ABSTRACT
The global increase in marine transportation of dilbit (diluted bitumen) can increase the risk of spills, and the application of chemical dispersants remains a common response practice in spill events. To reliably evaluate dispersant effects on dilbit biodegradation over time, we set large-scale (1,500 mL) microcosms without nutrient addition using a low dilbit concentration (30 ppm). Shotgun metagenomics and metatranscriptomics were deployed to investigate microbial community responses to naturally and chemically dispersed dilbit. We found that the large-scale microcosms could produce more reproducible community trajectories than small-scale (250 mL) ones based on the 16S rRNA gene amplicon sequencing. In the early-stage large-scale microcosms, multiple genera were involved in the biodegradation of dilbit, while dispersant addition enriched primarily Alteromonas and competed for the utilization of dilbit, causing depressed degradation of aromatics. The metatranscriptomic-based metagenome-assembled genomes (MAG) further elucidated early-stage microbial antioxidation mechanism, which showed that dispersant addition triggered the increased expression of the antioxidation process genes of Alteromonas species. Differently, in the late stage, the microbial communities showed high diversity and richness and similar compositions and metabolic functions regardless of dispersant addition, indicating that the biotransformation of remaining compounds can occur within the post-oil communities. These findings can guide future microcosm studies and the application of chemical dispersants for responding to a marine dilbit spill.
IMPORTANCE In this study, we employed microcosms to study the effects of marine dilbit spill and dispersant application on microbial community dynamics over time. We evaluated the impacts of microcosm scale and found that increasing the scale is beneficial for reducing community stochasticity, especially in the late stage of biodegradation. We observed that dispersant application suppressed aromatics biodegradation in the early stage (6 days), whereas exerting insignificant effects in the late stage (50 days), from both substance removal and metagenomic/metatranscriptomic perspectives. We further found that Alteromonas species are vital for the early-stage chemically dispersed oil biodegradation and clarified their degradation and antioxidation mechanisms. These findings help us to better understand microcosm studies and microbial roles for biodegrading dilbit and chemically dispersed dilbit and suggest that dispersant evaluation in large-scale systems and even through field trails would be more realistic after marine oil spill response.
KEYWORDS: Corexit 9500A, hydrocarbon degradation, metagenomics, metatranscriptomics, microbial communities, microcosm, oil spill
INTRODUCTION
Global energy demand boosts the daily unrefined petroleum use worldwide to nearly 100 million barrels per day. Meanwhile, the production of unconventional oils, such as bitumen, has also been increased (1, 2). In Canada, there are large oil sand deposits (e.g., Athabasca, Cold Lake, and Peace River regions) for bitumen recovery, which are projected to increase to 3.8 million barrels per day by 2022 (3). For transportation in pipelines or by rail, bitumen blends are diluted with lower-density hydrocarbons to form dilbit (diluted bitumen) oils. The increase in marine dilbit transportation increases the risk of a potential oil spill, causing harmful impacts on marine ecosystems and coastal environments (4). As an important marine oil spill treating agent, chemical dispersant can transfer floating oil slicks into the water column by lowering the oil-water interfacial tension and emulsifying oils into tiny droplets, which increases their bioavailability for degradation. The oil concentrations in the seawater could be increased to thousands of ppm in the first few hours of dispersant application and rapidly diluted to a few parts per million (5). Dispersant application can thus increase oil dilution in seawater, minimizing oil delivery to shorelines (6) and is a well-recognized response option for any potential marine dilbit spill.
During natural attenuation, a set of weathering processes, including evaporation, dissolution, photooxidation, and biodegradation, take place and affect the physicochemical properties of the dispersed oil at sea. In these processes, the volatile compounds and lighter weight aromatics can be easily evaporated (7), and highly polar compounds can be dissolved into the seawater (8), whereas photooxidation will increase the oxygen content in oils for producing solubilized carbons (9). In contrast, biodegradation depends on hydrocarbonoclastic bacteria, which will grow quickly, deriving energy and carbon from the degradation of petroleum hydrocarbons, especially alkanes and aromatics (10). Application of chemical dispersant will increase the bioavailability of spilled oils to hydrocarbonoclastic bacteria, change the composition of microbial communities, and ultimately affect the degradation of spilled oils.
Natural attenuation field trails, though reflecting the real marine environment with the real wave and current impact, are expensive and logistically and legislatively complicated. Therefore, laboratory microcosm studies have been widely adopted to identify dispersant effects on the natural attenuation of spilled oils in well-controlled and comparable conditions. Recent advancements in metagenomic and metatranscriptomic analyses, together with the oil composition characterization, have been applied as emerging tools to reveal the whole picture of microbial functions and activities and oil biodegradation processes, elucidate the dispersant effects on oil biodegradation, and predict their transport in marine environments. To date, controversial results on the biodegradation of chemically dispersed oil have been reported. They could be explained by the variations in types of spilled oils and seawaters and the microcosm conditions. The enhanced oil bioavailability, as a result of dispersant application, can stimulate biodegradation (11, 12), while the increased oil concentration in the water column may cause short-term inhibitory effects toward biodegradation (13, 14). Besides, compounds present in dispersant formulations can also act as carbon and energy sources and may be preferred by some microorganisms for competing for oil biodegradation (15). However, previously adopted microcosms for oil spill investigations can be improved by decreasing the concentration of oil used and enlarging the scale/size of the microcosms.
High oil concentrations (more than 100 ppm) in small-scale microcosms (a few hundred milliliters) have been widely applied (11, 12, 15, 16). In the field, however, dispersed oils will be rapidly diluted in the open ocean due to the high wave energies and diffuse apart and drop to concentrations well below 100 ppm within a few hours of dispersant application (17). The exponentially increased oil concentrations applied in closed systems will limit biodegradation due to the depletion of nutrients (17, 18). The small-scale microcosms, therefore, were supplemented with external nutrients (i.e., nitrogen and phosphate) to facilitate oil biodegradation (11, 16, 19–21). However, the increased nutrients were reported to eliminate some essential degraders like Cycloclasticus while accelerating the growth of others such as Alcanivorax (22), which will affect the actual evaluation of microbial community dynamics for oil biodegradation.
In addition, a small-scale system can hardly include a sufficient inoculum of hydrocarbonoclastic bacteria (5) and fails to have an in situ reflection of microbial communities in marine environments (19, 23). Further, microbial populations in a small-scale microcosm are more susceptible to stochastic effects over time, especially for species at the low diversity bottleneck, causing the biased subsequent microbial recovery trajectory (24, 25). Though it is challenging to design and interpret microcosm studies for monitoring in situ conditions, increasing the microcosm scale may help provide more reproducible community results. Such a hypothesis has been demonstrated in previous nutrient cycle microcosm studies (24) but not for oil spill investigations. Taking all of these together, an in-depth understanding of the scale-up effect on oil biodegradation is of great importance to understand the response of microbes to the dispersed oils and their ultimate fate in a natural marine environment (i.e., low dilbit concentration and no additional nutrients).
Here, we employed two scale microcosm systems (i.e., small, 250 mL, and large, 1,500 mL) without nutrient addition using a low dilbit concentration (30 ppm) to track microbial community dynamics during a hypothetical marine dilbit (i.e., Cold Lake Blend [CLB]) spill with and without a chemical dispersant (i.e., Corexit 9500A). The experimental design for the microcosm studies is presented in Fig. 1. Through 16S rRNA gene amplicon sequencing, we examined the impact of microcosm scale on community dynamics over time. To further explore microbial functions and activities, shotgun metagenomic and metatranscriptomic sequencing with combined contig-based and binning-centric analysis was applied for the large-scale microcosms.
FIG 1.
Overview of the microcosm setups in this study. The 30 ppm dilbit was used. The dispersant/oil ratio (DOR) (vol/vol) was 1:10. For each microcosm, large-scale means 1,500 mL and small-scale means 250 mL. The biotic microcosms were performed using fresh seawater, while the abiotic microcosms were performed using sterilized seawater.
RESULTS AND DISCUSSION
Effects of increasing microcosm scale on microbial community dynamics evaluation over time.
We performed two sets (i.e., small and large scale) of microcosms using fresh seawater sampled from the same place at the same time with incubation for 6 days (early stage) and 50 days (late stage), respectively. The DNA was harvested from the initial seawater and each microcosm for 16S rRNA gene amplicon sequencing. Four samples (one small-scale seawater sample and three early-stage small-scale microcosms amended with dilbit only) produced invalid results due to the low extractable DNA concentrations (see Fig. S1 in the supplemental material). In all, the amplicon data from a total of 26 samples were processed to generate operational taxonomic units (OUTs) for community profiling.
The microbial richness (i.e., based on Chao 1 and observed species indexes) and diversity (i.e., based on Shannon and Simpson indexes) metrics of the initial seawater samples for the large volume (1,500 mL) were higher than those for the small volume (250 mL) (Fig. 2A). The different microbial richness and diversity results of the same raw seawater could be explained by the limited DNA extracted from the small-scale communities (Fig. S1). Some species with extremely low abundance may be under the detection limit due to unsuccessful PCR amplification for sequencing. This indicates that large-scale microcosms could better represent in situ microbial communities of the marine environment due to the increased biomass for deep sequencing. The rarefaction curve also showed that the large volume provided a better estimate of the alpha diversity and a reduced stochasticity between replicates of each treatment (see Fig. S2 in the supplemental material). In the early-stage large-scale microcosms, microbial richness and diversity of microcosms amended with dilbit plus dispersant were significantly lower than those amended with dilbit only (Fig. 2A). In the late-stage large-scale microcosms, microbial communities were equally rich and diverse regardless of dispersant addition (Fig. 2A). In contrast, in the late-stage small-scale microcosms, dispersant addition apparently still had a negative effect on microbial richness but not on microbial diversity (Fig. 2A). Overall, this indicates that dispersant addition played a significant role in shaping the early-stage communities regardless of the microcosm scale.
FIG 2.
Microbial community dynamics in large-scale and small-scale microcosms over time based on 16S rRNA gene amplicon sequencing. (A) Alpha diversity analysis for microbial richness (Chao1 and observed species indexes) and diversity (Shannon and Simpson indexes). (B) Beta diversity analysis using PCoA analysis using Bray Curtis dissimilarity matrix. (C) Microbial community taxa at the class level. Large-scale microcosms (Top); small-scale microcosms (Bottom). T0, microbial community of the initial seawater; T6-Do and T6-DoD9500, microbial community in the early stage (6 days) with dilbit only and dilbit plus dispersant, respectively; T50-Do and T50-DoD9500, microbial community in the late stage (50 days) with dilbit only and dilbit plus dispersant, respectively.
The beta diversity analysis (i.e., principal-coordinate analysis [PCoA] for Bray Curtis dissimilarity matrix) showed that microbial populations become less and less similar to the initial community over time (Fig. 2B; see also Fig. S3 in the supplemental material). This finding is well-aligned with that of Schreiber et al. (19), in which the incubation time is identified as an essential factor in shaping community composition. In the early stage, dilbit plus dispersant communities are more dissimilar to the initial community than dilbit-only communities (Fig. 2B), similar to what was reported by Tremblay et al. (16) for small-scale crude oil microcosms. In the late-stage microcosms, microbial communities of both treatments gradually clustered together except for those in small-scale microcosms amended with dilbit plus dispersant (Fig. 2B). Both alpha and beta diversity analyses showed that late-stage large-scale microcosms were more similar, suggesting that dispersant application in the marine environment would have only a short-term effect on microbial communities after a dilbit spill.
The microbial taxa at the class level showed that the initial seawater (T0) was dominated by the phyla of Proteobacteria (Gammaproteobacteria and Alphaproteobacteria classes) and Bacteroidetes (Flavobacteriia class) (Fig. 2C). Proteobacteria, especially Gammaproteobacteria were stimulated in the early stage, while Alphaproteobacteria were promoted in the late stage in both scale microcosms, regardless of dispersant addition (Fig. 2C). The class Gammaproteobacteria contains many important members and a diversity of hydrocarbonoclastic bacteria involved in oil degradation (26), and Alphaproteobacteria persist more in polycyclic aromatic hydrocarbon (PAH)-contaminated sites (27). In the late stage, the abundance of microbial classes (e.g., Verrucomicrobiae, Betaproteobacteria, Cytophagia, Woesearchaeota, Planctomycetacia, Phycisphaerae), which were rare or even undetectable (<1%) in the initial seawater, were stimulated in large-scale microcosms of both treatments (Fig. 2C). However, these classes were not significantly enriched in the small-scale microcosms (Fig. 2C). Besides, in the early-stage microcosms amended with dilbit plus dispersant, the similarity between replicates representing the percentage of shared operational taxonomic units (OTUs) displayed similar values in both large-scale (46%) and small-scale (49%) microcosms. This may be because dispersant was deterministic in shaping the early-stage community composition regardless of the microcosm scale. In contrast, in the late stage, the similarity decreased for small-scale microcosms (36%) but increased for large-scale microcosms (69%) (see Fig. S4 in the supplemental material). Our small-scale microcosms displayed a similar trend as the previous study using 100-mL microcosms (19) that showed the community similarity decreased over time. The increased similarity observed in our late-stage large-scale microcosms of both treatments indicates that increased population size may decrease the effects of stochastic elimination for community assembly after long-term incubation.
In the large-scale microcosms, the early-stage microbial community at the family level (see Fig. S5 in the supplemental material) showed that the order of Alteromonadales, specifically the Alteromonadaceae family, made up the highest relative abundance in the bacterial communities of microcosms amended with dilbit plus dispersant. In contrast, the communities in oil-only microcosms showed an enrichment of the orders Oceanospirillales (dominated by the families Oceanospirillaceae and Alcanivoracaceae), Thiotrichales (dominated by Piscirickettsiaceae), and Rhodobacterales (dominated by Rhodobacteraceae) compared to communities in the initial seawater. Moreover, the families Cryomorphaceae and Thiotrichaceae showed a decreased relative abundance in microcosms of both treatments. The decreased abundance was possibly due to the effect of aromatics presented in the dilbit: the Cryomorphaceae family is reported to be inhibited entirely in the presence of pyrene and phenanthrene (28), and there is no evidence showing Thiotrichaceae can tolerate or degrade aromatics.
Microbial functions and activities toward dilbit degradation using the contig-based shotgun metagenomic approach.
To obtain a broad overview of metabolic functions and activities for the degradation of dilbit only and chemically dispersed dilbit, large-scale microcosms were analyzed by contig-based shotgun metagenomic and metatranscriptomic sequencing. All of the genes were annotated based on the KEGG database to reveal microbial functions. PCoA analysis, based on assembled contigs, confirmed the results of 16S rRNA gene amplicon sequencing. In detail, dilbit plus dispersant communities were significantly dissimilar to dilbit-only ones in the early stage but clustered together in the late stage (see Fig. S6 in the supplemental material). Taxonomic profiles at the species level showed that in the early-stage dilbit-only microcosms, species belonging to the genera Methylophaga, Marinobacterium, Alcanivorax, Phaeobacter, and Cycloclasticus were mostly enriched (Fig. 3). Inclusion of dispersant resulted in the enrichment of Alteromonas species, and there was a substantial increase in species related to Alteromonas australica (Fig. 3). Tremblay et al. (16) also showed that Alteromonas was the dispersant-associated genus in the early stage. In the late stage, similar taxonomic profiles were observed in both treatments, mostly with species belonging to Alphaproteobacteria (Fig. 3).
FIG 3.
Metagenomic sequencing showing the dominant microbial taxa at the species level using the contig-based approach in the large-scale microcosm communities. CPM, counts per million; T0, microbial community of the initial seawater; T6-Do and T6-DoD9500, microbial community in the early stage (6 days) with dilbit only and dilbit plus dispersant, respectively; T50-Do and T50-DoD9500, microbial community in the late stage (50 days) with dilbit only and dilbit plus dispersant, respectively.
Notably, microbial communities in both treatments appeared to strongly enrich pathways related to carbohydrate, energy, lipid, nucleotide, amino acid, cofactor and vitamin, and xenobiotic metabolism; genetic information processing related to translation, replication, and repair; environmental information processing related to membrane transport and signal transduction; as well as cellular processing related to cellular community, cell motility, and drug resistance over time (see Fig. S7 in the supplemental material). Metatranscriptomic data showed that with dispersant, microbial communities further enriched the transcripts of these metabolic functions in the early stage compared with the dilbit-only microcosms (see Fig. S8 in the supplemental material). This indicated that the presence of dispersant with dilbit might facilitate certain metabolic activities for using dispersant components or the relatively bioavailable dispersed dilbit (19).
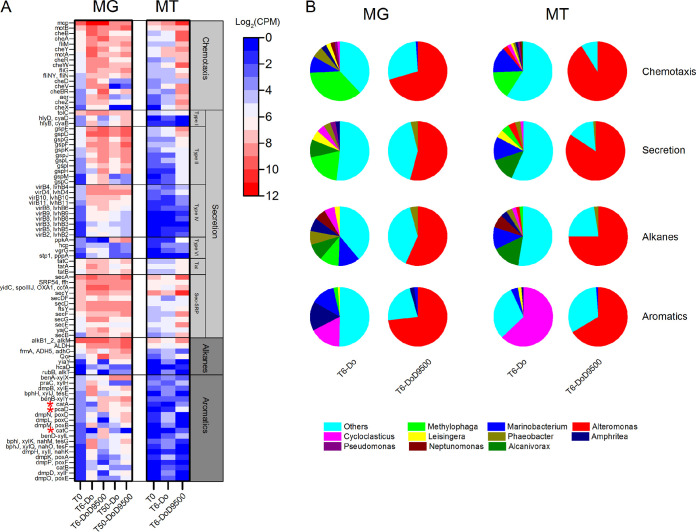
The abundance and expression levels of genes involved in accessing and degradation of hydrocarbons were further analyzed. Bacterial chemotaxis (KEGG pathway number ko02030) that mediates bacterial affinity and motility in response to gradients of chemical substrates and secretion systems (KEGG pathway number ko03070) that influence extracellular surface-active compounds (i.e., biosurfactants) for bioavailability of oils are both described as upstream pathways for hydrocarbon degradation (16). The metagenomic data showed enrichment of bacterial methyl-accepting (mcp) chemotaxis in both treatments compared with the initial seawater in the early stage (Fig. 4A). However, abundance decreased in the late stage. The abundance of genes for the type II and sec-SRP secretion systems was enriched over time, while virB series genes for the type IV secretion system decreased in the late stage (Fig. 4A). The type IV secretion system has been described as being responsible for the horizontal gene transfer of toxin resistance genes (29), indicating a potential role of these genes in the early-stage microbial survival and proliferation mechanisms toward dilbit-only or chemically dispersed dilbit. We suggest that both the chemotaxis and type IV general secretion systems are the primary induced strategies for sensing and accessing dilbit in the short term. The sustained high abundance of type II and sec-SRP secretion systems over time indicates their involvement in the relatively complex chemical components that remained for the long term (i.e., 50 days). The metatranscriptomic data also showed that dispersant addition increased the transcripts of the associated microbial accessing genes compared with the dilbit-only communities in the early stage (Fig. 4A). The associated microbial accessing mechanism toward dilbit and dispersed dilbit is in accordance with the findings of Tremblay et al. (16).
FIG 4.
Broad overview of microbial functions and activity for hydrocarbon access and degradation in the large-scale microcosms using the shotgun metagenomic contig-based approach. (A) The abundance and expression of corresponding genes for microbial access (i.e., chemotaxis and secretion) and degradation (i.e., alkanes and aromatics) pathways. Genes are annotated based on the KEGG database and are plotted with abundance Log2 CPM > 0. Genes involved in the degradation of catechol are labeled with red asterisks. (B) The dominant contributors (higher than 1%) toward the functions and activities. MG, metagenomic analysis; MT, metatranscriptomic analysis; T0, microbial community of the initial seawater; T6-Do and T6-DoD9500, microbial community in the early stage (6 days) with dilbit-only and dilbit plus dispersant, respectively; T50-Do and T50-DoD9500, microbial community in the late stage (50 days) with dilbit-only and dilbit plus dispersant, respectively. Mean values of three replicates are plotted.
Genes involved in alkane and aromatic degradation pathways were also analyzed. Alkanes are initially oxidized through alkane monooxygenase (alkB) to alcohols, which are further oxidized to aldehydes and then fatty acids, which enter the beta-oxidation pathway (30). The corresponding genes (as shown in Materials and Methods) in the KEGG pathway for the degradation of both fatty acids (KEGG pathway number ko00071) and aromatic compounds (KEGG pathway number ko01220) are then integrated and summarized. Generally, dispersant addition suppressed the abundance of genes for aromatics degradation in the early stage compared with the dilbit-only communities. Due to the similarly shaped communities for the degradation of residual aromatics, communities of both treatments showed increased gene abundance for aromatics degradation in the late stage (Fig. 4A). The metatranscriptomic data further indicated that, in the early stage, dispersant addition depressed the expression of most aromatics-degrading genes (Fig. 4A). In contrast, abundance and expression of genes like pcaD (KEGG entry k01055), catC (KEGG entry k03464), and catA (KEGG entry k03381), which are responsible for degradation of the intermediate catechol, were enriched (31). These findings suggest that dispersant addition may depress the degradation of aromatics.
Microbial contributions toward dilbit biodegradation in the early stage showed that multiple species were present in the dilbit-only microcosms, while Alteromonas was the most dominant contributor when dispersant was added (Fig. 4B). In dilbit-only communities, many genera, dominated by Methylophaga, carry the genes coding for the accessing system. The Methylophaga (15.4%) and Marinobacterium (14.1%) account for the high transcripts for chemotaxis, while Alcanivorax (13.2%) and Marinobacterium (12.7%) had a high expression in the secretion system. The degradation genes were identified within a diverse community, and their expression was primarily associated with Alcanivorax (15.3%) and Marinobacterium (12.1%) for alkane degradation and Cycloclasticus (62.9%) for aromatics degradation. These genera are widely reported for the degradation of alkanes and aromatics in previous studies (32–36). Interestingly, the genes coding for the accessing system were rarely detected and expressed in Cycloclasticus (Fig. 4B). This suggests that Cycloclasticus may not have an affinity toward low-molecular-weight (LMW) aromatics, which have higher solubility and bioavailability in the aqueous phase (37). In contrast, in the dilbit plus dispersant microcosms, Alteromonas was the predominant genus, possessing and expressing the accessing and degrading genes in the early stage (Fig. 3 and 4B). It is reported that coordinated microbial degradation (i.e., multiple microbes involved in the biodegradation) can achieve a higher degradation efficiency toward aromatics compared to a single taxon (38, 39). Since dispersant application suppressed microbial richness and diversity and primarily enriched Alteromonas, the early-stage degradation of aromatics may be inhibited.
Chemical analysis of percentage loss of n-alkanes and aromatics in abiotic and biotic microcosms.
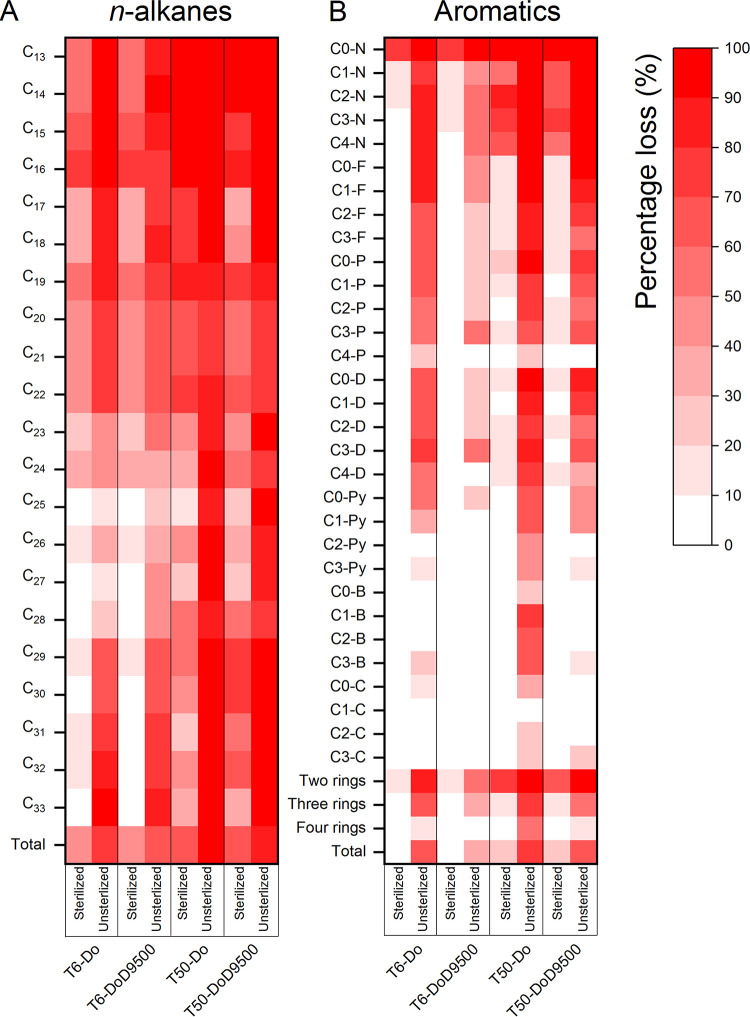
Chemical analysis was further applied to reveal the loss of n-alkanes and aromatics and changes of specific dispersant components in the abiotic and biotic microcosms. The gas chromatography-mass spectrometry (GC-MS) results proved a similar abiotic loss of n-alkanes and aromatics in the dilbit-only and dilbit plus dispersant microcosms. Nearly 41 to 42% of n-alkanes and 2 to 3% of aromatics were lost in the early stage, and the ratios reached 65 to 70% and 21 to 23%, respectively, in the late stage (Fig. 5). Obviously, the biotic microcosms could realize much higher removal of these hydrocarbons over time, evidencing that biodegradation played an indispensable role in dilbit removal (Fig. 5). However, dispersant addition had different effects on the loss of n-alkanes and aromatics over time. For n-alkanes, dispersant addition did not cause obvious change compared to the dilbit-only microcosms, with nearly 69 to 77% and 90 to 92% lost in the early stage and late stage, respectively (Fig. 5A). In contrast, dispersant addition dropped the percentage loss of aromatics from 61% to 33% and 80% to 62%, in the early stage and late stage, respectively (Fig. 5B). These results confirmed our observations from the metagenomic and metatranscriptomic analysis that dispersant addition exerted limited impacts on the removal of n-alkanes but inhibited the degradation of aromatics, especially in the early stage.
FIG 5.
Chemical analysis showing percentage loss of n-alkanes (A) and aromatics (B) after 6 days and 50 days incubation of both treatments in abiotic and biotic microcosms. C13-C33, n-alkanes with corresponding carbon atoms; C0-N, C1-N, C2-N, C3-N, and C4-N, naphthalene and its alkylated homologs; C0-F, C1-F, C2-F, and C3-F, fluorene and its alkylated homologs; C0-P, C1-P, C2-P, C3-P, and C4-P, phenanthrene and its alkylated homologs; C0-D, C1-D, C2-D, C3-D, and C4-D, dibenzothiophene and its alkylated homologs; C0-Py, C1-Py, C2-Py, and C3-Py, pyrene and its alkylated homologs; C0-B, C1-B, C2-B, and C3-B, benzonaphthothiophene and its alkylated homologs; C0-C, C1-C, C2-C, and C3-C, chrysene and its alkylated homologs. Two rings, naphthalene and its alkylated homologs; three rings, fluorene, phenanthrene, and dibenzothiophene, and their alkylated homologs; four rings, pyrene, benzonaphthothiophene, chrysene, and their alkylated homologs. Mean values between three replicates are plotted.
It has been widely acknowledged that LMW components, such as the short-chain n-alkanes, two-ring naphthalene, and its alkylated homologs, were more easily degraded. Our study further verified the degradation of multiple-ring aromatics like fluorene, pyrene, and dibenzothiophenes and their alkylated homologs in the early stage (Fig. 5B). Such results were different from the ones generated by Schreiber et al. (19) that used 150 ppm dilbit in their study. It is believed that lower oil concentrations could be more rapidly biodegraded in the marine environment than higher concentrations (40). Due to the lower initial concentration of dilbit (30 ppm) applied in our study, these multiple-ring aromatics may serve as carbon or energy sources after depletion of alkanes and LMW aromatics.
We also observed that the dispersant components, e.g., dioctyl sodium sulfosuccinate (DOSS), were under the limit of detection (LOD) in the early-stage biotic microcosms (see Fig. S10 in the supplemental material). The finding is in agreement with Gofstein et al. (21), who proved that Corexit 9500A could be biodegraded rapidly, with a concentration dropped to below the LOD within 5 days. In addition, we further estimated the cell density using the extractable DNA concentration. It showed that dispersant addition strongly enriched cell density in the early stage (see Fig. S11 in the supplemental material). Since the presence of dispersant in dilbit decreased the hydrocarbon utilization but enriched cell density in the early stage, we suggest that dispersant components could be primarily utilized for competing for microbial degradation of dilbit.
Dispersant application inducing microbial antioxidation in the early stage.
In our large-scale microcosms, the presence of dispersant in dilbit would depress microbial community richness and diversity and dispersant components can be primarily degraded for enriching cell density in the early stage. Apart from that, dispersant addition can also increase the aquatic exposure to xenobiotics, including aromatics, for inducing the overproduction of reactive oxygen species (ROS), causing potential inhibition to microorganisms (14).
To answer whether dispersant application could cause the inhibition effects in our large-scale microcosms with the low dilbit concentration, we applied a binning-centric approach to evaluate dominated bacterial responses. Based on a total of 62,437 assembled contigs, 3,850 contigs were binned to generate 39 metagenomic bins, representing an integration rate of 6.17%. The heatmap (see Fig. S12 in the supplemental material) presents the relative abundances. The 2-Thiotrichales (Methylophaga sp. 42_25_T18), 16-Oceanospirillales (Marinobacterium jannaschii), and bin-16 belonging to Gammaproteobacteria dominated the dilbit-only microcosms. In contrast, 1-Alteromonadales and 7-Alteromonadales (Alteromonas australica), which belong to the Alteromonas genus, were dominant in microcosms with added dispersant in the early stage. In the late stage, microbial communities became more diverse, with bin-14 and bin-6 showing the dominance of Proteobacteria. All of these highly abundant metagenome-assembled genomes (MAGs) were rare (<1%) in the initial seawater, indicating that these hydrocarbonoclastic bacteria can quickly respond and grow after a dilbit spill. However, based on the metagenomic binning approach, some important species for the degradation of alkanes (e.g., Alcanivorax) and aromatics (e.g., Cycloclasticus) were missing.
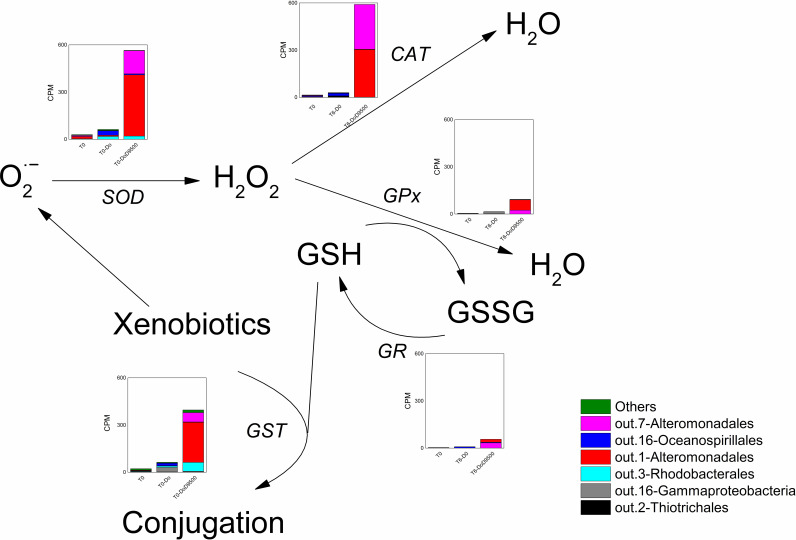
Microbial antioxidation can be utilized for evaluating the microbial responses to ROS. The mechanisms were recently reported in an anaerobic system based on the recovered MAGs (41). However, there is little information on the microbial antioxidation response toward naturally and chemically dispersed dilbit in a microbial community. We thus employed a metatranscriptomic-based MAGs approach to elucidate microbial antioxidation potential in response to the dilbit and dispersant application. We focused on antioxidant enzymes consisting of superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), glutathione reductase (GR), and glutathione S-transferases (GST) that can directly catalyze the conjugation of xenobiotics through glutathione (GSH) for detoxification (40, 42). Increased concentrations of xenobiotics can induce high concentrations of superoxide, which will be degraded by SOD to generate hydrogen peroxide (H2O2). There are two mechanisms for the removal of H2O2. The first one is directly through CAT, and the second one is via the GSH redox reaction. We found that the dilbit-only communities did not increase the transcripts of these genes compared to the initial seawater, while dispersant addition particularly enriched the transcripts of the GST, SOD, and CAT genes (Fig. 6). Transcripts of these genes were primarily increased from the two Alteromonas species, indicating that Alteromonas species may directly conjugate xenobiotics and quench the induced ROS mainly via the first mechanism to address the short-term exposure to chemically dispersed dilbit.
FIG 6.
Microbial antioxidation mechanism of dominated MAGs in the early-stage large-scale microcosms based on the shotgun metatranscriptomic analysis. O2·−, superoxide radical; H2O2, hydrogen peroxide; GSH, glutathione; GSSG, glutathione disulfide; SOD, superoxide dismutase; CAT, catalase; GPx, glutathione peroxidase; GR, glutathione reductase; GST, glutathione S-transferases. Mean values between three replicates are plotted.
It is well established that the expression of hydrocarbon-degrading genes will upregulate microbial antioxidant enzymes (43, 44). In our study, the expressions of alkanes- and aromatics-degrading genes were highly expressed in both the dilbit-only and dilbit plus dispersant microcosms (Fig. 4A). However, the expression of antioxidation genes was not stimulated in the dilbit-only microcosms as in the dilbit plus dispersant ones (Fig. 6). It was also reported that inefficient aromatics degradation or the presence of toxic compounds would generate more ROS to enhance the expression of antioxidation genes (43). These indicated that the increased toxicity effects rather than the increased expression of hydrocarbon-degrading genes might account for the stimulated antioxidation processes.
Evaluation of the chemical dispersant as a dilbit spill-treating agent.
Dispersant application was previously reported to enhance microbial degradation activity toward oils with a large proportion of alkanes and limited aromatics in marine microcosms (11, 45). However, as there is a large proportion of aromatics in dilbit (see Fig. S8), our microcosm results showed that dispersant addition inhibited the degradation of aromatics in the early stage. Based on the chemical and biological analyses in this study, we suggest that in the early stage, the presence of dispersant in dilbit would (i) depress microbial richness and diversity, (ii) compete for the utilization of hydrocarbons in dilbit, and (iii) induce potential toxicity.
In our microcosm study, we used a high dispersant/oil ratio (DOR = 1:10 [vol/vol]). A DOR of 1:20 or lower (i.e., 1:100) is the typical target ratio in an oil spill response, while for extremely heavy oils, it can be increased to 1:10 to enhance effectiveness (46). Due to the high viscosity of dilbit, the dispersant Corexit 9500A showed limited effectiveness below a DOR of 1:20 (see Fig. S13 in the supplemental material). It could only achieve 7.5% effectiveness after a 200-min retention time, as many dispersed oil droplets tended to resurface (see Fig. S10). Fortunately, under the DOR ratio of 1:10, it showed high dispersion effectiveness (41.7%) (Fig. S10). Hence, a DOR of 1:10 could be more applicable in a dilbit spill response. Efficiently dispersed dilbit can significantly increase early-stage concentrations of xenobiotics like aromatics in the aqueous phase, although under the high DOR, our marine microcosms still demonstrated strong dilbit attenuation ability. In the early stage, specific species from the Alteromonas genus increased and degraded both n-alkanes and aromatics.
The goal of dispersant application is to rapidly emulsify the spilled oil slicks, leading to decreased deposition on the shorelines and producing smaller oil droplets that are more readily attacked by the microbial degraders. As marine environments do have a large dilution capacity, the short-term adverse effects of dispersant application on aromatics degradation would be alleviated to some extent. Furthermore, in the late stage, our microcosms showed similar community composition and metabolic functions irrespective of dispersant addition. The transformation of the remaining high-molecular-weight compounds may be within the capabilities of the post-oil communities that contain higher microbial diversity and species richness.
Conclusions.
In summary, our study demonstrated that increasing the microcosm scale could produce more producible microbial communities during a simulated marine dilbit spill. The hydrocarbonoclastic bacteria initially present at a low abundance in the seawater responded rapidly, increasing their activity for dilbit degradation. The presence of dispersant in dilbit had different effects over time. In the early stage, dispersant addition inhibited dilbit degradation mainly because of (i) decreased microbial richness and diversity and predominantly enriched Alteromonas for degrading both alkanes and aromatics, (ii) microbial utilization of dispersant components for competing for the degradation of dilbit, and (iii) the induced potential toxicity. In the late stage, the inhibition effects could be relieved, and microbial communities showed similar compositions and metabolic functions regardless of dispersant addition. Although it is challenging to mimic a natural marine environment in laboratory microcosms, results generated from our large-scale microcosms without nutrient addition using low dilbit concentrations could increase our understanding of the microbial role in natural attenuation and help evaluate the impact of a chemical dispersant as a dilbit spill treating agent.
MATERIALS AND METHODS
Microcosm setup.
All biotic and abiotic microcosms were performed at identical settings with continual shaking at 150 rpm at room temperature (20 to 25°C). A total of 80 L of seawater (∼10 m depth, 31.24 ppt) was collected from Ocean Science Centre at Memorial University (47.6248°N, 52.6627°W) in the spring of 2019. This seawater was well mixed and used immediately for the preparation of all of the microcosms throughout this study to ensure that they were homogenous. All flasks were acid-washed and autoclaved prior to use, and all microcosms were conducted in triplicate. The biotic microcosms were set up with freshly collected seawater, while the abiotic microcosms were set up using the presterilized seawater. For small-scale microcosms, each 500 mL flask was filled with 250 mL seawater. For large-scale microcosms, each 4,000 mL flask was filled with 1,500 mL seawater. The dilbit (Cold Lake Blend [CLB]) and chemical dispersant (Corexit 9500A) were obtained from Fisheries and Oceans Canada, Dartmouth, NS. The dilbit was fully dispersed at room temperature and added directly using a pipette (Gilson Microman E) to achieve a final dilbit concentration of 30 ppm (i.e., 7.5 mg in small-scale microcosms and 45 mg in large-scale microcosms). To select the dispersant dose, the dispersion effectiveness was tested beforehand at dispersant/oil ratios (DOR) (vol/vol) of 1:10, 1:20, 1:50, and 1:100 using the baffled flask test (47). A DOR of 1:10 was selected due to the best performance. The dispersant was then directly pipetted to oil slick (i.e., 0.82 μL in small-scale microcosms and 4.92 μL in large-scale microcosms).
Chemical analysis.
Determination of n-alkanes and aromatics of dilbit and dispersant components was performed by sacrificing the complete microcosm content (including the flask wall) after 6 and 50 days of incubation to avoid the potential errors caused by sorption (48). Liquid-liquid extraction was performed for extraction (49). Briefly, approximately 30 mL dichloromethane was added three times into each large-scale flask to extract the hydrocarbons. The three organic solvent fractions were combined, and residual water was removed using anhydrous sodium sulfate. The solvent fraction was concentrated to 1 mL. An Agilent 7890A gas chromatography-mass spectrometer (GC-MS) equipped with a DB-5MS column was applied for oil analysis based on our previous study (37). Helium was the carrier gas at a flow rate of 1.0 mL min−1, and the GC oven temperature was set at 50°C for 2 min, then ramped up at 6°C min−1 to 300°C for 20 min. MS detection was conducted in the electron ionization (EI) mode with 70 eV and an ion source temperature of 300°C. Individual compounds were identified from their mass spectra. All hydrocarbon data were normalized to the conserved internal biomarker 17α(H), 21β(H) - hopane. Figure S10 in the supplemental material indicated the dispersant components did not affect the analysis of dilbit components.
Sample preparation for DNA and RNA sequencing.
Experimental procedures for sample preparation and DNA and RNA sequencing followed the same protocol. At each time point, 6 and 50 days, the microcosms for genomics analysis were harvested by filtering the entire contents of each flask through 0.22-μm polyethersulfone membranes. The initial seawater (250 mL for small-scale and 1,500 mL for large-scale microcosms) at T = 0 was directly filtered. Microbes were collected on the membranes, which were transferred into 50-mL Falcon tubes and immediately flash frozen by submersion in liquid nitrogen and stored at −80°C. The DNA and RNA were extracted simultaneously from the filtered membranes following the Qiagen RNeasy PowerWater kits. The extracted DNA samples were treated with RNase If (New England BioLabs) to remove RNA for downstream 16S rRNA gene amplicon (for both small and large scales microcosms) and shotgun metagenomic sequencing (only for large-scale microcosms). The RNA samples were treated with the Turbo DNA-free kit to remove all of the remaining DNA. The rRNA-depleted RNA was then subjected to reverse-transcription to produce cDNA for shotgun metatranscriptomic sequencing for large-scale microcosms.
16S rRNA gene amplicon sequencing for both small and large scales microcosms.
The V4 and V5 hypervariable region of the 16S rRNA gene amplified with 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAG TTT-3′) was used to detect both archaea and eubacteria. The 16S rRNA gene libraries for sequencing were prepared according to Illumina’s “16S Metagenomic Sequencing Library Preparation” guide (part number 15044223 Rev. B). Amplification of the DNA was performed in 25 μL containing 0.5 to 15 ng of DNA template, 0.4 μM each primer, and 0.5 mg/mL bovine serum albumin and Kapa HiFi HS RM polymerase (Roche). PCR amplification reaction conditions involved an initial denaturation at 96°C for 3 min, followed by 25 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, and final elongation for 5 min at 72°C. PCR products were evaluated by gel electrophoresis and purified with the Macherey-Nagel NucleoMag NGS Clean-up and Size Select kit (D-MARK Biosciences). Equal amounts of each purified PCR product were pooled and sequenced using the Illumina MiSeq platform.
Sequencing data were analyzed using AmpliconTagger (50). Briefly, the raw reads were quality checked, assembled, and clustered at 97% (VSEARCH) to form the final clusters and OTUs (51). Clusters having abundances lower than 25 were discarded. The OTUs were assigned to a taxonomic lineage with the RDP classifier software using an in-house training set containing the complete Silva release 128 database (52, 53). With taxonomic lineages in hand, an OTU table limited to bacterial and archaeal microorganisms was generated. A consensus rarefied OTU table was generated as described (50). Taxonomic summaries were generated using QIIME v1.9.1 (54). Community richness and diversity were estimated with 10 times rarefaction analysis to 10,765 sequences to improve data robustness. Alpha diversity (i.e., community richness and diversity metrics) and beta diversity (i.e., Bray Curtis dissimilarity matrix) were computed using the R package “vegan.” Statistical significance of diversity and richness differences was assessed using Student's t test.
Shotgun metagenomic and metatranscriptomic analysis for large-scale microcosms.
Metagenomic and metatranscriptomic libraries were sequenced on an Illumina HiSeq 4000 system with a rapid mode 2 × 100 bp configuration. A total of 15 samples were submitted for metagenomic sequencing, and 9 samples (i.e., the initial seawater at T = 0 and microcosms after 6 days of incubation with enough RNA) were used for metatranscriptomic sequencing. The resulting data (9.8 Gb for metagenome and 55.9 Gb for metatranscriptome) were processed through bioinformatic pipelines described previously (11). Shotgun metagenomic sequencing reads were quality controlled (QC) (Trimmomatic v0.39) and coassembled using Megahit v1.2.9 (55). Taxonomy of each contig was determined using CAT v5.0.3. Genes were ab initio predicted on each assembled contig using Prodigal v2.6.2 (56) and annotated following the guidelines of Joint Genome Institute (JGI), including the assignment of KEGG orthologs (57, 58). The QC-passed reads were mapped against contigs to assess the quality of assembly and to obtain contig abundance profiles. Alignment files in bam format were sorted by reading coordinates using SAMtools v1.9 (59), and only properly aligned read pairs were kept for downstream steps. Each bam file (containing properly aligned paired reads only) was analyzed for coverage of contigs and predicted genes using bedtools v2.17.0 (27) using a bed file having each gene coordinate on each contig. Only paired reads both overlapping their contig or gene were considered for gene and contig abundance computation. These latter matrices were normalized to obtain CPM (counts per million) using edgeR v3.10.2 (60). Shotgun metagenomic data were further processed to generate MAGs using MetaBAT v2.12.1 (61). Genes annotated with KEGG entries associated with dilbit accessing and degradation were chosen for a focused analysis. For the microbial chemotaxis and secretion systems, genes in bacterial chemotaxis (KEGG pathway number ko02030) and bacterial secretion system (KEGG pathway number ko03070) were integrated. For the degradation of alkanes, genes in fatty acid degradation (KEGG pathway number ko00071) that were responsible for the degradation of alkanes were analyzed. For the degradation of aromatics, genes in modules (KEGG pathway numbers M00538, M00537, M00419, M00547, M00548, M00551, M00568, M00569, M00539, M00543, M00544, M00418, M00541, M00540, M00534, M00624, M00623, M00636, M00545) in degradation of aromatic compounds (KEGG pathway number ko01220) were summarized.
Shotgun metatranscriptomic sequencing data were controlled for quality (Trimmomatic) and aligned on metagenomic coassembled contigs generated as described above to obtain a gene expression matrix of each sample readily comparable with the gene abundance matrix obtained with metagenomic data. The gene expression matrix was normalized with edgeR to generate normalized CPM counts.
Data availability.
All raw sequence reads, including 16S rRNA gene amplicon sequencing, metagenome, and metatranscriptome, have been submitted to NCBI's SRA and are available under the BioProject PRJNA704368. Other associated information is available upon request.
ACKNOWLEDGMENTS
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), Fisheries and Oceans Canada (DFO), Canada Foundation for Innovation (CFI), and Canada Research Chair (CRC) program.
We acknowledge Nathalie Fortin, Christine Maynard, Miria Elias, and Sylvie Sanschagrin from the NRC for helping with genomic sample treatments and data analysis. We thank Lars Schreiber (NRC) for helpful comments regarding the study design and for providing feedback on the manuscript. We acknowledge Compute Canada for access to the Graham high performance computing system.
Footnotes
Supplemental material is available online only.
Contributor Information
Baiyu Zhang, Email: bzhang@mun.ca.
Hideaki Nojiri, University of Tokyo.
REFERENCES
- 1.Nduagu EI, Gates ID. 2015. Unconventional heavy oil growth and global greenhouse gas emissions. Environ Sci Technol 49:8824–8832. 10.1021/acs.est.5b01913. [DOI] [PubMed] [Google Scholar]
- 2.Sleep S, Laurenzi IJ, Bergerson JA, MacLean HL. 2018. Evaluation of variability in greenhouse gas intensity of Canadian oil sands surface mining and upgrading operations. Environ Sci Technol 52:11941–11951. 10.1021/acs.est.8b03974. [DOI] [PubMed] [Google Scholar]
- 3.Nimana B, Canter C, Kumar A. 2015. Energy consumption and greenhouse gas emissions in upgrading and refining of Canada's oil sands products. Energy 83:65–79. 10.1016/j.energy.2015.01.085. [DOI] [Google Scholar]
- 4.Johannessen SC, Greer CW, Hannah CG, King TL, Lee K, Pawlowicz R, Wright CA. 2020. Fate of diluted bitumen spilled in the coastal waters of British Columbia, Canada. Mar Pollut Bull 150:110691. 10.1016/j.marpolbul.2019.110691. [DOI] [PubMed] [Google Scholar]
- 5.Lewis A, Prince RC. 2018. Integrating dispersants in oil spill response in Arctic and other icy environments. Environ Sci Technol 52:6098–6112. 10.1021/acs.est.7b06463. [DOI] [PubMed] [Google Scholar]
- 6.King TL, Robinson B, McIntyre C, Toole P, Ryan S, Saleh F, Boufadel MC, Lee K. 2015. Fate of surface spills of Cold Lake Blend diluted bitumen treated with dispersant and mineral fines in a wave tank. Environ Eng Sci 32:250–261. 10.1089/ees.2014.0459. [DOI] [Google Scholar]
- 7.Stout SA, Payne JR, Emsbo-Mattingly SD, Baker G. 2016. Weathering of field-collected floating and stranded Macondo oils during and shortly after the Deepwater Horizon oil spill. Mar Pollut Bull 105:7–22. 10.1016/j.marpolbul.2016.02.044. [DOI] [PubMed] [Google Scholar]
- 8.Stanford LA, Kim S, Klein GC, Smith DF, Rodgers RP, Marshall AG. 2007. Identification of water-soluble heavy crude oil organic-acids, bases, and neutrals by electrospray ionization and field desorption ionization Fourier transform ion cyclotron resonance mass spectrometry. Environ Sci Technol 41:2696–2702. 10.1021/es0624063. [DOI] [PubMed] [Google Scholar]
- 9.Vaughan PP, Wilson T, Kamerman R, Hagy ME, McKenna A, Chen H, Jeffrey WH. 2016. Photochemical changes in water accommodated fractions of MC252 and surrogate oil created during solar exposure as determined by FT-ICR MS. Mar Pollut Bull 104:262–268. 10.1016/j.marpolbul.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Zhang B, Zhu Z, Rostami M, Dong G, Ling J, Lee K, Greer CW, Chen B. 2021. Access-dispersion-recovery strategy for enhanced mitigation of heavy crude oil pollution using magnetic nanoparticles decorated bacteria. Bioresour Technol 337:125404. 10.1016/j.biortech.2021.125404. [DOI] [PubMed] [Google Scholar]
- 11.Tremblay J, Yergeau E, Fortin N, Cobanli S, Elias M, King TL, Lee K, Greer CW. 2017. Chemical dispersants enhance the activity of oil-and gas condensate-degrading marine bacteria. ISME J 11:2793–2808. 10.1038/ismej.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prince RC, McFarlin KM, Butler JD, Febbo EJ, Wang FC, Nedwed TJ. 2013. The primary biodegradation of dispersed crude oil in the sea. Chemosphere 90:521–526. 10.1016/j.chemosphere.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Kleindienst S, Paul JH, Joye SB. 2015. Using dispersants after oil spills: impacts on the composition and activity of microbial communities. Nat Rev Microbiol 13:388–396. 10.1038/nrmicro3452. [DOI] [PubMed] [Google Scholar]
- 14.Rahsepar S, Smit MP, Murk AJ, Rijnaarts HH, Langenhoff AA. 2016. Chemical dispersants: oil biodegradation friend or foe? Mar Pollut Bull 108:113–119. 10.1016/j.marpolbul.2016.04.044. [DOI] [PubMed] [Google Scholar]
- 15.Kleindienst S, Seidel M, Ziervogel K, Grim S, Loftis K, Harrison S, Malkin SY, Perkins MJ, Field J, Sogin ML, Dittmar T, Passow U, Medeiros PM, Joye SB. 2015. Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc Natl Acad Sci USA 112:14900–14905. 10.1073/pnas.1507380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremblay J, Fortin N, Elias M, Wasserscheid J, King TL, Lee K, Greer CW. 2019. Metagenomic and metatranscriptomic responses of natural oil degrading bacteria in the presence of dispersants. Environ Microbiol 21:2307–2319. 10.1111/1462-2920.14609. [DOI] [PubMed] [Google Scholar]
- 17.Lee K, Nedwed T, Prince RC, Palandro D. 2013. Lab tests on the biodegradation of chemically dispersed oil should consider the rapid dilution that occurs at sea. Mar Pollut Bull 73:314–318. 10.1016/j.marpolbul.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Prince RC, Butler JD. 2014. A protocol for assessing the effectiveness of oil spill dispersants in stimulating the biodegradation of oil. Environ Sci Pollut Res 21:9506–9510. 10.1007/s11356-013-2053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber L, Fortin N, Tremblay J, Wasserscheid J, Elias M, Mason J, Sanschagrin S, Cobanli S, King T, Lee K, Greer CW. 2019. Potential for microbially mediated natural attenuation of diluted bitumen on the coast of British Columbia (Canada). Appl Environ Microbiol 85:e00086-19. 10.1128/AEM.00086-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortmann AC, Cobanli SE, Wohlgeschaffen G, Thamer P, McIntyre C, Mason J, King TL. 2019. Inorganic nutrients have a significant, but minimal, impact on a coastal microbial community's response to fresh diluted bitumen. Mar Pollut Bull 139:381–389. 10.1016/j.marpolbul.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Gofstein TR, Perkins M, Field J, Leigh MB. 2020. The interactive effects of crude oil and Corexit 9500 on their biodegradation in Arctic seawater. Appl Environ Microbiol 86:e01194-20. 10.1128/AEM.01194-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh AK, Sherry A, Gray ND, Jones DM, Bowler BF, Head IM. 2014. Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front Microbiol 5:160. 10.3389/fmicb.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kujawinski EB, Reddy CM, Rodgers RP, Thrash JC, Valentine DL, White HK. 2020. The first decade of scientific insights from the Deepwater Horizon oil release. Nat Rev Earth Environ 1:237–250. 10.1038/s43017-020-0046-x. [DOI] [Google Scholar]
- 24.Pagaling E, Vassileva K, Mills CG, Bush T, Blythe RA, Schwarz‐Linek J, Strathdee F, Allen RJ, Free A. 2017. Assembly of microbial communities in replicate nutrient‐cycling model ecosystems follows divergent trajectories, leading to alternate stable states. Environ Microbiol 19:3374–3386. 10.1111/1462-2920.13849. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Ning D. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sierra-Garcia IN, Belgini DR, Torres-Ballesteros A, Paez-Espino D, Capilla R, Neto EVS, Gray N, de Oliveira VM. 2020. In depth metagenomic analysis in contrasting oil wells reveals syntrophic bacterial and archaeal associations for oil biodegradation in petroleum reservoirs. Sci Total Environ 715:136646. 10.1016/j.scitotenv.2020.136646. [DOI] [PubMed] [Google Scholar]
- 27.Kuppusamy S, Thavamani P, Megharaj M, Venkateswarlu K, Lee YB, Naidu R. 2016. Pyrosequencing analysis of bacterial diversity in soils contaminated long-term with PAHs and heavy metals: implications to bioremediation. J Hazard Mater 317:169–179. 10.1016/j.jhazmat.2016.05.066. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues EM, Morais DK, Pylro VS, Redmile-Gordon M, de Oliveira JA, Roesch LFW, Cesar DE, Tótola MR. 2018. Aliphatic hydrocarbon enhances phenanthrene degradation by autochthonous prokaryotic communities from a pristine seawater. Microb Ecol 75:688–700. 10.1007/s00248-017-1078-8. [DOI] [PubMed] [Google Scholar]
- 29.Sgro GG, Oka GU, Souza DP, Cenens W, Bayer-Santos E, Matsuyama BY, Bueno NF, Dos Santos TR, Alvarez-Martinez CE, Salinas RK, Farah CS. 2019. Bacteria-killing type IV secretion systems. Front Microbiol 10:1078. 10.3389/fmicb.2019.01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojo F. 2009. Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490. 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 31.Ghosal D, Ghosh S, Dutta TK, Ahn Y. 2016. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front Microbiol 7:1369. 10.3389/fmicb.2016.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Wang L, Shao Z. 2018. Polycyclic aromatic hydrocarbon (PAH) degradation pathways of the obligate marine PAH degrader Cycloclasticus sp. strain P1. Appl Environ Microbiol 84:e01261-18. 10.1128/AEM.01261-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasai Y, Kishira H, Harayama S. 2002. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl Environ Microbiol 68:5625–5633. 10.1128/AEM.68.11.5625-5633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park C, Park W. 2018. Survival and energy producing strategies of alkane degraders under extreme conditions and their biotechnological potential. Front Microbiol 9:1081. 10.3389/fmicb.2018.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregson BH, Metodieva G, Metodiev MV, McKew BA. 2019. Differential protein expression during growth on linear versus branched alkanes in the obligate marine hydrocarbon‐degrading bacterium Alcanivorax borkumensis SK2T. Environ Microbiol 21:2347–2359. 10.1111/1462-2920.14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neufeld JD, Schäfer H, Cox MJ, Boden R, McDonald IR, Murrell JC. 2007. Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J 1:480–491. 10.1038/ismej.2007.65. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y, Zhang B, Zhu Z, Song X, Cai Q, Chen B, Dong G, Ye X. 2020. Microbial eco-physiological strategies for salinity-mediated crude oil biodegradation. Sci Total Environ 727:138723. 10.1016/j.scitotenv.2020.138723. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, Hu Z, Wang H. 2019. Metagenomic analysis exhibited the co-metabolism of polycyclic aromatic hydrocarbons by bacterial community from estuarine sediment. Environ Int 129:308–319. 10.1016/j.envint.2019.05.028. [DOI] [PubMed] [Google Scholar]
- 39.Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ. 2016. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol 1:16057. 10.1038/nmicrobiol.2016.57. [DOI] [PubMed] [Google Scholar]
- 40.Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, Higuchi Y, Masai E. 2017. Bacterial catabolism of lignin‐derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep 9:679–705. 10.1111/1758-2229.12597. [DOI] [PubMed] [Google Scholar]
- 41.Wu Z, Nguyen D, Lam TY, Zhuang H, Shrestha S, Raskin L, Khanal SK, Lee P-H. 2021. Synergistic association between cytochrome bd-encoded Proteiniphilum and reactive oxygen species (ROS)-scavenging methanogens in microaerobic-anaerobic digestion of lignocellulosic biomass. Water Res 190:116721. 10.1016/j.watres.2020.116721. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Wang Y, Li Y, Santschi PH, Quigg A. 2017. Response of photosynthesis and the antioxidant defense system of two microalgal species (Alexandrium minutum and Dunaliella salina) to the toxicity of BDE-47. Mar Pollut Bull 124:459–469. 10.1016/j.marpolbul.2017.07.038. [DOI] [PubMed] [Google Scholar]
- 43.Mohapatra B, Phale PS. 2021. Microbial degradation of naphthalene and substituted naphthalenes: metabolic diversity and genomic insight for bioremediation. Front Bioeng Biotechnol 9:602445. 10.3389/fbioe.2021.602445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikel PI, Pérez‐Pantoja D, de Lorenzo V. 2016. Pyridine nucleotide transhydrogenases enable redox balance of Pseudomonas putida during biodegradation of aromatic compounds. Environ Microbiol 18:3565–3582. 10.1111/1462-2920.13434. [DOI] [PubMed] [Google Scholar]
- 45.Techtmann SM, Zhuang M, Campo P, Holder E, Elk M, Hazen TC, Conmy R, Santo Domingo JW. 2017. Corexit 9500 enhances oil biodegradation and changes active bacterial community structure of oil-enriched microcosms. Appl Environ Microbiol 83:e03462-16. 10.1128/AEM.03462-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFarlin KM, Prince RC, Perkins R, Leigh MB. 2014. Biodegradation of dispersed oil in arctic seawater at −1°C. PLoS One 9:e84297. 10.1371/journal.pone.0084297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang M, Chen B, Xin X, Song X, Liu J, Dong G, Lee K, Zhang B. 2021. Interactions between microplastics and oil dispersion in the marine environment. J Hazard Mater 403:123944. 10.1016/j.jhazmat.2020.123944. [DOI] [PubMed] [Google Scholar]
- 48.Cierniak D, Woźniak-Karczewska M, Parus A, Wyrwas B, Loibner AP, Heipieper HJ, Ławniczak Ł, Chrzanowski Ł. 2020. How to accurately assess surfactant biodegradation-impact of sorption on the validity of results. Appl Microbiol Biotechnol 104:1–12. 10.1007/s00253-019-10202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song X, Zhang B, Chen B, Lye L, Li X. 2018. Aliphatic and aromatic biomarkers for fingerprinting of weathered chemically dispersed oil. Environ Sci Pollut Res Int 25:15702–15714. 10.1007/s11356-018-1730-y. [DOI] [PubMed] [Google Scholar]
- 50.Tremblay J, Yergeau E. 2019. Systematic processing of ribosomal RNA gene amplicon sequencing data. GigaScience 8:giz146. 10.1093/gigascience/giz146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li D, Luo R, Liu C-M, Leung C-M, Ting H-F, Sadakane K, Yamashita H, Lam T-W. 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11. 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 56.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Palaniappan K, Szeto E, Pillay M, Chen I-MA, Pati A, Nielsen T, Markowitz VM, Kyrpides NC. 2015. The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v. 4). Stand Genomic Sci 10:86. 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S13. Download aem.02151-21-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)
Data Availability Statement
All raw sequence reads, including 16S rRNA gene amplicon sequencing, metagenome, and metatranscriptome, have been submitted to NCBI's SRA and are available under the BioProject PRJNA704368. Other associated information is available upon request.