ABSTRACT
Algae and heterotrophic bacteria have close and intricate interactions, which are regulated by multiple factors in the natural environment. Phages are the major factor determining bacterial mortality rates. However, their impacts on the alga-associated bacteria and thus on the alga-bacterium interactions are poorly understood. Here, we obtained a diatom-associated bacterium, Stappia indica SNL01, that could form a biofilm and had an inhibitory effect on the growth of the diatom Thalassiosira pseudonana. Meanwhile, phage SI01, with a double-stranded circular DNA genome (44,247 bp), infecting S. indica SNL01 was isolated. Phylogenetic analysis revealed that phage SI01 represents a novel member of the Podoviridae family. The phage contained multiple lysis genes encoding cell wall-lysing muramidase and spore cortex-lysing SleB, as well as depolymerase-like tail spike protein. By lysing the host bacterium and inhibiting the formation of biofilm, this phage could indirectly promote the growth of the diatom. Our results provide new insights into how phages indirectly regulate algal growth by infecting bacteria that are closely associated with algae or in the phycosphere.
IMPORTANCE The impact of phage infection on the alga-bacterium relationship in the ocean is poorly understood. Here, a novel phage infecting the diatom-associated bacterium Stappia indica SNL01 was isolated. This bacterium could form a biofilm and had a negative effect on diatom growth. We revealed that this phage contained multiple lysis genes and could inhibit the formation of the bacterial biofilm, thus indirectly promoting diatom growth. This study suggests that phages not only are important regulators of bacteria but also have substantial indirect effects on algae and the alga-bacterium relationship.
KEYWORDS: alga-bacterium interaction, bacteriophage lysis, biofilm-forming bacteria, diatom-associated bacteria, phage
INTRODUCTION
The marine ecosystem is driven by a network of interactions between diverse organisms. Of these, the interactions between autotrophic algae and heterotrophic bacteria are of significant importance. It is estimated that algae contribute about 50% of global photosynthesis and, together with bacteria, they are the key drivers of the marine carbon cycle (1). Bacteria, on the other hand, are found in all realms of the ocean, regulating all biogeochemical cycles of the marine ecosystem supplying inorganic nutrients for the growth of algae (2). Moreover, algae and heterotrophic bacteria are in constant interaction with each other, sharing various relationships shaped by themselves and/or the surrounding environment, which further intensifies their role in regulating marine biogeochemistry (1, 2).
The relationships of algae and bacteria are not stable and are influenced by multiple factors. The major biotic influencers are the marine viruses (phages), which are highly abundant in the oceans, with an average concentration of 107 per milliliter of surface seawater (3). In other words, there are at least 10 phages per bacterial cell (3). It is estimated that phages lyse about 20% of the marine microbial biomass daily, demonstrating their influential grip on the marine nutrient and energy cycle (4). Moreover, phage infection can have a significant impact on the phytoplankton-bacterium interactions and the surrounding environment (5–8). The organic nutrient-rich environment (phycosphere) surrounding algal cells serves as a hub for interactions between algae and bacteria (1), with bacterial concentrations in such environments frequently being multiple orders of magnitude higher than in surrounding waters (1), which makes them more susceptible to phage lysis (9). Also, it has been observed that the algal phycosphere is frequently dominated by a few specific bacterial lineages that are actively involved in symbiotic chemical interactions with the host algae (1, 10). According to the “kill the winner”/“piggyback the winner” hypothesis (8, 11–15), in this scenario phages would more likely infect the dominant bacteria, which could significantly affect the alga-bacterium interaction and might substantially affect the algal fitness. However, phages can also benefit the fitness of the host algae by regulating the abundance of or completely eliminating the harmful bacteria associated with the algae (16). Furthermore, phage lysis of bacterial cells would also increase organic nutrient load within the phycosphere (8, 17), which would boost noninfected bacterial metabolism (conversion of organic nutrients to inorganic nutrients), altering bacterial community structure (11, 18, 19) and providing more inorganic nutrients to the host algae (3, 20, 21).
During our investigation of the heterotrophic bacterial community associated with a model diatom, Thalassiosira pseudonana CCAP 1085, we observed that one particular bacterium was tightly associated with the diatom and could not be eliminated with mechanical and antibiotic treatments. Isolation and 16S rRNA gene identification suggested that the bacterium belonged to the genus Stappia and could form biofilms, which might have aided in its tight association with the diatom (22, 23). Biofilm formation by heterotrophic bacteria can aid in host colonization (24, 25) and essential nutrient supplementation (26), enhance algal aggregation (27–29), inhibit the entry of other bacteria within the phycosphere (30), and can even regulate host algal fitness (31). Moreover, phages are known to inhibit/disrupt bacterial biofilms (32, 33). Given that bacterial biofilms have such a profound effect on algae, phage modulation of bacterial biofilms or the bacteria within the phycosphere will have an indirect impact on the host algae. Isolates belonging to the genus Stappia were previously reported to be associated with algae (27, 34–37) and in various biofilm environments (38–42). The unique genomic capability of genus Stappia, such as the presence of biofilm-forming genes (43), as well as a wide variety of functionally important genes such as rhizobactin-like siderophore (44) for iron scavenging, could promote their interaction with the algae. Despite such evident signs of tight association with algae, little is known about their influence on algae or their phages. In fact, to date there has been no report of any lytic bacteriophage infecting strains of the genus Stappia. Thus, we hypothesized that phage infection of Stappia strains associated with algae would have a substantial impact on the host algal growth. To verify our hypothesis, we isolated and characterized a novel phage, SI01, and revealed that it could significantly promote the growth of diatoms by infecting the associated bacterium Stappia indica SNL01.
RESULTS
Stappia indica SNL01 was closely associated with diatom T. pseudonana.
A laboratory-grown culture of the diatom Thalassiosira pseudonana CCAP 1085, after physical and commonly used antibiotic treatments (45, 46), was found to be closely associated with the bacterium Stappia indica SNL01. From the Kirby-Bauer disc diffusion test (see Table S1 in the supplemental material) and depending on the mode of action (see Table S2), two S. indica SNL01-targeting antibiotic combinations (see Table S3) were prepared and tested to eliminate SNL01 colonization of the T. pseudonana culture. However, both antibiotic combinations failed to eliminate SNL01 colonization at an optimum antibiotic concentration (without inhibiting T. pseudonana growth). For detailed results, see the supplemental material. Further investigations revealed that S. indica SNL01 could form biofilms (see Fig. S1).
S. indica SNL01 inhibits diatom T. pseudonana growth at higher concentrations.
The influence of S. indica SNL01 on diatom T. pseudonana growth was evaluated. From chlorophyll intensity data, it was found that S. indica SNL01 at higher concentrations (106 to 107 cells mL−1) could significantly (P = 0.0008) inhibit the growth of T. pseudonana, compared to that of the control group (Fig. 1a).
FIG 1.
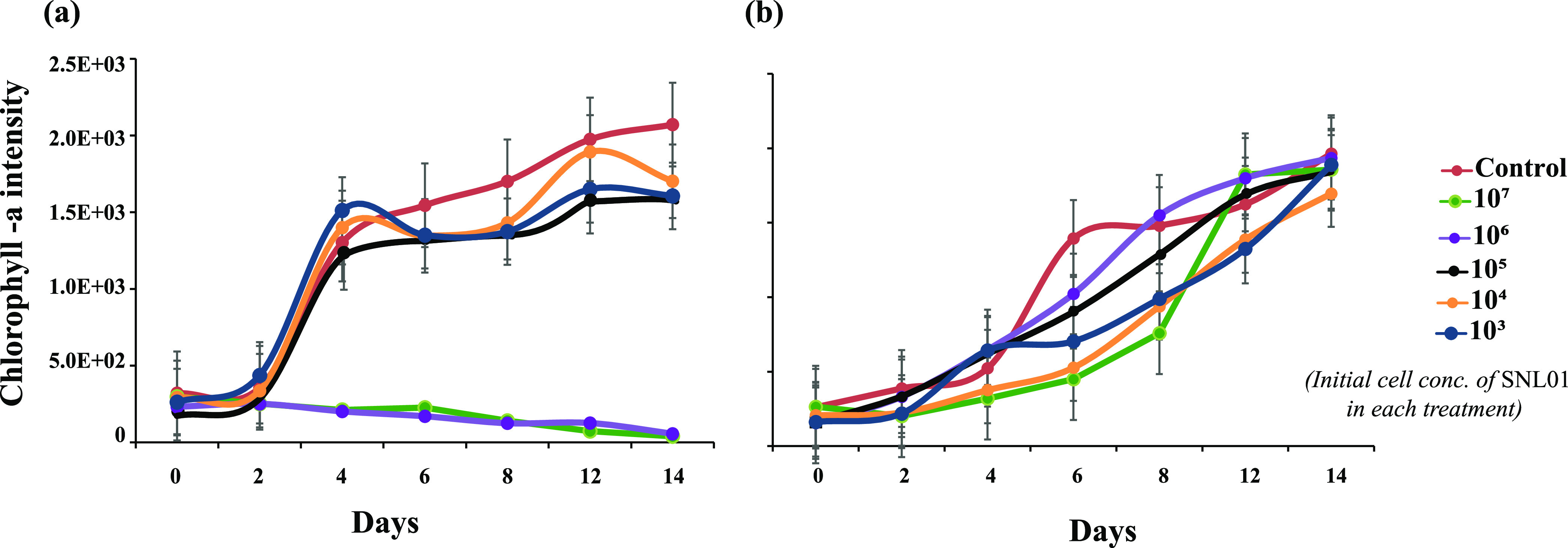
Effect of S. indica SNL01 on the diatom T. pseudonana. (a) Set I, in the absence of phage SI01. (b) Set II, in the presence of phage SI01. The key indicates the initial concentration of SNL01 for each treatment. The initial concentration of diatom in all treatments was constant (105 cells mL−1), while that of the phage in set II was an MOI of 10. The control in set I consisted of only T. pseudonana, while the control in set II contained T. pseudonana and phage SI01. Each treatment is a replicate three of samples.
Phage SI01 promotes diatom T. pseudonana growth by lysing S. indica SNL01 bacteria.
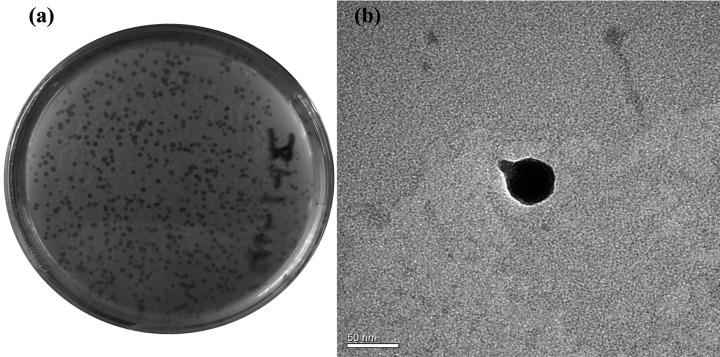
Phage SI01 was enriched and isolated from coastal seawater collected from Qingdao, China. It was observed that phage SI01 could actively lyse the S. indica SNL01 lawn, producing 0.5- to 1-mm clear spots within 12 h of inoculation on a soft agar plate (Fig. 2a). Furthermore, phage SI01 could also significantly disrupt the biofilms of S. indica SNL01 (P = 0.0004) (see Fig. S1). When it was introduced in a T. pseudonana-S. indica SNL01 interaction system, the S. indica SNL01 growth-inhibiting effect (at higher concentrations) was substantially suppressed (Fig. 1b).
FIG 2.
Phage morphology. (a) Plaque morphology of phage SI01 on a SNL01 lawn using a double-layer agar plating method. (b) TEM micrograph of phage SI01, showing a short noncontractile tail (∼17 nm in length) and an icosahedral head (∼45 nm in diameter). Scale bar, 50 nm.
Morphological, genomic, and phylogenetic characteristics of phage SI01.
From transmission electron microscopy (TEM) analysis, the phage showed the presence of a short noncontractile tail of ∼17 nm (Fig. 2b), a characteristic feature of the Podoviridae family (47), and an icosahedral capsid ∼45 nm in length and similar in diameter (Fig. 2b). Furthermore, we analyzed and characterized the genome of phage SI01. The phage genome consists of double-stranded DNA with a 44,247-bp genome and an average GC content of 58.3% (Fig. 3), which is similar to closely related phages (as suggested by NCBI-BLAST and the ViPTree algorithm) (see Table S5). PhageTerm and PCR analyses showed that the phage has a circular genome with short direct terminal repeats of 207 bases. Furthermore, the phage SI01 genome contains a total of 50 predicted open reading frames (ORFs), with 17 of them showing high similarity to previously defined functions (see Table S4). Interestingly, of the 50 predicted genes, 49 were located on the reverse strain, suggesting an unbalanced topology. The functionally annotated proteins covered modules for phage structure and packaging, DNA replication and metabolism, and lysis.
FIG 3.
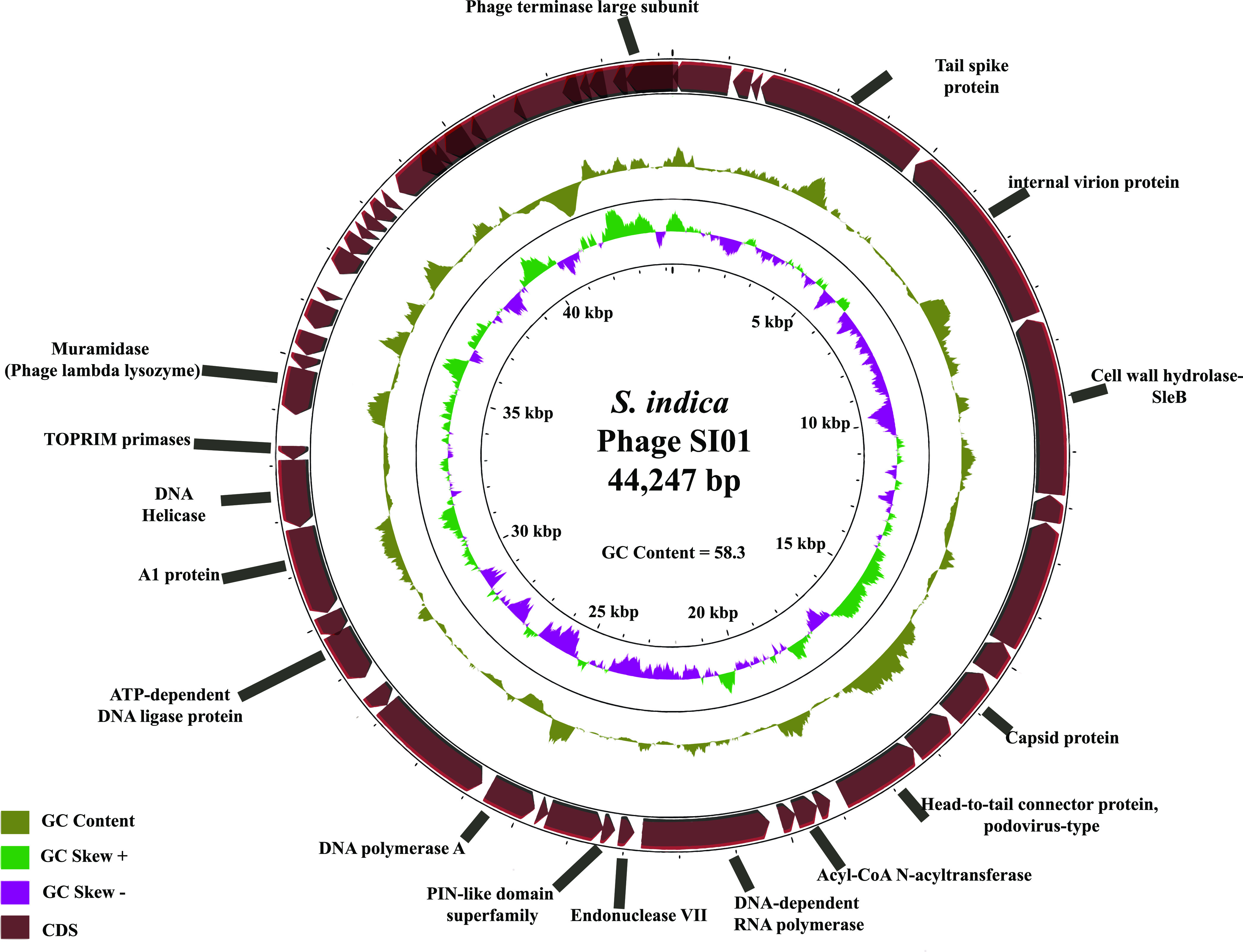
Genomic characteristics of phage SI01. In this circular representation of phage SI01, the rings (from outer to inner) represent predicted ORFs, with arrows (maroon) pointing in the strand direction, GC content (khaki green), positive GC skew (light green), and negative GC skew (magenta).
Five genes among the annotated genes represented the structure and packaging module, namely, those for phage terminase large subunit, tail spike/fiber protein, capsid protein, internal virion protein, and head-to-tail connector protein, all determining the structure and packaging mechanism of phage SI01. The DNA replication/metabolism module included eight genes, i.e., those for DNA polymerase A, TOPRIM primase, recombination endonuclease VII, a PIN-like domain superfamily protein, a putative ATP-dependent DNA ligase protein, acyl-coenzyme A (CoA) N-acyltransferase, DNA helicase, and A1 protein.
Finally, the lysis module was represented by three genes, i.e., two muramidase (peptidoglycogen-targeting lysozyme) genes, namely, those for SleB (gene 6) and lambda phage-like lysozyme (gene 29), and one depolymerase-like tail spike protein gene (gene 4). The two muramidases are bacterial cell wall-targeting lysins that work during the initial phage infection and at the end of the phage lytic cycle (48). While SleBs are lytic transglycosylase proteins capable of cleaving bacterial spore cortex (49), lambda phage-like lysozyme is a peptidoglycogen-targeting bacterial cell wall-cleaving protein (48). The third lysis gene is the tail spike protein gene (gene 4), showing high similarity to T7 tail fiber protein and 30.8% similarity (BLASTp E value of 1e−06) to the depolymerase protein of Klebsiella phage K5-4 (see Fig. S2). Additionally, no tRNAs were found within the SI01 genome, which is in contrast to the assumption that the presence of tRNAs is a characteristic of a more highly virulent phage (50).
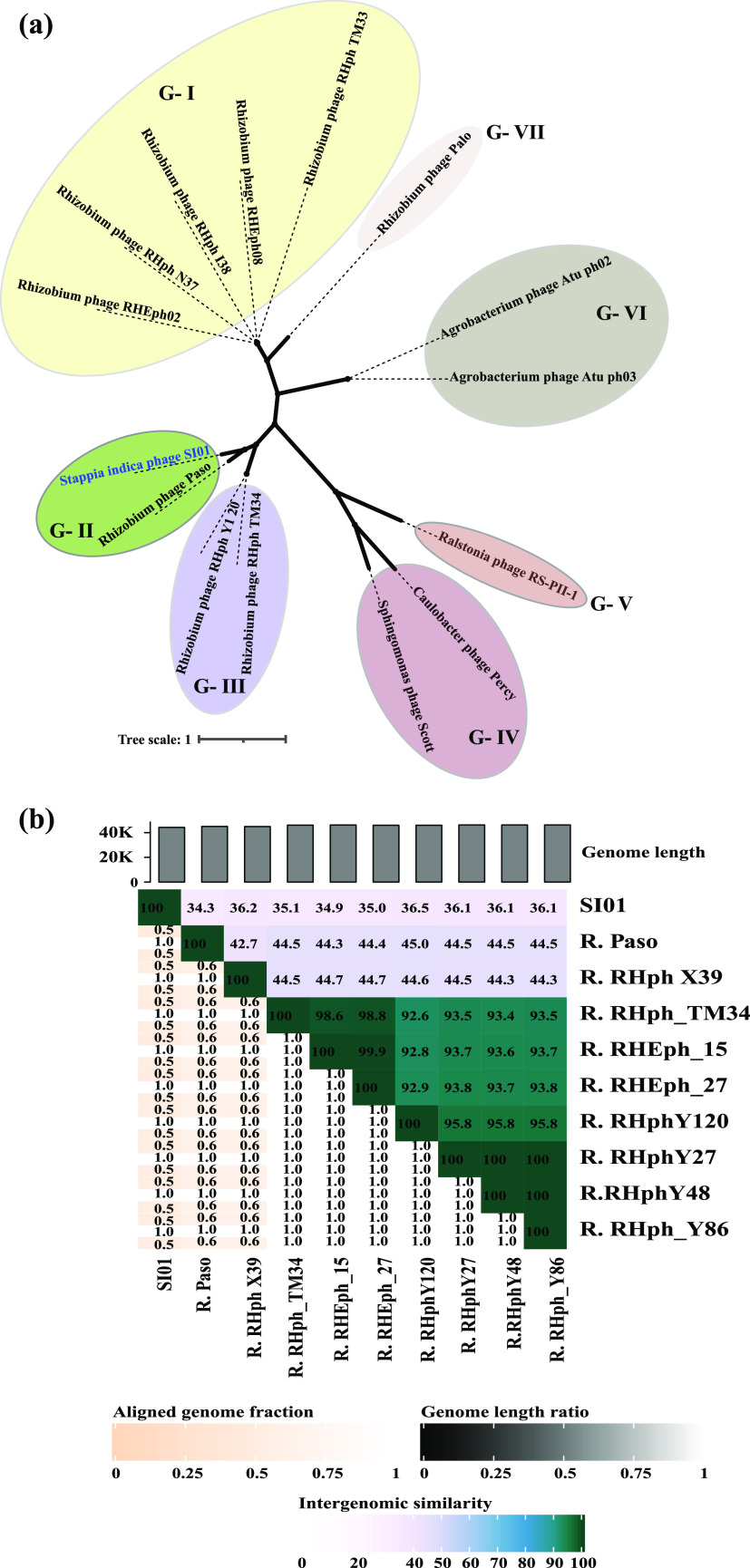
Moreover, the amino acid sequences of the phage terminase large subunit of 14 related phages and of SI01 were used for phylogenetic analysis. A nonrooted tree was constructed, grouping the 15 organisms into seven groups (Fig. 4a). Phage SI01 shared a node with Rhizobium phage Paso (node G-II) (Fig. 4a). Intergenomic distance calculations by VIRIDIC (which uses the same algorithm as that used by the International Committee on Taxonomy of Viruses [ICTV], Bacterial, and Archaeal Viruses Subcommittee) revealed that phage SI01 is less than 35% similar to Rhizobium phage Paso and other closely related phages (Fig. 4b). Furthermore, the genome similarity algorithm of ViPTree (51) suggested an SG score of less than 0.25 for phage SI01. Thus, from the phylogenetic analysis and intergenomic distance calculations, it is clear that phage SI01 represents a novel phage belonging to the family Podoviridae.
FIG 4.
Phylogenetic relationship between phage SI01 and closely related phages. (a) Unrooted tree drawn by aligning the phage terminase large subunits of 14 related phages via Clustal Omega and the neighbor-joining method with 1,000 bootstrap subsamples. (b) Heatmap representation of the intergenomic similarity between the complete genomic sequences of closely related phages, as calculated by the VIRIDIC tool.
Infection and biophysical stability characteristics of phage SI01.
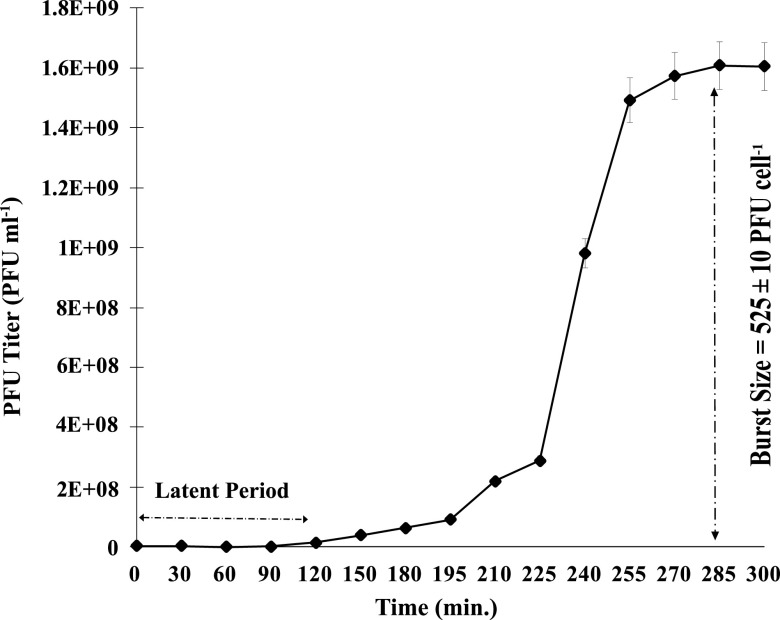
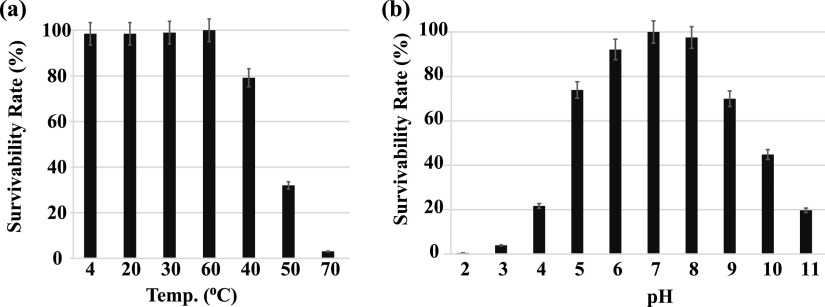
In the one-step growth experiment conducted to analyze the infection pattern of phage SI01 (Fig. 5), the phage showed a longer latent period of 120 min, compared with a few other marine Podoviridae phages (52–54), followed by a burst period of 150 min. The growth was stabilized after 255 min from the time of inoculation, with a burst size of 525 ± 10 PFU cell−1. The relatively large burst size is not surprising and might be attributed to the longer latent period (55). The host specificity of S. indica SNL01 was evaluated by using 30 strains related to different genera (Table 1). It was observed that phage SI01 was highly specific in its infection activity and could not lyse any tested strains except S. indica SNL01. Furthermore, to check the phage’s ability to cope with environmental changes, we tested phage SI01’s stability with a series of temperature and pH profiles. It was observed that phage SI01 was highly stable in a temperature range of 4 to 40°C, with a gradual decrease in survival ability between 50 and 70°C. Beyond 80°C, the phage was completely inactivated (Fig. 6a).
FIG 5.
One-step growth curve for phage SI01 on host S. indica SNL01 at 37°C and pH 8.0. Each point is an average of two replicates, with standard error bars.
TABLE 1.
Host range of phage SI01 against 30 selected bacterial strains
| Species | Strain | Isolation source | SI01 lysis effect |
|---|---|---|---|
| Stappia indica | SNL01 | Diatom culture | + |
| Stappia indica | 7002-212 | Cyanobacterial culture | − |
| Stappia indica | 7002-240 | Cyanobacterial culture | − |
| Stappia stellulata | HHTE138 | Seaweed culture | − |
| Stappia stellulata | HHTE115-1 | Seaweed culture | − |
| Alcanivorax xenomutans | SN006 | Cyanobacterial culture | − |
| Mesorhizobium sediminum | SN007 | Cyanobacterial culture | − |
| Georhizobium profundi | SN035 | Cyanobacterial culture | − |
| Nitratireductor aquibiodomus | SN008 | Cyanobacterial culture | − |
| Pseudomonas stutzeri | SN018 | Cyanobacterial culture | − |
| Erythrobacter citreus | SN021 | Cyanobacterial culture | − |
| Rhizobium aethiopicum | SN019 | Cyanobacterial culture | − |
| Marinobacter algicola | SN025 | Cyanobacterial culture | − |
| Bacillus mycoides | SN033 | Cyanobacterial culture | − |
| Aureimonas altamirensis | SN036 | Cyanobacterial culture | − |
| Gordonia alkanivorans | SN039 | Cyanobacterial culture | − |
| Vibrio alginolyticus | VB-288 | Collection | − |
| Vibrio harveyi | VB-645 | Collection | − |
| Vibrio campbellii | VB-18-1 | Collection | − |
| Vibrio scophthalmi | HS018 | Seawater | − |
| Vibrio azureus | HS031 | Seawater | − |
| Vibrio alginolyticus | HS058 | Seawater | − |
| Vibrio alfacsensis | HS062 | Seawater | − |
| Vibrio gigantis | HS128 | Seawater | − |
| Vibrio zhuhaiensis | HS171 | Seawater | − |
| Vibrio atlanticus | HS179 | Seawater | − |
| Vibrio scophthalmi | HS191 | Seawater | − |
| Vibrio zhuhaiensis | HS218 | Seawater | − |
| Vibrio chagasii | HS262 | Seawater | − |
| Vibrio scophthalmi | HS282 | Seawater | − |
FIG 6.
Biophysical characteristics of phage SI01. (a) Effect of temperature on the survivability rate. (b) Effect of pH on the survivability rate. Each point is an average of three replicates, with standard error bars.
Moreover, phage SI01 showed efficient lysis activity in a pH range of 6 to 8 (optimum, 7 to 8), well in line with the average seawater pH of 8.2 ± 0.3 (56). The phage survival ability was reduced to 70% at pH 5 and 9. A further drop in pH below 5 and an increase above 9 showed drastic reductions in phage activity (Fig. 6b). Thermal and pH profiles showed that phage SI01 could easily cope up with the seasonal and geographical temperature and pH fluctuations in the ocean, as well as that of changing climate (57). Phage SI01 was also found to be insensitive to chloroform, suggesting the absence of lipids in the phage capsid, which is also in line with the results for previously isolated Podoviridae phages (47).
DISCUSSION
Algae and bacteria are in constant interaction with each other in the ocean, and the fate of their interaction greatly influences carbon and other nutrient fluxes, eventually affecting the marine food web (1, 58). Phages, being the most abundant organisms in the ocean, are the main factor determining bacterial mortality rates and can infect about 20 to 30% of bacteria at any given time (3). Phage infection undoubtedly affects the interaction between bacteria and algae. However, to date little is known about the influence of phages on alga-bacterium interactions.
Here, S. indica SNL01 was closely associated with a laboratory-grown culture of the diatom T. pseudonana and could hardly be eliminated with mechanical and antibiotic treatments (see Table S3 in the supplemental material). Intriguingly, pure S. indica SNL01 isolates were susceptible to several tested antibiotics when tested on agar plates (see Table S1) but they could not be removed from T. pseudonana culture using the same antibiotics (see Table S3). Further investigations revealed that S. indica SNL01 could form biofilms (see Fig. S1), which might have helped it in tightly binding to diatom cells and evading the antibacterial effect of antibiotics in T. pseudonana culture. Biofilm-forming bacteria are typically resistant to most available antibiotics and disinfection methods (23, 59, 60), aiding in the survival of the bacteria within the biofilm environment (23). In fact, strains of Stappia indica have been found to be unaffected by antibiotic treatments due to their biofilm-forming nature (39). However, bacteriophages can infect bacteria within the biofilm environment (32, 33, 61, 62). Thus, we isolated and characterized phage SI01 infecting S. indica strain SNL01 and verified its efficiency in disrupting S. indica biofilms.
Moreover, we revealed that, at higher cell concentrations of S. indica SNL01 (compared with the T. pseudonana cell concentration), the bacterium could inhibit the growth of T. pseudonana (Fig. 1a). However, in the presence of phage SI01 (multiplicity of infection [MOI] of 10), the growth inhibitory effect of S. indica SNL01 was significantly suppressed (Fig. 1b) and the T. pseudonana growth was comparable to that of the control culture. This might be due to the complete or partial eradication of S. indica SNL01 (resulting in the suppression of SNL01 cell abundance) from the coculture system. Previously, it was seen that phages could promote algal growth by infecting the growth-inhibiting bacteria (16). Additionally, phage lysis of bacterial cells releases organic nutrients, which by microbial transformation increases the inorganic nutrient pool, ultimately boosting the algal growth (3, 20, 21). Here, phage SI01 could efficiently lyse the planktonic form as well as the biofilm of S. indica SNL01 (Fig. 2a; also see Fig. S1) within 12 h of infection, indicating its consequential role in altering S. indica SNL01 and the associated phytoplankton relationship.
This is the first report of isolation and characterization of a phage infecting strains of the Stappia genus. Phage SI01 is a novel phage belonging to the Podoviridae family, with little relatedness to other Podoviridae phages (Fig. 4a and b); it is stable at an optimum temperature and pH range, with a relatively long latent period and large burst size. The latent period of bacteriophages depends primarily on the host's growth rate (63, 64). If the host's growth is faster, then phages will shorten their latent period and vice versa (65). The host SNL01 takes 6 to 7 h to enter the exponential growth phase (data not shown), making the bacteriophage lengthen its latent period. Reports suggest that a longer latent period maximizes phage progeny generation, reflected in a large burst size (65), which might be the reason for the larger burst size of the phage SI01 (Fig. 5). Intriguingly, phage SI01 and other closely related phages (as suggested by NCBI-BLAST and the ViPTree algorithm) have hosts that are known to be closely associated with autotrophs (see Table S5) and need further detailed investigations.
Furthermore, we explored the genomic features of phage SI01 that might be contributing to the disruption of the host SNL01. Phage SI01 possesses a double-stranded circular DNA with a 44,247-bp genome, which is comparable to other Podoviridae phages (66, 67). On investigation for genes that could aid in lysing host bacteria, we identified two genes coding for cell wall-lysing enzymes (muramidases), namely, SleB and lambda phage-like lysozyme (Fig. 3; also see Table S4). Although muramidases and lysozymes are known to lyse bacterial cell walls postinfection to release the virions outside the bacterial cell, they may also have negative effects on bacterial aggregation, adhesion, and biofilm formation (68). It was reported that reduced secretion of cell wall-hydrolyzing/lysing enzymes increased cell aggregation and sedimentation and biofilm formation in bacteria (69, 70). Similarly, the application of muramidase enzyme has been shown to reduce the adhesive efficiency of biofilms (71). A subpopulation of bacteria in the biofilm are known to go into dormant inactive mode, the same as spore formation (23). Activation of SleB proteins aids in degrading the spore cortex (outer layer protecting dormant bacterial spores) and is highly regulated in some spore-forming bacteria (49, 72). This suggests that the presence of SleB and SleB like genes within the bacteriophage genome might be advantageous for penetrating dormant bacterial populations within biofilm environments. Although SleB-like genes have been observed in diverse phages (73–75), their exact role in phage lysis has not yet been reported, and detailed studies are needed to assess this hypothesis.
A peculiar feature of Podoviridae phages is the presence of depolymerase proteins, i.e., tail spike/fiber proteins that are capable of cleaving bacterial capsular polysaccharides (CPS), exopolysaccharides (EPS), or lipopolysaccharide (LPS) (which are the main components of biofilms) and are currently gaining wide attention as antibiofilm agents (32, 76–78). Given that phage SI01 belongs to the Podoviridae family, encodes a tail spike protein that is similar to known depolymerase proteins (see Fig. S2), and lyses S. indica SNL01 biofilm with high efficiency (see Fig. S1), SI01’s tail spike protein might possess depolymerase activity and might play an important role in disrupting SNL01 biofilm.
The tail spike protein depolymerase enzyme might aid in cleaving the bacterial CPS/EPS/LPS layer, followed by phage attachment and replication. Once the phage progenies are replicated, the phage would secret cell wall-lysing enzymes to rupture the bacterial cell wall, eventually killing the bacteria. This highly specific and effective lysis of S. indica SNL01 bacteria by phage SI01 eventually suppresses the bacterial growth, aiding in the weakening of the negative impact of SNL01 bacteria and promoting the healthy growth of the diatoms.
MATERIALS AND METHODS
Isolation and identification of alga-associated bacteria.
A laboratory-grown culture system of Thalassiosira pseudonana CCAP 1085 was found to be colonized by a single type of bacteria after treatment with commonly used antibiotic treatments (ampicillin, gentamicin sulfate, kanamycin, streptomycin, and ciprofloxacin) and mechanical axenization methods (45, 46). The bacteria were then isolated by spreading 100 μl of a serially diluted aliquot from the culture on solid marine agar 2216E medium (Hope Bio-Technology, Qingdao, China). The isolated strain was purified three times and employed for DNA extraction via a TIANamp bacteria DNA kit (Tiangen). The 16S gene from the extracted DNA was amplified using primers 27F and 1492R (79) under the following conditions: denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min 30 s with 30 cycles. Amplified PCR product was sequenced at Sangon Biotech (Shanghai, China) and identified as Stappia indica SNL01 by aligning the high-quality sequence with the EzBioCloud database (80). In addition, the antibiotic susceptibility of strain S. indica SNL01 was assessed as detailed in the supplemental material.
Effect of S. indica SNL01 on the growth of the diatom T. pseudonana CCAP 1085.
To evaluate the influence of S. indica SNL01 on the diatom T. pseudonana CCAP 1085, the diatom and the bacterium were cultured to the exponential growth phase and later inoculated in a multiwell plate in triplicate. In brief, S. indica SNL01 at various initial inoculum concentrations (103 to 107 cells mL−1) was added to the diatom (105 cells mL−1) in F/2 medium to a final volume of 1 mL. Prior to inoculation, the bacterial cells were washed twice with sterile F/2 solution. A control group was prepared with diatom (105 cells mL−1) in F/2 medium without the addition of SNL01. The culture setup was incubated for 15 days in an illumination chamber with a 12-h/12-h day/night cycle at a temperature of 28°C. Chlorophyll a intensity was taken as a measure of the growth of T. pseudonana CCAP 1085 and was quantified every 2 days with a Biotek Synergy HT reader (USA) at excitation and emission wavelengths of 440 nm and 680 nm, respectively.
Isolation and purification of phage infecting S. indica SNL01.
For phage isolation, water samples were collected from the coast of the Yellow Sea, China, and filtered through 0.22-µm-pore-size polycarbonate filters to remove microbial cells and larger particles. Five milliliters of the filtrate was inoculated with 100 mL of an overnight liquid culture of Stappia indica SNL01 and was incubated for 24 h at 30°C in a shaker. Plaque assays were performed by the double-layer agar plating method at 8 h, 24 h, and 48 h to determine phage infection (55). In brief, 1 mL of the suspension was filtered through a 0.22-µm polycarbonate filter, serially diluted with SM buffer, and inoculated with 1 mL of log-phase Stappia indica SNL01 in sterile polycarbonate tubes. The tubes were gently mixed and incubated for 20 min at 30°C in the dark in a shaker. Later, the suspension was mixed with 5 mL of sterile soft 2216E agar (0.5%) and spread on a solid 2216E agar plate (1.8%). The plates were incubated at 30°C overnight and observed for cleared patches (putative plaques). A single phage plaque was picked, resuspended in 100 μl of SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5]), and purified three times following the double-layer agar plating method described above. A high-titer phage lysate was obtained by enriching the phage suspension with log-phase Stappia indica SNL to 1 L.
The enriched phage suspension was centrifuged at 10,000 × g at 4°C for 15 min, filtered through 0.22-µm-pore-size polycarbonate filters, and treated with DNase I and RNase A (2 ng L−1) for 1 h at room temperature to remove/digest bacterial contamination. The pure phage lysate was precipitated overnight at 4°C by adding polyethylene glycol 8000 (final concentration, 10% [wt/vol]) and NaCl (final concentration, 1 M). The lysate was centrifuged at 10,000 × g at 4°C for 90 min, and the resulting precipitate was collected in 5 mL of SM buffer. The concentrated phage lysate was purified by CsCl gradient (gradient density of 1.7, 1.5, and 1.3 g mL−1) ultracentrifugation at 200,000 × g for 8 h at 4°C (CP-100WX; Hitachi Ltd., Tokyo, Japan). The purified phage particles (blue band) were collected and dialyzed three times in SM buffer via a 30-kDa superfilter (UFC5030; Millipore).
Morphological characterization of phage infecting S. indica SNL01.
Phage morphology was observed by TEM. In brief, samples were negatively stained with 2% (wt/vol) aqueous uranyl acetate (pH 7.0) on a 200-mesh copper grid and observed using an H-7650 transmission electron microscope (Hitachi). Micrographs were taken at an accelerating voltage of 80 kV via a charge-coupled-device (CCD) image transmission system (Gatan Inc., Pleasanton, CA, USA).
Infection pattern and biological characterization of phage SI01.
A one-step growth assay was performed as described previously (55) to investigate the phage infection pattern. In brief, 1 mL of exponentially growing S. indica SNL01 (107 CFU mL−1) and phage lysate were mixed at a MOI of 0.01 and incubated for 20 min at 37°C. The mixture was centrifuged at 6,000 × g for 10 min at 4°C. The supernatant was discarded to remove the nonadsorbed phages, and the pellet was washed twice in sterile 22I6E medium and resuspended in an equal volume of 22I6E medium. An aliquot of 100 µL of this resuspension was transferred to 100 mL of 2216E medium and incubated at 30°C in a shaker. The experiment was set up in duplicate. Samples were taken every 30 min for the first 3 h, followed by every 15 min until the optical density at 600 nm (OD600) had stabilized. The sampling interval (every 30 min for the first 3 h) was determined by monitoring the phage infection pattern of SNL01 by OD600 prior to the actual one-step growth curve experiment. The burst size was calculated by dividing the average titers for the postburst time points by the average initial titers.
The phage host range was determined by spot assay with the bacterial strains listed in Table S1 in the supplemental material. For this, 200 µL of exponentially growing bacterial culture was spread on 1.8% 2216E agar, and 2-µL spots of concentrated phage lysate were spotted in multiple places. The plates were incubated for 24 to 72 h and checked for any clear patches. A control was set by spotting phage lysate on the S. indica SNL01 spread plate.
The thermal stability of SI01 was evaluated by incubating the phage lysate in SM buffer at 4, 20, 30, 40, 50, 60, 70, 80, and 90°C for 2 h. For the pH stability test, SI01 lysate was inoculated in SM buffer with pH values ranging from 2 to 12 at 37°C for 2 h. The double-layer plate method was used to assess the survival rate. Additionally, for lipid detection, 1 mL of phage lysate was mixed with 20 µL (i.e., 2% [vol/vol]) and 200 µL (i.e., 20% [vol/vol]) of pure chloroform, mixed vigorously for 1 min, and incubated at room temperature for 30 min. The suspension was centrifuged at a low speed (3,000 × g), and the supernatant containing phages was spotted on an S. indica SNL01 plate for assessment of phage activity.
Phage SI01 DNA extraction, sequencing, and bioinformatics.
DNA from the CsCl-purified phage lysate was extracted as described previously (55). In brief, the lysate was mixed with 1 mL of extraction buffer (2× cetyltrimethylammonium bromide [CTAB], 100 mM Tris-HCl, 20 mM EDTA [pH 8.0], and 1.4 M NaCl, in 100 mL of double-distilled water), 0.4% SDS (4 µL), and 10 µL of 20 mM proteinase and EDTA (0.5 mol L−1 [pH 8.0]) and incubated at 60°C for 2 h with occasional shaking. The extracted DNA was separated from cell debris by phenol/chloroform/isoamyl alcohol (25:24:1) separation. Further, the DNA-containing phase was precipitated overnight at −20°C by adding 2/3 volume of ice-cold absolute ethanol and 250 µL of 10 M ammonium acetate. Precipitated DNA was pelleted by centrifugation at 12,000 × g, washed twice with ice-cold 70% ethanol, and resuspended in 60 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). DNA quality was checked via NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and integrity was checked via gel electrophoresis before sequencing.
DNA libraries were prepared using TruSeq DNA library preparation kits (Illumina, San Diego, CA, USA), and sequencing was performed using the Illumina HiSeq X 10 platform (Illumina, USA) to obtain paired-end reads (150 bp each). The reads were quality filtered and assembled into scaffolds using SPAdes v3.14.1 (81). Phage termini and the packaging mechanism were determined via PhageTerm v1.0.11 (82), and the genome type was determined via PCR amplification (83). A consensus set of ORFs (detected by at least two gene prediction tools) were filtered from Prodigal v2.6.3 (84), GeneMarkS v2 (85), and GLIMMER v3.02b (86). Genes were annotated via eggNOG-mapper v2.13 (87), InterProScan v5.51-85.0 (88), and ViPTree server v1.9 (51) using default settings. The annotations were manually checked using BLASTp against the NCBI nonredundant protein database and NCBI RefSeq viral database with an E value of <10−4. Transmembrane domains were identified with TMHMM (89) and tRNA genes with tRNAscan-SE v2.0 (90) with default settings.
Phylogenetic tree construction.
The protein sequences of the phage terminase large subunit of 14 related phages, as identified by the ViPTree server (51) and NCBI BLAST, were pulled into a single fasta file along with the phage SI01 genome and aligned via Clustal Omega v1.2.2 (91). The phylogenetic tree was then generated via MEGA v7.026 (92) by the neighbor-joining method (93) with 1,000 bootstrap samples. Furthermore, intergenomic distances between the complete genomic sequences of closely related phages (as suggested by NCBI BLAST) were calculated via the VIRIDIC tool (94) using recommended settings for prokaryotic viruses. The results were then used to infer a distance similarity heatmap.
Influence of phage SI01 on the relationship between the diatom and the bacterium.
To evaluate the influence of phage SI01 on the relationship between T. pseudonana and S. indica SNL01, the diatoms and the bacteria were cultured until the exponential growth phase and later inoculated in a multiwell plate in triplicate as described above. Here, each diatom-bacterium treatment (different concentrations) was inoculated with phage SI01 at an MOI of 10. The control group contained the diatom with phage (MOI of 10). The culture conditions were kept the same as described above, and chlorophyll a intensity was measured every 2 days.
Statistical analysis.
All results are reported as the means ± standard deviations (SDs) from triplicate samples. Student's t test was used to establish statistical significance, with P values of <0.05 indicating significance.
Data availability.
The sequences reported in this paper have been deposited in the NCBI GenBank database under accession number MZ462995.
ACKNOWLEDGMENTS
This work was funded by NSFC projects 41876174, 31700104, 42106107, 41806172, and 42006093, the Shandong Provincial Key Research and Development Plan (2021ZDSYS29), and the Senior User Project of RV KEXUE (grant KEXUE2019GZ03) supported by the Center for Ocean Mega-Science, Chinese Academy of Sciences.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Zenghu Zhang, Email: zhang_zh@qibebt.ac.cn.
Yongyu Zhang, Email: zhangyy@qibebt.ac.cn.
Hideaki Nojiri, University of Tokyo.
REFERENCES
- 1.Seymour JR, Amin SA, Raina JB, Stocker R. 2017. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065. 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 2.Amin SA, Parker MS, Armbrust EV. 2012. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76:667–684. 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitbart M, Bonnain C, Malki K, Sawaya NA. 2018. Phage puppet masters of the marine microbial realm. Nat Microbiol 3:754–766. 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. 2007. Marine viruses: major players in the global ecosystem. Nat Rev Microbiol 5:801–812. 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 5.Sheik AR, Brussaard CPD, Lavik G, Lam P, Musat N, Krupke A, Littmann S, Strous M, Kuypers MMM. 2014. Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J 8:212–225. 10.1038/ismej.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang J, Park JS, Jung SW, Kim HJ, Joo HM, Kang D, Seo H, Kim S, Jang MC, Lee KW, Jin Oh S, Lee S, Lee TK. 2021. Zooming on dynamics of marine microbial communities in the phycosphere of Akashiwo sanguinea (Dinophyta) blooms. Mol Ecol 30:207–221. 10.1111/mec.15714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutinho FH, Gregoracci GB, Walter JM, Thompson CC, Thompson FL. 2018. Metagenomics sheds light on the ecology of marine microbes and their viruses. Trends Microbiol 26:955–965. 10.1016/j.tim.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Weitz JS, Wilhelm SW. 2012. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep 4:17. 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baudoux AC. 2007. The role of viruses in marine phytoplankton mortality. PhD thesis. Royal Netherlands Institute for Sea Research, Texel, Netherlands. [Google Scholar]
- 10.Zhang Z, Nair S, Tang L, Zhao H, Hu Z, Chen M, Zhang YY, Kao S-J, Jiao N, Zhang YY. 2021. Long-term survival of Synechococcus and heterotrophic bacteria without external nutrient supply after changes in their relationship from antagonism to mutualism. mBio 12:e01614-21. 10.1128/mBio.01614-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Ma R, Yang Y, Jiao N, Zhang R. 2019. Viral regulation on bacterial community impacted by lysis-lysogeny switch: a microcosm experiment in eutrophic coastal waters. Front Microbiol 10:1763. 10.3389/fmicb.2019.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue C, Goldenfeld N. 2017. Coevolution maintains diversity in the stochastic “kill the winner” model. Phys Rev Lett 119:268101. 10.1103/PhysRevLett.119.268101. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Weinbauer MG, Jiao N, Zhang R. 2021. Revisiting marine lytic and lysogenic virus-host interactions: kill-the-winner and piggyback-the-winner. Sci Bull 66:871–874. 10.1016/j.scib.2020.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Korytowski DA, Smith H. 2017. Permanence and stability of a kill the winner model in marine ecology. Bull Math Biol 79:995–1004. 10.1007/s11538-017-0265-6. [DOI] [PubMed] [Google Scholar]
- 15.Silveira CB, Rohwer FL. 2016. Piggyback-the-winner in host-associated microbial communities. npj Biofilms Microbiomes 2:16010. 10.1038/npjbiofilms.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai W, Wang H, Tian Y, Chen F, Zheng T. 2011. Influence of a bacteriophage on the population dynamics of toxic dinoflagellates by lysis of algicidal bacteria. Appl Environ Microbiol 77:7837–7840. 10.1128/AEM.05783-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pourtois J, Tarnita CE, Bonachela JA. 2020. Impact of lytic phages on phosphorus- vs. nitrogen-limited marine microbes. Front Microbiol 11:221. 10.3389/fmicb.2020.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevallereau A, Pons BJ, van Houte S, Westra ER. 2022. Interactions between bacterial and phage communities in natural environments. Nat Rev Microbiol 20:49–62. 10.1038/s41579-021-00602-y. [DOI] [PubMed] [Google Scholar]
- 19.Braga LPP, Spor A, Kot W, Breuil MC, Hansen LH, Setubal JC, Philippot L. 2020. Impact of phages on soil bacterial communities and nitrogen availability under different assembly scenarios. Microbiome 8:52. 10.1186/s40168-020-00822-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shelford EJ, Middelboe M, Møller EF, Suttle CA. 2012. Virus-driven nitrogen cycling enhances phytoplankton growth. Aquat Microb Ecol 66:41–46. 10.3354/ame01553. [DOI] [Google Scholar]
- 21.Ankrah NYD, May AL, Middleton JL, Jones DR, Hadden MK, Gooding JR, Lecleir GR, Wilhelm SW, Campagna SR, Buchan A. 2014. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J 8:1089–1100. 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fei C, Ochsenkühn M, Shibl A, Isaac A, Wang C, Amin S. 2020. Quorum sensing regulates ‘swim-or-stick’ lifestyle in the phycosphere. Environ Microbiol 22:4761–4778. 10.1111/1462-2920.15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma D, Misba L, Khan AU. 2019. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control 8:76. 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao D, Webb JS, Kjelleberg S. 2006. Microbial colonization and competition on the marine alga Ulva australis. Appl Environ Microbiol 72:5547–5555. 10.1128/AEM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Lyu Y, Richlen ML, Anderson DM, Cai Z. 2016. Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. CRC Crit Rev Plant Sci 35:81–105. 10.1080/07352689.2016.1172461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Xu A, Elmerich C, Ma LZ. 2017. Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions. ISME J 11:1602–1613. 10.1038/ismej.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vuong TT, Kwon BR, Eom JI, Shin BK, Kim SM. 2020. Interaction between marine bacterium Stappia sp. K01 and diatom Phaeodactylum tricornutum through extracellular fatty acids. J Appl Phycol 32:71–82. 10.1007/s10811-019-01931-5. [DOI] [Google Scholar]
- 28.Grossart HP, Allgaier M, Passow U, Riebesell U. 2006. Testing the effect of CO2 concentration on the dynamics of marine heterotrophic bacterioplankton. Limnol Oceanogr 51:1–11. 10.4319/lo.2006.51.1.0001. [DOI] [Google Scholar]
- 29.Caruso G. 2020. Microbial colonization in marine environments: overview of current knowledge and emerging research topics. J Mar Sci Eng 8:78. 10.3390/jmse8020078. [DOI] [Google Scholar]
- 30.Dobretsov S, Abed RMM, Teplitski M. 2013. Mini-review: inhibition of biofouling by marine microorganisms. Biofouling 29:423–441. 10.1080/08927014.2013.776042. [DOI] [PubMed] [Google Scholar]
- 31.Mayali X, Azam F. 2004. Algicidal bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol 51:139–144. 10.1111/j.1550-7408.2004.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 32.Harper DR, Parracho HMRT, Walker J, Sharp R, Hughes G, Werthén M, Lehman S, Morales S. 2014. Bacteriophages and biofilms. Antibiotics 3:270–284. 10.3390/antibiotics3030270. [DOI] [Google Scholar]
- 33.Gutiérrez D, Rodríguez-Rubio L, Martínez B, Rodríguez A, García P. 2016. Bacteriophages as weapons against bacterial biofilms in the food industry. Front Microbiol 7:825. 10.3389/fmicb.2016.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharifah EN, Eguchi M. 2011. The phytoplankton Nannochloropsis oculata enhances the ability of Roseobacter clade bacteria to inhibit the growth of fish pathogen Vibrio anguillarum. PLoS One 6:e26756. 10.1371/journal.pone.0026756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J, Park BS, Wang P, Patidar SK, Kim JH, Kim SH, Han MS. 2017. Phycospheric native bacteria Pelagibaca bermudensis and Stappia sp. ameliorate biomass productivity of Tetraselmis striata (KCTC1432BP) in co-cultivation system through mutualistic interaction. Front Plant Sci 8:289. 10.3389/fpls.2017.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higashi A, Fujitani Y, Nakayama N, Tani A, Ueki S. 2016. Selective growth promotion of bloom-forming raphidophyte Heterosigma akashiwo by a marine bacterial strain. Harmful Algae 60:150–156. 10.1016/j.hal.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Kamalanathan M, Chiu MH, Bacosa H, Schwehr K, Tsai SM, Doyle S, Yard A, Mapes S, Vasequez C, Bretherton L, Sylvan JB, Santschi P, Chin WC, Quigg A. 2019. Role of polysaccharides in diatom Thalassiosira pseudonana and its associated bacteria in hydrocarbon presence. Plant Physiol 180:1898–1911. 10.1104/pp.19.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villemur R, Payette G, Geoffroy V, Mauffrey F, Martineau C. 2019. Impact of NaCl, nitrate and temperature on the microbial community of a methanol-fed, denitrifying marine biofilm. bioRxiv 607028. 10.1101/607028. [DOI] [PMC free article] [PubMed]
- 39.Yamada S, Sonoda Y, Sugimachi K, Toya H, Uehara K, Shinagawa Y, Tsuchimoto A, Nakano T, Kitazono T. 2021. A case of Stappia indica-induced relapsing peritonitis confirmed by 16S ribosomal RNA gene sequencing analysis in a patient undergoing continuous ambulatory peritoneal dialysis. CEN Case Rep 10:402–408. 10.1007/s13730-021-00579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jopia P, Ruiz-Tagle N, Villagrán M, Sossa K, Pantoja S, Rueda L, Urrutia-Briones H. 2010. Biofilm growth kinetics of a monomethylamine producing Alphaproteobacteria strain isolated from an anaerobic reactor. Anaerobe 16:19–26. 10.1016/j.anaerobe.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Blanco Y, Rivas LA, García-Moyano A, Aguirre J, Cruz-Gil P, Palacín A, Van Esta H, Parro V. 2014. Deciphering the prokaryotic community and metabolisms in South African deep-mine biofilms through antibody microarrays and graph theory. PLoS One 9:e114180. 10.1371/journal.pone.0114180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao Y, Zheng Y, Wu S, Zhang EH, Chen Z, Liang P, Huang X, Yang ZH, Ng IS, Chen BY, Zhao F. 2015. Pyrosequencing reveals a core community of anodic bacterial biofilms in bioelectrochemical systems from China. Front Microbiol 6:1410. 10.3389/fmicb.2015.01410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vick SHW, Greenfield P, Willows RD, Tetu SG, Midgley DJ, Paulsen IT. 2020. Subsurface Stappia: success through defence, specialisation and putative pressure-dependent carbon fixation. Microb Ecol 80:34–46. 10.1007/s00248-019-01471-y. [DOI] [PubMed] [Google Scholar]
- 44.Weber CF, King GM. 2007. Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl Environ Microbiol 73:1266–1276. 10.1128/AEM.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller DG, Gachon CMM, Küpper FC. 2008. Axenic clonal cultures of filamentous brown algae: initiation and maintenance. Cah Biol Mar 49:59–65. [Google Scholar]
- 46.Han J, Wang S, Zhang L, Yang G, Zhao L, Pan K. 2016. A method of batch-purifying microalgae with multiple antibiotics at extremely high concentrations. Chin J Ocean Limnol 34:79–85. 10.1007/s00343-015-4288-2. [DOI] [Google Scholar]
- 47.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2012. Podoviridae, p 63–85. In Virus taxonomy. Elsevier, London, United Kingdom. 10.1016/B978-0-12-384684-6.00003-3. [DOI] [Google Scholar]
- 48.Oliveira H, São-José C, Azeredo J. 2018. Phage-derived peptidoglycan degrading enzymes: challenges and future prospects for in vivo therapy. Viruses 10:292. 10.3390/v10060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Jin K, Setlow B, Setlow P, Hao B. 2012. Crystal structure of the catalytic domain of the Bacillus cereus SleB protein, important in cortex peptidoglycan degradation during spore germination. J Bacteriol 194:4537–4545. 10.1128/JB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailly-Bechet M, Vergassola M, Rocha E. 2007. Causes for the intriguing presence of tRNAs in phages. Genome Res 17:1486–1495. 10.1101/gr.6649807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S. 2017. ViPTree: the viral proteomic tree server. Bioinformatics 33:2379–2380. 10.1093/bioinformatics/btx157. [DOI] [PubMed] [Google Scholar]
- 52.Hardies SC, Hwang YJ, Hwang CY, Jang GI, Cho BC. 2013. Morphology, physiological characteristics, and complete sequence of marine bacteriophage ϕRIO-1 infecting Pseudoalteromonas marina. J Virol 87:9189–9198. 10.1128/JVI.01521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun X, Huang S, Long L. 2019. Characterization, complete genome and proteome of a bacteriophage infecting a coral-derived Vibrio strain. Mar Genomics 47:100674. 10.1016/j.margen.2019.03.009. [DOI] [Google Scholar]
- 54.Kalatzis PG, Bastías R, Kokkari C, Katharios P. 2016. Isolation and characterization of two lytic bacteriophages, φst2 and φgrn1: phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds. PLoS One 11:e0151101. 10.1371/journal.pone.0151101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Zhao J, Wang L, Li C, Liu J, Zhang L, Zhang Y. 2019. A novel benthic phage infecting Shewanella with strong replication ability. Viruses 11:1081. 10.3390/v11111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traving SJ, Clokie MRJ, Middelboe M. 2014. Increased acidification has a profound effect on the interactions between the cyanobacterium Synechococcus sp. WH7803 and its viruses. FEMS Microbiol Ecol 87:133–141. 10.1111/1574-6941.12199. [DOI] [PubMed] [Google Scholar]
- 57.Danovaro R, Corinaldesi C, Dell'anno A, Fuhrman JA, Middelburg JJ, Noble RT, Suttle CA. 2011. Marine viruses and global climate change. FEMS Microbiol Rev 35:993–1034. 10.1111/j.1574-6976.2010.00258.x. [DOI] [PubMed] [Google Scholar]
- 58.Amin SA, Hmelo LR, Van Tol HM, Durham BP, Carlson LT, Heal KR, Morales RL, Berthiaume CT, Parker MS, Djunaedi B, Ingalls AE, Parsek MR, Moran MA, Armbrust EV. 2015. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522:98–101. 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 59.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 60.Verderosa AD, Totsika M, Fairfull-Smith KE. 2019. Bacterial biofilm eradication agents: a current review. Front Chem 7:824. 10.3389/fchem.2019.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Łusiak-Szelachowska M, Weber-Dąbrowska B, Górski A. 2020. Bacteriophages and lysins in biofilm control. Virol Sin 35:125–133. 10.1007/s12250-019-00192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian F, Li J, Nazir A, Tong Y. 2021. Bacteriophage: a promising alternative measure for bacterial biofilm control. Infect Drug Resist 14:205–217. 10.2147/IDR.S290093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nabergoj D, Modic P, Podgornik A. 2018. Effect of bacterial growth rate on bacteriophage population growth rate. Microbiologyopen 7:e00558. 10.1002/mbo3.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abedon ST, Herschler TD, Stopar D. 2001. Bacteriophage latent-period evolution as a response to resource availability. Appl Environ Microbiol 67:4233–4241. 10.1128/AEM.67.9.4233-4241.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang YY, Huang CX, Yang J, Jiao NZ. 2011. Interactions between marine microorganisms and their phages. Chin Sci Bull 56:1770–1777. 10.1007/s11434-011-4503-2. [DOI] [Google Scholar]
- 66.Santamaría RI, Bustos P, Sepúlveda-Robles O, Lozano L, Rodríguez C, Fernández JL, Juárez S, Kameyama L, Guarneros G, Dávila G, González V. 2014. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl Environ Microbiol 80:446–454. 10.1128/AEM.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Limor-Waisberg K, Carmi A, Scherz A, Pilpel Y, Furman I. 2011. Specialization versus adaptation: two strategies employed by cyanophages to enhance their translation efficiencies. Nucleic Acids Res 39:6016–6028. 10.1093/nar/gkr169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. 2019. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10:331. 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Machata S, Hain T, Rohde M, Chakraborty T. 2005. Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol 187:8385–8394. 10.1128/JB.187.24.8385-8394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Renier S, Chagnot C, Deschamps J, Caccia N, Szlavik J, Joyce SA, Popowska M, Hill C, Knøchel S, Briandet R, Hébraud M, Desvaux M. 2014. Inactivation of the SecA2 protein export pathway in Listeria monocytogenes promotes cell aggregation, impacts biofilm architecture and induces biofilm formation in environmental condition. Environ Microbiol 16:1176–1192. 10.1111/1462-2920.12257. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Tang B, Gu Q, Yu X. 2014. Elimination of the formation of biofilm in industrial pipes using enzyme cleaning technique. MethodsX 1:e130–e136. 10.1016/j.mex.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sayer CV, Popham DL. 2019. YpeB dimerization may be required to stabilize SleB for effective germination of Bacillus anthracis spores. BMC Microbiol 19:169. 10.1186/s12866-019-1544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turner D, Ackermann HW, Kropinski AM, Lavigne R, Sutton JM, Reynolds DM. 2017. Comparative analysis of 37 Acinetobacter bacteriophages. Viruses 10:5. 10.3390/v10010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amarillas L, Rubí-Rangel L, Chaidez C, González-Robles A, Lightbourn-Rojas L, León-Félix J. 2017. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol 8:1355. 10.3389/fmicb.2017.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han F, Li M, Lin H, Wang J, Cao L, Khan MN. 2014. The novel Shewanella putrefaciens-infecting bacteriophage Spp001: genome sequence and lytic enzymes. J Ind Microbiol Biotechnol 41:1017–1026. 10.1007/s10295-014-1438-z. [DOI] [PubMed] [Google Scholar]
- 76.Latka A, Drulis-Kawa Z. 2020. Advantages and limitations of microtiter biofilm assays in the model of antibiofilm activity of Klebsiella phage KP34 and its depolymerase. Sci Rep 10:20338. 10.1038/s41598-020-77198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma R, Bajpai P, Sayyed U, Ahmad IZ. 2019. Approaches towards microbial biofilm disruption by natural bioactive agents, p 233–261. In Kumar S, Chandra N, Singh L, Hashmi M, Varma A (ed), Biofilms in human diseases: treatment and control. Springer, Cham, Switzerland. 10.1007/978-3-030-30757-8_15. [DOI] [Google Scholar]
- 78.Topka-Bielecka G, Dydecka A, Necel A, Bloch S, Nejman-Faleńczyk B, Węgrzyn G, Węgrzyn A. 2021. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics 10:175. 10.3390/antibiotics10020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 82.Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M. 2017. PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292. 10.1038/s41598-017-07910-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li C, Wang Z, Zhao J, Wang L, Xie G, Huang J, Zhang Y. 2021. A novel vibriophage vB_VcaS_HC containing lysogeny-related gene has strong lytic ability against pathogenic bacteria. Virol Sin 36:281–290. 10.1007/s12250-020-00271-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes: implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641. 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, Von Mering C, Bork P. 2019. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 90.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 94.Moraru C, Varsani A, Kropinski AM. 2020. VIRIDIC: a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. 10.3390/v12111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Fig. S1 and S2, and legends of Tables S1 to S5. Download aem.02138-21-s0001.pdf, PDF file, 0.2 MB (191.8KB, pdf)
Table S1. Download aem.02138-21-s0002.xlsx, XLSX file, 0.01 MB (10.7KB, xlsx)
Table S2. Download aem.02138-21-s0003.xlsx, XLSX file, 0.01 MB (9.9KB, xlsx)
Table S3. Download aem.02138-21-s0004.xlsx, XLSX file, 0.01 MB (10.4KB, xlsx)
Table S4. Download aem.02138-21-s0005.xlsx, XLSX file, 0.01 MB (12.8KB, xlsx)
Table S5. Download aem.02138-21-s0006.xlsx, XLSX file, 0.01 MB (11.5KB, xlsx)
Data Availability Statement
The sequences reported in this paper have been deposited in the NCBI GenBank database under accession number MZ462995.