Abstract
Waikikiamides A-C (1–3), structurally complex diketopiperazine derivatives, and putative biogenic precursors, (+)-semivioxanthin (4), notoamide F (5), and (−)-notoamide A (6), were isolated from Aspergillus sp. FM242. 1 and 2, bearing a hendecacyclic ring system, represent a novel skeleton. 3 features the first unique heterodimer of two notoamide analogs with an N–O-C bridge. Compounds 1 and 3 exhibit antiproliferative activity with IC50 values in the range of 0.56 to 1.86 μM. The gene clusters mined from the sequenced genome support their putative biosynthetic pathways.
Graphical Abstract
Natural products (NPs) have played a critical role in drug discovery.1 Marine-derived materials, for example, sponges, coral, bacteria, and fungi, are well recognized as a unique but less explored source for the discovery of structurally novel and biologically active secondary metabolites.2 For example, the antitumor agents ET-743,3 salinosporamide A,4 didemnin A,5 norhalichondrin A,6 and dolastatin 107 and the antituberculosis agent ilamycin E8 were discovered from different marine origins. These clinically valuable agents propel the continuous investigation of marine NPs for new drug discovery.
Aspergillus is a large genus that is well recognized to produce a wide array of bioactive chemicals.9 Indeed, the Aspergillus Secondary Metabolites Database (A2MDB) currently records 807 unique compounds produced by different Aspergillus species.9 We have previously identified a unique benzodiazepine circumdatin M from Aspergillus sp. FM242 that was isolated from a sample collected at Waikiki beach in Oahu, Honolulu, Hawaii in May 2014.10 In our ongoing research on this marine fungal strain, we isolated three novel complex prenylated indole diketopiperazine (DKP) derivatives, waikikiamides A-C (1–3), along with their potential biosynthetic precursors (+)-semivioxanthin (4), notoamide F (5), and (−)-notoamide A (6). Compounds 1 and 2 bear a hendecacyclic ring system formed from a prenylated indole DKP and polyketide 4, representing an unprecedented skeleton, whereas 3 was the first prenylated indole DKP heterodimer with an N-O-C bridge. Herein we report the isolation, structure elucidation, proposed biosynthetic pathways, and biological evaluation of waikikiamides A-C (1–3).
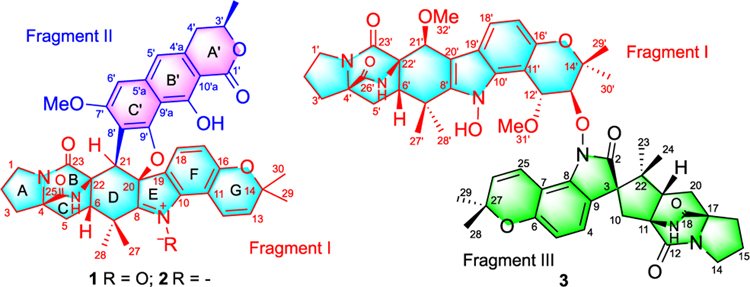
Waikikiamide A (1, Figure 1) was isolated as an optically active powder with a molecular formula of C41H39N3O9 (m/z 718.2743 [M + H]+, Calcd for C41H40N3O9 718.2759) that was established by high-resolution electrospray ionization mass spectrometry (HRESIMS) and required 24 degrees of unsaturation. The nuclear magnetic resonance (NMR)-based structural analysis of 1 was challenging due to the low H/C ratio. The correlation spectroscopy (COSY) spectrum of 1 showed five short spin systems: CH2-CH2-CH2 in ring A, CH2-CH in ring C, CH?CH (aromatic) in ring F, CH?CH (olefinic) in ring G, and CH2-CH-CH3 in ring A' (Figure 2). In the heteronuclear multiple bond correlation (HMBC) spectrum of 1, correlations from H-2 to C-4; from H-21 to C-6, C-20, C-22, and C-23; from H-5 to C-4 and C-25; and from H3-27/H3-28 to C-6 and C-8 could establish rings A-D in fragment I, as shown in Figure 2. On the contrary, HMBCs from H-18 to C-10, C-16, and C-20; from H-17 to C-11 and C-19; from H-12 to C-10, C-14, and C-16; and from H3-29/ H3-30 to C-13 and C-14 indicated that rings F and G were a 5,6-disubstututed 2,2-dimethyl-2H-chromene. Although there was no HMBC to determine the connectivity of the three nitrogen atoms in fragment I, we assumed that ring E was a penta-substituted pyrrole 1-oxide and rings B/C had a 2,5-diazabicyclo[2.2.2]octane-3,6-dione core. The detailed NMR analysis revealed that fragment I of 1 was a Pro-Trp 2,5-DKP with two isoprenyl groups, which was the same as (+)-avrainvillamide, except for the 20- and 21-positions. The highly aromatic character of another fragment (II) was apparent from the presence of only 4 sp3 carbons, including 1 methoxy and the CH2-CH-CH3 spin system, and 11 sp2 carbons, including 9 nonprotonated and 2 tertiary aromatic carbons. When using DMSO-d6 and pyridine-d5 as solvents, the HMBC analysis clearly demonstrated the correlation from 10'-OH to C-9a', C-10', and C-10a'. NMR analysis further revealed that fragment II, a 3',7',8',9',10'-pentasubstituted benzoisochroman-1-one, was similar to (+)-semivioxanthin (4), except for the 8'- and 9'-positions, which was also isolated in this study. The connectivity between C-21 and C-8' was established on the basis of HMBCs from H-21 to C-6, C-20, C-22, C-23, C-7', C-8', and C-9'. According to the molecular formula of 1, one additional ring and an oxygen atom had to be assigned. Because only the valences at C-20 and C-9' remained unsatisfied, the additional ring and oxygen could result only from an ether linkage between C-20 and C-9'. Hence, the planar structure of 1 (Figure 2) was determined, as shown.
Figure 1.
Chemical structures of compounds 1–3
Figure 2.
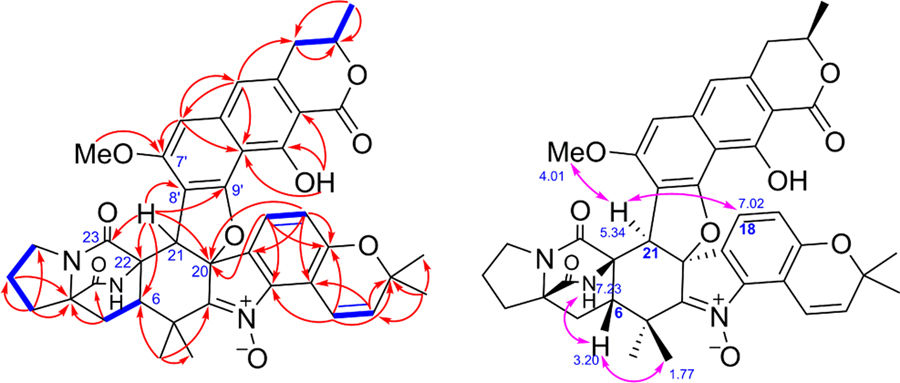
Key COSY correlations (bold, blue) and HMBCs (single-headed, red) of 1. Key NOESY correlations (double-headed, pink) of 1.
In the nuclear Overhauser effect spectroscopy (NOESY) spectrum of 1 (Figure 2), H-6 correlated to 26-NH, and H-21 showed correlations to 7'-OMe and H-18. These data indicated that H-6 and 26-NH had the same orientation, whereas H-21 and the C20-C19 bond had opposite orientations. To further determine the relative configuration of fragment I, we undertook quantum calculations of NMR shifts for correlation with experimental data, one leading strategy to facilitate the structural elucidation of complex molecules.11 The continuous advances in the field enable the in silico predictions to complement the standard NMR spectroscopy nicely, emerging as a new tool when dealing with challenging architectures featuring a low H/C ratio.12 Among the different methods of data correlation,13 the DP4+ probability stands out for its good performance over a wide variety of systems.14 The diastereoisomers resulting from varying the configurations at C-6, C-20, and C-21 were generated, leading to eight structures (1a to 1h, SI Section S10) including 1a (all β H-6, 26-NH, C-19-C-20, and H-21) and 1h (β H-6 and 26-NH; α C-19-C-20 and H-21). After a conformational sampling at the Merck molecular force field (MMFF) level (using both Spartan and Macromodel), the NMR shifts of all conformers found with a 10 kcal/mol cutoff for each configuration were computed at the PCM/ mPW1PW91/6–31+G**//B3LYP/6–31G* level.14 Among all of the isomers, 1h showed the closest match with the experimental values, demonstrating the smallest CMAE (corrected mean absolute error) values of 1.2 and 0.09 ppm for carbon and proton data, respectively (SI Section S14). Therefore, the DP4+ quantum calculations strongly supported 1h to represent the real structure of fragment I (>99.9% overall probability), in good agreement with the NOESY correlations observed for waikikiamide A (1). Connecting the relative configurations of the fragment I and fragment II (C-3' position) was, on the contrary, much more difficult given the large separation of both stereoclusters by a plane aromatic core. Given the challenge to obtain suitable crystals for X-ray analysis, we computed the NMR shifts of 3'epi-1h in an attempt to suggest a sound full stereochemical assignment of 1. As expected, the NMR shifts of 3'epi-1h were similar to those computed for 1h, with slightly larger CMAE values (1.26 and 0.093 ppm vs 1.21 and 0.087 ppm, respectively), and DP4+ values favored 1h in moderate probability (80.7%) (SI Section S16). Hence, the structure including the relative configuration of compound 1 was proposed, as shown.
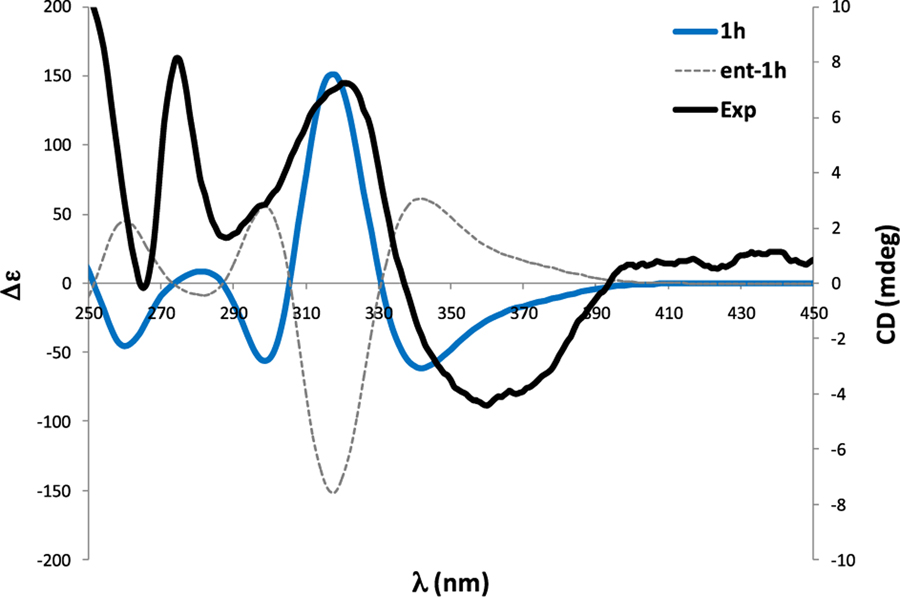
To define the absolute configuration of 1, the time-dependent density functional theory-electronic circular dichroism (TDDFT-ECD) method was applied to the most likely candidates (1h and 3'epi-1h).15 The ECD spectra experimentally collected (Figure 3) showed positive Cotton effects at 225 and 275 nm and a strong negative Cotton effect at ~350 nm. The ECD calculations were carried out with the most stable conformers found for 1h and 3'epi-1h at the B3LYP/6–31G* level. The Boltzmann-weighted ECD spectra of both 1h candidates matched well with the experimental ECD spectrum (Figure 3), allowing us to assign the absolute configuration of 1, as shown. The calculated ECD spectrum of 3'epi-1h (SI Section S26) was almost identical to that of 1h, suggesting that the ECD spectrum seems to be governed mainly by the handedness of fragment I. Similar results were obtained at other levels of theory (including ωB97XD and CAM-B3LYP).
Figure 3.
Experimental and calculated ECD of 1.
Waikikiamide B (2) had a molecular formula of C41H39N3O8 (m/z 702.2845 [M + H]+, Calcd for C41H40N3O8 702.2810), with one oxygen less than that of 1. The detailed NMR analysis revealed that 2 was almost the same as 1, except that the oxygen at the 9-position in 1 was missing in 2. It is also supported by the chemical shift of C-8 (186.5) (SI Sections S27 and S54).16 Its NOESY correlations and CD spectrum were almost the same as those of 1 (SI Sections S50, S56, and S59). Hence, the structure of 2 was determined, as shown.
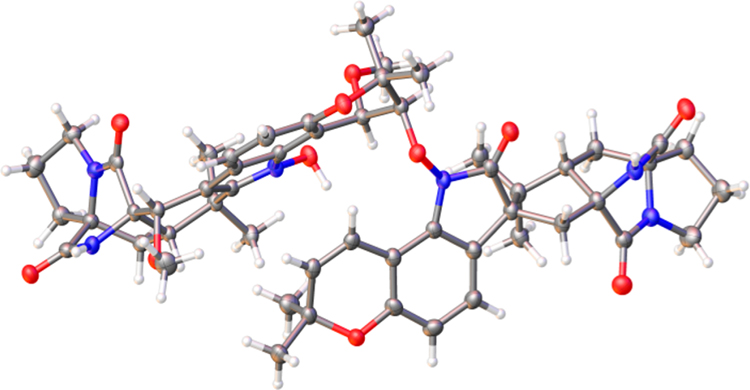
The HRESIMS data of compound 3 provided a molecular formula of C54H62N6O11 (m/z 971.4545 [M + H]+, Calcd for C54H63N6O11 917.4549). NMR analyses revealed that 3 was also composed of two fragments (I and III). Fragment I was assigned as the prenylated indole DKP notoamide B, whereas fragment III was 12,13-dihydro-13-hydroxy-12-methoxy-notoamide G (SI Section S61). There was no HMBC between these two fragments, which precluded the connectivity of fragments I and III of the molecule. Suitable crystals (0.16 × 0.10 × 0.08 mm) for an X-ray crystallographic study were obtained from acetonitrile at room temperature after repeated recrystallization. The final structure including the connectivity of fragments I and III, and the absolute configuration of 3 were then unambiguously determined by single-crystal X-ray diffraction (Figure 4, CCDC 1994347) with Flack(x) = 0.02(6), Hooft(y) = 0.02(5), p3(true) = 1.000, p3(false) = 0.3 × 10−82, and p3(racemic twin) = 0.9 × 10−20 using PLATON/ BIJVOETPAIR.17
Figure 4.
X-ray crystal structure of 3.
The structures of known compounds 4 ((+)-semivioxanthin)18 and 5 (notoamide F (21-methoxy stephacidin A))19 (SI Section S6, CCDC 1994346; 0.18 × 0.12 × 0.10 mm crystals were obtained from methanol at room temperature) and 6 ((−)-notoamide A)20 were identified by the comparison of their physical data with the reported values in the literature and X-ray analysis.
Multiple prenylated indole DKP dimers (e.g., stephacidin B21 and waikialoids A and B22) as well as the hybrids of DKP and polyketide (e.g., variecolortines A-C23 and versicoamides F-H24) have been previously reported. To our knowledge, compound 3 was the first DKP heterodimer with an N-O-C bridge. Furthermore, compounds 1 and 2 represent a rare class of fungal secondary metabolites, which are hybrids of DKPs and polycyclic aromatic polyketides.
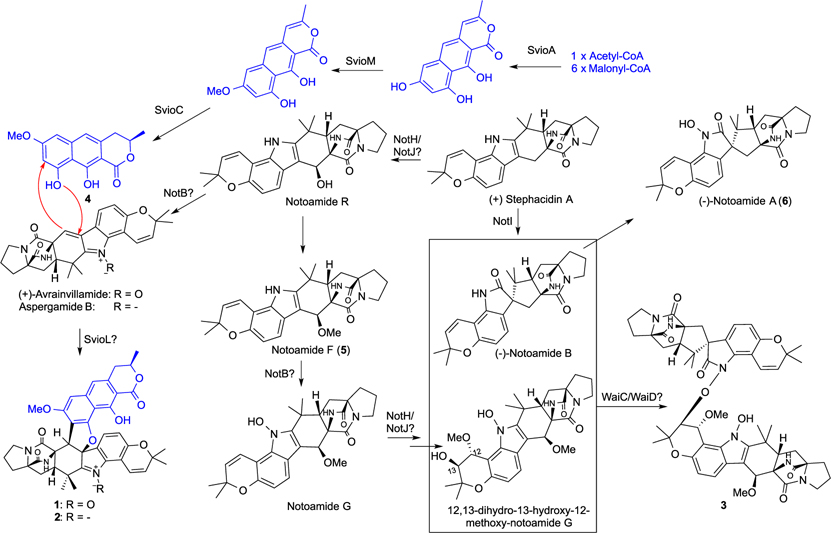
The biosynthesis of waikikiamides A and B (1 and 2) can be derived from the polyketide (+)-semivioxanthin (4) and the prenylated indole DKP (+)-avrainvillamide and aspergamide B, respectively, whereas the coupling of notoamide B and a derivative of notoamide G can produce compound 3 (Scheme 1). Co-isolation of 4–6 together with the LC/MS detection of notoamides R and G supported the proposed biosynthetic pathway of waikikiamides. Indeed, (+)-stephacidin A is a known common biosynthetic intermediate of (+)-avrainvillamide, aspergamide B, and notoamides25 (SI Section S4). Furthermore, the genome of Aspergillus sp. FM242 was sequenced and assembled to provide 36.6 Mbp (395 contigs, 49.1% GC). The antiSMASH analysis revealed a gene cluster highly similar to the notoamide (not) cluster known in other fungal species and can be responsible for the biosynthesis of multiple prenylated indole DKPs for the production of waikikiamides.25–27 It is of note that the N-O-C bridge in 3 is a unique feature among known dimeric DKPs, and the enzymatic formation of this bridge remains unstudied. Compared with known notoamide gene clusters,26,27 the cluster identified in this work encodes five additional proteins: WaiA1, WaiA2, WaiB, WaiC, and WaiD (Figure 5). Among them, WaiD is predicted to be a benzoate 4-monooxygenase P450, whereas WaiC is an uncharacterized 740-amino-acid protein. The vast catalytic versatility of P450s28 and the unknown function of WaiC make them potential candidates for the formation of the N-O-C bridge (Scheme 1). The sequenced genome also carries eight predicted polyketide synthases but only one nonreducing polyketide synthase (NRPKS, SvioA) at a different chromosomal region withthe not cluster. The set of SvioA, an O-methyltransferase (SvioM), and a short-chain dehydrogenase/reductase (SvioC) can synthesize the naphtho-α-pyrone semivioxanthin (4) from one acetyl-CoA and six malonyl-CoAs, whereas the pathway-specific laccase (SvioL) may fuse 4 with (+)-avrainvillamide and aspergamide B29 to produce 1 and 2, respectively (Scheme 1, Figure 5). SvioL homologues are known to catalyze phenol coupling in the biosynthesis of polyketide dimers.29
Scheme 1.
Proposed Biosynthetic Pathways of 1 –3
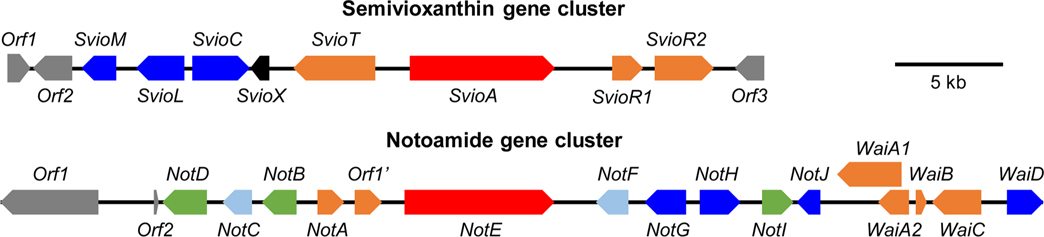
Figure 5.
Waikikiamide (wai) biosynthetic gene cluster derived from the complete sequencing and bioinformatic mining of the Aspergillus sp. FM 242 genome. The mined wai cluster comprises two separate subclusters, the semivioxanthin (svio) gene cluster (~34 kb, Genbank accession number: MT457560) and the notoammide (not) gene cluster (~47 kb, Genbank accession number: MT457561), located at two different chromosomal regions. The svio cluster (top) contains all genes required for the biosynthesis of semivioxanthin, and the putative laccase SvioL may catalyze the enzymatic fusion of semivioxanthin and (+)-avrainvillamide or aspergamide B to produce waikikiamides A and B (1 and 2). A total of 11 proteins encoded by genes from the not cluster (bottom) share high amino acid sequence similarities with those of the not cluster from notoamide-producing Aspergillus sp. MF297–2 and can be responsible for the biosynthesis of notoamides (+)-avrainvillamide and aspergamide B via the common biosynthetic intermediate (+)-stephacidin A. The not cluster from Aspergillus sp. FM292 also encodes an uncharacterized protein (WaiC) and an additional P450 (WaiD), which could be the candidates for the formation of a N-O-C bridge between (−)-notoamide B and 12,13-dihydro-13-hydroxy-12-methoxy-notoamide G to produce waikikiamide C (3).
Compounds 1–6 were evaluated for their antiproliferative activity against four different types of cancer cell lines, HT1080 (a human fibrosarcoma cell line), PC3 (a human prostatic tumor cell line), Jurkat (an immortalized T lymphocyte cell line), and A2780 (a human ovarian cancer cell line) (Table 1). Interestingly, compounds 1 and 3, each with an N-O bond, were active with IC50 values between 0.56 and 1.86 μM, whereas compound 2 without an N-O bond and monomers (4–6) were inactive at 20 and 40 μM (the highest concentration tested) for 2 and 4–6, respectively. The more rigid DKP-polyketide hybrid (1) was more active than the heterodimer of two DKPs (3).
Table 1.
Antiproliferative Activity (IC50) of Compounds 1 and 3 (μM)a
| cell lines | 1 | 3 | taxol |
|---|---|---|---|
| HT1080 | 0.519 | 1.135 | 0.0075 |
| PC3 | 1.855 | 1.805 | 0.0136 |
| Jurkat | 0.62 | 1.79 | 0.0087 |
| A2780S | 0.78 | 1.127 | 0.0081 |
Note: all in triplicate.
In summary, three novel complex compounds (1–3) were isolated from Aspergillus sp. FM242 and characterized as either DKP-polyketide hybrids (1 and 2) or a heterodimer of two DKPs (3). The hybrids of DKP and polyketide (1 and 2) are quite rare secondary metabolites, and the DKP heterodimer (3) with an N-O-C bridge is unprecedented. Clearly, to determine the structure of 3 without X-ray analysis was impossible. The discovery of 1–3 expands the chemical diversity of the known DKP scaffolds and furnishes intriguing templates for synthetic and biosynthetic chemists. Furthermore, 1, a DKP-polyketide hybrid with a nitrogenated methoxy group, showed the most antiproliferative activity, which may attract wide attention from cancer biologists, biochemists, medicinal chemists, and pharmacologists for new anticancer drug discovery and development.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by start-up funding from the Daniel K. Inouye College of Pharmacy (S.C.), Seed Grants from the University of Hawaii at Hilo (S.C.), the Victoria S. and Bradley L. Geist Foundation (15ADVC-74420 and 17CON-86295 to S.C.), NIH INBRE (5P20GM103466 to S.C.), NIH 1R35GM128742 (Y.D.), ANPCyT (PICT-2016-0116), and UNR (BIO 500) (A.M.S.). We thank Dr. Peilan Zhang at the University of Florida for her technical support.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.orglett.0c01411.
Experimental procedures and spectroscopic data for all new compounds (PDF)
Accession Codes
CCDC 1994346–1994347 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.orglett.0c01411
Contributor Information
Fuqian Wang, Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo, Hilo, Hawai'i 96720, United States; Department of Pharmacy, Wuhan No. 1 Hospital, Wuhan 430022, China.
Ariel M. Sarotti, Instituto de Química Rosario (CONICET), Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario, Rosario 2000, Argentina.
Guangde Jiang, Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, College of Pharmacy, University of Florida, Gainesville, Florida 32610, United States.
José C. Huguet-Tapia, Department of Plant Pathology, University of Florida, Gainesville, Florida 32611, United States
Shao-Liang Zheng, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138, United States.
Xiaohua Wu, Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo, Hilo, Hawai'i 96720, United States.
Chunshun Li, Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo, Hilo, Hawai'i 96720, United States.
Yousong Ding, Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, College of Pharmacy, University of Florida, Gainesville, Florida 32610, United States.
Shugeng Cao, Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo, Hilo, Hawai’i 96720, United States; Cancer Biology Program, University of Hawaii Cancer Center, Honolulu, Hawai’i 96813, United States.
REFERENCES
- (1).(a) Newman DJ; Cragg GMJ Nat. Prod. 2016, 79, 629–661. [DOI] [PubMed] [Google Scholar]; (b) Cragg GM; Newman DJ Biochim. Biophys. Acta, Gen. Subj 2013, 1830, 3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mathur S; Hoskins C Biomed. Rep. 2017, 6, 612–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).(a) Blunt JW; Carroll AR; Copp BR; Davis RA; Keyzers RA; Prinsep MR Nat. Prod. Rep. 2018, 35, 8–53 [DOI] [PubMed] [Google Scholar]; (b) Deshmukh SK; Prakash V; Ranjan N Front. Microbiol. 2018, 8 (2536), 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Blunt JW; Copp BR; Keyzers RA; Munro MH; Prinsep MR Nat. Prod. Rep. 2016, 33, 382–431. [DOI] [PubMed] [Google Scholar]; (d) Blunt JW; Copp BR; Keyzers RA; Munro MH; Prinsep MR Nat. Prod. Rep. 2015, 32, 116–211. [DOI] [PubMed] [Google Scholar]
- (3).Damia G; Silvestri S; Carrassa L; Filiberti L; Faircloth GT; Liberi G; Foiani M; D’Incalci M Int. J. Cancer 2001, 92, 583–588. [DOI] [PubMed] [Google Scholar]
- (4).Feling RH; Buchanan GO; Mincer TJ; Kauffman CA; Jensen PR; Fenical W Angew. Chem., Int. Ed. 2003, 42, 355–357. [DOI] [PubMed] [Google Scholar]
- (5).Crampton SL; Adams EG; Kuentzel SL; Li LH; Badiner G; Bhuyan BK Cancer. Res. 1984, 44, 1796–1801. [PubMed] [Google Scholar]
- (6).Uemura D; Takahashi K; Yamamoto T; Katayama C; Tanaka J; Okumura Y; Hirata YJ Am. Chem. Soc. 1985, 107, 4796–4798. [Google Scholar]
- (7).Schwartsmann G; da Rocha AB; Berlinck RG; Jimeno J Lancet Oncol. 2001, 2, 221–225. [DOI] [PubMed] [Google Scholar]
- (8).Ma J; Huang H; Xie Y; Liu Z; Zhao J; Zhang C; Jia Y; Zhang Y; Zhang H; Zhang T; Ju J Nat. Commun. 2017, 8, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Vadlapudi V; Borah N; Yellusani KR; Gade S; Reddy P; Rajamanikyam M; Vempati LNS; Gubbala SP; Chopra P; Upadhyayula SM; Amanchy R Sci. Rep. 2017, 7, 7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wang F; Hu Z; Li C; Wu X; Cao S Tetrahedron Lett. 2019, 60, 1724–1726. [Google Scholar]
- (11).Lodewyk MW; Siebert MR; Tantillo D Chem. Rev. 2012, 112, 1839–1862. [DOI] [PubMed] [Google Scholar]
- (12).Kutateladze AG; Krenske EH; Williams CM Angew. Chem., Int. Ed. 2019, 58, 7107–7112. [DOI] [PubMed] [Google Scholar]
- (13).Grimblat N; Sarotti AM Chem. - Eur. J. 2016, 22, 12246–12261. [DOI] [PubMed] [Google Scholar]
- (14).Grimblat N; Zanardi MM; Sarotti AM J. Org. Chem. 2015, 80, 12526–12534. [DOI] [PubMed] [Google Scholar]
- (15).Li XC; Ferreira D; Ding Y Curr. Org. Chem. 2010, 14, 1678–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li F; Zhang Z; Zhang G; Che Q; Zhu T; Gu Q; Li D Org. Lett. 2018, 20, 1138–1141. [DOI] [PubMed] [Google Scholar]
- (17).Spek AL Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65 (Pt 2), 148–155. [Google Scholar]
- (18).(a) Zeeck A; Ruß P; Laatsch H; Loeffler W; Wehrle H; Zähner H; Holst H Chem. Ber. 1979, 112, 957–978. [Google Scholar]; (b) Yada H; Sato H; Toshima H; Deura M; Ichihara A Biosci., Biotechnol., Biochem. 2001, 65, 484–486. [DOI] [PubMed] [Google Scholar]
- (19).Tsukamoto S; Kato H; Samizo M; Nojiri Y; Onuki H; Hirota H; Ohta TJ Nat. Prod. 2008, 71, 2064–2067. [DOI] [PubMed] [Google Scholar]
- (20).Kato H; Yoshida T; Tokue T; Nojiri Y; Hirota H; Ohta T; Williams RM; Tsukamoto S Angew. Chem., Int. Ed. 2007, 46, 2254–2256. [DOI] [PubMed] [Google Scholar]
- (21).Qian-Cutrone J; Huang S; Shu Y; Vyas D; Fairchild C; Menendez A; Krampitz K; Dalterio R; Klohr SE; Gao QJ Am. Chem. Soc. 2002, 124, 14556–14557. [DOI] [PubMed] [Google Scholar]
- (22).Wang X; You J; King JB; Powell DR; Cichewicz RH J. Nat. Prod. 2012, 75, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhong W; Wang J; Wei X; Chen Y; Fu T; Xiang Y; Huang X; Tian X; Xiao Z; Zhang W; Zhang S; Long L; Wang F Org. Lett. 2018, 20, 4593–4596. [DOI] [PubMed] [Google Scholar]
- (24).Liu L; Wang L; Bao L; Ren J; Bahadur Basnet B; Liu R; He L; Han J; Yin W; Liu H Org. Lett. 2017, 19, 942–945. [DOI] [PubMed] [Google Scholar]
- (25).Klas KR; Kato H; Frisvad JC; Yu F; Newmister SA; Fraley AE; Sherman DH; Tsukamoto S; Williams RM Nat. Prod. Rep. 2018, 35, 532–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ding Y; Wet J. R. d.; Cavalcoli J; Li S; Greshock TJ; Miller KA; Finefield JM; Sunderhaus JD; McAfoos TJ; Tsukamoto S; Williams RM; Sherman DH J. Am. Chem. Soc. 2010, 132, 12733–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Li S; Srinivasan K; Tran H; Yu F; Finefield JM; Sunderhaus JD; McAfoos TJ; Tsukamoto S; Williams RM; Sherman DH MedChemComm 2012, 3, 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).(a) Zhang X; Li S Nat. Prod. Rep. 2017, 34, 1061–1089. [DOI] [PubMed] [Google Scholar]; (b) Rudolf JD; Chang CY; Ma M; Shen B Nat. Prod. Rep. 2017, 34, 1141–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Podust LM; Sherman DH Nat. Prod. Rep. 2012, 29, 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).(a) Fürtges L; Obermaier S; Thiele W; Foegen S; Müller M ChemBioChem 2019, 20, 1928–1932. [DOI] [PubMed] [Google Scholar]; (b) Su J; Fu J; Wang Q; Silva C; Cavaco-Paulo A Crit. Rev. Biotechnol. 2018, 38, 294–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.