Abstract
Coronavirus disease, also known as COVID-19, is an infectious disease caused by SARS-CoV-2. It has a direct impact on the upper and lower respiratory tract and threatened the health of many people around the world. The latest statistics show that the number of people diagnosed with COVID-19 is growing exponentially. Diagnosing positive cases of COVID-19 is important for preventing further spread of the disease. Currently, Coronavirus is a serious threat to scientists, medical experts and researchers around the world from its detection to its treatment. It is currently detected using reverse transcription polymerase chain reaction (RT-PCR) analysis at the most test centers around the world. Yet, knowing the reliability of a deep learning based medical diagnosis is important for doctors to build confidence in the technology and improve treatment. The goal of this study is to develop a model that automatically identifies COVID-19 by using chest X-ray images. To achieve this, we modified the DarkCovidNet model which is based on a convolutional neural network (CNN) and plotted the experimental results for two scenarios: binary classification (COVID-19 versus No-findings) and multi-class classification (COVID-19 versus pneumonia versus No-findings). The model is trained on more than 10 thousand X-ray images and achieved an average accuracy of 99.53% and 94.18% for binary and multi-class classification, respectively. Therefore, the proposed method demonstrates the effectiveness of COVID-19 detection using X-ray images. Our model can be used to test the patient via cloud and also be used in situations where RT-PCR tests and other options aren't available.
Keywords: Coronavirus disease, COVID-19, X-ray, Deep learning, CNN, Chest radiography
Introduction
In Wuhan Province of China, Coronavirus Disease or COVID-19 was reported first in December, 2019 and confirmed to the World Health Organization (WHO) on December 31, 2019 [1]. The cause of this disease is found to be Severe Acute Respiratory Syndrome Coronavirus2 (SARS-CoV-2). It became a pandemic around the world within just three months after its discovery and it is being compared to the Spanish flu of 1920 [2] that took millions of lives. The spread primarily occurs during close contact between people, via small droplets produced by coughing, sneezing, and talking [2]. COVID-19 transmission is faster when people are closer to each other. Thus, the restrictions of travelling help to control the spread of the disease, and frequent hand washing and hand sanitizing are highly recommended to prevent the infection. Fast transmission, the ever-increasing number of deaths, and shortage of production of vaccine made it one of the biggest problems on earth right now. Even though the fatality rate of the disease is only 2–3%, the more significant concern is its rapid spreading among humans [3]. Fever and cough are the most common infection symptoms with other mild symptoms, including chest discomfort, sputum development, and sore throat. COVID-19 may progress to viral pneumonia which has a 5.8% mortality risk [4]. As of November 20, 2020, the total number of people infected with COVID-19 worldwide is 57,531,513, while the confirmed deaths and recoveries are 1,370,616 and 39,929,010, respectively [5]. Most cases have been registered in the United States, India, Brazil, France, Russia, Spain, the United Kingdom, Argentina and Italy [5]. At present, one of the most accurate ways of diagnosing COVID-19 is by a test called Reverse Transcription Polymerase Chain Reaction (RT-PCR) [6]. The coronavirus is an RNA virus and to get complementary DNA; its genetic material is transcribed inversely. This can then be amplified by polymerase chain reaction or PCR, making it easier to measure. It is however a complex and time-consuming operation, taking almost 2–3 h and needing a specialist to be involved. Even then, there have been studies showing that RT-PCR can yield false negatives [6]. Moreover, RT-PCR only detects the presence of viral RNA. It cannot prove that the virus is alive and transmissible [7]. Owing to the massive number of cases in the pandemic, the testing material is also limited, contributing to higher expenses. It has become critical for radiologists to be confident with the imaging appearance of the disease as the world is dealing with the burden of the disease. The main modality for the diagnosis and treatment of respiratory diseases is thoracic imaging [8] with chest X-ray and CT [8]. Chest imaging in patients with suspected or probable COVID-19 disease where RT-PCR is not available or results are delayed has been regarded as part of the diagnostic workup. Thus, automatic diagnosis system is needed. In this paper, a COVID-19 diagnosis method using deep CNN is presented. We modified the existing DarkCovidNet [9] model which is based on DarkNet-19 and achieved high classification accuracy.
Section II describes the literature review where as in section III the proposed model is explained. Experimental results and discussion are presented in section IV. Finally, Conclusion is presented in the last section.
Literature review
Several studies have recently been conducted based on CNN models related to COVID-19 analysis and detection using chest X-ray images. Narin et al. [10] proposed a CNN-based model for identifying Covid-19 patients using 100 chest X-ray images. ResNet-50, Inception-v3, and Inception-ResNetv2 were tested using fivefold cross-validation and found that ResNet-50 had the highest detection performance (98%). Hemdan et al. [11] have suggested the COVIDX-Net model to aid radiologists in diagnosing COVID-19 patients using X-ray images. They used a dataset of 50 X-ray images separated into two groups: 25 positive COVID-19 images and 25 negative COVID-19 images. The image was resized to 224 × 224 pixels. MobileNet, ResNet-v2, Inception-ResNet-v2, Xception, Inception-v3, DenseNet, and revised VGG19 were among the deep learning models used in the COVIDX-Net model. According to their findings, the VGG19 and DenseNet models performed similarly, with an F-score of 91 percent for COVID-19 cases. Sethy and Behera [12] extracted features from various pre-trained CNN architectures. With 50 X-ray images (25 normal and 25 COVID-19 cases), the ResNet50 together with the Support Vector Machine (SVM) classifier achieved the maximum accuracy of 95.38%. Tulin Ozturk [13] also introduced DarkCovidNet, a deep-learning model built on the DarkNet model. It is used to extract data from X-ray images in order to detect COVID-19. They divided the experiment into two categories: binary classification and multi-class classification. They used 125 COVID-19 positive results in total. For binary classification, they achieved an accuracy of 98.08%, while for multi-class classification, they achieved an accuracy of 87.02%. Ucar and Korkmaz [14] proposed a COVIDiagnosis-Net model that combined SqueezeNet and Bayesian optimizers and achieved 98.3% test accuracy across three classification groups. Apostolopoulos et al. [15] tested a number of known CNN models for classification of COVID-19 cases and found the best accuracy of 98.75% and 93.48% for binary and multi-class classification cases. Shrivastava et al. [16] has used various state-of-the-art deep CNN models with fine tuning approach to perform experiment on the same dataset. They recorded the classification accuracy of 98.56% using InceptionResNetV2 model for binary classification and 88.44% using Xception model for multiclass classification. Recently, Farooq and Hafeez [17] developed COVID-ResNet. It is a ResNet-based CNN model for classification of COVID-19 and three other instances (normal, bacterial pneumonia and viral pneumonia). They achieved a 96.23% accuracy rate using a freely accessible dataset (COVIDx); however, in this study, only 68 COVID-19 X-ray images were used.
The most challenging aspect of using deep learning models is collecting enough number of correctly recorded samples for successful training. According to the literature, earlier models are validated with smaller samples and in most cases the data is unbalanced. In addition, in order to detect COVID-19, the majority of the above studies used standard techniques such as VGG and ResNet. However, DarkCovidNet used DarkNet model and achieved better accuracy compared to other studies. So, instead of starting our model from scratch, this paper uses DarkCovidNet as starting model and modifies it to get better accuracy and also, we managed to develop a dataset which consists more than 3000 Covid-19 X-ray images.
The following are the paper's key contributions:
A modified DarkCovidNet model has been presented to achieve high classification performance.
We have obtained over 10,000 Covid-19 X-ray images from various sources, addressing the lack of X-ray images in various literatures.
As compared to the base DarkCovidNet model, the obtained classification accuracy is very high.
Methodology
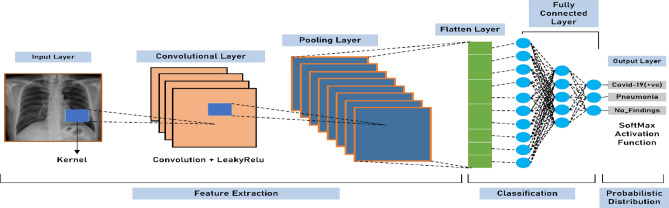
CNN mainly consists of three layers, a convolutional, pooling, and fully connected layer. These layers are sequentially explained below and Fig. 1 illustrates a standard CNN model with these layers.
Fig. 1.
A standard architecture of convolutional neural network
In general, a deep CNN model has alternate stacking of multiple convolution and pooling layers. Deep learning is a subset of machine learning techniques. It is a particular form of representation-based computing in which the network learns and establishes the inherent features for every subsequent hidden layer of neurons [15]. The term "deep" is derived from several hidden layers within the structure of the Artificial Neural Network (ANN). Among various deep learning models, CNNs showed excellent performance in various domains such as object detection [16], image classification [17], medical image analysis [14], agriculture [18], remote sensing [19] and so on.
Convolution layer
The convolution layer utilizes filters that lead convolution tasks as it checks the input image with its dimensions. Its hyperparameters incorporate the size of the stride and the filter. Stride is the size of the step passed each time by the convolution filter. Normally, a stride scale is 1, indicating that the filter slides pixel by pixel. The output is called a feature map or an activation map. We use an activation mechanism to make our output non-linear, much like every other neural network. The yield of the convolution will be gone through the activation mechanism. Mathematically, the 2D convolution operation can be defined using Eq. (1) for kernel (Z) and input image (Y). Further, Fig. 2 shows a basic convolution operation using 3 × 3 filter with stride of 1.
| 1 |
sliding convolutional filters are added to the input by a 2D convolutional layer. The layer compresses the input by vertically and horizontally shifting the filters on the input and measuring the scalar product for weights, thereby adding a bias term.
Fig. 2.
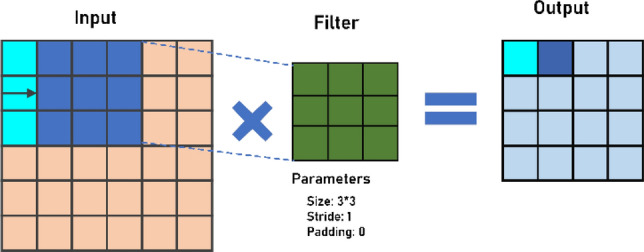
An example of Convolution Operation
Pooling layer
The pooling operation reduces the spatial dimension of input sample while retaining substantial information. Average and max pooling are commonly used pooling operations. Typically, the max-pooling function is used. Figure 3 depicts the max pooling operation with pool size of 2 × 2 and stride of 2.
Fig. 3.
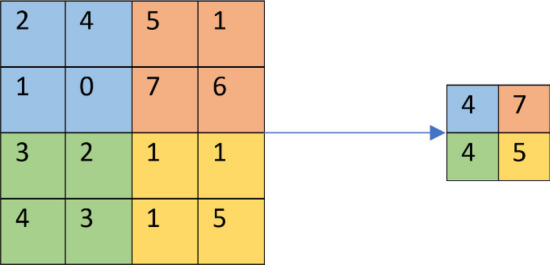
Max pooling with 2 × 2 filter size and strides of 2
Fully connected layer
It signifies the completely connected i.e., each neuron of the current layer is linked to all the neurons of the previous layer. Fully connected layer "flattens" and converts the output of the previous layers to a single vector that can be used to input the next level. There may be more than one fully connected layers. In the last fully connected layer, the total number of neurons is equal to the number of classes. Softmax function is generally used at the last layer that provides probability distribution of predicted classes. The Fig. 4 shows a graphical representation of fully connected layer.
Fig. 4.
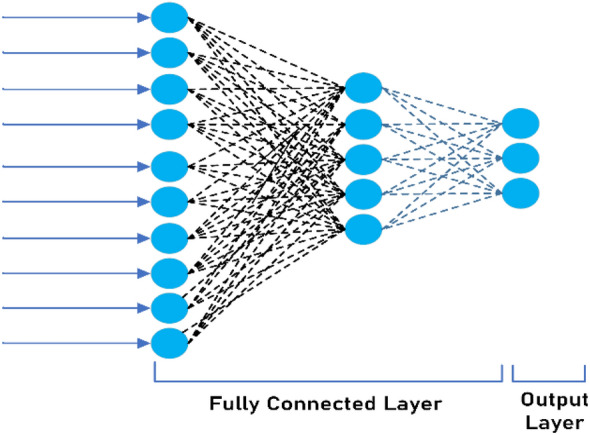
Fully Connected Layer
Modified DarkCovidNet
CNN has played a significant role in detecting and classifying images particularly for medical purposes. The layers and internal parameters of CNN can be adjusted to achieve better performance in a particular task. Along this line, we have modified the DarkCovidNet [9] model to achieve improved performance. DarkCovidNet uses DarkNet-19 [20] as a base model which is constructed using YOLO (You only look once) real-time object identification system. Let, CL denotes a convolutional layer, and MP denotes a Maxpool layer. Then, the structure of DarkNet-19 can be represented in Eq. (2). It uses Leaky rectified linear unit (LeakyReLU) as activation function that is represented in Eq. (3).
| 2 |
| 3 |
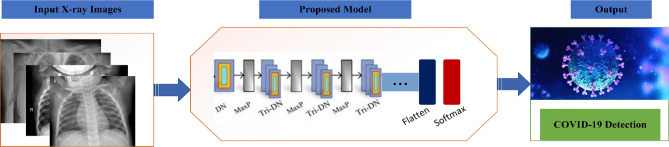
Compared to the original DarkNet architecture, fewer layers and filters were used in DarkCovidNet. DarkCovidNet model has 17 convolutional layers and 5 max pool layers. We modified the model by increasing the number of convolutional layers from 17 to 19 and keeping the number of pooling layers as it is which is 5. Hence, our modified DarkCovidNet model can be represented using Eq. (4). Figure 5 indicates the general demonstration of the proposed model where DN represent DarkNet Layers and MaxP denotes Max pool layer. Likewise, each Tri-DN denotes three DN blocks.
| 4 |
Fig. 5.
General demonstration of the proposed model
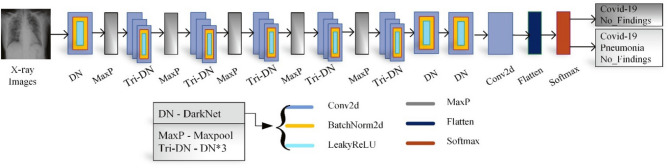
Figure 6 represents the architecture of proposed modified DarkCovidNet model. It has input layer, DarkNet (DN) layers, convolution layers (Conv2d), max pooling layers (MaxP), fully connected layers, and output layer. X-ray images are fed to DN layer of the model with each DN blocks consisting a Conv2d, BatchNorm layer and LeakyReLU operation. Likewise, each Tri-DN denotes three DN blocks. To standardize the inputs and reduce the training time, the batch normalization process is used. Thus, it increases the model stability. The LeakyReLU operation is the modification of ReLU which allows a small negative value when the input is less than zero.
Fig. 6.
Architecture of proposed modified DarkCovidNet model
The details of layers and model parameters are given in Table 1. The layer type shows the type of layer used and output shape represents the size of the output image of corresponding layer. The # Parameters indicates the number of learnable parameters.
Table 1.
The details of layers and parameters of the proposed model
| # Layer | Layer type | Output shape | # Parameters |
|---|---|---|---|
| 1 | DN | [8, 256, 256] | 216 |
| 2 | MaxP | [8, 128, 128] | 0 |
| 3 | DN | [16, 128, 128] | 1,152 |
| 4 | DN | [8, 130, 130] | 128 |
| 5 | DN | [16, 130, 130] | 1,152 |
| 6 | MaxP | [16, 65, 65] | 0 |
| 7 | DN | [32, 65, 65] | 4,608 |
| 8 | DN | [16, 67, 67] | 512 |
| 9 | DN | [32, 67, 67] | 4,608 |
| 10 | MaxP | [32, 33, 33] | 0 |
| 11 | DN | [64, 33, 33] | 18,432 |
| 12 | DN | [32, 35, 35] | 2,048 |
| 13 | DN | [64, 35, 35] | 18,432 |
| 14 | MaxP | [64, 17, 17] | 0 |
| 15 | DN | [128, 17, 17] | 73,728 |
| 16 | DN | [64, 19, 19] | 8,192 |
| 17 | DN | [128, 19, 19] | 73,728 |
| 18 | MaxP | [128, 9, 9] | 0 |
| 19 | DN | [256, 9, 9] | 294,912 |
| 20 | DN | [128, 11, 11] | 32,768 |
| 21 | DN | [256, 11, 11] | 294,912 |
| 22 | DN | [128, 13, 13] | 32,768 |
| 23 | DN | [256, 13, 13] | 294,912 |
| 24 | Conv2d | [3, 13] | 6,912 |
| 25 | Flatten | [507] | 0 |
| 26 | Softmax | [3] | 1,524 |
Results and discussion
Dataset description
X-ray images obtained from various sources were used in this study for the diagnosis of COVID-19. First source is Cohen JP [21] who developed a COVID -19 X-ray image database with images from different sources and it is continuously updated with images from other researchers studying various areas. There are currently 468 X-ray images in the database, which have been diagnosed with COVID-19. In addition, from COVID-19 Radiography Database [22], we have used 1200 COVID-19 positive X-ray images, 1341 X-ray images of normal, and 1345 X-ray images of Pneumonia found in the Database. Furthermore, for the normal and pneumonia images, we have used Chest X-Ray Images (Pneumonia) [23]. From this database, 1583 X-ray images of normal, and 4273 X-ray images of Pneumonia are found. In total, our dataset includes 1648 X-ray images of Covid-19 positive cases, 5618 X-ray images of Pneumonia cases and 2924 X-ray images of normal cases. We have prepared around 10,000 X-ray images of dataset. The overall conclusion of our database is presented in Tables 2 and 3. The sample for the X-ray images of each case are shown in Fig. 7.
Table 2.
Detailed description of number of X-ray images used for binary class
| X-ray image Category | Train | Testing |
|---|---|---|
| Covid-19 | 1120 | 280 |
| No-Findings | 3600 | 900 |
Table 3.
Detailed description of number of X-ray images used for multi-class classification
| X-ray image Category | Train | Testing |
|---|---|---|
| Covid-19 | 1120 | 280 |
| No-Findings | 3600 | 900 |
| Pneumonia | 3600 | 900 |
Fig. 7.
Sample x-ray images from the database: a Covid-19 positive, b No-findings, c Pneumonia
Experimental analysis
The input chest X-ray image is reshaped to 256 × 256 pixels in size and separated into batches. Following that, the normalization operation would take place. The tests are conducted in two cases to diagnose COVID-19 using X-ray images. In the first case, the model was trained to differentiate between COVID-19 and No Findings classes using binary classification. In the second case, the model has been trained to divide the X-ray images into three classes: COVID-19, Pneumonia, and No Findings. The dataset has been split into two parts: 80 percent for training and 20 percent for testing. The Five-fold cross-validation is used to assess the proposed model's performance. We used 100 epochs for binary classification and 50 epochs for multiclass classification. Due to the fact that the number of X-ray images used for multiclass classification was more than 10,000, the training time was too high. That is the reason why we have used a smaller number of epochs for multiclass classification. In addition, to prevent overfitting, we used early stopping parameters. The model's success was assessed using a variety of evaluation metrics such as F1 score, precision, specificity, sensitivity, and accuracy.
Our model has shown an average accuracy of 99.53% for a binary classification with F1 score, precision, specificity, and sensitivity shown in Table 4. Similarly, for multiclass classification our model has achieved an average accuracy of 94.13% with F1 score, precision, specificity, and sensitivity shown in Table 5.
Table 4.
Classification results obtained using proposed model for binary classification
| Folds | Performance Metrics (%) | ||||
|---|---|---|---|---|---|
| F1 score | Precision | Specificity | Sensitivity | Accuracy | |
| First Fold | 98 | 100 | 100 | 98.57 | 99.58 |
| Second Fold | 99 | 100 | 100 | 97.50 | 99.49 |
| Third Fold | 99 | 100 | 100 | 97.86 | 99.41 |
| Fourth Fold | 99 | 100 | 100 | 98.21 | 99.66 |
| Fifth Fold | 99 | 100 | 100 | 97.86 | 99.49 |
| Average | 98.8 | 100 | 100 | 98 | 99.53 |
Table 5.
Classification results obtained using proposed model for multiclass classification
| Folds | Performance Metrics (%) | ||||
|---|---|---|---|---|---|
| Specificity | Sensitivity | F1 score | Precision | Accuracy | |
| First Fold | 93.66 | 95.71 | 89 | 82 | 94.09 |
| Second Fold | 93.81 | 95.71 | 90 | 84 | 93.94 |
| Third Fold | 94 | 96.07 | 89 | 83 | 94.28 |
| Fourth Fold | 93.66 | 95.71 | 89 | 82 | 94.23 |
| Fifth Fold | 94.05 | 94.64 | 88 | 83 | 94.13 |
| Average | 93.84 | 95.57 | 89 | 82.8 | 94.13 |
Confusion matrix
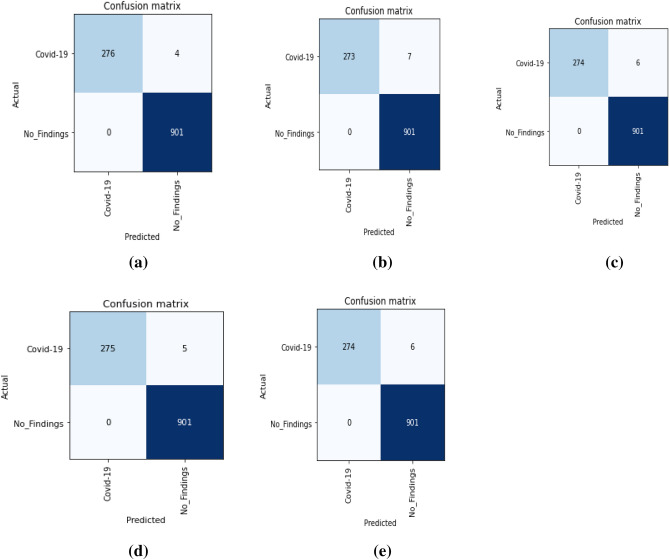
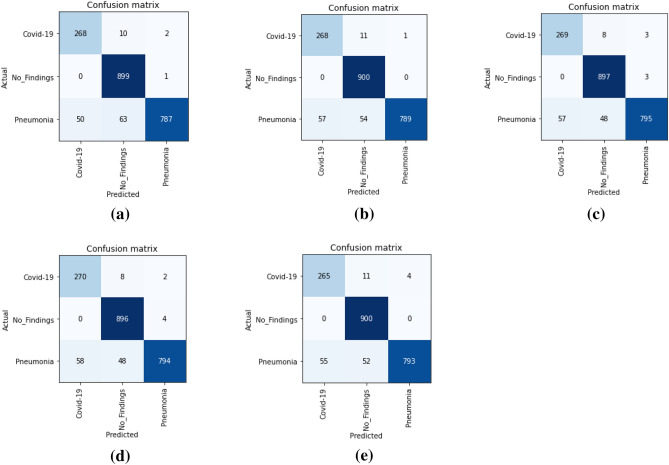
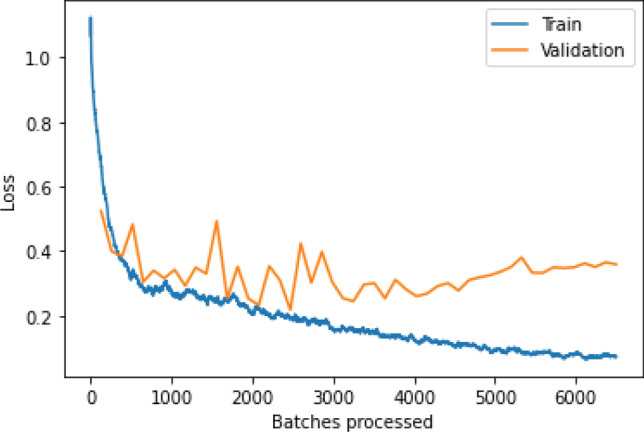
The counts of test records correctly and incorrectly estimated by a classification model are used to determine the model's accuracy. The confusion matrix (CM) gives a more detailed image of not only a predictive model's results, but also which classes are being predicted correctly and incorrectly, as well as the types of errors that are occurring. Figure 8 and Fig. 9 shows the confusion matrix obtained for binary and multiclass classification, respectively for five folds. In confusion matrix, the diagonal elements show the correct classification and non-diagonal elements represent incorrect classification. From Fig. 8, it can be observed that the non-diagonal elements have very low numbers for all five folds which demonstrate that the misclassification rate is very low in case of binary classification. On the other hand, Fig. 9 shows the low misclassification rate in all the classes except the Pneumonia class in case of multiclass classification. In all the five folds, few of the test images which belongs to Pneumonia class were classified as either COVID-19 or No-finding class. In addition, the number of batches processed vs. validation loss for multiclass classification were depicted in Fig. 10. To validate our model’s performance, we have presented the comparative performance analysis along with existing models on diagnosis of COVID-19 using X-ray images in Table 6. It can be observed that, compared to other literature, the proposed model has a better accuracy.
Fig. 8.
Confusion matrix for binary classification: a First-fold CM, b Second-fold CM, c Third-fold CM, d Fourth-fold CM, e Fifth-fold CM
Fig. 9.
Confusion matrix for multiclass classification: a First-fold CM, b Second-fold CM, c Third-fold CM, d Fourth-fold CM, e Fifth-fold CM
Fig. 10.
The number of batches processed vs. validation loss for multiclass classification
Table 6.
Comparative performance analysis of the proposed diagnostic method of COVID-19 with other research papers established using X-ray images
| Authors | Modality | # Classes | Model Used | Accuracy (%) |
|---|---|---|---|---|
| Hemdan et al. [13] | X-ray | Infected (COVID-19(+ ve)–25 non-infected- 25 | COVIDX-Net | 90.0 |
| Wang and Wong [24] | X-ray | Infected ( +)–53 COVID-19 ( −)–5526 No findings—8066 | COVID-Net | 92.4 |
| Sethy and Behra [10] | X-ray | Infected ( +)–25 Non-Infected ( −)–25 | ResNet50 + SVM | 95.38 |
| Ioannis et al. [25] | X-ray | Infected ( +)–224 Pneumonia–700 Healthy–504 | VGG-19 | 93.48 |
| Ozturk et al. [9] | X-ray | Infected ( +)–125 No-Infected–500 | DarkCovidNet | 98.08 |
| Infected ( +)–125 Pneumonia–500 No-Infected–500 | 87.02 | |||
| Proposed Model | X-ray | Infected ( +)–1300 No-Infected–4500 | Modified DarkCovidNet | 99.53 |
| Infected ( +)–1300 Pneumonia–4500 No-Infected—4500 | 94.13 |
We plan to add more X-ray radiology images to our model in the future to further validate it. This model can be deployed in the cloud and used to easily identify and rehabilitate patients. The workload for clinicians could be greatly reduced as a result of this. We'll use CT images to detect COVID-19 and compare the findings to the proposed model trained on X-ray images. We'll also try to gather local radiology images for COVID-19 cases from Bhubaneswar sites and compare them to our model. Once the requisite experiments are performed, we expect to use the built model to screen patients in local hospitals.
Conclusion
To avoid spreading of illness to numerous people, early prediction of COVID-19 patients is crucial. In this study, a convolutional neural network-based model has been proposed using chest X-ray images obtained from COVID-19 patients, viral pneumonia and normal. Our designed model is capable of performing binary class tasks with 99.53% accuracy and 94.13% accuracy in multi-class cases. Due to these promising performances, it may be used in hospitals or through cloud to help medical doctors and prioritize the patients in need. Moreover, in rural areas of countries that are heavily impacted by COVID-19, our model can be used to resolve problems such as delayed outcomes and fewer radiologists. However, the limited number of COVID-19 X-ray images is a downside of this work. By using more of these images, we aim to make our model more consistent and reliable in future.
COVID-19 is already a threat to the global healthcare system, and thousands of people have already lost their lives. The loss of breathing, failing other organs, has caused deaths. Physician time is dedicated to outdoor care or emergencies, and computer-aided analysis can save lives with early screening. In a pandemic where the burden of disease and the need for prevention steps do not match the availability of services, this automatic model could be particularly useful.
Funding
No funding has been received for this research work.
Declarations
Conflict of interest
All authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dawit Kiros Redie, Email: dawitkiros1922@gmail.com.
Abdulhakim Edao Sirko, Email: abdulhakim.edao1@gmail.com.
Tensaie Melkamu Demissie, Email: tensaiemelkamu@gmail.com.
Semagn Sisay Teferi, Email: semagn2712@gmail.com.
Vimal Kumar Shrivastava, Email: vimal.shrivastavafet@kiit.ac.in.
Om Prakash Verma, Email: vermaop@nitj.ac.in.
Tarun Kumar Sharma, Email: taruniitr1@gmail.com.
References
- 1.World Health Organization website (2020) Pneumonia of unknown cause, China. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/
- 2.Centers for Disease Control and Prevention (CDC) (2020) How COVID-19 Spreads. Centers for Disease Control and Prevention (CDC). https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html
- 3.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. New England J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alazab M, Awajan A, Mesleh A, Abraham A, Jatana V, Alhyari S. COVID-19 prediction and detection using deep learning. Int J Comput Inf Syst Ind Manag Appl. 2020;12:168–181. [Google Scholar]
- 5.Worldometers. (2020) Coronavirus Cases. Available: https://www.worldometers.info/coronavirus/
- 6.Zhang J, Cao Y, Dong X, Wang B, Liao M, Lin J et al. (2020) Distinct characteristics of COVID-19 patients with initial rRT-PCR-positive and rRT-PCR-negative results for SARS-CoV-2. Allergy [DOI] [PMC free article] [PubMed]
- 7.Joynt GM, Wu WK. Understanding COVID-19: what does viral RNA load really mean? Lancet Infect Dis. 2020;20(6):635–636. doi: 10.1016/S1473-3099(20)30237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zohra A, Parul D, Deepjyoti DC, Satabdi K, Zohaib H, Zainab A. Chest x-ray findings in covid-19 pneumonia: a pictorial review. International J Sci Res. 2020;2:1069. [Google Scholar]
- 9.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;2:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sethy PK, Santi KB (2020) Detection of coronavirus disease (covid-19) based on deep features. Preprints 2020030300
- 11.Song Y, Shuangjia Z, Liang L, Xiang Z, Xiaodong Z, Ziwang H, Jianwen C et al. (2020) Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv [DOI] [PMC free article] [PubMed]
- 12.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 13.Hemdan EED, Shouman MA, Karar ME (2020) COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv preprint arXiv:2003.11055
- 14.Shrivastava VK, Monoj KP (2020) Deep convolutional neural network-based diagnosis of COVID-19 using X-ray images. IOP Publishing Ltd
- 15.Oludare IA, Aman J, Abiodun EO, Kemi VD, Nachaat AEM, Humaira A. State-of-the-art in artificial neural network applications: a survey. Heliyon. 2018;4(11):e00938. doi: 10.1016/j.heliyon.2018.e00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y-H, Kim Y. Comparison of CNN and YOLO for object detection. J Semicond Disp Technol. 2020;19(1):85–92. [Google Scholar]
- 17.Sanjana S, Shriya VR, Vaishnavi G, Ashwini K. A review on various methodologies used for vehicle classification, helmet detection and number plate recognition. Evol Intel. 2021;14(2):979–987. doi: 10.1007/s12065-020-00493-7. [DOI] [Google Scholar]
- 18.Pattnaik G, Shrivastava VK, Parvathi K. Transfer learning-based framework for classification of pest in tomato plants. Appl Artif Intell. 2020;34(13):981–993. doi: 10.1080/08839514.2020.1792034. [DOI] [Google Scholar]
- 19.Bera S, Shrivastava VK. Effect of pooling strategy on convolutional neural network for classification of hyperspectral remote sensing images. IET Image Proc. 2020;14(3):480–486. doi: 10.1049/iet-ipr.2019.0561. [DOI] [Google Scholar]
- 20.Redmon J, Ali F (2017) YOLO9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7263–7271
- 21.Cohen JP (2020) COVID-19 Image Data Collection. https://github.com/ieee8023/COVID-chestxray-dataset
- 22.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal MBI, Al-Emadi N, Reaz MBI (2020) Can AI help in screening Viral and COVID-19 pneumonia? arXiv preprint, 29 March 2020. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database
- 23.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: Hospital scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106
- 24.Wang L, Zhong QL, Alexander W. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannis D, Apostolopoulos, TB (2003) COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks.11617 [DOI] [PMC free article] [PubMed]