Abstract
Phenotype-driven chemical genetic screens in zebrafish have become a proven approach for both dissection of developmental mechanisms and discovery of potential therapeutics. A library of small molecules can be arrayed into multiwell plates containing zebrafish embryos. The embryo becomes a whole organism in vivo bioassay that can produce a phenotype upon treatment. Screens have been performed that are based simply on the morphology of the embryo. Other screens have scored complex phenotypes using whole mount in situ hybridization, fluorescent transgenic reporters, and even tracking of embryo movement. The availability of many well-characterized zebrafish mutants has also enabled the discovery of chemical suppressors of genetic phenotypes. Importantly, the application of chemical libraries that already contain FDA-approved drugs has allowed the rapid translation of hits from zebrafish chemical screens to clinical trials.
INTRODUCTION
The zebrafish has emerged as an ideal vertebrate model organism for in vivo phenotype-based screens. The high degree of gene conservation between zebrafish and mammals has allowed the rapid translation of screen results to humans. There is also a growing collection of mutants and transgenics in zebrafish that can be used to ask more complex questions. The large number of embryos that adult zebrafish can spawn each week enables them to support high-throughput studies. The optically clear embryos can be easily observed under a light microscope, facilitating scoring of morphology or fluorescent transgene expression, and more recently adoption of automated screening platforms. Many chemicals can be easily diluted to a physiological dose in the embryos’ water. Perhaps most importantly, chemicals can be added conditionally; treatment during a specific developmental window adds a temporal dimension not possible with a genetic mutant, which is essentially constitutive. With advancing screen design, chemicals that were first used to simply induce a morphological change, are now being used to rescue genetic mutations, modulate gene expression patterns and levels, and produce subtle phenotypes at the cellular level that can be detected with transgenic reporter lines. Together, the intersection of chemical screening and zebrafish development is emerging as a powerful approach to acquire complex phenotypic data.
THE DEVELOPMENT OF PHENOTYPE-DRIVEN CHEMICAL SCREENS USING THE ZEBRAFISH EMBRYO
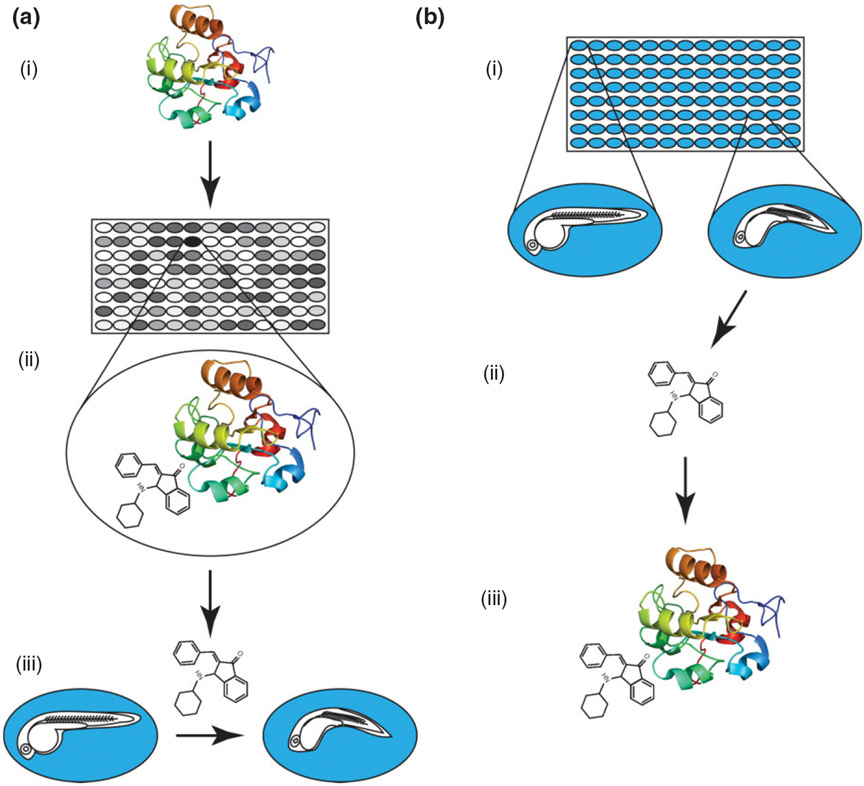
The traditional approach for chemical screening that still dominates the pharmaceutical industry is target-oriented drug discovery. The goal is to first find a specific molecular target relevant to a disease and then, using either a cell-based reporter assay or in silico design, identify a compound that can activate or inhibit that target. If a potential ‘hit’ is found in the primary screen of a large chemical library, the compound must translate to an in vivo model before being tested and developed as a potential new therapeutic (Figure 1(a)). Most target-based screens have a very clear readout and compounds of interest can be identified quickly. However, compounds can fail in the following stages of verification for a number of reasons. For example, a compound can behave differently in vivo due to pharmacokinetic properties or have off-target effects that were not detected in a primary screen. Perhaps most importantly, the action of a compound on a single target or pathway may not be enough to improve a disease state. One way to overcome these pitfalls of target-oriented drug discovery is to screen for compounds based on phenotype (Figure 1(b)). Using a phenotype-driven screen, a therapeutic compound can be identified without knowing the exact mechanism of the disease. Even if a compound acts on multiple pathways or has off-target effects, a positive phenotypic outcome becomes the only important result.
FIGURE 1 ∣.
(a) A target-based chemical screen. (i) A protein of interested is selected. (ii) High-throughput screening is used to find a small molecule that produces a positive result in a bioassay. (iii) The compound must be tested to determine if it produces a desired phenotype in vivo—in this example zebrafish embryos are treated. (b) A phenotype-based chemical screen. (i) Zebrafish embryos are arrayed in multiwell plates containing compounds from a library. Stage-matched wild-type embryos (left) are compared to those with a phenotype (right). (ii) The small molecule that produces the phenotype is retrieved from the library and retested. (iii) The protein target and mechanism of action must be determined.
In the 1960s, the NIH began screening a large number of molecules against the growth of human cancer cell lines to identify compounds that might be clinically relevant.1 In these studies, a growth inhibition phenotype was the major focus, with less emphasis on the mechanisms of action. Although cell-based screens have proven useful, they do not allow for the capture of complex phenotypes that require in vivo and often heterotypic cellular interactions. The idea of small molecule screening in a whole organism emerged from this need for phenotypic assays with a more complex readout. The application of zebrafish embryos specifically came from observations made during toxicology screens; chemical contaminants in water were producing complex phenotypic effects. As early as the 1950s researchers began to describe the benefits of the zebrafish embryo as a bioassay for chemical treatments.2-4 It was not until more than four decades later that these early observations would become the foundation for large-scale in vivo screens of drug libraries.
STATE-OF-THE-ART CHEMICAL SCREENING WITH ZEBRAFISH EMBRYOS
The first large-scale application of small molecules to zebrafish embryos was described by Peterson and colleagues in 2000.5 They examined the developmental effects of 1100 molecules on the central nervous system, ear, pigment, and heart, and found molecules that selectively perturbed their development. They recognized that discrete windows of chemical treatment allowed for temporal refinement of the resulting phenotypes. Peterson et al. followed up on these observations by asking whether a small molecule could rescue the aortic coarctation phenotype in hey2 (formerly known as gridlock) mutant embryos. By screening for restoration of blood flow in the tail, they discovered compounds that upregulate vascular endothelial growth factor (VEGF), which is sufficient to suppress the hey2 mutant phenotype.6 Similarly, Stern et al. utilized a 16,000 chemical library to identify suppressors of the mybl2 (formerly known as bmyb) mutation, which leads to an increase in phospho-H3 positive mitotic cells.7 They identified one molecule, persynthamide, that not only rescued the phospho-H3 abnormality but also apoptotic and cell cycle defects; other small molecules were found that modulated the cell cycle in wild-type embryos.8 After further testing of persynthamide, its effects on the cell cycle were found to be zebrafish-specific; this demonstrated that screen hits must be carefully validated before assuming they will translate directly from fish to mammalian systems. Together, these screens paved the way for small molecule approaches to elucidate complex underlying biology. This concept is often encapsulated in the term ‘chemical biology’ or ‘chemical genetics’ because of the obvious parallels with classical genetic screens. A comparison of chemical and genetic screens is presented in Table 1.
TABLE 1 ∣.
Advantages of Chemical Methods Compared with Genetic Methods
| Questions | Genetic Approaches | Chemical Approaches |
|---|---|---|
| Genes involved in later biological processes | Suitable for studying early onset phenotypes | A conditional approach, feasible for studying specific gene functions at different stages |
| Genes with multiple copies or redundant isoforms | Genetic redundancy is often undetected and compound mutants may be needed | Small molecules can bind to different isoforms simultaneously and therefore target the function of multiple genes Both maternal and zygotic products can be targeted |
| The effect of gene dosage | Need to generate an allelic series | Pathway disruption can be easily modulated with dosage |
| Suppressor/enhancer screens | For recessive mutations, only 1/16 of the offspring from parents heterozygous for two mutations will have the phenotype | Small molecules can affect all 1/4 mutant embryos uniformly, increasing the penetrance of phenotype |
The next important step was to bring models of genetic disease onto the platform of chemical screening in zebrafish. A small molecule that can modify or suppress a disease phenotype in the zebrafish embryo has the potential for translation into a novel therapy. This approach was applied to a common and life-threatening human genetic disorder called polycystic kidney disease (PKD). There are two zebrafish mutations that have been mapped to the PKD-related genes pkd2 and ift172. Cao and colleagues performed a chemical modifier screen on these mutants and found that the pan-histone deacetylase (HDAC) inhibitor trichostatin A (TSA) could suppress the embryos’ gross morphological defects.9 Further validation revealed similar effects using another structurally unrelated HDAC inhibitor, valproic acid (VPA). Importantly, treatment of a PKD mouse model with VPA showed a dramatic improvement in kidney function and a reduction in cyst formation. Another notable example of a zebrafish disease model that responds to drug treatment is a transgenic line carrying the acute myeloid leukemia associated fusion gene AML1-ETO.10 Interestingly, its phenotype, which includes accumulation of granulocytic cells, is also suppressed by the HDAC inhibitor TSA. As more zebrafish mutants and transgenics are found that effectively model human disease, so will the potential to match these models with successful drug therapies.
Hematopoietic stem cell (HSC) transplantation is one of the most important life-saving treatments for leukemia and blood-borne disease. However, there is a need to improve the efficiency and success rate of this procedure. To address this, North and colleagues undertook a zebrafish chemical screen to find compounds that expanded the emerging pool of HSCs in the embryo.11 Embryos were treated during the period of HSC specification, and in situ hybridization was used to detect changes in hematopoietic progenitor markers cmyb and runx1. By screening a library of known bioactive compounds, many of which were already FDA-approved, novel links to HSC specification were identified, including the prostaglandin E2 (PGE2) and nitric oxide pathways. Remarkably, the small molecules from the zebrafish screen translated well to HSC assays in the mouse. This strong conservation of function between fish and mammals, and the identification of drugs already clinically approved for other applications, allowed rapid progression to a phase I clinical trial in humans. This trial is currently ongoing and will determine if PGE2 promotes long-term engraftment of HSCs after transplantation, and therefore improved patient survival. There are currently other chemical screens underway in our laboratory that we hope will, independently or synergistically with PGE2, improve the homing and engraftment of HSCs.
The last 10 years have seen a major expansion of zebrafish as a cancer model (reviewed in Ref 12). Mutation or overexpression of various oncogenes has generated zebrafish models of melanoma,13-16 leukemia,10,17-21 and more recently pancreatic cancer22 and rhabdomyosarcoma.23,24 Zebrafish cancer models are now being applied in chemical screens and will potentially reveal how drugs can suppress oncogenic transformation and tumor formation. In 2011, White and Zon used a BRAFV600E transgenic zebrafish that exhibits an expansion of neural crest cells during development.25 By isolating chemical suppressors of the embryonic neural crest lineage, they hypothesized that they could also identify suppressors of adult melanoma. They found that an inhibitor of transcriptional elongation, leflunomide, suppressed not only embryonic neural crest and melanocyte development, but also diminished melanoma growth in vitro and in mouse xenografts. Whether this molecule will have utility in the treatment of human melanoma awaits the planned phase II trial, but again highlights the rapidity with which observations in zebrafish embryos can move to clinical application.
A unique and emerging niche for zebrafish embryos is its capacity for high-throughput behavioral screens. Recently, two landmark studies found that there was a high correlation between complex phenotypes and certain classes of psychoactive drugs. With embryos arrayed into 96-well plates, an automated tracking system was used to record movements in response to changes in light. These data were assembled into a multifactoral behavioral ‘fingerprint’ or ‘barcode’. The screens by Rihel et al. and Kokel et al. had coverage of approximately 4000 or 14,000 unique molecules, and 60,000 or 250,000 behavioral profiles, respectively.26,27 Multidimensional clustering was used to group compounds by particular behaviors, which allowed for predictions of mechanism for previously uncharacterized molecules. Although this screen focused on locomotor activity, the same approach could potentially be applied to other behaviors, and the discovery of new applications for psychotropic drugs.
CHEMICAL GENETIC SCREENS ANSWER DEVELOPMENTAL QUESTIONS
The many strengths of the zebrafish as a model for developmental biology, and for chemical screening, are demonstrated in the discovery of dorsomorphin. Yu and colleagues designed a screen to find a compound that would selectively inhibit bone morphogenetic protein (BMP) receptors.28 This was a daunting task in vitro because the ideal chemical would selectively perturb a BMP receptor without affecting the many other similar receptor kinases. Without an in vivo readout it would be difficult to assay for this level of specificity. Yu and colleagues applied the concept of a forward genetic screen to a chemical screen; they started with a morphological phenotype and worked backwards to find its cause. When conducted in this manner, the embryo becomes a highly sensitive assay for signal pathway output, and rapid reading of the embryonic morphology can lead directly to the affected pathway.
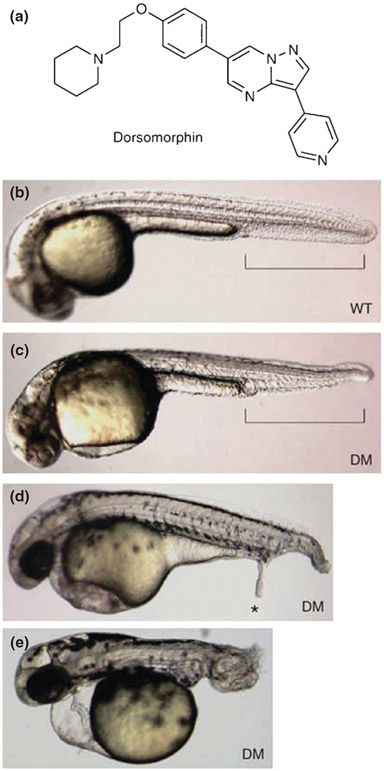
After screening over 7500 compounds, Yu and colleagues identified one that reproducibly dorsalized zebrafish embryos, which they called dorsomorphin (Figure 2). What was striking about this compound was how addition at discrete times and doses produced such specific BMP-related effects; this illustrates the advantage of a chemical over a genetic mutant because the early requirement for BMP in patterning the embryo can be bypassed and requirements for BMP in other contexts can be studied. The addition of the drug at 24 h.p.f, thereby bypassing any gastrulation patterning defects, was used to disrupt BMP-mediated bone mineralization. Systemic injection of dorsomorphin into adult fish was effective at inhibiting BMP-dependent SMAD activation, which is a necessary signaling component in iron homeostasis. The discovery of dorsomorphin illustrates how a phenotypic assay can lead directly to a pathway-specific chemical that can be applied conditionally to study multiple processes throughout the development of the organism.
FIGURE 2 ∣.
Dorsomorphin (DM) induces dorsalization in zebrafish embryos. (a) Structure of DM. (b) Vehicle-treated wild-type zebrafish embryo 36 h.p.f. Ventral tail fin is highlighted in brackets. (c) Zebrafish embryo treated with 10 μM DM at 6–8 h.p.f. and photographed at 36 h.p.f. (d) Zebrafish embryos treated with 10 μM DM at 6 h.p.f. occasionally develop ectopic tails (*) at 48 h.p.f. (e) Embryos treated with 10 μM DM at 1–2 h.p.f. show severe dorsalization at 48 h.p.f. Embryos (b–e) are shown on lateral view. (Adapted with permission from Ref 28. Copyright 2008 Macmillan Publishers Ltd.)
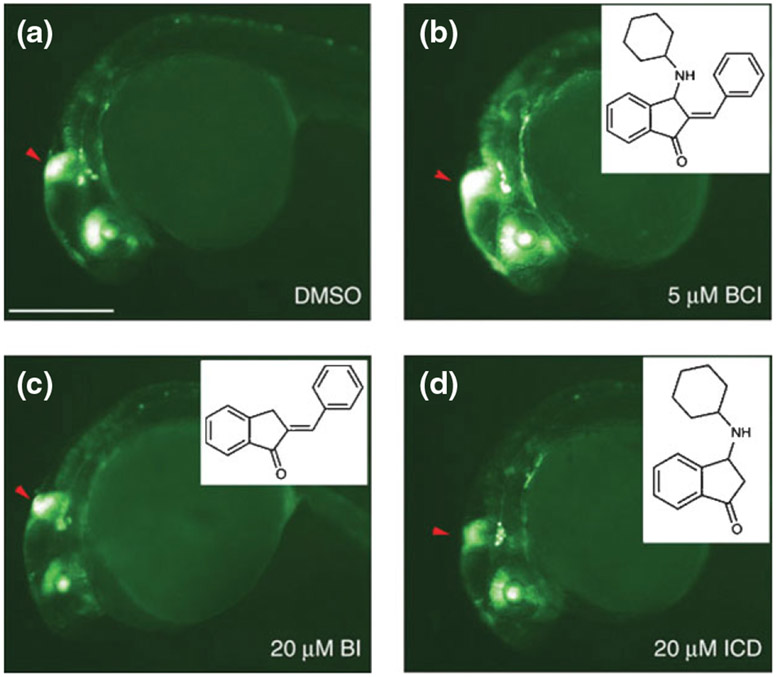
Molina and colleagues used the zebrafish to find a chemical modulator of another important pathway—fibroblast growth factor (FGF) signaling via extracellular signal-regulated kinase (ERK).29 Using a highly sensitive transgenic FGF signaling reporter, embryos were arrayed into 96-well plates and treated from 24 h.p.f for 6–8 h with a 5000 compound library. An increase in intensity of destabilized green fluorescent protein was scored by visual analysis. One small molecule was found that dose-dependently increased FGF reporter signal and was named BCI. A structure–function relationship study was performed to determine which of the chemical side groups were required for enhancement of FGF signaling (Figure 3).
FIGURE 3 ∣.
BCI structure–activity relationship studies. (a–d) Lateral views of 30 h.p.f. Tg(dusp6:EGFP) embryos treated with dimethyl sulfoxide (DMSO) (a), BCI (b), BI (c), and ICD (d). GFP fluorescence was enhanced in BCI-treated embryos (b), whereas related analogs, shown in inner panels, had no effect, even at fourfold higher concentrations (c,d). Red arrowheads mark the mid-hindbrain boundary. Scale bar, 250 μM. (Adapted with permission from Ref 29. Copyright 2009 Macmillan Publishers Ltd.)
As in the study of dorsomorphin, a combination of genetics and chemical treatment allowed a more detailed dissection of the role of BCI in FGF signaling. First, loss of BCI-mediated signal enhancement in an fgf8a (formerly known as ace) null mutant background demonstrated that the FGF ligand is required for BCI to take effect. Next, to test the hypothesis that BCI functions by blocking an endogenous FGF inhibitor, thereby resulting in a net increase of FGF signal, a number of inhibitors were overexpressed. Strikingly, the only FGF inhibitor that could be rescued by BCI was the dual specificity phosphatase 6 (dusp6). Further characterization of this interaction showed that BCI binds an allosteric site on the inactive Dusp6 enzyme, thereby blocking the ERK2-mediated activation of the enzyme that allows dephosphorylation of ERK2 at the catalytic site. Again, it was the ability to conditionally apply the small molecule at various developmental stages that revealed a peak phenotypic dependency; in this case, the phenotype assayed was the FGF-dependent expansion of the cardiac progenitor pool in the anterior lateral plate mesoderm. Although FGF signal enhancement can have an effect on cardiac progenitors at various stages of embryonic development, it was found that BCI treatment at the one-somite stage produced the most prominent result.
Together, the discovery of dorsomorphin and BCI demonstrates the power of using a chemical approach to modulate signaling pathways during development. While an in vitro assay may show an interaction between a chemical and a protein, it cannot bring to light the complex phenotypic result of adding a small molecule to a whole organism at various stages of development. The in vivo chemical genetic screen can quickly read out if a compound produces non-specific effects or toxicity, while often pointing directly to a signaling pathway of interest.
CURRENT TECHNIQUES IN ZEBRAFISH CHEMICAL SCREENING
Although zebrafish chemical genetic screens are gaining wider acceptance, the logistics of designing and performing a screen are still being developed. This section will briefly describe the most current techniques available and how they can be adapted to optimize screening for a specific project (we have previously published detailed protocols elsewhere30,31).
Embryo Collection
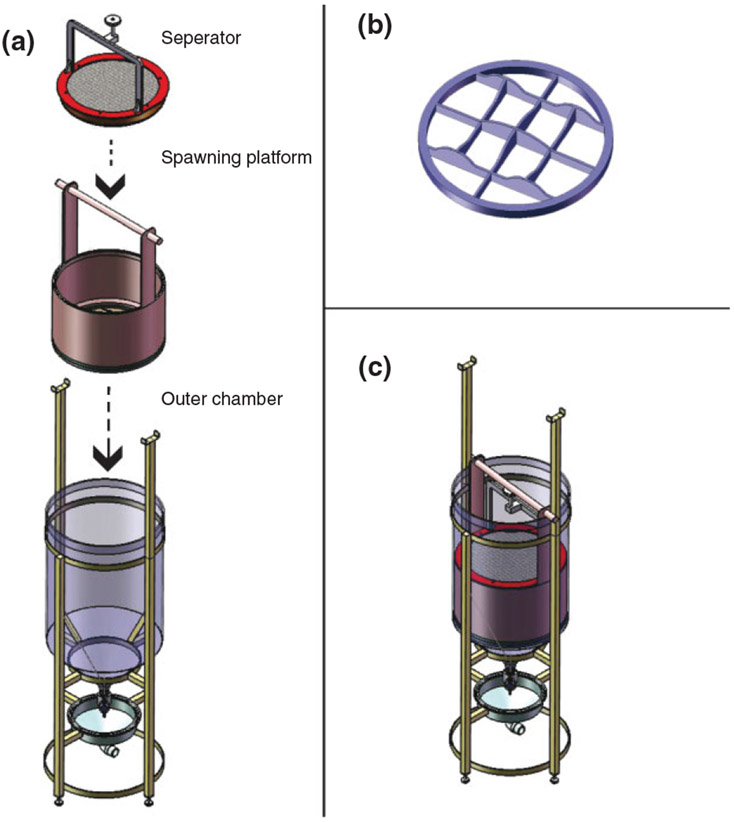
The high fecundity and rapid development of zebrafish is key to their suitability for chemical screening. To achieve high throughput, large numbers of developmentally synchronized embryos are required; particularly at early stages, even a few minutes can shift the embryo into the next discrete developmental stage. Setting up many mating pairs at once is possible but has a number of important limitations; not only is it labor intensive, but collecting increasing numbers of embryos extends spawning intervals and leads to loss of staging synchronization. To address these issues, we have developed a mass spawning vessel that optimizes embryo production (Figure 4).32 Briefly, this large vessel allows for overnight separation of typically 50 male and 50 female adult fish with a shared water supply. In the morning, the fish are mixed in shallow water above a contoured mesh surface that provides an optimal spawning environment. Within a short spawning interval of 10 min or less many thousands of embryos can collected.
FIGURE 4 ∣.
Zebrafish mass breeding vessel. (a) The three primary components of the breeding vessel. (b) Framework of the bottom or ‘floor’ of the spawning platform, showing variation in topography. (c) The breeding vessel, with all three primary components engaged and ready for operation. (Reprinted with permission from Ref 32. Copyright 2011 PLoS ONE)
Embryo Distribution and Handling
After collecting thousands of developmentally synchronized embryos, the challenge becomes removing dead or unfertilized embryos and arraying healthy embryos into plate format for treatment (e.g., 48-well or 96-well plates). For chemical treatments at early gastrula stages (~5.25 h), this requires rapid processing that, at present, does not seem achievable with automated methods. With adequate staff, this process, although tedious, can be performed by hand with high reliability and speed. Automated embryo handlers such as the ZebraFactor (CSEM) or BioSorter (Union Biometrica) may find utility for later chemical treatments when speed is less critical and when embryo quality can be verified by eye.
Another consideration is removal of the protective chorion that surrounds the embryo. The chorion often interferes with downstream processing such as whole mount in situ hybridization. The embryos do not hatch from the chorion until between 48 and 72 h. Unfortunately, at gastrula stages and earlier the embryos are very fragile and cannot come into contact with air or plastic. Processing of dechorionated embryos at early stages must be performed on glass or agarose-coated plastic dishes. Chemical treatments are often performed with the chorions intact but there is concern that this impedes penetration of some small molecules. Small numbers of embryos can be dechorionated by hand with forceps, but for large numbers the chorions are removed using a pronase treatment. It is recommended that chorions be removed before embryos are transferred to mesh bottom plates because they will clog the mesh and block transfer of liquids.
Chemical Application
Small molecules from a master chemical library plate, often in 384-well format, are diluted into embryo media and then distributed onto previously arrayed embryos. Application of chemicals is easily achieved using a standard liquid handling robot, similar to what is often used for cell culture-based applications. Given the small size of the zebrafish embryo, even in the chorion >10 embryos can be arrayed in standard 48-well or 96-well formats to provide multiple biological replicates. Choosing the chemical library depends on the specific goal of the screen. Library size can range from a few dozen to over 100,000. Some libraries focus on specific pathways or targets, such as the BIOMOL Kinase Inhibitor Library, and are useful for a hypothesis-driven study. There are libraries that encompass all FDA-approved drugs and others that are enriched for compounds with known bioactivity (e.g., ICCB). Natural product libraries contain chemically diverse compounds with good potential for drug discovery. There are also libraries (e.g., Chembridge DIVERSet) that have been computationally filtered for pharmacophores that represent a large chemical space but have not been prescreened for bioactivity. The million-compound libraries used by pharmaceutical research companies have not yet been used for zebrafish screening, but with large-scale embryo spawning and robotic sorting this could become a possibility in the future.
Readouts and Scoring
The scoring of chemical effects in a screen is usually the most time-consuming aspect of the process. Usually readout of morphology, whole mount in situ hybridization, or a fluorescent transgene is assessed by eye. The behavioral screens described above made significant advancements in automated recording of dynamic locomoter activity in 96-well plate format. Recently, Yanik and colleagues have developed a microfluidics system that can rapidly load embryos from multiwell plates, orient them correctly in all three axes, and then perform imaging and even laser microsurgery.33 Although the throughput of approximately 30 seconds per embryo in this system is impressive, drift in stage would still be prohibitive for large-scale applications, unless embryos were fixed and the fluorescent signal was preserved. Advances in high-throughput screening of transgenic embryos have also been achieved using multiwell plate readers, with automated image analysis that takes advantage of changes in fluorescent reporter expression.34-36
The Final Step: Linking a Phenotypic Chemical Hit to a Target Protein
Finally, when a screen is complete and hits have been found, a compound and its resultant phenotype must be linked to a pathway and target. If a library of known bioactives was used for the screen sometimes the mechanism can be readily identified based on annotation and existing data. However, there is high potential for small molecules to act on uncharacterized targets, and any annotation will be biased toward the focus of previous screens.
The more common situation is one in which either the protein target is unknown or the annotated target protein is not linked to the phenotype of interest. In this case, chemoinformatic algorithms like Discoverygate can provide mechanistic clues. These approaches all rely on structural similarities between a lead compound and those within ‘chemical space’ for which some mechanistic information is known. The chemical space in this setting can be very large, for example, that found within the MDL Drug Data Report utilized by the Discoverygate algorithm. This can incorporate not only published papers about mechanism, but perhaps more importantly patent applications from pharmaceutical companies. The latter provides a vast trove of mechanistic data that is otherwise difficult to access in a systematic manner. Other algorithms, such as PubChem, ChemBank and SEA, provide similar functionality.
If the chemoinformatic approaches do not provide clues, unbiased microarray-based experiments can be useful. In this case, the chemical is applied to elicit the phenotype of interest, and a microarray ‘signature’ is generated. This signature can then be used as a query into large repositories of microarray drug signatures, such as the Connectivity Map database. When a similarity between the signature of a novel compound and a known compound is found, that may offer a potentially conserved mechanism that can then be tested. Conceptually, any microarray signature database can be queried, not just the Connectivity Map. For example, we treated zebrafish embryos with a molecule called leflunomide, generated a microarray signature, and then utilized Gene Set Enrichment Analysis to compare our signature to that of known genetic mutants.25 This strongly suggested that leflunomide acted to suppress transcriptional elongation in a manner analogous to the supt5h (formerly known as spt5) elongation mutant, which can then be proven with downstream assays. This approach will improve over time as an increasing number of microarray signatures of zebrafish embryos are generated.
As mass spectrometry becomes better optimized for the fish, measurement of drug-induced changes in protein levels and post-translational modifications will complement transcriptome analysis.37 Sometimes a small molecule hit in a library can be successfully modified with a tag for pulldown of associated proteins. Unfortunately, this approach can be difficult because tagging a small molecule often blocks or reduces its function. Successful tagging of a compound often requires extensive structure–activity relationship analysis. One approach that has proved successful is tagging of an entire library before performing the phenotypic screen, with the caveat that functional inhibition by a tag could result in a false negative. The advantage of screening with a tagged library is that the hit can be used directly for pulldown of a protein target.38 Lastly, as more chemical genetic screens are performed in zebrafish, there is a growing need for a centralized database that would collate important data; for example: phenotypic results, treatment windows, wild-type or mutant background, target associations, toxicity, and linking tables between differently annotated chemical libraries. There are already a growing number of online resources that can be used to follow up hits from chemical screens (Table 2). Undoubtedly, there will soon be a convergence of rapid screening techniques, automated scoring, and target protein identification that will allow larger and more productive chemical genetic screens in zebrafish.
TABLE 2 ∣.
Examples of Web-Based Resources for Chemoinformatics31
| Web Address | Information Provided by this Resource; Free or Site License |
|---|---|
| http://www.discoverygate.com/ | Information on published literature and patents; site license |
| http://sea.bkslab.org/ | Relates protein targets based on chemical similarity of their ligands; free |
| http://pubchem.ncbi.nlm.nih.gov/ | Information on all publicly available molecules; free |
| http://chembank.broadinstitute.org/ | Public access repository of screens done at the Broad Institute and elsewhere; free |
| http://www.broadinstitute.org/cmap/ | Database connecting chemicals to phenotypes to gene expression; free |
| http://www.ingenuity.com/ | Relates defined biological pathways to chemicals which perturb the pathway; site license |
CONCLUSION
It is now just over 10 years since the first large-scale small molecule screens in zebrafish have been performed, and several themes are emerging. First, small molecules can be highly useful tools in understanding basic aspects of developmental, stem cell and cancer biology. Second, these screens can yield selective, if not specific, phenotypes in virtually any organ system. Third, these in vivo and phenotypically complex screens complement, rather than compete with, similar screens performed in cell culture; for example, a molecule that gives an interesting phenotype in fish may have its mechanism of action more easily worked out in cell culture systems. Finally, advances in throughput and automated analysis are needed if the application of much more structurally diverse and potentially interesting chemical libraries are to be achieved in the next few years.
Footnotes
WEB RESOURCES
HHMI’s BioInteractive—What is chemical genetics? (http://www.hhmi.org/biointeractive/genomics/poster_a2.html)
Harvard Medical School Screening Facility—Chemical Libraries http://iccb.med.harvard.edu/screening/compound_libraries/index.htm
REFERENCES
- 1.Driscoll JS. The preclinical new drug research program of the National Cancer Institute. Cancer Treat Rep 1984, 68:63–76. [PubMed] [Google Scholar]
- 2.Hisaoka K, Hopper A. Some effects of barbituric and diethylbarbituric acid on the development of the zebra fish, Brachydanio rerio. Anat Rec 1957, 129:297–307. [DOI] [PubMed] [Google Scholar]
- 3.Jones R, Gibson W, Nickolls C. Factors influencing mitotic activity and morphogenesis in embryonic development. I. The effects of thyroxine and thiouracil on the development of Brachydanio rerio (zebrafish). Anat Rec 1951, 111:440–586. [Google Scholar]
- 4.Jones R, Huffman M. Fish embryos as bio-assay material in testing chemicals for effects on cell division and differentiation. Trans Am Microsc Soc 1957, 76:177–183. [Google Scholar]
- 5.Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A 2000, 97:12965–12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, Macrae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol 2004, 22:595–599. [DOI] [PubMed] [Google Scholar]
- 7.Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, Weber G, Tallarico JA, King RW, Zon LI. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol 2005, 1:366–370. [DOI] [PubMed] [Google Scholar]
- 8.Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Design 2006, 68:213–219. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Semanchik N, Lee SH, Somlo S, Barbano PE, Coifman R, Sun Z. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc Natl Acad Sci U S A 2009, 106:21819–21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh JR, Munson KM, Chao YL, Peterson QP, Macrae CA, Peterson RT. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development 2008, 135:401–410. [DOI] [PubMed] [Google Scholar]
- 11.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang I-H, Grosser T, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 2007, 447:1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Leach SD. Zebrafish models for cancer. Annu Rev Pathol 2011, 6:71–93. [DOI] [PubMed] [Google Scholar]
- 13.Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A 2005, 102:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol 2005, 15:249–254. [DOI] [PubMed] [Google Scholar]
- 15.Dovey M, White RM, Zon LI. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 2009, 6:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anelli V, Santoriello C, Distel M, Koster RW, Ciccarelli FD, Mione M. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish 2009, 6:417–424. [DOI] [PubMed] [Google Scholar]
- 17.Kalev-Zylinska ML, Horsfield JA, Flores MVC, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development 2002, 129:2015–2030. [DOI] [PubMed] [Google Scholar]
- 18.Onnebo SM, Condron MM, McPhee DO, Lieschke GJ, Ward AC. Hematopoietic perturbation in zebrafish expressing a tel-jak2a fusion. Exp Hematol 2005, 33:182–188. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia 2007, 21:462–471. [DOI] [PubMed] [Google Scholar]
- 20.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 2003, 299:887–890. [DOI] [PubMed] [Google Scholar]
- 21.Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2006, 103:15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology 2008, 134:2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci U S A 2007, 104:9410–9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev 2007, 21:1382–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 2011, 471:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kokel D, Bryan J, Laggner C, White R, Cheung CYJ, Mateus R, Healey D, Kim S, Werdich AA, Haggarty SJ, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol 2010, 6:231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010, 327:348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 2008, 4:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol 2009, 5:680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trompouki E, Zon LI. Small molecule screen in zebrafish and HSC expansion. Methods Mol Biol 2010, 636:301–316. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman CK, White RM, Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protocols 2009, 4:1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adatto I, Lawrence C, Thompson M, Zon LI. A new system for the rapid collection of large numbers of developmentally staged zebrafish embryos. PLoS One 2011, 6:e21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardo-Martin C, Chang TY, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. High-throughput in vivo vertebrate screening. Nat Methods 2010, 7:634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt A, Codore H, Day BW, Hukriede NA, Tsang M. Development of automated imaging and analysis for zebrafish chemical screens. J Vis Exp 2010, 40:e1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt A, Cholewinski A, Shen X, Nelson SG, Lazo JS, Tsang M, Hukriede NA. Automated image-based phenotypic analysis in zebrafish embryos. Dev Dyn 2009, 238:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran TC, Sneed B, Haider J, Blavo D, White A, Aiyejorun T, Baranowski TC, Rubinstein AL, Doan TN, Dingledine R, et al. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res 2007, 67:11386–11392. [DOI] [PubMed] [Google Scholar]
- 37.Forné I, Abián J, Cerdà J. Fish proteome analysis: model organisms and non-sequenced species. Proteomics 2010, 10:858–872. [DOI] [PubMed] [Google Scholar]
- 38.Ahn Y-H, Chang Y-T. Tagged small molecule library approach for facilitated chemical genetics. Acc Chem Res 2007, 40:1025–1033. [DOI] [PubMed] [Google Scholar]