Abstract
Background
The past few years have seen a rapid emergence of artificial intelligence (AI)-enabled technology in the field of sleep medicine. AI refers to the capability of computer systems to perform tasks conventionally considered to require human intelligence, such as speech recognition, decision-making, and visual recognition of patterns and objects. The practice of sleep tracking and measuring physiological signals in sleep is widely practiced. Therefore, sleep monitoring in both the laboratory and ambulatory environments results in the accrual of massive amounts of data that uniquely positions the field of sleep medicine to gain from AI.
Method
The purpose of this article is to provide a concise overview of relevant terminology, definitions, and use cases of AI in sleep medicine. This was supplemented by a thorough review of relevant published literature.
Results
Artificial intelligence has several applications in sleep medicine including sleep and respiratory event scoring in the sleep laboratory, diagnosing and managing sleep disorders, and population health. While still in its nascent stage, there are several challenges which preclude AI’s generalizability and wide-reaching clinical applications. Overcoming these challenges will help integrate AI seamlessly within sleep medicine and augment clinical practice.
Conclusion
Artificial intelligence is a powerful tool in healthcare that may improve patient care, enhance diagnostic abilities, and augment the management of sleep disorders. However, there is a need to regulate and standardize existing machine learning algorithms prior to its inclusion in the sleep clinic.
Keywords: Polysomnogram, Artificial intelligence, Machine learning, Disorders of excessive somnolence, Sleep apnea
Introduction
In recent times, artificial intelligence (AI) has entered our everyday lives, for example through hyper-personalized product suggestions based on our data and virtual assistants (i.e., “Alexa” and “Siri”) in our households. Tracing the history of AI in medicine (Fig. 1) demonstrates the rapid advancements over the past decade, due to a number of changes, which include the accrual of massive amounts of health data, greater computing power and storage capacity, and highly sophisticated algorithms powering AI applications.
Fig. 1.
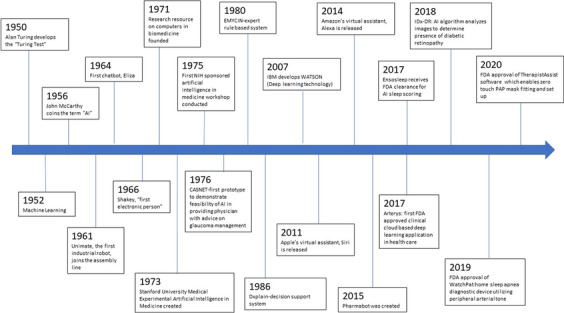
Artificial Intelligence timeline
AI refers to the capability of computer systems to perform tasks conventionally considered to require human intelligence, such as speech recognition, decision-making, and visual recognition of patterns and objects. While AI has gained popularity in several fields of medicine including radiology and oncology, the field of sleep medicine stands to greatly benefit from AI [1, 2]. Sleep is a physiological state marked by dynamic changes in a variety of organ systems, which is reflected by our use of the polysomnogram, which records various physiological signals across the night. Additionally, sleep tracking over long durations is ubiquitous given the availability and popularity of fitness trackers and smart watches. Therefore, sleep monitoring in both the laboratory and ambulatory environments results in the accrual of massive amounts of data. Large and complex datasets are amenable to analysis with AI algorithms, which uniquely positions the field of sleep medicine to gain from AI. Sleep medicine is expected to benefit from artificially intelligent computer programs to effectively score polysomnograms. However, use cases transcend automation and include improved diagnosis of sleep disorders, identification of the mechanisms underlying sleep disorders, treatment selection, and prediction of sleep disorder sequela. The greater insight provided by AI will have applications at both the level of individual patients and in population health.
The emergence of AI is well timed, as we start to realize the constraints of traditional medicine in bridging some of the knowledge gaps which challenge our ability to provide optimal patient care. The heterogeneity of endotypes, interindividual variability in treatment response, and the over-reliance on the identification and quantification of specific “events” occurring during sleep studies have been widely discussed [3]. Researchers have successfully leveraged “big data” to offer new insights into sleep physiology, improve accuracy of diagnosis of sleep disorders, predict response and adherence to treatment, define endotypes, and use sleep parameters as predictors of future physical and mental health [4–13]. This holds promise for the future of sleep medicine, where AI will usher in an era of precision medicine with the advent of “Sleep-Omics” [14]. Integrating sleep physiological data with genetic/imaging markers will provide insight far beyond our clinic walls.
As AI rapidly evolves, it is important to demystify AI for the sleep clinician. This article reviews the basic principles that underlie machine learning algorithms and how to assess the performance of such algorithms in sleep study scoring. Additionally, we will review other applications of AI in sleep medicine and research and identify the challenges of implementing AI tools in clinical practice.
What is machine learning?
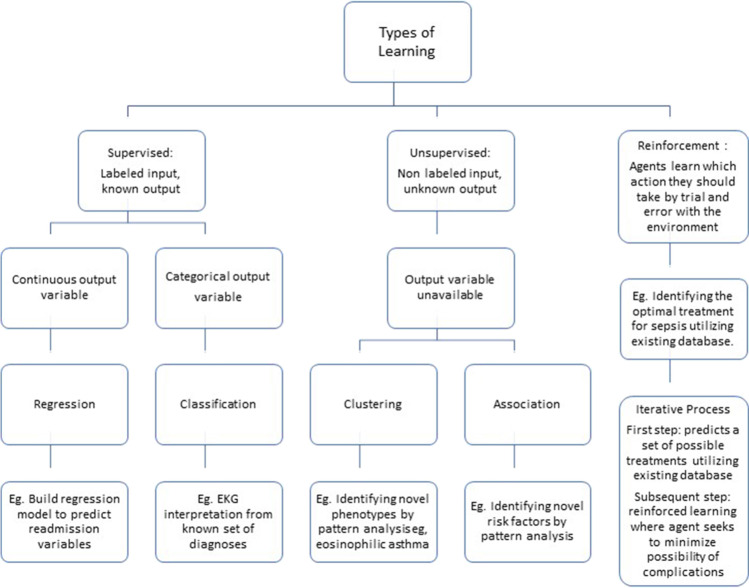
Broadly speaking, machine learning (ML) uses computer algorithms that improve with experience and prior data, without intervention from direct programming commands. Most ML tasks can be divided into supervised learning (learning to map an input x to an output y, based on a set of input–output examples [e.g., predicting human scored sleep stages from polysomnogram signals]), unsupervised learning (finding patterns or clusters in a set of inputs, without labeled output variables provided), or reinforcement learning (algorithms learn based on interacting with the environment and receiving penalties and rewards) (Fig. 2). Recent advances utilize combinations of these strategies to develop new algorithms that may not clearly fit in one of these categories. Additionally, due to the intricacies of the mathematical or statistical model used, various control systems have been designed. Control systems regulate the behavior of other systems using control loops. The efficiency and precision of control systems can be improved with the help of machine learning algorithms utilizing innovative statistical and mathematical models [15]. For instance, the random forest techniques utilizes the principle of simple regression [16]. The details of control systems are beyond the scope of this review, but awareness of this technique is beneficial as the reader appraises the available literature regarding machine learning and sleep.
Fig. 2.
Types of machine learning
Multiple learning models are utilized for training the machine [17]. They are categorized in the following four types:
Supervised learning is used when there is a training dataset that has a well-defined relationship between each input and expected output. Weights are adjusted in this training process to reduce the error of the predicted output from the expected output.
Unsupervised training involves the use of unlabeled inputs, given to the machine for training without known outputs. During training process, the machine is expected to identify patterns or grouping of the data.
Semi-supervised learning is a combination of the supervised learning and the unsupervised learning.
Reinforcement learning, like supervised learning, involves the use of a measurable outcome to guide training process rather than a predefined expected outcome for each input. This model is typically used when the input is stimuli from the environment rather than a compiled dataset.
These processes are analogous to the biological process of learning, where we would repeatedly study and memorize facts (most like supervised learning model), learn from observation (unsupervised learning model), or learn from trial and error (reinforcement learning model). Conventional machine learning algorithms involve feature extraction and classification. Feature extraction is a process by which an initial set of data is reduced by identifying key features of the data for machine learning. The inputs obtained through feature extraction would then be classified based on predetermined criteria.
In recent times, deep learning has emerged as one of the popular modalities of machine learning. Deep learning is inspired by the way a human brain works. Such biologically inspired computational networks which facilitate deep learning are known as neural networks. Unlike conventional machine learning, neural networks do not have to rely on feature extraction and can sometimes utilize raw signals as input. Thus, neural network is a step towards computers being able to perform tasks without explicit programming.
How are machine learning algorithms developed?
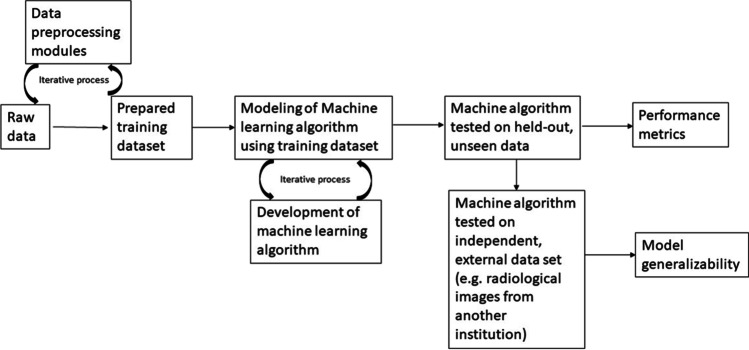
The development and optimization of ML algorithms is an iterative process involving a training dataset and previously unseen or “held-out” test data. Because ML algorithms learn from the provided data, models may overfit the training dataset. Therefore, use of a held-out test dataset is required to avoid biased (usually inflated) estimates of how well a model performs. Next, to ensure generalizability of the model, it is deployed on a completely independent test dataset (i.e., data obtained from a different study cohort or clinical population that uses different data acquisition methods). A simplified depiction of the process of ML algorithm development and testing is depicted in Fig. 3.
Fig. 3.
Development of machine algorithms
AI sleep staging algorithms have utilized training datasets comprised of both healthy and unhealthy subjects. Notably, an algorithm trained on healthy subjects may demonstrate reduced performance when validated in an unhealthy population, for example, patients with neurodegenerative disease who may display electroencephalogram and electromyogram findings uncharacteristic for a given sleep stage [18]. Most studies utilize existing datasets for training and testing including sleep-EDF, sleep-EDF (expanded), Montreal Archive of Sleep Studies, Sleep Heart Health Study (SHHS), Massachusetts General Hospital Sleep Laboratory, Apnea-ECG database, Multi-Ethnic Study of Atherosclerosis (MESA), University College Dublin Sleep Apnea Database, Seoul National University Bundang Hospital (SNUBH), Sleep Center of Samsung Medical Center in Seoul, Korea, and Osteoporotic Fractures in Men Study (MrOS) [19–21]. These datasets are easy to access (some are publicly available) and provide adequate data (even smaller cohorts may provide sufficient data as 800 30-s epochs are available per night of PSG recording). Disadvantages include variability in signal preprocessing and sampling rate and study subject characteristics that may not translate to the heterogeneity of patients and disease presentations seen in real-world clinical populations.
Artificial intelligence in polysomnogram sleep staging and respiratory event scoring
The rapid expansion of AI in sleep is evident from a Pubmed search on “Artificial Intelligence” and “polysomnogram” that shows more than 360 articles, with 204 articles (57%) published in the past 5 years.
Evolution of artificial intelligence-based sleep staging
One of the first evaluations of AI sleep scoring was the use of a learning vector quantizer and the induction of decision trees to stage polygraphic data in eight infants and demonstrated overall recognition accuracy of 75% [22]. An early use of neural networks specifically was presented by Schaltenbrand and colleagues in 1996 [23]. The automatic scoring of 61,949 epochs from 60 subjects with a neural network model achieved comparable agreement to human experts, with expert-model agreement and inter-expert agreement of 82.3% and 87.5%, respectively. This agreement improved to 90% with expert supervision for unknown or ambiguous epochs.
Over the next few years, several studies used neural network models to score sleep studies in patients with obstructive sleep apnea (OSA), epilepsy, Cheyne Stokes respirations, and Parkinson’s disease [24–28]. While some of the studies focused on analyzing sleep spindles and power spectra of sleep for staging, others focused on integrating cardiorespiratory events to diagnose sleep-related breathing disorders while a few focused on snore signal [29–34]. Interestingly, some of the earlier studies concentrated on use of AI to score sleep studies in infants, particularly those at risk for sudden infant death syndrome (SIDS) [35, 36].
Additionally, during this time period, different methods to improve AI sleep scoring were explored; for example, a neural network model to identify sleep-disordered breathing events was iteratively refined with use of a supervised approach [37]. This entailed input from clinicians each time a new pattern was found. Whenever a clinician demonstrated good self-agreement, the neural network model was retrained. Over the next few years, there was a progression towards development of models which relied less on expert supervision. This led to newer approaches such as the fuzzy set theory which allowed modification of the morphological detection criteria and performed a detailed characterization of the identified events to approximate human intuition [38].
More recently, different techniques were explored to develop ML polysomnogram scoring models (Table 1). Conventional ML for sleep staging utilizes two main components: feature extraction and classification. The traditional styles of feature extraction relied on raw signal input through a variety of methods, including Fourier transforms, wavelet analysis, and Hilbert transform [39–41]. Feature extraction can also utilize time–frequency images, generated by short-time Fourier transform or wavelet transform instead of raw signal inputs [18, 42]. Use of spectrograms has also been reported as a processing method prior to input of polysomnogram data [43]. With advances in ML, feature extraction techniques have evolved to reduce the number of features in a dataset by creating new features from existing ones. A thorough review can be found here [44].
Table 1.
Examples of studies using machine learning algorithms for sleep stage and respiratory scoring
| Author, year | Population studied | Dataset source | Channels and sensors | Data preprocess | Classifier used | Performance measures | Other findings |
|---|---|---|---|---|---|---|---|
| Perslev [87], 2021 | 15,660 participants | 16 clinical datasets | Single channel EEG and EOG sugnal | U Sleep software | Convoluted Neural Network(CNN) | U sleep performed as well as other algorithms, even though U sleep was not trained on similar datasets | It predicts sleep stages in a single forward pass |
| Sharma [88] 2021 | 80 subjects comprising of healthy controls as well as various sleep disorders | Cyclic Alternating Pattern (CAP) database | Dual channel EEG | Optimized wavelet filters | Bagged tree (EBT) classifier with tenfold cross validation |
Accuracy 85.3% κ = 0.786 |
Accuracy improved in a balanced dataset created using over-sampling and under-sampling techniques |
| Sun, 2020 [6] | 8682 PSG | N/A | ECG and respiratory signals | 270 s time windows used | CNN |
κ = 0.585 (sleep stages) κ = 0.76 (wake vs NREM vs REM) |
Performance is better for younger ages |
| Jaoude 2020 [89] | 6431 patients |
MGH-PSG dataset; Ambulatory scalpEEG dataset |
4 EEG channels | Bandpass filter, downsampling to 100 Hz, generated bipolar montage to make it “reference channel-free” | CNN followed by RNN |
κ = 0.74 (MGH held out dataset) κ = 0.78 (scalpEEG dataset) |
Performance was consistent across common EEG background abnormalities |
| Zhange et al., 2020 [90] | 294 sleep studies (122 training data set, 20 validation dataset, 152 testing data set) | Prospectively collected | 2 channel EEG, EOG, EMG, ECG, airflow | Filtered signal at 66 Hz, downsampled signal sampling frequency to 66 Hz | CNN |
Accuracy 81.81% κ = 0.7276 |
Number of arousals affected model’s performance |
| Peter-Derex 2020 [91] | 23 patients with insomnia, 24 patients with idiopathic hypersomnia, 24 patients with narcolepsy, 24 patients with OSA | Lyon sleep database | Single channel EEG | ASEEGA software | ASEEGA software | Agreement between software and consensual scorer was: insomnia 85.5% (κ = 0.80), narcolepsy 83.8% (κ = 0.78), idiopathic hypersomnia 86.1% (κ = 0.68), and obstructive sleep disorder 87.2% (κ = 0.82) | |
| Sridhar, 2020 [92] |
800 (561 subjects); 993 nights (993 subjects) |
Sleep Heart Health Study; Multi-ethnic study of Atherosclerosis; Physionet Computing in Cardiology (CinC) |
ECG | Normalize ECG signal, interbeat interval time series computed | CNN |
Accuracy 77% κ = 0.66 (SHHS) Accuracy 72% κ = 0.55 (CinC) |
|
| Zhu, 2020 [93] | 8 recordings (4 healthy, 4 sleep disorders); 20 recordings (healthy) | Sleep EDF, Sleep EDFX | Single channel EEG | Z score normalization of data | CNN + attention mechanism |
Accuracy 93.7% F1 score 84.5% |
Attention mechanism helped in learning inter and intra-epoch features |
| Xu, 2020 [94] | 5793 participants (sleep disorders) | Sleep Heart Health Study | Multichannel EEG, EOG | Time frequency spectra | LSTM (Long short term memory)/RNN |
Accuracy 87.4% κ = 0.8216 |
RNNtakes temporal information into account |
| Zhang et al. Sleep, 2019 [95] | 5213 patients | Sleep Heart Health Study | Multichannel EEG, EOG, EMG | Raw signal, short term Fourier transform for spectogram | Recurrent and convolutional neural networks |
κ = 0.82 Validation MrOS κ = 0.68 Validation SOF κ = 0.70 |
|
| Yildrim, 2019 [96] |
8 recordings; 61 recordings (healthy and insomnia) |
Sleep EDF, Sleep EDFX | Single channel EEG, Single channel EOG | Raw PSG signal | CNN | Accuracy 98.06% | |
| Phan, 2019 [97] | 200 recordings | MGH sleep lab | Single channel EEG, EOG, EMG | Time frequency images | CNN + RNN |
Accuracy 87% κ = 0.81 |
Trained network in end to end fashion |
| Zhang and Wu, 2018 [98] |
25 recordings (sleep disorders) 16 recordings |
MIT-BIH database, Sleep EDF | Single channel EEG | Phase encoder, unsupervised training | CNN |
Accuracy 87% κ = 0.81 |
|
| Stephansen, 2018 [57] | 3000 recordings (healthy and sleep disorders) | 10 databases | Multichannel EEG, EOG, | Filter + octave encoding | CNN + RNN | Accuracy = 87% | Automates type 1 narcolepsy diagnosis |
| Sors et al., 2018 [99] | 5793 recordings (sleep disorders) | Sleep Heart Healthy Study | Single channel EEG | raw | CNN |
Accuracy = 87% κ = 0.81 |
|
| Biswal et al. 2018 [50] |
1000 recordings 5804 recordings |
Sleep Heart Health study; ISRUC-sleep |
Multichannel EEG | Spectogram | Recurrent and convolutional neural networks |
accuracy 87.5% κ = 0.805 Validation SHHS accuracy 77.7% κ = 0.732 |
Minimal reduction in accuracy noted when working on single channel |
| Patanaik et al. Sleep, 2018 [18] |
1046 recordings (healthy adolescents) 284 recordings (healthy young adults) 210 recordings(sleep disorders) 77 recordings (Parkinson disease adults) |
CNL lab, CSL lab, Singapore; UCSD sleep lab, UC database | Multichannel EEG, EOG, | Spectral images | Convolutional neural network |
accuracy 89.2% κ = 0.86 Validation set 1- accuracy 81.4% κ = 0.740 Validation set 2 (PD)- accuracy 72.1% κ = 0.597 |
Faster compared to human experts (F sec compared to 30–60 min) |
| Olesen 2018 [7] | 2310 recordings (healthy and sick) | CNL lab, Singapore | Multichannel EEG, EOG, EMG | Raw data | CNN | κ = 0.75 | Most errors made in stage N1 and N3 |
| Malafeev, 2018 [56] |
54 recordings (healthy); 43 recordings (22 PSG and 21 MSLT, hypersomnia patients) |
Warsaw (healthy); Wisconsin Sleep Cohort (hypersomnia) |
Single channel EEG, EMG, EOG | Spectrogram | CNN | κ = 0.8 (except stage N1) | Performance in healthy subjects were better compared to those on hypersomnia patients |
| Cui 2018 [46] | 116 recordings including healthy and sick population | University of Zurich | Multichannel EEG, EOG, EMG | Time series | CNN | Accuracy = 92.2% | |
| Chambon, 2018 [100] | 61 recordings from healthy adults | MGH sleep lab |
Multichannel EEG Three chin EMG |
Linear spatial filtering | CNN | Sensitivity = 52% | Utilized 1 min of data before and after each data segment which offered the strongest improvement |
| Vilamala, 2017 [101] | 40 recordings from 20 healthy adults | Montreal archive | Single channel EEG | Multitaper spectral analysis | CNN | Acc = 84–88% | |
| Supratak,2017 [55] | 62 healthy recordings | MGH sleep lab, Montreal Archive | Single channel EEG | Raw data | CNN + RNN |
Acc = 86.2% K = 0.8 |
Used multiple datasets |
| Tsinalis, 2016 [42] | 40 recordings from 20 Healthy young adults | Montreal archive | Single channel EEG | Class balanced random sampling | CNN and 2D stack of frequency-specific activity in time |
Accuracy = 71–76% Per stage Accuracy = 80–84% |
Performance balanced across classes |
CNN convolutional neural network, RNN recurrent neural network, LSTM long-short-term memory, EBT ensemble bagged tree, Κ kappa
In addition to improvements in feature extraction, advances were also made in the realm of classification. Most of the published AI sleep staging to date has utilized convolutional or recurrent neural networks or a combination of both [45]. In general, neural networks utilize a network of filters and subsampling layers. While convolutional neural networks consider only the current input, recurrent neural networks consider the current input together with the previously received inputs and, therefore, are well-suited for sequential data.
Information sources for AI-based sleep staging and respiratory event scoring
Multiple considerations are needed regarding the data substrates for AI scoring of sleep and respiratory events. The type and number of channels used, the input of raw or processed signal, and artifact are factors in the development of ML algorithms deployed on polysomnogram data.
Sleep staging channels
While most studies utilize electroencephalogram (EEG) signal for sleep staging, several studies have combined EEG channels with electromyogram (EMG) and electro-oculogram (EOG channels) [32]. General consensus is that use of multi-channel EEG improves performance.
Given the utility of home monitoring, there is growing interest in AI sleep staging from one or a few easily recorded physiological signals [46]. Electrocardiogram (ECG), respiratory effort, and photoplethysmography (PPG) have all demonstrated promise as alternative signals that can be leveraged for sleep staging [6, 8]. For example, a deep neural network that utilized both ECG and respiratory signals performed well in the classification of sleep stages and was not impacted by patient age or comorbid sleep disordered breathing. However, accuracy was lower compared to networks trained on EEG models [6]. Another group demonstrated accurate estimation of sleep time and differentiation between sleep stages with use of PPG signal, obtained from pulse oximetry [8].
The ability of ML algorithms to estimate sleep stages from limited channels facilitates data acquisition in the ambulatory environment, particularly given the ubiquity of PPG in consumer facing technologies.
Respiratory event channels
Similarly, for ML scoring of respiratory events, analysis often uses signal from traditional sensors (nasal oral thermistor, nasal pressure transducer, abdominal and thoracic respiratory inductance plethysmography, and oximetry). However, to automate scoring of data collected in the home, investigators have used ECG or PPG signal in isolation, or combined with a limited complement of traditional respiratory parameters [47–50]. For example, studies have utilized ECG inter-beat intervals or heart rate variability (HRV) to detect respiratory events [6, 31]. Because PPG can estimate HRV and is widely available, ML algorithms may allow obstructive sleep apnea detection from consumer facing technologies.
Signal type
ML algorithms can process raw or pre-processed signal. One of the popular options is to extract features from raw EEG signals for sleep stage classification [51]. However, an EEG spectogram can also act as input by first calculating the power spectral density. Power spectral density is the measure of the signal’s power content versus frequency. The importance of power spectral density has been highlighted in studies which have shown an increase in delta and beta EEG activity in certain sleep disorders [30].
For respiratory event scoring, several studies use raw airflow, respiratory effort, and oximetry signals as inputs [50]. However, another approach utilizes raw signals normalized based on the mean and standard deviation of the normal samples for each subject or employs a combination of raw input signals to reshape it into a matrix for classifier use [52].
Filtering
Raw signals can be contaminated with noise that can affect the classifier’s performance. Basic band pass filters, as recommended by the technical specifications in the American Academy of Sleep Medicine (AASM) Manual for the Scoring of Sleep and Associated Events, can diminish this noise [53]. Several approaches have been utilized when using filtered signal as information source. For example, one group of researchers employed artifact reduction approaches by using several different options including Butterworth filters, weight decay, or adaptive normalization [46, 54, 55]. In another example, a group demonstrated that deep learning algorithms can learn key information from epochs with artifact [56].
Performance of artificial Intelligence sleep staging algorithms
AI algorithms require testing on unseen data to evaluate their performance, which is often achieved by a cross-fold validation process where the dataset is partitioned into several equal groups. A single group is retained to test model performance while the other groups are utilized for algorithm training.
Several performance metrics are available to describe performance of AI algorithms. Sleep stages and obstructive sleep apnea severity classes (no, mild, moderate, or severe disease) are categorical constructs, and therefore, results can be represented as percent agreement with gold standard (visual scoring by a human expert). Use of Cohen’s kappa instead of percent agreement is more stringent as it mitigates the effect of agreement occurring by chance.
Accuracy values should be approached with caution if used in isolation to describe algorithm performance. Specifically, accuracy can be misleading if there is an unequal number of observations in each class or more than two classes in the dataset. Use of the accuracy metric can lead to a situation where the model is completely and consistently misidentifying one class, but this misidentification is missed because on average, performance is good. A confusion matrix can overcome these issues. The confusion matrix identifies when the algorithm confuses two classes by counting the number of instances data is misclassified. Each row in a confusion matrix represents a predicted class, while each column represents an actual class. The number of correct and incorrect predictions for each class is calculated and represented in the confusion matrix. Therefore, the confusion matrix may provide a better gauge of performance than accuracy alone (Table 2).
Table 2.
Confusion matrix: confusion matrix for predicted sleep stage displays agreement between human experts and the prediction by the dataset (example-2 stage classification of sleep and wake)
| Dataset scored wake (predicted class positive) | Dataset scored sleep (predicted class negative) | ||
|---|---|---|---|
| Human expert scored wake (actual class positive) | True positive (TP) | False negative (FN) | Sensitivity TP/(TP + FN) |
| Human expert scored sleep (actual class negative) | False positive (FP) | True negative (TN) | Specificity TN/(TN + FP) |
|
Precision TP/(TP + FP) |
Negative predictive value TN/(TN + FN) |
Accuracy (TP + TN)/(TP + TN + FP + FN) |
Traditional two by two tables can also provide descriptive statistics when comparing a binary outcome (i.e., obstructive sleep apnea versus no obstructive sleep apnea) between algorithm and human. In this case, algorithm-identified cases are compared to cases based on visual scoring of respiratory events and described by true positive (TP, obstructive sleep apnea cases correctly identified by the algorithm), false positive (FP, healthy subjects incorrectly identified as obstructive sleep apnea cases by the algorithm), true negative (TN, healthy subjects correctly identified as normal by the algorithm), and false negative (FN, obstructive sleep apnea cases incorrectly identified as normal by the algorithm) values. Table 3 lists commonly used performance metrics.
Table 3.
Performance metrics
| Performance metric | Formula |
|---|---|
| Accuracy | (TP + TN)/(total population) |
| Specificity | TN/(TN + FP) |
| Sensitivity | TP/(TP + FN) |
| PPV | TP/(TP + FP) |
| F1 | 2 * [(PPV*sensitivity)/(PPV + sensitivity)] |
| AUC | Area under ROC curve |
Commonly used metrics to quantify algorithm performance against gold standard
PPV, positive predictive value; AUC, area under the curve; TP, true positive; TN, true negative; FP, false positive; FN, false negative; ROC, receiver operating characteristic curve (y axis = sensitivity, x axis = 1-specificity)
One of the commonly encountered problems in classification predictive modeling is imbalanced classification. Most machine learning algorithms used for classification are designed around the assumption of an equal number of examples for each class. When classes are imbalanced, this results in models that have poor predictive performance, specifically for the minority class. This holds true in the realm of sleep studies, given that most of the nighttime period is sleep when healthy participants are used. Additionally, imbalance is present among sleep stages and N1 sleep can be misclassified since the percentage of N1 sleep is less compared to other stages of sleep. This can be overcome by balancing classes in the training dataset or by improving classification algorithms. For imbalance classification, how well the positive class was predicted or sensitivity (TP/(TP + FN)) may be of more interest than how well the negative class was predicted or specificity (TN/(FP + TN)).
Other challenges in appraising the performance of AI sleep staging and respiratory event scoring stem from characteristics of training and testing datasets. Training datasets are often derived from healthy populations or convenience samples. To diagnose sleep disorders, training datasets should consist of patients with heterogeneous sleep problems to facilitate deep learning.
Highlighting the need for diverse data sets, researchers found that having more data sources significantly improved classification performance and generalizability. Specifically, the group noted that using 75% of the PSGs available yielded just as high performance compared to using 100% once they included PSGs from five different sources [7]. This underscores the importance of availability of public datasets from multiple heterogenous populations.
If a test dataset comes from the same sleep center, acquired with the same equipment and scored by the same human scorers, performance metrics may be falsely elevated, even with use of held-out, unseen data. Therefore, testing with use of an external, independent database is typically considered more reliable [18, 57]. There is considerable value in standardizing testing data from various sleep laboratories as well as standardizing performance metrics, which can help users compare different algorithms.
Notably, pediatric populations have been underrepresented and expansion of pediatric sleep datasets for algorithm development and testing is required.
Other use cases for artificial intelligence and sleep medicine
Although the first obvious use for AI in sleep medicine is to automate the staging of sleep and scoring of respiratory events to reduce technician burden and decrease time from PSG recording to interpretation, other use cases will deepen our understanding of sleep disorders and the role of sleep in health and disease.
Improved phenotyping, endotyping, and treatment response prediction in sleep disordered breathing
There is growing evidence that the underlying etiology (i.e., endotype) and clinical manifestation (i.e., phenotype) of OSA in an individual are not well described by the traditionally used AHI [58]. Artificial intelligence has paved the way for a better understanding of the various endotypes and phenotypes of OSA to form the foundation of personalized treatment for OSA. AI-assisted graphical models of chemoreflex feedback loop have been used to identify ventilatory instability in OSA patients which can guide treatment selection [59]. Routine polysomnographic characteristics and clinical data have been utilized to estimate upper airway collapsibility and arousal threshold using AI-assisted data-driven models [60]. Endotyping OSA through PSG is increasingly recognized as vastly important to our field and there is an increasing interest in making OSA endotyping algorithms accessible, inexpensive, and, ultimately, scalable [12]. Even adherence to treatment may be better predicted with use of ML. Compliance classifiers with CPAP therapy have enabled early prediction of compliant patients.
In addition to ramifications for personalized treatment, the use of AI in sleep disordered breathing is relevant for outcomes. Unsupervised and supervised clustering models were used to cluster 2277 OSA patients into sic phenotypes based on their polysomnogram data. The phenotypes show different risk for the development of cardio-neuro-metabolic comorbidity, unlike the conventional single-metric apnea–hypopnea index-based phenotype [61].
Tools to improve risk stratification will also benefit from AI. Support vector machine-based models have been created utilizing clinical data for early identification of patients at risk for OSA presenting to a primary care clinic which may potentially prioritize them for sleep studies [9].
Hypersomnia
While AI has made significant strides in the realm of sleep-disordered breathing, this innovative technology has been investigated for the evaluation and management of other sleep disorders including suspected central disorders of hypersomnolence. The objective confirmation of a central disorder of hypersomnolence requires a PSG followed by a multiple sleep latency test (MSLT). An MSLT entails 4–5 nap opportunities with recording of EEG, EOG, EMG, and EKG leads. Sleep onset latency for each nap (averaged as the mean sleep latency) and the presence of sleep onset stage REM (R) sleep are recorded. Completion of the overnight PSG and daytime series of nap opportunities is burdensome for the patient, and manual review of PSG and MSLT data is time-consuming, expensive, and subjective.
The central disorder of hypersomnolence, narcolepsy, type I (narcolepsy with cataplexy), is confirmed by reduced mean sleep latency on MSLT and at least 2 sleep onset stage R periods across overnight PSG and daytime MSLT. However, poor nocturnal sleep consolidation is also a characteristic feature of narcolepsy, type I. After development of an automatic classifier capable of separating sleep and wakefulness epochs with single channel EEG, individuals with narcolepsy with cataplexy were observed to have significantly more sleep–wake transitions during night than patients with narcolepsy without cataplexy and normal controls [62]. In subsequent work, Olsen et al. used a linear discriminant analysis (LDA) model which utilized 38 features from EOG, EMG, and EEG to identify features that differentiated wake, stage N1, N2, N3, and REM sleep in control subjects [63]. Next, the derived 2-dimensional sleep state space projection was used to distinguish patients with narcolepsy, type I from controls by leveraging the known sleep state dissociation in narcolepsy patients.
More recently, Stephansen and colleagues utilized deep learning to diagnose narcolepsy, type 1 from overnight PSG alone [57]. First, a hypnodensity graph was generated from PSG signal, which does not enforce a single sleep stage label, but instead assigns a membership function to each of the sleep stages. Therefore, use of neural networks not only automated sleep staging but allowed for more detailed representation of sleep trends over the course of the night. Next, deep learning was used to identify features of sleep state dissociation predictive of narcolepsy, type 1. Analysis of a single night of PSG was able to identify narcolepsy, type 1 with high sensitivity (91%) and specificity (96%) compared to the more laborious PSG-MSLT.
Narcolepsy, type 2 presents a different diagnostic challenge given the lack of cataplexy and poor test–retest reliability of the MSLT for this condition [64]. A stochastic gradient boosting (SGB) model was used to explore the features characteristic of type 1 and type 2 narcolepsy based on a dataset of individuals in the European Narcolepsy Network (EU-NN) [65]. The SGB model allowed for selection of features independent from existing diagnostic criteria and demonstrated the capacity to classify narcolepsy subtypes with high accuracy. Furthermore, the model can use a mixture of clinical features and identifies the most important features. Therefore, machine learning may identify novel potential candidates for future diagnostic criteria for narcolepsy, type 1 and 2.
To employ data sources beyond polysomnogram in the evaluation of excessive daytime sleepiness (EDS), Liu and colleagues utilized an artificial neural network of modified adaptive resonance theory to differentiate subjects with and without sleep disorders that cause EDS from normal control subjects based on EEG and pupil size [66].
Insomnia
Insomnia can also benefit from AI analytic techniques, and one of the initial investigations in this area assessed singular spectrum analysis (SSA) of sleep EEG to differentiate paradoxical insomnia, psychophysiological insomnia, and control groups [67]. In 2016, Chaparro-Vargas et al. used 3 tandem models to distinguish insomnia patients from controls [68]. First, a preprocessing module was used that utilized state-space time-varying autoregressive moving average (TVARMA) processes to identify the features that characterize sleep onset. Next, a hypnogram generation module used a fuzzy inference system to infer sleep stages and the macrostructure of sleep architecture. Lastly, the characterization module compared hypnograms with similarity distances and used logistic regression to distinguish controls from insomnia patients. Another group trained deep neural network classifiers with features extracted from a maximum of two EEG channels and accurately differentiated patients with insomnia from controls [69]. When compared with manual scoring, the classifier had excellent discrimination accuracy between patients and controls using both (92%) or only one EEG channel (86%).
While most of these studies use PSG signal as a substrate for machine learning algorithms, other sleep data sources outside of the laboratory have been explored. For example, natural language processing techniques were used to extract causality from twitter messages that included stress, headache, and insomnia content [70]. Additionally, unsupervised learning has been applied to wearable data and identified 5 different clusters of insomnia activity [71].
The use of AI in insomnia has expanded to include intervention. During the COVID-19 pandemic, a group of researchers devised a smartphone app called KANOPEE that allowed users to interact with a virtual agent that screened for sleep disturbances and delivered digital behavioral interventions. The program used decision tree architecture and interacted with users through natural body motion and voice [72]. AI digital screening and intervention tools, easily deployed through smart phone applications, confer the ability to provide behavioral interventions remotely, at scale.
Circadian rhythm sleep–wake disorders
The circadian timing system regulates a variety of biological processes in addition to the sleep–wake cycle. Therefore, misalignment of behavioral, light–dark, sleep–wake, and peripheral rhythms can produce detrimental impacts on human health. Data that demonstrate circadian oscillation can be derived from numerous sources and the level, degree, and impact of circadian disruption may vary; therefore, AI provides a unique opportunity to improve our understanding of circadian rhythms.
For example, researchers built an expert system that identifies the characteristics that contribute to negative effects of shift work and then selects mitigation efforts according to their importance in preventing these negative effects. With a fuzzy analytic hierarchy process model, the shift “expert” prioritizes prevention advice to shift workers at the individual and organization level [73].
Additionally, given the difficulties in measuring circadian rhythms, AI has also been used to understand and predict circadian states. The cyclic ordering by periodic structure (CYCLOPS) algorithm uses machine learning to identify circadian rhythms at a molecular level including rhythmic transcripts in human liver and lung [74]. Another group of researchers utilized machine learning to predict circadian phase within 2 h from gene expression in peripheral blood samples [75]. A particular strength of this study was excellent predictive performance with use of an independent test set, suggesting generalizability of this circadian measurement.
Utilization of machine learning to predict circadian timing from gene expression has ramifications beyond sleep disorders. An application that has drawn considerable attention is precision timing of cancer treatment based on AI estimates of circadian timing. Chemotherapy timed in accordance with the patient’s internal time may reduce toxicity and improve outcomes [76].
Machine learning has not only allowed circadian timing predictions from peripheral blood samples, but also from data collected by ubiquitous wearable devices [77]. Real-time circadian tracking in the ambulatory environment from wearable devices may hold promise as an easy to use, inexpensive adjunct to expert clinical evaluation and management.
REM sleep behavior disorder
Appropriate diagnosis of REM sleep behavior disorder (RBD), which includes dream enactment behavior and loss of normal atonia of stage REM sleep during PSG, is crucial given its association with both co-morbid and incident alpha-synucleinopathy neurodegenerative disease. Furthermore, identifiable characteristics that separate individuals with idiopathic RBD (RBD in the absence of a neurodegenerative disorder) from patients with RBD in the setting of alpha-synucleinopathy (e.g., Parkinson’s disease, dementia with Lewybodies, and multiple systems atrophy) could assist with the development of prediction tools. Christensen et al. utilized data driven topic modeling and unsupervised learning to characterize sleep EEG and EOG among controls, patients with periodic limb movements of sleep (PLMS), idiopathic RBD, and Parkinson’s disease [28]. A Lasso regularized regression model was then used to differentiate patient groups. The most salient features were the number and stability of EEG topics linked to REM and N3, respectively, and the model was able to distinguish patients with idiopathic RBD from individuals with Parkinson’s disease with a sensitivity of 91.4% and a specificity of 68.8%.
Another dilemma in RBD is the determination of REM sleep without atonia (RSWA). Scoring criteria and quantification metrics have been delineated, but the implementation of these rules in the context of manual, visual scoring is laborious [53]. Oftentimes, qualitative assessment of EMG tone in REM sleep is made, which results in a lack of standardization across sleep laboratories. Therefore, automation of the process is an area of active research which may benefit from AI [78]. For example, a random forest classifier was developed that used established RSWA metrics along with an EMG fractal exponent ratio between sleep stages and sleep architecture measures [79]. The random forest classifier that supplemented traditional computerized metrics with novel features related to sleep architecture was able to automate RSWA scoring and identify RBD with accuracy, sensitivity, and specificity of 0.96, 0.98, and 0.94, respectively, and outperformed automated scoring that uses traditional measures in isolation (atonia index, motor activity, and STREAM).
Apart from PSG data, machine learning that incorporates other clinical features, such as olfactory loss, cerebrospinal fluid measurements, and the results of functional imaging, with a diagnosis of RBD may allow model prediction of early, or even preclinical Parkinson’s disease [80]. The ability to use clinical or PSG characteristics related to RBD combined with other features to identify individuals at risk for neurodegenerative disease is essential to the development of primary prevention therapeutics.
Sleep-related movement disorders
Movements during sleep may be incidental findings during PSG or may present clinically if troublesome to patients or their bedpartners. Periodic limb movements of sleep (PLMS) are highly prevalent among patients with restless legs syndrome but rarely seen as an isolated finding causing daytime symptoms (periodic limb movement disorder). PLMS are typically scored with use of the anterior tibialis EMG lead and deep learning has been used to automate this process with 85% accuracy; however, with use of a K-nearest neighbors algorithm, investigators could identify PLMS without use of EMG [50, 81]. Additionally, with use of machine learning analysis, novel data sources that do not contact the patient such as 3D cameras and infrared sensors were able to detect 75% of PSG confirmed PLMS [82].
AI has been used outside the sleep laboratory in sleep-related movement disorders in the diagnosis of restless legs syndrome by analyzing bed acceleration sensors with deep learning [83].
Population health-predicting morbidity and mortality
An important use of AI beyond the diagnosis and treatment of defined sleep disorders is its application in population health, with emphasis on the relationship between disturbed sleep and morbidity and mortality. Sleep health is a multidimensional construct influenced by inherent, person-specific characteristics and external social and environmental demands. Optimal sleep health has been characterized by satisfactory subjective quality, alertness during desired wakefulness, appropriate timing, adequate duration, sufficient continuity, robust rhythmicity, and high regularity [84]. This comprehensive definition of sleep health provides a more inclusive description than isolated aspects of sleep such as duration and has relevance for individuals without diagnosable sleep disorders.
A multidimensional definition of sleep health has the potential to influence large-scale public health initiatives by informing screening programs and interventions that are more precise and comprehensive with the ultimate aim of improving not only sleep but other aspects of health and wellness. Wallace and colleagues applied three multivariable approaches to determine which sleep characteristics increased mortality risk in the osteoporotic fractures in men cohort [85]. Across multivariable approaches, lower sleep–wake rhythmicity, and continuity (assessed by actigraphy) increased the risk for all-cause mortality even after considering other important sleep, demographic, health, and behavioral risk factors. Notably, use of a random forest model, which is more flexible than traditional statistical models, allowed for the simultaneous consideration of potentially correlated variables and identified which facets of sleep health were the greatest driver of outcomes [85].
AI also confers the ability to conduct scalable research, as evident by the over 11 million nights of wearable activity characterizing sleep duration and timing data by age and gender [5]. With a focus on younger populations, the application of structural equation modeling to almost 5000 children allowed researchers to assess repeated data and showed a bidirectional association between behavioral sleep problems and health related quality of life [13].
In addition to ambulatory sleep information, PSG findings that may not be traditionally considered in the quantification of OSA severity, such as sleep fragmentation, oxygen desaturation magnitude, and the percentage of stage REM sleep, are independently related to mortality risk [67]. Therefore, PSG datasets can also inform population health with use of novel measures beyond the AHI. New insights on sleep microarchitecture were already obtained through automated detection of cyclic alternating pattern in older men and women from two community cohorts [4].
AI algorithms alone will not fully delineate the role of sleep in health and disease, and a combined approach of advanced analytics, novel sensors, and measurement of sleep both in and outside of the laboratory is likely required. The Sleep and Obstructive Sleep Apnoea Monitoring with Non-Invasive Applications (SOMNIA) project helps support this goal as in addition to recording the usual signals, sensors not typically monitored as part of PSG are simultaneously recorded including suprasternal pressure monitoring, multielectrode electromyography of the diaphragm, wrist worn accelerometry and optical photoplethysmography, and mattress embedded sensors. Therefore, in addition to providing a data source that can be analyzed with machine learning algorithms to provide novel insight from data typically recorded in PSG, new sensors may demonstrate utility, and some are even adaptable for ambulatory use [86].
Challenges of artificial intelligence in sleep medicine
Despite the huge advances made, there are some critical challenges to consider in the implementation of AI in clinical sleep medicine and sleep research. These include (1) logistics of creating datasets, (2) standardization of commercial algorithms, (3) limited data available for research, (4) regulation, and (5) integration of “omics” data.
One of the biggest challenges is creating training datasets. Most of the existing datasets using polysomnogram data are research datasets collected from a subgroup meeting certain inclusion criteria. Hence, they are not generalizable and not representative of what the clinician encounters in real practice. Another challenge is ensuring optimal data quality by reducing external noise and artifact. Finally, algorithm validation requires independent data sets that are sequestered and not available for training purposes.
With multiple commercial companies developing FDA cleared algorithms, there is a need to standardize commercial algorithms through certification by an accredited regulatory body. While FDA approval ensures that the algorithms are safe to use, the approval does not ensure clinical validity. This can be overcome by creating standardized certification programs, which will test the algorithms and disclose performance metrics on independent test sets. For appropriate use and generalization, the circumstances in which the data was collected and characteristics of the population the data were derived from should be well described.
There is an acute need for larger-scale research trials which can corroborate machine algorithm generated measures to clinically significant outcomes. This prompts the need for research datasets with heterogeneity in signals, patient demographics, sleep disorders, and clinical outcomes. Projects like SOMNIA are strides towards that direction.
There is a strong need for policies and best clinical practices regarding use of AI in sleep medicine.
There is a need to integrate data obtained through “omics” technology (transcriptomics, proteomics, metabolomics) with traditional health and demographic data with polysomnographically derived data [14]. This further emphasizes the need for a universal database formed by collaborative efforts across the sleep community.
In addition to concerns development, testing, and certification, clinical implementation of AI tools for sleep staging and respiratory event scoring will also require user interface improvements to streamline use [57]. Additionally, as many programs require upload of sleep data to external servers, security of protected health information is required. Issues regarding bias and health disparities require continued evaluation and mitigation to avoid scaling inequities.
Future possibilities for artificial intelligence in sleep medicine
Artificial intelligence in sleep medicine undoubtedly holds promise. There are currently FDA-cleared AI Scoring software available in the market. With regulation and careful standardization, these softwares can facilitate scoring. However, in its present form, it will still require health care provider oversight and clinical correlation will be strongly recommended. As the machines continue to learn, it will be imperative to continuously regulate these scoring systems.
With continued advancement in technology, AI scoring can be further utilized to identify polysomnogram features which are not easily identified by humans or are time/labor-intensive. Examples include microspindles, sleep–wake transitions, and thoracoabdominal asynchrony. These features may assist in diagnosis as well as monitoring progression of several sleep disorders. Big data analysis of wearable/nearable devices can be a very useful tool in the hands of the sleep clinician in determining an individual’s sleep health. This can be utilized at the population health level to generate ideas on how to improve health issues including sleep deprivation. AI can improve clinic flow by voice-assisted documentation and automated organization of available clinical information from multiple sources, thereby allowing more time for physician–patient interaction. This is turn will augment physician–patient relationship.
Conclusion
In summary, AI has made considerable advancements in sleep medicine. Polysomnograms result in the acquisition of robust data, and AI applications will allow for improved understanding, screening, diagnosis, and management of sleep disorders. AI augmentation of the polysomnogram scoring process will allow for diversion of human effort and time from repetitive, laborious tasks to face-to-face patient care. Wearable technology and large-scale clinical databases can supplement the novel information extracted from polysomnograms with AI to improve our understanding of the role of sleep in human health and disease. However, there are certain challenges which preclude AI’s generalizability and wide-reaching clinical application.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
This study does not contain any human or animal subjects. Institutional IRB or informed consent was not required.
Conflict of interest
Author Anuja Bandyopadhyay declares that she has no conflict of interest. Author Cathy Goldstein is on the medical advisor boards of Huxley medical and eviCore. She receives royalties from UpToDate. She is 5% inventor of a circadian mobile application licensed to Arcascope, LLC.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anuja Bandyopadhyay, Email: anubandy@iupui.edu.
Cathy Goldstein, Email: cathygo@med.umich.edu.
References
- 1.Watson NF, Fernandez CR. Artificial intelligence and sleep advancing sleep medicine. Sleep Med Rev. 2021;59:101512. doi: 10.1016/j.smrv.2021.101512. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein CA, Berry RB, Kent DT, Kristo DA, Seixas AA, Redline S, et al. Artificial intelligence in sleep medicine: background and implications for clinicians. J Clin Sleep Med. 2020;16(4):609–618. doi: 10.5664/jcsm.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malhotra A, Ayappa I, Ayas N, Collop N, Kirsch D, McArdle N, et al. Metrics of sleep apnea severity: beyond the apnea-hypopnea index. Sleep. 2021;44(7). [DOI] [PMC free article] [PubMed]
- 4.Hartmann S, Bruni O, Ferri R, Redline S, Baumert M. Characterization of cyclic alternating pattern during sleep in older men and women using large population studies. Sleep. 2020;43(7). [DOI] [PMC free article] [PubMed]
- 5.Jonasdottir SS, Minor K, Lehmann S. Gender differences in nighttime sleep patterns and variability across the adult lifespan: a global-scale wearables study. Sleep. 2021;44(2). [DOI] [PubMed]
- 6.Sun H, Ganglberger W, Panneerselvam E, Leone MJ, Quadri SA, Goparaju B, et al. Sleep staging from electrocardiography and respiration with deep learning. Sleep. 2020;43(7). [DOI] [PMC free article] [PubMed]
- 7.Olesen AN, Jørgen Jennum P, Mignot E, Sorensen HBD. Automatic sleep stage classification with deep residual networks in a mixed-cohort setting. Sleep. 2021;44(1). [DOI] [PubMed]
- 8.Korkalainen H, Aakko J, Duce B, Kainulainen S, Leino A, Nikkonen S, et al. Deep learning enables sleep staging from photoplethysmogram for patients with suspected sleep apnea. Sleep. 2020;43(11). [DOI] [PMC free article] [PubMed]
- 9.Huang WC, Lee PL, Liu YT, Chiang AA, Lai F. Support vector machine prediction of obstructive sleep apnea in a large-scale Chinese clinical sample. Sleep. 2020;43(7). [DOI] [PMC free article] [PubMed]
- 10.Lechat B, Hansen K, Catcheside P, Zajamsek B. Beyond K-complex binary scoring during sleep: probabilistic classification using deep learning. Sleep. 2020;43(10). [DOI] [PMC free article] [PubMed]
- 11.Wickwire EM, Jobe SL, Oldstone LM, Scharf SM, Johnson AM, Albrecht JS. Lower socioeconomic status and co-morbid conditions are associated with reduced continuous positive airway pressure adherence among older adult medicare beneficiaries with obstructive sleep apnea. Sleep. 2020;43(12). [DOI] [PubMed]
- 12.Finnsson E, Ólafsdóttir GH, Loftsdóttir DL, Jónsson S, Helgadóttir H, Ágústsson JS, et al. A scalable method of determining physiological endotypes of sleep apnea from a polysomnographic sleep study. Sleep. 2021;44(1). [DOI] [PMC free article] [PubMed]
- 13.Williamson AA, Zendarski N, Lange K, Quach J, Molloy C, Clifford SA, et al. Sleep problems, internalizing and externalizing symptoms, and domains of health-related quality of life: bidirectional associations from early childhood to early adolescence. Sleep. 2021;44(1). [DOI] [PMC free article] [PubMed]
- 14.Redline S, Purcell SM. Sleep and Big Data: harnessing data, technology, and analytics for monitoring sleep and improving diagnostics, prediction, and interventions-an era for Sleep-Omics? Sleep. 2021;44(6). [DOI] [PMC free article] [PubMed]
- 15.Moe S, Rustad AM, Hanssen KG, editors. Machine learning in control systems: an overview of the state of the art. International Conference on Innovative Techniques and Applications of Artificial Intelligence; 2018: Springer.
- 16.Willetts M, Hollowell S, Aslett L, Holmes C, Doherty A. Statistical machine learning of sleep and physical activity phenotypes from sensor data in 96,220 UK Biobank participants. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-26174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayyadevara VK. Basics of machine learning. Pro Machine Learning Algorithms: Springer; 2018. pp. 1–15. [Google Scholar]
- 18.Patanaik A, Ong JL, Gooley JJ, Ancoli-Israel S, Chee MWL. An end-to-end framework for real-time automatic sleep stage classification. Sleep. 2018;41(5). [DOI] [PMC free article] [PubMed]
- 19.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circ. 2000;101(23):e215–e20. doi: 10.1161/01.CIR.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 20.O’reilly C, Gosselin N, Carrier J, Nielsen T. Montreal Archive of Sleep Studies: an open-access resource for instrument benchmarking and exploratory research. J Sleep Res. 2014;23(6):628–635. doi: 10.1111/jsr.12169. [DOI] [PubMed] [Google Scholar]
- 21.Quan SF, Howard BV, Iber C, Kiley JP, Nieto FJ, O’Connor GT, et al. The sleep heart health study: design, rationale, and methods. Sleep. 1997;20(12):1077–1085. [PubMed] [Google Scholar]
- 22.Kubat M, Pfurtscheller G, Flotzinger D. AI-based approach to automatic sleep classification. Biol Cybern. 1994;70(5):443–448. doi: 10.1007/BF00203237. [DOI] [PubMed] [Google Scholar]
- 23.Schaltenbrand N, Lengelle R, Toussaint M, Luthringer R, Carelli G, Jacqmin A, et al. Sleep stage scoring using the neural network model: comparison between visual and automatic analysis in normal subjects and patients. Sleep. 1996;19(1):26–35. doi: 10.1093/sleep/19.1.26. [DOI] [PubMed] [Google Scholar]
- 24.El-Solh AA, Mador MJ, Ten-Brock E, Shucard DW, Abul-Khoudoud M, Grant BJ. Validity of neural network in sleep apnea. Sleep. 1999;22(1):105–11. doi: 10.1093/sleep/22.1.105. [DOI] [PubMed] [Google Scholar]
- 25.Espie CA, Paul A, McFie J, Amos P, Hamilton D, McColl JH, et al. Sleep studies of adults with severe or profound mental retardation and epilepsy. Am J Ment Retard. 1998;103(1):47–59. doi: 10.1352/0895-8017(1998)103<0047:SSOAWS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 26.El-Solh AA, Magalang UJ, Mador MJ, Dmochowski J, Veeramachaneni S, Saberi A, et al. The utility of neural network in the diagnosis of Cheyne-Stokes respiration. J Med Eng Technol. 2003;27(2):54–58. doi: 10.1080/0309190021000043693. [DOI] [PubMed] [Google Scholar]
- 27.Almazaydeh L, Elleithy K, Faezipour M. Obstructive sleep apnea detection using SVM-based classification of ECG signal features. Annu Int Conf IEEE Eng Med Biol Soc. 2012;2012:4938–4941. doi: 10.1109/EMBC.2012.6347100. [DOI] [PubMed] [Google Scholar]
- 28.Christensen JA, Zoetmulder M, Koch H, Frandsen R, Arvastson L, Christensen SR, et al. Data-driven modeling of sleep EEG and EOG reveals characteristics indicative of pre-Parkinson’s and Parkinson’s disease. J Neurosci Methods. 2014;235:262–276. doi: 10.1016/j.jneumeth.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Huupponen E, Saastamoinen A, Niemi J, Virkkala J, Hasan J, Värri A, et al. Automated frequency analysis of synchronous and diffuse sleep spindles. Neuropsychobiology. 2005;51(4):256–264. doi: 10.1159/000085821. [DOI] [PubMed] [Google Scholar]
- 30.Redmond SJ, Heneghan C. Cardiorespiratory-based sleep staging in subjects with obstructive sleep apnea. IEEE Trans Biomed Eng. 2006;53(3):485–496. doi: 10.1109/TBME.2005.869773. [DOI] [PubMed] [Google Scholar]
- 31.Suhas SR, Behbehani K, Vijendra S, Burk JR, Lucas EA. ECG biomarkers for simultaneous detection of obstructive sleep apnea and Cheyne-Stokes breathing. Annu Int Conf IEEE Eng Med Biol Soc. 2007;2007:1047–1050. doi: 10.1109/IEMBS.2007.4352474. [DOI] [PubMed] [Google Scholar]
- 32.Lewicke A, Sazonov E, Corwin MJ, Neuman M, Schuckers S. Sleep versus wake classification from heart rate variability using computational intelligence: consideration of rejection in classification models. IEEE Trans Biomed Eng. 2008;55(1):108–118. doi: 10.1109/TBME.2007.900558. [DOI] [PubMed] [Google Scholar]
- 33.Azarbarzin A, Moussavi ZM. Automatic and unsupervised snore sound extraction from respiratory sound signals. IEEE Trans Biomed Eng. 2011;58(5):1156–1162. doi: 10.1109/TBME.2010.2061846. [DOI] [PubMed] [Google Scholar]
- 34.Willemen T, Van Deun D, Verhaert V, Vandekerckhove M, Exadaktylos V, Verbraecken J, et al. An evaluation of cardiorespiratory and movement features with respect to sleep-stage classification. IEEE J Biomed Health Inform. 2014;18(2):661–669. doi: 10.1109/JBHI.2013.2276083. [DOI] [PubMed] [Google Scholar]
- 35.Burke MJ, Downes R. A fuzzy logic based apnoea monitor for SIDS risk infants. J Med Eng Technol. 2006;30(6):397–411. doi: 10.1080/03091900600590140. [DOI] [PubMed] [Google Scholar]
- 36.Gerla V, Paul K, Lhotska L, Krajca V. Multivariate analysis of full-term neonatal polysomnographic data. IEEE Trans Inf Technol Biomed. 2009;13(1):104–110. doi: 10.1109/TITB.2008.2007193. [DOI] [PubMed] [Google Scholar]
- 37.Fontenla-Romero O, Guijarro-Berdiñas B, Alonso-Betanzos A, Moret-Bonillo V. A new method for sleep apnea classification using wavelets and feedforward neural networks. Artif Intell Med. 2005;34(1):65–76. doi: 10.1016/j.artmed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Otero A, Felix P, Alvarez MR, Zamarron C. Fuzzy structural algorithms to identify and characterize apnea and hypopnea episodes. Annu Int Conf IEEE Eng Med Biol Soc. 2008;2008:5242–5245. doi: 10.1109/IEMBS.2008.4650396. [DOI] [PubMed] [Google Scholar]
- 39.Dimitriadis SI, Salis C, Linden D. A novel, fast and efficient single-sensor automatic sleep-stage classification based on complementary cross-frequency coupling estimates. Clin Neurophysiol. 2018;129(4):815–828. doi: 10.1016/j.clinph.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 40.Procházka A, Kuchyňka J, Vyšata O, Cejnar P, Vališ M, Mařík V. Multi-class sleep stage analysis and adaptive pattern recognition. Appl Sci. 2018;8(5):697. doi: 10.3390/app8050697. [DOI] [Google Scholar]
- 41.Rahman MM, Bhuiyan MIH, Hassan AR. Sleep stage classification using single-channel EOG. Comput Biol Med. 2018;102:211–220. doi: 10.1016/j.compbiomed.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 42.Tsinalis O, Matthews PM, Guo Y, Zafeiriou S. Automatic sleep stage scoring with single-channel EEG using convolutional neural networks. arXiv preprint arXiv:161001683. 2016.
- 43.Zhang L, Fabbri D, Upender R, Kent D. Automated sleep stage scoring of the sleep heart health study using deep neural networks. Sleep. 2019;42(11):zsz159. doi: 10.1093/sleep/zsz159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chriskos P, Frantzidis CA, Nday CM, Gkivogkli PT, Bamidis PD. Kourtidou-Papadeli C. A review on current trends in automatic sleep staging through bio-signal recordings and future challenges. Sleep medicine reviews. 2021;55:101377. doi: 10.1016/j.smrv.2020.101377. [DOI] [PubMed] [Google Scholar]
- 45.Fiorillo L, Puiatti A, Papandrea M, Ratti P-L, Favaro P, Roth C, et al. Automated sleep scoring: a review of the latest approaches. Sleep medicine reviews. 2019;48:101204. doi: 10.1016/j.smrv.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Cui Z, Zheng X, Shao X, Cui L. Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity. 2018;2018.
- 47.Erdenebayar U, Kim YJ, Park J-U, Joo EY, Lee K-J. Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram. Comput Methods Programs Biomed. 2019;180:105001. doi: 10.1016/j.cmpb.2019.105001. [DOI] [PubMed] [Google Scholar]
- 48.Calderón JM, Álvarez-Pitti J, Cuenca I, Ponce F, Redon P. Development of a minimally invasive screening tool to identify obese pediatric population at risk of obstructive sleep apnea/hypopnea syndrome. Bioeng. 2020;7(4):131. doi: 10.3390/bioengineering7040131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ElMoaqet H, Eid M, Glos M, Ryalat M, Penzel T. Deep recurrent neural networks for automatic detection of sleep apnea from single channel respiration signals. Sensors. 2020;20(18):5037. doi: 10.3390/s20185037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswal S, Sun H, Goparaju B, Westover MB, Sun J, Bianchi MT. Expert-level sleep scoring with deep neural networks. J Am Med Inform Assoc. 2018;25(12):1643–1650. doi: 10.1093/jamia/ocy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Wu Y. A new method for automatic sleep stage classification. IEEE Trans Biomed Circuits Syst. 2017;11(5):1097–1110. doi: 10.1109/TBCAS.2017.2719631. [DOI] [PubMed] [Google Scholar]
- 52.Haidar R, McCloskey S, Koprinska I, Jeffries B, editors. Convolutional neural networks on multiple respiratory channels to detect hypopnea and obstructive apnea events. 2018 International Joint Conference on Neural Networks (IJCNN); 2018: IEEE.
- 53.Berry RB, Brooks R, Gamaldo CE, Harding SM, Marcus C, Vaughn BV. The AASM manual for the scoring of sleep and associated events Rules, Terminology and Technical Specifications, Darien, Illinois. Am Acad Sleep Med. 2012;176:2012. [Google Scholar]
- 54.Banluesombatkul N, Rakthanmanon T, Wilaiprasitporn T, editors. Single channel ECG for obstructive sleep apnea severity detection using a deep learning approach. TENCON 2018–2018 IEEE Region 10 Conference; 2018: IEEE.
- 55.Supratak A, Dong H, Wu C, Guo Y. DeepSleepNet: a model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans Neural Syst Rehabil Eng. 2017;25(11):1998–2008. doi: 10.1109/TNSRE.2017.2721116. [DOI] [PubMed] [Google Scholar]
- 56.Malafeev A, Laptev D, Bauer S, Omlin X, Wierzbicka A, Wichniak A, et al. Automatic human sleep stage scoring using deep neural networks. Front Neurosci. 2018;12:781. doi: 10.3389/fnins.2018.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephansen JB, Olesen AN, Olsen M, Ambati A, Leary EB, Moore HE, et al. Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy. Nat Commun. 2018;9(1):1–15. doi: 10.1038/s41467-018-07229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edwards BA, Redline S, Sands SA, Owens RL. More than the sum of the respiratory events: personalized medicine approaches for obstructive sleep apnea. Am J Respir Crit Care Med. 2019;200(6):691–703. doi: 10.1164/rccm.201901-0014TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miaskowski C, Barsevick A, Berger A, Casagrande R, Grady PA, Jacobsen P, et al. Advancing symptom science through symptom cluster research: expert panel proceedings and recommendations. JNCI: Journal of the National Cancer Institute. 2017;109(4). [DOI] [PMC free article] [PubMed]
- 60.Dutta R, Delaney G, Toson B, Jordan AS, White DP, Wellman A, et al. A novel model to estimate key obstructive sleep apnea endotypes from standard polysomnography and clinical data and their contribution to obstructive sleep apnea severity. Ann Am Thorac Soc. 2021;18(4):656–667. doi: 10.1513/AnnalsATS.202001-064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma E-Y, Kim J-W, Lee Y, Cho S-W, Kim H, Kim JK. Combined unsupervised-supervised machine learning for phenotyping complex diseases with its application to obstructive sleep apnea. Sci Rep. 2021;11(1):1–15. doi: 10.1038/s41598-021-84003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen JB, Sorensen HB, Kempfner J, Sørensen GL, Knudsen S, Jennum P. Sleep-Wake transition in narcolepsy and healthy controls using a support vector machine. J Clin Neurophysiol. 2014;31(5):397–401. doi: 10.1097/WNP.0000000000000074. [DOI] [PubMed] [Google Scholar]
- 63.Olsen AV, Stephansen J, Leary E, Peppard PE, Sheungshul H, Jennum PJ, et al. Diagnostic value of sleep stage dissociation as visualized on a 2-dimensional sleep state space in human narcolepsy. J Neurosci Methods. 2017;282:9–19. doi: 10.1016/j.jneumeth.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 64.Trotti LM. Twice is nice? Test-retest reliability of the Multiple Sleep Latency Test in the central disorders of hypersomnolence. J Clin Sleep Med. 2020;16(S1):17–18. doi: 10.5664/jcsm.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Mayer G, Dauvilliers Y, Plazzi G, Pizza F, Fronczek R, et al. Exploring the clinical features of narcolepsy type 1 versus narcolepsy type 2 from European Narcolepsy Network database with machine learning. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-28840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D, Pang Z, Lloyd SR. A neural network method for detection of obstructive sleep apnea and narcolepsy based on pupil size and EEG. IEEE Trans Neural Networks. 2008;19(2):308–318. doi: 10.1109/TNN.2007.908634. [DOI] [PubMed] [Google Scholar]
- 67.Aydın S, Saraoǧlu HM, Kara S. Singular spectrum analysis of sleep EEG in insomnia. J Med Syst. 2011;35(4):457–461. doi: 10.1007/s10916-009-9381-7. [DOI] [PubMed] [Google Scholar]
- 68.Chaparro-Vargas R, Ahmed B, Wessel N, Penzel T, Cvetkovic D. Insomnia characterization: from hypnogram to graph spectral theory. IEEE Trans Biomed Eng. 2016;63(10):2211–2219. doi: 10.1109/TBME.2016.2515261. [DOI] [PubMed] [Google Scholar]
- 69.Shahin M, Ahmed B, Hamida ST, Mulaffer FL, Glos M, Penzel T. Deep learning and insomnia: assisting clinicians with their diagnosis. IEEE J Biomed Health Inform. 2017;21(6):1546–1553. doi: 10.1109/JBHI.2017.2650199. [DOI] [PubMed] [Google Scholar]
- 70.Doan S, Yang EW, Tilak SS, Li PW, Zisook DS, Torii M. Extracting health-related causality from twitter messages using natural language processing. BMC Med Inform Decis Mak. 2019;19(Suppl 3):79. doi: 10.1186/s12911-019-0785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park S, Lee SW, Han S, Cha M. Clustering insomnia patterns by data from wearable devices: algorithm development and validation study. JMIR Mhealth Uhealth. 2019;7(12):e14473. doi: 10.2196/14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Philip P, Dupuy L, Morin CM, de Sevin E, Bioulac S, Taillard J, et al. Smartphone-based virtual agents to help individuals with sleep concerns during COVID-19 confinement feasibility study. J Med Internet Res. 2020;22(12):e24268. doi: 10.2196/24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esen H, Hatipoğlu T, Cihan A, Fiğlali N. Expert system application for prioritizing preventive actions for shift work: shift expert. Int J Occup Saf Ergon. 2019;25(1):123–137. doi: 10.1080/10803548.2017.1350392. [DOI] [PubMed] [Google Scholar]
- 74.Anafi RC, Francey LJ, Hogenesch JB, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc Natl Acad Sci U S A. 2017;114(20):5312–5317. doi: 10.1073/pnas.1619320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Braun R, Kath WL, Iwanaszko M, Kula-Eversole E, Abbott SM, Reid KJ, et al. Universal method for robust detection of circadian state from gene expression. Proc Natl Acad Sci U S A. 2018;115(39):E9247–E9256. doi: 10.1073/pnas.1800314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hesse J, Malhan D, Yalҫin M, Aboumanify O, Basti A, Relógio A. An optimal time for treatment-predicting circadian time by machine learning and mathematical modelling. Cancers (Basel). 2020;12(11). [DOI] [PMC free article] [PubMed]
- 77.Huang Y, Mayer C, Cheng P, Siddula A, Burgess HJ, Drake C, et al. Predicting circadian phase across populations: a comparison of mathematical models and wearable devices. Sleep. 2021. [DOI] [PMC free article] [PubMed]
- 78.Högl B, Santamaria J, Iranzo A, Stefani A. Precision medicine in rapid eye movement sleep behavior disorder. Sleep Med Clin. 2019;14(3):351–362. doi: 10.1016/j.jsmc.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 79.Cooray N, Andreotti F, Lo C, Symmonds M, Hu MT, De Vos M. Detection of REM sleep behaviour disorder by automated polysomnography analysis. Clin Neurophysiol. 2019;130(4):505–514. doi: 10.1016/j.clinph.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 80.Prashanth R, Roy SD, Mandal PK, Ghosh S. High-accuracy detection of early Parkinson’s disease through multimodal features and machine learning. Int J Med Informatics. 2016;90:13–21. doi: 10.1016/j.ijmedinf.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 81.Umut I, Çentik G. Detection of periodic leg movements by machine learning methods using polysomnographic parameters other than leg electromyography. Computational and mathematical methods in medicine. 2016;2016. [DOI] [PMC free article] [PubMed]
- 82.Veauthier C, Ryczewski J, Mansow-Model S, Otte K, Kayser B, Glos M, et al. Contactless recording of sleep apnea and periodic leg movements by nocturnal 3-D-video and subsequent visual perceptive computing. Sci Rep. 2019;9(1):16812. doi: 10.1038/s41598-019-53050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou P, Huang L, Zhao Q, Xiao W, Li S. A domestic diagnosis system for early restless legs syndrome based on deep learning. Zhongguo Yi Liao Qi Xie Za Zhi. 2019;43(2):79–82. doi: 10.3969/j.issn.1671-7104.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 84.Buysse DJ. Sleep health: can we define it? Does it matter? Sleep. 2014;37(1):9–17. doi: 10.5665/sleep.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wallace ML, Stone K, Smagula SF, Hall MH, Simsek B, Kado DM, et al. Which sleep health characteristics predict all-cause mortality in older Men? An application of flexible multivariable approaches. Sleep. 2018;41(1). [DOI] [PMC free article] [PubMed]
- 86.van Gilst MM, van Dijk JP, Krijn R, Hoondert B, Fonseca P, van Sloun RJG, et al. Protocol of the SOMNIA project an observational study to create a neurophysiological database for advanced clinical sleep monitoring. BMJ Open. 2019;9(11):e030996. doi: 10.1136/bmjopen-2019-030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perslev M, Darkner S, Kempfner L, Nikolic M, Jennum PJ, Igel C. U-Sleep: resilient high-frequency sleep staging. NPJ Digit Med. 2021;4(1):72. doi: 10.1038/s41746-021-00440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma M, Tiwari J, Acharya UR. Automatic sleep-stage scoring in healthy and sleep disorder patients using optimal wavelet filter bank technique with EEG signals. Int J Environ Res Public Health. 2021;18(6). [DOI] [PMC free article] [PubMed]
- 89.Abou Jaoude M, Sun H, Pellerin KR, Pavlova M, Sarkis RA, Cash SS, et al. Expert-level automated sleep staging of long-term scalp electroencephalography recordings using deep learning. Sleep. 2020;43(11). [DOI] [PMC free article] [PubMed]
- 90.Zhang X, Xu M, Li Y, Su M, Xu Z, Wang C, et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020;24(2):581–590. doi: 10.1007/s11325-019-02008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peter-Derex L, Berthomier C, Taillard J, Berthomier P, Bouet R, Mattout J, et al. Automatic analysis of single-channel sleep EEG in a large spectrum of sleep disorders. J Clin Sleep Med. 2021;17(3):393–402. doi: 10.5664/jcsm.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sridhar N, Shoeb A, Stephens P, Kharbouch A, Shimol DB, Burkart J, et al. Erratum: Author Correction: Deep learning for automated sleep staging using instantaneous heart rate. NPJ Digit Med. 2020;3:131. doi: 10.1038/s41746-020-00337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu T, Luo W, Yu F. Convolution-and attention-based neural network for automated sleep stage classification. Int J Environ Res Public Health. 2020;17(11). [DOI] [PMC free article] [PubMed]
- 94.Xu Z, Yang X, Sun J, Liu P, Qin W. Sleep stage classification using time-frequency spectra from consecutive multi-time points. Frontiers in Neuroscience. 2020;14(14). [DOI] [PMC free article] [PubMed]
- 95.Zhang L, Fabbri D, Upender R, Kent D. Automated sleep stage scoring of the Sleep Heart Health Study using deep neural networks. Sleep. 2019;42(11). [DOI] [PMC free article] [PubMed]
- 96.Yildirim O, Baloglu UB, Acharya UR. A deep learning model for automated sleep stages classification using PSG signals. Int J Environ Res Public Health. 2019;16(4). [DOI] [PMC free article] [PubMed]
- 97.Phan H, Andreotti F, Cooray N, Chen OY, De Vos M. SeqSleepNet: end-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans Neural Syst Rehabil Eng. 2019;27(3):400–410. doi: 10.1109/TNSRE.2019.2896659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Wu Y. Complex-valued unsupervised convolutional neural networks for sleep stage classification. Comput Methods Programs Biomed. 2018;164:181–191. doi: 10.1016/j.cmpb.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 99.Sors A, Bonnet S, Mirek S, Vercueil L, Payen J-F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed Signal Process Control. 2018;42:107–114. doi: 10.1016/j.bspc.2017.12.001. [DOI] [Google Scholar]
- 100.Chambon S, Galtier MN, Arnal PJ, Wainrib G, Gramfort A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans Neural Syst Rehabil Eng. 2018;26(4):758–769. doi: 10.1109/TNSRE.2018.2813138. [DOI] [PubMed] [Google Scholar]
- 101.Vilamala A, Madsen KH, Hansen LK, editors. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. 2017 IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP); 2017 25–28 Sept. 2017.