Highlights
-
•
This study provides a novel insight into the mechanism of degradation of E6 and E7 caused by punicalagin-induced autophagy. Therefore, our results will offer new strategy for treatment of HPV-infected cervical cancer.
Keywords: Punicalagin, E6, E7, Cervical cancer, Autophagy
Abstract
Punicalagin, which is derived from pomegranate peel, is reported to exert growth-inhibitory effects against various cancers. However, the underlying mechanisms have not been elucidated. Human papillomavirus (HPV), a major oncovirus, utilizes the host autophagic machinery to support its replication. Here, punicalagin markedly downregulated the levels of the major HPV oncoproteins E6 and E7 in cervical cancer cells through the autophagy-lysosome system. Additionally, punicalagin activated the reactive oxygen species (ROS)-JNK pathway and promoted the phosphorylation of BCL2, which led to the dissociation of BCL2 from BECN1 and the induction of autophagy. Treatment with autophagy and JNK inhibitors or ROS scavengers mitigated the punicalagin-induced degradation of E6 and E7. Moreover, the knockout of ATG5 using the clustered regularly interspaced palindrome repeat/Cas 9 system mitigated the punicalagin-induced downregulation of E6/E7. This indicated that punicalagin-induced degradation of E6 and E7 was dependent on autophagy. The results of in vivo studies demonstrated that punicalagin efficiently inhibits cervical cancer growth. In conclusion, this study elucidated a mechanism of punicalagin-induced autophagic degradation of E6 and E7. It will enable the future applications of punicalagin as a therapeutic for HPV-induced cervical cancer.
Introduction
Human papillomavirus (HPV), a double-stranded DNA virus, is the major etiological agent for cervical cancer, which is one of the most fatal cancers among women worldwide. Based on their pathogenicity, HPVs can be classified into high-risk and low-risk types. Type 16 and 18 HPVs, which are high-risk types, are the etiological agents for approximately 70% of invasive cervical cancers [1]. The severe consequences of type 16 and 18 HPV infections can be attributed to the two transforming oncoproteins E6 and E7. E6 mainly targets and degrades p53. Meanwhile, E7 regulates the expression of genes involved in modulating cell cycle arrest, apoptosis, and differentiation through the degradation of retinoblastoma (Rb) that interacts with E2F1 (E2F transcription factor 1) and other protein factors [2, 3]. HPV promotes cervical carcinogenesis through the modulation of p53 and Rb. Additionally, E6 promotes cell proliferation and cervical carcinogenesis by activating STAT3 (signal transducer and activator of transcription 3) [4]. The expression of E6 and E7 is sustained in HPV-positive cervical cancer and the continuous expression of E6 and E7 is required to sustain the immortality of HPV-positive cervical cancer cells and their xenografts in vivo [2]. Several E6 and E7-based therapeutic strategies for cervical cancer have been developed. For example, transfection with short-interfering RNA (siRNA) targeting E7 simultaneously downregulated the expression of E6 and E7 and induced senescence in HeLa cells. Additionally, specific ligands and blocking peptides that bind to E6 restore the p53-mediated cellular signaling pathway [5], [6], [7].
Recently, several novel small molecular weight compounds have been developed for HPV-positive cancer therapy. Docosahexaenoic acid and antimycin analogs derived from a marine Streptomyces sp. are considered novel therapeutics for cervical cancer as they promote the degradation of E6 and E7 in HPV-positive cervical cancer cells and induce cell apoptosis [8, 9].
Punicalagin is a polyphenolic tannin derived from pomegranate. Previous studies have suggested that punicalagin exhibits various biological activities, including anti-cancer, anti-inflammatory, antioxidant, and antiviral activities [10], [11], [12]. Punicalagin suppresses proliferation and induces apoptosis or autophagic cell death in various types of cancer cells in vitro, including colon cancer, lung cancer, cervical cancer, and breast cancer cells [13], [14], [15], [16], [17], [18], [19], [20]. Additionally, punicalagin is involved in immune system regulation and antiviral processes. Previous studies have reported that punicalagin suppresses inflammation in RAW264.7 cells, modulates murine macrophage polarization, and exerts immunosuppressive activities through the inhibition of NFAT (nuclear factor of activated T cells). Furthermore, punicalagin can effectively inhibit influenza and hepatitis B viruses by targeting viral neuraminidase and covalently closed circular DNA, respectively [11, 12, [21], [22], [23]]. The mechanisms underlying the anti-tumor, anti-inflammatory, and antiviral activities vary. However, studies on the biological functions of punicalagin will provide the basis for its future clinical applications.
Previous studies have demonstrated that punicalagin exhibits growth-inhibitory activities against cervical cancer. However, the underlying mechanisms have not been elucidated. This study aimed to examine the mechanism underlying the growth-inhibitory activities of punicalagin against cervical cancer with a special focus on its effect on the main oncoproteins. The findings of this study will enable the development of novel therapeutic strategies for cervical cancer.
Materials and methods
Cell culture
The cervical cancer cell lines (HeLa and SiHa cells) were purchased from American Type Culture Collection. STR profiling for HeLa and SiHa cells was performed by Biowing Applied Biotechnology company (Shanghai, China) to confirm that these two cell lines were authenticated. The cells were cultured in Dulbecco's modified Eagle's medium (Hyclone, USA) and minimal essential medium/Earle's balanced salt solution (Hyclone, USA), respectively, supplemented with 10% fetal bovine serum (FBS; Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Yeasen, Shanghai, China). All cells were cultured at 5% CO2 and 37°C and passaged every three days using 0.25% trypsin (Gibco, USA).
Regents and antibodies
Punicalagin was purchased from Macklin (purity ≥ 98% determined using high-performance liquid chromatography; Shanghai, China). The anti-HPV type 16/18 E6 and anti-HPV type 16/18 E7 antibodies were purchased from Novus and Genetex, respectively. The horseradish peroxidase (HRP)-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies were purchased from Zhongshan Goldenbridge Biotechnology (Beijing, China). All other antibodies were purchased from Cell Signaling Technology. LipoRNAi transfection reagent, MG-132, and radioimmunoprecipitation assay (RIPA) lysis buffer were purchased from Beyotime (Shanghai, China). TRIzol was purchased from Invitrogen. E64d, pepstatin A, SP600125 (JNK inhibitor), and 3-methyladenine (3-MA) (PI3KC3 inhibitor) were purchased from Sigma-Aldrich. The reactive oxygen species (ROS) scavenger N-acetyl-L-cysteine (NAC) was obtained from Selleck. Cycloheximide (CHX) was purchased from Absin. The details of the antibodies and reagents are listed in Table S1 and Table S2.
siRNA transfection
All siRNAs were synthesized by RiboBio (Guangzhou, China). The anti-sense sequence used to target JNK was 5′-UCACAGUCCUGAAACGAUA-3′. The HeLa cells were transfected with siRNAs against JNK1 using the lipoRNAi transfection reagent, following the manufacturer's instructions. Briefly, the cells were seeded in 6-well plates one day before transfection. Transfection was performed after the cells achieved 70%–80% confluency. The medium was replaced with fresh complete medium supplemented with 10% FBS and penicillin-streptomycin. In individual wells of the six-well plates, the cells were incubated with a mixture of 125 μL Opti-MEM, 100 nM siRNA, and 5 μL transfection reagent for 48 h. The knockdown efficiency was evaluated using western blotting.
Co-immunoprecipitation and western blotting
The cells were lysed for 30 min in cell lysis buffer (Beyotime). and for ubiquitin detection, cells were treated with 50 μM MG 132 for 6 h before lysis. The lysates were centrifuged at 14000 g for 30 min. The protein concentration in the supernatant was quantified. Next, the proteins in the supernatant were incubated with the antibodies overnight at 4°C, followed by incubation with protein A/G plus agarose beads (Santa Cruz, USA) for 4 h. The beads were washed thrice with ice-cold phosphate-buffered saline (PBS, pH 7.4). The whole-cell lysates were incubated with 2× loading buffer at 100°C for 10 min. The samples were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Western blotting was performed as described previously. Briefly, the cells were scraped from the wells and lysed in RIPA containing 10 mM phenylmethylsulfonyl fluoride for 30 min on ice. The lysate was centrifuged at 12000 rpm and 4°C for 20 min. The supernatant was boiled and the concentration of protein was determined using a bicinchoninic acid protein assay kit (Takara, Japan). Equal amounts of proteins were subjected to SDS-PAGE. The resolved proteins were transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% skimmed milk in Tris-buffered saline with Tween-20 for 1 h at room temperature, followed by incubation with the primary antibodies at 4℃ overnight. Next, the membrane was washed thrice with Tris-buffered saline with Tween-20 (10 min/step) and incubated with the corresponding HRP-conjugated secondary antibodies. Further, the membrane was washed thrice (10 min/step) and the immunoreactive signals were detected using enhanced chemiluminescence.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from the HeLa cells using TRIzol, following the manufacturer's instructions. The isolated RNA was reverse transcribed into complementary DNA (cDNA) using the PrimeScript RT Master Mix (Takara, Japan) under the following thermal cycling conditions: 37°C for 30 min and 85°C for 5 s and hold at 4°C. The qRT-PCR analysis was performed using the SYBR Green Ex Taq mix (Takara, Japan), following the manufacturer's instructions. Each sample was analyzed in triplicate. The PCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, and 60°C for 45 s. The mRNA levels of E6 and E7 were quantified using qRT-PCR. GAPDH and beta-actin were used as an internal control. The following primers were used for qRT-PCR analysis: HPV type 18 E6, 5′-ATAAGGTGCCTGCGGTGCC-3′ (forward) and 5′-TGCGTCGTTGGAGTCGTTC-3′ (reverse); HPV type 18 E7, 5′-GAGCACGACAGGAACGACT-3′ (forward) and 5′-GGGCTGGTAAATGTTGATGAT-3′ (reverse); GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse); beta-actin, 5′-GACGACATGGAGAAAATCTG-3′ (forward) and 5′-ATGATCTGGGTCATCTTCTC-3′ (reverse). The fold change of target mRNA expression was analyzed with the 2-ΔΔCt method.
Autophagic flux
The autophagy flux was analyzed using the AdPlus-mCherry-green fluorescent protein (GFP)-LC3B adenovirus (Beyotime), following the manufacturer's instructions. The HeLa cells were infected with the adenovirus for 24 h and treated with various drugs. The cells positive for GFP, mCherry, or both were visualized using a confocal fluorescence microscope (Leica Microsystems). Autophagic flux was determined by analyzing the punctate patterns of GFP and mCherry.
Colony formation assay
The cells plated in a 6-well plate (1.5×103 cells/well) for 24 h were incubated with punicalagin. On day 14 post-treatment, the cells were washed twice with PBS, fixed with methanol for 30 min, and subjected to crystal violet staining.
Cell counting kit-8 assay
The proliferation of HeLa and SiHa cells was determined using the cell counting kit-8 (CCK-8) (Dojindo, Japan). Briefly, the cells plated in a 96-well plate (5×103 cells/well) were incubated with different concentrations of punicalagin for different durations. Cell viability was determined using CCK-8, following the manufacturer's instructions.
Transmission electron microscopy
For transmission electron microscopy analysis, HeLa cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer at room temperature for 30 min, followed by incubation at 4°C overnight. The samples were embedded, sectioned, and observed under a transmission electron microscope (FEI Tecnai Spirit120kV).
In vivo study
The mouse studies were performed according to the regulations of the Institute of Microbiology, Chinese Academy of Sciences of Research Ethics Committee. The study protocol was approved by the Research Ethics Committee (permit number PZIMCAS2011001). All animal experiments were performed according to the Institutional Guidelines on the Handling of Laboratory Animals. BALB/c nude mice (female; aged 5–6 weeks) were purchased from SPF Biotechnology company (Beijing, China) and housed under SPF conditions. To establish the HeLa cell xenograft model, 5 × 106 HeLa cells were subcutaneously injected into the right flank of the nude mice. The mice were randomly divided into the following two groups after the mean diameter of the xenograft reached 5 mm (6 mice/ group), punicalagin-treated group, mice intraperitoneally administered with punicalagin at a dose of 20 mg/kg bodyweight; PBS-treated group, mice intraperitoneally administered with PBS. The tumor size was monitored once every two days. On day 18 post-punicalagin/PBS administration, the mice were sacrificed. Western blotting and immunohistochemical analyses were performed.
ROS assessment
Punicalagin-treated cells were incubated with the oxidation-sensitive fluorescent probe 2′,7′-dichlorofluorescein diacetate (10 mM; Beyotime) for 20 min at 37°C. The cells were washed thrice with culture medium, harvested, and subjected to flow cytometry analysis.
Cell apoptosis analysis
The cells were treated with punicalagin for 24 h, trypsinized, and subjected to Annexin V-fluorescein isothiocyanate/propidium iodide staining using the cell apoptosis kit (Invitrogen, USA), following the manufacturers’ instructions. The stained cells were subjected to flow cytometry analysis.
Immunohistochemistry
The tumor tissues were fixed with formalin, dehydrated, embedded in paraffin blocks, sliced into thin sections, and transferred to glass slides. The sections were incubated with 3% H2O2 in methanol for 10 min to inactivate endogenous peroxidase activity. Next, the sections were incubated in a 100°C water bath with ethylenediaminetetraacetic acid antigen retrieval buffer for 10 min and cooled to room temperature to unmask the antigenic epitope. The sections were then blocked with 5% bovine serum albumin at 37°C for 30 min and incubated with specific primary and secondary antibodies for 30 min at 37°C. Further, the sections were incubated with streptavidin-biotin complex and 3,3′-diaminobenzidine and counterstained with hematoxylin. Finally, the sections were subjected to immunohistochemical and image analyses.
Establishment of ATG5 knockout HeLa cell lines using the clustered regularly interspaced palindrome repeat/Cas 9 system
The pSpCas9 (BB)-2A-GFP plasmid was a kind gift from Professor Cuihua Liu. Single guide RNA (sgRNA) oligonucleotides targeting ATG5 were annealed and ligated to the vector digested with BbsI (NEB, USA). The sgRNA sequences used for targeting ATG5 were as follows: 5′-CACCGTTCCATGAGTTTCCGATTGA-3′ (forward) and 5′-AAACTCAATCGGAAACTCATGGAAC-3′ (reverse). HeLa cells were transfected with the vector containing the ATG5 sgRNA using lipofectamine 3000 reagent (Invitrogen, USA). At 24 h post-transfection, GFP positive single cells were sorted into 96-well plates using an AriaIII flow cytometer (BD, USA). The expression levels of ATG5 in cell clones were assessed using western blotting.
Statistical analysis
Data are expressed as mean ± standard deviation (shown as error bars). The data were analyzed using the Student's t-test. All statistical analyses were performed using GraphPad Prism 8 software (CA, USA). Densitometry analysis of the protein bands in immunoblots was performed using ImageJ. Differences were considered significant at *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Punicalagin suppresses the proliferation of cervical cancer cells, induces caspase-dependent cell apoptosis, and inhibits carcinogenesis
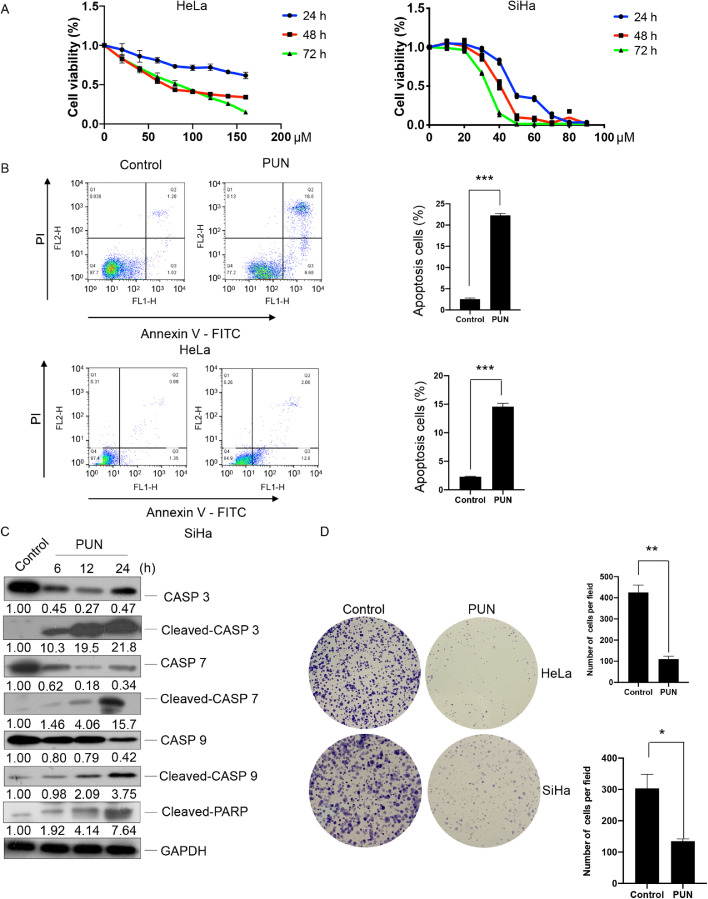
The effect of punicalagin on the proliferation of HeLa and SiHa cells was examined. As shown in Fig. 1A, punicalagin dose-dependently and time-dependently inhibited the proliferation of HeLa and SiHa cells. The half-maximal inhibitory concentration values of punicalagin against HeLa and SiHa cells were 67 and 40 μM, respectively, at 48 h post-treatment. This indicated that SiHa cells were highly sensitive to punicalagin treatment. Flow cytometric analysis revealed that apoptosis in HeLa and SiHa cells treated with 140 or 90 μM punicalagin for 24 h was approximately 8.5-fold and 6.3-fold, respectively, higher than that in control cells (both P < 0.001) (Fig. 1B). Western blotting results indicated that punicalagin-induced apoptosis was dependent on caspase. The expression levels of CASP3, CASP7, and CASP9 were downregulated, whereas those of cleaved CASP3, CASP7, and CASP9 were upregulated in HeLa cells upon treatment with 140 μM punicalagin for 6, 12, or 24 h (Fig. 1C). Moreover, the number and size of cell colonies markedly decreased in HeLa and SiHa cells upon treatment with punicalagin (Fig. 1D).
Fig. 1.
Effect of punicalagin (PUN) on the proliferation, apoptosis, and colony-forming ability of HeLa and SiHa cells. (A) HeLa and SiHa cells were treated with different doses of punicalagin for 24, 48, or 72 h. Cell viability was evaluated using the cell counting kit-8. (B) Apoptosis in cells treated with 140 or 90 μM PUN or dimethyl sulfoxide (DMSO) (control) for 24 h was analyzed using flow cytometry. (C) The levels of apoptosis-related caspase proteins in HeLa cells treated with 140 μM PUN for 6, 12, or 24 h were examined using western blotting. (D) HeLa and SiHa cells treated with 15 or 10 μM PUN for 14 days were stained with crystal violet. Representative images of colonies are shown. Values represent mean ± standard deviation. All experiments were performed in three replicates. * p<0.05, ** p<0.01.
Punicalagin promotes the lysosomal degradation of HPV E6 and E7 proteins
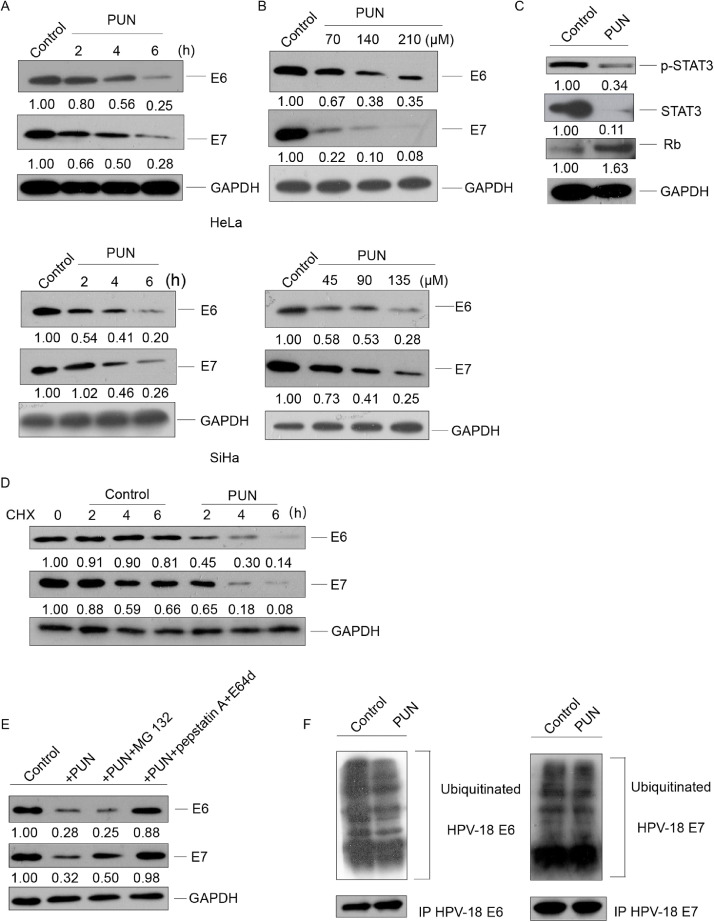
Next, the effect of punicalagin on E6 and E7, which are the two main oncoproteins of HPVs that regulate the proliferation, apoptosis, and carcinogenesis of cervical cancer cells, was examined. As shown in Fig. 2A–B, punicalagin dose-dependently and time-dependently downregulated the levels of E6 and E7 in HeLa and SiHa cells. Consistently, the expression levels of the E6 related targeted proteins STAT3 and phosphorylated STAT3 were markedly downregulated, whereas E7 targeted protein Rb was markedly upregulated in the punicalagin (140 μM)-treated HeLa cells (Fig. 2C). However, treatment with 140 μM punicalagin did not downregulate the mRNA levels of E6/E7 (Fig. S2). This indicated that punicalagin did not regulate the expression levels of E6 and E7 at the transcriptional level. The stability of E6 and E7 protein markedly decreased in the punicalagin (140 μM)-treated HeLa cells (Fig. 2D). Next, the pathways through which punicalagin mediates the degradation of E6 and E7 were examined. HeLa cells were co-treated with 140 μM punicalagin and MG 132 (a proteasome inhibitor) or pepstatin A (a protease inhibitor)/ E64d (a lysosome inhibitor). Co-treatment with E64d and pepstatin A almost recovered the punicalagin-mediated downregulation of E6/E7 (Fig. 2E). Meanwhile, MG 132 partially mitigated the punicalagin-mediated downregulation of E7. Treatment with 140 μM punicalagin did not affect the ubiquitination levels of E7 and led to a moderate decrease of E6 ubiquitination in HeLa cells (Fig. 2F). Together, these data indicate that besides lysosomal degradation, proteasome degradation may have a role in punicalagin-mediated E7 loss.
Fig. 2.
Punicalagin promotes the degradation of E6 and E7 through the lysosomal pathway. (A and B) HeLa or SiHa cells were treated with 140 or 90 μM punicalagin for 2, 4, and 6 h (A) or with the indicated concentrations of punicalagin for 6 h (B). Cells treated with dimethyl sulfoxide (DMSO) served as a control. The total cellular E6 or E7 levels were determined using western blotting. (C) The levels of E6-related and E7-related proteins in HeLa cells treated with 140 μM punicalagin or DMSO (control) for 6 h were examined using western blotting. (D) The levels of E6 and E7 in HeLa cells treated with 50 μg/mL cycloheximide (CHX) or the combination of 50 μg/mL CHX for 1 h and 140 μM punicalagin for 2, 4, and 6 h were examined using western blotting. (E) The levels of E6 and E7 in HeLa cells treated with 140 μM punicalagin for 6 h or pretreated with 50 μM MG 132 or 20 μM pepstatin A and 20 μM E64d for 1 h, followed by treatment with punicalagin for 6 h were examined using western blotting. (F) HeLa cells were pretreated with 50 μM MG 132 for 1 h, followed by treatment with 140 μM punicalagin or MG 132 for 6 h. The cells were lysed and E6 or E7 was immunoprecipitated with the corresponding antibody. The levels of ubiquitinylated proteins were analyzed using ubiquitin antibody by western blotting. All experiments were performed in three replicates.
Punicalagin induces autophagy in cervical cancer cells
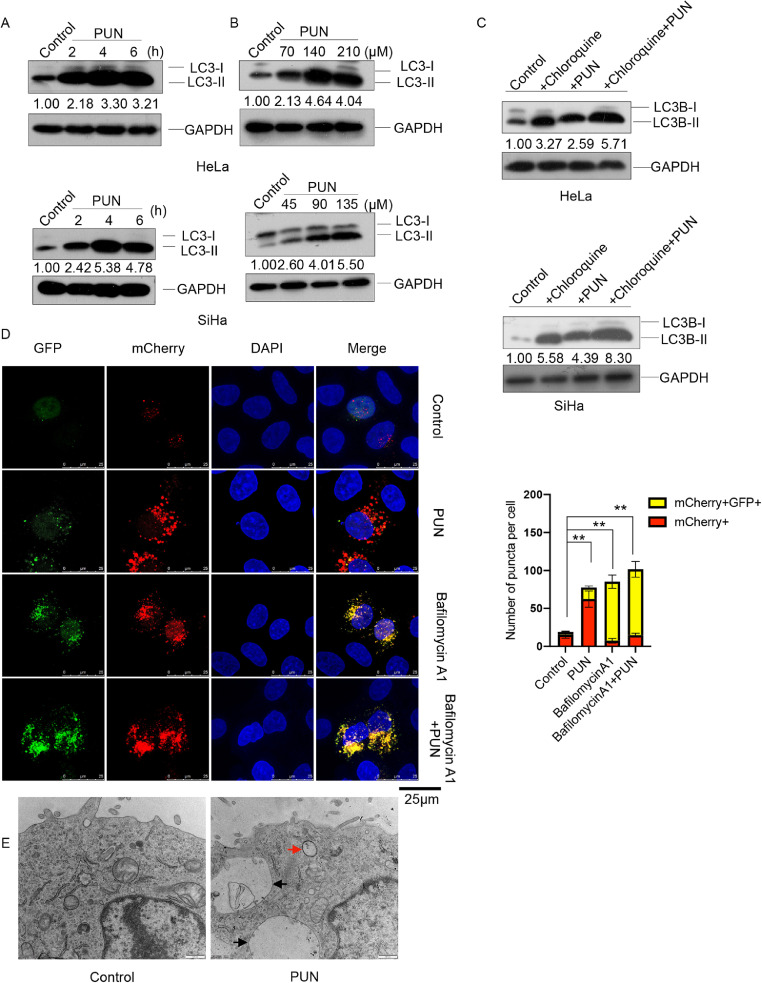
To elucidate the underlying mechanisms of punicalagin-induced lysosomal degradation of E6 and E7, the effect of punicalagin on autophagy in HeLa and SiHa cells was examined. The expression of the microtubule-associated protein 1 light chain 3 (LC3-II), a marker of autophagy [24], was analyzed in punicalagin-treated cells. Treatment with 140 or 90 μM time-dependently upregulated the LC3-II levels and punicalagin treatment for 6 h also exhibited a dose-dependent upregulation of LC3-II levels in HeLa and SiHa cells (Fig. 3A–B). Next, the cells were treated with chloroquine, an autophagosome/lysosome fusion inhibitor [25], to determine whether the accumulation of LC3-II resulted from increased autophagosome formation and/or inhibition of autophagosome/lysosome fusion. Compared with those in the untreated control HeLa and SiHa cells, either punicalagin or chloroquine markedly upregulated LC3-II level. Meanwhile, the LC3-II levels in the punicalagin (140 or 90 μM)/chloroquine-co-treated cells were upregulated when compared with those in the punicalagin (140 or 90 μM)-treated and chloroquine-treated cells (Fig. 3C). This indicated that punicalagin induces autophagy in HeLa and SiHa cells.
Fig. 3.
Effect of punicalagin on autophagy. (A–B) The LC3-I and LC3-II levels in HeLa or SiHa cells treated with 140 or 90 μM punicalagin for 2, 4, and 6 h (A) or the indicated concentrations of punicalagin for 6 h (B) were determined using western blotting. (C) The LC3-I and LC3-II levels in HeLa or SiHa cells pretreated with 100 μM chloroquine for 1 h, followed by treatment with 140 or 90 μM punicalagin for 6 h were examined using western blotting. (D) HeLa cells were infected with AdPlus-mCherry-GFP-LC3B adenovirus at the multiplicity of infection of 200 for 24 h, followed by pretreatment with 100 nM bafilomycin A1 for 1 h and treatment with 140 μM punicalagin for 24 h. The fluorescent images were captured using a confocal fluorescence microscope. The red and yellow LC3-II puncta were quantitatively analyzed in 100 cells per group and the results were reported as mean ± standard deviation. (E) HeLa cells treated with 140 μM punicalagin for 2 h were examined under a transmission electron microscope. The representative autophagosomes and autolysosomes are indicated by red and black arrows, respectively. ** P<0.01. All experiments were repeated three times.
Moreover, a tandem fluorescence probe-tagged adenovirus system (AdPlus-mCherry GFP-LC3II) was used to evaluate autophagic flux in punicalagin-treated HeLa cells (Fig. 3D). The red and yellow puncta (merged from mCherry and GFP) indicate fused autophagosome-lysosome and unfused autophagosomes, respectively. The number of red and yellow puncta was counted after treatment with 140 μM punicalagin for 24 h. The autophagy flux in the punicalagin-treated HeLa cells was higher than that in the control HeLa cells. Compared with that in the punicalagin-treated cells, the autophagic flux was higher in the cells co-treated with punicalagin and bafilomycin A1, which inhibits both lysosomal acidification and autophagosome-lysosome fusion [26]. Transmission electron microscopy analysis provided direct insights into autophagosomes and autolysosomes. The formation of autophagosomes and autolysosomes in the punicalagin-treated HeLa cells was higher than that in the control HeLa cells (Fig. 3E). Additionally, treatment with 3-MA, a PIK3C3 (phosphatidylinositol 3-kinase catalytic subunit type 3) inhibitor that inhibits the formation of autophagosomes, mitigated the punicalagin-induced downregulation of E6 and E7. The levels of E6 and E7 in the 3-MA/punicalagin-co-treated HeLa and SiHa cells were similar to those in the control HeLa and SiHa cells, respectively. This indicated that punicalagin promotes the degradation of E6 and E7 through the upregulation of autophagy (Fig. S3).
Punicalagin induces autophagy in cervical cancer cells through the ROS-JNK-BCL2 pathway
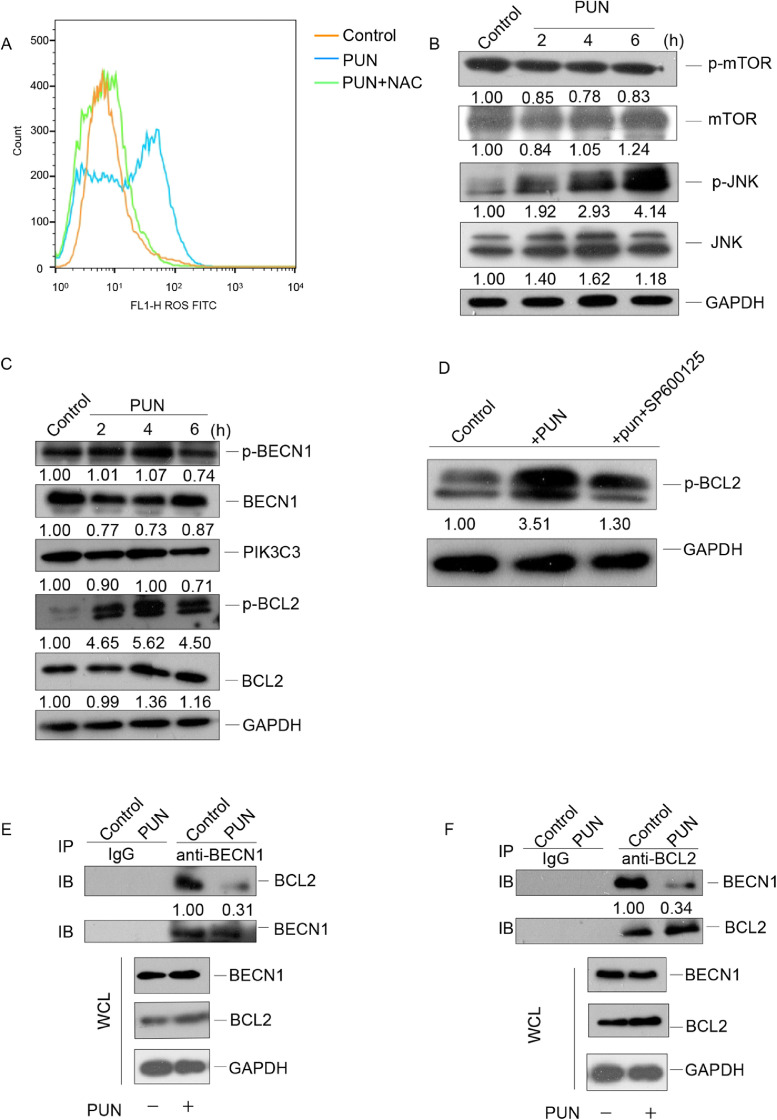
Several studies have demonstrated that punicalagin promotes mitochondrial dysfunction and consequently promotes the production of ROS [14, 27]. Additionally, the ROS-mediated PI3K/Akt/mTOR (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta, PI3K/serine/threonine kinase, Akt /mechanistic target of rapamycin kinase, mTOR) pathway activation and/or activation of the JNK (mitogen-activated protein kinase 8, MAPK8) pathway are critical for ROS-mediated autophagy induction [28], [29], [30], [31]. Therefore, the ROS levels were examined in the punicalagin-treated cervical cancer cells. As shown in Fig 4A, treatment with 140 μM punicalagin upregulated the ROS levels in HeLa cells. The mTOR pathway negatively regulates autophagy [32]. Hence, the mTOR and phosphorylated mTOR (p-mTOR) levels were examined in the punicalagin-treated HeLa cells. Treatment with 140 μM punicalagin did not markedly affect the mTOR and p-mTOR levels (Fig. 4B), suggesting that punicalagin-induced autophagy does not involve the mTOR pathway. The total and phosphorylated JNK (p-JNK) levels were examined in HeLa cells treated with 140 μM punicalagin for different durations. As shown in Fig. 4B, punicalagin time-dependently upregulated the p-JNK levels. JNK-mediated BCL2 (BCL2 apoptosis regulator) phosphorylation disrupts the binding of BCL2 to BECN1 (beclin 1SQ). This process plays an important role in regulating cellular autophagy and apoptosis [33], [34], [35].. Hence, the total and phosphorylated BCL2 (p-BCL2) levels were examined in punicalagin-treated HeLa cells. As shown in Fig. 4C, treatment with 140 μM punicalagin for 6 h markedly increased the p-BCL2 levels but did not affect the levels of phosphorylated BECN1 (p-BECN1) and PIK3C3, another important binding partner of BECN1. Treatment with SP100625 (JNK inhibitor) significantly reduced the punicalagin-induced upregulation of p-BCL2, and this indicated that this induction was dependent on JNK (Fig. 4D). As shown in Fig. 4E–F, treatment with 140 μM punicalagin markedly inhibited the interaction between BCL2 and BECN1 in HeLa cells. These results demonstrate that punicalagin activates the ROS-JNK-BCL2 pathway and it promotes the dissociation of the BCL2-BECN1 complex and the subsequent activation of autophagy.
Fig. 4.
Punicalagin promotes BCL2 phosphorylation and consequently the dissociation of the BCL2-BECN1 complex through the ROS-JNK pathway. (A) HeLa cells treated with 140 μM punicalagin or dimethyl sulfoxide (DMSO; control) and 10 mM N-acetyl-L-cysteine (NAC) for 4 h were stained with 10 μM of 2′,7′-dichlorofluorescein diacetate and subjected to flow cytometric analysis. (B) The mTOR, phosphorylated mTOR (p-mTOR), JNK, and phosphorylated JNK (p-JNK) levels in HeLa cells treated with 140 μM punicalagin for 2, 4, or 6 h were detected using western blotting. (C) The phosphorylated and total BECN1, BCL2, and total PIK3C3 levels in HeLa cells treated as in B were determined using western blotting. (D) The p-BCL2 levels in HeLa cells treated with 140 μM punicalagin or DMSO (control) or the combination of 20 μM SP600125 treatment for 1 h and punicalagin treatment for 6 h were detected using western blotting. (E and F) The lysates of HeLa cells treated with 140 μM punicalagin or DMSO (control) for 6 h were subjected to immunoprecipitation with anti-BECN1 (E) or anti-BCL2 (F) antibodies. Additionally, the levels of BECN1 and BCL2 in the immunoprecipitate were analyzed using western blotting. All experiments were repeated three times.
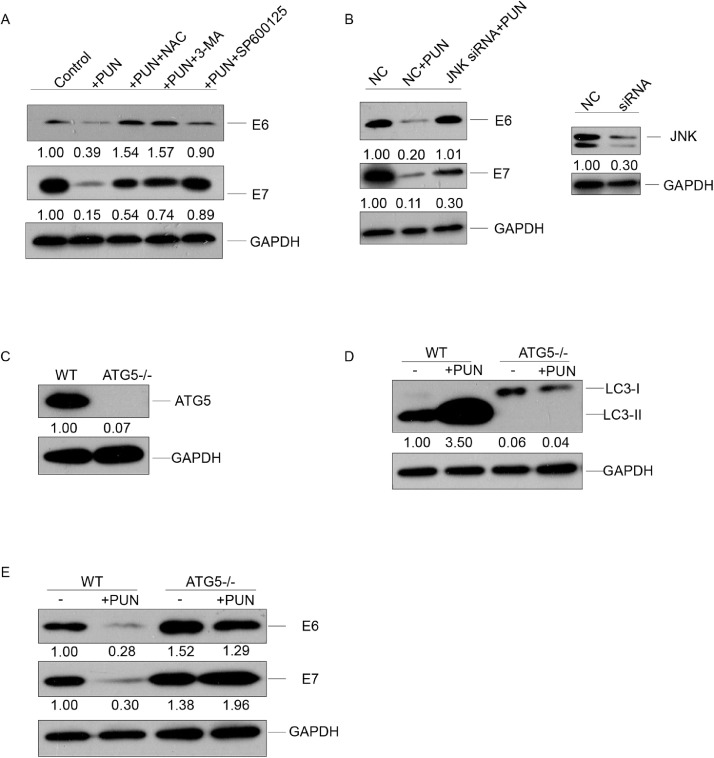
JNK-mediated autophagy is involved in the punicalagin-mediated degradation of E6 and E7 proteins
To evaluate the role of the punicalagin-induced JNK pathway in autophagic E6/E7 degradation, the HeLa cells were co-treated with punicalagin and NAC (ROS scavenger), SP100625, or 3-MA. As shown in Fig. 5A, the E6 and E7 levels in the cells co-treated with punicalagin (140 μM) and NAC, SP100625, or 3-MA for 6 h were similar to those in the control cells. Similar results were obtained for E6 protein levels and partially for E7 protein levels after knocking down JNK (MAPK8, gene ID: 5599) using siRNA (Fig. 5B).
Fig. 5.
Role of JNK pathway-induced autophagy in punicalagin-mediated degradation of E6 and E7. (A) The E6 and E7 levels in HeLa cells treated with 140 μM punicalagin or dimethyl sulfoxide (DMSO; control) or pretreated with N-acetyl-L-cysteine (NAC; 10 mM), 3-MA (5 mM) or SP600125 (20 μM) for 1 h, followed by treatment with punicalagin for 6 h were assessed using western blotting. (B) HeLa cells were transfected with 100 nM short-interfering RNA (siRNA) against JNK or control siRNA. At 48 h post-transfection, the cells were treated with 140 μM punicalagin for 6 h. The E6 and E7 protein levels and JNK knockdown efficiency were analyzed using western blotting. (C–E) The levels of ATG5 in wild-type (WT) and ATG5 knockout (KO) HeLa cells were examined using western blotting (C). The levels of LC3-II (D), E6, and E7 (E) in WT and ATG5 KO cells treated with 140 μM punicalagin for 6 h were analyzed using western blotting. All experiments were repeated three times.
ATG5 (gene ID: 9474) plays a vital role in autophagosome formation by promoting the conversion of LC3-I to LC3-II [36, 37]. Next, the role of autophagy in punicalagin-mediated E6/E7 degradation was examined. The ATG5 knockout HeLa cell line was established using the CRISPR/Cas9 system (Fig. 5C). The wild-type (WT) and ATG5 knockout HeLa cells were treated with 140 μM punicalagin for 6 h. The knockout of ATG5 inhibited the conversion of LC3-I to LC3-II in HeLa cells (Fig 5D). The E6 and E7 levels in the punicalagin-treated WT cells were markedly downregulated when compared with control cells and those in the punicalagin-treated ATG5 knockout cells (Fig. 5E). ATG5 knockout slightly increased the E6 and E7 levels. This indicated that the basal levels of autophagy may play a role in the degradation of E6 and E7 proteins.
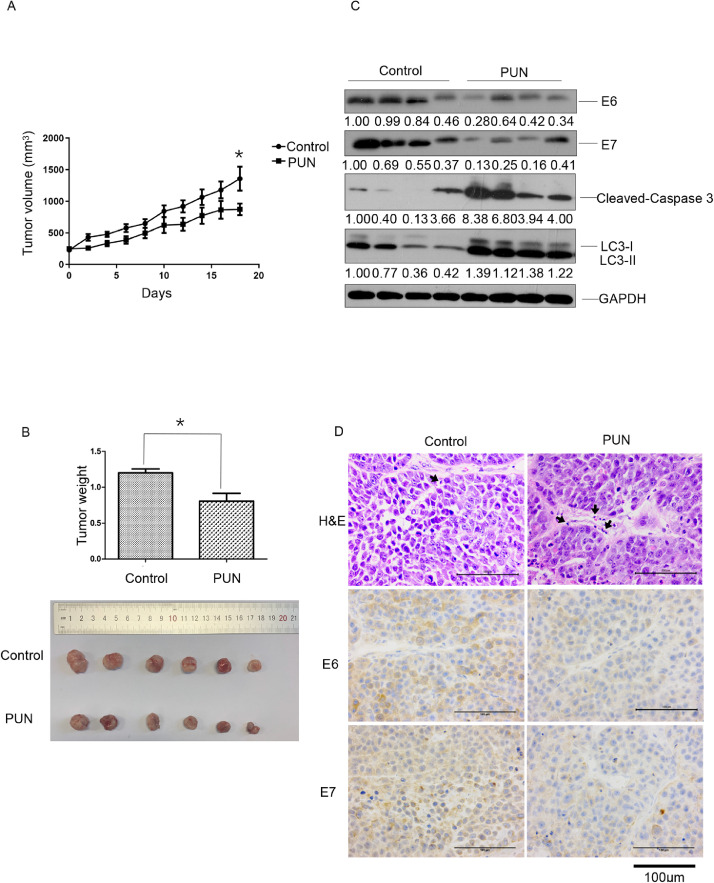
Punicalagin downregulates E6/E7 levels and suppresses cervical tumor growth in vivo
Finally, the therapeutic efficacy of punicalagin in the HeLa cell xenograft nude mouse model was examined. Consistent with the in vitro results, the tumor growth in the punicalagin (20 mg/kg bodyweight)-treated group was significantly slower than that in the PBS-treated group (P < 0.05) (Fig. 6A–B). Additionally, the tumor weight in the punicalagin (20 mg/kg bodyweight)-treated group significantly decreased by approximately 32.7% when compared with that in the PBS-treated group (P < 0.05). The downregulation of E6 and E7 in the punicalagin-treated group was verified using western blotting (Fig. 6C) and immunohistochemical analyses (Fig. 6D). Compared with those in the PBS-treated group, the levels of LC3-II and cleaved CASP3 (the apoptosis-related proteins) were upregulated in the punicalagin (20 mg/kg bodyweight)-treated group (Fig. 6C). In addition, increased infiltration of inflammatory cells was observed in the punicalagin-treated group and the PBS-treated group (Fig. 6D).
Fig. 6.
Therapeutic efficiency of punicalagin against cervical tumors. BALB/c-nu/nu mice with the HeLa cell xenograft tumor of diameter 5 mm were injected with punicalagin (20 mg/kg bodyweight) or phosphate-buffered saline (PBS) (control) once every two days. (A) Tumor diameter was measured once every two days for 18 days. (B) Tumor weight in the two groups was measured (upper panel) and representative images of the tumors were captured on day 18 when all mice were sacrificed (low panel). (C) The E6, E7, LC3-II, and cleaved CASP3 levels in the tumor tissues were examined using western blotting. (D) Representative images of tumor sections stained with hematoxylin and eosin and tumor infiltration of inflammatory cells were indicated by black arrows and immunohistochemical analysis of E6 and E7. * p<0.05. Data are expressed as mean ± standard deviation (6 mice/group). The experiments were performed twice with similar results.
Discussion
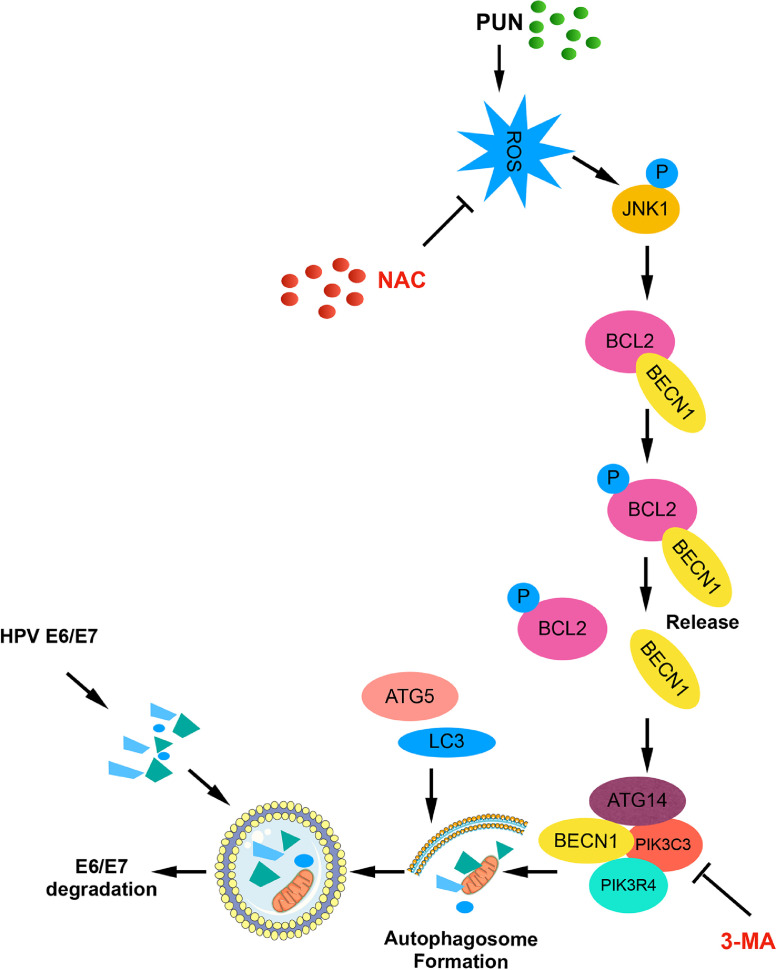
The malignant phenotype of HPV-infected cervical cancer cells is mainly maintained by E6 and E7. In this study, punicalagin time-dependently and dose-dependently downregulated the E6 and E7 levels in cervical cancer cells through the autophagy lysosome system. However, the role of the proteasome pathway in the punicalagin-mediated regulation of E6 and E7 cannot be completely ruled out because treatment with the proteasome inhibitor MG 132 partially mitigated the punicalagin-mediated downregulation of E7. The autophagy-lysosome pathway majorly contributed to the regulation of E6 and E7 levels as the inhibitors of autophagy and lysosome almost completely inhibited the punicalagin-induced downregulation of E6 and E7. Further investigation revealed that punicalagin activated the ROS-JNK pathway and that treatment with NAC (ROS scavenger) and SP600125 (JNK inhibitor) mitigated the punicalagin-induced downregulation of E6 and E7 levels. This indicated that the ROS-JNK pathway is involved in the degradation of E6 and E7. Activated JNK promoted BCL2 phosphorylation, which led to the dissociation of the BECN1/BCL2 complex and the formation of the BECN1-PIK3C3 complex, which is important for autophagic phagophore assembly and recruitment of accessory proteins to induce autophagy. The results of co-immunoprecipitation and ATG5 knockout experiments confirmed our hypothesis that punicalagin promotes the dissociation of the BECN1-BCL2 complex and promoted autophagy. As shown in Figure 7, punicalagin-mediated E6/E7 degradation was largely dependent on autophagy. This model provided a novel mechanism of autophagy induced degradation of the two oncoproteins E6 and E7 in HPV positive cervical cancer cells.
Fig. 7.
Schematic of the pathways involved in punicalagin-induced autophagic degradation of E6 and E7. In human papillomavirus (HPV)-infected cervical cancer cells, the binding of BCL2 to BECN1 blocks its interaction with other components of the PIK3C3 complex that regulates autophagosome nucleation for membrane formation. Treatment with punicalagin promotes the production of reactive oxygen species (ROS), which leads to the activation and phosphorylation of JNK. Activated JNK directly phosphorylates BCL2. Phosphorylated BCL2 promotes the dissociation of the BECN1/BCL2 complex. BECN1 binds to the PIK3C3 complex and initiates autophagy. Accelerated autophagosome formation and autophagosome-lysosome fusion lead to the lysosomal degradation of its targets E6 and E7. This results in the suppression of cervical cancer cell proliferation and survival and the inhibition of tumor growth.
SQSTM1/p62 (sequestosome 1), an adaptor of selective autophagy, interacts with ubiquitinylated proteins and transports them to the autophagosome for degradation. During this process, SQSTM1/p62 also undergoes degradation. Treatment with punicalagin did not affect the levels of SQSTM1/p62 (Fig. S4). However, the possibility of punicalagin promoting SQSTM1/p62-induced selective autophagy cannot be excluded. The SQSTM1/p62 levels can be affected by other factors, such as NRF2 (nuclear factor, erythroid 2 like 2) after ROS exposure [38]. Autophagy induction can result in subtle changes in SQSTM1/p62 levels. Therefore, the SQSTM1/p62 levels may be regulated by multiple regulators. The role of punicalagin in inducing selective or non-selective autophagy in cervical cancer was not fully determined in this study. It awaits further investigation whether punicalagin causes degradation of E6 and E7 through specific or generalized mechanisms of the autophagic pathway.
Autophagy can be induced through various pathways. AMP-activated protein kinase (AMPK) and mTOR regulate several autophagy-related pathways. Punicalagin upregulated AMPK phosphorylation in HeLa cells (Fig. S4). This is consistent with the results of a previous study, which reported that punicalagin upregulates the levels of AMPK [20].. However, punicalagin did not affect the levels of mTOR and p-mTOR (Fig 4B), which must be explored in future studies.
Several studies demonstrate that autophagy is a host defense mechanism against pathogens, including HPV. HPVs are reported to counteract this protective system and exploit cellular autophagy to facilitate their replication, which significantly contributes to cervical cancer progression. Griffin et al. reported that the ability of type 16 HPV pseudovirion to infect primary human keratinocytes is limited when compared with its ability to infect established keratinocyte cell lines. This may be attributed to the decreased levels of basal autophagy in established keratinocyte cells lines. The knock down of autophagy related genes or treatment with an autophagy inhibitor enhances the infectivity of virions [39, 40]. To establish a successful infection, HPV utilizes several strategies to inhibit host autophagy. E6 and E7 may inhibit autophagy by modulating the Akt/mTOR pathway [41, 42]. Conversely, the depletion of E6 and E7 induces autophagy in a cervical carcinoma model. This suggested that autophagy and HPV are inversely correlated. Cervical carcinogenesis results from compromised autophagy and successful HPV invasion. Recently, the clinical significance of autophagy in cervical cancer has piqued the interest of the scientific community. The downregulation of BECN1 and LC3-II is associated with poor survival in patients with cervical cancer [43], [44], [45], [46], [47]. Autophagy can function as a tumor suppressor mechanism by inhibiting tumor initiation, proliferation, invasion, and development, especially at the early stages of tumorigenesis [48], [49], [50]. However, at the late stage of tumor progression, autophagy can promote the pathogenesis of tumor cells, as well as tumor growth, invasion, and metastasis [51], [52], [53], [54], [55]. Several autophagy inhibitors, including chloroquine, 3-MA, bafilomycin A1, LY294002, SB202190, and SB203580, exert growth-inhibitory effects against tumors. These inhibitors mainly target the nucleation and extension of the phagophore or inhibit the endosomal-lysosomal acidification process. Additionally, various natural compounds have been developed to inhibit or activate autophagy. Natural compounds, such as artemisinin, dihydroartemisinin, curcumin, and resveratrol are reported as autophagy by modulating different pathways or proteins related to autophagy [56], [57], [58], [59]. The findings of this study demonstrated that punicalagin efficiently induced autophagy-mediated degradation of E6 and E7, and inhibited cervical cancer both in vitro and in vivo. In addition, induction of tumor cell autophagy and apoptosis, as well as increased infiltration of inflammatory cells in tumors were also observed under punicalagin treatment. It may be exploited to develop novel therapeutic strategies for cervical cancer.
In conclusion, this study provided novel insight into the mechanism underlying the punicalagin-induced autophagic degradation of E6 and E7. Punicalagin promoted autophagy in cervical cancer by activating the ROS-JNK-BCL2 pathway and promoted the release of BECN1 from BCL2. The growth-inhibitory effects of punicalagin against cervical cancer were verified both in vitro and in vivo. Therefore, these findings will enable the development of a novel therapeutic strategy for patients with HPV-infected cervical cancer.
Authors’ Contributions
Designed the experiments: XLX, CFL, HAA, and SDM; Performed the experiments: XLX, LYH, LLL, and JRW; Analyzed the data: XLX and CFL; Prepared the manuscript: XLX, CFL, and SDM. All authors discussed the results and critically commented on the manuscript.
Ethical statement
The mouse studies were performed according to the regulations of the Institute of Microbiology, Chinese Academy of Sciences of Research Ethics Committee. The study protocol was approved by the Research Ethics Committee (permit number PZIMCAS2011001). All animal experiments were performed according to the Institutional Guidelines on the Handling of Laboratory Animals.
Funding information
This work was supported by a grant from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29040000), One Belt and One Road International Science and Technology Cooperation of the Chinese Academy of Sciences (153211KYSB20170001), the Industrial Innovation Team grant from Foshan Industrial Technology Research Institute, Chinese Academy of Sciences, and grants from the National Natural Science Foundation of China (32070163, 81761128002, 81871297).
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgment
We are grateful to Professor Cuihua Liu (Key Laboratory of Pathogenic Microbiology and Immunology, Chinese Institute of Microbiology, Chinese Academy of Sciences) for providing the pSpCas9 (BB)-2A-GFP plasmid (Key Laboratory of Pathogenic Microbiology and Immunology, Chinese Institute of Microbiology, Chinese Academy of Sciences) for their technical assistance with flow cytometry and confocal microscopy. Additionally, we would like to thank Zhongshuang Lv, Xueke Tan, and Xixia Li (Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences) for their technical assistance with sample preparation and image acquisition for electron microscopy. We would like to thank Editage (www.editage.com) for English language editing. This work was supported by a grant from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29040000), One Belt and One Road International Science and Technology Cooperation of the Chinese Academy of Sciences (153211KYSB20170001), the Industrial Innovation Team grant from Foshan Industrial Technology Research Institute, Chinese Academy of Sciences, and grants from the National Natural Science Foundation of China (32070163, 81761128002, 81871297).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101388.
Contributor Information
Changfei Li, Email: lichangfei2006@163.com.
Haji Akber Aisa, Email: haji@ms.xjb.ac.cn.
Songdong Meng, Email: mengsd@im.ac.cn.
Appendix. Supplementary materials
References
- 1.de Sanjose S., Quint W.G.V., Alemany L., Geraets D.T., Klaustermeier J.E., Lloveras B., Tous S., Felix A., Bravo L.E., Shin H.-R., Vallejos C.S., de Ruiz P.A., Lima M.A., Guimera N., Clavero O., Alejo M., Llombart-Bosch A., Cheng-Yang C., Tatti S.A., Kasamatsu E., Iljazovic E., Odida M., Prado R., Seoud M., Grce M., Usubutun A., Jain A., Suarez G.A.H., Lombardi L.E., Banjo A., Menéndez C., Domingo E.J., Velasco J., Nessa A., Chichareon S.C.B., Qiao Y.L., Lerma E., Garland S.M., Sasagawa T., Ferrera A., Hammouda D., Mariani L., Pelayo A., Steiner I., Oliva E., Meijer C.J.L.M., Al-Jassar W.F., Cruz E., Wright T.C., Puras A., Llave C.L., Tzardi M., Agorastos T., Garcia-Barriola V., Clavel C., Ordi J., Andújar M., Castellsagué X., Sánchez G.I., Nowakowski A.M., Bornstein J., Muñoz N., Bosch F.X. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. The Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 2.Hoppe-Seyler K., Bossler F., Braun J.A., Herrmann A.L., Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26:158–168. doi: 10.1016/j.tim.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 3.van den Heuvel S., Dyson N.J. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 4.Morgan E.L., Wasson C.W., Hanson L., Kealy D., Pentland I., McGuire V., Scarpini C., Coleman N., Arthur J.S.C., Parish J.L., Roberts S., Macdonald A. STAT3 activation by E6 is essential for the differentiation-dependent HPV18 life cycle. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall A.H., Alexander K.A. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J. Virol. 2003;77:6066–6069. doi: 10.1128/JVI.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez J., Poirson J., Foltz C., Chebaro Y., Schrapp M., Meyer A., Bonetta A., Forster A., Jacob Y., Masson M., Deryckere F., Trave G. Targeting the two oncogenic functional sites of the HPV E6 oncoprotein with a high-affinity bivalent ligand. Angew. Chem. Int. Ed Engl. 2015;54:7958–7962. doi: 10.1002/anie.201502646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sterlinko Grm H., Weber M., Elston R., McIntosh P., Griffin H., Banks L., Doorbar J. Inhibition of E6-induced degradation of its cellular substrates by novel blocking peptides. J. Mol. Biol. 2004;335:971–985. doi: 10.1016/j.jmb.2003.10.079. [DOI] [PubMed] [Google Scholar]
- 8.Jing K., Shin S., Jeong S., Kim S., Song K.S., Park J.H., Heo J.Y., Seo K.S., Park S.K., Kweon G.R., Wu T., Park J.I., Lim K. Docosahexaenoic acid induces the degradation of HPV E6/E7 oncoproteins by activating the ubiquitin-proteasome system. Cell Death. Dis. 2014;5:e1524. doi: 10.1038/cddis.2014.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W., Che Q., Tan H., Qi X., Li J., Li D., Gu Q., Zhu T., Liu M. Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin-proteasome system. Sci. Rep. 2017;7:42180. doi: 10.1038/srep42180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulkarni A.P., Mahal H.S., Kapoor S., Aradhya S.M. In vitro studies on the binding, antioxidant, and cytotoxic actions of punicalagin. J. Agric. Food Chem. 2007;55:1491–1500. doi: 10.1021/jf0626720. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y., Chen J., Ren G., Zhang Y., Tan X., Yang L. Punicalagin prevents inflammation in LPS-induced RAW264.7 macrophages by inhibiting foxo3a/autophagy signaling pathway. Nutrients. 2019;11 doi: 10.3390/nu11112794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C., Cai D., Zhang L., Tang W., Yan R., Guo H., Chen X. Identification of hydrolyzable tannins (punicalagin, punicalin and geraniin) as novel inhibitors of hepatitis B virus covalently closed circular DNA. Antiviral Res. 2016;134:97–107. doi: 10.1016/j.antiviral.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larrosa M., Tomas-Barberan F.A., Espin J.C. The dietary hydrolysable tannin punicalagin releases ellagic acid that induces apoptosis in human colon adenocarcinoma Caco-2 cells by using the mitochondrial pathway. J. Nutr. Biochem. 2006;17:611–625. doi: 10.1016/j.jnutbio.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Berkoz M., Krosniak M. Punicalagin induces apoptosis in A549 cell line through mitochondria-mediated pathway. Gen. Physiol. Biophys. 2020;39:557–567. doi: 10.4149/gpb_2020024. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Chinnathambi A., Alharbi S.A., Veeraraghavan V.P., Mohan S.K., Zhang G. Punicalagin promotes the apoptosis in human cervical cancer (ME-180) cells through mitochondrial pathway and by inhibiting the NF-kB signaling pathway. Saudi J. Biol. Sci. 2020;27:1100–1106. doi: 10.1016/j.sjbs.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J., Li B., Hong S., Liu C., Min J., Hu M., Li Y., Liu Y., Hong L. Punicalagin suppresses the proliferation and invasion of cervical cancer cells through inhibition of the beta-catenin pathway. Mol. Med. Rep. 2017;16:1439–1444. doi: 10.3892/mmr.2017.6687. [DOI] [PubMed] [Google Scholar]
- 17.Pan L., Duan Y., Ma F., Lou L. Punicalagin inhibits the viability, migration, invasion, and EMT by regulating GOLPH3 in breast cancer cells. J. Recept. Signal Transduct. Res. 2020;40:173–180. doi: 10.1080/10799893.2020.1719152. [DOI] [PubMed] [Google Scholar]
- 18.Tang J.M., Min J., Li B.S., Hong S.S., Liu C., Hu M., Li Y., Yang J., Hong L. Therapeutic effects of punicalagin against ovarian carcinoma cells in association with beta-catenin signaling inhibition. Int. J. Gynecol. Cancer. 2016;26:1557–1563. doi: 10.1097/IGC.0000000000000805. [DOI] [PubMed] [Google Scholar]
- 19.Li J., Wang G., Hou C., Li J., Luo Y., Li B. Punicalagin and ellagic acid from pomegranate peel induce apoptosis and inhibits proliferation in human HepG2 hepatoma cells through targeting mitochondria. Food Agric. Immunol. 2019;30:897–912. [Google Scholar]
- 20.Wang S.G., Huang M.H., Li J.H., Lai F.I., Lee H.M., Hsu Y.N. Punicalagin induces apoptotic and autophagic cell death in human U87MG glioma cells. Acta Pharmacol. Sin. 2013;34:1411–1419. doi: 10.1038/aps.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X., Guo Y., Zhao J., He S., Wang Y., Lin Y., Wang N., Liu Q. Punicalagin, a PTP1B inhibitor, induces M2c phenotype polarization via up-regulation of HO-1 in murine macrophages. Free Radic. Biol. Med. 2017;110:408–420. doi: 10.1016/j.freeradbiomed.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Lee S.I., Kim B.S., Kim K.S., Lee S., Shin K.S., Lim J.S. Immune-suppressive activity of punicalagin via inhibition of NFAT activation. Biochem. Biophys. Res. Commun. 2008;371:799–803. doi: 10.1016/j.bbrc.2008.04.150. [DOI] [PubMed] [Google Scholar]
- 23.Li P., Du R., Chen Z., Wang Y., Zhan P., Liu X., Kang D., Chen Z., Zhao X., Wang L., Rong L., Cui Q. Punicalagin is a neuraminidase inhibitor of influenza viruses. J. Med. Virol. 2020 doi: 10.1002/jmv.26449. [DOI] [PubMed] [Google Scholar]
- 24.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. In search of an "autophagomometer". Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 25.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauvezin C., Neufeld T.P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang L., Wang H., Zhang J., Fang X. Punicalagin induces ROS-mediated apoptotic cell death through inhibiting STAT3 translocation in lung cancer A549 cells. J. Biochem. Mol. Toxicol. 2021:e22771. doi: 10.1002/jbt.22771. [DOI] [PubMed] [Google Scholar]
- 28.Li H.Y., Zhang J., Sun L.L., Li B.H., Gao H.L., Xie T., Zhang N., Ye Z.M. Celastrol induces apoptosis and autophagy via the ROS/JNK signaling pathway in human osteosarcoma cells: an in vitro and in vivo study. Cell Death. Dis. 2015;6:e1604. doi: 10.1038/cddis.2014.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poillet-Perez L., Despouy G., Delage-Mourroux R., Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox. Biol. 2015;4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo C., Yang M., Jing L., Wang J., Yu Y., Li Y., Duan J., Zhou X., Li Y., Sun Z. Amorphous silica nanoparticles trigger vascular endothelial cell injury through apoptosis and autophagy via reactive oxygen species-mediated MAPK/Bcl-2 and PI3K/Akt/mTOR signaling. Int. J. Nanomed. 2016;11:5257–5276. doi: 10.2147/IJN.S112030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X.L., Zhang X.W., Zhai H.J., Zhang D., Ma S.Y. Magnoflorine inhibits human gastric cancer progression by inducing autophagy, apoptosis and cell cycle arrest by JNK activation regulated by ROS. Biomed. Pharmacother. 2020;125 doi: 10.1016/j.biopha.2019.109118. [DOI] [PubMed] [Google Scholar]
- 32.Jung C.H., Ro S.H., Cao J., Otto N.M., Kim D.H. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vega-Rubin-de-Celis S. The role of Beclin 1-dependent autophagy in cancer. Biology (Basel) 2019;9 doi: 10.3390/biology9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain A., Lamark T., Sjottem E., Larsen K.B., Awuh J.A., Overvatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffin L.M., Cicchini L., Pyeon D. Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes. Virology. 2013;437:12–19. doi: 10.1016/j.virol.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishii Y. Electron microscopic visualization of autophagosomes induced by infection of human papillomavirus pseudovirions. Biochem. Biophys. Res. Commun. 2013;433:385–389. doi: 10.1016/j.bbrc.2013.02.130. [DOI] [PubMed] [Google Scholar]
- 41.Spangle J.M., Munger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J. Virol. 2010;84:9398–9407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menges C.W., Baglia L.A., Lapoint R., McCance D.J. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 2006;66:5555–5559. doi: 10.1158/0008-5472.CAN-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H.Y., Yang G.F., Huang Y.H., Huang Q.W., Gao J., Zhao X.D., Huang L.M., Chen H.L. Reduced expression of autophagy markers correlates with high-risk human papillomavirus infection in human cervical squamous cell carcinoma. Oncol. Lett. 2014;8:1492–1498. doi: 10.3892/ol.2014.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z.H., Xu L., Wang Y., Cao M.Q., Li L., Bai T. Clinicopathologic correlations between human papillomavirus 16 infection and Beclin 1 expression in human cervical cancer. Int. J. Gynecol. Pathol. 2011;30:400–406. doi: 10.1097/PGP.0b013e31820f874a. [DOI] [PubMed] [Google Scholar]
- 45.Zhu W., Pan X., Li F., Zhang Y., Lu X. Expression of Beclin 1 and LC3 in FIGO stage I-II cervical squamous cell carcinoma and relationship to survival. Tumour Biol. 2012;33:1653–1659. doi: 10.1007/s13277-012-0421-4. [DOI] [PubMed] [Google Scholar]
- 46.Hu Y.F., Lei X., Zhang H.Y., Ma J.W., Yang W.W., Chen M.L., Cui J., Zhao H. Expressions and clinical significance of autophagy-related markers Beclin1, LC3, and EGFR in human cervical squamous cell carcinoma. Oncol. Targets Ther. 2015;8:2243–2249. doi: 10.2147/OTT.S86844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng H.Y., Zhang Y.N., Wu Q.L., Sun X.M., Sun J.R., Huang X. Expression of beclin 1, an autophagy-related protein, in human cervical carcinoma and its clinical significance. Eur. J. Gynaecol. Oncol. 2012;33:15–20. [PubMed] [Google Scholar]
- 48.Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R.S., Karantza-Wadsworth V., White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnard R.A., Regan D.P., Hansen R.J., Maycotte P., Thorburn A., Gustafson D.L. Autophagy inhibition delays early but not late-stage metastatic disease. J. Pharmacol. Exp. Ther. 2016;358:282–293. doi: 10.1124/jpet.116.233908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., He S., Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J.M., Dell'antonio G., Mautner J., Tonon G., Haigis M., Shirihai O.S., Doglioni C., Bardeesy N., Kimmelman A.C. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y.F., Shi Y.H., Ding Z.B., Ke A.W., Gu C.Y., Hui B., Zhou J., Qiu S.J., Dai Z., Fan J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9:2056–2068. doi: 10.4161/auto.26398. [DOI] [PubMed] [Google Scholar]
- 53.Macintosh R.L., Timpson P., Thorburn J., Anderson K.I., Thorburn A., Ryan K.M. Inhibition of autophagy impairs tumor cell invasion in an organotypic model. Cell Cycle. 2012;11:2022–2029. doi: 10.4161/cc.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lock R., Kenific C.M., Leidal A.M., Salas E., Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–479. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White E. The role for autophagy in cancer. J. Clin. Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Efferth T. From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin. Cancer Biol. 2017;46:65–83. doi: 10.1016/j.semcancer.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Hu W., Chen S.S., Zhang J.L., Lou X.E., Zhou H.J. Dihydroartemisinin induces autophagy by suppressing NF-kappaB activation. Cancer Lett. 2014;343:239–248. doi: 10.1016/j.canlet.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J., Wang J., Xu J., Lu Y., Jiang J., Wang L., Shen H.M., Xia D. Curcumin targets the TFEB-lysosome pathway for induction of autophagy. Oncotarget. 2016;7:75659–75671. doi: 10.18632/oncotarget.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Y., Chang H., Peng X., Bai Q., Yi L., Zhou Y., Zhu J., Mi M. Resveratrol inhibits breast cancer stem-like cells and induces autophagy via suppressing Wnt/beta-catenin signaling pathway. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.