Abstract
Combination therapies are superior to monotherapy for many cancers. This advantage was historically ascribed to the ability of combinations to address tumor heterogeneity, but synergistic interaction is now a common explanation as well as a design criterion for new combinations. We review evidence that independent drug action, described in 1961, explains the efficacy of many practice-changing combination therapies: it provides populations of patients with heterogeneous drug sensitivities multiple chances of benefit from at least one drug. Understanding response heterogeneity could reveal predictive or pharmacodynamic biomarkers for more precise use of existing drugs and realize the benefits of additivity or synergy.
Significance:
The model of independent drug action represents an effective means to predict the magnitude of benefit likely to be observed in new clinical trials for combination therapies. The “bet-hedging” strategy implicit in independent action suggests that individual patients often benefit from only a subset—sometimes one—of the drugs in a combination. Personalized, targeted combination therapy, consisting of agents likely to be active in a particular patient, will increase, perhaps substantially, the magnitude of therapeutic benefit. Precision approaches of this type will require a better understanding of variability in drug response and new biomarkers, which will entail preclinical research on diverse panels of cancer models rather than studying drug synergy in unusually sensitive models.
Introduction
The introduction of targeted anticancer drugs and the use of these drugs in various combinations have broadly and substantially improved rates and durability of response to therapy; like blood cancers, many advanced solid cancers are now treated with multidrug combinations (in addition to surgery and radiation where applicable; refs. 1–3). For optimal use of existing combination therapies and discovery of new ones, it is important to understand precisely why combining some but not all drugs is successful. Contemporary clinical trial reports make few if any claims about mechanism of action, and most scientific understanding of combination cancer therapy derives from preclinical studies (in cell culture or mouse models)—often performed after a drug or combination is evaluated clinically. The theoretical rationale for combination therapy—as historically understood—derives from the heterogeneity of cancer (4). The explanation more commonly advanced today is drug synergy (1), and screening for synergy is the focus of many ongoing research programs (5–17). However, whereas pharmacologic synergy is well defined in the case of preclinical experiments, particularly in cell lines, it has no precise definition in the context of survival data collected in cancer clinical trials.
In this review, we discuss historical and contemporary perspectives on combination cancer therapy, particularly the hypothesis that combinations can be highly effective in patient populations in the absence of drug interaction (either additivity or synergy). The underlying mechanism in this case is independent drug action. We review evidence from trials run by the founders of combination therapy, including the Acute Leukemia Group B (ALGB) and its chairs Emil Frei III and James Holland (18), that led to this hypothesis. We then review contemporary evidence collected from solid tumors that provides further support for this independent action in the context of targeted therapies and solid tumors. We close by describing an alternative framework for thinking about combination therapies and discuss its impact on future trials and preclinical mechanism-of-action studies.
This perspective does not call into question the results of any published randomized clinical trial (RCT), and it is not intended to guide clinical practice. It therefore differs from systematic reviews and meta-analyses (19, 20), the primary formats in which multiple clinical trials are compared retrospectively. The goal of a systematic review or meta-analyses is to guide clinical practice using scientific evidence, whereas this perspective primarily aims to inform the design of future trials and preclinical studies.
Combination Therapy and Cancer Heterogeneity
The proteins targeted by anticancer drugs are commonly part of multicomponent mitogenic networks and drug resistance pathways (acquired, adaptive, and innate; refs. 21–23). By analogy with synthetic lethality in genetic screens (24), the existence of parallel and converging molecular mechanisms in oncogenic and drug resistance networks provides a molecular rationale for the use of combination therapy (25–27). These arguments also have historical antecedents in cytotoxic chemotherapies that target multiple metabolic pathways (28). Identifying examples of synergy has now become an explicit goal for preclinical development of combination therapies (5, 6, 29–32). Clinically successful combination therapies are, in turn, often described as arising from synergy and explained in terms of activity against overlapping response and resistance networks (33–35).
Intra- and intertumor heterogeneity is evident at the level of genetics, histology, and disease progression, and single-cell sequencing has revealed rapid and diverse cancer genome evolution in response to therapy (36, 37). Moreover, heterogeneity and evolvability, via genetic or nongenetic mechanisms, are two of the greatest obstacles to the successful treatment of cancers. Variability in drug response is observed in clinical trials of even highly successful precision therapies that target mutant or amplified oncogenes (the drugs that are ideal Ehrlich “magic bullets”; ref. 38). For example, fewer than half of biomarker-positive patients respond initially to trastuzumab (Herceptin) in HER2-overexpressing breast cancers or to MEK/BRAF inhibitors in BRAFV600E/K-mutant melanomas (39, 40). However, such responses can be of sufficient magnitude in the patients in whom they occur to make trastuzumab and MEK/BRAF inhibitors practice-changing (41, 42). Studies in genetically homogeneous mouse models of cancer suggest that, were all cancers biologically similar (and tumor evolution relatively limited), a single therapeutic vulnerability might be sufficient for tumor eradication. However, heterogeneity among cells within a single human cancer commonly promotes drug resistance and disease progression, and heterogeneity between cancers limits clinically meaningful responses (with rare exception) to a subset of patients. The molecular origins of variability could include patient-to-patient (or tumor-to-tumor) differences in cancer cell genetics, drug pharmacokinetics, or patient immune function (43). Among these possibilities, patient-specific differences in pharmacokinetics remain the least studied or understood.
Since some of the earliest preclinical (mouse model; ref. 44) and clinical (45) studies of chemotherapies, combination therapy has been understood as a way of addressing intra- and intertumor heterogeneity, whatever its origins. Whereas a single drug might not be effective in killing every cancer cell in a heterogeneous tumor, drug combinations have the potential to kill different subsets of cells, improving the likelihood and durability of response (46). Indeed, this logic applies to antibacterial and antiviral drug combinations (47–50), and the ability of combination therapy to cure tuberculosis inspired early studies on combination cancer therapy (51). The same reasoning also applies to interpatient heterogeneity: any single therapy will not be effective in every patient, but combination therapies provide patients with several opportunities for a clinically meaningful response.
Here, we focus on the role that interpatient heterogeneity has on the effectiveness of combination therapies. We revisit historical theories in light of contemporary clinical trial data and a molecular understanding of cancer and discuss the implications for the modern concept of precision medicine. In particular, we review evidence that many combination therapies used for solid tumors, including all combination therapies with immune checkpoint inhibitors reported up to early 2020 (52), provide a benefit to a patient population equal to that expected from independent drug action. In this case, different patients benefit from the independent activities of different drugs (exhibiting “highest single-agent” response), without enhanced drug activity from the other constituents of the combination (either additive or synergistic pharmacologic interaction). Because patients exhibit differences in response, some patients benefit from one drug and others from a different drug, increasing response rates in the population as a whole. We describe how such a benefit can be quantified and discuss how independent drug action can be used to predict the likely benefit of new combinations (53). We also discuss why independent action is not sufficient to explain curative regimens for lymphoma, leukemia, and germ-cell tumors; in these cases, drugs exhibit additivity (54). We close by discussing how increasing the precision of cancer therapy may allow us to realize the substantial benefits of pharmacologic interactions.
Historical Clinical Evidence for Bet-Hedging by Combinations of Cancer Therapies
Sidney Farber's 1948 report that aminopterin induced temporary remissions in a fraction of children with acute lymphocytic leukemia (ALL; ref. 55) was the first demonstration of a successful cancer chemotherapy. Achieving any degree of cancer control in these patients was remarkable at the time, but two limitations were immediately evident, each of which arose from heterogeneity in drug response. First, only a subset of patients with disease responded to therapy. Second, among responders, remissions were temporary, with a duration of around three months, consistent with the hypothesis that some cancer cells survived therapy and caused recurrent disease. Fortunately, childhood ALL was subsequently found to be responsive to multiple chemotherapies having different mechanisms of action and lacking cross-resistance, which allowed the ALGB [today Cancer and Leukemia Group B (CALGB); ref. 18] to develop an increasingly effective series of multidrug regimens.
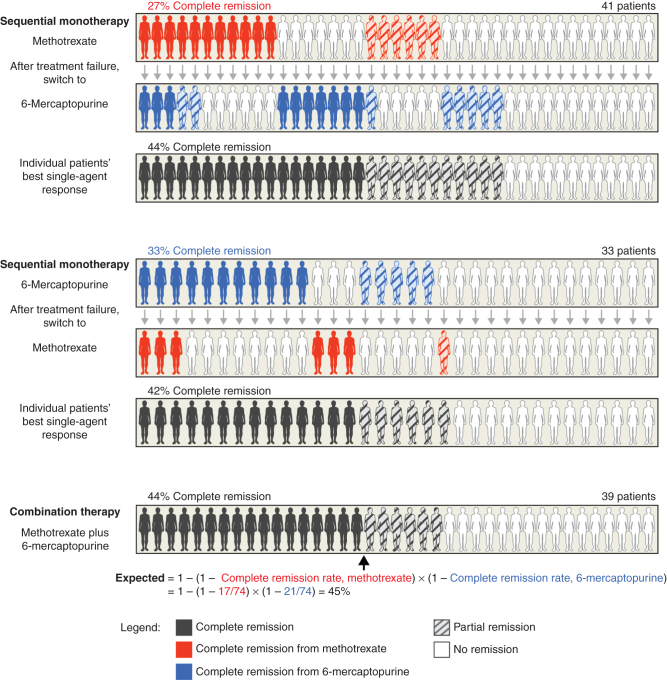
In a 1961 clinical trial on ALL by the ALGB, Frei and colleagues (45) compared sequential monotherapy with combination therapy and identified pharmacologic principles that remain relevant today. ALGB “Protocol 2” randomized patients to either 6-mercaptopurine (6MP) combined with methotrexate (MTX) or sequential monotherapy with the same agents—switching from one agent to the other when the first failed. Sequential treatment made it possible to compare each of 74 individual patient responses to the first therapy (6MP or MTX) with the responses of the same patients to the second (MTX or 6MP, respectively; Fig. 1). It was found that responses of patients (partial or complete remission) to the first and second stages of therapy were not correlated, and remission rates were not significantly different between the first and second phases of treatment. Frei and colleagues (45) concluded that MTX and 6MP exhibited neither “cross-resistance” nor “collateral sensitivity.”
Figure 1.
The earliest clinical trials of combination cancer therapy found that therapeutic benefit was due to independent drug action. Clinical trials of drug combinations for ALL repeatedly observed that a model of independent drug action could accurately explain the superior remission rates achieved by drug combinations (45, 56). In this example from 1961, trial arms evaluating sequential monotherapy found that MTX and 6MP were not cross-resistant (no correlation in response), and that the proportion of patients experiencing a complete remission was identical whether the drugs were given sequentially or concurrently in a combination. Furthermore, the complete remission rate of the combination therapy was consistent with that expected from independent drug action.
In addition, no difference in survival was observed among the sequential and combination therapy arms (42% or 44% complete remission with sequential therapy as compared with 44% with simultaneous combination therapy). Thus, patients treated with combination therapy effectively had two unbiased chances of responding to one of the two agents (Fig. 1). The probability of responding to either of two agents present in a combination individually having probabilities PA and PB was calculated by Frei and colleagues as:
 |
That is, the probability of responding to the combination, PAB is the sum of the probability of response from drug A (PA) and probability of response from drug B in patients who did not respond to drug A ((1 – PA) × PB). This calculation was found to be accurate to within 1% of observed response rates, which excluded the possibility that therapies each became more effective when used in combination (that is, that PA, PB became larger, as expected for pharmacologic interaction; ref. 45). A scenario in which individual drug efficacies (PA, PB) do not change when combined was described by Frei and colleagues (45) as independent drug action (Box 1), and was subsequently observed for multiple drug pairs for ALL (56).
The ALGB's insights into independent action in ALL came from a comparison of sequential monotherapy with combination therapy, but such trials are uncommon in solid cancers. A notable exception is the 2003 trial (E1193; ref. 57) by the Eastern Cooperative Oncology Group (ECOG), which randomized patients with metastatic breast cancer to either doxorubicin plus paclitaxel, or sequential monotherapy with the same agents. Patients treated with the combination experienced a higher rate of response (complete or partial reduction in lesion size; ref. 58) than those treated with a single drug, but no differences in overall survival were observed between arms in which the drugs were given sequentially or simultaneously. Echoing Emil Frei's conclusion 40 years prior, Sledge and colleagues interpreted E1193 as showing that different patients respond best to different drugs and the “composite response rate” of two therapies given sequentially (calculated as 49%) approximated that of the simultaneous combination (47%; ref. 57). As a result of this trial, sequential mono-therapy remains the internationally recognized approach to chemotherapy for metastatic breast cancer. Sequential therapy can, in principle, deliver the same therapeutic benefit as a combination with less toxicity and higher quality of life, provided that patients are able to receive the second therapy and the therapies are not cross-resistant (59–61). Sequential therapy is not appropriate when there is an urgent need for tumor control or for cancers where progressing patients may be too ill for a second line: use of up-front combination therapy allows the benefits of multiple drugs to be realized immediately (59).
What general lessons emerge from these studies? First, trials with drugs administered sequentially clearly show that different patients respond best to different drugs. Second, because of patient-to-patient variability, combinations of individually active therapies can increase the number of positive responses overall simply by providing several opportunities for benefit from monotherapies. Because it was not possible then, nor is it possible now, to predict which drug will be superior at the level of an individual patient, using combination therapy provides a therapeutic advantage in the form of “bet-hedging.” Third, when it is observed in a clinical trial that a population of patients responds better to combination therapy A + B as compared with monotherapies A or B given individually, it is not necessarily true that any individual patient will experience a superior response to the two drugs given together. As discussed below, data from a limited number of sequential human trials as well as patient-derived xenograft (PDX) studies involving panels of mice carrying PDXs make it possible to compare drug responses at the level of individual patients. These data confirm that drug combinations can improve outcomes in solid tumors without any pharmacologic interaction. Thus, the clinical superiority of a combination therapy does not demonstrate (or require) synergistic or additive interaction among the constituent drugs.
Assessing the Role of Synergy, Additivity, and Independent Action in the Lab and the Clinic
Drug “additivity” and “synergy” have rigorous definitions for preclinical experiments used to inform—or at least rationalize—which combinations proceed to clinical trials (Box 1; ref. 62). Pharmacologic interaction among two drugs was evaluated using isobologram analysis as early as 1872 (63) and defined mathematically in 1928 (64). As described by Loewe, two (equipotent) drugs are additive if combining a half-dose of each drug is as effective as a full dose of one drug; the drugs are synergistic if the effect is greater than additivity (this is often quantified by the combination index). Synergy is therefore assessed against a null hypothesis of additivity. In “Bliss Independence,” the expected effect of noninteracting drugs or toxins (the null model) is computed by assuming that each drug has a statistically independent chance of killing a target cell or organism; for example, if each of two drugs kills 90% of cancer cells alone, Bliss Independence predicts that the combination will kill 99% of cancer cells (that is, 10% of 10% survive). Synergy is demonstrated when this level of killing is exceeded. For a more thorough discussion of drug interactions in preclinical studies, see reviews by Foucquier and Guedj (65) and Twarog and colleagues (66) as well as extensions to pharmacodynamic (67) and toxicologic (68) interactions. Meyer and colleagues (69) have recently proposed an elegant reconciliation of different definitions of additivity and synergy.
Drug additivity, synergy, and antagonism are evaluated using measures of potency (e.g., IC50) and efficacy (e.g., fractional cell kill) from dose–response measurements that are common only in preclinical settings (Box 1). As yet, no rigorous definition exists for drug synergy using survival or other data commonly obtained from patients in oncology trials. This reflects the fact that sufficiently resolved dose–response data are rarely available from human trials. It is also noteworthy that assessing pharmacologic interaction using laboratory data is more complex than the definitions in Box 1 imply, in part due to experimental noise and in part because dose–response data vary in parameters other than IC50 (e.g., maximum effect and slope of the dose–response curve; ref. 70).
Box 1: Definitions of synergy, additivity, and independent action by combination therapies.
“Bliss Independence” is a null model for efficacy based on the statistically independent probability of a drug-induced death; if a combination exceeds the expected level of cytotoxicity, the drugs are judged to be synergistic.
“Loewe Additivity” is a null model for potency based on a principle of “dose equivalence.” For example, drugs are additive if combining half the IC50 of drug A and half the IC50 of drug B also achieves a 50% effect; this principle applies to any threshold of effect, e.g., IC90. A drug combination is synergistic if it is more potent than the null model of additivity.
Independent drug action—“Frei Independence”—is a null model for progression-free survival (PFS) in clinical trials. It assumes that PFS for each patient is equal to the longer of the two possible PFS times conferred by one or the other drug (see Box 2 for details). Combinations whose activity equals that expected from independent action are neither additive nor synergistic: the effect is the “highest single agent” per patient. Because of interpatient variability in drug response, combination therapy can confer substantial benefit, relative to monotherapy, via independent drug action and “bet-hedging.”
Regardless of which null model is used, a drug combination exhibiting synergy allows the same level of efficacy to be achieved at lower drug doses (or superior efficacy to be achieved with the same doses) as compared with an additive combination, and is therefore desirable (2). In principle, an efficacious drug combination exhibiting strong synergy could be achieved using individual agents that are inactive individually. Retrospective analysis has shown, however, that high single-agent activity is usually associated with good activity in a combination (71). Moreover, if two drugs are each highly effective, a combination can be weaker than additive and still provide a substantial level of benefit (Fig. 2A). This is particularly true when patients vary in their responses to the agents individually. This observation motivates a definition of drug interaction first explored by Gaddum in 1940, developed by Frei, and now known as “independent drug action” (or “Frei Independence” to distinguish it from Bliss Independence; Box 1).
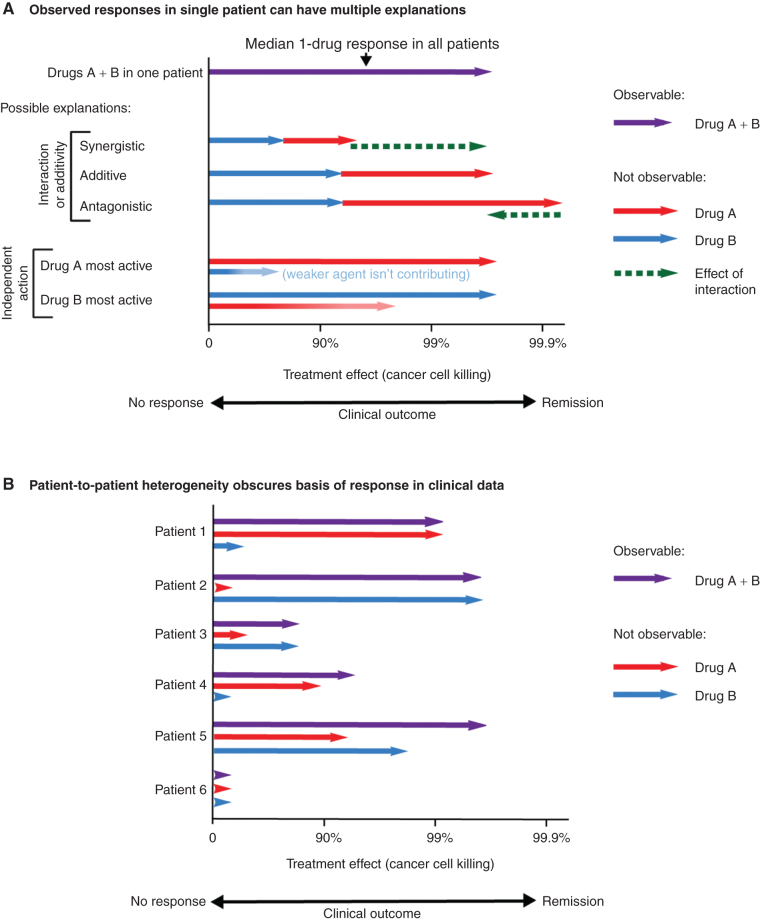
Figure 2.
The basis for combinatorial efficacy is difficult to discern in clinical trial data because of patient heterogeneity in single-drug response. A, Clinical benefit from combination therapy can be mediated both by high single-agent efficacy (enabling many log-kills of tumor cells) and by combined effects that could be affected by positive or negative pharmacologic interactions (additive, synergistic, or antagonistic). The observation that a combination response in a single patient is superior to the population median single-drug response could therefore have multiple possible explanations. B, Patient responses to a single therapy are heterogeneous across a population. Variation in drug response among patients makes it challenging to understand the precise nature of drug interaction across a population. It is therefore necessary to formulate appropriate null models and then determine whether the data in aggregate exceed the predictions of the null model. Note that this scenario is purely illustrative because current technology does not make it possible to observe single-drug effects in individual patients who are treated with combination therapy.
Independent drug action is the benefit provided by a combination when the effect is equal to the stronger of the effects of the two drugs considered individually (72). In this case, neither additivity nor synergy is present at the level of individual tumor (or patient in the case of survival data), and responses are scored as though the less effective agent is neither present nor active (Fig. 2A). The key insight is that the most effective single agent typically varies across patients, and thus drug independence can be sufficient for a combination to provide substantial clinical benefit: some patients benefit from one drug and others from a different drug in the combination (because we do not know this information a priori, this is a bet-hedging approach). The level of efficacy observed for a drug combination exhibiting independent action is therefore less than the null model for either Loewe additivity or Bliss Independence. Independent action is of interest only when response is measured in a heterogeneous population of samples, as in a human clinical trial or a panel of PDXs (73, 74).
Drug synergy evaluated by either Loewe or Bliss criteria is most commonly assessed in cell lines and sometimes in animal models. However, translating simple mathematical formulations of additivity and synergy into reliable experimental methodologies to detect these mechanisms is not straightforward even in cell lines, and a series of papers dating back to at least 1977 (65) has raised concerns about the widespread misuse of experiments and definitions associated with this terminology. All too often, “synergy” is used as a substitute for “more active” in cell line studies, and there is reason to approach the concept with caution even in preclinical studies (75).
Measuring Loewe additivity or Bliss Independence in patients is generally impossible because dose–response measurements comparing monotherapy and combination therapy are required (76). Were it possible to dose a population of patients with varying levels of a drug (as in a phase I trial, for example), differences in pharmacokinetics would mean that the effective drug concentration in each patient's tumor would be unknown. In a population of patients, interpreting responses to combination therapy is even more challenging, as each patient is likely to experience different magnitudes of antitumor efficacy from different therapies (Fig. 2A and B). Thus, any single patient's response to a combination therapy will have competing explanations in terms of best single-agent response, additivity, or synergy (Fig. 2A).
Independent Action Explains the Clinical Activity of Many Drug Combinations
In contrast to a Loewe or Bliss null, evaluating the null model of independent drug action is straightforward using data from RCTs. When this model is exceeded, either additivity or synergy is present, and these cannot currently be distinguished. Estimating the magnitude of independent action in the precomputing era of 1960s oncology, as undertaken by Frei and others (56), involved rates of response but did not consider time-series survival data or correlation (ρ) in the probabilities of response [Box 2; Eq. (B)]; such correlations can arise from shared mechanism, cross-resistance, or prognostic factors. Today, independent action is evaluated using a simple algorithm whose inputs are the clinically observed distributions of responses to each of two treatments given individually, commonly progression-free survival (PFS) from Kaplan–Meier survival curves and an estimate of correlation in response [Box 2; Eqs. (C) and (D)]. Computing this null model is most conveniently implemented as the computational procedure shown in Supplementary Fig. S1. This procedure provides another way of thinking about independence: for each of a series of simulated patients, a response to therapy A and to therapy B is chosen at random from a joint distribution of PFS values, constructed from PA(t), PB(t), and the correlation in response ρ. Then, the better of the two responses for each simulated patient to either drug A or B is selected under the assumption that the less effective drug does not enhance response to the more effective drug. Because empirical survival data are used to compute the magnitude of independent action, the affect of prognostic factors, tumor heterogeneity, and time-dependent treatment effects is accounted for to the same extent as in monotherapy data from the original phase III trials.
The null model for independent drug action is applicable to a population of patients, or potentially to animal models (e.g., panels of diverse PDX tumors; refs. 73, 74), when individuals differ in response. If all individuals had the same drug sensitivities, independent drug action would predict no benefit relative to monotherapy. Benefit from independent action also requires that drugs be active as monotherapies, at least in some patients. A complication in determining if a particular drug combination exhibits benefit consistent with or exceeding drug independence is that efficacy data must be available for the constituent monotherapies at the same dosage, ideally from the same trial or one with closely matched patient characteristics.
Box 2: The independent drug action model.
-
In the simple case in which responses to drugs A and B are not correlated, the expected PFS at time t from A + B combination therapy is calculated similarly to Eq. (A):

Eq. (B) shows that the probability of tumor control, or PFS, from the combination (PAB(t)) is the sum of the probability of tumor control by drug A (PA(t)) and the probability of tumor control by drug B if drug A fails (PB(t) × (1 – PA(t))). Here, “independence” means that the activity of drug A (PA(t)) does not change the activity of drug B (PB(t)) or vice versa.
-
Drug responses are often partially correlated (partially cross-resistant; ref. 74). To account for this, Eq. (B) is extended with a correlation coefficient ρ:

where A is the more effective of the two drugs. When ρ = 0 (no cross-resistance), Eqs. (B) and (C) are the same, and the benefit of independence is maximized. When ρ = 1, the drugs are completely cross-resistant, and the less active drug provides no additional benefit.
-
Chen and colleagues (53) recently derived a version of this equation that is symmetric for drugs A and B:

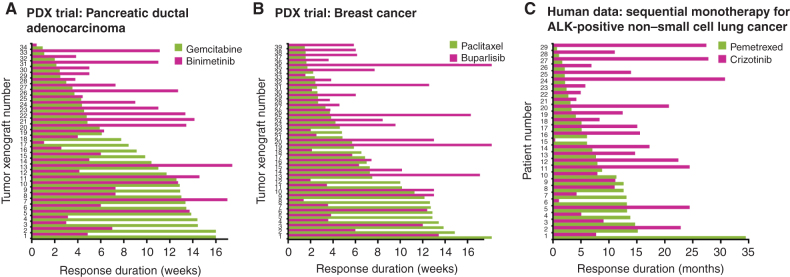
The correlation in drug responses [ρ in Eqs. (C) and (D)] is the only parameter in these calculations. Clinical trials of sequential therapy and studies with PDX panels provide an opportunity to estimate this value. For example, Fig. 3A and B show data from a study by Gao and colleagues (73) of approximately 1,000 PDXs from 277 patients and six different types of tumors exposed to one of 62 different monotherapies or combination therapies (73, 74). For each type of therapy, 30 to 40 different human tumors were propagated in mice and exposed to drugs individually or in combination, and response duration was measured. The data show that only a subset of tumors responds strongly to a single drug and even fewer respond strongly to both drugs. However, tumors responding poorly to one drug (e.g., the green bars at the top of each graph) frequently exhibited strong responses to the other drug (depicted in magenta), so that animals receiving both drugs are predicted to benefit from a combination simply because they have two chances at a substantial monotherapy response; this is the essence of independent action. Figure 3C shows that the same reasoning can be applied to human patients receiving sequential therapy with pemetrexed and crizotinib for ALK-positive non–small cell lung cancer (77). Correlation values used to compute the magnitude of independent action can be estimated from these data and vary from Spearman ρ of 0.1 to 0.5 for mechanistically dissimilar drugs (e.g., cytotoxic agents vs. oncogene inhibitors), up to ≈ 0.7 for drugs with related targets (e.g., inhibitors of signaling kinases). Data from sequential human clinical trials are consistent with low correlations between dissimilar drugs, and ranges for benefits attributable to independent action have been found to lie within the range of ρ = 0.3 ± 0.2 (52, 74). Because responses are correlated to this degree, as opposed to truly independent in a statistical sense, the term “independent action” refers to pharmacologic, not statistical, independence (72).
Figure 3.
Different cancer drugs benefit different patients in a population. Responses to different single drugs can be measured in the same tumor (as duration of PFS) through PDXs (A and B; ref. 73) or human patients (C) treated with sequential monotherapy (77). None of these drug pairs have a statistically significant correlation in response duration (as determined by a Spearman rank test). Patients and xenografts are sorted based on response to the single agent depicted in green.
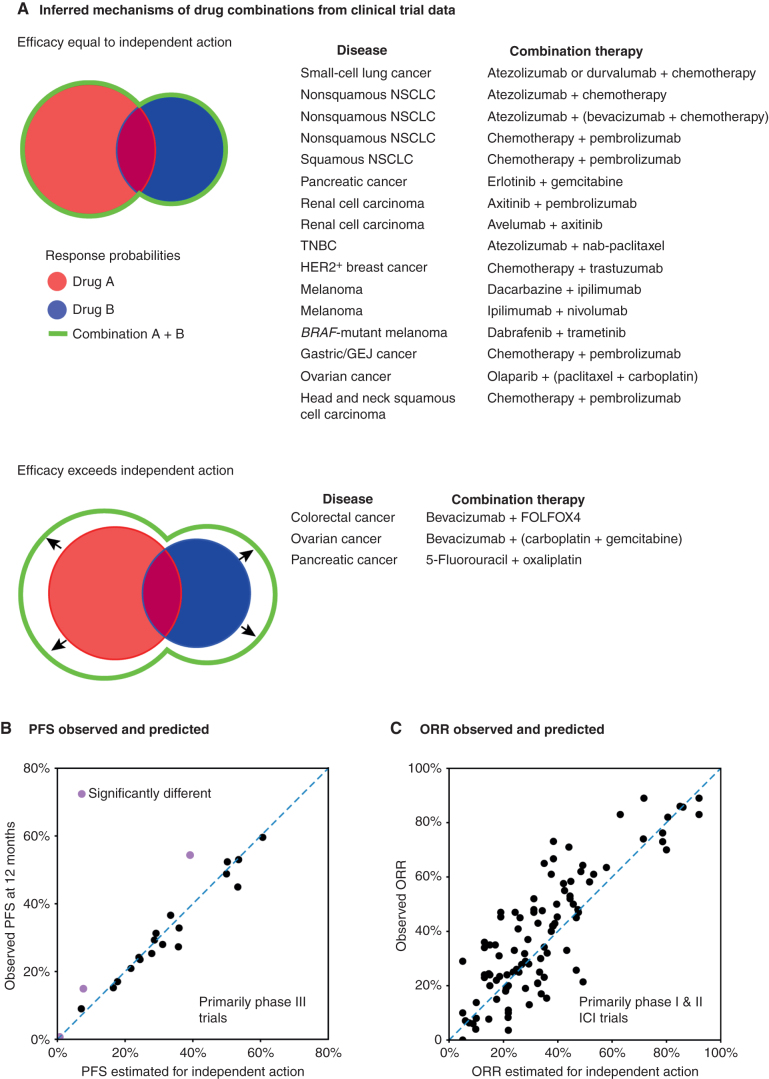
It is currently possible to evaluate the magnitude of independent action using PFS data for a total of 21 published phase III clinical trial combination results (the great majority of which led to FDA approvals), including 13 recent trials involving immune checkpoint inhibitors (ICI), which in aggregate involve 11 types of advanced solid cancers. The key requirement for such an evaluation is the availability of sufficient data on responsiveness to monotherapies. Roughly three quarters of approved combinations analyzed, including all combinations involving ICIs, exhibit clinical activity very close to that predicted by independent action (Fig. 4A and B; Pearson r = 0.98, P < 10–8, n = 21 comparisons; data from 13,689 patients in 38 clinical trials; refs. 52, 74). These conclusions are supported in biomarker-stratified and nonstratified RCTs across a variety of advanced cancers (cancers of the lung, breast, skin, head and neck, ovary, pancreas, stomach, and kidney) and treatment modalities (including chemotherapies, molecularly targeted therapies, and ICIs).
Figure 4.
Independent drug action explains the survival benefit of many drug combinations. A, Combination therapies with efficacy equal to or greater than independent action simulations, based on analysis of clinical trial PFS data (52, 74). Parentheses denote combinations tested as a single arm of a clinical trial. GEJ, gastroesophageal junction; NSCLC, non–small cell lung cancer; TNBC, triple-negative breast cancer. B, Observed combination PFS at 12 months (mostly from phase III clinical trials) correlates with 12-month PFS estimated by independent action simulations (n = 21 combinations). Note that differences from the independent action predictions (black or purple color) were calculated using longitudinal data over the total trial length. C, Observed combination ORR (mostly from phase I and II trials) correlates with ORR estimated by independent action (n = 100 combinations; ref. 78).
Similar approaches have recently been used to estimate the magnitude of independent action using overall response rate (ORR) data from 98 early trials (primarily phase I and II) performed by Merck & Co. on combination therapies involving ICIs (78, 79). The Merck group found that independent drug action was also sufficient to explain response rates observed for the majority of drug combinations in this data set (Fig. 4C; Pearson r = 0.84, P < 10–26, n = 100 comparisons). Unfortunately, for some standard-of-care combination therapies, it is not currently possible to assess the magnitude of either drug independence or synergy: most commonly, the efficacy of at least one drug has not been measured in the appropriate patient population at the appropriate dose for a formal comparison with the combination. It is also unfortunate that there exist relatively few phase III trials involving multiple targeted agents for which preclinical research has provided molecular hypotheses about expected mechanistic interactions. However, the fact that the independent action model can only rarely be rejected when analysis of suitable RCTs is possible implies that it is not reasonable to presume synergy whenever data are lacking.
The most effective drug combinations in oncology have historically been those that combine active single agents (71), particularly those having nonoverlapping mechanisms of drug resistance (28, 80). In these cases, calculation of independence provides a quantitively accurate explanation for disease-specific differences in observed activity. For example, in advanced non–small cell lung cancer, ICIs and chemotherapy (81, 82) are active as single agents and are superior in a combination. Similarly, ICIs and multitargeted receptor tyrosine kinase inhibitors are effective in advanced renal cancer, and a combination of the two is superior to either one alone (83, 84). Benefit from independent drug action is possible only when both agents in a combination are active. The magnitude of this benefit falls as cross-resistance (correlation) between drugs increases (Box 2). In this regard, it is important to note that “no single-agent activity” has two different meanings in the clinical setting. The first meaning is that an agent truly has no measurable ability to shrink tumors or delay progression, and the second is that a drug has some antitumor activity, but it is insufficient for approval as a single agent (typically this means the new drug is inferior to standard of care). Very few approved combination therapies contain drugs in the first category, whereas many successful combination therapies contain a drug in the second category, which is expected according to independent drug action. Ambiguity about the meaning of “no single-agent activity” has contributed to the erroneous perception that synergy is commonly observed in clinical trial data.
Conversely, among trials analyzed to date, independent action is most commonly exceeded when at least one drug is relatively inactive on its own; this is the case in the metastatic setting for combinations involving bevacizumab, which targets VEGF and appears to enhance the effects of chemotherapies by affecting tumor vasculature (ref. 85; Fig. 4).
Two notable cases of coinhibiting one pathway exhibit synergistic (supra-additive) interaction, because at least one agent is devoid of single-agent activity in the given disease but improves the activity of a combination. The first is fluorouracil plus leucovorin (86), which is approved in many indications, and the second is EGFR plus BRAF inhibition for BRAF-mutant colorectal cancer (ref. 87; these effects are not quantified in Fig. 4 due to lack of monotherapy data). These examples demonstrate that synergy does not ensure high response rates, and analysis of 18 approved combinations involving one agent with limited activity (~40% being VEGF inhibitors; ref. 71) has found that median overall survival increased only by ~1.6 months.
Whether coinhibition of one pathway or even a single target (e.g., a small-molecule and biological drug against the same protein) is more likely to result in supra-additive efficacy than drugs inhibiting multiple pathways is not yet known. Regardless, synergy is not guaranteed: combinations of antibacterial drugs targeting a single protein complex, the ribosome, have been shown to exhibit synergy, no interaction (independence), and even antagonism (88). Pathway-specific models are needed to understand such effects, and additional mechanistic analysis could help us understand what principles distinguish independence from interaction in these scenarios. Higher-order drug combinations can also provide benefit using a mixture of effects, including independent action against different targets and pharmacologic interaction for one target or pathway. For example, the triplet combination of trastuzumab, pertuzumab, and docetaxel (two HER2 antibodies and a microtubule inhibitor) may confer benefit by both “bet-hedging” and pharmacologic interaction (74). Whether interacting or independent, it is nevertheless the case that combinations of inactive agents rarely succeed in the clinic, which is cause for caution in the development of agents that lack single-agent antitumor effect.
Some combination therapies that have been tested in RCTs exhibit activity less than predicted by independence (52). This could arise because drugs are strongly cross-resistant, such that patients nonresponsive to therapy A almost never benefit from therapy B. Alternatively, the drugs could have a strong antagonistic interaction, which can render a combination less active than monotherapy (89). A third possibility is that the combination induces adverse side effects that either worsen patient survival directly or require dose reductions or interruptions. In the latter case, the use of lower doses of each drug results in a weaker antitumor effect than full-dose monotherapy. This appears to explain the underperformance of dabrafenib plus trametinib plus pembrolizumab in the treatment of metastatic BRAF-mutant melanoma; the management of adverse effects necessitated dose reduction or interruption in nearly all patients (90, 91).
In summary, the available evidence provides strong support for independent action as the mechanism underlying the majority of approved combination therapies for advanced cancers as well as many combinations in current development. “Bet-hedging” is therefore an effective strategy for increasing ORRs in a heterogeneous population of patients, precisely as the ALGB first observed in 1961 (45). Merck investigators have recently suggested that the ability of independent action to estimate the outcomes of future combination trials will be particularly valuable for ICIs (78), because more than 3,000 ICI trials are currently under way, and historical experience suggests that many will fail (based on data in ClinicalTrials.gov).
The Role of Drug Additivity in Curative Combination Therapies
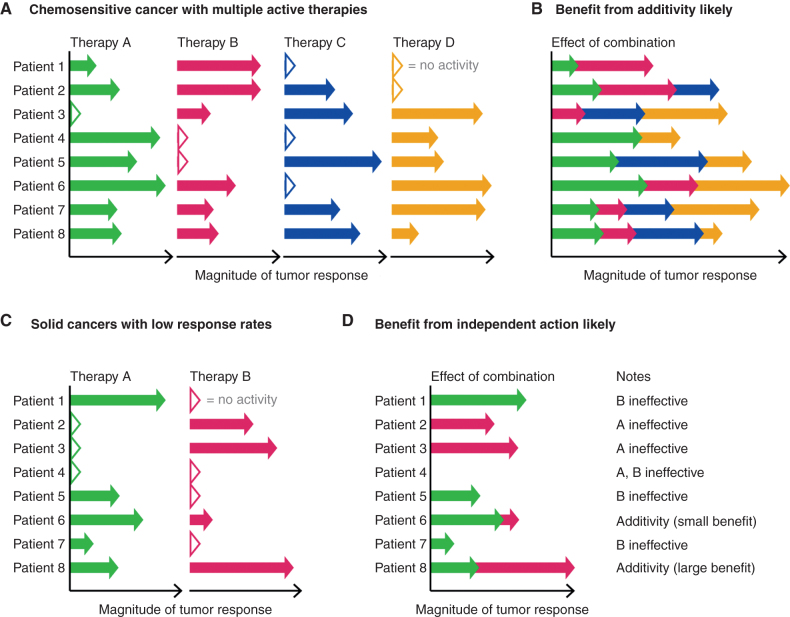
In multiple hematologic malignancies, it is possible to administer combination therapies that achieve long-lasting responses and even cures. Independent drug action is inadequate as an explanation for such therapies. The classic “fractional kill hypothesis,” as well as recent preclinical experiments (54), suggests that drug additivity but not synergy is involved in cures. For example, the five-drug R-CHOP regimen cures most patients with diffuse large B-cell lymphoma (DLBCL) and is strictly additive (or even slightly antagonistic) in cell lines by both Loewe and Bliss criteria, as demonstrated by clone-tracing experiments that measure a 106-fold range of tumor cell killing (54); a similar conclusion is supported by murine models of B-cell lymphoma (25). The mechanistic basis for curative therapies for advanced cancers, which remain largely restricted to chemosensitive liquid tumors and a small number of solid tumors, involves a high degree of cell killing through the use of multiple drugs, each potent on its own in the majority of patients and having low cross-resistance. Intratumor heterogeneity and tumor evolution are a still a challenge in these therapies, and it appears that many tumors are initially resistant to one or more drugs that make up the combination (Fig. 5A). Resistance is also acquired in the course of therapy. However, when cross-resistance is low, the likelihood that a single resistance mechanism will cause multiple drugs to be ineffective initially is correspondingly low. When the components of a combination are each highly effective (e.g., >99% tumor cell killing each), additivity results in many-orders-of-magnitude improvements in fractional cell kill (Fig. 5B). Hematologic oncologists have long been taught (4) that drug additivity of this type is responsible for the efficacy of curative combination therapies, and recent evidence supports this interpretation (54). The fractional kill of individual therapies (illustrated as arrow length in Fig. 5) will depend on the chemosensitivity of a given cancer type, and may explain why some curative regimens involve five to eight therapies, whereas germ-cell tumors can often be cured with fewer therapies [e.g., the bleomycin, etoposide, and cisplatin (BEP) combination for testicular cancer; ref. 92].
Figure 5.
Clinical context affects the prevalence of benefit from drug additivity or from bet-hedging via independent action. A, In chemosensitive cancers treated with high-order combinations (e.g., DLBCL), multiple agents are typically active per patient, although to varying extents. B, Additive interactions among many highly active therapies are sufficient for strong tumor response and cure. C, In many solid tumors, single therapies have low response rates and heterogeneous effects. Low single-drug response rates imply that few patients are highly responsive to multiple therapies. D, Bet-hedging confers substantial advantage in solid tumors without the need for drug interaction. Additivity or synergy may arise in some patients but are statistically expected to be uncommon, or small in effect because one therapy is usually the principal contributor to tumor response (e.g., patient 6).
Sequential, Concurrent, and Biomarker-Guided Combination Therapy to Address Interpatient Heterogeneity in Advanced Cancers
Many advanced solid malignancies are more difficult to treat than blood cancers, and response rates to single drugs are typically lower than in hematologic malignancies. Low response rates alone are a reason to expect that drug additivity will be less often observed in solid tumors, because few patients will experience strong responses to multiple agents (Fig. 5C). These are the scenarios in which independent drug action is expected to provide the greatest benefit relative to monotherapy: even if additivity is possible with these drugs, it would be limited to too few “doubly responsive” patients to substantially affect survival in a trial arm (Fig. 5D).
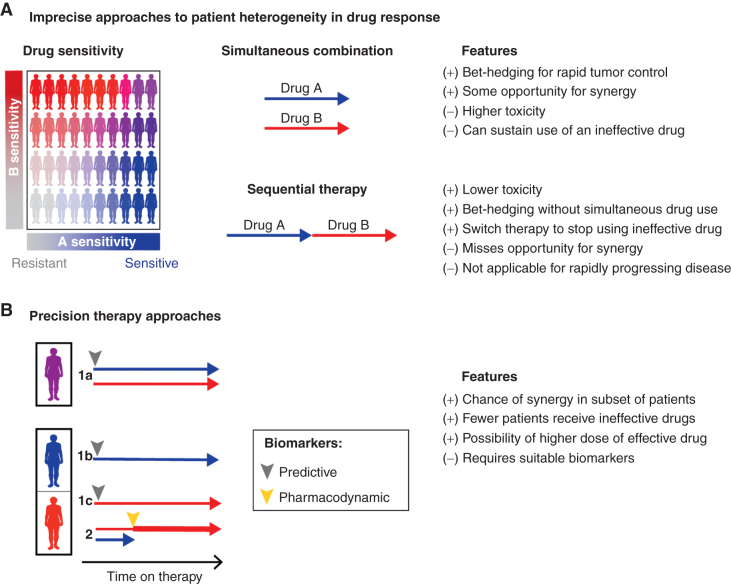
There are two main therapeutic strategies for exploiting independently acting combinations in the clinic (Fig. 6A): (i) concurrent combination therapy (commonly used for new drug approvals, for example, Checkmate 067 for ipilimumab and nivolumab in advanced melanoma, ref. 93; and the combinations shown in Fig. 4B and C), and (ii) sequential administration of different single agents, switching on a predetermined schedule or on evidence of disease progression (e.g., ECOG 1193 showing benefit of sequential chemotherapy in metastatic breast cancer, ref. 57; a strategy described in detail below). Because benefit consistent with independence assumes that each patient benefits from the best monotherapy, the efficacy conferred by concurrent or sequentially administered combinations could in principle be matched by a third strategy: treating each patient with the therapies most likely to be active in that specific patient (Fig. 6B). This strategy could use biomarkers predictive of therapeutic response, such that combination therapy A + B is restricted to patients predicted to benefit from both drugs (standard therapy A alone would be assigned to patients whose biomarker status predicts no benefit from B, as in KEYNOTE-355 with pembrolizumab and chemotherapy combined for PD-L1–positive advanced triple-negative breast cancer; ref. 94). A variant of this approach is to start all patients on A + B and then use pharmacodynamic biomarkers to measure response at a molecular level; the inactive agent could then be withdrawn and the dose of the active agent increased or a new drug introduced (Fig. 6B). These approaches have a critical limitation: we currently lack the biomarkers to make accurate treatment assignments at the level of individual patients. A corollary of independent action is that, were response biomarkers available, benefit could be increased using existing drugs. On the basis of data from PDX panels in which drug additivity or synergy is occasionally demonstrable (73, 95), the magnitude of improvement relative to independent action could be dramatic and might even extend to cures (as in Fig. 5B).
Figure 6.
Possible strategies for combining multiple cancer drugs. Patient heterogeneity in drug response presents a fundamental challenge in designing combination cancer therapies. A, Existing, imprecise strategies for use of multiple drugs include first-line combination therapies for all patients and sequential therapy. B, Precision-based approaches can use pretreatment predictive biomarkers to select one or more agents likely to be active in a given patient (patients labeled 1a, 1b, and 1c), or use on-treatment pharmacodynamic biomarkers (patient 2) to identify which agents are active or inactive in a patient to stop use of an inactive agent and increase dose of an active agent (particularly if combination therapy required dose reduction). Here, treatment “arrows” are to compare strategies and are not intended to evaluate response duration.
Is Identifying the Mechanisms Underlying Successful Combination Therapy Important?
The distinction between independent action and additivity or synergy is not simply semantic and has profound implications for improving patient care with existing drug combinations and for developing new and improved combination therapies. In the immediate term, clinical research on drug combinations would benefit from better predicting the efficacy expected in new clinical trials by simulating the likely benefits conferred by independent action (53, 79). In the longer term, the focus of preclinical and translational cancer pharmacology needs to include understanding, measuring, and predicting variability in drug response and in developing better response biomarkers, including pharmacodynamic biomarkers (96, 97).
The Benefits Conferred by New Drug Combinations Are Largely Predictable
One of the great advantages of independent drug action is that it provides a simple methodology for predicting the likely benefits of a drug combination based on knowledge of responses to monotherapy in similar patient populations. Critically, accurate prediction does not require making any assumptions about targets or mechanisms of action. Criteria for combinations likely to provide substantial benefit via independent action include drugs with high single-agent activity and uncorrelated responses (correlation is lowest when mechanisms of action differ). As discussed below, this does not mean that drugs can be combined at random. One substantial limitation with respect to the design of new combinations is that we cannot currently predict treatment-related adverse events from drugs individually or in combination; we do not view this as a limitation of the model of independent action per se, but as a reflection of generally poor understanding of drug toxicity. With further empirical data, adverse effects could be scored for independence, additivity, synergy, or antagonism, and their relationships to therapeutic effects could be ascertained. This may be particularly important in the case of immune-related adverse events (irAE), which appear to be closely related in mechanism to the therapeutic effects of ICIs (98).
Sequential Therapy May Be as Good as a Combination in Some Cancers
Intensive combination therapy is vital to most treatments with curative intent, and coadministration is necessary for drug synergy to manifest (Fig. 6). However, among noncurative combination therapies exhibiting independent action, drugs need not be given at the same time to benefit patients. This implies that combinations in current clinical practice could be modified to involve sequential monotherapy, as currently practiced for metastatic breast cancer, where patients switch to a different chemotherapy when the first one fails (60). This approach has the potential to reduce drug toxicities while delivering equivalent duration of tumor control (and possibly better control if dose escalation of the monotherapy is possible; ref. 59). Of course, sequential therapy is possible only when a disease does not progress so rapidly that it is necessary to maximize the probability of response up front. Sequential therapy requires that patients are healthy enough to switch therapy upon progression. First-line combination therapy will also remain optimal for cancer types, or specific patients, where there is a sizable risk that patients will be unable to receive a second line of therapy, for example, in many cases of advanced non–small cell lung cancer (99).
Prioritize Rather Than Penalize Single-Agent Efficacy in a Preclinical Setting
One of the practical problems in identifying drug combinations involving synergy by either Loewe Additivity or Bliss Independence criteria in preclinical studies is that the greatest deviation from additivity occurs over a relatively narrow range of drug doses typically centered on the midpoint (IC50) of a sigmoidal dose–response curve. At these concentrations, target coverage is incomplete, and responses are suboptimal by definition. In contrast, the goal with many oncology drugs, particularly biologics, is to achieve a dose that results in substantially more than 50% of target engagement (100). Screens for synergistic drug combinations therefore penalize highly active drugs and the dose ranges at which target coverage is optimal. Furthermore, drug combinations with meager activity can score as “synergistic” if the constituent drugs are inactive alone (12) and therefore much less likely to be useful clinically (71). In contrast, efficacy via independent action (as well as the additivity required for successful curative therapy) benefits from maximal tolerable dosing and high single-agent activities—the hallmarks of most clinically useful combinations. Matching preclinical to clinical drug concentration ranges has also been shown to facilitate accurate prediction of combination drug efficacy from in vitro data (101). Preclinical studies should therefore shift to studying drugs at dose ranges that provide target coverage similar to that achieved in patients (we note that these might not be the same drug concentrations in vivo and in vitro due to pharmacokinetic effects). Under these conditions, additivity should be considered a very promising finding and could be sufficient for the design of curative therapies; for example, clinical and experimental data show that rituximab is additive with the CHOP regimen for DLBCL (54).
Deprioritize Screening for Synergy and Emphasize Screening to Avoid Cross-Resistance
Data from curative drug combinations suggest that preclinical studies in cell lines should also shift away from searching for synergy to screening for single-agent activity, drug additivity, and identification of drugs having nonoverlapping resistance mechanisms. As mentioned above, pharmacologic interaction is scored near drug IC50 values, but the primary obstacle to cure in most settings is thought to be acquired drug resistance caused by outgrowth of rare resistant cells. Systematic analysis of cross-resistance is challenging using conventional cell culture techniques, but clone tracing and genome-wide CRISPR screening make it feasible to empirically measure cross-resistance for any combination therapy. We propose that screening for drug cross-resistance be performed alongside screens for collateral drug sensitivities (102). Resistance screens are best performed one drug at a time, with cross-resistance identified in a subsequent computational comparison, making it possible to build systematic resources relevant to many therapeutic approaches (54, 103).
Common Misconceptions and Limitations in the Analysis of Combination Therapy
Given the long history of the drug independence model (45), we have been surprised by resistance to the idea that it likely explains the benefits conferred by many approved combination therapies. We believe that this resistance arises from several misunderstandings. Most importantly, in saying that a particular drug combination does not involve synergy (or pharmacologic interaction more generally) we are not saying that the combination is not clinically superior to monotherapy. Independent action can explain practice-changing improvements in patient outcomes without the need to invoke any specific (and possibly poorly understood) molecular mechanism. In contrast, concluding that drugs interact synergistically is a statement about a specific mechanism of action; it is not a measure of the quality or importance of a combination therapy. As noted above, primary articles that report clinical trial results rarely make specific claims about drug interactions. These claims typically arise in reviews and preclinical studies. By equating clinical benefit with synergistic drug interaction, these claims make it difficult to determine how outcomes might best be improved in the clinic and where preclinical studies are likely to be most informative.
Another common misconception is that the model of independent action implies that any two (or more) drugs can be combined to improve outcomes. This is not true. Instead, benefit by independent action occurs only when both drugs are individually sufficiently active and their responses weakly correlated. In an analysis of all possible drug pairs in PDX data from Gao and colleagues (73), fewer than 5% of combinations were predicted to improve PFS when compared with the best observed monotherapy for that tumor type (74). This low rate of success is consistent with evidence that drug combinations superior to standard monotherapies are challenging to identify and develop clinically.
The model of independent drug action, like that of Loewe Additivity or Bliss Independence, represents a null model against which evidence of additivity or synergy is scored. If the null model cannot be rejected by an appropriate statistical test, then it is considered the most likely explanation. By analogy, if we cannot reject the null model that survival is no greater in the presence of therapy than a placebo, as evaluated by Cox proportional hazards regression, we conclude that a therapy provides no benefit (104). However, this is not the same as saying that drugs in a combination cannot interact. On the contrary, there is ample evidence from preclinical models and clinical samples that cells exposed to multiple therapies adopt states distinct from monotherapy-treated cells (29, 105). An additive or synergistic response to therapy might also be attributable to different tumor cell subpopulations responding to different drugs in a combination (106). However, if survival distributions are consistent with independent action, then any pharmacologic interactions that do occur must not measurably extend response duration across a patient cohort.
The simplest way to reconcile preclinical and clinical data on drug interactions is to note that variability of response in patients is so large that it is statistically unlikely that a patient will experience a similar magnitude of response to two agents, and one agent therefore has the dominant effect (Figs. 5 and 6). Large-scale preclinical experiments (107, 108) have shown that synergy at the level of cytostasis or cytotoxicity is relatively uncommon when drugs are combined and tested across panels of cell lines, and in many cases, additivity is not observed either (101). Moreover, computational methods attempting to predict synergy in cancer cells have found that the magnitude of drug interaction varies across cell line identity (17). Finally, it is not true that most approved combination therapies have been demonstrated to exhibit better-than-additive efficacy in cell lines or animal models using Bliss or Loewe criteria (109).
Moving Beyond Independent Drug Action: Replacing Bet-Hedging with Precision
Patient heterogeneity in drug response is a fundamental challenge in oncology that has often been addressed with concurrent or sequential use of multiple therapies to maximize chance of response (bet-hedging; Fig. 6A). Because only a small minority of solid tumors (10%–15% in PDX studies or in sequential human trials) actually respond to both drugs, it is uncommon for favorable pharmacologic interaction by drug combinations to have a significant impact. Were it routinely achievable, positive pharmacologic interaction between drugs in a combination (additivity or synergy) by definition provides a magnitude of benefit that exceeds independent action. How might this be achieved? The key is likely to be greater precision in the use of drugs individually and in combination (Fig. 6B). However, combination therapies generally cannot be used in a personalized manner, because for many individual therapies, biomarkers are presently not available to identify responsive patients. To generate such biomarkers, we need to measure and then understand determinants of drug response at the level of individual patients, including the possibility that differences in response involve patient-to-patient variability in pharmacokinetics and optimal dosing, which are not areas of current emphasis in publicly funded translational cancer research. At a molecular level, the development of sensitive methods for transcriptional profiling (110, 111) and multiplexed imaging (112–114) of biopsies, along with increasing acceptance of sequential biopsies as a means to monitor patients, may provide an opportunity to develop sensitive new pharmacodynamic assays. Patients could potentially be started on a combination followed by use of a biopsy to see if their tumor is responding at a molecular level to both drugs; if not, then the inactive drug might be withdrawn (in advance of radiologic evidence of tumor progression) and a new combination tried, or the active agent dose escalated. In the intermediate term, drug responses might be measured in patient-derived primary tumor cells using BH3 profiling (115), microfluidic technologies (116), or image-based assays (117), or assessed in situ using implantable drug-delivery microdevices (118). Some of these approaches are already being used to guide therapy in programs such as the SMMART trial initiative at Oregon Health and Science University (Portland, OR; ref. 119).
Improving rates of response outside of academic medical centers, where multiple biopsies are unlikely to be feasible, will ultimately require diagnostics predictive of drug response (or lack of resistance) based on genetic, histologic, and transcriptional features of tumors pretreatment (95, 120–124). The I-PREDICT trial recently demonstrated proof of concept for personalized combination therapies in advanced metastatic cancers using pretreatment genomic and histologic profiling (125), and such approaches should improve with advances in biomarker discovery. In cases in which an evaluation is possible (e.g., BRAF-mutant melanomas), existing biomarkers significantly improve average response, as expected, but they do not appear to reduce variability in response as measured by the coefficient of variation in PFS distributions (74). This is most easily understood as reflecting the presence of multiple additional and unknown drug response determinants. Identifying these unknown determinants must become a priority for preclinical and translational research, even in the case of successful therapies. Conversely, it will also be important to understand why a small minority of univariate biomarkers are so effective at predicting some drug responses, for example, NTRK fusions and sensitivity to entrectinib and larotrectinib in multiple cancer types (126).
Prioritizing Heterogeneity in the Preclinical Setting
Molecular oncology has been well served by a focus in preclinical studies on outliers in response that reveal the presence of genes and mutations conferring high drug sensitivity or strong acquired resistance (127, 128). However, the resulting emphasis on drug-sensitive cell lines and murine models is the likely cause of consistent overestimation of drug efficacy (whether of individual agents or combinations) in the preclinical setting as compared with observed benefit in human trials. We do not yet fully understand the molecular origins of diversity in drug response even in panels of cancer lines carrying a known driving oncogene, such as BRAFV600E in cutaneous melanoma or EGFRL858R in non–small cell lung cancer; therefore, it is not surprising that we do not understand the phenomenon in human patients. The problem becomes more acute as the models get more complex; PDX panels such as those used by Gao and colleagues (73) show promise, but in-depth molecular analysis is required as a complement to what is most commonly a phenomenologic study of tumor volume. In the case of preclinical study of immunotherapies, genetically engineered mice and syngeneic models have been valuable, but the current focus on a relatively small number of responsive models should be complemented by a mechanistic study of variability in response in a set of models having different drug sensitivities. Relatively simple experiments have the potential to be informative in the context of a panel of heterogenous preclinical models. We might then ask: if a panel of cell lines or tumor explants (129) is exposed to a drug over a concentration range, is target coverage similar in all cases (as measured using a pharmacodynamic assay)? If the target is equally covered (e.g., inhibited) in multiple lines, are the phenotypic consequences the same (130, 131)? Drug–response studies using large panels of cell lines and PDX explants have become increasingly common (70, 129, 132, 133), and what is now needed is a molecular understanding of observed differences in response. Bringing the full power of preclinical cancer biology to bear on these questions, first in cell lines and then in PDX studies, would almost certainly yield information useful in the more challenging problem of understanding variability in drug response in patients. In humans, variability in drug pharmacokinetics also needs more investigation.
Making the Data from Clinical Trials Accessible to Promote Mechanistic Studies
The analysis of clinical trial data reviewed here, and any assessment of pharmacologic mechanism in clinical trials, would greatly benefit from access to the individual participant data (IPD) used to create the Kaplan–Meier curves and compute survival functions. Analysis of IPD is widely regarded as the gold standard for formal meta-analysis (134, 135), and the International Committee of Medical Journal Editors (ICMJE) in fact required a data-sharing statement from papers reporting clinical trials effective July 1, 2018; as of April 2020, only two of 487 relevant articles in the Journal of the American Medical Association, the Lancet, and the New England Journal of Medicine had actually made IPD publicly accessible (136). Figures showing Kaplan–Meier estimators are universally part of oncology clinical trial reports, and the absence of numerical data has little to do with patient privacy or intellectual property and more with the continuing view of computational biologists as data parasites (137). As a result, much of the analysis described above comes from papers in which image processing was used to extract data from published figures (52, 74). One positive development is the depiction of individual patients’ tumor responses in “waterfall” and “swimmer” plots in phase I and II trials (138). Such data are rarely reported in phase III trials, but their publication could provide rich insights into interpatient heterogeneity. It may nonetheless be necessary to amend the requirements for data deposition on ClinicalTrials.gov (per U.S. Public Law 110-85: Food and Drug Administration Amendments Act of 2007, Title VIII, Section 801) so that trial reporting includes IPD and not just summary statistics. Journals should also enforce the ICMJE standards that already exist.
Conclusion
The (re)realization that many approved and effective combination therapies perform in a manner consistent with independent drug action and therefore represent a necessary and beneficial form of bet-hedging, in the face of continued ignorance about many of the determinants of drug response and resistance, should be viewed as a positive development. Not only does it provide a realistic way to design and predict the probable benefit conferred by new drug combinations, it suggests that a renewed focus on precision medicine will yield very substantial benefits because it will unlock the potential for pharmacologic interaction. In the immediate future, assays that reduce the number of ineffective therapies received by each patient could also reduce toxicity and cost without compromising effective tumor control. Moreover, data from combinations used in liquid tumors suggest that additivity among active agents (not necessarily synergy) will be sufficient to elicit the degree of curative control over solid cancers currently possible in some hematologic malignancies.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grant U54-CA225088. D. Plana is also supported by NIGMS grant T32-GM007753 and NCI grant F30-CA260780. We thank Emmett Schmidt and David Weinstock for discussions.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
A.C. Palmer reports personal fees from Merck and grants from Prelude Therapeutics outside the submitted work. P.K. Sorger reports grants from NCI during the conduct of this work, as well as other support from Glencoe Inc. and personal fees from Applied Biomath, RareCyte, NanoString, Flagship Pioneering, and Merck outside the submitted work. No disclosures were reported by the other authors.
References
- 1. Mokhtari RB, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das Bet al. Combination therapy in combating cancer. Oncotarget 2017;8:38022–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cavalli F. Textbook of medical oncology. Boca Raton, FL: CRC Press; 2010. [Google Scholar]
- 3. Frei E, Eder JP. Principles of dose, schedule, and combination therapy. In: Kufe DW, Pollock RE, Weichselbaum RR, editors. Holland-Frei Cancer Medicine. 6th ed. Ontario, Canada: BC Decker; 2003. [Google Scholar]
- 4. Bast RC, Hait WN, Kufe DW, Weichselbaum RR, Holland JF, Croce CMet al. , editors. Holland-Frei Cancer Medicine [Internet]. 9th ed. Hoboken (NJ): Wiley; 2016. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9781119000822. [Google Scholar]
- 5. Menden MP, Wang D, Mason MJ, Szalai B, Bulusu KC, Guan Yet al. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat Commun 2019;10:2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang Het al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol 2014;32:1213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rationalizing combination therapies. Nat Med 2017;23:1113. [DOI] [PubMed] [Google Scholar]
- 8. Pemovska T, Bigenzahn JW, Superti-Furga G. Recent advances in combinatorial drug screening and synergy scoring. Curr Opin Pharmacol 2018;42:102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He L, Kulesskiy E, Saarela J, Turunen L, Wennerberg K, Aittokallio Tet al. Methods for high-throughput drug combination screening and synergy scoring. Methods Mol Biol 2018;1711:351–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol 2012;30:679–92. [DOI] [PubMed] [Google Scholar]
- 11. Chou T-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 12. Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 2017;35:463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lehár J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ERet al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol 2009;27:659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun X, Vilar S, Tatonetti NP. High-throughput methods for combinatorial drug discovery. Sci Transl Med 2013;5:205rv1. [DOI] [PubMed] [Google Scholar]
- 15. Sidorov P, Naulaerts S, Ariey-Bonnet J, Pasquier E, Ballester PJ. Predicting synergism of cancer drug combinations using NCI-ALMANAC data. Front Chem 2019;7:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma A, Rani R. An integrated framework for identification of effective and synergistic anti-cancer drug combinations. J Bioinform Comput Biol 2018;16:1850017. [DOI] [PubMed] [Google Scholar]
- 17. Julkunen H, Cichonska A, Gautam P, Szedmak S, Douat J, Pahikkala Tet al. Leveraging multi-way interactions for systematic prediction of pre-clinical drug combination effects. Nat Commun 2020;11:6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schilsky RL, McIntyre OR, Holland JF, Frei E III. A concise history of the cancer and leukemia group B. Clin Cancer Res 2006;12:3553s–5s. [DOI] [PubMed] [Google Scholar]
- 19. Haidich AB. Meta-analysis in medical research. Hippokratia 2010;14:29–37. [PMC free article] [PubMed] [Google Scholar]
- 20. Sutton AJ, editor. Methods for meta-analysis in medical research. Chichester, NY: Wiley; 2000. [Google Scholar]
- 21. Sun C, Wang L, Huang S, Heynen GJJE, Prahallad A, Robert Cet al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014;508:118–22. [DOI] [PubMed] [Google Scholar]
- 22. Bell CC, Gilan O. Principles and mechanisms of non-genetic resistance in cancer. Br J Cancer 2020;122:465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gerosa L, Chidley C, Fröhlich F, Sanchez G, Lim SK, Muhlich Jet al. Receptor-driven ERK pulses reconfigure MAPK signaling and enable persistence of drug-adapted BRAF-mutant melanoma cells. Cell Syst 2020;11:478–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat Rev Drug Discov 2020;19:23–38. [DOI] [PubMed] [Google Scholar]
- 25. Pritchard JR, Bruno PM, Gilbert LA, Capron KL, Lauffenburger DA, Hemann MT. Defining principles of combination drug mechanisms of action. Proc Natl Acad Sci U S A 2013;110:E170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao B, Pritchard JR, Lauffenburger DA, Hemann MT. Addressing genetic tumor heterogeneity through computationally predictive combination therapy. Cancer Discov 2014;4:166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao B, Hemann MT, Lauffenburger DA. Modeling tumor clonal evolution for drug combinations design. Trends Cancer 2016;2:144–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeVita VT, Schein PS. The use of drugs in combination for the treatment of cancer: rationale and results. N Engl J Med 1973;288:998–1006. [DOI] [PubMed] [Google Scholar]
- 29. Diaz JE, Ahsen ME, Schaffter T, Chen X, Realubit RB, Karan Cet al. The transcriptomic response of cells to a drug combination is more than the sum of the responses to the monotherapies. eLife 2020;9:e52707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuenzi BM, Park J, Fong SH, Sanchez KS, Lee J, Kreisberg JFet al. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell 2020;38:672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Preuer K, Lewis RPI, Hochreiter S, Bender A, Bulusu KC, Klambauer G. DeepSynergy: predicting anti-cancer drug synergy with Deep Learning. Bioinformatics 2018;34:1538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Narayan RS, Molenaar P, Teng J, Cornelissen FMG, Roelofs I, Menezes Ret al. A cancer drug atlas enables synergistic targeting of independent drug vulnerabilities. Nat Commun 2020;11:2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang RX, Wong HL, Xue HY, Eoh JY, Wu XY. Nanomedicine of synergistic drug combinations for cancer therapy – strategies and perspectives. J Control Release 2016;240:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nowak AK, Robinson BWS, Lake RA. Synergy between chemotherapy and immunotherapy in the treatment of established murine solid tumors. Cancer Res 2003;63:4490–6. [PubMed] [Google Scholar]
- 35. Fan W, Yung B, Huang P, Chen X. Nanotechnology for multimodal synergistic cancer therapy. Chem Rev 2017;117:13566–638. [DOI] [PubMed] [Google Scholar]
- 36. Kuipers J, Jahn K, Beerenwinkel N. Advances in understanding tumour evolution through single-cell sequencing. Biochim Biophys Acta Rev Cancer 2017;1867:127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DLet al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 2020;182:1232–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strebhardt K, Ullrich A. Paul Ehrlich's magic bullet concept: 100 years of progress. Nat Rev Cancer 2008;8:473–80. [DOI] [PubMed] [Google Scholar]
- 39. Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher Let al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002;20:719–26. [DOI] [PubMed] [Google Scholar]
- 40. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin Jet al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene 2007;26:3637–43. [DOI] [PubMed] [Google Scholar]
- 42. Zhu Z, Liu W, Gotlieb V. The rapidly evolving therapies for advanced melanoma—towards immunotherapy, molecular targeted therapy, and beyond. Crit Rev Oncol Hematol 2016;99:91–9. [DOI] [PubMed] [Google Scholar]
- 43. Lim Z-F, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol 2019;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Law LW. Effects of combinations of antileukemic agents on an acute lymphocytic leukemia of mice. Cancer Res 1952;12:871–8. [PubMed] [Google Scholar]
- 45. Frei E, Freireich EJ, Cehan E, Pinkel D, Holland JF, Selawry Oet al. Studies of sequential and combination antimetabolite therapy in acute leukemia: 6-mercaptopurine and methotrexate. Blood 1961;18:431–54. [Google Scholar]
- 46. Schmitt MW, Loeb LA, Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol 2016;13:335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Worthington RJ, Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 2013;31:177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Richman DD. HIV chemotherapy. Nature 2001;410:995–1001. [DOI] [PubMed] [Google Scholar]
- 49. Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev 2012;25:450–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kerantzas CA, Jacobs WR. Origins of combination therapy for tuberculosis: lessons for future antimicrobial development and application. mBio 2017;8:e01586–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Frei E, Holland JF, Schneiderman MA, Pinkel D, Selkirk G, Freireich EJet al. A comparative study of two regimens of combination chemotherapy in acute leukemia. Blood 1958;13:1126–48. [PubMed] [Google Scholar]
- 52. Palmer AC, Izar B, Hwangbo H, Sorger PK. Predictable clinical benefits without evidence of synergy in trials of combination therapies with immune-checkpoint inhibitors. Clin Cancer Res 2022;28:368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen C, Liu F, Ren Y, Suttner L, Sun Z, Shentu Yet al. Independent drug action and its statistical implications for development of combination therapies. Contemp Clin Trials 2020;98:106126. [DOI] [PubMed] [Google Scholar]
- 54. Palmer AC, Chidley C, Sorger PK. A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity. eLife 2019;8:e50036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Farber S, Diamond L, Mercer R, Sylvester R, Wolff R. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid (aminopterin). N Engl J Med 1948;238:787–93. [DOI] [PubMed] [Google Scholar]
- 56. Frei E, Karon M, Levin RH, Freireich EJ, Taylor RJ, Hananian Jet al. The effectiveness of combinations of antileukemic agents in inducing and maintaining remission in children with acute leukemia. Blood 1965;26:642–56. [PubMed] [Google Scholar]
- 57. Sledge GW, Neuberg D, Bernardo P, Ingle JN, Martino S, Rowinsky EKet al. Phase III trial of doxorubicin, paclitaxel, and the combination of doxorubicin and paclitaxel as front-line chemotherapy for metastatic breast cancer: an intergroup trial (E1193). J Clin Oncol 2003;21:588–92. [DOI] [PubMed] [Google Scholar]
- 58. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein Let al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst 2000;92:205–16. [DOI] [PubMed] [Google Scholar]
- 59. Dear RF, McGeechan K, Jenkins MC, Barratt A, Tattersall MH, Wilcken N. Combination versus sequential single agent chemotherapy for metastatic breast cancer. Cochrane Database Syst Rev 2013;12:CD008792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cardoso F, Bedard PL, Winer EP, Pagani O, Senkus-Konefka E, Fallowfield LJet al. International guidelines for management of metastatic breast cancer: combination vs sequential single-agent chemotherapy. J Natl Cancer Inst 2009;101:1174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Conlin AK, Seidman AD. Point: combination versus single-agent chemotherapy: the argument for sequential single agents. J Natl Compr Canc Netw 2007;5:766–70. [DOI] [PubMed] [Google Scholar]
- 62. Subbiah V, Baik C, Kirkwood JM. Clinical development of BRAF plus MEK inhibitor combinations. Trends Cancer 2020;6:797–810. [DOI] [PubMed] [Google Scholar]
- 63. Fraser TR. 5. An experimental research on the antagonism between the actions of physostigma and atropia. Proc R Soc Edinb 1872;7:506–11. [Google Scholar]
- 64. Loewe S. Die quantitativen probleme der pharmakologie. Ergeb Physiol 1928;27:47–187. [Google Scholar]
- 65. Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 2015;3:e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Twarog NR, Connelly M, Shelat AA. A critical evaluation of methods to interpret drug combinations. Sci Rep 2020;10:5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sudalagunta P, Silva MC, Canevarolo RR, Alugubelli RR, DeAvila G, Tungesvik Aet al. A pharmacodynamic model of clinical synergy in multiple myeloma. EBioMedicine 2020;54:102716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ianevski A, Timonen S, Kononov A, Aittokallio T, Giri AK. SynToxProfiler: an interactive analysis of drug combination synergy, toxicity and efficacy. PLoS Comput Biol 2020;16:e1007604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Meyer CT, Wooten DJ, Paudel BB, Bauer J, Hardeman KN, Westover Det al. Quantifying drug combination synergy along potency and efficacy axes. Cell Syst 2019;8:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW, Sorger PK. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol 2013;9:708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gyawali B, Prasad V. Drugs that lack single-agent activity: are they worth pursuing in combination? Nat Rev Clin Oncol 2017;14:193–4. [DOI] [PubMed] [Google Scholar]
- 72. Gaddum JH. Pharmacology. 1st ed. London: Oxford University Press; 1940. [Google Scholar]
- 73. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh Met al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015;21:1318–25. [DOI] [PubMed] [Google Scholar]
- 74. Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 2017;171:1678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tang J, Wennerberg K, Aittokallio T. What is synergy? The Saariselkä agreement revisited. Front Pharmacol 2015;6:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wittes RE, Goldin A. Unresolved issues in combination chemotherapy. Cancer Treat Rep 1986;70:105–25. [PubMed] [Google Scholar]
- 77. Berge EM, Lu X, Maxson D, Barón AE, Gadgeel SM, Solomon BJet al. Clinical benefit from pemetrexed before and after crizotinib exposure and from crizotinib before and after pemetrexed exposure in patients with anaplastic lymphoma kinase-positive non–small-cell lung cancer. Clin Lung Cancer 2013;14:636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schmidt EV, Chisamore MJ, Chaney MF, Maradeo ME, Anderson J, Baltus GAet al. Assessment of clinical activity of PD-1 checkpoint inhibitor combination therapies reported in clinical trials. JAMA Netw Open 2020;3:e1920833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun LZ, Wu C, Li X, Chen C, Schmidt EV. Independent action models and prediction of combination treatment effects for response rate, duration of response and tumor size change in oncology drug development. Contemp Clin Trials 2021;106:106434. [DOI] [PubMed] [Google Scholar]
- 80. Law LW. Differences between cancers in terms of evolution of drug resistance. Cancer Res 1956;16:698–716. [PubMed] [Google Scholar]
- 81. Ettinger DS, Aisner DL, Wood DE, Akerley W, Bauman J, Chang JYet al. NCCN guidelines insights: non–small cell lung cancer, version 5.2018. J Natl Compr Canc Netw 2018;16:807–21. [DOI] [PubMed] [Google Scholar]
- 82. Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen Let al. Immunotherapy in non–small cell lung cancer: facts and hopes. Clin Cancer Res 2019;25:4592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov Det al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1116–27. [DOI] [PubMed] [Google Scholar]
- 84. Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MTet al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kazazi-Hyseni F, Beijnen JH, Schellens JHM. Bevacizumab. Oncologist 2010;15:819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rustum YM. Biochemical rationale for the 5-fluorouracil leucovorin combination and update of clinical experience. J Chemother 1990;2:5–11. [DOI] [PubMed] [Google Scholar]
- 87. Tabernero J, Grothey A, Van Cutsem E, Yaeger R, Wasan H, Yoshino Tet al. Encorafenib plus cetuximab as a new standard of care for previously treated BRAF V600E-mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol 2021;39:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kavčič B, Tkačik G, Bollenbach T. Mechanisms of drug interactions between translation-inhibiting antibiotics. Nat Commun 2020;11:4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Osborne CK, Kitten L, Arteaga CL. Antagonism of chemotherapy-induced cytotoxicity for human breast cancer cells by antiestrogens. J Clin Oncol 1989;7:710–7. [DOI] [PubMed] [Google Scholar]
- 90. Ribas A, Lawrence D, Atkinson V, Agarwal S, Miller WH, Carlino MSet al. Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat Med 2019;25:936–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ascierto PA, Ferrucci PF, Fisher R, Vecchio MD, Atkinson V, Schmidt Het al. Dabrafenib, trametinib and pembrolizumab or placebo in BRAF-mutant melanoma. Nat Med 2019;25:941. [DOI] [PubMed] [Google Scholar]
- 92. Nichols CR, Catalano PJ, Crawford ED, Vogelzang NJ, Einhorn LH, Loehrer PJ. Randomized comparison of cisplatin and etoposide and either bleomycin or ifosfamide in treatment of advanced disseminated germ cell tumors: an Eastern Cooperative Oncology Group, Southwest Oncology Group, and Cancer and Leukemia Group B Study. J Clin Oncol 1998;16:1287–93. [DOI] [PubMed] [Google Scholar]
- 93. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CDet al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. [DOI] [PubMed] [Google Scholar]
- 94. Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im S-A, Yusof MMet al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020;396:1817–28. [DOI] [PubMed] [Google Scholar]
- 95. Palmer AC, Plana D, Gao H, Korn JM, Yang G, Green Jet al. A proof of concept for biomarker-guided targeted therapy against ovarian cancer based on patient-derived tumor xenografts. Cancer Res 2020;80:4278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sarker D, Workman P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res 2007;96:213–68. [DOI] [PubMed] [Google Scholar]
- 97. Gainor JF, Longo DL, Chabner BA. Pharmacodynamic biomarkers: falling short of the mark? Clin Cancer Res 2014;20:2587–94. [DOI] [PubMed] [Google Scholar]
- 98. König D, Läubli H. Mechanisms of immune-related complications in cancer patients treated with immune checkpoint inhibitors. Pharmacology 2020;106:67–80. [DOI] [PubMed] [Google Scholar]
- 99. Lazzari C, Bulotta A, Ducceschi M, Viganò MG, Brioschi E, Corti Fet al. Historical evolution of second-line therapy in non-small cell lung cancer. Front Med 2017;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sachs JR, Mayawala K, Gadamsetty S, Kang SP, de Alwis DP. Optimal dosing for targeted therapies in oncology: drug development cases leading by example. Clin Cancer Res 2016;22:1318–24. [DOI] [PubMed] [Google Scholar]
- 101. Ling A, Huang RS. Computationally predicting clinical drug combination efficacy with cancer cell line screens and independent drug action. Nat Commun 2020;11:5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McDonald ER, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GRet al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017;170:577–92. [DOI] [PubMed] [Google Scholar]
- 103. Bhang HC, Ruddy DA, Krishnamurthy Radhakrishna V, Caushi JX, Zhao R, Hims MMet al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 2015;21:440–8. [DOI] [PubMed] [Google Scholar]
- 104. Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care 2004;8:389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wei SC, Anang N-AAS, Sharma R, Andrews MC, Reuben A, Levine JHet al. Combination anti-CTLA-4 plus anti–PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. Proc Natl Acad Sci U S A 2019;116:22699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chang C-H, Liu W-T, Hung H-C, Gean C-Y, Tsai H-M, Su C-Let al. Synergistic inhibition of tumor growth by combination treatment with drugs against different subpopulations of glioblastoma cells. BMC Cancer 2017;17:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lehar J, Krueger AS, Zimmermann GR, Borisy AA. Therapeutic selectivity and the multi-node drug target. Discov Med 2009;8:185–90. [PubMed] [Google Scholar]
- 108. O'Neil J, Benita Y, Feldman I, Chenard M, Roberts B, Liu Yet al. An unbiased oncology compound screen to identify novel combination strategies. Mol Cancer Ther 2016;15:1155–62. [DOI] [PubMed] [Google Scholar]
- 109. Ocana A, Amir E, Yeung C, Seruga B, Tannock IF. How valid are claims for synergy in published clinical studies? Ann Oncol 2012;23:2161–6. [DOI] [PubMed] [Google Scholar]
- 110. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJet al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto Het al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lin J-R, Izar B, Wang S, Yapp C, Mei S, Shah PMet al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 2018;7:e31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf Bet al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014;11:417–22. [DOI] [PubMed] [Google Scholar]
- 114. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky ADet al. Multiplexed ion beam imaging of human breast tumors. Nat Med 2014;20:436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Del Gaizo Moore V, Letai A. BH3 profiling–measuring integrated function of the mitoch ondrial apoptotic pathway to predict cell fate decisions. Cancer Lett 2013;332:202–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Stockslager MA, Malinowski S, Touat M, Yoon JC, Geduldig J, Mirza Met al. Functional drug susceptibility testing using single-cell mass predicts treatment outcome in patient-derived cancer neurosphere models. Cell Rep 2021;37:109788. [DOI] [PubMed] [Google Scholar]
- 117. Kornauth C, Pemovska T, Vladimer GI, Bayer G, Bergmann M, Eder Set al. Functional precision medicine provides clinical benefit in advanced aggressive hematological cancers and identifies exceptional responders. Cancer Discov 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jonas O, Landry HM, Fuller JE, Santini JT, Baselga J, Tepper RIet al. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci Transl Med 2015;7:284ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mitri ZI, Parmar S, Johnson B, Kolodzie A, Keck JM, Morris Met al. Implementing a comprehensive translational oncology platform: from molecular testing to actionability. J Transl Med 2018;16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Nowakowski GS, Feldman T, Rimsza LM, Westin JR, Witzig TE, Zinzani PL. Integrating precision medicine through evaluation of cell of origin in treatment planning for diffuse large B-cell lymphoma. Blood Cancer J 2019;9:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Coussy F, El-Botty R, Château-Joubert S, Dahmani A, Montaudon E, Leboucher Set al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med 2020;12:eaax2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bernicker EH, Allen TC, Cagle PT. Update on emerging biomarkers in lung cancer. J Thorac Dis 2019;11:S81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hodgson DR, Dougherty BA, Lai Z, Fielding A, Grinsted L, Spencer Set al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br J Cancer 2018;119:1401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sicklick JK, Kato S, Okamura R, Patel H, Nikanjam M, Fanta PTet al. Molecular profiling of advanced malignancies guides first-line N-of-1 treatments in the I-PREDICT treatment-naïve study. Genome Med 2021;13:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Karrila S, Lee JHE, Tucker-Kellogg G. A comparison of methods for data-driven cancer outlier discovery, and an application scheme to semisupervised predic tive biomarker discovery. Cancer Inform 2011;10:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ochs MF, Farrar JE, Considine M, Wei Y, Meshinchi S, Arceci RJ. Outlier analysis and top scoring pair for integrated data analysis and biomarker discovery. IEEE/ACM Trans Comput Biol Bioinform 2014;11:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RNet al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016;167:260–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods 2016;13:521–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Fallahi-Sichani M, Becker V, Izar B, Baker GJ, Lin J-R, Boswell SAet al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol 2017;13:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Daemen A, Griffith OL, Heiser LM, Wang NJ, Enache OM, Sanborn Zet al. Modeling precision treatment of breast cancer. Genome Biol 2013;14:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu Xet al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017;171:1437–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Stewart LA, Tierney JF. To IPD or not to IPD?: advantages and disadvantages of systematic reviews using individual patient data. Eval Health Prof 2002;25:76–97. [DOI] [PubMed] [Google Scholar]
- 135. Riley RD, Lambert PC, Abo-Zaid G. Meta-analysis of individual participant data: rationale, conduct, and reporting. BMJ 2010;340:c221. [DOI] [PubMed] [Google Scholar]
- 136. Danchev V, Min Y, Borghi J, Baiocchi M, Ioannidis JPA. Evaluation of data sharing after implementation of the international committee of medical journal editors data sharing statement requirement. JAMA Netw Open 2021;4:e2033972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Longo DL, Drazen JM. Data sharing. N Engl J Med 2016;374:276–7. [DOI] [PubMed] [Google Scholar]
- 138. Chia PL, Gedye C, Boutros PC, Wheatley-Price P, John T. Current and evolving methods to visualize biological data in cancer research. J Natl Cancer Inst 2016;108:djw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.