Abstract
Mycobacterium smegmatis DSM43756 inactivates rifampin, and the inactivated antibiotic product recovered from culture medium was ribosylated on the 23-OH group. To study this process, the gene responsible for the inactivation was expressed at high levels by the lac promoter in Escherichia coli conferring resistance to >500 μg of antibiotic per ml. Cell homogenates generated a novel derivative designated RIP-TAs; in this study, we determined that RIP-TAs is 23-(O-ADP-ribosyl)rifampin. Our results indicated that RIP-TAs is an intermediate in the pathway leading to ribosylated rifampin and that the previously characterized gene encodes a mono(ADP-ribosyl)transferase which, however, shows no sequence similarity to other enzymes of this class.
Rifampin is a valuable antibiotic and one of the principal chemotherapeutic agents used to combat tuberculosis and other pathogenic mycobacteria. Its antimicrobial activity is due to inhibition of prokaryotic DNA-dependent RNA polymerases, and most rifampin-resistant Mycobacterium tuberculosis and Mycobacterium leprae strains have been reported to have an alteration in the β-subunit of this enzyme (9, 18). However, most rifampin-resistant clinical isolates of Mycobacterium avium and Mycobacterium intracellulare do not have any mutations in the rpoB gene (6), which suggests that there are other genes which are able to confer some degree of resistance. Hetherington et al. (8) showed that Mycobacterium smegmatis, which is naturally resistant to the antibiotic, had no identifiable rpoB mutations and suggested that another mechanism of resistance is at work in this species.
During our studies on the mechanisms of rifampin resistance in acid-fast bacteria, we found that the antibiotic was modified to give a glucosylated or phosphorylated derivative in pathogenic Nocardia spp. (23–25). We also found that M. smegmatis DSM43756 has the ability to inactivate rifampin by ribosylation (3), the first reported case of such a mechanism. Gene disruption experiments showed that ribosylative inactivation of rifampin is a major contributor to the low susceptibility of M. smegmatis to rifampin and that this is the principal rifampin inactivation mechanism in this bacterium (20). We have also reported that many mycobacteria including M. chelonae subsp. abscessus, M. flavescens, M. vaccae, and M. parafortuitum as well as Gordona and Tsukamurella strains inactivate rifampin by ribosylation (23). The ribosylated antibiotic was purified from culture broth of M. smegmatis or Rhodococcus rhodochrous carrying the cloned gene responsible (20), so in this work, we determined the inactivated form of the antibiotic produced by cell homogenates in vitro when the gene was expressed at a high level by using the lac promoter in Escherichia coli.
Rifampin was generously provided by CIBA-Geigy Pharmaceuticals. MICs were determined by an agar dilution method with brain heart infusion agar (Difco Laboratories, Detroit, Mich.) medium. Strains, plasmids, and other methods were as described previously (20).
The cloned mycobacterial gene was found in E. coli MM294-4 to confer a twofold increase in rifampin resistance when expressed off its own promoter, but in vitro antibiotic inactivation by homogenates was not detected with this transformant. By using the previously sequenced 1.1-kb SalI fragment (20) as starting material, various subclones were obtained in E. coli plasmid pGEM3Zf(−). One construct, in which a 600-bp BstNI fragment blunt ended with Klenow enzyme was cloned into the HindII site of the vector, conferred resistance to >500 μg of rifampin per ml and was used for in vitro studies.
E. coli carrying this clone, pGEM3Z-Bst49, was grown in a 500-ml Erlenmeyer flask containing 200 ml of Luria broth (LB) with 100 μg of ampicillin per ml on a Brunswick rotary shaker for 18 h at 37°C. Cells were collected by centrifugation and resuspended in 5 ml of extraction buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 5 mM dithiothreitol). The cell suspension on ice was sonicated for 30 s three times each time (Bioruptor; Cosmo Bio, Tokyo, Japan). To 4 ml of this cell homogenate, 4 ml of reaction mixture containing 60 mg of rifampin and 100 mg of NADH was added. After incubation at 37°C for 3 h, the reaction mixture was centrifuged and the supernatant was freeze-dried. Dried samples were extracted with 8 ml of methanol, and the extract was chromatographed on a LH-20 Sephadex column. From 60 mg of rifampin, 43 mg of purified inactivated compound (designated RIP-TAs) was obtained. The purification state of RIP-TAs was checked by reverse-phase high-pressure liquid chromatography [LiChrospher 100, RP-18(e), Cica-Merck; column dimensions, 4.6 by 150 mm; 38% acetonitrile (MeCN) with 0.05% trifluoroacetic acid) with a detection system at UV 270 nm. Reverse-phase thin-layer chromatography (KC18F; J. T. Baker, Inc.) with a development solvent of 0.2 M NaCl–dimethyl sulfoxide–MeCN (4:1.5:4) was also useful for the identification of rifampin, ribosylated rifampin (RIP-Mb), and ADP-ribosylated rifampin (RIP-TAs), and their Rf values on the thin-layer chromatographic plate were 0.37, 0.55, and 0.80, respectively.
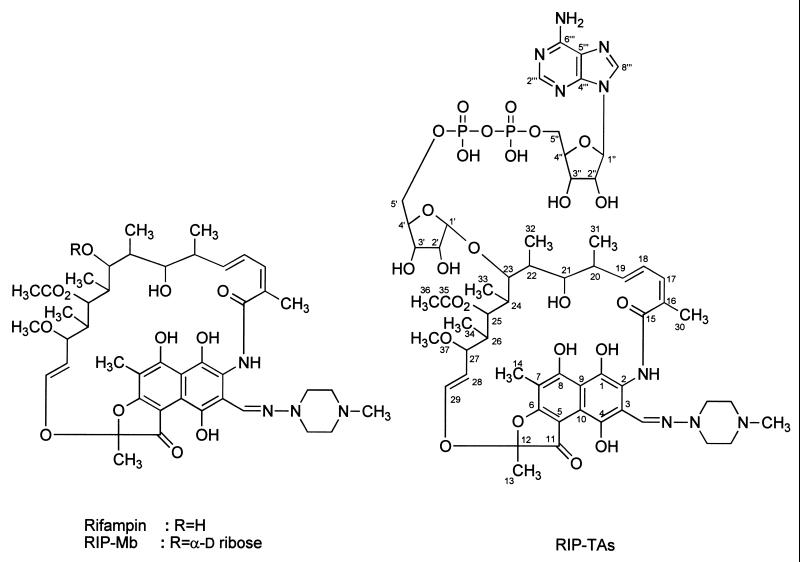
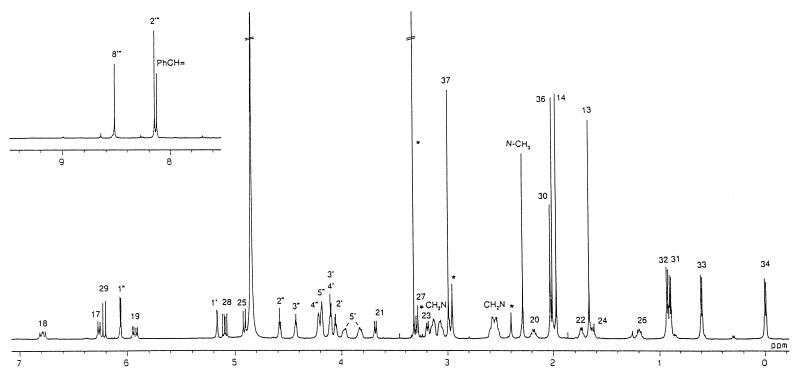
Structure assignment of RIP-TAs was mainly based on 1H and 13C nuclear magnetic resonance (NMR) analyses; 1H-1H correlated spectroscopy, pulsed-field gradient heteronuclear multiquantum coherence, and pulsed-field gradient heteronuclear multiple-bond correlation. The molecular weight and molecular formula of RIP-TAs were determined to be C58H79N9O25P2 (molecular weight, 1,363) by positive- and negative-ion fast atom bombardment mass spectrometry and high-resolution fast atom bombardment mass spectrometry. Comparison of the 1H and 13C NMR signals measured in CD3OD and of the molecular formula of RIP-TAs with that of RIP-Mb (14) suggested that RIP-TAs is the 5′-adenosine diphosphate of RIP-Mb (Fig. 1). The presence of adenosine phosphate was determined by signals at δC 89.2 (C-1"), 76.4 (C-2"), 71.8 (C-3"), 85.5 (C-4"), 66.3 (C-5"), 153.8 (C-2′"), 150.7 (C-4′"), 120.1 (C-5′"), 157.3 (C-6′"), and 141.3 (C-8′") ppm and at δH 6.08 (H-1"), 4.60 (H-2"), 4.45 (H-3"), 4.23 (H-4"), 4.20 (H-5"), 8.17 (H-2′"), and 8.54 (H-8′") ppm (1, 21). Phosphorylation of 5′-OH and 5"-OH was confirmed by couplings of C-4′ (δC 85.1 ppm), C-4", C-5′ (δC 66.8 ppm) and C-5" with the phosphorous atom; JC-O-P of the former two carbons is 9.3 Hz, and that of the latter two carbons is 3.7 Hz (21). The H-5′ signals of RIP-TAs (δH 3.85 and 3.99 ppm) were shifted to lower field relative to those of RIP-Mb (δH 3.51 ppm) (14) by phosphorylation (15, 16). The signals due to the other protons and carbons in the spectra of RIP-TAs were similar to those of RIP-Mb, indicating that the rest of the structure was unchanged. The glycosylation site was confirmed by a pulsed-field gradient heteronuclear multiple-bond correlation experiment to correlate H-23 with C-1′ and H-1′ with C-23. The broad-band proton-decoupled 31P NMR spectrum of RIP-TAs showed signals at δP −8.88 and −8.89 ppm of AB spin system for the P-O-P unit of diphosphate (JP-O-P = 18.3 Hz) (17). Chemical shifts of 1H and 13C NMR were recorded in δ units relative to internal tetramethylsilane (δ = 0), and chemical shifts of 31P NMR were relative to external potassium phosphate (δ = 0). Consequently, the structure of RIP-TAs was determined as shown in Fig. 1: RIP-TAs is 23-(O-ADP-ribosyl)rifampin. The 1H NMR spectrum of RIP-TAs and the complete assignment of each signal are shown in Fig. 2. Details of structure determination will be given elsewhere.
FIG. 1.
Structures of rifampin, RIP-Mb, and RIP-TAs.
FIG. 2.
1H NMR spectrum of RIP-TAs in CD3 OD. Peaks caused by impurity or methanol are indicated by asterisks.
The antimicrobial activity of RIP-TAs was compared with that of rifampin. MICs of RIP-TAs and rifampin for Bacillus subtilis PCl 219 were 25 and 0.02 μg/ml, respectively, and for M. smegmatis DSM43756 they were >100 and >50 μg/ml, respectively. We recently found that M. smegmatis homogenates acting upon rifampin also produce RIP-TAs as an intermediate. The nature of the inactivated antibiotic demonstrates that the previously characterized gene (20) is a mono(ADP-ribosyl)transferase (11); this enzyme transfers the ADP-ribose moiety of NADH to acceptor molecules, usually proteins (7). Many bacterial mono(ADP-ribosyl)transferases are toxins, such as those of diphtheria (2), cholera (22), Bordetella pertussis, or Clostridium botulinum C3 (13, 22). ADP-ribosylation of the nitrogen-fixing enzyme dinitrogenase reductase in the photosynthetic bacterium Rhodospirillum rubrum mediates the reversible inactivation of the enzyme and is influenced by light (12). T4 and related bacteriophages code for two ADP-ribosylating activities, the Alt and Mod gene products, whose main target is the host DNA-dependent RNA polymerase altering promoter specificity (10).
Endogenous mono(ADP-ribosyl)ation has been demonstrated in several bacteria including Pseudomonas aeruginosa (11, 19), but little is known about the physiological role of this modification process (11). In these cases, the acceptors were proteins. However, in the present study the acceptor was a low-molecular-weight antibiotic, rifampin, and to our knowledge, this is the first case of ADP-ribosylation as a mechanism of antibiotic inactivation. Moreover, the ADP-ribosyl moiety is joined to an oxygen atom, in contrast to the examples cited above, where it is a nitrogen atom.
We find that NAD is the preferred source of the ADP-ribosyl group; Domenighini and Rappuoli (5) have analyzed the protein sequences from a number of ADP-ribosyltransferases and based on the crystal structures have suggested a plausible NAD binding consensus motif present in all the sequences they have examined. They propose two groups of these enzymes, the CT and DT group. The CT group have a conserved atom-His/Arg motif and a conserved motif in a β-strand forming part of the scaffold of the catalytic cavity with an arom-ph-Ser-Thr-Ser-ph consensus sequence. An additional consensus sequence is centered around the key glutamic acid residue and is (Glu/Gln)-X-Glu. In the DT group, the NAD-binding motif is Tyr-X10-Tyr. The predicted amino acid sequence of the rifampin ADP-ribosylating enzyme does not have an arom-His/Arg motif or the Ser-Thr-Ser NAD-binding motif; a Glu-X-Glu motif was present.
Studies on the detailed enzymology of conversion of ADP-ribosylated rifampin (RIP-TAs) to ribosylated rifampin (RIP-Mb) is in progress. We have identified phosphoribosylated rifampin both in vivo and in vitro, so the ADP-ribosylated rifampin (RIP-TAs) may be hydrolyzed by an ADP-ribose phosphohydrolase, as previously described (4).
Acknowledgments
This work was partly supported by a Grant-in-Aid for Scientific research (C) from the Ministry of Education, Sciences, Sports, and Culture of Japan to Y.M.
REFERENCES
- 1.Breitmaier E, Voelter W. A 13C nuclear-magnetic-resonance study of the enzyme cofactor flavin-adenine dinucleotide. Eur J Biochem. 1972;31:234–238. doi: 10.1111/j.1432-1033.1972.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 2.Carrol S F, Collier R J. NAD binding site of diphtheria toxin: identification of a residue within the nicotinamide subsite by photochemical modification with NAD. Proc Natl Acad Sci USA. 1984;81:3307–3311. doi: 10.1073/pnas.81.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dabbs E R, Yazawa K, Mikami Y, Miyaji M, Morisaki N, Iwasaki S, Furihata K. Ribosylation by mycobacterial strains as a new mechanism of rifampin inactivation. Antimicrob Agents Chemother. 1995;39:1007–1009. doi: 10.1128/aac.39.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doherty M D, Morrison J F. The hydrolysis of adenosine diphosphate ribose by a specific phosphohydrolase of rabbit-muscle extracts. Biochim Biophys Acta. 1962;65:364–366. doi: 10.1016/0006-3002(62)91061-2. [DOI] [PubMed] [Google Scholar]
- 5.Domenighini M, Rappuoli R. Three conserved consensus sequences identify the NAD-binding site of the ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol Microbiol. 1996;21:667–674. doi: 10.1046/j.1365-2958.1996.321396.x. [DOI] [PubMed] [Google Scholar]
- 6.Guerrero C, Stockeman L, Marchesi F, Bodmer T, Roberts G D, Telenti A. Evaluation of rpoB gene in rifampicin-susceptible and -resistant Mycobacterium avium and Mycobacterium intracellulare. J Antimicrob Chemother. 1994;33:661–663. doi: 10.1093/jac/33.3.661-a. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi O, Ueda K. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 8.Hetherington S V, Watson A S, Patrick C C. Sequence and analysis of rpoB gene of Mycobacterium smegmatis. Antimicrob Agents Chemother. 1995;39:2164–2166. doi: 10.1128/aac.39.9.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honore N, Cole S T. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother. 1993;37:414–418. doi: 10.1128/aac.37.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch T, Ruger W. The ADP-ribosyltransferase (gpAlt) of bacteriophages T2, T4, and T6: sequencing of the genes and comparison of their products. Virology. 1994;203:294–298. doi: 10.1006/viro.1994.1487. [DOI] [PubMed] [Google Scholar]
- 11.Lowery R G, Ludden P W. Endogenous ADP ribosylation in procaryotes. In: Moss J, Vaughan M, editors. ADP-ribosylating toxins and G proteins: insights into signal transduction. Washington, D.C: American Society for Microbiology; 1990. pp. 459–468. [Google Scholar]
- 12.Ludden P W, Roberts G P. Regulation of nitrogenase activity by reversible ADP-ribosylation. Curr Topics Cell Regul. 1989;30:23–56. doi: 10.1016/b978-0-12-152830-0.50004-9. [DOI] [PubMed] [Google Scholar]
- 13.Middlebrook J L, Dorland R B. Bacterial toxins: cellular mechanisms to action. Microbiol Rev. 1984;48:199–221. doi: 10.1128/mr.48.3.199-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morisaki N, Kobayashi H, Iwasaki S, Furihata K, Dabbs E R, Yazawa K, Mikami Y. Structure determination of ribosylated rifampicin and its derivative: new inactivated metabolites of rifampicin by mycobacterial strains. J Antibiot. 1995;48:1299–1303. doi: 10.7164/antibiotics.48.1299. [DOI] [PubMed] [Google Scholar]
- 15.Morisaki N, Iwasaki S, Yazawa K, Mikami Y, Maeda A. Inactivated products of rifampicin by pathogenic Nocardia spp.: structures of glycosylated and phosphorylated metabolites of rifampicin and 3-formylrifamycin SV. J Antibiot. 1994;46:1605–1610. doi: 10.7164/antibiotics.46.1605. [DOI] [PubMed] [Google Scholar]
- 16.Morisaki N, Iwasaki S, Furihata K, Yazawa K, Mikami Y. Structural elucidation of rokitamycin, midecamycin and erythromycin metabolites formed by pathogenic Nocardia. Magn Reson Chem. 1995;33:481–489. [Google Scholar]
- 17.Nonaka Y, Fujii S, Yamano T. Phosphorus-31 nuclear magnetic resonance and electronic spectroscopic studies of adrenodoxin reductase and its binary complex with NADP+ J Biochem. 1985;97:1263–1271. doi: 10.1093/oxfordjournals.jbchem.a135177. [DOI] [PubMed] [Google Scholar]
- 18.Ohno K, Koga H, Kohno S, Tashiro T, Hara K. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother. 1996;40:1053–1056. doi: 10.1128/aac.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penyige A, Deak E, Kalmanczhelyi A, Barabas G. Evidence of a role for NAD+ glycohydrolase and ADP-ribosyltransferase in growth and differentiation of Streptomyces griseus NRRL B-2682: inhibition by m-aminophenylboronic acid. Microbiology. 1996;142:1937–1944. doi: 10.1099/13500872-142-8-1937. [DOI] [PubMed] [Google Scholar]
- 20.Quan S, Venter H, Dabbs E R. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob Agents Chemother. 1997;41:2456–2460. doi: 10.1128/aac.41.11.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rag E, Scallion L, Mindless R, Carol I, Casino A, Tortorella S. 1H-, 13C-, 31P-NMR studies and conformational analysis of NADP+, NADP coenzymes and of dimers from electrochemical reduction of NADP+ Biochim Biophys Acta. 1991;1076:49–60. doi: 10.1016/0167-4838(91)90218-o. [DOI] [PubMed] [Google Scholar]
- 22.Spangler D B. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol Rev. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka Y, Yazawa K, Dabbs E R, Nishikawa K, Komaki H, Mikami Y, Miyaji M, Morisaki N, Iwasaki S. Different rifampicin inactivation mechanisms in Nocardia and related taxa. Microbiol Immunol. 1996;40:1–4. doi: 10.1111/j.1348-0421.1996.tb03303.x. [DOI] [PubMed] [Google Scholar]
- 24.Yazawa K, Mikami Y, Maeda A, Akao M, Morisaki N, Iwasaki S. Inactivation of rifampin by Nocardia brasiliensis. Antimicrob Agents Chemother. 1993;37:1127–1135. doi: 10.1128/aac.37.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazawa K, Mikami Y, Maeda A, Morisaki N, Iwasaki S. Phosphorylative inactivation of rifampicin by Nocardia otitidiscaviarum. J Antimicrob Chemother. 1994;33:1127–1135. doi: 10.1093/jac/33.6.1127. [DOI] [PubMed] [Google Scholar]