Abstract
Despite limited reports on glutamine methylation, methylated glutamine is found to be highly conserved in a "GGQ" motif in both prokaryotes and eukaryotes. In bacteria, glutamine methylation of peptide chain release factors 1/2 (RF1/2) by the enzyme PrmC is essential for translational termination and transcript recycling. Two PrmC homologs, HEMK1 and HEMK2, are found in mammals. In contrast to those of HEMK2, the biochemical properties and biological significance of HEMK1 remain largely unknown. In this study, we demonstrated that HEMK1 is an active methyltransferase for the glutamine residue of the GGQ motif of all four putative mitochondrial release factors (mtRFs)—MTRF1, MTRF1L, MRPL58, and MTRFR. In HEMK1-deficient HeLa cells, GGQ motif glutamine methylation was absent in all the mtRFs. We examined cell growth and mitochondrial properties, but disruption of the HEMK1 gene had no considerable impact on the overall cell growth, mtDNA copy number, mitochondrial membrane potential, and mitochondrial protein synthesis under regular culture condition with glucose as a carbon source. Furthermore, cell growth potential of HEMK1 KO cells was still maintained in the respiratory condition with galactose medium. Our results suggest that HEMK1 mediates the GGQ methylation of all four mtRFs in human cells; however, this specific modification seems mostly dispensable in cell growth and mitochondrial protein homeostasis at least for HeLa cells under fermentative culture condition.
Subject terms: Biochemistry, Cell biology, Genetics, Molecular biology
Introduction
Post-translational modifications (PTMs) such as phosphorylation, acetylation, methylation, SUMOylation, can alter protein functions. Among these PTMs, research on protein methylation has attracted increasing attention due to its role in epigenetic regulations1. A large number of studies have been conducted on the methylation of lysine and arginine residues owing to their specific roles in histone PTMs, which are highly relevant in chromatin regulation and gene expression2,3. Besides adenosine triphosphate (ATP), S-adenosylmethionine (SAM)—the donor of methyl moiety for protein methylation—is the second most used cosubstrate in biological processes, suggesting that protein methylation is a pervasive biochemical event involved in different cellular processes4,5.
Glutamine methylation, although rarely studied, is one of the essential protein modifications in prokaryotes6. The Gly-Gly-Gln (GGQ) motif of release factors (RF1 and RF2) is a remarkable substrate for glutamine methylation. Generally, at the end of translation, the stop codons in transcripts are read by RF1 (UAA and UAG) and RF2 (UAA and UGA). Subsequently, these factors cleave the nascent polypeptide chain from the last linked tRNA in the peptidyl transferase center (PTC) of the ribosome. The GGQ motif in RF1 and RF2 plays a critical role in orienting a water molecule and facilitating a nucleophilic attack on the carbonyl carbon of the ester bond between the tRNA and the attached polypeptide. Glutamine methylation driven by the N5-glutamine methyltransferase (PrmC) at the GGQ motif in both bacterial RF1 and RF2 can stabilize the GGQ motif at the PTC through hydrophobic interactions, thus enhancing the polypeptide-releasing activity by about 5-fold and 20 fold, respectively7–10. Sequence and structural analyses revealed that PrmC is a typical seven-stranded β-sheet methyltransferase containing a GxGxG type SAM-binding motif and an Asn-Pro-Pro-Tyr (NPPY) glutamine/cystine-binding motif. Deletion of the SAM-binding motif of PrmC causes a loss of methylation in both RF1 and RF2 and results in an impaired growth phenotype in rich medium and lethality in nutrient-depleted medium6. In the E. coli genome, the prmC gene is positioned immediately downstream of its substrate (prfA gene, encoding RF1), ensuring in-time methylation of bacterial RF16.
PrmC evolved divergently beyond the prokaryotic lineage. It evolved into a homolog protein Mtq2 in archaea (archaeal PrmC), with a slight difference in the molecular basis. A recent study published by van Tran et al.11 demonstrated that in H. volcanii, a PrmC homolog HvoMtq2 could methylate HvoaRF1, with HovaRF3 and GTP acting as cofactors. In addition, unlike the bacterial PrmC, HvoMtq2 forms a heterodimer with a protein called multifunctional methyltransferase subunit 112 (HvoTrm112). Although this dimerization is not essential for the methyltransferase activity of HvoMtq2, the enzymatic activity of the HvoMtq2-HvoTrm112 heterodimer is enhanced11. Similar to the phenotypes induced by prmC inactivation in bacteria, the deletion of the HvoMtq2 gene also causes growth retardation in H. volcanii11.
In eukaryotic cells, two PrmC homologs, HEMK1 (also known as MTQ1 or MPRMC) and HEMK2 (also known as MTQ2 or N6AMT1), are localized in the mitochondria and cytosol, respectively12,13. HEMK2 exhibits similar enzymatic properties as those of the archaeal HvoMtq2. It requires the protein TRM112 as a binding partner and the eukaryotic peptide chain release factor subunit 3 (eRF3) and GTP as cofactors for efficient methylation of the eukaryotic peptide chain release factor subunit 1 (eRF1)14. The deletion of scMtq2, an HEMK2 homolog, in S. cerevisiae yielded a strain with multiple defects, including growth retardation, cold-sensitivity, and hypersensitivity to translational fidelity drugs12. It has been reported that the loss of HEMK2 leads to early embryonic lethality in mice15. In contrast to bacterial PrmC, mammalian HEMK2 has a broader range of substrate-binding activity, as it can also catalyze the methylation of lysine residues and nucleotides16–18.
Similarly, scMtq1, another PrmC homolog found in S. cerevisiae, can methylate one of the mitochondrial peptide chain release factors, Mrf1, required for translation termination inside the organelle19. The loss of methylation in Mrf1 shows no significant impact when solid media is used, in contrast to the PrmC knockout (KO) phenotype in prokaryotes. The growth rate is about 15% slower when non-fermentable carbon sources are used in liquid media, suggesting that the methylation in Mrf1 affects mitochondrial function and energy metabolism12. Considering the high sequence identity of the enzyme and substrates, it is possible that the mammalian HEMK1 also possesses a methyltransferase activity against human mitochondrial peptide chain release factors (mtRFs). To date, four potential mtRFs have been identified in humans, namely MTRF1, MTRF1L, MRPL58 (also known as ICT1), and MTRFR (also known as C12orf65)20–23. As with other class I release factors, all four mtRFs contain the universally conserved GGQ motif. Although direct evidence from LC–MS/MS is missing, previous studies have demonstrated that HEMK1 could cause a 14 Da increase in molecular mass in a GGQ-containing peptide fragment derived from MTRF1L, implying the addition of a methyl group on the glutamine residue of the GGQ motif13. Recently, Desai et al. demonstrated that MTRFR is methylated at position Q73 in its GGQ motif24. Glutamine methylation on MTRF1 and MTRL58 has so far been reported yet. Furthermore, it remains unclear how glutamine methylation of the GGQ motif affects the functions of mtRFs and mitochondrial translation. Thus, using proteomic analysis and genomic manipulation, we assessed the methyltransferase activity of HEMK1 towards the glutamine residue in GGQ motif of all potential mtRFs and examined possible biological consequences of HEMK1 depletion on mitochondrial properties.
Results
Domain and motif highlights of HEMK1 and mtRFs
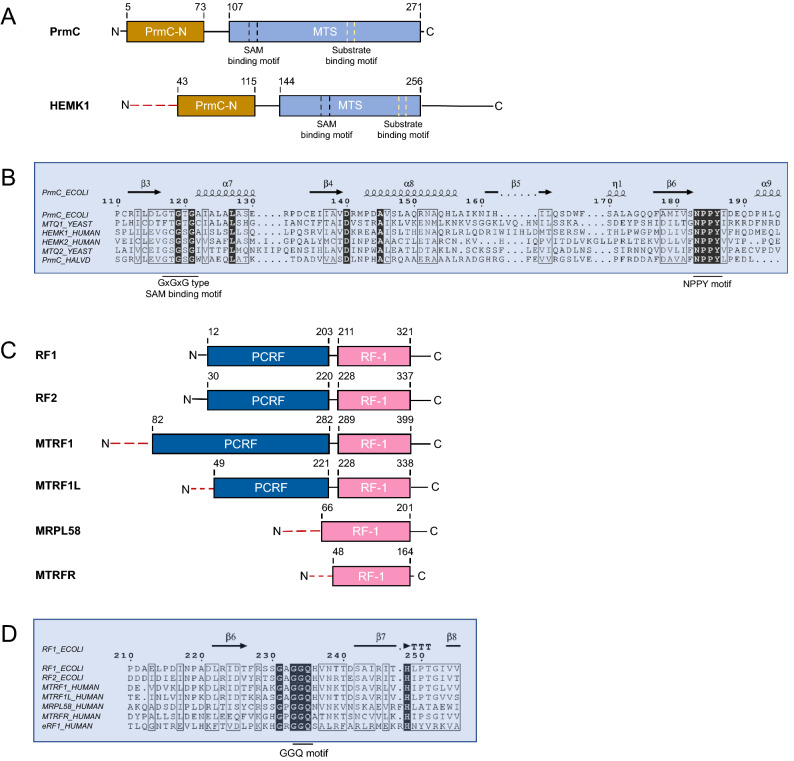
Human HEMK1 is a 338 amino acid residue protein that shows high sequence and structural similarity to bacterial PrmC (Fig. 1A,B, Figure S1). Similar to PrmC, HEMK1 belongs to the seven beta-strand class of methyltransferases. It possesses two distinct domains, a PrmC-N terminal domain (Pfam: PrmC_N, PF17827) and a methyltransferase small domain (Pfam: MTS, PF05175). The MTS domain contains a GxGxG type SAM-binding motif (position 117–121, referring to the bacterial PrmC sequence) and a NPPY glutamine/cystine-binding motif (position 183–186) (Fig. 1B, Figure S1). Sequence-based algorithms, such as DeepMito, MitoFate, and DISOPRED, predicted that the first 40 residues of the HEMK1 N-terminal constitute a disordered, amphiphilic region containing a mitochondrial localization signal (MLS) (Figure S2A; Table S1), suggesting its localization in the mitochondria.
Figure 1.
Domain architecture and sequence alignment of HEMK1 and mtRFs. (A) Domain architecture of PrmC and HEMK1. The red dash line indicated the predicted mitochondrial localization signal. PrmC-N: PrmC N-terminal domain (PF17827); MTS: methyltransferase small domain (PF05175). (B) Sequence alignments of various PrmC family proteins. The amino acid position is numbered according to the PrmC sequence. PCRF: peptide chain release factor domain, bacterial Class 1 (PF03462). RF-1: peptide chain release factor, bacterial Class I, PTH domain (PF00472). (C) Domain architecture of bacterial RF1, RF2, and mtRFs. The red dash lines indicated the predicted mitochondrial localization signal. (D) Sequence alignments of bacterial RF1, RF2, mtRFs, and the human eRF1. The amino acid position is numbered according to the bacterial RF1 sequence.
Amino acid sequence comparison of HEMK1 with another human Mtq2 homolog, HEMK2, revealed a radical amino acid replacement at the − 1 position of the NPPY motif towards the N terminus. At this position, a large and hydrophobic residue, phenylalanine, was found in HEMK2, yeast Mtq2, and HvoPrmC (HvoMtq2), but a small residue, serine/glycine, was found in HEMK1, yeast Mtq1, and PrmC (Fig. 1B). Considering the critical role of the NPPY motif in the PrmC methyltransferase family14, it is possible that HEMK1 and HEMK2 have different preferences for substrate binding.
Given the potential difference in substrate recognition, we thought that HEMK1 might modify GGQ-containing proteins other than eRF1 (targeted by HEMK2). The apparent candidates were four potential mtRFs—MTRF1, MTRF1L, MTPL58, and MTRFR20–22. All four mtRFs contained the conserved GGQ motif, while only MTRF1 and MTRF1L possessed a peptide chain release factor domain (Pfam: PCRF, PF03462) (Fig. 1C,D). Permutation array of enzyme–substrate binding, using the sequence of eRF1 (position 179–192), revealed that G-Q-X3-R was the minimal recognition element for HEMK2, with the + 4 position of the GGQ motif (R189) being absolutely essential16 (Figure S2B). However, the arginine residue was not found at the + 4 position of the GGQ motif in any of the four human mtRFs (Figures S2B,S3), suggesting it is unlikely that HEMK2 is the methyltransferase responsible for human mtRFs.
HEMK1 is responsible for glutamine methylation of mtRFs
Although serval reports have demonstrated the mitochondrial localization of HEMK1 and mtRFs13,20–22,25, co-localization of HEMK1 and mtRFs has yet been shown. In order to determine whether HEMK1 methylates the mtRFs, we first confirmed the subcellular localization of both exogenously expressed enzymes and substrates. We co-expressed HA-tagged mtRFs individually with FLAG-tagged HEMK1 and performed immunofluorescence imaging to track their localization. As expected and consistent with previous reports, HEMK1 and all four mtRFs were localized in the mitochondria (Fig. 2A).
Figure 2.
Glutamine methylation in the “GGQ” motif of four mtRFs mediated by HEMK1. (A) Subcellular localization of transiently expressed exogenous HEMK1 and mtRFs in HeLa cells. Representative images of the cell expressing both HEMK1-cFLAG and individual mtRF-cHA were shown. The number of cells showing the colocalization of mtRF, HEMK1, and MitoTracker per the number of analyzed cells are shown in the right side of the panels. Cells with both HEMK1-cFLAG and individual mtRF-cHA co-expressed were analyzed. Note that, although some cells showed mtRFs-cHA and/or HEMK1-cFLAG in the nucleus possibly because of too-much expression of transgene(s), we excluded these cells for quantification to avoid collecting abnormal phenotypes. HEMK1-cFLAG, magenta; mtRFs-cHA, green; MitoTracker Red, orange. Scale 20 μm in total length. (B) Approximate methylation efficiency. Stably expressed exogenous mtRFs-cHA in naïve HeLa, HEMK1 KO clone #1, and HEMK1 KO clone #1 complemented with stably expressed HEMK1-cFlag. For fragment counts and methylation ratio of each methylated peptide, see Figure S5A. (C) MS1 spectra of mtRFs. Theoretical mass corresponds to specific methylated mtRFs peptides. See Table S2 for detailed MS2 spectra and chromatograms of methylation of each sites.
The purified recombinant PrmC could induce the methylation of bacterial RF1 in vitro7. Therefore, we attempted to detect the catalytic activity of recombinant HEMK1 against mtRFs in vitro; however, we did not observe any methyltransferase activity (data not shown). Hence, we decided to monitor the methyltransferase activity of HEMK1 in cells. We generated HEMK1 KO HeLa cells via CRISPR-Cas9-mediated gene deletion. This deletion removed both the SAM-binding and substrate-binding motifs of HEMK1, resulting in a non-functional enzyme (Figure S4). To monitor the impact of HEMK1 depletion on the methylation of mtRFs, we expressed HA-tagged mtRFs individually in the naïve or HEMK1-KO HeLa cells and immune-purified the proteins for mass spectrometry analysis to detect methylated peptides. In naïve HeLa cells, we found glutamine methylation at Q252 of MTRF1L, Q313 of MTRF1, Q90 of MRPL58, and Q73 of MTRFR, within their GGQ motifs (Fig. 2B,C; Table S2). In contrast, these glutamine methylation events were not detected in the HEMK1 KO cells (Fig. 2B, Figure S5A). Exogenous expression of HEMK1-cFLAG in the HEMK1 KO cells rescued and in fact, enhanced the methylation of all four mtRFs, compared to that in the naïve cells (Fig. 2B, Figure S5A).
To further clarify whether glutamine methylation of mtRFs is directly caused by the enzymatic activity of HEMK1, we generated a mutant HEMK1 with a Y242E substitution at the NPPY substrate-binding motif, which was predicted to inactivate the enzymatic activity. HEMK1Y242E displayed the same mitochondrial localization as the wild-type HEMK1 (Figure S5C). However, the glutamine methylation of MTRF1L in HEMK1 KO HeLa cells was not induced by the HEMK1Y242E mutant (Figure S5A). These data further support that HEMK1 is a glutamine methyltransferase for all the four mtRFs.
Loss of HEMK1 has minimal impact on HeLa cell physiology
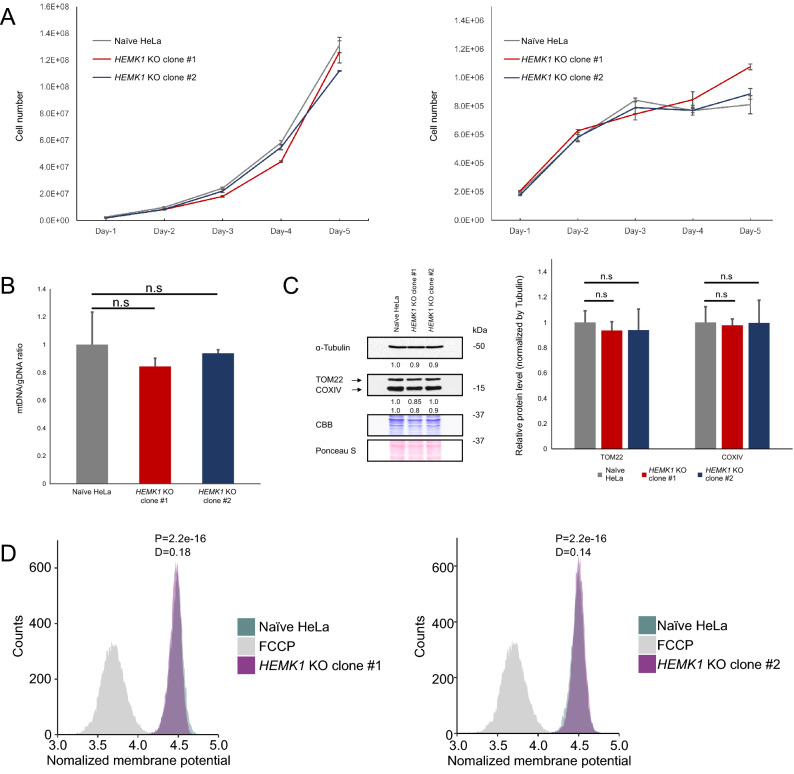
Since the loss of PrmC in K-12 E. coli causes growth retardation6, we assessed the impact of the loss of HEMK1 on cell growth. Under the fermentative culture condition with glucose as a carbon source, we monitored the growth of naïve and HEMK1 KO HeLa cells, but did not find a clear difference between them (Fig. 3A, left). We also assayed under the respiratory condition with galactose as a carbon source, but HEMK1 depletion did not show clear impact on the cell growth (Fig. 3A, right).
Figure 3.
Impacts of HEMK1 depletion on cell growth and mitochondrial properties. (A) Cell growth in high-glucose and galactose media. Growth curve of naïve HeLa and HEMK1 KO clones #1 and #2 in different media. Left panel: high-glucose media; right panel: galactose media. The numbers represent a mean ± SEM from three independent samples. (B) Assessment of relative mtDNA amounts (mtDNA/gDNA) by qPCR. Data were normalized to gDNA abundance. Data represent a mean ± SEM from three independent samples. The Student’s t-test, two-tail, was used for testing the statistical significance. n.s not significant. (C) Western blot of mitochondrial outer membrane protein TOM22, and inner membrane protein COXIV from naïve HeLa and HEMK1 KO clones (#1 and #2). Left panel shows representative western blot result. α-Tubulin was used as a loading control. CBB and Ponceau S staining were shown as loading and transfer indicators. Numbers shown at the bottom of each panel indicated the normalized intensity referring to signals from naïve HeLa cells. Right panel: quantitation of mitochondrial membrane protein TOM22 and COXIV Western blot signals with a mean ± SEM from four independent samples. The Student’s t-test, two-tail, was used for testing the statistical significance. n.s not significant. Biological repeats were shown in Figures S6C and S9A–F. (D) Assessment of mitochondrial properties by FACS analysis. Mitotracker Red CMXRos signal was normalized with Mitotracker Green FM signal (see “Materials and methods” section for the details). Data from 10,000 cells are depicted. The Mann–Whitney U-test, two-tail, was used for testing the statistical significance (P value). The Cohen’s d-test was used for testing the effect size (D value).
It has been shown that mitochondria DNA (mtDNA) replication is a highly dynamic process that is linked to mitochondrial properties including total mass and activities26. We thus examined the potential impact of HEMK1 depletion on mtDNA copy number. To this end, we performed a quantitative polymerase chain reaction using total DNA extracts to assess mtDNA amounts (as a ratio to genomic DNA [gDNA]). Again, we failed to detect an apparent difference between naïve and HEMK1 KO cells regarding the mtDNA-gDNA ratios, suggesting that the mtDNA replication is likely not affected by HEMK1-mediated glutamine methylation on mtRFs (Fig. 3B).
We then examined the impact of HEMK1 depletion on mitochondrial membrane protein contents. Potential changes in one of the mitochondrial outer membrane proteins, TOM22, and one of the mitochondrial inner membrane proteins, COXIV, were investigated. However, the western blot analysis of these mitochondrial proteins revealed no changes in protein expression in HEMK1 KO cells (Fig. 3C, Figure S6C).
It has been shown that mitochondria DNA (mtDNA) replication is a highly dynamic process that is linked to mitochondrial properties including total mass or activities26. Therefore, we measured mitochondrial membrane potential via a flow cytometry-based assay. Here, we used MitoTracker Red, a membrane potential sensitive dye27. To control the effect of mitochondrial volume, we used membrane potential insensitive MitoTracker Green, which stains the mitochondrial membrane. Although the loss of HEMK1 significantly affected (P value = 2.2e−16) the signals of MitoTracker Red (Fig. 3D), the change was nearly negligible (D value < 0.2) in both clones. Thus, the impact on the net mitochondrial membrane potential (normalized MitoTracker Red signal) was marginal (Fig. 3D).
HEMK1-mediated mtRF methylation could be dispensable in mitochondrial translation under standard culture conditions
Even with the limited physiological influence of HEMK1 depletion in mitochondria, we extended our analysis to mitochondrial translation. To directly examine the impact of HEMK1 depletion and the role of HEMK1-mediated mtRFs methylation in mitochondrial translation, we performed fluorescent noncanonical amino acid tagging for mitochondrial translation (mito-FUNCAT)28,29 to assess nascent protein synthesis in HEMK1 KO cells. Out of four HEMK1 KO clones, only KO clone #1 showed a decrease in nascent protein synthesis (Fig. 4A,B, Figure S6A,B), suggesting that this reduction stemmed from clonal variation and not HEMK1 depletion. To further ascertain this, we performed a rescue experiment using the wild-type HEMK1 and the HEMK1Y242E mutant. As expected, the exogenous expression of neither wild-type HEMK1 nor HEMK1Y242E mutant could reverse the changes in nascent protein synthesis in HEMK1 KO clone #1 (Figure S7A,B). Furthermore, we performed western blot of an mtDNA-encoding protein MT-CO1. Similar to that in the mito-FUNCAT assay, HEMK1 KO clone #1 showed lower MT-CO1 levels, and this phenotype was unrelated to HEMK1 expression (Figure S7C,D). Therefore, we concluded that HEMK1 inactivation does not have a clear impact on mitochondrial translation.
Figure 4.

The assessment of mitochondrial translation in HEMK1 KO cells. (A) Representative gel images of total and nascent mitochondrial proteins. Infrared (IR)-800 dye was conjugated to HPG-containing newly synthesized proteins. Total protein was stained with CBB. A black line is inserted between lanes 4 and 5 (counting from the left-hand side) because they were not adjacent lanes, although these lanes are originated from the same gel. The original image is shown in Figure S6A. (B) Bar graph of HPG signal/CBB signal for (A). Error bars represented the mean ± SEM calculated from samples shown in Figure S6A (see detailed measurements in Figure S6A). The Student’s t test, two-tail, was used for testing the statistical significance.
Discussion
Ishizawa et al. reported HEMK1-mediated methylation (14 Da addition) of the MTRF1L-derived peptide13; here, we extended their findings onto other mtRFs and demonstrated that HEMK1 could methylate glutamine in the GGQ motif of all the four mtRFs. However, our analysis, which utilized exogenously expressed mtRFs, posed an analytic hurdle in the assessment of the methylation level of endogenous mtRFs. Since we could not access to good antibody to detect HEMK1 protein, we compared RNA expression level of HEMK1 in naïve Hela (endogenous one) vs. the HEMK1 KO clone #1 complemented with exogenous HEMK1 (exogenous HEMK1 cDNA only), which was used for exogenous mtRFs GGQ motif methylation analysis shown in Fig. 2B,C and Fig. S5A. Exogenous HEMK1 was highly expressed (> 700 times higher than endogenous one in naïve Hela, Fig S5B) in this complemented HEMK1 KO cells. However, with such high HEMK1 expression, mtRFs methylation was hardly reaching 50%. Given that the exogenously expressed mtRFs in naïve HeLa cells were methylated at a low frequency (Fig. 2B, Figure S5A), the expression levels of HEMK1 in HeLa cells might be insufficient for full methylation of endogenous mtRFs. Another possibility for the incomplete methylation of mtRFs is that although HEMK1 is the enzyme responsible for glutamine methylation, other cofactor(s) may be needed for the catalytic reaction, as seen in HEMK2 homologs14, which might also explain why recombinant HEMK1 did not show enzymatic activity in the in vitro assay. Therefore, even if a sufficient amount of HEMK1 was exogenously supplied (Figure S5B), the amount of such putative endogenous cofactor(s) might restrict this enzymatic reaction in cells. In any case, if any approach for investigating GGQ methylation of endogenous mtRFs is accessible, such as, specific antibodies capable of immunoprecipitation of mtRFs or the epitope tagging to the endogenous mtRFs, it will be possible to enrich the endogenous ones and clarify their GGQ methylation status. Furthermore, as described, we have not succeeded in detecting the methylation of Q by HEMK1 in vitro. If the HEMK1-mediated Q methylation reaction can be established in vitro, the details of the enzymatic reaction of HEMK1 and its regulatory mechanism could be further elucidated.
Given that the loss of PrmC in E. coli K-12 strain leads to growth retardation6,7,30, it is possible that the loss of methylation of mtRFs could cause similar biological consequences. Unfortunately, we could not identify any clear mitochondria-associated phenotypes in terms of cell growth, mitochondrial DNA replication, membrane potential, or even mitochondrial translation, a biological reaction directly supported by mtRFs. Methylated bacterial RFs can just accelerate the termination reaction, and this methylation is not essential at the termination step, as hydrolysis still occurs, although at a slower rate9. Again, the lower rate of mtRF methylation seen in HeLa cells suggests the limited role of mtRFs in mitochondrial protein synthesis. The restricted function of GGQ methylation in mtRFs is also supported by the Genome Aggregation Database31, which shows the probability of "being the loss-of-function intolerant" score of HEMK1 as 0, suggesting that HEMK1 is unlikely an essential gene.
Although our results showed that HEMK1 depletion plays a limited role in translation, further analysis is required for clarifying any biological role of HEMK1-mediated GGQ methylation, especially a comprehensive molecular understanding of the four putative mtRFs, which has been long contested. mtRF1L is a canonical release factor with peptidyl-tRNA hydrolysis activity in ribosomes22. Furthermore, such activity has also been reported in MRPL58 (ICT1)32, but the role in termination is still a matter of debate33,34. A recent structural study showed that MRPL58 recognizes the vacant A site in ribosomes translating truncated mRNA23, suggesting that MRPL58 may rescue the stalled ribosomes on the ends of aberrant mRNAs, rather than participate in the standard termination reaction on stop codons. Moreover, Ramakrishnan and colleagues reported that MTRFR, associated with a split large subunit of the mitoribosome, serves as a quality control factor and cleaves the peptidyl-tRNAs halted in the middle of elongation24. In addition to termination and ribosome recycling, MRPL58 may also plays an important role in mitoribosome biogenesis or basal stability, as this protein is an integral constituent of the large subunit of the mitoribosome20,35,36. Further functional analysis of mtRFs in mitochondrial translation will help in enhancing the understanding of HEMK1-mediated methylation.
Materials and methods
Cell culture
HeLa cells were obtained from RIKEN BioResource Research Center (BRC) and cultured in standard Dulbecco's modified Eagle' medium (DMEM high-glucose 4.5 g/l, Nacalai) with 10% FBS (Biosera), 1 × glutamine (Gibco), and 1 × Penicillin–Streptomycin (Gibco) at 37 °C humidified incubators with 5% CO2 supplied. For cell growth experiment under the respiratory condition, we used modified Dulbecco's modified Eagle' medium (No Glucose, Nacalai) with the addition of 4.5 g/l D(+)-galactose (Wako), 10% dialyzed FBS (Thermo Fisher Scientific), and 1 × Penicillin–Streptomycin (Gibco). The medium was changed routinely every 48 h. For the cell counting experiment, 0.5 million cells were seeded on a 10-cm dish for cell counting experiment. Cell counts started the next day after seeding for five consecutive days.
Sequence alignment, structure analysis, and phylogenetic analysis
Amino acid sequences were retrieved from UniProt (https://www.uniprot.org/). Alignments were generated using ClustalOmega (https://www.ebi.ac.uk/Tools/msa/clustalo/) with default settings. Alignments editing, color-coding, and phylogenetic analysis (by BLOUSUM62 matrix) were performed using the Jalview program. Domain organizations were retrieved from Pfam37 database (http://pfam.xfam.org/). HEMK1/2 sequence-specific substrate binding sites were generated by WebLogo (http://weblogo.threeplusone.com/)38. DeepMito, deep-learning approaches trained with dataset comprising known mitochondrial proteins, (http://busca.biocomp.unibo.it/deepmito/)39 and MitoFates, amino acid composition and physiochemical properties-based analysis, (http://mitf.cbrc.jp/MitoFates/cgi-bin/top.cgi)40 were used for predictions of mitochondrial targeting sequences. Disordered region and secondary structure prediction were conducted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/)41. Sequence alignments with secondary structure elements were generated by ESPript (http://espript.ibcp.fr/ESPript/ESPript/)42. All UniProt protein IDs used in this study are listed in Table S1.
DNA construction and establishment of HEMK1 KO cells
HEMK1 KO cell lines were generated by CRISPR-Cas9 mediated gene knockout. In brief, two sgRNAs (targeting HEMK1 exon 5 and exon 8) were cloned into pL-CRISPR.EFS.tRFP (Addgene #57819)43 and pKLV2-U6gRNA5(BbsI)-PGKpuro2ABFP-W (Addgene #67974)44, respectively. After verification by Sanger sequencings, naïve HeLa cells were transfected with both sgRNA containing plasmids by Lipofectamine 2000 (Thermo Fisher Scientific) as instructed by the manufacturer. At 48 h post-transfection, HeLa cells were sorted by flow cytometry for RFP and BFP dual positive populations. Generally, 200–300 cells were seeded onto a 6-cm dish and cultured until visible colonies were seen. Single clones were then expanded in a 24-well dish. HEMK1 KO clones were confirmed by PCR. For HEMK1 KO clone #3 and #4, Sanger sequencing was also performed because large genomic deletion was only found on one allele. See Table S1 for the sequencing results.
DNA fragments encoding four mtRFs were PCR-amplified from HeLa total cDNA using specific primers. DNA fragments encoding mtRFs were cloned into EcoRI-linearized pQCXIP vector (Clontech) by In-Fusion cloning (Clontech) with HA tag at the C-terminal of mtRFs. DNA fragment encoding HEMK1 cDNA was PCR-amplified from HeLa total cDNA by specific primers, and the PCR fragments was cloned into pcDNA3-cFLAG (Thermo Fisher Scientific) using EcoRI and NotI double digestion. HEMK1 mutant was generated by site-directed mutagenesis with Phusion polymerase (Thermo Fisher Scientific). For transient expression (HEMK1, HEMK1Y242E, and mtRF1L), HeLa cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) followed the protocol from the manufacturer. At 72 h post-transfection, cells were harvested with ice-cold PBS, and then lysed in RIPA buffer for SDS-PAGE analysis.
Stable cell line establishment
HEMK1 wild-type and the mutants were subcloned from pcDNA3-cFLAG (Thermo Fisher Scientific) constructs into EcoRI-linearized pQCXIN (Clontech) by In-Fusion cloning (Clontech). Viral particles were produced in HEK293T cells (cultured at the same conditions as HeLa cells) by pQCXIN/P and the Retroviral Expression System (Clontech). The medium was replaced on the next day after transfection. Viral particles were collected at 48 h post-transfection and filtered by 0.45-μm membrane (EMD). HeLa cells were infected by the viral particles (with 8 μg/ml polybrene, Sigma). At 24 h post-infection, cells were selected with neomycin (500 μg/ml for up to 14 days). Stable cell lines expressing mtRFs were generated similarly using the former established HEMK1 wild-type/mutants cell lines (selected with 2 μg/ml puromycin for 3 days). Primers and vectors used in this study were listed in Table S1.
Western blot analysis
Cells harvested using ice-cold PBS (with 1 × protease inhibitor cocktail [Nacalai] and 1 mM PMSF), were lysed by RIPA buffer (with 1 × protease inhibitor cocktail [Nacalai] and 1 mM PMSF) on ice for 5 min. Sonication was performed on ice subsequently. The lysate was centrifuged at 10,000g, 4 °C for 10 min. The supernatant was transferred to a new tube, and protein concentration was measured using the Bradford Protein Assay Kit (BioRad Laboratories). The lysate was denatured by NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) with 1 mM 2-mercaptoethanol at room temperature. Proteins were separated by SDS-PAGE and transferred onto PVDF membrane (Immobilon-FL-PVDF, Millipore). For imaging, both X-ray film (FUJIFILM) with chemiluminescence and ODYSSEY CLx (LI-COR Biosciences) with infrared fluorescence were used. In brief, PVDF membrane was blocked using TBS-T (1 × TBS and 0.1% Tween-20) with skimmed milk at room temperature for 1 h, followed by incubation with the primary antibody in TBS-T with 3% BSA at 4 °C overnight with mild shaking. The membrane was washed with TBS-T for 10 min twice before secondary antibody incubation. In the case when infrared fluorescence was used for detection, Tween-20 was omitted from the blocking buffer, and secondary antibody was used in the dark for 1 h at room temperature. Antibodies used in this study are listed in Table S1.
Immunofluorescence microscopy analysis
For the mtRFs and HEMK1 co-localization analysis in mitochondria, 5 × 105 of HeLa cells were reverse transfected using Fugene HD (Promega) with 0.5 μg each of HEMK1-cFLAG WT and individual mtRFs-cHA expressing plasmids followed the protocol from the manufacturer. For the HEMK1Y242E Mut cellular localization experiment, 0.5 μg HEMK1-cFLAG WT or HEMK1Y242E Mut expressing plasmid was used for transfection. The transfected cells were grown on sterilized cover glasses for 24 h. Then, the cells were first washed with PBS then fixed using 4% PFA at 37 °C for 10 min. Cells were washed with PBS twice, followed by permeabilization using PBS with 0.2% Triton X-100 for 10 min at room temperature. Then, cells were blocked with 3% goat serum in PBS at room temperature for 30 min. Cells then incubated with primary antibodies at 4 °C, overnight. Secondary antibodies were administered as guided by manufacturers. MitoTracker Red CMXRos reagent (200 nM, Thermo Fisher Scientific) was added directly into the media at 30 min before PFA fixation for mitochondria staining. Slides were mounted on slide glasses (Matsunami) with Prolong with DAPI mounting reagent (Thermo Fisher Scientific). Images were captured with 60× objective lens and processed by Olympus FV3000 system and its suite software. Exogenous HEMK1-cFLAG and mtRFs-cHA Double-expressing cells were selected for quantitation. Quantitation was described as x/y, where x was the number of cells showing the colocalized phenotype, y was the total number of the examined cells in which (1) both HEMK1-cFLAG and individual mtRF-cHA were co-expressed and (2) none of mtRFs-cHA and HEMK1-cFLAG were not detected in the nucleus in order to exclude abnormal phenotypes by too much expression of transgene(s). Antibodies used in this study are listed in Table S1.
Detection of mitochondrial membrane potential by flow cytometry
Experimental procedures, signal visualization, and normalization were adapted from previous studies with minor modifications on the cell seeding step45–47. Cells (naïve HeLa, HEMK1 KO clone #1, and HEMK1 KO clone #2) were seeded on 6-cm dishes at ~ 70% confluency. Then, after 16 h cell culture (not confluent), the cells were first washed with PBS once and then replaced with Mitotracker Red CMXRos (200 nM, Thermo Fisher Scientific) and Mitotracker Green FM (150 nM, Thermo Fisher Scientific) containing PBS with 5% FCS. Cells were incubated at 37 °C for 25 min. Then, cells were dissociated from the dishes by 0.5% trypsin and neutralized by PBS with 5% FCS. Cells were pelleted and resuspended in 0.3 ml of PBS with 5% FCS for flow cytometry analysis (BD FACSAria II, BD Biosciences). The value of Mitotracker Green FM was standardized by the mean of naïve HeLa, HEMK1 KO clone #1, and HEMK1 KO clone #2 experiments (10,000 cells data each). Then the value of Mitotracker Red CMXRos in each cell was normalized by the standardized value of Mitotracker Green FM from the cell. The statistical significance (P value) was calculated by the Mann–Whitney U-test, two-tail. The effect size (D value) was calculated by the Cohen's D test.
Immunoprecipitation for mass spectrometry (MS) analysis
Stable mtRFs and mtRFs with HEMK1 (co-)overexpressing cells were first lysed on ice for 10 min using lysis buffer A (50 mM Hepes pH 7.5, 140 mM KCl, 10% glycerol, 0.5% NP40, 1 × protease inhibitor cocktail [Nacalai], and 1 mM PMSF). The lysate was then sonicated with Ultrasonic Disruptor (TOMY UR-21P) with 70% output for 10 pluses on ice and precleared using Protein G Sepharose 4 Fast Flow beads (GE Healthcare) at 4 °C for 30 min with mild rotation. The supernatant was collected by 10,000 g spinning at 4 °C for 10 min and subsequently incubated with anti-HA antibody at 4 °C for 1 h and then with Protein G beads for 2 h. Then, beads were collected at 500g centrifugation and washed once with lysis buffer A and three times with bead wash buffer (20 mM Tris 6.8, 150 mM NaCl, 10% glycerol, and 1 mM PMSF). The beads were boiled in 1 × Laemmli buffer at 95 °C for 5 min for elution. Eluted proteins were separated on 15% Tris–glycine gel with MOPS buffer. Gels were stained using the Pierce Silver Stain for Mass Spectrometry kit (Thermo Fisher Scientific), and regions corresponding to different mtRFs were excised for mass spectrometry.
MS analysis
The excised gels were destained as instructed by the manufacturer. The proteins were digested in gel with trypsin (TPCK-treated, Worthington Biochemical Corporation). The peptide mixture was separated on a nanoflow LC (Easy nLC 1200; Thermo Fisher Scientific, Waltham, MA) using a nano-spray column (NTCC analytical column, C18, φ75 μm × 150 mm, 3 μm, Nikkyo Technos) with a linear gradient of 0–100% buffer B (80% aqueous acetonitrile containing 0.1% formic acid) at a flow rate of 300 nl/min over 20 min. The elution was sprayed online to a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific) equipped with a nano-spray ion source. The MS/MS data were acquired in a Top10 data-dependent manner. Proteins were identified using Proteome Discoverer 2.2 (Thermo Fisher Scientific) with MASCOT program 2.6 (Matrix science) using an in-house database. The mass spectra and mass chromatograms were drawn by Qual Browser 4.1.50 (Thermo Fisher Scientific).
qPCR analysis
MT-ND2 and ALU repeats were selected for quantification of mtDNA and gDNA content, respectively. Total DNA was extracted with the Nucleospin DNA isolation kit (Macherey Nagel). One nanogram of total DNA was used for qPCR with Power SYBR Green PCR master mix (Thermo Fisher Scientific). Step one plus real-time PCR (Applied Biosystems) was performed according to the manufacturer's instruction. Data were processed and plotted with Microsoft Excel. For primers used in qPCR, see Table S1.
On-gel mito-FUNCAT assay
Cells (naïve HeLa, HEMK1 KO clone #1, clone #2, and HEMK1 KO clone #1 rescued lines) were grown on 6-well dish in standard culture media (-antibiotics) until the day of assay. Cells were washed with protein labelling media (DMEM [ThermoFisher Scientific], 10% FBS, 48 mg/ml l-cystine dihydrochloride [Nacalai], and 862 mg/ml l-alanyl-l-glutamine [Nacalai]) once and then incubated in methionine-free medium with L-homopropargylglycine 50 μM (Jena Bioscience) and anisomycin 100 μg/ml (Alomone Labs) for 4 h at 37 °C humidified incubators with 5% CO2 supplied. Cells were lysed with ice-chilled lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100). Lysates were cleared by centrifugation for 10 min at 20,000g, 4 °C. The supernatants were used for CLICK reaction with Click-iT Cell Reaction Buffer Kit (ThermoFisher Scientific) and IRdye800CW Azide (LI-COR Biosciences), according to the manufacturer's instruction. After the CLICK reaction, free IRdye800CW Azide was removed using illustra MicroSpin G-25 Column (GE Healthcare) equilibrated with lysis buffer containing 1 mM DTT. Samples were denatured at 50 °C with 4 × Protein Loading Buffer (LI-COR Biosciences) with 10% 2-mercaptoethanol (Nacalai). Samples were loaded onto the polyacrylamide gel (NuPAGE, 4 to 12%, Bis–Tris [ThermoFisher Scientific]) with 5 μl protein ladder marker (NIPPON Genetics). Electrophoresis was conducted in 1 × MES buffer (ThermoFisher Scientific) for 22 min at 200 V constant. The gel was fixed with fixation reagent (40% ethanol, 10% acetic acid, and 50% ddH2O) for 15 min at room temperature. After fixation, the gel was destained with MilliQ water and subsequently imaged by Odyssey CLx (LI-COR Biosciences) with IR800 channel for the newly synthesized mitochondrial polypeptides. For the total protein imaging, the same gel (after IR800-channel imaging) was incubated with GelCode Blue reagent (ThermoFisher) for 15–60 min then destained with MilliQ water. The destained gel was imaged by Odyssey CLx (LI-COR Biosciences) with IR700 channel.
Supplementary Information
Acknowledgements
We thank the staff of the Support Unit for Bio-Material Analysis (BMA) at RIKEN Center for Brain Science (CBS) Research Resources Division (RRD) for DNA sequencing and flow cytometry. We are also grateful to Mingming Chen for technical help and Hironori Saito for statistical analysis. This study used the following materials: pL-CRISPR.EFS.tRFP from Benjamin Ebert and pKLV2-U6gRNA5(Bbsl)-PGKpuro2ABFP from Kosuke Yusa. We thank Asce Zheng for typesetting. We would also like to thank our colleagues at Shinkai laboratory and members of Iwasaki laboratory for their support and valuable comments.
Author contributions
Q.F., S.I., and Y.S. wrote the original and revised manuscript. Q.F., Y.K., and A.Y. performed the experiments other than MS analysis. T.S. and N.D. performed MS analysis. T.S. produced some experimental materials and performed initial experiments. S.I., T.S., and Y.S. guided entire experiments.
Funding
This work was supported by a RIKEN internal research fund for Y.S. Q.F. was funded by the International Program Associate (IPA) Fellowship and an International Joint Graduate School Program of RIKEN. S.I. was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (a Grant-in-Aid for Transformative Research Areas [B], JP20H05784), the Japan Society for the Promotion of Science (JSPS) (a Grant-in-Aid for Young Scientists [A], JP17H04998; a Challenging Research [Exploratory], JP19K22406), the Japan Agency for Medical Research and Development (AMED) (AMED-CREST, JP21gm1410001), and RIKEN (Pioneering Projects "Biology of Intracellular Environments" and Ageing Project). Y.K. was supported by a Grant-in-Aid for JSPS Fellows (JP20J10665) from JSPS. Y.K. was a RIKEN Junior Research Associate Program recipient and a JSPS Research Fellow (DC2).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08061-y.
References
- 1.Murn J, Shi Y. The winding path of protein methylation research: Milestones and new frontiers. Nat. Rev. Mol. Cell Biol. 2017;18:517–527. doi: 10.1038/nrm.2017.35. [DOI] [PubMed] [Google Scholar]
- 2.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell. Mol. Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 6.Heurgue-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graille M, et al. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005;20:917–927. doi: 10.1016/j.molcel.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 8.Pierson WE, et al. Uniformity of peptide release is maintained by methylation of release factors. Cell Rep. 2016;17:11–18. doi: 10.1016/j.celrep.2016.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng F, Jin H. Conformation of methylated GGQ in the peptidyl transferase center during translation termination. Sci. Rep. 2018;8:2349. doi: 10.1038/s41598-018-20107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakahigashi K, et al. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. USA. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Tran N, et al. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018;46:8483–8499. doi: 10.1093/nar/gky638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polevoda B, Span L, Sherman F. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 2006;281:2562–2571. doi: 10.1074/jbc.M507651200. [DOI] [PubMed] [Google Scholar]
- 13.Ishizawa T, Nozaki Y, Ueda T, Takeuchi N. The human mitochondrial translation release factor HMRF1L is methylated in the GGQ motif by the methyltransferase HMPrmC. Biochem. Biophys. Res. Commun. 2008;373:99–103. doi: 10.1016/j.bbrc.2008.05.176. [DOI] [PubMed] [Google Scholar]
- 14.Liger D, et al. Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein. Nucleic Acids Res. 2011;39:6249–6259. doi: 10.1093/nar/gkr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu P, et al. Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality. Mol. Cell. Biol. 2010;30:4245–4253. doi: 10.1128/MCB.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusevic D, Kudithipudi S, Jeltsch A. Substrate specificity of the HEMK2 protein glutamine methyltransferase and identification of novel substrates. J. Biol. Chem. 2016;291:6124–6133. doi: 10.1074/jbc.M115.711952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodcock CB, Yu D, Zhang X, Cheng X. Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112. Cell Discov. 2019;5:50. doi: 10.1038/s41421-019-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao CL, et al. N(6)-Methyladenine DNA modification in the human genome. Mol. Cell. 2018;71:306–318e307. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Kummer E, Ban N. Mechanisms and regulation of protein synthesis in mitochondria. Nat. Rev. Mol. Cell Biol. 2021;22:307–325. doi: 10.1038/s41580-021-00332-2. [DOI] [PubMed] [Google Scholar]
- 20.Richter R, et al. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antonicka H, et al. Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am. J. Hum. Genet. 2010;87:115–122. doi: 10.1016/j.ajhg.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soleimanpour-Lichaei HR, et al. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol. Cell. 2007;27:745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kummer E, Schubert KN, Schoenhut T, Scaiola A, Ban N. Structural basis of translation termination, rescue, and recycling in mammalian mitochondria. Mol. Cell. 2021;81:2566–2582 e2566. doi: 10.1016/j.molcel.2021.03.042. [DOI] [PubMed] [Google Scholar]
- 24.Desai N, et al. Elongational stalling activates mitoribosome-associated quality control. Science. 2020;370:1105–1110. doi: 10.1126/science.abc7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zorkau M, Albus CA, Berlinguer-Palmini R, Chrzanowska-Lightowlers ZMA, Lightowlers RN. High-resolution imaging reveals compartmentalization of mitochondrial protein synthesis in cultured human cells. Proc. Natl. Acad. Sci. USA. 2021 doi: 10.1073/pnas.2008778118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009;37:749–761. doi: 10.1093/nar/gkn993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteiro LB, Davanzo GG, de Aguiar CF, Moraes-Vieira PMM. Using flow cytometry for mitochondrial assays. MethodsX. 2020;7:100938. doi: 10.1016/j.mex.2020.100938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon BC, et al. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148:752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mora L, Heurgue-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 2007;282:35638–35645. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- 31.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akabane S, Ueda T, Nierhaus KH, Takeuchi N. Ribosome rescue and translation termination at non-standard stop codons by ICT1 in mammalian mitochondria. PLoS Genet. 2014;10:e1004616. doi: 10.1371/journal.pgen.1004616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrzanowska-Lightowlers ZM, Lightowlers RN. Response to "ribosome rescue and translation termination at non-standard stop codons by ict1 in mammalian mitochondria". PLoS Genet. 2015;11:e1005227. doi: 10.1371/journal.pgen.1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi N, Nierhaus KH. Response to the formal letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers regarding our article "ribosome rescue and translation termination at non-standard stop codons by ICT1 in mammalian mitochondria". PLoS Genet. 2015;11:e1005218. doi: 10.1371/journal.pgen.1005218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greber BJ, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 36.Desai N, Brown A, Amunts A, Ramakrishnan V. The structure of the yeast mitochondrial ribosome. Science. 2017;355:528–531. doi: 10.1126/science.aal2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savojardo C, Bruciaferri N, Tartari G, Martelli PL, Casadio R. DeepMito: Accurate prediction of protein sub-mitochondrial localization using convolutional neural networks. Bioinformatics. 2020;36:56–64. doi: 10.1093/bioinformatics/btz512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukasawa Y, et al. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones DT, Cozzetto D. DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heckl D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzelepis K, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puleston D. Detection of mitochondrial mass, damage, and reactive oxygen species by flow cytometry. Cold Spring Harbor Protoc. 2015;2015:pdb prot086298. doi: 10.1101/pdb.prot086298. [DOI] [PubMed] [Google Scholar]
- 46.Hoshino S, et al. A stalled-ribosome rescue factor Pth3 is required for mitochondrial translation against antibiotics in Saccharomyces cerevisiae. Commun. Biol. 2021;4:300. doi: 10.1038/s42003-021-01835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.