Abstract
An open reading frame (ORF) homologous to norA was insertionally inactivated with cat in a fluoroquinolone-resistant pneumococcus with an efflux phenotype; this inactivation caused reversion to drug sensitivity. The ORF product has 24% amino acid sequence identity each to NorA and Bmr, which suggests that it is a major facilitator system pump of the 12-transmembrane-segment class.
Recent studies in our laboratories and elsewhere have described a putative efflux mechanism as a cause of low-level fluoroquinolone resistance in pneumococci (1–3, 13). As for other well-described multidrug efflux pumps, such as NorA and Bmr (9), concomitant reduced susceptibility to several diverse compounds is observed, and the pump is inhibited by reserpine. However, the genetic basis of this mechanism of resistance is not known. We therefore undertook this study to determine whether a homologue of norA or bmr is responsible for efflux-mediated resistance in Streptococcus pneumoniae.
Nucleotide sequence data from the Institute of Genome Research (Rockville, Md.; http://www.tigr.org) S. pneumoniae genome project were screened against the EMBL prokaryote library by using FASTA (10). One genomic sequence showed some homology with norA. An open reading frame (ORF) of 1,200 bp within this sequence showed 52% homology to a 430-nucleotide overlap with norA. By using PCR, DNA from 53 bp 5′ of the start codon to 1,186 bp into this ORF was amplified from the chromosome of S. pneumoniae R6. The primer sequences were 5′GGATCCGATATCAGAACCAGCTTCTTA3′ (forward; BamHI/EcoRV linker underlined) and 5′GGTACCAAGCTTTTAATAATGTTCGAAA3′ (reverse; KpnI/HindIII linker underlined). The conditions used were an initial extended denaturation cycle of 94°C for 5 min followed by 30 cycles of 1 min at 50°C, 1 min at 72°C, and 1 min at 94°C.
The ORF PCR product was cloned into pBluescript SK− (Stratagene, La Jolla, Calif.) and transformed into Escherichia coli XL1-Blue (Stratagene). cat excised from pR326 (4) was ligated into the cloned ORF. The resultant plasmid (pNBT5CAT), maintained in E. coli XL1-Blue, contained cat cloned into the AccI site of the ORF, flanked by 478 and 764 bp of ORF sequence. pNBT5CAT was used to transform (6) S. pneumoniae R6N. Strain R6N is a strain with an efflux phenotype derived by transformation of strain R6 with DNA from strain 1N27; strain R6 is an easily transformed laboratory strain; strain 1N27 is spontaneous mutant (selected by norfloxacin) of S. pneumoniae ATCC 49619 (3) with an efflux phenotype and is not easily transformable. Transformants of R6N were selected on Columbia agar supplemented with 5% horse blood and containing chloramphenicol (8 μg/ml). The antibiotic susceptibility of transformants was tested by an agar dilution method (3) using Iso-Sensitest agar (Oxoid, Basingstoke, United Kingdom) supplemented with 5% horse blood and an inoculum of 104 CFU per spot. Following incubation at 37°C in 4 to 6% CO2 for 20 h, the MIC was taken as the lowest concentration completely inhibiting growth.
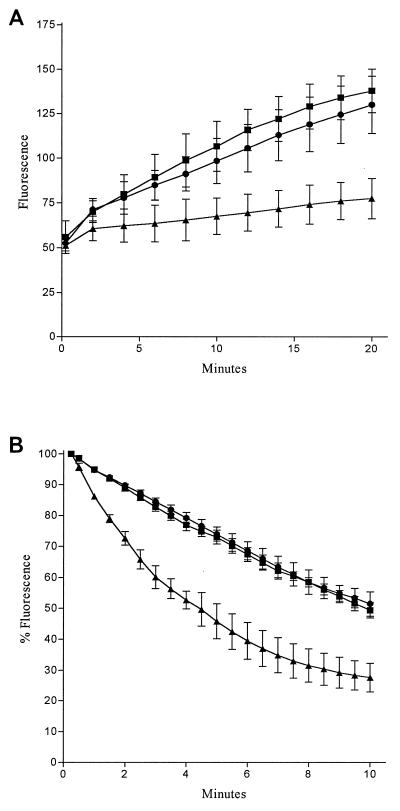
Table 1 shows the antibiotic susceptibilities of isogenic derivatives of R6. The efflux phenotype of R6N (increased MICs of norfloxacin, ciprofloxacin, acriflavine, and ethidium bromide [EtBr]; fourfold reduction of norfloxacin MIC in the presence of reserpine) (2, 3) was reversed to the fully susceptible phenotype in R6N-cat. Confirmation that these changes in susceptibility were due to altered efflux was made by demonstrating an increase in accumulation and a decrease in efflux of EtBr (1, 3) in R6N-cat in comparison to those of R6N. Briefly, for accumulation, bacteria were suspended in uptake buffer (NaCl, 110 mM; KCl, 7 mM; NH4Cl, 50 mM; Na2HPO4, 0.4 mM; glucose, 0.2%; Tris base, 52 mM adjusted to pH 7.5 with HCl) and then exposed to EtBr (2 μg/ml). The increase in fluorescence as EtBr entered the cells was recorded fluorometrically (excitation λ = 530 nm; emission λ = 600 nm) at 30°C. EtBr accumulation at 20 min was significantly greater (Student’s t test, P < 0.02) in R6N-cat than in R6N (Fig. 1A). For determining EtBr loss, bacteria were first exposed to EtBr (2 μg/ml) in the presence of reserpine (1 μg/ml) for 20 min at 37°C. The cells were then pelleted by centrifugation and resuspended in fresh uptake buffer. The loss of EtBr from the cells was measured as a decrease in fluorescence. EtBr efflux was significantly lower in R6N-cat than in R6N (Fig. 1B). There was no significant difference in the accumulation of efflux of EtBr between R6 and R6N-cat.
TABLE 1.
Characteristics and antibiotic susceptibilities of S. pneumoniae strains
| Strain | Characteristic | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Nor | Nor + R | Cip | Moxi | Spar | EtBr | Acri | Cm | ||
| R6 | Transformation recipient | 2 | 2 | 0.5 | 0.12 | 0.25 | 2 | 4 | 4 |
| R6N | R6 transformed with 1N27 DNA (3) | 16 | 4 | 2.0 | 0.12 | 0.25 | 16 | 16 | 2 |
| R6N-cat | R6N transformed with pNBT5CAT | 2 | 2 | 0.5 | 0.12 | 0.25 | 2 | 4 | 32 |
Nor, norfloxacin; R, reserpine; Cip, ciprofloxacin; Moxi, moxifloxacin (Bay 12-8039); Spar, sparfloxacin; Acri, acriflavine; Cm, chloramphenicol.
FIG. 1.
EtBr accumulation (A) and efflux (B) in isogenic pneumococci. Fluorescence is directly proportion to EtBr concentration. For accumulation (A), fluorescence is in arbitrary units; for efflux (B), fluorescence is expressed as a percentage of that at the first time point (15 s) for each strain. Each point represents the mean of three separate determinations; error bars show standard deviations. Symbols: ■, R6, susceptible strain; ▴, R6N, efflux strain; •, R6N-cat, strain with putative efflux pump gene insertionally inactivated by cat.
PCR of chromosomal DNA using the above primers and ClaI restriction digest analysis of the resultant amplimers showed that the putative efflux pump gene was present in R6, R6N, and R6N-cat. In addition, PCR product digestion with ClaI showed that the cat orientation in the pump gene in R6N-cat was such that its transcription was in the opposite direction to that predicted for the pump gene. Southern blotting was performed on PstI- and HindIII-digested chromosomal DNA (from R6, R6N, and R6N-cat) (13). The Southern blot was probed with an efflux gene probe (made by using the above PCR product) or a cat probe (derived by SmaI and HindII digestion from p326). Probes were labelled with the ECL random prime labelling system (Amersham Life Science Ltd., Little Chalfont, United Kingdom). Hybridizations were carried out for 18 h at 60°C; the membrane was washed three times for 15 min at 60°C in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate and then washed three times for 15 min at 60°C in 0.1× SSC containing 0.1% sodium dodecyl sulfate. Probe signal generation was detected on chemiluminescence film in accordance with the manufacturer’s instructions (Amersham Life Science Ltd.). Probing with the efflux gene demonstrated that there was only one copy of the efflux pump gene on the chromosome of R6 and its isogenic derivatives. The efflux pump gene probe and the cat probe both hybridized to the same 1.8-kb PstI fragment of R6N-cat chromosomal DNA. Finally, DNA sequencing of PCR products generated from chromosomal DNA of R6N and R6N-cat also confirmed that cat had inserted into the AccI site of the putative efflux gene in the latter strain.
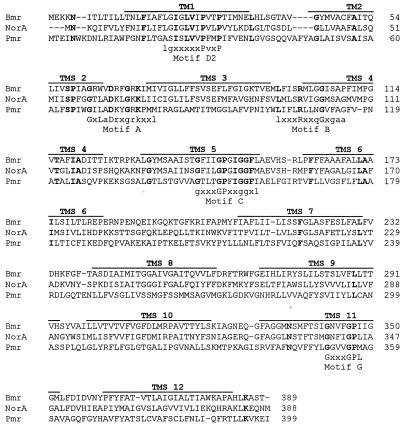
Alignment of the putative pneumococcal efflux pump sequence with the amino acid sequences of Bmr and NorA is shown in Fig. 2. Overall, it has 24% sequence identity each with Bmr and NorA. Transmembrane segment (TMS) predictions using SOSUI (7) indicate that the pneumococcal efflux pump has 12 TMSs. The TMSs are in positions comparable to those in other 12-TMS proton-dependent efflux pumps (9). Further evidence that it is an efflux pump comes from the finding that it contains sequences similar to those of motifs A, B, C, D2, and G (Fig. 2) that have been described in other 12-TMS proton-dependent efflux pumps (8). These motifs are thought to be essential for protein structure and function but are unlikely to be involved in pump substrate specificity (9). Analysis of sequence found at the 5′ end of the ORF revealed the presence of promoter, transcription start, and Shine-Dalgarno sequences consistent with those found in pneumococci (11). In common with other efflux pumps, the pneumococcal pump gene was found in all strains examined by PCR. It is therefore likely that the associated drug resistance phenotype results from increased pump expression.
FIG. 2.
Amino acid sequence alignment of B. subtilis Bmr (EMBL accession no. M33768), S. aureus NorA (EMBL accession no. D90119), and the pneumococcal efflux pump (Pmr). Alignments were performed by using Multalin (5). Amino acids in bold are those common to the three proteins. Motifs A, B, C, D2, and G are those described by Paulsen and Skurray (8). TMS predictions are from SOSUI predictions (7) of the pneumococcal sequence.
In summary, we have insertionally inactivated an ORF that had sequence homology with norA in a pneumococcus with a drug efflux phenotype. This inactivation resulted in the loss of the efflux phenotype. The corresponding amino acid sequence of this ORF is consistent with it encoding a 12-TMS proton-dependent multidrug efflux pump of the major facilitator family. Since there may be other multidrug efflux pumps in pneumococci, we propose calling this pump PmrA (pneumococcal multidrug resistance protein).
Nucleotide sequence accession number.
The efflux gene sequence is deposited in the EMBL sequence database (accession no. AJ007367; 16 June 1998).
Acknowledgments
We are grateful to A. Dahlhoff of Bayer AG for financial support.
We also thank C. G. Dowson and V. Barcus for their advice on pneumococcal transformation and the gift of competence-stimulating peptide. J.-P. Claverys kindly sent us pR326.
REFERENCES
- 1.Baranova N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenwald N P, Gill M J, Wise R. The effect of reserpine, an inhibitor of multi-drug efflux pumps, on the in-vitro susceptibilities of fluoroquinolone-resistant strains of Streptococcus pneumoniae to norfloxacin. J Antimicrob Chemother. 1997;40:458–460. doi: 10.1093/jac/40.3.458. [DOI] [PubMed] [Google Scholar]
- 3.Brenwald N P, Gill M J, Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverys J-P, Dintilhac A, Pestova E K, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 5.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haverstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 8.Paulsen D, Skurray R A. Topology, structure and evolution of two families of proteins involved in antibiotic and antiseptic resistance in eukaryotes and prokaryotes—an analysis. Gene. 1993;124:1–11. doi: 10.1016/0378-1119(93)90755-r. [DOI] [PubMed] [Google Scholar]
- 9.Paulsen I T, Brown M H, Skurry R A. Proton-dependent multi-drug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 11.Sanford A L. Cloning and expression of pneumococcal genes in Streptococcus pneumoniae. Microb Drug Res. 1997;3:327–337. doi: 10.1089/mdr.1997.3.327. [DOI] [PubMed] [Google Scholar]
- 12.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 13.Zeller V, Janoir C, Kitzis M, Gutmann L, Moreau N J. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1973–1978. doi: 10.1128/aac.41.9.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]