The asthmatic airway inflammation is regulated by an elaborate network of cytokines (1). Among these, interleukin (IL)-4 and IL-5 are thought to be central players in perpetuating a Th2-type immune response. In an autocrine fashion, IL-4 regulates the expansion and activation of Th2 lymphocytes, a subset of CD4+ T helper cells. IL-4 also stimulates the expression of IL-5, the cytokine specifically responsible for differentiation, activation and survival of eosinophil granulocytes in the inflamed tissue. Although their exact role is still subject to debate, eosinophils have been regarded to have a major pathogenic importance in allergic inflammation and asthma. Effective targeting of this pathway has been therefore the subject of intensive research in the past two decades. The IL-4-driven signaling pathway has been well mapped: upon binding to the IL-4 receptor, IL-4 activates the receptor-associated kinases JAK-1 and JAK-3 (Fig. 1A). Tyrosine phosphorylation of STAT-6 by JAK-1 and JAK-3 induces dimerization and nuclear translocation of STAT-6 (Fig. 1B). STAT-6 then activates transcription of a number of proinflammatory genes (Fig. 1C) including that of GATA-3 (GATA binding protein-3), a master transcriptional regulator of IL-5 expression [reviewed in (1)]. Datta et al. (2) reported in this issue of Allergy that the nuclear protein PARP-1 [poly(ADP-ribose) polymerase-1] is a novel regulator of the IL-4-driven signaling pathway leading to IL-5 expression. They proposed a mechanism of action through enhancement of GATA-3 expression by PARP-1 mediated stabilization of STAT-6 protein (Fig. 1D).
Figure 1.
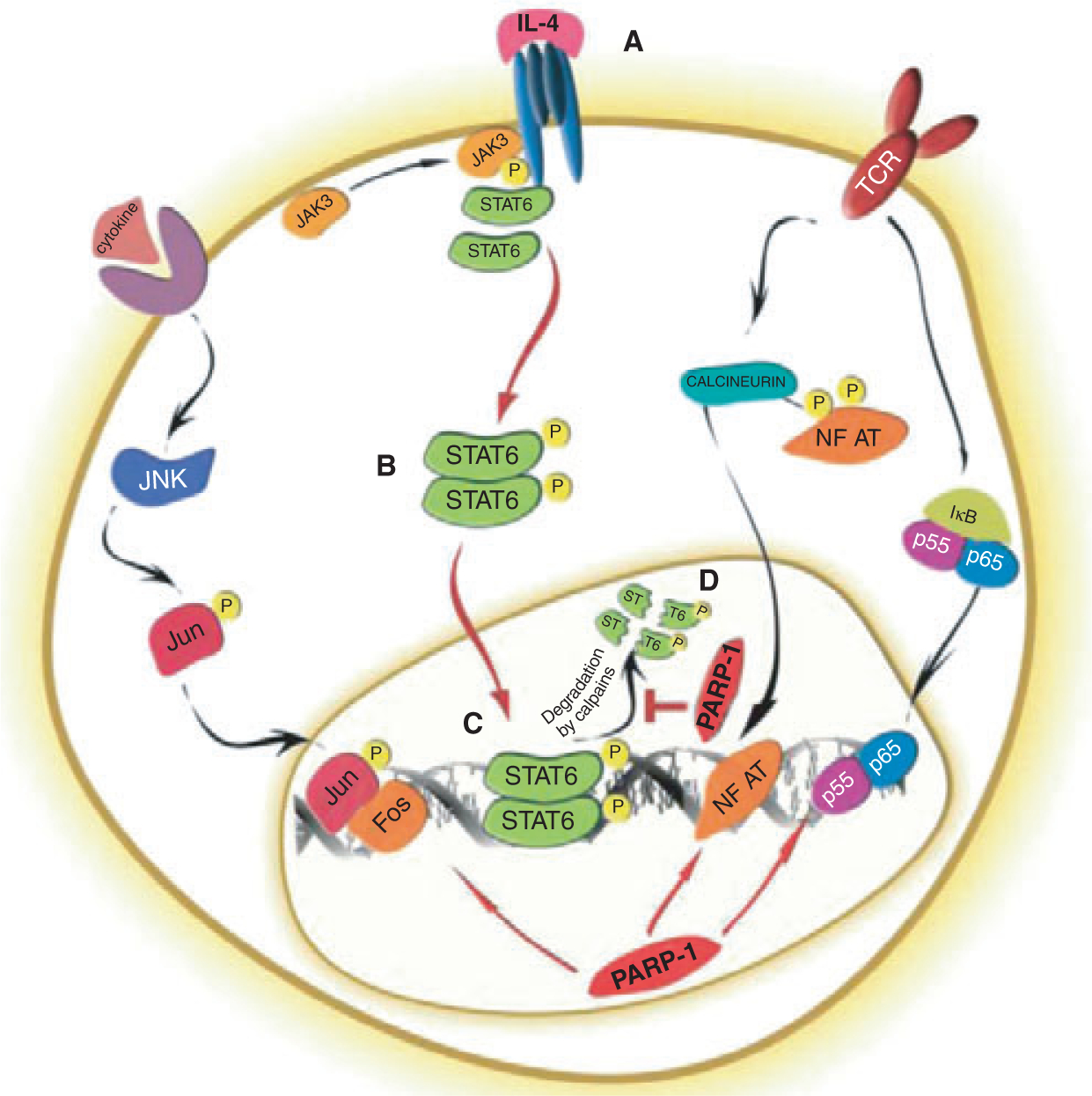
Role of PARP-1 in regulating the STAT-6 signaling cascade. PARP-1 regulates the AP-1, NFκB, NF-AT as well as STAT-6 transcription pathways during proinflammatory T-cell responses. The putative role of PARP-1 is illustrated in the STAT-6 signaling pathway. (A) Binding of IL-4 induces IL-4R heterodimerization and activates JAK1 (constitutively associated with the α chain) and JAK3 (constitutively associated with the γ chain of the IL-4R). (B) JAK1 and JAK3 are activated by trans-phosphorylation of the specific and conserved tyrosine residues found in their activation loops. The activated JAK kinases phosphorylate multiple tyrosine residues in the cytoplasmic domain of IL-4Rα, which facilitates the recruitment of the IL-4Rα-specific transcription factor STAT-6 via its SH2 domain. STAT-6 then homodimerizes upon phosphorylation by JAK kinases and translocates into the nucleus. (C) Nuclear STAT-6 binds to and activates the promoter of various proinflammatory genes (such as GATA3, not shown here). (D) PARP-1 stabilizes STAT6 by preventing its calpain-mediated cleavage. PARP-1 acts downstream of JAK1 and JAK3 by stabilizing STAT-6 in activated T cells.
Poly(ADP-ribosyl)ation
PARP-1 is the founding member of the PARP enzyme family consisting of 17 homologues. (Of note, the enzyme family members share a conserved 50 aa sequence found in the catalytic domain of PARP1, but most members may function as mono- or oligo-ADP-ribosyltransferases rather than poly-ADP-ribosyl-transferases.) PARP enzymes covalently modify target proteins by cleaving NAD+ into nicotinamide and ADP-ribose and attaching the latter onto glutamate or aspartate residues of target proteins (3). In turn, PARPs cleave further NAD+ molecules and elongate the ADP-ribose chain into a branched poly(ADP-ribose) (PAR) polymer consisting of a maximum of approximately 250 monomers. PARylation is transient as the polymer is quickly degraded by PARG enzymes [poly(ADP-ribose) glycohydrolases]. PARG activity results in ADP-ribose monomers and also in free polymer as PARG can cleave off bigger chunks because of endoglycosidase activity. Free polymer may also function as a signaling molecule by non-covalent binding to PAR binding motives found in various proteins (3). The PAR polymer confers negative charge to the acceptor proteins and thereby changes their physico-chemical behavior usually leading to inhibition of function. For example, auto-PARylation of PARP-1 during recognition of DNA breaks leads to inhibition of the enzymatic activity and detachment from the DNA ends (4). PARylation is a multifunctional regulatory mechanism that has been implicated in chromatin organization and remodeling, DNA repair, replication, transcription, differentiation and cell death, only to name a few (4).
PARylation in allergic inflammation
PARP-1 has been shown to mediate inflammation in a wide range of animal models (4). Using pharmacological inhibitors and/or mice deficient in PARP-1, it has been demonstrated that inhibition of the enzyme or genetic ablation of its gene protected against tissue injury in colitis, arthritis, uveitis, contact dermatitis as well as septic or endotoxin shock. Treatment with PARP inhibitors or lacking a functional PARP-1 gene resulted in suppressed migration of granulocytes, reduced expression of inflammatory cytokines, chemokines, adhesion molecules, inducible nitric oxide synthase, and matrix metalloproteinases in animals (4).
The question whether PARP-1/PARylation may also contribute to Th2-mediated diseases such as asthma was investigated by several different groups (5–9). Increased PARP-1 protein expression and activation has been reported in the ovalbumin-induced airway inflammation model (5) suggesting direct involvement of the enzyme in allergic inflammation. Using mouse and guinea pig models and ovalbumin as allergen, pharmacological inhibition by 3-aminobenzamide (3-AB), PJ-34 and 5-aminoisoquinoline-1(2H)-one or the PARP-1 knockout phenotype attenuated allergen-induced lung injury. Improvement of the respiratory abnormalities (including cough, dyspnea, airway obstruction, air space enlargement, leukocyte infiltration, mast cell degranulation, oxidative and nitrosative stress, and inflammatory cytokine and chemokine release) indicated the effectiveness of PARP inhibition/deficiency in these models. The potent PARP inhibitor thieno[2,3-c]isoquinolin-5-one (TIQ-A) was also found to exert anti-inflammatory effects (2) even when administered after the antigen challenge (9). Thus, the involvement of PARP-1 and PARylation in animal models of asthma has been unequivocally demonstrated, but the molecular pathways need further elucidation.
Role of PARP-1 in inflammatory signaling
The key molecular component of the anti-inflammatory role of PARP inhibition is the connection between PARP-1 and NFκB, the master transcriptional regulator of inflammatory mediators. PARP-1 has been identified as a coactivator of NFκB (10). Interestingly, this coactivator function of PARP-1 does not appear to require enzymatic activity (11) raising the possibility that the suppressed inflammation caused by PARP inhibitors or the PARP-1 knockout phenotype is because of a different mechanism. Other transcription factors such as AP1 and NFAT that are important mediators of inflammatory diseases have also been shown to be regulated by PARP-1 (12–14). However, the importance of the PARP-1-AP-1 and PARP-1-NFAT connections has not yet been investigated in vivo. As NFκB, AP-1 and NFAT also play a role in allergic inflammation, it is plausible to hypothesize that their interaction with PARP-1 may be important in the effector phase of asthma.
Datta et al. probed the IL-4R → JAK1/3 → STAT-6 → GATA-3 → IL-5 mRNA pathway to unravel the mechanisms underlying the anti-inflammatory effects of PARP-1 inhibition or deficiency in an allergen-induced mouse model. The study stemmed from a previous observation of the authors that PARP-1 regulated eosinophil recruitment downstream of IL-4 and upstream of IL-5. The authors here found that PARP-1 deficiency was associated with reduction in IL-5 mRNA upon allergen exposure after IL-4 receptor engagement but independently of JAK1/JAK3 activation. STAT-6 protein (but not mRNA) was severely suppressed in spleens of PARP-1−/− mice and this PARP-1 effect required enzymatic activity. The dowregulation of STAT-6 coincided with reduced GATA-3 mRNA and protein expression and occupancy of its binding site on the IL-5 gene-promoter. IL-4 was sufficient to induce STAT-6 downregulation in both PARP-1−/− mice and isolated splenocytes. These results by Datta et al. (2) demonstrated that PARP-1 acts downstream of JAK1/3 by stabilizing STAT-6 protein activation in an allergen-stimulation-dependent manner.
Role of PARP-1 in protein stabilization or degradation
Another interesting finding in this study was that STAT-6 degradation was mediated by calpains rather than the proteasome system. Calpains have been implicated in the degradation of nuclear transcription factors such as nuclear IkBα (15). However, the involvement of the nuclear proteasome cannot be ruled out entirely as a potential mechanism in the degradation of nuclear STAT-6. Transcription factor translocation into the nucleus and nuclear action is usually followed by inactivation and degradation, often mediated by the proteasomal system. This system contains many components, including the 20S and 26S proteasomes, numerous regulators and the ubiquitination machinery and is the major proteolytic mechanism that catalyzes 70–80% of the intracellular protein degradation (16). As it is potentially dangerous for a cell or for cellular proteins to have an active protease in the cytosol or nucleus, proteasomal degradation is strongly controlled by specific pathways mostly through ubiqitination. In the nucleus, several (nuclear-)specific proteasomal regulators as the PA28γ, the PA200 and, interestingly, PARP-1 mediated PARylation are known (16). Indeed PARP-1 mediated PARylation is able to activate the proteasome itself (17), whereas PARylation of potential substrate proteins can inhibit proteasomal degradation (18). Therefore, PARP-1 mediated PARylation can contribute to either the degradation (via proteasome activation), or the stabilization of substrate proteins. Such a PARP-1 mediated stabilization was already demonstrated for HIF1α (19) or p53 (20).
Therapeutic considerations
Similarly to Th1-mediated inflammations, allergic diseases such as asthma now join the growing list of disease models where PARP inhibitors may have therapeutic benefit. It is important however to consider the involvement of this enzyme in DNA strand break repair and because of this effect special precautions need to be taken before PARP inhibitors are tested in humans. So far most clinical trials with PARP inhibitors targeted patients with cancer. Moreover, acute life-threatening diseases such as myocardial or brain ischemia-reperfusion injury are also among the conditions where clinical trials test the potential therapeutic value of PARP inhibition. Whether patients suffering from chronic diseases such as asthma or arthritis may also benefit from this novel therapeutic modality, remains to be seen.
Acknowledgements
Work in the authors’ laboratory is supported by the following grants OTKA, K73003, K60780, K82009, TAMOP-4.2.2-08/1-2008-0019 and TÁMOP 4.2.1./B-09/1/KONV-2010-0007. TG was supported by DFG. AH is supported by R01AI072197, RC1ES018505, CEET.
References
- 1.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest 2008;118:3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datta R, Naura AS, Zerfaoui M, Oumouna M, Kim H, Ju J et al. PARP-1 deficiency blocks allergen-induced IL-5 expression by promoting a calpain-dependent degradation of STAT-6. Allergy 2011; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 2008;13:3046–3082. [DOI] [PubMed] [Google Scholar]
- 4.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev 2002;54:375–429. [DOI] [PubMed] [Google Scholar]
- 5.Havranek T, Aujla PK, Nickola TJ, Rose MC, Scavo LM. Increased poly(ADP-ribose) polymerase (PARP)-1 expression and activity are associated with inflammation but not goblet cell metaplasia in murine models of allergen-induced airway inflammation. Exp Lung Res 2010;36:381–389. [DOI] [PubMed] [Google Scholar]
- 6.Boulares AH, Zoltoski AJ, Sherif ZA, Jolly P, Massaro D, Smulson ME. Gene knockout or pharmacological inhibition of poly(ADP-ribose) polymerase-1 prevents lung inflammation in a murine model of asthma. Am J Respir Cell Mol Biol 2003;28:322–329. [DOI] [PubMed] [Google Scholar]
- 7.Virag L, Bai P, Bak I, Pacher P, Mabley JG, Liaudet L et al. Effects of poly(ADP-ribose) polymerase inhibition on inflammatory cell migration in a murine model of asthma. Med Sci Monit 2004;10:BR77–BR83. [PubMed] [Google Scholar]
- 8.Suzuki Y, Masini E, Mazzocca C, Cuzzocrea S, Ciampa A, Suzuki H et al. Inhibition of Poly(ADP-Ribose) Polymerase Prevents Allergen-Induced Asthma-Like Reaction in Sensitized Guinea Pigs. J Pharmacol Exp Ther 2004;311:1241–1248 [DOI] [PubMed] [Google Scholar]
- 9.Naura AS, Hans CP, Zerfaoui M, You D, Cormier SA, Oumouna M et al. Post-allergen challenge inhibition of poly(ADP-ribose) polymerase harbors therapeutic potential for treatment of allergic airway inflammation. Clin Exp Allergy 2008;38:839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver FJ, Menissier-De MJ, Nacci C, Decker P, Andriantsitohaina R, Muller S et al. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J 1999;18: 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem 2001;276:45588–45597. [DOI] [PubMed] [Google Scholar]
- 12.Andreone TL, O’Connor M, Denenberg A, Hake PW, Zingarelli B. Poly(ADP-ribose) polymerase-1 regulates activation of activator protein-1 in murine fibroblasts. J Immunol 2003;170:2113–2120. [DOI] [PubMed] [Google Scholar]
- 13.Valdor R, Schreiber V, Saenz L, Martinez T, Munoz-Suano A, Dominguez-Villar M et al. Regulation of NFAT by poly(ADP-ribose) polymerase activity in T cells. Mol Immunol 2008;45:1863–1871. [DOI] [PubMed] [Google Scholar]
- 14.Olabisi OA, Soto-Nieves N, Nieves E, Yang TT, Yang X, Yu RY et al. Regulation of transcription factor NFAT by ADP-ribosylation. Mol Cell Biol 2008;28:2860–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcilhac A, Raynaud F, Clerc I, Benyamin Y. Detection and localization of calpain 3-like protease in a neuronal cell line: possible regulation of apoptotic cell death through degradation of nuclear IkappaBalpha. Int J Biochem Cell Biol 2006;38:2128–2140. [DOI] [PubMed] [Google Scholar]
- 16.Jung T, Catalgol B, Grune T. The proteasomal system. Mol Aspects Med 2009;30:191–296. [DOI] [PubMed] [Google Scholar]
- 17.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc NatlAcadSci USA 1999;96:6223–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalgol B, Wendt B, Grimm S, Breusing N, Ozer NK, Grune T. Chromatin repair after oxidative stress: role of PARP-mediated proteasome activation. Free Radic Biol Med 2010;48:673–680. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Romero R, Martinez-Lara E, Aguilar-Quesada R, Peralta A, Oliver FJ, Siles E. PARP-1 modulates deferoxamine-induced HIF-1alpha accumulation through the regulation of nitric oxide and oxidative stress. J Cell Biochem 2008;104:2248–2260. [DOI] [PubMed] [Google Scholar]
- 20.Won J, Chung SY, Kim SB, Byun BH, Yoon YS, Joe CO. Dose-dependent UV stabilization of p53 in cultured human cells undergoing apoptosis is mediated by poly(ADP-ribosyl)ation. Mol Cells 2006;21: 218–223. [PubMed] [Google Scholar]


