Abstract
Background
This study aimed to screen and identify potential immune biomarker to predict the prognosis of oral squamous cell carcinoma (OSCC).
Methods
Data of OSCC patient from The Cancer Genome Atlas (TCGA) database were downloaded, and the ESTIMATE algorithm was used to calculate stromal and immune scores. Differentially expressed genes (DEGs) between the high and low immune score groups were screened, and Kaplan-Meier survival analysis was performed to identify the DEGs linked to the overall survival (OS) time of OSCC patients. Then, those DEGs were validated in anther cohort. A correlation analysis was used to further screen the prognostic genes which were tightly linked to the expression of programmed cell death 1 (PD-1)/programmed death-ligand 1 (PD-L1). The expression profiles of the candidate genes interleukin 12 receptor subunit beta 1 (IL-12RB1), cytotoxic T-lymphocyte associated protein 4 (CTLA4), and G protein-coupled receptor 25 (GPR25) were identified in the single-cell RNA sequence OSCC dataset from GSE103322. Finally, immunohistochemistry (IHC) and immunofluorescence (IF) were applied to confirm the expression pattern of IL-12RB1 in OSCC tissue microarray. Kaplan-Meier analysis was used to assess the prognostic significance of IL-12RB1 staining score in the malignant and non-malignant cells among the patients.
Results
The high immune score group showed better OS compared with that of the low immune scores group. Among 339 DEGs, 90 were identified as being tightly linked to OS time. In the validation set, 23 genes were confirmed to be closely associated with survival prognosis, and the expression levels of IL-12RB1, CTLA4, and GPR25 were commonly associated with the expression of PD-1/PD-L1. The RNA-sequencing showed that IL-12RB1 was expressed in epithelial and immune cells, whereas CTLA4 and GPR25 were relatively poorly expressed in the OSCC tissue. IHC showed that IL-12RB1 was positively expressed in both malignant and non-malignant cells. IF showed that IL-12RB1 was co-expressed with CD3, CD68, PD-1, and PD-L1 on the cytomembrane. Additionally, high score of IL-12RB1 expression in the non-malignant cells was a prognostic risk factor for OS of OSCC.
Conclusions
IL-12RB1 was tightly associated with survival of OSCC and with the expression levels of PD-1/PD-L1 in the tumor immune microenvironment.
Keywords: Oral squamous cell carcinoma (OSCC), tumor microenvironment (TME), interleukin 12 receptor subunit beta 1 (IL-12RB1), programmed cell death 1 (PD-1), programmed death-ligand 1 (PD-L1)
Introduction
Immuno-oncology (I-O) has been an active and growing field ever since Dr. William Coley observed in 1893 that cancer symptoms could be relieved by acute bacterial infections (1). Immunoediting includes 3 distinct but connected phases (2). In the first step, “immune-surveillance”, tumor-associated antigens (TAAs) activate the immune system and precancerous cells are eliminated; in the second step, “immune equilibrium”, a few abnormal cells achieve a balance between evading immune surveillance and being destroyed by the immune system, which manifests clinically as a dormant occult tumor; in the third phase, “immune escape”, the mutated cells evade the immune system’s elimination more efficiently and the tumor begins to grow, invading and metastasizing (3). Based on these theories, activation of the immune system to treat cancer has become the central topic of I-O in the past decades. However, due to serious systemic toxicity, the clinical results of simple immunotherapies such as IL-2 treatment were not as positive as hoped (4). As we have learned more about I-O, it has become clear that the tumor microenvironment (TME) formed by the extracellular matrix, immune cells, cancer cells, and other tumor-associated cells play a key role in tumorigenesis, invasion, metastasis and recurrence (5,6). Recent studies have shown that functional changes of tumor-infiltrating lymphocytes (TILs) in the TME obviously affect tumor growth and the outcome of immune checkpoint blockade therapy (7,8). Programmed cell death 1 (PD-1) engaged its ligands, programmed death-ligand 1 (PD-L1), was considered as a very important way to disable the TILs. Thus, efficiently reactivating TILs which have been disabled in the TME is a key goal of immune checkpoint blockade therapy (1). Blockade therapy especially in targeting the PD-1/PD-L1 axis had been widely applied to treat a variety of tumors (9).
Head and neck squamous cell carcinoma (HNSCC) is in the top 10 most common cancers worldwide (10). In 2016, the United States Food and Drug Administration (FDA) first approved anti-PD-1/PD-L1 therapy for advanced HNSCC, and by 2019 it was regarded as first-line drug treatment for patients with metastatic/recurrent/inoperable HNSCC (11,12). Oral squamous cell carcinoma (OSCC) is the most common type of HNSCC, typically occurring in the oral mucosa, palate, and throat and characterized by squamous differentiation (13,14). In the past few years, hampered by tumor heterogeneity and various immune conditions of patients, the efficacy of PD-1/PD-L1 axis blockade therapies has been limited for OSCC (15). TILs, especially cytotoxic CD8+ T cells, are pivotal for immune therapy due to complex mechanisms (16). Studies have attempted to accurately distinguish the characteristics of TME and predict the therapeutic effect of anti-PD-1/PD-L1 inhibitors. For instance, Tsakiroglou et al. found that OSCC patients with high frequencies of proximal CD8+ and PD-L1+ cells had poor prognosis in overall survival (OS) (17). Foy et al. further showed that OSCC patients with a higher CD8+ T cell infiltration received a significantly higher clinical benefit from PD-1 blockade treatment (18). However, many patients still show no clinical response to anti-PD-1/PD-L1 therapy.
Therefore, it is crucial to identify more novel immune prognostic biomarkers for patients with OSCC and to improve the efficacy of anti-PD-1/PD-L1 therapies. With this aim in mind, we used a series of methods, including ESTIMATE algorithm, gene expression correlation analysis, single-cell RNA-sequencing (single-cell RNA-seq) of OSCC, immunohistochemistry (IHC), and immunofluorescence (IF), to identify potential predictive indicators for OSCC and anti-PD-1/PD-L1 therapy. We present the following article in accordance with the REMARK reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6915/rc).
Methods
Data collection
The gene expression profiles of 144 patients with OSCC were obtained from The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/). Only those patients whose histological diagnosis was OSCC and whose primary sites were buccal, tongue, gingival, and oropharynx were included in our analysis. An additional Gene Expression Omnibus (GEO) cohort, GSE41613, was downloaded to validate our results. The “GEOquery” R package was used to obtained gene expression profiles and GPL data (19). Clinical data of survival and outcome were also downloaded from the TCGA and GEO data portals.
Calculation of immune and stromal scores, identification of differentially expressed genes (DEGs), and function enrichment analysis
Immune and stromal scores were calculated by the ESTIMATE algorithm from the “limma” package (20). A fold change of >2 and an adjusted P value of <0.05 were set as the threshold to screen for DEGs. Heatmaps and clustering were generated by the web tool ClustVis (21). Functional enrichment analysis of DEGs was conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (22) to identify Gene Ontology (GO) categories, including cellular components (CCs), biological processes (BPs) and molecular functions (MFs). Pathway enrichment analysis from the Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed though the DAVID database. A false discovery rate (FDR) of <0.05 was considered the cut-off.
Identification of prognostic genes in DEGs extracted from the TCGA database and validation of those genes in the GEO database
A Kaplan-Meier survival analysis was performed to explore the relationship between the OS of patients with OSCC and the gene expression levels of the DEGs extracted from the TCGA database. The relationship was calculated by log-rank test. Then, gene expression data of a cohort of 97 patients with OSCC were downloaded from GEO, an independent cancer database. To verify the value of the prognostic genes identified from the TCGA database, Kaplan-Meier plots were used to illustrate the relationship between OS and gene expression levels in the validation set.
Correlation analysis between the expression of the prognostic genes and PD-1/PD-L1 in the GSE41613 cohort
The prognostic genes screened from the previous step and PD-1/PD-L1 expression levels were downloaded from the validation cohort. A correlation analysis of the expression of the prognostic genes and PD-1/PD-L1 was applied and a P value of <0.05 was considered statistically significant.
Screening of the cluster of cells expressing interleukin 12 receptor subunit beta 1 (IL-12RB1), G protein-coupled receptor 25 (GPR25), and cytotoxic T-lymphocyte associated protein 4 (CTLA4) by the single-cell RNA-seq of OSCC in GSE103322
The single-cell RNA sequence dataset (GSE103322), which consisted of 5,902 single cells from 18 patients with oral cavity tumors, was downloaded from the GEO database and analyzed according to the pipeline of the “Seurat” R package (23). In brief, the data were normalized and scaled. Then, principal component analysis (PCA) was performed to approach linear dimensional reduction. After determining the dimensionality of the dataset, non-linear dimensional reduction was applied with the Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) algorithm (24). Cells were successfully classified into 17 separate clusters. Differential expression analysis was applied to find the marker genes of these clusters. Afterwards, each cluster was annotated by singleR and CellMarker (25,26). The top 3 marker genes of each cluster were visualized by dotplot. Expression of IL-12RB1, GPR25 and CTLA4 in these clusters were visualized with UMAP projection and violin plot.
Evaluation of IL-12RB1 expression pattern by hematoxylin and eosin (H&E), IHC, and IF staining of the histologic microarray of patients with OSCC
Human tissue microarrays were attained from the First Affiliated Hospital of Sun Yat-sen University. The clinical characteristics of 92 patients with OSCC were downloaded from the medical record database. All patients had no preoperative radiation or chemotherapy and were informed of the purpose of our study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Sun Yat-sen University (No. 2017-173), and informed consent was taken from all the patients. All tissues were embedded in paraffin, and 4 µm sections were prepared.
First, malignant and non-malignant cells in the section was confirmed by H&E staining. IHC and IF was performed as reported before (27,28). In brief, paraffin sections were routinely deparaffinized, rehydrated, and processed for antigen retrieval in pH6.0 citrate antigen retrieval solution. For IHC, slides were incubated with rabbit anti-IL-12RB1 antibodies (1:150, Abcam) at 4 °C overnight. Then, sections were incubated with horseradish peroxide (HRP)-labeled goat anti-rabbit secondary antibodies for 60 min at 37 °C, followed by 2% 3,3'-diaminobenzidine (DAB) chromogenic solution for 10 seconds at room temperature. Nuclei were then counter-stained with Mayer’s hematoxylin before slides were sealed. For IF, slides were co-incubated anti-IL-12RB1 (1:150, Abcam) with anti-CD3 (1:150, Abcam), anti-CD68 (1:150, Abcam), anti-PD-1 (1:150, Abcam), or anti-PD-L1 (1:150, Abcam) at 4 °C overnight. Then, all slides were incubated with fluorescence conjugated secondary antibody for 60 minutes at room temperature, protected from light. Finally, nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) before slides were sealed with an antifade mounting medium. Images were visualized with a fluorescence microscope.
With respect to the IL-12RB1 expressed in malignant cells and tumor-infiltrating immune cells, the IHC scores were detected in malignant and non-malignant cells in the tissues, which were calculated by multiplying the proportion of positively stained cells by the staining intensity (29). A Kaplan-Meier curve was used for the survival analysis, and univariate and multivariate Cox regression analyses were used to assess the prognostic factors linked to OS. P values of <0.05 were considered significant.
Statistical analysis
All data were shown as mean ± standard deviation. Data were analyzed and plotted by the GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, CA, USA) and SPSS Statistics 25.0 software (IBM Corp., Armonk, NY, USA). The data were evaluated using Student’s t-test to determine significance between two groups. The Kaplan-Meier curve was used to evaluate OS, and the log-rank test was used to estimate differences between the survival curves. P value of <0.05 was considered statistically significant.
Results
Association of immune scores with OS of OSCC in the TCGA database and identification of the DEGs profile with the group of immune scores
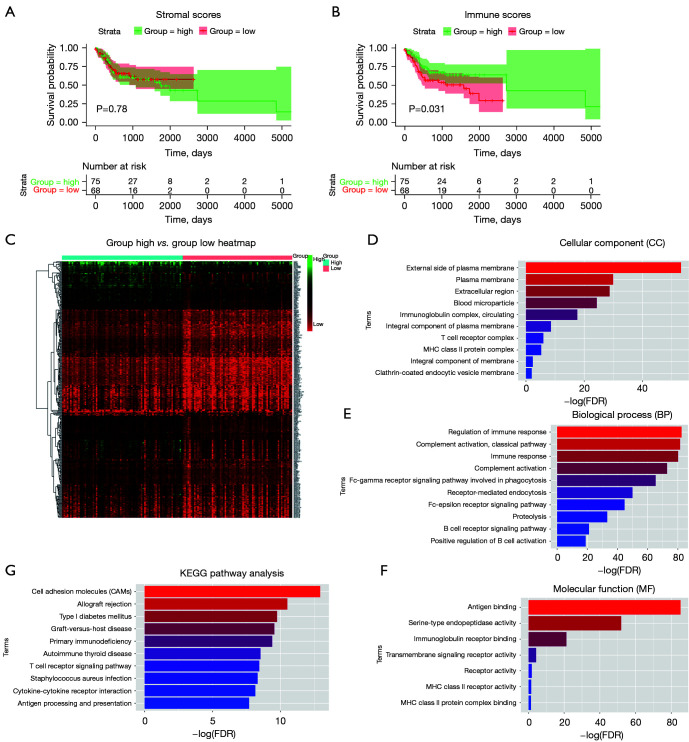
The gene expression profiles of 144 patients with OSCC were downloaded from the TCGA database. Using the ESTIMATE algorithm, we found that stromal scores ranged from 15,102.03 to 22,822.44, and immune scores ranged from 17,834.73 to 21,900.78. After dividing these cases into high and low score groups, we found that there was no significant difference between the high and low stromal score groups (Figure 1A). However, the Kaplan-Meier survival analysis showed that the high immune score group had better OS than that of the low immune scores group (Figure 1B). To further reveal the DEGs profiles in the high and low immune score groups, we analyzed Affymetrix microarray data of the 144 patients. As shown in the heatmaps in Figure 1C, 331 genes were upregulated and 8 genes were downregulated in the high score group compared to those of the low score group. After applying GO and KEGG analyses, we found that the top GO terms were mainly concentrated in the external side of the plasma membrane and involved in BPs such as immune response regulation and classical pathway complement activation, and in MFs such as antigen binding (Figure 1D-1F). Additionally, all the pathways that were identified from the KEGG analysis were deeply involved with cell adhesion molecules and other immune responses (Figure 1G).
Figure 1.
Stromal scores and immune scores were associated with OS in OSCC, and the profile of the DEGs was identified from the immune score groups. (A) Kaplan-Meier curves of the high and low groups based on their stromal scores with OS time. (B) Kaplan-Meier curves of the high and low groups based on their immune scores with OS time. (C) Heatmap of the DEGs between the high and low immune scores, P<0.05. (D-F) GO analysis of the DEGs in the two groups based on immune score, including CCs, BPs, and MFs. (G) KEGG pathway enrichment analysis of the DEGs. OS, overall survival; OSCC, oral squamous cell carcinoma; DEGs, differentially expressed genes; GO, Gene Ontology; CCs, cellular components; BPs, biological processes; MFs, molecular functions; KEGG, Kyoto Encyclopedia of Genes and Genomes; MHC, major histocompatibility complex; FDR, false discovery rate.
Identification of prognostic genes from the DEGs and further validation in the GSE41613 cohort
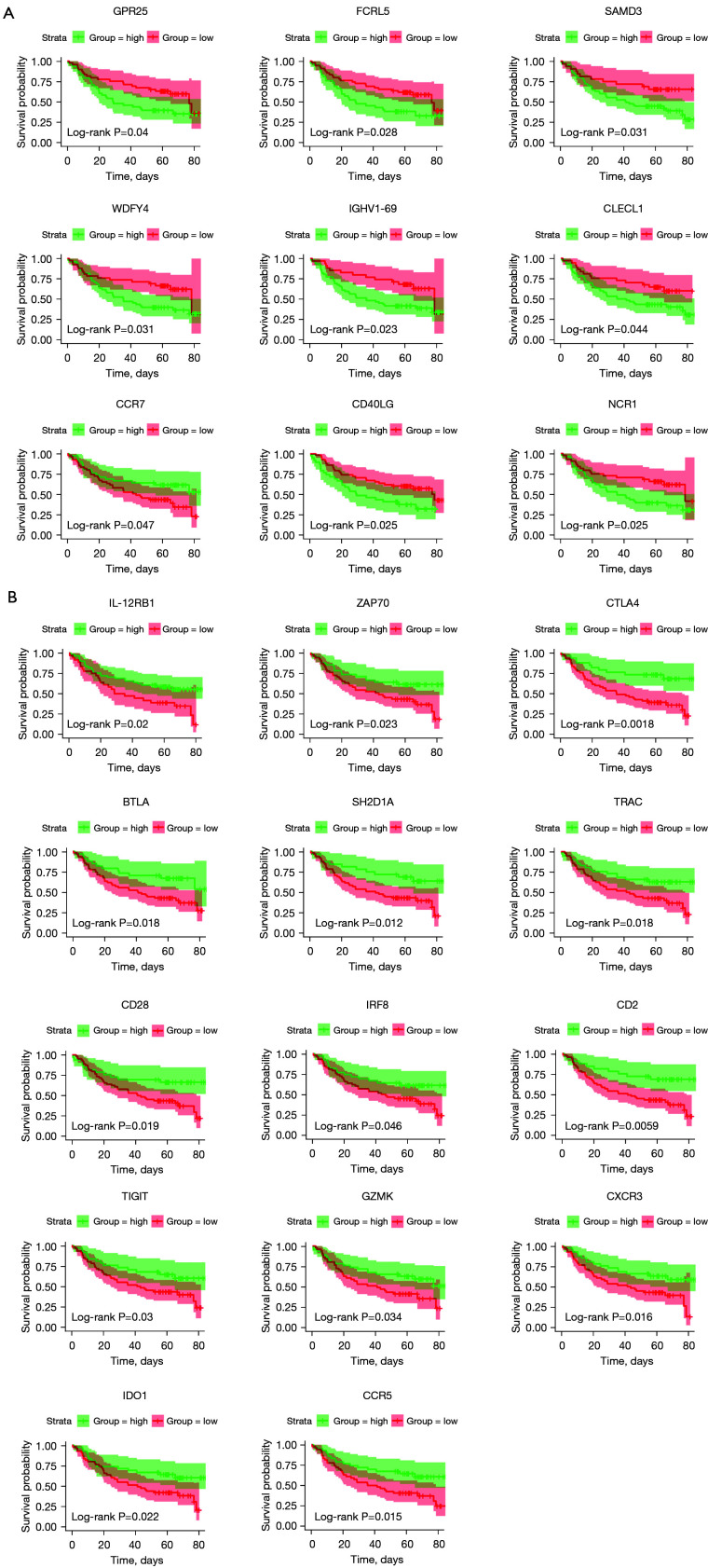
To identify the key genes that effect OS time, we performed a Kaplan-Meier curve analysis. As shown in Table S1, 90 genes from the 339 DEGs extracted from the TCGA database had a significant effect on prognosis. To detect the genes with the best predictive value, we downloaded and analyzed the data of gene expression from an additional cohort of OSCC cases in the GEO database. Thus, 23 genes among the 90 prognostic genes were further confirmed to be tightly linked to OS time (Figure 2).
Figure 2.
Validation of the prognostic genes extracted from the TCGA database with OS in GSE41613. Kaplan-Meier survival curves were used to select the DEGs though the comparison of high (green line) and low (red line) groups of gene expression. (A) Nine low expression genes such as GPR25 and FCRL5 etc. were shown to predict better 5-year OS. (B) Fourteen high expression genes such as IL-12RB1 and CTLA4 etc. were shown to predict better 5-year OS. P<0.05 in log-rank test. TCGA, The Cancer Genome Atlas; OS, overall survival; DEGs, differentially expressed genes; GPR25, G protein-coupled receptor 25; FCRL5, Fc receptor like 5; IL-12RB1, interleukin 12 receptor subunit beta 1; CTLA4, cytotoxic T-lymphocyte associated protein 4; SMAD3, SMAD family member 3; WDFY4, WDFY family member 4; IGHV1-69, immunoglobulin heavy variable 1-69; CLECL1, C-type lectin like 1; CCR7, C-C motif chemokine receptor 7; CD40LG, CD40 antigen ligand; NCR1, natural cytotoxicity triggering receptor 1; ZAP70, zeta chain of T cell receptor associated protein kinase 70; BTLA, B and T lymphocyte associated; SH2D1A, SH2 domain containing 1A; TRAC, T cell receptor alpha constant; IRF8, interferon regulatory factor 8; TIGIT, T cell immunoreceptor with Ig and ITIM domains; GZMK, granzyme K; CXCR3, C-X-C motif chemokine receptor 3; IDO1, indoleamine 2,3-dioxygenase 1; CCR5, C-C motif chemokine receptor 5.
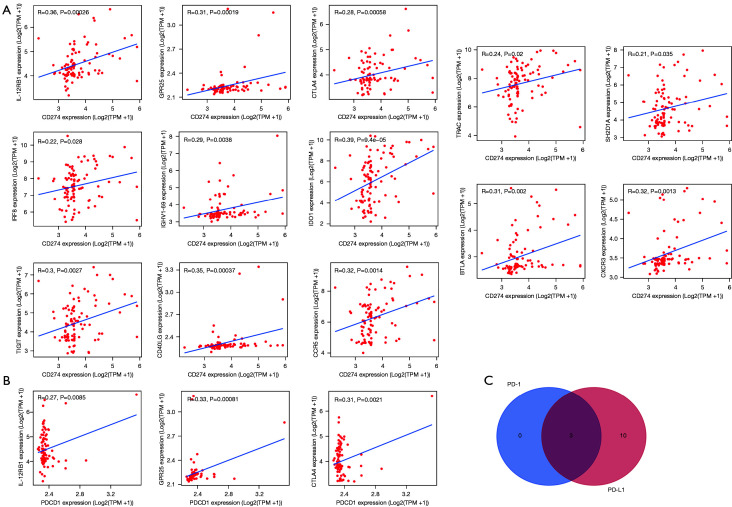
Correlation analysis showed that the expression of 13 genes were related to PD-L1 expression, while 3 genes were linked to the level of PD-1
We further downloaded the expression profile of PD-L1 (CD274) and PD-1 (PDCD1) in the GSE41613 cohort. According to the correlation analysis, the expression of 13 genes (IL-12RB1, GPR25, CTLA4, CCR5, CD40LG, CXCR3, IDO1, IGHV1-69, IRF8, SH2D1A, TIGIT, TRAC, and BTLA) was closely related to PD-L1 level, while 3 genes (IL-12RB1, GPR25, and CTLA4) were linked to the expression of PD-1 (Figure 3A,3B). Moreover, the Venn diagram indicated that these 3 genes (IL-12RB1, GPR25, and CTLA4) were commonly related to the expression of PD-1 and PD-L1 (Figure 3C).
Figure 3.
Screening the validated genes related to expression of PD-1 and PD-L1 in the GSE41613 cohort. (A) The genes positively correlated with PD-L1 expression. (B) The genes positively correlated with PD-1 expression. (C) Venn diagram showing the 3 genes (IL-12RB1, GPR25, and CTLA4) related to the expression of PD-1 and PD-L1. PD-1 (PDCD1), programmed cell death 1; PD-L1 (CD274), programmed death-ligand 1; IL-12RB1, interleukin 12 receptor subunit beta 1; GPR25, G protein-coupled receptor 25; CTLA4, cytotoxic T-lymphocyte associated protein 4; CCR5, C-C motif chemokine receptor 5; CD40LG, CD40 antigen ligand; CXCR3, C-X-C motif chemokine receptor 3; IDO1, indoleamine 2,3-dioxygenase 1; IGHV1-69, immunoglobulin heavy variable 1-69; IRF8, interferon regulatory factor 8; SH2D1A, SH2 domain containing 1A; TIGIT, T cell immunoreceptor with Ig and ITIM domains; TRAC, T cell receptor alpha constant; BTLA, B and T lymphocyte associated.
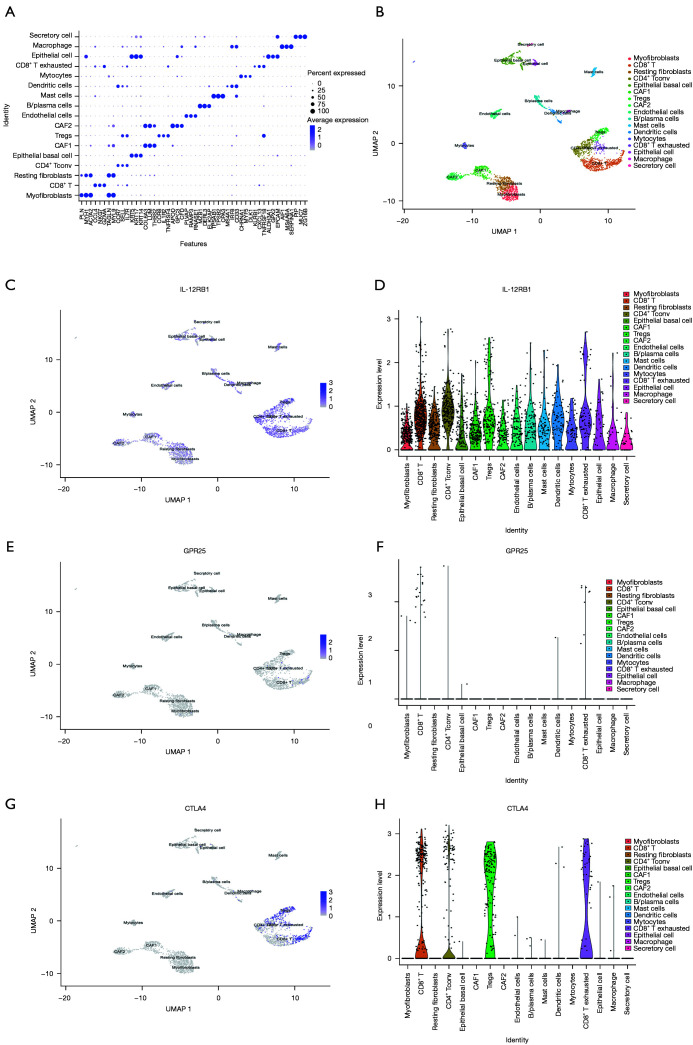
The single-cell RNA-seq of OSCC in the GSE103322 cohort showed that IL-12RB1 was mainly expressed in the cluster of epithelial cells and immune cells
As the composition of malignant and non-malignant cells in the TME was found to be heterogeneous in HNSCC by Puram et al. (30), we wanted to investigate the expression pattern of IL-12RB1, GPR25, and CTLA4 among these heterogeneous cells. Thus, we downloaded a single-cell RNA sequence dataset (GSE103322) from the GEO database. This dataset consisted of 5,902 singles cells from 18 patients with oral cavity tumors. We reanalyzed the data following the pipeline of the R package “Seurat” and clustered the cells into 17 clusters as visualized by UMAP projection. We annotated these 17 clusters according to the marker genes (Figure 4A). Our results showed similar cells clusters to those found by Puram et al. As shown in Figure 4B, the tumor cells consisted of 17 kinds of cells, including cancer cells and immune cells. Interestingly, we found that IL-12RB1 was mainly expressed in epithelial cells and immune cells, especially in CD8+ T cells and the CD4+ T conv cluster (Figure 4C,4D). However, GPR25 was poorly expressed in OSCC (Figure 4E,4F) and CTLA4 was only expressed in T cells (Figure 4G,4H).
Figure 4.
The expression pattern of IL-12RB1, GPR25, and CTLA4 in malignant and non-malignant cells from the single-cell RNA sequence dataset of GSE103322. (A) Dot plot showing the top 3 marker genes of indicated clusters cells. (B) UMAP projection of the OSCC cells colored by clusters, which were annotated based on the expression of known marker genes. (C) UMAP plot showing the expression level of IL-12RB1 among clusters. (D) Violin plot showing the expression level of IL-12RB1 in distinct clusters. (E) UMAP plot showing the expression level of GPR25 in distinct clusters. (F) Violin plot showing the expression level of GPR25 among clusters. (G) UMAP plot showing the expression level of CTLA4 in distinct clusters. (H) Violin plot showing the expression level of CTLA4 among clusters. IL-12RB1, interleukin 12 receptor subunit beta 1; GPR25, G protein-coupled receptor 25; CTLA4, cytotoxic T-lymphocyte associated protein 4; UMAP, Uniform Manifold Approximation and Projection for Dimension Reduction; OSCC, oral squamous cell carcinoma; CAF1, cancer associated fibroblast 1; CAF2, cancer associated fibroblast 2.
The expression pattern of IL-12RB1 and colocalization staining of IL-12RB1 and CD3, CD68, PD-1, and PD-L1 in OSCC tissue
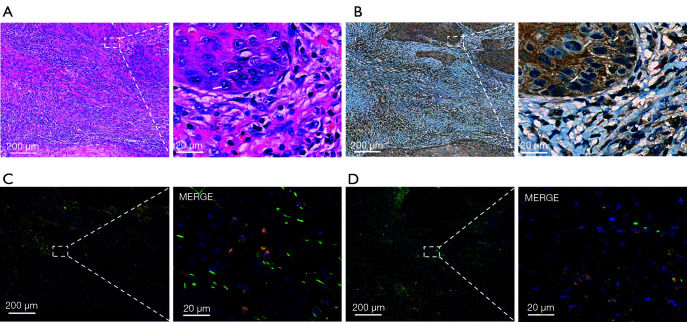
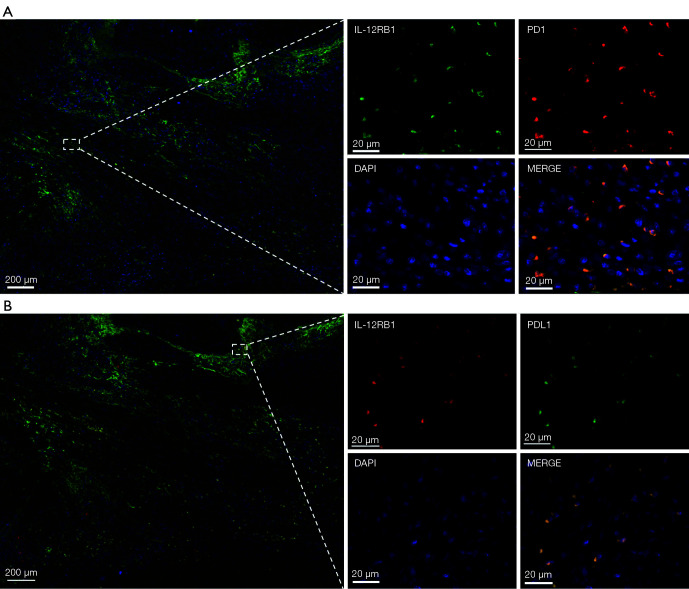
As the expression of CTLA4 and GPR25 was relatively low in the OSCC tissue, we focused on IL-12RB1. IHC and IF were performed to confirm the expression pattern of IL-12RB1 in patients with OSCC. As shown in Figure 5A,5B, compared with H&E staining, we found that IL-12RB1 was expressed in both malignant and non-malignant cells. The results of IF staining showed that IL-12RB1 was colocalized with CD3 and CD68, the biomarker protein of T cells, and macrophages (Figure 5C,5D). Furthermore, IF staining showed that IL-12RB1 and PD-1 or PD-L1 were also co-expressed in those cells (Figure 6A,6B).
Figure 5.
IL-12RB1 is highly expressed in OSCC tissue. (A) H&E staining of the OSCC samples. (B) IHC indicated the expression of IL-12RB1 in malignant and non-malignant cells of OSCC tissues. (C) IF indicated the co-expression of IL-12RB1 and CD3 in OSCC tissues. (D) IF indicated the co-expression of IL-12RB1 and CD68 in OSCC tissues. White and red arrows indicate the malignant and non-malignant cells, respectively. Scale bar: 5×, 200 µm; 40×, 20 µm. IL-12RB1, interleukin 12 receptor subunit beta 1; OSCC, oral squamous cell carcinoma; H&E, hematoxylin and eosin; IHC, immunohistochemical; IF, immunofluorescence.
Figure 6.
IL-12RB1 co-expressed with PD-1 and PD-L1 in OSCC tissue. (A) Representative images of IL-12RB1 (red) and PD-1 (green) in OSCC samples. (B) Representative IF images of IL-12RB1 (green) and PD-L1 (red) in OSCC samples. Scale bar: 5×, 200 µm; 40×, 20 µm. IL-12RB1, interleukin 12 receptor subunit beta 1; PD-1, programmed cell death 1; PD-L1, programmed death-ligand 1; OSCC, oral squamous cell carcinoma; IF, immunofluorescence.
The degree of IL-12RB1 expression was tightly associated with OS of OSCC in the tissue microarray
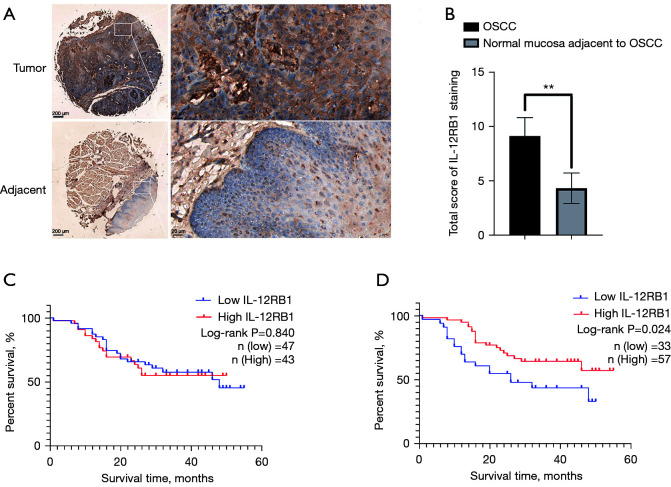
In the microarray comprising 92 OSCC samples and 60 samples of adjacent normal oral epithelia, the IHC score for each tissue was calculated by multiplying the proportion of positively stained cells by the staining intensity to evaluate the IHC staining. As shown in Figure 7A, IL-12RB1 was mainly expressed in the cytomembrane of malignant and non-malignant cells; meanwhile, significantly higher expression of IL-12RB1 was found in OSCC tissues than that in normal oral epithelial tissues (Figure 7B). To further elucidate the prognostic role of IL-12RB1 in patients with OSCC, we investigated the relationship between IL-12RB1 expression and patient outcome using the long-term follow-up period as the variable. The Kaplan-Meier curve showed that a higher score of IL-12RB1 in the non-malignant cells indicated a better prognosis of OSCC, but the high and low IHC scores of malignant cells had no significantly effect on OS time (Figure 7C,7D). Furthermore, the IL-12RB1 expression levels were significantly associated with clinical stage and node metastasis but not with age, gender, or pathologic differentiation (Table 1).
Figure 7.
IL-12RB1 was upregulated in OSCC. Kaplan-Meier plots of the OS time of the patient based on IHC score of IL-12RB1 in the tissue microarray. (A) Representative IHC staining images of OSCC tissue microarrays in tumor tissues and paired adjacent tissues. (B) Analysis of the scores of OSCC and the normal mucosa adjacent to OSCC in tissue microarray according to IL-2RB1 staining results. (C) Kaplan-Meier curves of the high and low groups based on the IHC scores of malignant cells with OS time. (D) Kaplan-Meier curves of the high and low groups based on IHC scores of non-malignant cells with OS time. **P<0.01. Scale bar: 200 and 20 μm. IL-12RB1, interleukin 12 receptor subunit beta 1; OSCC, oral squamous cell carcinoma; OS, overall survival; IHC, immunohistochemical.
Table 1. Univariate and multivariate analysis of factors affecting the OS of patients with OSCC.
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| IL-12RB1 expression in malignant cells (low vs. high) | 1.07 (0.57–2.01) | 0.842 | 1.10 (0.58–2.07) | 0.781 | |
| IL-12RB1 expression in non-malignant cells (low vs. high) | 0.50 (0.26–0.93) | 0.029* | 0.45 (0.24–0.86) | 0.015* | |
| Age (≤60 vs. >60) | 1.61 (0.84–3.07) | 0.150 | – | – | |
| Gender (male vs. female) | 1.12 (0.56–2.20) | 0.754 | – | – | |
| T classification (T1+2 vs. T3+4) | 1.16 (0.53–2.53) | 0.714 | 1.342 (0.60–3.00) | 0.473 | |
| LN metastasis (LN− vs. LN+) | 2.13 (1.11–4.10) | 0.023* | 2.34 (1.20–4.56) | 0.013* | |
| Histological (well vs. moderate) | 0.71 (0.26–1.95) | 0.503 | 0.74 (0.27-2.05) | 0.563 | |
| Histological (well vs. poor) | 0.74 (0.37–1.46) | 0.381 | 0.70 (0.34–1.34) | 0.255 | |
*P<0.05. OS, overall survival; OSCC, oral squamous cell carcinoma; HR, hazard ratio; CI, confidence interval; IL-12RB1, interleukin 12 receptor subunit beta 1; LN, lymph node.
Discussion
TME has been confirmed to play a critical role in the tumorigenesis and development of OSCC. Immune cells, an important component of TME, have a significant effect on the invasion and therapeutic resistance of tumors (31). In 2016, as a response to the encouraging clinical results of immune checkpoints inhibitors, the FDA approved anti-PD-1/PD-L1 therapy for patients with advanced HNSCC (11). In current clinical practice, PD-1/PD-L1 therapy is selected based on the intensity of PD-L1 expression by IHC. However, due to the heterogeneity of tumors, there are still a considerable number of patients for whom this treatment has poor curative effect (15). Therefore, it is important to identify the biomolecules that can predict or even affect the accuracy and efficacy of anti-PD-1/PD-L1 treatment for OSCC.
In the present study, we found that immune scores were associated with the OS of OSCC, and that the GO terms of 339 DEGs were mainly involved in the regulation of the immune response. Then, through validation in an independent cohort of OSCC, 23 genes were found to be tightly linked to OS time, of which 13 genes were positively correlated with PD-L1 expression and 3 genes were positively correlated with the expression of PD-1. Interestingly, we found that IL-12RB1, CTLA4 and GPR25 were positively associated with PD-1/PD-L1 expression. Furthermore, we analyzed the single-cell RNA-seq of OSCC in GSE103322 and found that only IL-12RB1 was highly expressed in epithelial cells and immune cells, especially in CD8+ T cells and the CD4+ T conv cluster. Finally, we used IHC and IF to confirm that IL-12RB1 was expressed in both cancer cells and immune cells, such as T cells and macrophages, and also co-expressed with PD-1/PD-L1 on the cytomembrane. The tissue microarray also confirmed that IL-12RB1 was highly expressed in OSCC tissue, and a high score of expression in stromal cells was a prognostic risk factor for OS in OSCC. Thus, we suggest that the activation of IL-12RB1 may have a potential effect on the OS of patients with OSCC and the expression of PD-1/PD-L1 in the tumor immune microenvironment, further affecting the efficacy of PD-1/PD-L1 axis inhibitor treatment.
IL-12RB1, encoded by the gene IL12RB1, is a type I transmembrane receptor that can bind to the P40-domain of IL-12/IL-23 and promote their immune response separately (32). IL-12, including 2 subunits (P35 and P40), was discovered from B cell lines and was regarded as a “natural killer-stimulatory factor” and “cytotoxic lymphocyte maturation factor” (33,34). As another member of the IL-12 family, IL-23 is also structured by 2 disulfide linked subunits, a light p19 chain (α chains), and a heavy P40 chain (β chains) (35). It is generally accepted that IL-12 and IL-23 are pro-inflammatory/pro-stimulatory cytokines, balancing with other negative feedback members in the IL-12 family, such as IL-27 or IL-35 (36). Consistent with its important role in regulating the immune response, IL-12 has been confirmed to be tightly associated with immune-related diseases (36). Epidemiological studies have found that IL-12 polymorphisms are closely related to cancers (37). Due to its ability to activate cytotoxic cells from the immune response, IL-12 is regarded as a potential antitumor drug (38). The antitumor mechanism of IL-12 involves various aspects, including increasing the synthesis and secretion of IFN-γ and enhancing the cytotoxicity of natural killer (NK) cells, CD4+ cells, and CD8+ T cells (38,39). The activity of IL-12 depends on the binding of its subunits to the receptors (P40 IL-12RB1 and P35 IL-12RB2). Therefore, the expression of IL-12RB1 affects the development of tumors. Additionally, Kundu et al. found that in prostate cancer patients, the serum level of P40 was much greater than that in healthy people. Meanwhile, excess P40 binding to IL-12RB1 reduces the antitumor effect of normal IL-12, resulting in immune evasion of the tumor cells (40). Therefore, it can be inferred that reducing the production of P40 and increasing the expression of IL-12RB1 might be beneficial to tumor therapy. In the current study, we found that the survival prognosis of the higher IL-12RB1 expression group was better than that of the low expression group.
Through correlation analysis of gene expression level, we found that IL-12RB1 was tightly positively related to the expression of PD-1 and PD-L1. This is a very interesting phenomenon that has not been reported before. Through the single-cell RNA-seq and IHC analyses, we confirmed that IL-12RB1 was highly expressed in the cancer cells and immune cells of OSCC. This differs from the results of previous studies, which have suggested that IL-12R is predominantly expressed in immune cells, especially on NK cells and T cells (41,42). Notably, we further found that IL-12RB1 was co-expressed with PD-1/PD-L1 in T cells. We hypothesized that activation of IL-12RB1 could affect the expression of PD-1/PD-L1 in cancer cells and immune cells via a previously unrecognized pathway, thus affecting the survival prognosis and efficacy of PD-1/PD-L1 axis therapy. This putative mechanism may include two processes: (I) direct activation of PD-1/PD-L1 expression through the crosstalk of intracellular signal pathways; and (II) regulation of PD-1/PD-L1 expression through the IL-12/interferon gamma (IFN-γ) axis and its downstream signaling pathway. Quatrini et al. found that IL-12 together with glucocorticoids increased PD-1 transcription and mRNA translation, eventually leading to an increased expression of PD-1 in NK cells (43). Previous studies have shown that the bioactive function of the IL-12 family is mainly mediated by the Janus kinase-signal transducers and activators of transcription (JAK-STAT) family (32,44). The STATs cause the nuclear translocation of the PD-L1 transcription factors IRF, S6K, and AP-1 (Jun/Fos) either in a direct way or through the PI3K/Akt/mTOR and MEK/Erk pathways, resulting in an effect on PD-L1 encoding (11). In addition, IL-12R activates the mediated mitogen-activated protein kinase 3/6 (MKK3/6)/p38/AP-1 axis, which upregulates the expression of PD-L1 (45). Various studies have shown that the physiological function of IL-12 is mainly realized by increasing IFN-γ production, which can affect the transcription of PD-1 or PD-L1 through multiple signaling pathways (46,47).
In conclusion, our study, which used a variety of data mining methods, found that IL-12RB1 was tightly associated with PD-1/PD-L1 expression and may have a potential effect on treatments targeting the PD-1/PD-L1 axis. This study has some shortcomings, and further in vivo and in vitro experiments are needed to verify the effect of IL-12RB1 on the prognosis of OSCC and the PD-1/PD-L1 expression. This will be the direction of our future research.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors gratefully acknowledge Dr. Changchang Jia and Xinlin Chen for their technical support and expert advice.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Sun Yat-sen University (No. 2017-173), and informed consent was taken from all the patients.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6915/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6915/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6915/coif). The authors have no conflicts of interest to declare.
(English Language Editor: C. Gourlay)
References
- 1.Kamta J, Chaar M, Ande A, et al. Advancing Cancer Therapy with Present and Emerging Immuno-Oncology Approaches. Front Oncol 2017;7:64. 10.3389/fonc.2017.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn GP, Bruce AT, Ikeda H, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002;3:991-8. 10.1038/ni1102-991 [DOI] [PubMed] [Google Scholar]
- 3.Messerschmidt JL, Prendergast GC, Messerschmidt GL. How Cancers Escape Immune Destruction and Mechanisms of Action for the New Significantly Active Immune Therapies: Helping Nonimmunologists Decipher Recent Advances. Oncologist 2016;21:233-43. 10.1634/theoncologist.2015-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbas AK. The Surprising Story of IL-2: From Experimental Models to Clinical Application. Am J Pathol 2020;190:1776-81. 10.1016/j.ajpath.2020.05.007 [DOI] [PubMed] [Google Scholar]
- 5.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett 2017;387:61-8. 10.1016/j.canlet.2016.01.043 [DOI] [PubMed] [Google Scholar]
- 6.Giancotti FG. Deregulation of cell signaling in cancer. FEBS Lett 2014;588:2558-70. 10.1016/j.febslet.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 2016;126:3447-52. 10.1172/JCI87324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348:56-61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 9.Lei Q, Wang D, Sun K, et al. Resistance Mechanisms of Anti-PD1/PDL1 Therapy in Solid Tumors. Front Cell Dev Biol 2020;8:672. 10.3389/fcell.2020.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vicente JC, Rodríguez-Santamarta T, Rodrigo JP, et al. PD-L1 Expression in Tumor Cells Is an Independent Unfavorable Prognostic Factor in Oral Squamous Cell Carcinoma. Cancer Epidemiol Biomarkers Prev 2019;28:546-54. 10.1158/1055-9965.EPI-18-0779 [DOI] [PubMed] [Google Scholar]
- 11.Lenouvel D, González-Moles MÁ, Talbaoui A, et al. An update of knowledge on PD-L1 in head and neck cancers: Physiologic, prognostic and therapeutic perspectives. Oral Dis 2020;26:511-26. 10.1111/odi.13088 [DOI] [PubMed] [Google Scholar]
- 12.Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915-28. 10.1016/S0140-6736(19)32591-7 [DOI] [PubMed] [Google Scholar]
- 13.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 14.Gleber-Netto FO, Yakob M, Li F, et al. Salivary Biomarkers for Detection of Oral Squamous Cell Carcinoma in a Taiwanese Population. Clin Cancer Res 2016;22:3340-7. 10.1158/1078-0432.CCR-15-1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao A, Pan X, Yang X, et al. Predictive factors in the treatment of oral squamous cell carcinoma using PD-1/PD-L1 inhibitors. Invest New Drugs 2021;39:1132-8. 10.1007/s10637-021-01082-w [DOI] [PubMed] [Google Scholar]
- 16.Le Saux O, Ray-Coquard I, Labidi-Galy SI. Challenges for immunotherapy for the treatment of platinum resistant ovarian cancer. Semin Cancer Biol 2021;77:127-43. 10.1016/j.semcancer.2020.08.017 [DOI] [PubMed] [Google Scholar]
- 17.Tsakiroglou AM, Fergie M, Oguejiofor K, et al. Spatial proximity between T and PD-L1 expressing cells as a prognostic biomarker for oropharyngeal squamous cell carcinoma. Br J Cancer 2020;122:539-44. 10.1038/s41416-019-0634-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foy JP, Bertolus C, Michallet MC, et al. The immune microenvironment of HPV-negative oral squamous cell carcinoma from never-smokers and never-drinkers patients suggests higher clinical benefit of IDO1 and PD1/PD-L1 blockade. Ann Oncol 2017;28:1934-41. 10.1093/annonc/mdx210 [DOI] [PubMed] [Google Scholar]
- 19.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. 10.1093/bioinformatics/btm254 [DOI] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 2015;43:W566-70. 10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W , Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 23.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888-902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hozumi Y, Wang R, Yin C, et al. UMAP-assisted K-means clustering of large-scale SARS-CoV-2 mutation datasets. Comput Biol Med 2021;131:104264. 10.1016/j.compbiomed.2021.104264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Lan Y, Xu J, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res 2019;47:D721-8. 10.1093/nar/gky900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163-72. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie GF, Zhao LD, Chen Q, et al. High ACTN1 Is Associated with Poor Prognosis, and ACTN1 Silencing Suppresses Cell Proliferation and Metastasis in Oral Squamous Cell Carcinoma. Drug Des Devel Ther 2020;14:1717-27. 10.2147/DDDT.S244516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng J, Chen Z, Liu C, et al. Bone mesenchymal stem cell-derived exosome-loaded injectable hydrogel for minimally invasive treatment of spinal cord injury. Nanomedicine (Lond) 2021;16:1567-79. 10.2217/nnm-2021-0025 [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Lu Z, Deng Y, et al. Up-regulation of INSR/IGF1R by C-myc promotes TSCC tumorigenesis and metastasis through the NF-κB pathway. Biochim Biophys Acta Mol Basis Dis 2018;1864:1873-82. 10.1016/j.bbadis.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 30.Puram SV, Tirosh I, Parikh AS, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017;171:1611-24.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat AA, Yousuf P, Wani NA, et al. Tumor microenvironment: an evil nexus promoting aggressive head and neck squamous cell carcinoma and avenue for targeted therapy. Signal Transduct Target Ther 2021;6:12. 10.1038/s41392-020-00419-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol 2012;13:722-8. 10.1038/ni.2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi M, Fitz L, Ryan M, et al. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med 1989;170:827-45. 10.1084/jem.170.3.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stern AS, Podlaski FJ, Hulmes JD, et al. Purification to homogeneity and partial characterization of cytotoxic lymphocyte maturation factor from human B-lymphoblastoid cells. Proc Natl Acad Sci U S A 1990;87:6808-12. 10.1073/pnas.87.17.6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreymborg K, Böhlmann U, Becher B. IL-23: changing the verdict on IL-12 function in inflammation and autoimmunity. Expert Opin Ther Targets 2005;9:1123-36. 10.1517/14728222.9.6.1123 [DOI] [PubMed] [Google Scholar]
- 36.Ullrich KA, Schulze LL, Paap EM, et al. Immunology of IL-12: An update on functional activities and implications for disease. EXCLI J 2020;19:1563-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Núñez-Marrero A, Arroyo N, Godoy L, et al. SNPs in the interleukin-12 signaling pathway are associated with breast cancer risk in Puerto Rican women. Oncotarget 2020;11:3420-31. 10.18632/oncotarget.27707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tait Wojno ED, Hunter CA, Stumhofer JS. The Immunobiology of the Interleukin-12 Family: Room for Discovery. Immunity 2019;50:851-70. 10.1016/j.immuni.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro F, Cardoso AP, Gonçalves RM, et al. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front Immunol 2018;9:847. 10.3389/fimmu.2018.00847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundu M, Roy A, Pahan K. Selective neutralization of IL-12 p40 monomer induces death in prostate cancer cells via IL-12-IFN-γ. Proc Natl Acad Sci U S A 2017;114:11482-7. 10.1073/pnas.1705536114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desai BB, Quinn PM, Wolitzky AG, et al. IL-12 receptor. II. Distribution and regulation of receptor expression. J Immunol 1992;148:3125-32. [PubMed] [Google Scholar]
- 42.Robinson RT. IL12Rβ1: the cytokine receptor that we used to know. Cytokine 2015;71:348-59. 10.1016/j.cyto.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quatrini L, Vacca P, Tumino N, et al. Glucocorticoids and the cytokines IL-12, IL-15, and IL-18 present in the tumor microenvironment induce PD-1 expression on human natural killer cells. J Allergy Clin Immunol 2021;147:349-60. 10.1016/j.jaci.2020.04.044 [DOI] [PubMed] [Google Scholar]
- 44.van der Heijden T, Bot I, Kuiper J. The IL-12 cytokine family in cardiovascular diseases. Cytokine 2019;122:154188. 10.1016/j.cyto.2017.10.010 [DOI] [PubMed] [Google Scholar]
- 45.Morinobu A, Gadina M, Strober W, et al. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A 2002;99:12281-6. 10.1073/pnas.182618999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon S, Park Y, Hyeon S, et al. Niche-specific MHC II and PD-L1 regulate CD4+CD8αα+ intraepithelial lymphocyte differentiation. J Exp Med 2021;218:e20201665. 10.1084/jem.20201665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song Q, Wang X, Wu X, et al. IL-22-dependent dysbiosis and mononuclear phagocyte depletion contribute to steroid-resistant gut graft-versus-host disease in mice. Nat Commun 2021;12:805. 10.1038/s41467-021-21133-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as