Abstract
In 2019 the COVID-19 pandemic, caused by SARS-CoV-2, demonstrated the urgent need for rapid, reliable, and portable diagnostics. The COVID-19 pandemic was declared in January 2020 and surges of the outbreak continue to reoccur. It is clear that early identification of infected individuals, especially asymptomatic carriers, plays a huge role in preventing the spread of the disease. The current gold standard diagnostic for SARS-CoV-2 is quantitative reverse transcription polymerase chain reaction (qRT-PCR) test based on the detection of the viral RNA. While RT-PCR is reliable and sensitive, it requires expensive centralized equipment and is time consuming (∼2 h or more); limiting its applicability in low resource areas. The FDA issued Emergency Use Authorizations (EUAs) for several COVID-19 diagnostics with an emphasis on point-of care (PoC) testing. Numerous RT-PCR and serological tests were approved for use at the point of care. Abbott's ID NOW, and Cue Health's COVID-19 test are of particular interest, which use isothermal amplification methods for rapid detection in under 20 min. We look to expand on the range of current PoC testing platforms with a new rapid and portable isothermal nucleic acid detection device.
We pair reverse transcription loop mediated isothermal amplification (RT-LAMP) with a particle imaging technique, particle diffusometry (PD), to successfully detect SARS-CoV-2 in only 35 min on a portable chip with integrated heating. A smartphone device is used to image the samples containing fluorescent beads post-RT-LAMP and correlates decreased diffusivity to positive samples. We detect as little as 30 virus particles per μL from a RT-LAMP reaction in a microfluidic chip using a portable heating unit. Further, we can perform RT-LAMP from a diluted unprocessed saliva sample without RNA extraction. Additionally, we lyophilize SARS-CoV-2-specific RT-LAMP reactions that target both the N gene and the ORF1ab gene in the microfluidic chip, eliminating the need for cold storage. Our assay meets specific target product profiles outlined by the World Health Organization: it is specific to SARS-CoV-2, does not require cold storage, is compatible with digital connectivity, and has a detection limit of less than 35 × 104 viral particles per mL in saliva. PD-LAMP is rapid, simple, and attractive for screening and use at the point of care.
Keywords: Particle diffusometry, Loop-mediated isothermal amplification, SARS-CoV-2, Diagnostic, Smartphone, rapid detection
Graphical abstract
1. Introduction
COVID-19, the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a pandemic in 2020 [1,2]. Many individuals infected with SARS-CoV-2 do not exhibit symptoms but are still carriers of the virus [3,4]. SARS-CoV-2 has an average incubation period of 6 days prior to the onset of symptoms and is more infectious than severe acute respiratory syndrome (SARS) or Middle East respiratory syndrome (MERS).
[5]. COVD-19 often mimics flu or pneumonia-like symptoms, making it difficult to diagnose [6,7]. Such ambiguous symptoms highlights the importance of rapid and specific testing to confirm and control the spread of COVID-19. Additionally, early detection of SARS-CoV-2 is a key factor to preventing the spread of the disease and aiding in proper quarantine and isolation safety protocols [6] While RT-PCR tests were quickly developed and commercialized for SARS-CoV-2 detection, the need for RNA extraction and thermocycler equipment limits its use in low resource settings [5].
Nucleic acid amplification tests (NAATs) and serological tests are the two types of diagnostic testing receiving FDA-EUA approval. The current gold standard test uses quantitative RT-PCR (a highly sensitive NAAT) [8]. However, RT-PCR is a laboratory-based technique that requires trained personnel, specialized equipment, expensive reagents, and is labor-intensive [6,9]. Some researchers have developed RT-PCR kits for self-collected salivary samples. However, the sample then must be sent back to the lab for analysis [10,11]. Therefore, there is a need for a rapid point of care molecular diagnostic due to continued high number of cases [6].
An alternative to conventional PCR methods are isothermal nucleic acid amplification (iNAAT) tests. INAAT-based diagnostics do not require expensive equipment or trained personnel due to the ability to amplify DNA without thermal cycling. Specifically, reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) allows for sensitive, rapid, and robust detection of nucleic acids in under an hour with fluorescent, colorimetric, or electrochemical read-outs [[12], [13], [14]]. Several research groups are developing LAMP-based assays for the detection of SARS-CoV-2 [[15], [16], [17], [18]]. RT-LAMP amplifies targeted RNA into 109 copies with more resistance to inhibitors than PCR [19]. This resistance to inhibition allows LAMP assays to be performed in crude sample matrices such as diluted saliva or urine, eliminating the need for costly and timely RNA extraction [15,20].
Some of the SARS-CoV-2 diagnostics approved for point of care testing include Mesa Biotech and Cepheid's RT-PCR tests which take less than 40 min [21]. Abbott's ID NOW [22], and Cue Health's COVID-19 test [23] both use isothermal methods for detection in less than 20 min. These tests use nasal or throat swabs for sample collection which may be difficult for patients to perform at home [15]. Alternatively, saliva has recently been found as a promising non-invasive sample for SARS-CoV-2 detection and contains a relatively high viral load in comparison to nasopharyngeal swabs [[24], [25], [26]]. When nasopharyngeal swabs and saliva samples were collected from the same individuals, saliva was found to provide more sensitive results for COVID-19 diagnosis [24,27]. Further, there have been studies confirming frequent complications such as chronic nose bleeds from nasal swabs, making saliva a safer alternative [28]. The FDA has approved saliva-based tests, but these tests still rely on RNA extraction followed by qRT-PCR and may take days to return results [9].
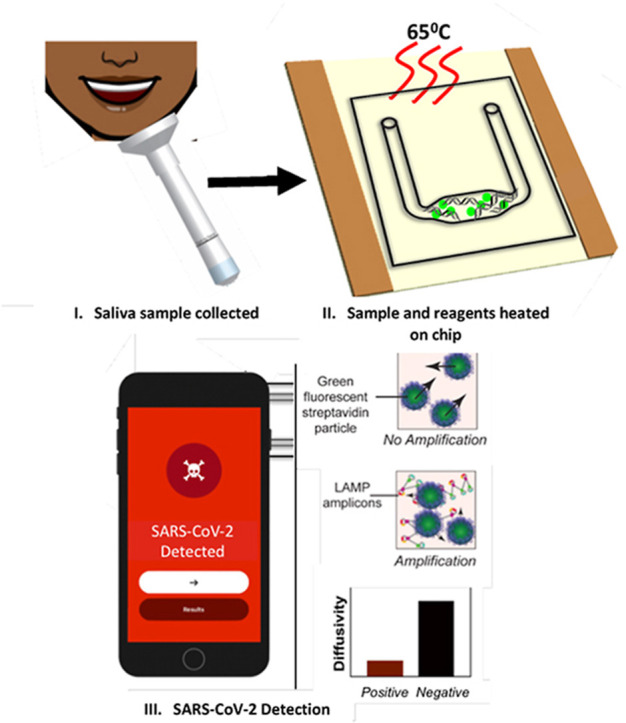
Here, we report the use of RT-LAMP paired with a smartphone imaging technique, particle diffusometry (PD), to detect SARS-CoV-2 that can be carried out in 35 min from crude saliva samples with a limit of detection (LOD) of 30 virus particles μL−1 (Fig. 1 ). We developed new primers targeting nsp3, a conserved region of the SARS-CoV-2 ORF1ab gene, and paired these with primers targeting the N2 region of the N gene for increased sensitivity [29]. Previously we have shown PD-LAMP to successfully detect pathogen nucleic acids from Vibrio cholerae (V. cholerae) in water [30] and Plasmodium parasites in unprocessed blood [31]. However, the testing in these studies were performed with LAMP reactions in tube and required the amplification product to be transferred to a simple test chip for measuring with PD. Here, we utilize a portable indium tin oxide (ITO) coated polyethylene terephthalate (PET) heater. ITO-PET has been used previously for continuous-flow PCR which requires multiple heating zones as the sample is flowed through and then analyzed outside of the flow through device [32,33]. Here, we developed a low power ITO-PET heater for isothermal amplification directly on chip. This method of on-chip PD-LAMP eliminates the need for sample transfer from tube and potential cross-contamination.
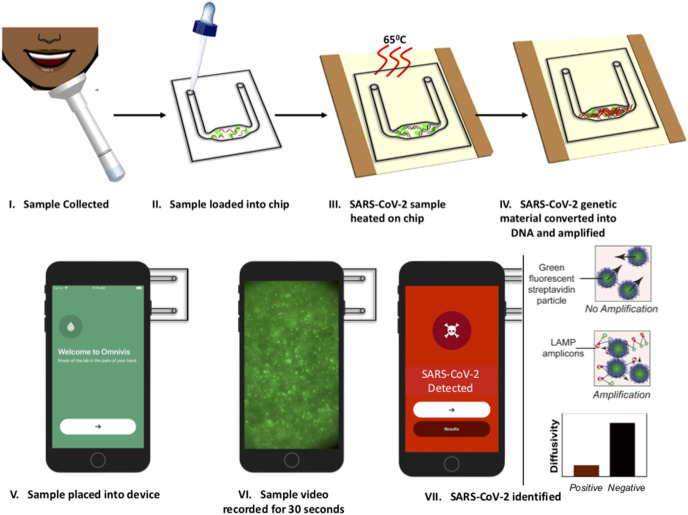
Fig. 1.
Illustration of PD-LAMP process. (I) The sample is collected and (II) loaded into the test chip containing lyophilized reagents and streptavidin coated particles for PD. (III) The chip is heated on an ITO-PET heater for 35 min and then (IV) the amplified product is placed into the (V) smartphone device. (VI) A 30 s video is taken of the sample and the results are analyzed (VII) indicating the presence or absence of SARS-CoV-2 based on particle diffusivity.
The RT-LAMP reagents and positive or negative sample are loaded onto the chip, amplified using the 1-inch square ITO-PET heater and analyzed in the smartphone device using PD. PD measures the diffusion coefficient of fluorescent particles suspended within the amplification reaction. As amplification occurs, the streptavidin-coated particles bind to the LAMP products via the biotinylated LF primer. The long strands of the amplified LAMP products increase the hydrodynamic radius of the particle as well as the overall solution viscosity, thereby reducing the diffusion coefficient of the particles in the solution. The smartphone images the particle motion and outputs the diffusion coefficient, which outputs lower for positive samples than negative samples in which LAMP has not occurred (Fig. 1). Within 35 min, we detected as few as 50 SARS-CoV-2 viral particles per reaction with up to 10% saliva in the reaction. The LOD of 50 particles per reaction was further obtained using lyophilized reagents stored on-chip targeting both nsp3 and N2 conserved regions in 8% (vv−1) saliva. Resuspending lyophilized reagents and heating on test chips increases ease-of-use and eliminates cold storage, both of which are necessary for a PoC device [34,35]. These detection limits meet the requirements outlined by the WHO (103 to 101 copies μL−1) for testing and have the advantage of minimal sample processing [36].
2. Methods
2.1. Viral targets
The following reagents were obtained through BEI Resources (Manassas, VA), NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, Heat Inactivated, NR-52286; Middle East Respiratory Syndrome Coronavirus (MERS CoV), EMC/2012, Irradiated Infected Cell Lysate, NR-50549; and SARS-CoV, Gamma-Irradiated and Sucrose-Purified, 1 × 109 PFU Equivalents per mL in PBS, NR-9547.
2.2. Reverse transcription loop mediated isothermal amplification (RT-LAMP)
RT-LAMP was performed using a primer set developed in-house to target the nsp3 region of ORF1ab (Table S1) and a primer set targeting the N2 region of the N gene developed by Butler et al. (Table S2) [37]. The reaction mixture contained the following components (also listed in Table S3): 1.6 μM FIP/BIP primers, 0.2 μM F3/B3 primers, 0.4 μM LF/LB primers, 1x Isothermal Amplification Buffer II (New England Biolabs, Ipswich, MA), 1.4 mM dNTPs (Agilent, Santa Clara, CA), 200 mM Betaine (Sigma-Aldrich, St. Louis, MO), 0.5x ROX Reference dye (Invitrogen, Carlsbad, CA), 0.4x Eva Green (Fisher, Hampton, NH), 6U of BST 3.0 DNA polymerase (New England Biolabs, Ipswich, MA) and 10U of SuperScript IV Reverse Transcriptase (Invitrogen, Carlsbad, CA) with 1.6 μL of template (8% saliva experiments; 2 μL of template for 10% saliva experiments) in a total volume of 20 μL. Pooled saliva (Innovative Research, Novi, MI) was used as the sample matrix at up to 10% (v v−1) (2 μL) of the final reaction volume for saliva experiments.
RNA or heat inactivated virus templates were serially diluted 10-fold with DEPC treated nuclease free water (Invitrogen, Carlsbad, CA). Isothermal incubation at 65 °C was performed for 35 min in a QuantStudio 5 Real-Time PCR machine (ThermoFisher, Waltham, MA) or on ITO-PET (see On-Chip Amplification).
For all on-chip experiments, 400 nm streptavidin coated Dragon Green fluorescent particles (Bangs Laboratories, Inc. Fishers, IN) were added to the reactions at a final concentration of 2.3 × 109 particles mL−1 prior to amplification. For initial in-tube amplification experiments, the fluorescent beads were added at the same final concentration prior to detection on-chip.
RT-LAMP products were visualized using an ethidium bromide stained 2% agarose gel at 100V for 50 min. The gel was imaged using an ultraviolet light gel system (c400, Azure Biosystems, Dublin, CA). Gel images were collected with an exposure time of 15 s using the Azure Series software at settings of UV302. Gel images were transferred from the Azure as.tiff files and have not been edited in this manuscript.
2.3. Chip fabrication
The chip is composed of 3 layers: 2 layers of 188 μm cyclic olefin polymer (COP) and, 1 layer of 50 μm COP (Zeonor, Tokyo, Japan). For the 2 layers of 188 μm COP, patterns are designed using Adobe Illustrator and transferred to the Silhouette Studio 2.0 Software (Silhouette America, Lindon, UT). The 188 μm COP layers were cut using the Silhouette Cameo Craft Cutter (Silhouette America, Lindon, UT) with two passes using a 10-blade, force of 29, and a speed of 10 (arbitrary units within the Silhouette Cameo software). The top imaging layer, made from 50 μm COP, was hand-cut to cover the channel cutout on the 188 μm COP layer (Fig. S1). The first two layers of 188 μm were bonded together using a Carver 4386 hydraulic press (Carver Inc., Wabash, IN) with 1.2 tons of pressure at 120 °C for 2 min. After bonding the two 188 μm layers together, the remaining 50 μm COP is bonded with the same pressure and temperature settings in the hydraulic press for 1 min. The final assembled chip has dimensions of 25.4 mm long by 22.10 mm wide and 0.426 mm thick, holding approximately 20 μL of sample volume.
Prior to loading the reactions, the completed COP chips were cleaned by flowing three separate rinsing solutions through the channel: 10% bleach, followed by 70% ethanol, and then MilliQ water using a pipette and capillary action. Compressed air was used to blow dry the chips in between each rinse. After rinsing and blowing with compressed air, the chips were filled with MilliQ water again without removal and re-annealed by heating in the Carver press for 60 s at 120 °C and 1 metric ton rotating 180° after 30 s.
2.4. Lyophilization in chip
For lyophilization experiments, the amount of betaine was reduced to 100 mM final concentration to ensure proper freeze drying of the RT-LAMP reagents in chip. 10% (w v−1) trehalose was added as a cryoprotectant. The chips (Fig. S1) were frozen for 30 min at −80 °C and then placed on a lyophilizer shelf (SP, Warminster, PA) at −40 °C and 100 mTorr overnight. Before removal, the shelf temperature was set to −25 °C for 1 h and then −5 °C for 1 h. The reaction was rehydrated with 20 μL of diluted saliva and SARS-CoV-2 sample or controls.
2.5. On-chip amplification
The heater component was made by using ITO-PET (MSE Supplies, Tucson, AZ) cut into 1.5 × 1.5 inch2 squares with two strips of 0.5-inch copper tape (McMaster-Carr, Elmhurst, IL) adhered to the ITO making the electrical connections needed to drive heating (Fig. S1).
To supply the power to the heating unit, a DC benchtop power supply (Model 1550, B&K Precision, Yorba Linda, CA) was initially set to 3.5 V. A thermocouple (RDXL4SD, Omega Engineering, Stamford CT) was used to monitor the temperature at 65 1 °C, while increasing or decreasing voltage accordingly (Fig. S8). RT-LAMP reactions were prepared as previously described with 20 μL reaction volumes embedded into the chip. The chip was then sealed with PCR tape (Bio-Rad, Hercules, CA) and placed on the ITO heater for a 35-min reaction time. After 35 min of amplification, chips were removed from the heater, left to cool under ambient temperature for 2 min allowing the chip to equilibrate to the room temperature, and then analyzed using the smartphone device.
2.6. Particle diffusometry (PD) theory
PD quantifies the Brownian motion of suspended particles within the liquid sample by calculating the statistical average diffusion coefficient using correlation analysis. Individual images are equipartitioned into smaller areas each containing, on average, 8–10 particles, and pair-wise correlation analysis is performed.
| (1) |
The first image is correlated with a second image that is separated by a time step, , for cross-correlation, (Sc), and the image is correlated with itself for an autocorrelation process, (Sa). Peak widths from both correlation analyses, Sa and Sc, are calculated, and Equation (1) is used to determine the average diffusion coefficient [38]. For statistical robustness and to reduce random error, each data point is comprised of an average of 100 diffusion coefficients. For recording the experimental data, sample is placed in chip with its overall dimension and the imaging chamber size requirements held by the recording device. Experimental data are imaged via 30 s videos collected using an iPhone6-driven custom microscope device and processed using an in-house smartphone application, fully described in previous work [30,39].
2.7. Statistical analysis
All PD measurements of SARS-CoV-2 RT-LAMP amplicons were compared to negative controls with a one-way ANOVA post-hoc Dunnett's test to determine statistical significance. Tukey's multiple comparison test was used in specificity experiments to compare each negative control to every positive concentration. Box and whisker plots were generated for the PD data where the upper and lower bounds of the boxes represent the 75th and 25th percentile about the mean, respectively, and the minimum and maximum values are represented by the upper and lower whiskers.
3. Results
3.1. RT-LAMP primer optimization
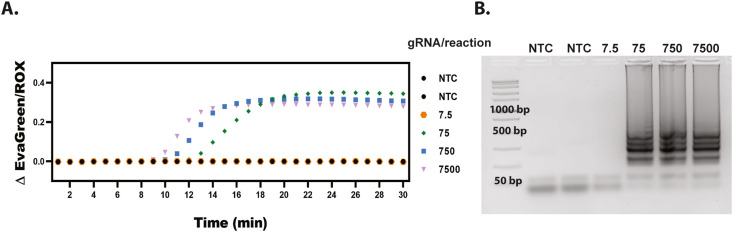
To ensure sensitive detection of SARS-CoV-2 we designed a primer set targeting the conserved nsp3 region of the ORF1ab gene (Table S1) and tested it in an in-tube RT-LAMP reaction with various concentrations of SARS-CoV-2 genomic RNA (gRNA). The in-house designed primers could amplify the genomic RNA in less than 30 min without any negative amplification occurring. We ran the assay for 30 min five times (Fig. S2) and found a LOD of 75 genomic RNA (gRNA) copies per reaction (representative results shown in Fig. 2 ). 7.5 gRNA copies per reaction did amplify 2 out of the 5 times. Therefore our limit of detection was determined to be 75 gRNA copies per reaction as this concentration amplified for every repeat (Fig. S2, N = 5).
Fig. 2.
SARS CoV-2 gRNA LAMP LOD. (A) Representative results of quantitative RT-PCR graph from SARS-CoV-2 genomic RNA showing amplification for 75, 750, and 7500, but not 7.5 gRNA copies per reaction. (B) Representative 2% agarose gel indicating LAMP banding down to 75 copies per reaction.
3.2. On-chip amplification of SARS-CoV-2 virus
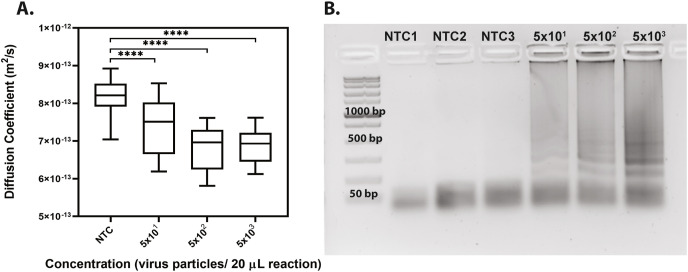
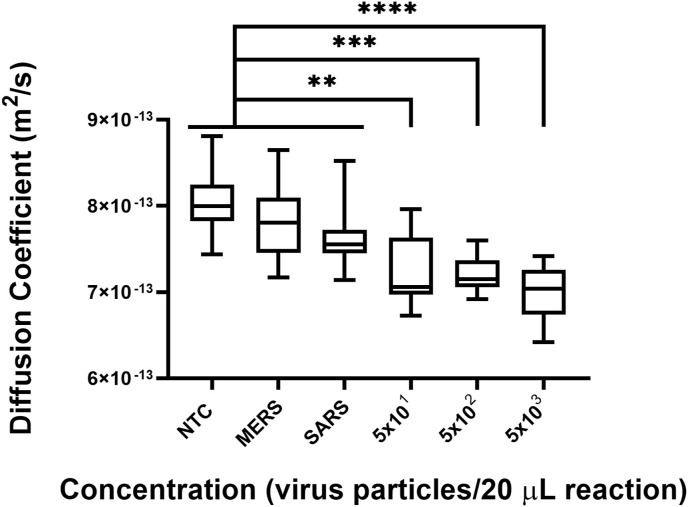
After optimizing the RT-LAMP in-tube, we performed further experiments on-chip using our in-house primer set and inactivated SARS-CoV-2 viral particles and analyzed the samples with PD-LAMP. A significant difference in the diffusion coefficient of fluorescent particles in SARS-CoV-2 samples relative to controls in which no genetic template was added (no template controls, NTC) was found for each repeat (N = 3) (Fig. 3 and Fig. S3). Each chip was analyzed for 30 s, and the resulting diffusion coefficient measurements were recorded and analyzed using a one-way ANOVA with post hoc Dunnett's against the NTC. The diffusion coefficients were all significantly different from the NTC (p < 0.0001 for 5 × 101, 5 × 102 and 5 × 103 virus particles per reaction). We determined a LOD for on-chip amplification to be 50 virus particles per reaction which was consistent with our detection limits off-chip of 75 gRNA copies per reaction and 50 virus particles per reaction. This detection limit is within the range recommended by the WHO [40]. Therefore, our ITO-PET heater can be used to drive RT-LAMP reactions in a portable, and compact manner.
Fig. 3.
LOD of SARS-CoV-2 detection on chip. (A) PD-LAMP diffusion coefficient measurements show a decreasing trend with higher concentration of virus (from 5 × 103 to 5 × 101 virus particles (20 μL reaction) −1). A significant difference from the NTC was seen for 5 × 101, 5 × 102 and 5 × 103 (∗∗∗∗p < 0.0001) virus particles per reaction using one-way ANOVA with Dunnett's post-hoc relative to NTC (N = 3). (B) A representative image of 2% agarose gel confirming amplification in the positive samples as seen by the LAMP banding pattern.
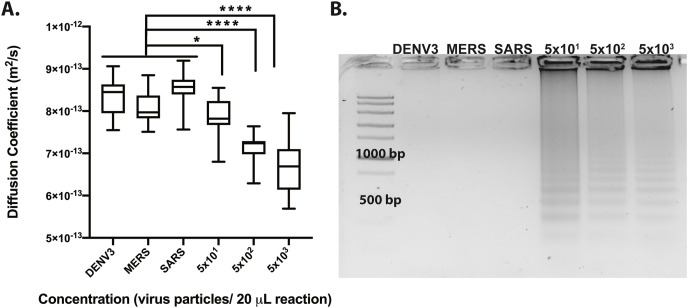
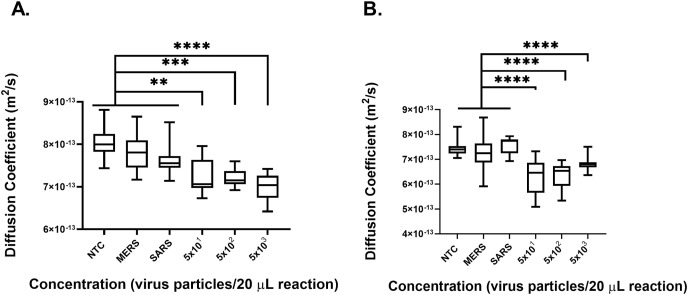
After confirming the LOD of on chip amplification, we investigated the cross-reactivity of the PD-LAMP assay against other viruses. 1.6 μL of Dengue Virus type III (DENV3), SARS, and MERS from an initial concentration of 1 × 104 virus particles μL−1 were added as control templates to a total reaction volume of 20 μL. DENV3 causes similar respiratory symptoms to SARS-CoV-2, and SARS and MERS are coronaviruses, also with similar symptoms to SARS-CoV-2. After 35 min of amplification on chip, it was confirmed PD-LAMP was indeed specific to SARS-CoV-2 and did not amplify DENV3, MERS, or SARS (Fig. 4 , N = 3). After the 35 min of reaction, samples were removed from the chip and run on gel electrophoresis to visually confirm amplification (Fig. 4B). Diffusion coefficients of SARS-CoV-2 at various concentrations were significantly different from each off-target virus (DENV3, MERS, SARS), using a Tukey's multiple comparison test (∗p < 0.05 for 5 × 101, ∗∗∗∗p < 0.0001 for 5 × 102 and 5 × 103 virus particles per reaction). Fig. S4 shows each individual run. Further, the diffusion coefficients of the off-target viruses did not exhibit a significant difference from each other (p > 0.05). These results indicate that PD-LAMP is specific to the SARS-CoV-2 coronavirus and no other common viruses, even coronaviruses.
Fig. 4.
On chip heating comparing cross-reactivity of SARS-CoV-2 nsp3 primer set against DENV3, MERS, and SARS inactivated virus. (A) Diffusion coefficient measurements indicate a significant difference between each off-target viruses and SARS-CoV-2 virus. Diffusion coefficients measurements trend toward lower values for the higher positive SARS-CoV-2 concentrations. While DENV3, MERS, and SARS were not statistically different from one another, significant differences were found for 5 × 101 (∗p < 0.05), 5 × 102 and 5 × 103 (∗∗∗∗p < 0.0001) SARS-CoV-2 viral particles per reaction concentrations compared to these off-target viruses via Tukey's multiple comparison test (N = 3). (B) 2% agarose gel confirmed PD results from samples extracted from each chip.
3.3. On chip amplification with saliva
Having confirmed that PD-LAMP for SARS-CoV-2 detection had little cross-reactivity to other viruses, we then investigated the robustness of PD-LAMP in saliva samples. The viscosity of saliva reduces the solution's initial diffusion coefficient and can introduce changes in RT-LAMP amplification. Prior to testing on-chip, we amplified SARS-CoV-2 gRNA in-tube and performed secondary confirmation with gel electrophoresis to determine the maximum concentration of saliva that could be used in an RT-LAMP assay (Fig. S5). Saliva was added at a final concentration of 8% (v v−1) (Fig. 5 A) and 10% (v v−1) (Fig. 5B) in the 20 μL reaction. SARS and MERS were again introduced as negative controls at a final concentration of 1.6 × 104 virus particles per 20 μL reaction while SARS-CoV-2 concentrations of 5 × 103 - 5 × 101 virus particles per 20 μL reaction were added to the reaction for on-chip testing. The chips were heated for 35 min with the ITO-PET heater and then analyzed via PD. There was a significant difference between the controls relative to the positive samples (∗∗p < 0.01). PD yielded a lower diffusion coefficient for the positive samples when compared to the negative samples for both 8% and 10% saliva (Fig. 5A and B), indicating that RT-LAMP successfully produced amplicons for positive samples on-chip. Thus, demonstrating that PD sensitively measured SARS-CoV-2 presence. Overall, diffusion coefficients presented lower values in the presence of saliva, which was expected due to its higher viscosity. Additionally, one of the three MERS samples amplified in 10% saliva. The MERS amplification may have been due to a pipetting error, carryover contamination, or because the diluted MERS template was contaminated. Due to this negative amplification event, MERS was significantly different from the other negative control SARS (∗p < 0.05). PD-LAMP was still found to be specific for SARS-CoV-2 viral particles in 8% and 10% saliva (Fig. 5). Detection of SARS-CoV-2 via PD-LAMP was found to have a LOD of 50 particles per 20 μL reaction in both 8 and 10% saliva.
Fig. 5.
On chip heating sensitivity and specificity of PD-LAMP for SARS-CoV-2 samples containing 8% and 10% saliva. MERS and SARS inactivated virus were diluted to a concentration of 1 × 104 particles μL−1. SARS-CoV-2 inactivated virus was diluted with DEPC treated nuclease free water at concentrations ranging from 5 × 103 to 5 × 101 virus particles per 20 μL reaction in 10-fold dilutions. (A) Diffusion coefficient measurements from reactions containing 8% saliva indicate significant differences from the NTC for 5 × 101, 5 × 102 (∗∗p < 0.01) and 5 × 103 (∗∗∗∗p < 0.0001) virus particles per 20 μL reaction. The same significance was found against MERS and then SARS. There was no significant difference between the negative controls. (B) Diffusion coefficient measurements from reactions containing 10% saliva indicate significant differences from the NTC, MERS and SARS for 5 × 101, 5 × 102, and 5 × 103 (∗∗∗∗p < 0.001). There was no significant difference between NTC and SARS or NTC and MERS. However, MERS was significantly different from SARS (∗p < 0.05). Statistical analysis was performed with Tukey's multiple comparison test (N = 3).
3.4. Amplification from lyophilized master mix on chip
Reactions were unable to lyophilize with the full concentration of Betaine, an anti-freezing agent. Therefore, the lyophilization protocol was optimized by reducing the amount of betaine in the reaction to allow for proper freezing. The master mix containing two primer sets (S1 and S2 Tables) and 2.3 × 109 fluorescent beads mL−1 particles in a total reaction volume of 20 μL was lyophilized in the chips. After at least one day of storage at room temperature the chips were rehydrated with 20 μL of a mixture containing positive or negative template, 8% (v v−1) saliva and water. MERS and SARS were again tested to evaluate cross-reactivity of the combined SARS-CoV-2 primer sets and the off-target viruses at a concentration of 1.6 × 104 virus particles per reaction volume. SARS-CoV-2 was tested at concentrations from 5 × 103 to 5 × 101 virus particles per reaction. PD-LAMP was successful in detecting positive samples from negative and off-target samples (Fig. 6 ). SARS-CoV-2 amplification resulted in lower diffusion coefficients for the positive samples relative to NTC and off-target samples. Each of the SARS-CoV-2 positive concentrations was found to be significantly different from each of the control samples using Tukey's multiple comparison test (∗∗p < 0.01 5 × 101 virus particles per reaction, ∗∗∗p < 0.001 5 × 102 virus particles per reaction, and ∗∗∗∗p < 0.0001 5 × 103 virus particles per reaction) (Fig. 6). Performing PD-LAMP from resuspending lyophilized reagents yielded a LOD of 30 particles μL−1 and was specific to SARS-CoV-2.
Fig. 6.
Performing PD-LAMP on-chip with lyophilized reagents. MERS and SARS inactivated virus were added at a concentration of 1.6 × 104 virus particles per 20 μL reaction. SARS-CoV-2 inactivated virus was diluted with DEPC treated nuclease free water at concentrations ranging from 5 × 103 to 5 × 101 viral particles per reaction in 10-fold dilutions. Diffusion coefficient measurements from reactions containing 8% saliva indicate significant differences from the controls (NTC, MERS, and SARS) for 5 × 101, 5 × 102 (∗∗p < 0.01) and 5 × 103 (∗∗∗∗p < 0.0001) virus particles per 20 μL reaction. Statistical analysis was performed using Tukey's multiple comparison test (N = 3).
4. Discussion
Tracing and rapidly diagnosing infected patients are key to controlling disease spread. The COVID-19 pandemic is a prime example of the importance of affordable and rapid diagnostics [34,41]. iNAAT-based methods offer an alternative to RT-PCR tests by eliminating the need for expensive temperature controlling units [17,42]. Furthermore, the ability to perform RT-LAMP with minimal sample prep in a portable platform would provide a highly sensitive PoC device to aid in rapid confirmation of cases and surveillance of the virus. RT-LAMP provides an alternative to current serological tests that require strenuous sample prep.
Smartphone devices have advantages such as a GPS, camera, and advanced computational processing power that can aid in medical diagnosis [43,44]. The pairing of PoC testing with automated sample processing is of great importance for scalability and rapid processing [21]. We were able to target SARS-CoV-2 by performing RT-LAMP in a microfluidic chip with a LOD of 50 virus particles per reaction. We achieved LODs with lyophilized reagents and on-chip heating that are equivalent to those from heating in the PCR machine. These comparable results are promising for the development of a portable point of care detection device for SARS-CoV-2.
We performed RT-LAMP in the presence of MERS cell lysate and SARS inactivated virus to assess the cross-reactivity of the reaction against other coronaviruses as well as with DENV3 inactivated virus, a virus with similar symptoms as SARS-CoV-2. We found SARS-CoV-2 PD-LAMP to be specific even when challenged with other coronaviruses and DENV3 (Fig. 4). Next, we performed RT-LAMP in various concentrations of saliva and determined the LOD in 8% and 10% saliva to be 50 virus particles per reaction (Fig. 5). Diffusion coefficients measured by PD in the presence of saliva were overall lower when compared to samples without saliva. This was expected due to higher viscosity of saliva. Regardless, there still may be room for further optimization of the assay to ensure no non-specific amplification occurs in the presence of saliva. In the MERS samples, the average diffusion coefficient-value was significantly different than the positive SARS-CoV-2 amplification, and not significantly different from other negative controls (Fig. 5), however in some repeats non-specific amplification and primer dimers may have occurred in some cases, as seen in the increased standard deviation of MERS sample in Fig. 5B. Comparing the 8% and 10% saliva samples we saw a greater statistical difference between positive SARS-CoV-2 samples and controls in the 10% saliva samples. This difference may be due to the larger volume of virus and saliva. Investigation of PD-LAMP in minimally processed saliva samples, moves the technology towards use at the PoC.
In order to realize these methods for PoC testing, the lyophilization of reagents in-chip allows for fewer sample processing steps and negates the need for cold-chain storage. Fewer sample processing steps reduces pipetting operations and possible contamination easing the usability for minimally trained personnel. Rehydration of the lyophilized reagents in a single step also helps to prevent contamination. The elimination of cold storage is significant as this reduces transportation costs and is key for use in developing areas where the cold chain may be difficult to maintain. In addition, drying down multiple primer sets for the targeting of two genes allows for greater sensitivity in the event of variants and mutations which are common for SARS-CoV-2 [6,45].
Various lyophilization techniques have been studied but have been limited by the difficulty in freezing the buffers and dyes of the LAMP reaction [46,47]. The addition of 10% (w v−1) trehalose as a cryoprotectant improves the performance of LAMP and aids in stabilization of proteins [48]. One group explored the creation of lyophilized LAMP buttons which were stable for at least 24 h at room temperature but found a degradation in the intensity of the dyes needed for their paper-based method [49]. Others have lyophilized the dyes in the cap of tubes separately from the remaining reagents to combat the loss in dye intensity [50]. In the current work dyes are not a necessary component of the assay, making the lyophilization process less complicated, however optimization of lyophilization conditions was still necessary. We found that the lyophilization process would not complete in concentrations of betaine greater than 200 mM due to its extremely low freezing point. The concentration was lowered to 100 mM with no reduction in assay sensitivity. We performed PD-LAMP on chip after at least 24 h of room temperature storage and saw a significant difference from the negative diffusion coefficients with a LOD of 30 virus particles μL−1 and the technique was specific to SARS-CoV-2 (Fig. 6). The performance of PD-LAMP reagents over a range of storage conditions remains to be studied.
Furthermore to truly make the device field ready, an analysis of more samples using the current setup and microfluidic chips will be needed to create proper cut-off values for positive and negative samples. Previously we have determined cut-offs for PD-LAMP of 150 blinded samples of V. Cholerae in pond water [30]. Coefficients less than 7.0 × 10−13 m2/s were considered positive and coefficients greater than 7.2 × 10−13 m2/s were negative with all coefficients in between yielding results which were inconclusive and would need to be retested. However, the chips used for the previous study were not the same design as the microfluidic chip described here and the differences in saliva vs water samples are expected to affect background solution viscosity. While all negative samples did maintain values greater than 7.0 × 10−13 there was a wider range for the positive samples. One limitation of the current chip design, is that it does not always create an airtight seal. This allows for the sample to flow which can interfere with the true Brownian motion of particles and in turn yield higher diffusion coefficients even in more complex matrices with higher viscosities than water. After lyophilization, the contact angle of liquid in the chip changes also inducing greater susceptibility to flow. This will need to be further explored to reduce flow or a change in our in-house processing application made to account for flow. This would aid in improving the robustness of the assay and readiness for use at the point-of-care.
5. Conclusions
In summary, we demonstrate the performing RT-LAMP on-chip using a portable ITO-PET heating platform with lyophilized reagents with detection of SARS-CoV-2 down to 50 virus particles per 20 μL reaction in 10% saliva via PD-LAMP. This assay could be used to selectively identify a sample that is negative for SARS-CoV-2 even when other pathogenic viruses are present in the sample. Performing this assay in less than 35 min is another key step to placing it within the realm of current FDA-EUA approved diagnostics that use isothermal amplification methods such as Abbot ID Now and the Cue Covid-19 test [22,23]. These approved tests have 100% sensitivity and specificity. The next steps towards full integration and use at the point of care for the PD-LAMP detection of SARS-CoV-2 would be verification with clinical samples.
Funding
This work was supported by Vodafone Americas Foundation Wireless Innovation Project Award, National Institutes of Health (NIH), National Institute of Allergy and Infection Disease (NIAID) Award Number R61AI140474, the Indiana Clinical and Translational Sciences Institute (IN-CTSI) which is funded in part by Award Number UL1TR002529 from the NIH, National Center for Advancing Translational Sciences (NCATS), Clinical and Translational Sciences Award, and the George Washington Carver Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and other funding institutions.
CRediT authorship contribution statement
Ashlee J. Colbert: Methodology, Visualization, Validation, Investigation, Writing – original draft, Formal analysis, Writing – review & editing. Dong Hoon Lee: Methodology, Visualization, Validation, Investigation, Writing – original draft, Formal analysis, Writing – review & editing. Katherine N. Clayton: Resources, Funding acquisition, Writing – review & editing. Steven T. Wereley: Resources, Supervision, Writing – review & editing, Funding acquisition. Jacqueline C. Linnes: Conceptualization, Supervision, Writing – review & editing, Funding acquisition. Tamara L. Kinzer-Ursem: Conceptualization, Supervision, Writing – review & editing, Funding acquisition.
Declaration of competing interest
Katherine N. Clayton, Steven T. Wereley, Jacqueline C. Linnes, and Tamara L. Kinzer-Ursem are co-founders of OmniVis Inc., a company licensing IP from Purdue University to translate the smartphone PD technology. Dr. Clayton is presently the CEO of OmniVis Inc. Ashlee J. Colbert and Dong Hoon Lee have declared that they have no competing interests.
Acknowledgement
We would like to recognize Melinda Lake for providing feedback throughout the manuscript writing process. We would also like to acknowledge Navaporn Sritong, Holly Witten, and Riley Brown for helpful insights into primer optimization.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aca.2022.339702.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.WHO Director-General’s opening remarks at the media briefing on COVID-19. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (n.d.),11 March 2020.
- 2.Novel coronavirus (2019-nCoV), situation report 11. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200131-sitrep-11-ncov.pdf?sfvrsn=de7c0f7_4 n.d.
- 3.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan. Euro Surveill. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization The selection and use of essential in vitro diagnostics. 2021. https://apps.who.int/iris/bitstream/handle/10665/339064/9789240019102-eng.pdf?sequence=1&isAllowed=y
- 6.World Health Organization . World Health Organization; 2021. COVID-19 Weekly Epidemiological Update 22; pp. 1–3.https://www.who.int/docs/default-source/coronaviruse/situation-reports/weekly_epidemiological_update_22.pdf [Google Scholar]
- 7.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization . 2021. Recommendations for National SARS-CoV-2 Testing Strategies and Diagnostic Capacities Interim Guidance. [Google Scholar]
- 9.Eftekhari A., Alipour M., Chodari L., Dizaj S.M., Ardalan M.R., Samiei M., Sharifi S., Vahed S.Z., Huseynova I., Khalilov R., Ahmadian E., Cucchiarini M. A comprehensive review of detection methods for SARS-CoV-2. Microorganisms. 2021;9:1–18. doi: 10.3390/microorganisms9020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi Y., Furuchi M., Kamimoto A., Honda K., Matsumura H., Kobayashi R. Saliva-based PCR tests for SARS-CoV-2 detection. J. Oral Sci. 2020;62:350–351. doi: 10.2334/josnusd.20-0267. [DOI] [PubMed] [Google Scholar]
- 11.Smithgall M.C., Dowlatshahi M., Spitalnik S.L., Hod E.A., Rai A.J. Types of assays for SARS-COV-2 testing: a review. Lab. Med. 2021;51 doi: 10.1093/LABMED/LMAA039. E59–E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/BBRC.2001.5921. [DOI] [PubMed] [Google Scholar]
- 13.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. : Official Journal of the Japan Society of Chemotherapy. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalli M.A., Chen X., Langmade S.J., Fronick C.C., Sawyer C.S., Burcea L.C., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP., MedRxiv. The Preprint Server for Health Sciences. 2020 doi: 10.1101/2020.05.07.20093542. 2020.05.07.20093542. [DOI] [Google Scholar]
- 16.González-González E., Lara-Mayorga I.M., Pablo Rodríguez-5 Sánchez I., Yee-De León F., García-Rubio A., Ezio Garciaméndez-6 Mijares C., Guerra-Alvarez G.E., García-Martínez G., Andrés J., Hernández A., Márquez-García E., Zhang Y.S., Martínez-8 Chapa S.O., Zúñiga J., Trujillo-De Santiago G., Moisés Alvarez M. Scaling diagnostics in times of COVID-19: colorimetric Loop-mediated Isothermal 1 Amplification (LAMP) assisted by a 3D-printed incubator for cost-effective and 2 scalable detection of SARS-CoV-2 3 4. 2020. [DOI]
- 17.Huang W.E., Lim B., Hsu C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microbial. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park G.S., Ku K., Baek S.H., Kim S.J., Il Kim S., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priye A., Bird S.W., Light Y.K., Ball C.S., Negrete O.A., Meagher R.J. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 2017;7(1):1–11. doi: 10.1038/srep44778. 7 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ID NOW COVID-19 | Abbott point of care testing. https://www.globalpointofcare.abbott/en/product-details/id-now-covid-19.html n.d.
- 23.Real-time COVID-19 testing | Cue. https://www.cuehealth.com/what-is-cue/how-cue-detects-covid-19/ (n.d.
- 24.To K.K.W., Tsang O.T.Y., Yip C.C.Y., Chan K.H., Wu T.C., Chan J.M.C., Leung W.S., Chik T.S.H., Choi C.Y.C., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W.S., Fung A.Y.F., Hung I.F.N., Cheng V.C.C., Chan J.F.W., Yuen K.Y. vol. 71. An Official Publication of the Infectious Diseases Society of America; 2020. pp. 841–843. (Consistent Detection of 2019 Novel Coronavirus in Saliva, Clinical Infectious Diseases). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C.Y., Cai J.P., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C.K., Poon R.W.S., Luo C.T., Cheng V.C.C., Chan J.F.W., Hung I.F.N., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lalli M.A., Chen X., Langmade S.J., Fronick C.C., Sawyer C.S., Burcea L.C., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP., MedRxiv. The Preprint Server for Health Sciences. 2020 doi: 10.1101/2020.05.07.20093542. 2020.05.07.20093542. [DOI] [Google Scholar]
- 27.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Geng B., Muenker M.C., Moore A., Vogels C., Petrone M., Ott I., Lu P., Lu-Culligan A., Klein J., Venkataraman A., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L., Valdez J., White E., Lapidus S., Kalinich C., Jiang X., Kim D., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C., Omer S., Dela Cruz C., Farhadian S., Martinello R., Iwasaki A., Grubaugh N., Ko A. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. Camila Odio. 2020;8(3):12. doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- 28.Koskinen A., Tolvi M., Jauhiainen M., Kekäläinen E., Laulajainen-Hongisto A., Lamminmäki S. JAMA Otolaryngology-- Head & Neck Surgery; 2021. Complications of COVID-19 Nasopharyngeal Swab Test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park G.S., Ku K., Baek S.H., Kim S.J., Il Kim S., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moehling T.J., Lee D.H., Henderson M.E., McDonald M.K., Tsang P.H., Kaakeh S., Kim E.S., Wereley S.T., Kinzer-Ursem T.L., Clayton K.N., Linnes J.C. A smartphone-based particle diffusometry platform for sub-Attomolar detection of Vibrio cholerae in environmental water. Biosens. Bioelectron. 2020;167:112497. doi: 10.1016/j.bios.2020.112497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colbert A.J., Co K., Lima-Cooper G., Lee D.H., Clayton K.N., Wereley S.T., John C.C., Linnes J.C., Kinzer-Ursem T.L. Towards the use of a smartphone imaging-based tool for point-of-care detection of asymptomatic low-density malaria parasitaemia. Malar. J. 2021;20(1):1–13. doi: 10.1186/S12936-021-03894-W. 20 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuba T., Yamamoto T., Naganuma T., Fujii T. Microfabricated flow-through device for DNA amplification - towards in situ gene analysis. Chem. Eng. J. 2004;101:151–156. doi: 10.1016/j.cej.2003.11.016. [DOI] [Google Scholar]
- 33.K. Sun, A. Yamaguchi, Y. Ishida, S. Matsuo, H. Misawa, A Heater-Integrated Transparent Microchannel Chip for Continuous-Flow PCR, n.d.
- 34.Nguyen T., Bang D.D., Wolff A. Novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines. 2019;11:1–7. doi: 10.3390/MI11030306. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen T., Andreasen S.Z., Wolff A., Bang D.D. From lab on a chip to point of care devices: the role of open source microcontrollers. Micromachines. 2018;9 doi: 10.3390/mi9080403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0. https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1 n.d.
- 37.Butler D.J., Mozsary C., Meydan C., Danko D., Foox J., Rosiene J., Shaiber A., Afshinnekoo E., MacKay M., Ivanov N.A., Sierra M., Pohle D., Zietz M., Ramlall V., Westover C.D., Ryon K., Young B., Bhattacharya C., Ruggiero P., Langhorst B.W., Tanner N., Meleshko D., Xu D., D Steel P.A., Shemesh A.J., Thierry-Mieg J., Thierry-Mieg D., Schwartz R.E., Bezdan D., Sipley J., Cong L., Craney A., Velu P., Tatonetti N., Imielinski M., Rennert H., Mason C.E. Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Shawn Levy. 2020;18:14. doi: 10.1101/2020.04.20.048066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen M.G., Adrian R.J. Brownian motion and correlation in particle image velocimetry. Opt Laser. Technol. 2000;32:621–627. doi: 10.1016/S0030-3992(00)00119-5. [DOI] [Google Scholar]
- 39.Clayton K.N., Moehling T.J., Lee D.H., Wereley S.T., Linnes J.C., Kinzer-Ursem T.L. Particle diffusometry: an optical detection method for Vibrio cholerae presence in environmental water samples. Sci. Rep. 2019;9 doi: 10.1038/s41598-018-38056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0. https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1 n.d.
- 41.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China — key questions for impact assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 42.Hou Tieying, Zengid W., Yang M., Chenid W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Guid B., Wang J., Xuid T. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. 2020. [DOI] [PMC free article] [PubMed]
- 43.Hernández-Neuta I., Neumann F., Brightmeyer J., Ba Tis T., Madaboosi N., Wei Q., Ozcan A., Nilsson M. Smartphone-based clinical diagnostics: towards democratization of evidence-based health care. J. Intern. Med. 2019;285:19–39. doi: 10.1111/joim.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon L., Long K.D., Wan Y., Yu H., Cunningham B.T. Medical diagnostics with mobile devices: comparison of intrinsic and extrinsic sensing. Biotechnol. Adv. 2016;34:291–304. doi: 10.1016/J.BIOTECHADV.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Rahimi A., Mirzazadeh A., Tavakolpour S. Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics. 2021;113:1221–1232. doi: 10.1016/j.ygeno.2020.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H.-W., Weissenberger G., Ching W.-M. Development of lyophilized loop-mediated isothermal amplification reagents for the detection of leptospira. Mil. Med. 2016;181:227. doi: 10.7205/MILMED-D-15-00149. [DOI] [PubMed] [Google Scholar]
- 47.Hayashida K., Kajino K., Hachaambwa L., Namangala B., Sugimoto C. Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for human African trypanosomiasis. 2015. [DOI] [PMC free article] [PubMed]
- 48.Mok E., Wee E., Wang Y., Trau M. Nature Publishing Group; 2016. Comprehensive Evaluation of Molecular Enhancers of the Isothermal Exponential Amplification Reaction OPEN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paper-based loop-mediated isothermal amplification and lateral flow (LAMP-LF) assay for identification of tissues of cattle origin | Elsevier Enhanced Reader. https://reader.elsevier.com/reader/sd/pii/S0003267021000465?token=60524D75B499F92E4307BF2997888215B3DDFFFD36240212A7D774CB038705EED4ACE1154ACEEE158B460534E5F85EDD&originRegion=us-east-1&originCreation=20210623120726 n.d. [DOI] [PubMed]
- 50.Advanced lyophilised loop mediated isothermal amplification (L-LAMP) based point of care technique for the detection of dengue virus | Elsevier enhanced reader. https://reader.elsevier.com/reader/sd/pii/S0166093421001075?token=DCD0CF6DD629433F3A2F7C2EDBCEB3D008A6E8567410504BE3D383C76B9FAABB36A4029C8CE92D66B98EDB450F3BB1BA&originRegion=us-east-1&originCreation=20210623122317 n.d. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.