Abstract
Therapeutic mRNA has the potential to revolutionize the treatment of myriad diseases and, in 2020, facilitated the most rapid vaccine development in history. Among the substantial advances in mRNA technology made in recent years, the incorporation of base modifications into therapeutic mRNA sequences can reduce immunogenicity and increase translation. However, experiments from our lab and others have shown that the incorporation of base modifications does not always yield superior protein expression. We hypothesized that the variable benefit of base modifications may relate to lipid nanoparticle chemistry, formulation, and accumulation within specific organs. To test this theory, we compared IV-injected lipid nanoparticles formulated with reporter mRNA incorporating five base modifications (ψ, m1ψ, m5U, m5C/ψ, and m5C/s2U) and four ionizable lipids (C12-200, cKK-E12, ZA3-Ep10, and 200Oi10) with tropism for different organs. In general, the m1ψ base modification best enhanced translation, producing up to 15-fold improvements in total protein expression compared to unmodified mRNA. Expression improved most dramatically in the spleen (up to 50-fold) and was attributed to enhanced protein expression in monocytic lineage splenocytes. The extent to which these effects were observed varied with delivery vehicle and correlated with differences in innate immunogenicity. Through comparison of firefly luciferase and erythropoietin mRNA constructs, we also found that mRNA modification-induced enhancements in protein expression are limited outside of the spleen, irrespective of delivery vehicle. These results highlight the complexity of mRNA-loaded lipid nanoparticle drug design and show that the effectiveness of mRNA base modifications depend on the delivery vehicle, the target cells, and the site of endogenous protein expression.
Keywords: mRNA, lipidoid, lipid nanoparticles, pseudouridine, non-viral gene delivery
Introduction
In vitro transcribed (IVT) mRNA has emerged as a powerful gene therapy platform for a wide range of therapeutic applications such as protein replacement,1,2 gene editing,3,4 vaccines,5-9 and immunotherapies.10,11 Indeed, the lipid nanoparticle mRNA vaccines produced by BioNTech/Pfizer and Moderna have been effectively combating the SARS-CoV-2 pandemic since late 2020.8,9,12 While traditional vaccines typically require years of development, time scales are reduced for mRNA vaccines because mRNA synthesis is relatively quick13,14 and mRNA therapeutics are inherently modular. These attributes confer mRNA therapeutics with theoretically unlimited clinical potential for existing and emerging viruses.15-17
The FDA-approved and other clinically advanced mRNA vaccines and therapeutics rely on a lipid nanoparticle-based delivery vehicle to protect the mRNA cargo and deliver it to the cytosol of target cells.8,9 Fortunately, numerous lipid nanoparticles have been discovered that potently deliver mRNA to select targets including muscle, immune cells, and the liver.3,4,18-22 Despite these successes, many therapeutic applications are hindered by immunogenicity due to both the lipid carrier and the exogenous mRNA molecules.
While the lipid carrier interfaces with immune cells outside of the target cell, the mRNA is primarily exposed to intracellular components of the immune system, which defend against viral genetic material.23 Many cell types express toll-like receptors (TLRs) that, upon binding to mRNA, trigger a signaling cascade. This cascade produces a cytokine and interferon inflammatory response that stalls mRNA translation.24-26 While TLR activation may enable vaccine immunogenicity and efficacy,10,27 it also hinders the potency of protein replacement therapies. Further, all applications of mRNA therapeutics are limited by similar inflammatory responses induced by cytoplasmic receptors such as RIG-I, PKR, and MDA5 upon binding exogenous mRNA.28
These immunostimulatory mRNAs are unmodified – meaning that they contain only unmodified nucleosides – adenosine (A), cytosine (C), uridine (U), and guanosine (G). Such mRNAs are recognized as foreign because they lack post-transcriptional modifications (e.g. pseudouridylation and methylation) that are characteristic of endogenous mRNAs.29,30 Karikó, Weissman, and others have found that the immunogenicity of exogenous mRNA can be mitigated through the incorporation of modified nucleosides.31-34 These nucleosides, including pseudouridine (ψ), 5-methylcytidine (m5C), 5-methyluridine (m5U), and 2-thiouridine (s2U), reduce activation by TLRs and RIG-I.32,35-37 Decreased immune recognition often results in an increase in mRNA translation. Early studies using ψ-modified mRNA achieved both greater mRNA translation and decreased inflammatory cytokine production in human dendritic cells and in mice following intraperitoneal administration relative to uridine-containing mRNA.37 Later studies found that the modification 1-methylpseudouridine (m1ψ) outperforms ψ in vitro and in mice following intramuscular and intradermal injection38 by decreasing immunogenicity and enhancing ribosome binding.35,38,39 The success of this modification led to its incorporation into the BioNTech/Pfizer and Moderna SARS-CoV-2 vaccines.8,9 Of note, while these clinically successful mRNA products used modified mRNA, there are also mRNA candidates without base modifications that have failed in clinical trials, including Translate Bio’s cystic fibrosis treatment40 and CureVac’s SARS-CoV-2 vaccine.41
Given the increasing interest in mRNA therapeutics, we anticipate a surge in related delivery research that will build upon previous discoveries of lipid nanoparticles that potently delivery mRNA. 3,4,10,18,19,21,38-49 Although mRNA base modifications are generally considered to improve efficacy, our lab and others have found that this is not always the case.54 As such, the field would benefit from a better understanding of how delivery vehicles work together with mRNA base modifications to impact protein expression in different tissues. We hypothesized that the variable benefit of base modifications may relate to the chemistry and formulation of mRNA delivery vehicles, and, therefore, to the distribution or translation of the RNA within specific cell or tissue types. To test this idea, we systematically studied a series of lipid nanoparticles incorporating six types of mRNA and four ionizable amino lipids and evaluated efficacy in the liver, spleen, and lungs. Our results show that the utility of mRNA base modifications is not universal; instead, it depends on the delivery material, the biology of target cells, and on the site of endogenous protein expression. Further, we show that, as anticipated, effects relate to the immunogenicity of the mRNA-loaded lipid nanoparticles. Together, these data underscore the importance of tailoring the nucleoside modification to both the delivery system, the target cells, and the application.
Results and Discussion
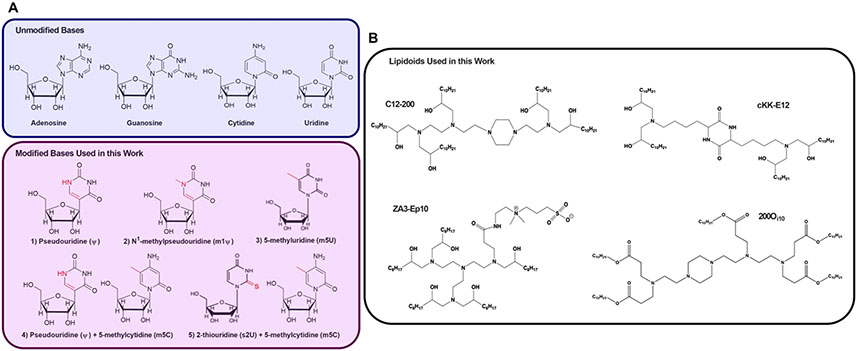
Our goal in this study was to elucidate the relationship between mRNA delivery vehicle, biological site of mRNA translation, and base modification performance. Therefore, we delivered a group of in vitro transcribed (IVT) mRNAs containing nucleoside modifications demonstrated to reduce immune activation and enhance translation using four lipid nanoparticles with differential tropism in vivo. Specifically, we tested unmodified mRNA along with five modified mRNAs (Figure 1A), each of which contained either modified uridine (ψ, m1ψ, and m5U) or modified uridine and cytidine (m5C/ψ and m5C/s2U). Because previous studies have described the mechanisms by which these modifications reduce immunogenicity,32,55,56 we focused on how they affected protein expression in vivo. These six mRNAs were delivered using lipid nanoparticles formulated with the four lipids shown in Figure 1B. These lipid nanoparticles were selected to allow comparison of tissue tropism as a function of mRNA modification. C12-200 and cKK-E12 (also known as MD-1) are five- and four-tailed ionizable lipids that predominately deliver mRNA to the liver.57,58 The ionizable lipid 200Oi10, which has the same amino core as C12-200 but contains ester linkages next to the alkyl tails with a branch on the terminal carbon, induces protein expression in the spleen and the liver.19 Finally, the zwitterionic amino lipid ZA3-Ep10 facilitates mRNA delivery predominantly to the lungs of mice.3
Figure 1. Four unique ionizable amino lipids materials were formulated into lipid nanoparticles containing base-modified mRNA.
(A) In addition to unmodified mRNA, we examined five modified mRNAs. The modifications were of either the uridine nucleoside (ψ, m1ψ, or m5U) or of both uridine and cytidine (m5C/ψ and m5C/s2U). The groups shown in red represent the difference between the modified and unmodified nucleoside. (B) The six mRNAs were formulated into nanoparticles using one of four different lipids: C12-200, cKK-E12, 200Oi10, and ZA3-Ep10.
Nanoparticle biodistribution does not correlate with protein expression
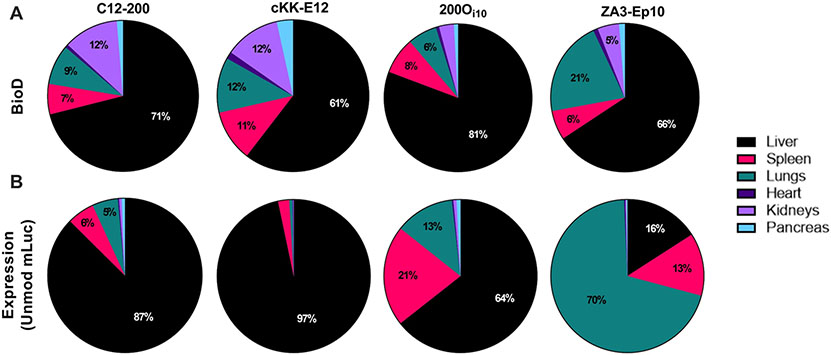
Before delivering modified mRNAs, we conducted a side-by-side comparison of the in vivo delivery properties of the four lipid nanoparticles. First, we examined the biodistribution by injecting mice intravenously with nanoparticles loaded with Cy5-labeled mRNA. One hour after injection, the mice were sacrificed, and the major organs were removed for ex vivo fluorescent imaging. This time point was chosen because our previous work demonstrates that lipid nanoparticles have a serum half-life of ~6 minutes with nearly 100% clearance by 1 hour post-injection with immediate uptake by hepatocytes.59 As shown in Figure 2A and Figure S1, most nanoparticles accumulated in the liver (>60%) and, to a lesser extent, the spleen, kidneys, and lungs.
Figure 2. mRNA biodistribution does not indicate sites of mRNA translation.
(A) To assess biodistribution, mice received IV injections of LNPs carrying Cy5-labeled mRNA at a dose of 0.75 mg/kg. LNPs predominantly distributed to the liver and to the spleen, lungs, and kidneys to a lesser extent. (B) Protein expression was determined by injecting mice via tail vein with LNPs carrying unmodified luciferase mRNA at a dose of 0.75 mg/kg. C12-200 and cKK-E12 facilitated protein expression almost entirely in the liver. 200Oi10 delivery resulted in a more diverse expression profile, as did ZA3-Ep10, with 70% of luciferase expression occurring in the lungs.
Next, we assessed protein expression for each delivery vehicle, as the relationship between nanoparticle biodistribution and protein expression does not always correlate. To do this, we formulated nanoparticles with unmodified firefly luciferase-encoding mRNA (mLuc) and delivered them intravenously at a dose of 0.75 mg/kg. Six hours after injection, we sacrificed the mice and imaged the major organs for bioluminescence (Figure 2B). This time point was chosen because our previous studies show that maximum luciferase expression occurs 6 hours after LNP injection, which allows sufficient time for the mRNA to undergo translation and resultant protein to accumulate and maximize detection sensitivity.4 Consistent with previous reports, C12-200 and cKK-E12 facilitated protein expression almost entirely in the liver (87 and 97%, respectively).57,58 Lipid nanoparticles formulated with 200Oi10 induced a more diverse protein expression pattern (64% liver, 21% spleen, and 13% lungs). Also consistent with previous studies, ZA3-Ep10 nanoparticles produced protein mostly in the lungs (70%).3
Interestingly, despite cKK-E12 having the most diverse biodistribution profile (Figure 2), it had the narrowest expression pattern with almost all translation occurring in the liver. We also noted that, despite having low accumulation in the spleen and lungs, 200Oi10 had significant levels of protein expression in these organs. These discrepancies between biodistribution and protein expression are commonly observed in mRNA delivery studies.19,60 While it isn’t always clear why these discrepancies occur, we postulate that a complex combination of cell-specific differences in nanoparticle internalization, endosomal escape, and translational kinetics likely contribute to this. Based on these results, we categorized the lipid nanoparticles into three categories based on the organ(s) in which they facilitated protein expression: liver (C12-200 and cKK-E12), hybrid (200Oi10), and lung (ZA3-Ep10).
Base modifications improve translation in the spleen in a nanoparticle-dependent manner
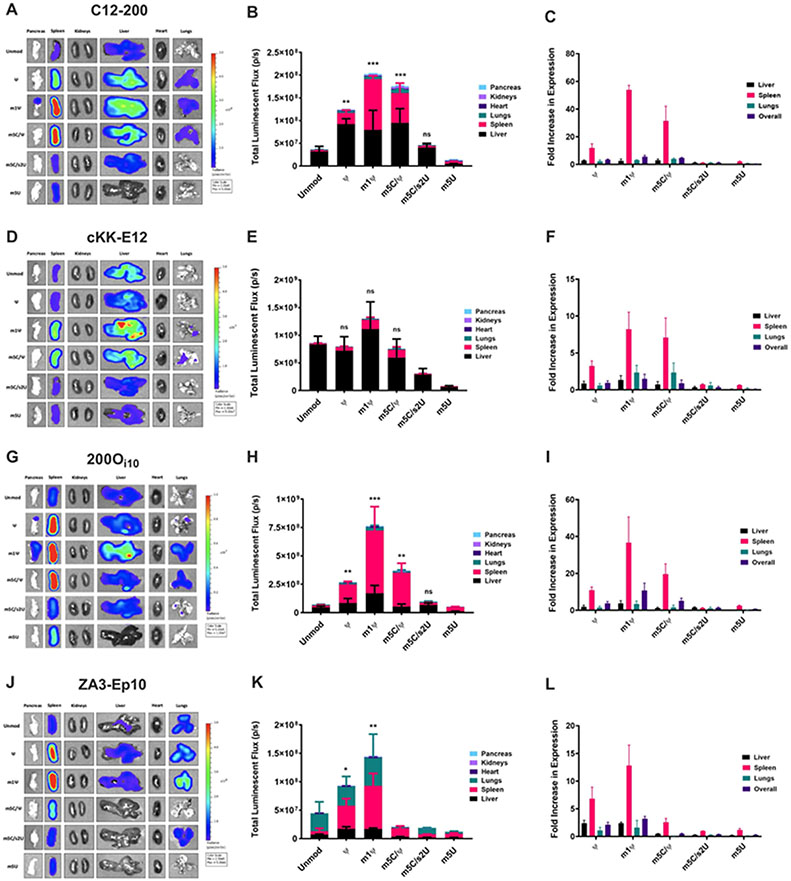
To determine the influence of mRNA modifications on protein translation, we combinatorially formulated nanoparticles with the four lipids and five modified mRNAs encoding mLuc. Nanoparticles were injected intravenously at an mRNA dose of 0.75 mg/kg. Six hours later, mice were sacrificed, and their major organs were imaged for bioluminescence (left column, Figure 3, S2). Luminescence was quantified in each organ using a region of interest analysis to determine relative protein expression (middle column, Figure 3, S2). We also calculated the fold increase in expression resulting from mRNA modifications for each organ by normalizing the bioluminescence for that modification by the bioluminescence for unmodified mLuc in that organ (right column, Figure 3, S2).
Figure 3. The m1ψ mRNA modification best enhanced in vivo luciferase expression for all delivery vehicles, with greatest improvement in the spleen.
Mice were IV-injected with LNPs carrying either unmodified or modified luciferase mRNA at a dose of 0.75 mg/kg. Six hours after injection, organs were harvested and imaged for bioluminescence by IVIS. (A-C) Protein expression resulting from C12-200 delivery was enhanced by the ψ, m1ψ, and m5C/ψ modifications, with the vast majority of the improvement in the spleen. (D-F) mRNA modifications did not improve overall expression for cKK-E12 mRNA delivery with statistical significance. Nonetheless, spleen expression improved with m1ψ and m5C/ψ modifications. (G-I) 200Oi10-mediated protein expression benefitted the most from nucleoside modifications, particularly with the m1ψ modification. Improvements in spleen expression were dramatic. (J-L) When formulated into ZA3-Ep10 LNPs, ψ and m1ψ modifications enhanced expression in the spleen, with very little improvement in the lungs. Error bars represent s.d. (n = 3; *p< 0.05, **p< 0.01, ***p<0.001; unpaired t-test relative to unmodified).
For nanoparticles incorporating C12-200, only ψ-, m1ψ-, and m5C/ψ-modified mRNAs significantly improved overall Luc expression (Figure 3A-B, S2). These improvements were driven by enhanced translation in the spleen. Of the mRNAs, m1ψ-modified mLuc was the most effective. It produced a 6-fold increase in overall expression and a marked 54-fold increase in spleen expression compared to unmodified mLuc (Figure 3C). Notably, the results for cKK-E12 differed from those for C12-200, even though these materials both deliver unmodified mRNA almost entirely to the liver. None of the five modified mLuc sequences resulted in a statistically significant increase in expression when delivered with cKK-E12 (Figure 3D-E, S2). However, even though overall efficacy was not enhanced, the m1ψ and m5C/ψ modifications nonetheless increased spleen expression 8-fold and 7-fold, respectively, for this delivery vehicle.
We then examined 200Oi10 lipid nanoparticles, which delivered unmodified mLuc to the liver, spleen, and lungs. As shown in Figure 3G-H, S2, the m1ψ modification performed best, producing an 11-fold increase in overall Luc expression. As with C12-200, this enhancement was attributable to a substantial increase in spleen expression (37-fold over unmodified mRNA, Figure 3I). Delivery of ψ- and m5C/ψ-modified mLuc with 200Oi10 also resulted in significant improvements in overall Luc expression (4-fold and 5-fold, respectively).
As for the lung-targeting lipid nanoparticle ZA3-Ep10, the m1ψ modification, once again, enhanced protein expression most effectively in mice, with a 3-fold increase in overall Luc expression (Figure 3J-K, S2). This, too, was driven by increased expression in the spleen (13-fold). Delivery of ψ-modified mLuc with this material also produced a 7-fold increase in spleen expression but no improvement in the lungs.
Although the performance of the IVT mRNAs varied significantly with delivery vehicle, there were several consistencies. For example, two of the modified mRNAs – m5C/s2U (25% substitution) and m5U – did not improve protein expression when incorporated into any of the four delivery vehicles. This confirms that there are many important factors in mRNA design, and that incorporations of base modifications known to decrease immune stimulation does not guarantee improved translation. The m5C/s2U modification had no impact on total expression when incorporated into C12-200 and 200Oi10 nanoparticles, and it decreased expression for cKK-E12 and ZA3-Ep10. The m5U modification was always detrimental to efficacy, inducing up to 90% reductions in total Luc expression.
Another consistency across vehicles was that the m1ψ modification was always the most effective. This nucleoside modification enhanced overall efficacy 1.5-fold to 11-fold, depending on the delivery vehicle. Although the m1ψ modification offered modest improvements in liver and lung expression, improvements in Luc expression were driven by exceptional increases in spleen expression (8- to 54-fold), depending on the delivery vehicle. The increased expression in the spleen extended to the other two pseudouridine-containing modifications (ψ and m5C/ψ). A previous study noted a similar phenomenon in which ψ-modified mRNA delivered to mice with a commercially available transfection agent produced protein almost entirely in the spleen.61 Together, the results in Figure 3 provide an important takeaway: the choice of nucleoside modification should be tailored to the delivery vehicle, the target organ, and the degree of specificity required.
m1ψ enhances mRNA translation in splenic monocytic lineage cells
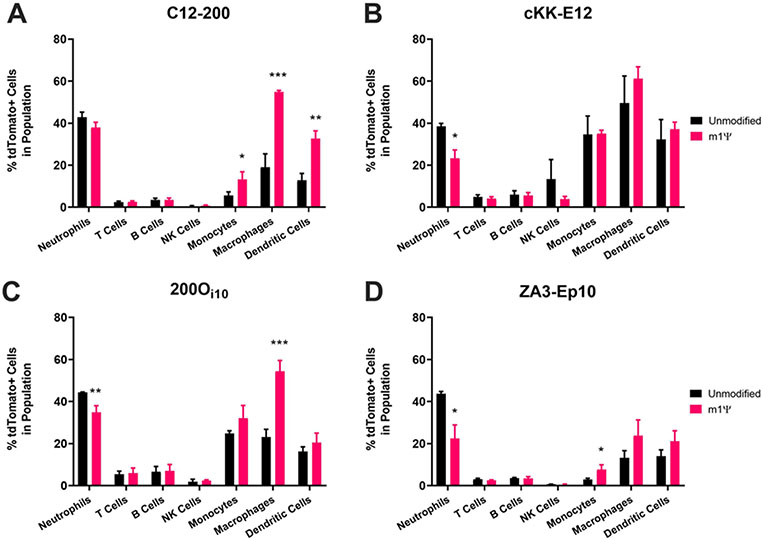
We next asked why m1ψ-modified mRNA improved protein expression when incorporated into some delivery vehicles but not others. We noted that 200Oi10, which was the most spleen-tropic of the lipids tested with unmodified mRNA, benefited the most from nucleoside modifications. Conversely, the performance of the least spleen-directed material, cKK-E12, was not augmented by any modification. To better understand the effect of base modifications on protein expression in the spleen, we used flow cytometry to identify the splenic cell types being transfected. We used all four lipids to deliver unmodified and m1ψ-modified Cre recombinase-encoding mRNA (mCre) to Ai9 mice, which express tdTomato upon Cre-mediated recombination.62 Importantly, this model reveals only which cells translate mCre, not the amount of protein expressed. Eighteen hours post-injection, spleens were removed, mechanically digested into single-cell suspensions, and analyzed by flow cytometry. The gating scheme for this experiment is shown in Figure S3. Results were similar for all four lipid nanoparticles in that modified mRNA produced the best improvement in percent of cells transfected for monocytic lineage cells (Figure 4). Generally, both modified and unmodified mRNA was delivered to splenic monocytes, macrophages, neutrophils, and dendritic cells, as evidenced by the fractions of tdTomato+ cells within these populations. Unmodified and modified mCre induced tdTomato activation in the highest percentage of neutrophils and macrophages, respectively, though splenic macrophages make up a considerably greater portion of the total splenocyte population than neutrophils. Overall, the data indicate that base modifications best improve transfection rates in monocytes and macrophages compared to other cell types in the spleen.
Figure 4. m1ψ modifications, when effective, improved mRNA translation in monocytic lineage immune cells.
Ai9 mice were injected with LNPs carrying unmodified or m1ψ-modifed Cre recombinase mRNA using either A) C12-200, B) cKK-E12, C) 200Oi10, or D) ZA3-EP10 at a dose of 0.5 mg/kg. Cells were isolated from spleens 18 hours post-injection and stained with antibodies to identify cell populations. Error bars represent s.d. (n = 3; *p< 0.05, **p< 0.01, ***p<0.001; unpaired t-test relative to unmodified mRNA for each cell type).
Because immune cells have evolved to recognize and degrade foreign nucleic acids, we hypothesized that the increased percentage of transfected monocytic splenocytes was due to the reduced immunogenicity of modified mRNAs.32,61 We tested this hypothesis by conducting a reporter assay for immunogenicity in cultured macrophages. Specifically, we used RAW Blue™ cells, which express a secreted alkaline phosphatase (SEAP) reporter inducible by NF-κB and AP-1. NF-κB and AP-1 are transcription factors that mediate cellular inflammatory responses downstream of endosomal and cytosolic receptors including TLRs, RIG-I, and MDA-5. Therefore, cells that secrete higher levels of SEAP are undergoing an inflammatory response.
For all nanoparticles except cKK-E12, m1ψ-mRNA significantly reduced innate immunogenicity compared to unmodified mRNA (Figure S4). Specifically, m1ψ decreased SEAP levels for 200Oi10, C12-200, and ZA3-Ep10 by 40, 30, and 70%, respectively. The lipid cKK-E12 was the only delivery material for which modifications did not significantly improve protein expression or reduce immunogenicity. These data suggest that improvements in splenic protein expression with modified mRNA correlate with reduced immunogenicity. While the SEAP reduction we observed in this study was significant, it was also fairly modest and may not completely account for the drastic increases in mRNA translation shown in Fig 3. A simple explanation for this may be that RAW Blue cells are a useful model for interrogating innate immunogenicity in vitro but may not fully recapitulate the signaling endogenous to monocytic splenocytes. Further, one study showed that m1ψ-modified mRNAs exhibit increased ribosome density through eIF2α-dependent and independent mechanisms, indicating another potential mechanism by which modified mRNAs undergo enhanced translation independent of innate immune activation.39
The site of endogenous protein expression affects the extent to which mRNA modifications enhance translation
Finally, we asked whether the results we observed for reporter proteins Luc and Cre recombinase also applied to mRNA encoding a therapeutic protein. As such, we delivered mRNA encoding murine erythropoietin (mEPO) synthesized with the same five modified nucleosides and delivered with the four lipid nanoparticles. EPO is produced predominantly in the kidneys and to a lesser extent in the liver. It is then secreted into the bloodstream to stimulate red blood cell production in the bone marrow.63 Because EPO is secreted (unlike Luc and Cre), we measured only overall changes in serum EPO levels instead of expression in individual organs. Despite our observations that modifications are most impactful in the spleen, we hypothesized that the modification of mEPO would best improve protein expression in the liver because it is a site of endogenous EPO production.
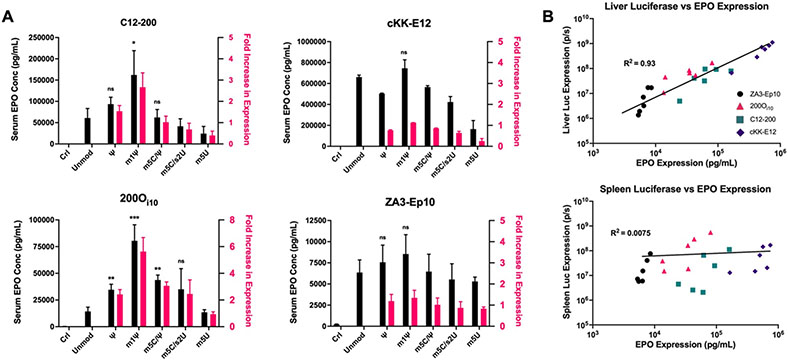
As expected, EPO expression was greatest using liver-targeting nanoparticles (Figure 5A). C12-200 lipid nanoparticles encapsulating m1ψ-modified mEPO produced a 2.6-fold increase in protein expression relative to unmodified mEPO. While this improvement did not match the overall increase in Luc expression for m1ψ (5.6-fold), it was equivalent to the enhancement in liver Luc expression for the m1ψ modification (2.5-fold). Results for cKK-E12 lipid nanoparticles were also consistent with mLuc delivery data. Indifferent to mRNA modification, this ionizable lipid nonetheless boasted the highest serum EPO concentrations of any nanoparticle given its exceptional potency in the liver. Next, 200Oi10 nanoparticles benefitted the most from mRNA base modifications, with m1ψ-modified mEPO producing a 5.6-fold increase in serum EPO levels relative to unmodified mRNA. Improvements observed from the various mRNA modifications were more consistent with our Luc expression data from the liver than with overall Luc expression. Finally, ZA3-Ep10 nanoparticles facilitated the lowest serum EPO concentrations of any delivery vehicle due to its relatively low tropism for the liver. As expected for a lung targeting formulation, ZA3-Ep10 nanoparticle potency was not influenced by mEPO modifications that would be most influential in the liver (Figure 5A).
Figure 5. The m1ψ modification best enhanced in vivo erythropoietin expression at the site of endogenous protein expression.
A) Unmodified or modified erythropoietin mRNA was delivered using either C12-200, cKK-E12, 200Oi10, or ZA3-EP10 lipid nanoparticles at a dose of 0.75 mg/kg. Serum EPO levels were determined by ELISA. m1ψ-modified mRNA facilitated a statistically significant increase in EPO expression when delivered with C12-200 and 200Oi10. The vehicle cKK-E12 mediated the highest serum levels of EPO, with the m1ψ modification facilitating a small but non-statistically significant increase in EPO concentrations. There was no statistically significant increase in EPO liver production with the m1ψ modification using lung-targeting ZA3-Ep10 lipid nanoparticles. Error bars represent s.d. (n = 3; *p< 0.05, **p< 0.01, ***p<0.001; unpaired t-test relative to unmodified mRNA). B) The expression of erythropoietin (EPO) correlates strongly with the expression of luciferase in the liver but not in the spleen for the delivery of modified mRNAs for all LNPs. The six different mRNAs were delivered using the four lipid nanoparticles: purple diamonds = cKK-E12, teal squares = C12-200, pink triangles= 200Oi10, black circles= ZA3-Ep10.
With these data in hand, we compared EPO expression to the expression of Luc in the liver and in the spleen for all four delivery vehicles, using all six mRNAs. As shown in Figure 5B, the EPO and liver Luc expression data collected for the 24 mRNA-lipid combinations correlated well (R2= 0.93), whereas the EPO and spleen Luc expression data did not (R2= 0.0075). This analysis indicates that EPO was produced predominantly in the liver. This is interesting because some of the delivery materials (e.g., 200Oi10) were expected to produce EPO in the spleen based on biodistribution and tropism data and, therefore, benefit significantly from mRNA modification. However, this was not the case – thus demonstrating that, at least for endogenous proteins, the site of endogenous protein expression must be considered in addition to delivery vehicle chemistry when assessing the potential benefits of modified mRNA inclusion in lipid nanoparticle formulations.
Conclusions
These results shed important light on the use of modified mRNAs to improve protein expression and decrease aberrant immunogenicity. While modified bases reduced inflammation in macrophages and improved protein translation in a variety of organs in an ionizable amino lipid nanoparticle-dependent manner, the greatest increases in translation occurred in monocytic lineage cells, which are plentiful in the spleen. However, in the case of proteins that are not endogenously produced in the spleen or in immune cells, mRNA modifications may increase splenic mRNA translation to a lesser extent. Together, our data indicate that the use and choice of modified mRNA in lipid nanoparticle formulations is a complex decision that must be made in the context of the native site of target protein expression (when applicable), the target cell type, and the delivery vehicle itself.
Materials and Methods
Materials.
Cy5-labeled mRNA, ψ- 5’-triphosphate, m1ψ- 5’-triphosphate, m5C- 5’-triphosphate, s2U- 5’-triphosphate, and m5U- 5’-triphosphate were purchased from Trilink Biotechnologies (San Diego, CA). MEGAscript™ T7 Transcription Kit was purchased from Thermo Fisher Scientific (Waltham, MA). ScriptCap™ 2’-O-Methyltransferase Kit was purchased from CellScript (Madison, WI). Cholesterol and 1,2-epoxyhexadecane (C12) were purchased from Sigma Aldrich (St Louis, MO). The phospholipid 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and C14-PEG2000 were purchased from Avanti Polar Lipids (Alabaster, AL). DMG-PEG2000 was purchased from NOF America (White Plains, NY). The isodecyl acrylate (Oi10) amine and the 2[4-2(2-aminoethyl)amino)ethylpiperazine-1-YL)ethan-1-amine (200) were purchased from Sartomer (Colombes, France) and Enamine (Princeton, NJ), respectively. XenoLight D-Luciferin Potassium Salt was purchased from PerkinElmer (Waltham, MA). The lipid cKK-E12 was generously donated by the Anderson Lab at the Massachusetts Institute of Technology (Cambridge, MA). The Mouse Erythropoietin Quantikine ELISA Kit was purchased from R&D Systems (Minneapolis, MN).
mRNA Synthesis.
mRNAs were synthesized as previously described.64,65 Linearized plasmids encoding firefly luciferase (pTEV-Luc-A101), Cre recombinase (pTEV-Cre-A101), mouse erythropoietin (pTEV-muEPO-A51), and eGFP (pTEV-eGFP-A101) were transcribed using the MEGAscript™ T7 Transcription Kit. IVT reactions were performed in the presence of 100% modified uridines (i.e. ψUTP, m1ψUTP, or m5UTP) except for the m5C/s2U modified mRNA where 5 methylcytosine and thiouridine were added at a 1:4 mol ratio (25%) in the NTP mixture to ensure efficient transcription and subsequent translation. mRNAs were capped using either the ScriptCap™ 2′-O-Methyltransferase Kit (eGFP, muEPO) or co-transcriptionally (mLuc, Cre) using the trinucleotide cap1 analog, CleanCap (TriLink). All mRNAs were purified by cellulose purification as described.66 All mRNAs were analyzed by electrophoresis using agarose gels, endotoxin tested, and dsRNA content assessed using dot blot.
Lipid Synthesis.
To synthesize the ionizable lipid C12-200, the amine 2[4-2(2-aminoethyl)amino)ethylpiperazine-1-YL)ethan-1-amine (200) was combined with the tail 1,2-epoxyhexadecane (C12) in a glass scintillation vial at a molar ratio of 1 : 5 and stirred for 3 days at 90°C without solvent. To synthesize the lipid 200Oi10, the amine 2[4-2(2 aminoethyl)amino)ethylpiperazine-1-YL)ethan-1-amine (200) was combined with the tail isodecyl acrylate (Oi10) in a glass scintillation vial at a molar ratio of 1 : 5 and stirred for 3 days at 90°C without solvent. The lipids were then purified using a Teledyne ISCO Chromatography system (Thousand Oaks, CA) to isolate the fully substituted lipid product. The zwitterionic amino lipid ZA3-Ep10 was synthesized as previously described.3
Lipid Nanoparticle Formulations.
The lipids cKK-E12, C12-200, and 200Oi10 were formulated into lipid nanoparticles as previously described.19,20 Specifically, ionizable lipid, DOPE, cholesterol, and C14-PEG2000 were combined at a molar ratio of 35 : 16 : 46.5 : 2.5 in 100% ethanol. The mRNA was diluted in 10 mM sodium citrate buffer (pH 3). For cKK-E12 and C12-200, the solutions were combined in a microfluidic device (Precision Nanosystems) at a flow ratio of 1 : 3 (ethanol : aqueous phase) with a total flow rate of 4 mL/min (cKK-E12) or 12 mL/min (C12-200). For 200Oi10, the lipid and mRNA solutions were combined at 1 : 1 volume ratio, and rapidly mixed by pulse vortexing. The final weight ratio of lipid : mRNA was 10 : 1. The zwitterionic lipid ZA3-Ep10 was formulated as previously described,3 by combining ZA3-Ep10, cholesterol, and DMG-PEG2000 at a molar ratio of 50 : 38.5 : 0.5 in 100% ethanol. The mRNA was diluted in 10 mM sodium citrate (pH 3). The solutions were combined by microfluidic mixing at a flow ratio of 1 : 3 (ethanol : aqueous phase) with a total flow rate of 12 mL/min. The final weight ratio of lipid : mRNA was 7.5 : 1. All lipid nanoparticles were dialyzed against PBS for 1 hour prior to use.
Biodistribution Studies.
All animal experiments were conducted using institutionally approved protocols (IACUC). Female C57BL/6 mice (Charles River Laboratories) were injected via tail vein with lipid nanoparticles carrying Cy5-labeled mRNA at a dose of 0.75 mg/kg (0.375 mg/kg for ZA3-Ep10). One hour after injection, the mice were sacrificed and the pancreas, spleen, liver, kidneys, heart, and lungs were harvested and imaged for Cy5 fluorescence at an excitation of 649 nm and an emission of 670 nm on an IVIS® imaging system (PerkinElmer). The total radiant efficiency was determined by a region of interest analysis using the Living Image® software (PerkinElmer). The percent biodistribution for each organ was determined by dividing the signal in that organ by the total signal.
In Vivo Luciferase mRNA Delivery.
Female C57BL/6 mice were injected via tail vein with each of the four lipid nanoparticles carrying either unmodified, ψ-modified, m1ψ-modifed, m5C/ψ-modified, m5C/s2U-modifed, or m5U-modified firefly mLuc at a dose of 0.75 mg/kg. Six hours after injection, mice received an intraperitoneal injection of 130 μL D-luciferin at a concentration of 30 mg/mL. Fifteen minutes later, the mice were sacrificed, and the pancreas, spleen, liver, kidneys, heart, and lungs were harvested and imaged for bioluminescence by IVIS®. The total luminescent flux was determined by a region of interest analysis using the Living Image® software.
In Vivo Erythropoietin mRNA Delivery.
Female C57BL/6 mice were injected via tail vein with each of the four lipid nanoparticles carrying either unmodified, ψ-modified, m1ψ-modifed, m5C/ψ-modified, m5C/s2U-modifed, or m5U-modified mouse erythropoietin mRNA at a dose of 0.75 mg/kg. Six hours after injection, mice were sacrificed, and blood was collected by cardiac puncture. Serum was isolated by centrifuging blood samples in BD Microtainer Serum Separator Tubes at 14,000 RPM for 10 minutes. Serum samples were diluted either 1 : 10 or 1 : 100, and erythropoietin concentrations were determined using the Mouse Erythropoietin Quantikine ELISA Kit according to the manufacturer’s instructions.
Flow Cytometry.
Female Ai9 mice received tail vein injections of lipid nanoparticles carrying either unmodified or m1ψ-modifed Cre mRNA at a dose of 0.5 mg/kg. Mice were sacrificed 18 hours post-injection, and the spleens was harvested. Cells were isolated by mashing the spleen through a 70 μm nylon mesh cell strainer (Thermo Fisher), followed by treatment with red blood cell lysis buffer (Thermo Fisher). Cells were then pelleted by centrifuging at 400 x g for 5 mins, followed by resuspension in DMEM supplemented with 10% FBS. 2x106 cells were suspended in blocking buffer (PBS + 1% FBS + 10% Fc block; Miltenyi Biotec) and stained with antibodies (BioLegend) diluted 1 : 100 against CD45, CD19, CD11c, CD11b, CD64, F4/80, CD31, Ly-6G, CD3, and MCHII for 30 minutes at 4 °C. Cells were washed by centrifugation, resuspended, and analyzed by flow cytometry using a NovoCyte 3000 (ACEA Biosciences). Flow cytometry data were compensated and analyzed using NovoExpress software (ACEA Biosciences).
In Vitro Immunogenicity.
Raw Blue™ cells were obtained from InvivoGen and maintained at 37°C, 5% CO2 in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, 100 μg/mL Normocin supplemented with Zeocin selective antibiotic according to the manufacturer’s instructions. Cells were seeded at 100,000 cells/well in 96-well plates and treated with 40 nM (0.5 μg/mL) modified or unmodified mRNA using each of the four lipid nanoparticles investigated in this work. 48 hours later, samples were analyzed for SEAP using a QUANTI-Blue™ assay (InvivoGen) performed according to the manufacturer’s instructions. Data were collected using a Synergy H1 plate reader (Biotek), and absorbance values for treated samples were normalized to that of untreated control cells.
Supplementary Material
Highlights:
The extent to which base modifications alter mRNA delivery depends on lipid nanoparticle chemistry.
Base modifications most effectively increase protein expression in the spleen.
Translation enhancement due to base modifications is greatest in monocytic splenocytes.
Acknowledgments
Funding was provided by the Defense Advanced Research Projects Agency (DARPA) (award number D16AP00143), The Shurl and Kay Curci Foundation, and National Institutes of Health (NIH, award number DP2-HD098860). J.R.M. was supported by an NIH F32 fellowship (award number 1F32EB029345), and M.L.A. was supported by an NSF Graduate Research Fellowship Program award (number DGE1745016). D.J.S. acknowledges support from the NIH (NIBIB R01 EB025192-01A1), the Cystic Fibrosis Foundation (CFF) (SIEGWA18XX0), the Cancer Prevention and Research Institute of Texas (CPRIT) (RP190251), and the American Cancer Society (ACS) (RSG-17-012-01). D.W. was supported by grants from NIAID (AI142596 and AI124429), IPCAVD (U19AI142596).
Footnotes
Conflicts of Interest
K.A.W. is an inventor on at least seven U.S. Patents that are related to the materials described here. D.J.S. is a co-founder of ReCode Therapeutics. D.W. and N.P. are named on U.S. patents describing the use of RNA containing modified nucleotides.
References
- 1.DeRosa F et al. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 23, 699–707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramaswamy S et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A 114, E1941–E1950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JB et al. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chemie - Int. Ed 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajj KA et al. A potent branched-tail lipid nanoparticle enables multiplexed mRNA delivery and gene editing in vivo. Nano Lett. 10.1021/acs.nanolett.0c00596 (2020) doi: 10.1021/acs.nanolett.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richner JM et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 168, 1114–1125.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardi N et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543, 248–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazzoli M et al. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol 90, JVI.01786–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulligan MJ et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586, 589–593 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Jackson LA et al. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberli MA et al. Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy. Nano Lett. 17, 1326–1335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billingsley M et al. Ionizable Lipid Nanoparticle Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. DOI: 10.1021/acs.nanolett.9b04246 (2020) doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin MD et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol (2020) doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 13.Pardi N, Hogan MJ & Weissman D Recent advances in mRNA vaccine technology. Curr. Opin. Immunol 65, 14–20 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Pardi N, Hogan MJ, Porter FW & Weissman D mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahin U, Karikó K & Türeci Ö mRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hajj KA & Whitehead KA Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater 2, 1–17 (2017). [Google Scholar]
- 17.Kowalski PS, Rudra A, Miao L & Anderson DG Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther 27, 710–728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenton OS et al. Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent in Vivo mRNA Delivery. Adv. Mater 28, 2939–2943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajj KA et al. Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small 1805097, 1805097 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Kauffman KJ et al. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 15, 7300–7306 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Hassett KJ et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. - Nucleic Acids 15, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trepotec Z, Lichtenegger E, Plank C, Aneja MK & Rudolph C Delivery of mRNA Therapeutics for the Treatment of Hepatic Diseases. Mol. Ther 27, 794–802 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitehead KA, Dahlman JE, Langer RS & Anderson DG Silencing or stimulation? siRNA delivery and the immune system. Annu. Rev. Chem. Biomol. Eng 2, 77–96 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Karikó K, Ni H, Capodici J, Lamphier M & Weissman D mRNA Is an Endogenous Ligand for Toll-like Receptor 3. J. Biol. Chem 279, 12542–12550 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Diebold SS, Kaisho T, Hemmi H, Akira S & Reis E Sousa C Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science (80-. ). 303, 1529–1531 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Heil F et al. Species-Specific Recognition of Single-Stranded RNA via Toll-like Receptor 7 and 8. Science (80-. ). 303, 1526–1529 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Zhang H et al. Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc. Natl. Acad. Sci 118, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson BR et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlile TM et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Karikó K & Weissman D Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: Implication for therapeutic RNA development. Curr. Opin. Drug Discov. Dev 10, 523–532 (2007). [PubMed] [Google Scholar]
- 32.Karikó K, Buckstein M, Ni H & Weissman D Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Karikó K et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karikó K, Muramatsu H, Keller JM & Weissman D Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther 20, 948–953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parr CJC et al. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 48, e35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andries O et al. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Karikó K, Muramatsu H, Keller JM & Weissman D Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther 20, 948–953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andries O et al. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Svitkin YV et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Translate Bio. Study to Evaluate the Safety & Tolerability of MRT5005 Administered by Nebulization in Adults With Cystic Fibrosis (RESTORE-CF). clinicaltrials.gov https://clinicaltrials.gov/ct2/show/NCT03375047 (2020). [Google Scholar]
- 41.Dolgin E CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature 10.1038/d41586-021-01661-0 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Ramishetti S et al. A Combinatorial Library of Lipid Nanoparticles for RNA Delivery to Leukocytes. Adv. Mater 1906128, 1–8 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Brito LA et al. Self-Amplifying mRNA Vaccines. Advances in Genetics vol. 89 (Elsevier Ltd, 2015). [DOI] [PubMed] [Google Scholar]
- 44.Paunovska K et al. Nanoparticles Containing Oxidized Cholesterol Deliver mRNA to the Liver Microenvironment at Clinically Relevant Doses. Adv. Mater 31, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sago CD et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci 201811276 (2018) doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Q et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol 10.1038/s41565-020-0669-6 (2020) doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel S et al. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 17, 5711–5718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riley RS et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv 7, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka H et al. In Vivo Introduction of mRNA Encapsulated in Lipid Nanoparticles to Brain Neuronal Cells and Astrocytes via Intracerebroventricular Administration. Mol. Pharm 15, 2060–2067 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Li B et al. An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo. Nano Lett. 15, 8099–8107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong Y et al. Poly(glycoamidoamine) Brushes Formulated Nanomaterials for Systemic siRNA and mRNA Delivery in Vivo. Nano Lett. 16, 842–848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarzębińska A et al. A Single Methylene Group in Oligoalkylamine-Based Cationic Polymers and Lipids Promotes Enhanced mRNA Delivery. Angew. Chemie - Int. Ed 55, 9591–9595 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Ball RL, Hajj KA, Vizelman J, Bajaj P & Whitehead KA Lipid Nanoparticle Formulations for Enhanced Co-delivery of siRNA and mRNA. Nano Lett. 18, acs.nanolett.8b01101 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Kauffman KJ et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109, 78–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karikó K & Weissman D Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: Implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel 10, 523–532 (2007). [PubMed] [Google Scholar]
- 56.Anderson BR et al. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 39, 9329–9338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love KT et al. Erratum: Lipid-like materials for low-dose, in vivo gene silencing (Proceedings of the National Academy of Sciences of the United States of America (2010) 107, 5 (1864–1869)) DOI: 10.1073/pnas.0910603106. Proc. Natl. Acad. Sci. U. S. A. 107, 9915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yizhou D et al. Erratum: Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates (Proceedings of the National Academy of Sciences of the United States of America (2014) 111:11 (3955–3960) DOI: 10.1073/pnas.1322937111). Proc. Natl. Acad. Sci. U. S. A. 111, 5753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitehead KA et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun 5, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaczmarek JC et al. Polymer–Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chemie - Int. Ed 55, 13808–13812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karikó K et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maxwell PH et al. Sites of erythropoietin production. Kidney Int. 51, 393–401 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Pardi N et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun 9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freyn AW et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther 28, 1569–1584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baiersdörfer M et al. A facile method for removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol. Ther. - Nucleic Acids 15, 26–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.