Over the past 2 decades, antimicrobics have become increasingly available for a broad range of pathogens. Due to the widespread use of these drugs, new forms of antimicrobial resistance have emerged. Detection of this resistance by conventional methods may not always be easy or possible.
The genetics of antimicrobial resistance has been elucidated completely or in part for many organism-antimicrobial agent combinations. As a result, genetic methods for assessing antimicrobial resistance have been developed, and many of these are being used or soon will become part of the standard testing menus in clinical microbiology laboratories. The intent of this review is to describe the principles and applications of the more frequently reported genetic methods for evaluating antimicrobial resistance.
CONVENTIONAL METHODS FOR SUSCEPTIBILITY TESTING
Conventional antimicrobial susceptibility testing methods require that pathogens are first isolated from human specimens by culture methods. In separate assays, isolated microorganisms are then exposed to various concentrations of antimicrobial agents under specified growth conditions, and the ability of these antimicrobics to inhibit growth is determined. Methods that are frequently used for testing cultivated bacteria and yeasts include disk diffusion, broth dilution, agar dilution, and gradient diffusion (Epsilometer test). Isolated bacteria can also be screened for antimicrobic-modifying enzymes. A commercially available chromogenic disk method (Cefinase; Becton Dickinson Microbiology Systems, Cockeysville, Md.) is frequently used as a rapid procedure to detect β-lactamase production in Staphylococcus spp., Haemophilus influenzae, and Bacteroides spp. This method relies on the visualization of a colored product that results from hydrolysis of the substrate β-lactam molecule, nitrocefin, contained in a paper disk.
Tissue culture methods, with either human or primate cells, have been used to determine the susceptibility of herpesviruses and human immunodeficiency virus (HIV) to antiviral agents. For herpesviruses, the effects of antiviral compounds are assessed by cytopathic (plaque) reduction assays or the quantitation of specific herpesvirus DNA. For HIV, inhibition of reverse transcriptase activity or p24 antigen production by antiretroviral agents is assessed after exposure of virus cultured in human lymphocytes.
Currently, published standards exist for media preparation, incubation parameters, and the interpretation of results for disk diffusion, broth dilution, and agar dilution methods. These standards are provided by the National Committee for Clinical Laboratory Standards (NCCLS) and are available and updated periodically for selected aerobic (44) and anaerobic bacteria (42) and yeasts (43). Tentative NCCLS guidelines exist for mycobacteria (41). No published standards have been developed for gradient diffusion or genetic testing for any microorganisms.
GENETIC METHODS FOR ASSESSING ANTIMICROBIAL RESISTANCE
Potential advantages of genetic susceptibility testing methods.
For several reasons, genetic methods, compared to conventional susceptibility methods, have the potential to provide a more rapid and reliable assessment of antimicrobial resistance. (i) Genetic susceptibility testing methods can be performed directly with clinical specimens obviating the need for isolation of the organism by culture. (ii) These methods assess the genotype of the organism, whereas conventional susceptibility techniques asses the phenotype or expression of the genotype under artificial or laboratory conditions. Although debate exists among authorities as to which of these assessments is more clinically relevant, it seems reasonable that the lowest-risk approach for the patient is to determine the genotype. This may be especially true if one is dealing with serious life-threatening infections such as meningitis or bacteremia or infections such as endocarditis or osteomyelitis, which require prolonged courses of antimicrobial therapy. (iii) In some cases, genotypes may be discerned long before phenotypes can be determined due to the slow growth of the organism. (iv) Some organisms cannot be cultured or are not easily cultured and so only genotypes can be determined in these cases. (v) Genetic methods may lessen the biohazard risk which may occur with the propagation by culture of a microorganism, a requirement for conventional test methods.
Disadvantages of genetic testing methods.
Several disadvantages exist for genetic methods compared to conventional phenotypic methods. (i) They may lack sensitivity when only a few organisms are present in a sample. Refinements in techniques for concentrating nucleic acids in large volumes of clinical specimens hopefully will address this problem. (ii) Different assays are required for each antimicrobial agent tested. (iii) Resistance of a microorganism to a specific antimicrobial agent may occur via different mechanisms associated with different resistance genes or a large array of single or coincidental mutations. Pertinent to this and the prior point, is the concept that “with genetic testing methods, one only gets what one specifically is looking for.” This is in contrast to culture-based methods which are more comprehensive in assessing antimicrobial resistance. That is, by using the same culture-based assay, different forms of resistance can be detected. By virtue of the capacity to survey for different forms of resistance, culture-based methods also are useful for detecting emerging or new forms of antimicrobial resistance. (iv) A genetic mechanism for resistance for some antimicrobics may not have been defined. (v) Genetic methods may detect resistance genotypes that are expressed at levels that may not be clinically relevant. Examples of this include low-level vancomycin resistance encoded by vanC genes and poorly expressed extended-spectrum β-lactamase resistance. (vi) False-positive results may occur due to contamination of the test sample with extraneous nucleic acid or residual nucleic acid from prior samples. This problem is of particular concern when nucleic acid amplification techniques such as PCR are used. However, the development of enzymatic and chemical sterilization methods for nucleic acid amplified by PCR has greatly enhanced the specificity for this technique (14). (vii) Unlike for conventional culture-based susceptibility test methods, no standards exist for performing genetic testing methods. (viii) Multicenter, controlled clinical trials to assess the accuracy, reproducibility, and clinical utility of these methods have only been conducted for a few of these tests.
Evaluation of genetic testing methods for introduction into the clinical laboratory.
Clinical microbiologists should critically evaluate the advantages and disadvantages of genetic versus conventional methods before changing test menus. Important in this process are assessments of performance characteristics; turnaround time for results, costs related to reagents, royalties or licensing fees; and personnel time. Genetic susceptibility tests may be “add-ons,” their introduction may not obviate conventional testing methods. An example of this is mecA gene detection for staphylococci. The mecA gene encodes a penicillin-binding protein, designated 2a or 2′, which has decreased affinity for methicillin and related compounds. Because methicillin resistance may result from mechanisms other than the mecA gene (e.g., hyperproduction of β-lactamase), conventional susceptibility testing is still required for mecA-negative staphylococcal isolates. Also, if the result for the mecA gene analysis is negative, testing for susceptibility to penicillin must still be performed, because for β-lactamase-negative strains, penicillin remains the therapy of choice. In this situation, determination of penicillin susceptibility is important as this compound is more effective therapeutically and is cheaper than β-lactamase stable penicillins (nafcillin, oxacillin, dicloxacillin, and cloxacillin). Thus, for each staphylococcal strain, both a genetic and conventional testing method may ultimately be required before a final susceptibility report can be issued.
The above is one example of how a genetic test for determining antimicrobial resistance could be incorporated into the testing repertoire of a clinical microbiology laboratory. It is not yet possible to determine how or if any of the genetic tests covered in the subsequent discussion in this review will fit into either routine or reference laboratory test protocols. As the technology continues to evolve, it is likely that more user-friendly testing formats based on the principles of some of these tests will be developed. This should make the technology more easily adaptable by small hospital-based laboratories as well as large reference laboratories.
Genetic mechanisms of antimicrobial resistance.
Antimicrobial resistance may occur as the result of intrinsic or acquired genetic material. Furthermore, intrinsic or acquired genetic material may undergo changes, usually single base mutations, which may affect the spectrum of antimicrobial resistance.
In Table 1, organism-antimicrobial agent combinations are shown for which genetic mechanisms for or genetic associations with antimicrobial resistance have been defined. This is not meant to be comprehensive summary, rather it is limited to those organism-antimicrobial agent combinations for which conventional phenotypic susceptibility test methods lack accuracy, are technically difficult to perform, and/or require significant time periods before results are available. Furthermore, for these organism-antimicrobic combinations, specific genetic testing methods have been reported to be useful. These methods are also provided in Table 1. For general bacteria, these combinations include the assessment of methicillin resistance in coaglase-negative Staphylococcus species (4, 18, 28, 29, 35, 36, 77, 78), low-level vancomycin resistance in Enterococcus species (46), and extended-spectrum β-lactamase resistance in gram-negative facultatively anaerobic bacteria such as Escherichia coli and Klebsiella pneumonia (2, 45). For Mycobacterium tuberculosis, genetic methods have been reported for detecting resistance to isoniazid (7, 19, 21, 34, 37, 39, 52, 61, 66, 69, 74, 76), rifampin (16, 39, 48, 51, 66, 67, 68, 79, 81, 82), streptomycin (39, 60) ethambutol (59), pyrazinamide (58), and the fluoroquinolones (65). Detection of mutations associated with resistance of herpes simplex virus and herpes zoster virus to acyclovir (54, 56) and of cytomegalovirus to ganciclovir (57) have also been described. Assessments of mutations in the reverse transcriptase (31, 50, 64) and protease (8, 22, 75) genes of HIV have been reported to be convenient means by which resistance to reverse transcriptase and protease inhibitors can be determined. One must be cautious, however, in attributing all mutations in these genes to antiretroviral resistance; further clinical studies are required to validate these associations. Although a direct mechanistic link between nucleotide sequences in hepatitis C virus (HCV) genomes and the effect of immunotherapy (interferon) has not been proven, such an association appears to predict response to this therapy (9, 10, 33, 62, 83) and so this combination is included in the Table 1. In the subsequent discussion, examples of genetic testing methods which have been developed for these purposes are described.
TABLE 1.
Application of genetic methods for assessing antimicrobial resistance
| Organism(s) | Antimicrobial agent(s) | Genetic material-/changes associated with resistance | Comments | Examples of reported molecular methods (reference)a |
|---|---|---|---|---|
| Staphylococcus spp., especially coagulate-negative staphylococci | Methicillin, (nafcillin, oxacillin, cloxacillin, dicloxacillin) | Acquisition of mecA gene | The mecA gene may be heterogeneously expressed and therefore not all methicillin-resistant staphylococcal strains may be detectable with standard culture-based methods | PCR-gel electrophoresis (18, 77, 78), PCR-dot blot hybridization (4), chemiluminescent DNA hybridization of cultured staphylococci (28), PCR-gel electrophoresis or Southern hybridization: dot-blot hybridization of DNA from cultivated staphylococcal strains (35), PCR-Southern hybridization (36), bDNA signal amplification assay (29) |
| Enterococcus spp. | Vancomycin | vanC genes | Current NCCLS breakpoints for culture-based methods may not identify low-level vancomycin resistance in all Enterococcus sp. strains that carry vanC genes (27); however, the clinical significance of low-level vancomycin resistance remains unanswered | PCR-RFLP (46) |
| Enterobacteriaceae, especially K. pneumoniae and E. coli | Ceftazidime, cefotaxime, and aztreonam | Acquisition of genes encoding extended-spectrum β-lactamases (ESBL) which are derivatives of TEM, SHV, and OXA-type β-lactmases | Current NCCLS breakpoints for culture-based methods may not identify low-level resistance encoded by some ESBL genes (23). If ESBL genes are identified, then penicillins, cephalosporins, or monobactams should not be used. | PCR-RFLP (2,45), PCR-SSCP (15, 38) |
| M. tuberculosis | Isoniazid | Mutations in the catalase-peroxidase (katG) gene account for 50–70% of resistant strains (30–65% due to a single mutation [S315T]) | Due to the slow growth of M. tuberculosis, standard culture-based methods frequently require several weeks. Due to the prolonged incubation of test media, activity of drugs within media may decrease | PCR-SSCP (21, 52, 66, 69, 76) PCR-RFLP (7, 19, 39, 74, 76), PCR-CFLP (5), PCR-DNA sequencing (34, 37) |
| Mutations in the inhA locus account for 5–10% of resistant strains | PCR-SSCP (52, 66), PCR-DNA sequencing (37) | |||
| Mutations in promoter region of ahpC encoding the alkylhydro-peroxide reductase gene account for 6–13% of resistant strains | PCR-SSCP (66), PCR-DNA sequencing (61) | |||
| Rifampin | Mutations in rpoB gene, which encodes the β subunit of RNA polymerase | PCR-SSCP (16, 55, 66, 67, 68, 79), PCR-ddF (16), PCR-HDP analysis (39, 81, 82), PCR-CFLP (20), PCR-LiPA (51), PCR-molecular beacon sequence analysis (48), PCR-DNA sequencing (24, 39, 51, 81, 82) | ||
| Ethambutol | Mutations in embB gene, which encodes polymerization of arabinose into arabino galactan, account for ∼70% of resistant strains | PCR-SSCP (59), PCR-DNA sequencing (59) | ||
| Streptomycin | Mutations in genes encoding 16S rRNA (rrs) and ribosomal protein S12 (rpsL) account for 64–68% of resistant strains | PCR-RFLP (39), PCR-DNA sequencing (39, 60) | ||
| Pyrazinamide | Mutations in the pncA gene, which encodes pyranzinamidase, account for ∼70% of resistance strains | PCR-DNA sequencing (58) | ||
| Fluoroquinolones | Mutations in gyrA gene, which encodes the A subunit of DNA gryase, account for ∼100% of resistant strains | PCR-SSCP (65), PCR-DNA sequencing (65) | ||
| M. avium complex | Macrolides | Mutations in the 23S rRNA gene account for ∼100% of resistant strains | Due to the slow growth of M. avium complex, standard cultured-based methods frequently require weeks. Due to prolonged incubation of test media, the activity of drugs in media may decrease | PCR-NIRCA (40) |
| Herpes simplex virus, herpes zoster virus | Acyclovir | Mutations frequently in the thymidine kinase (tk) gene and rarely in the DNA polymerase gene | Standard cytopathic (plaque) reduction assays or quantitation of specific herpesvirus DNA are technically difficult to perform and require 7–14 days | PCR-DNA sequencing (54, 56) |
| Cytomegalovirus | Ganciclovir | Mutations in the viral phosphotransferase gene (UL97) and the DNA polymerase gene (UL54) | PCR-DNA sequencing (57) | |
| HIV | Reverse transcriptase inhibitors | Mutations in polymerase (pol) gene | Standard reverse transcriptase or p24 antigen assays of HIV cultures are technically difficult and require 60–80 days | RT-PCR-LiPA (64), RT-PCR-DNA sequencing (50) |
| Protease inhibitors | Mutations in protease gene | Standard reverse transcriptase or p24 antigen assays of HIV cultures are technically difficult and require 60–80 days | RT-PCR-RFLP (75), RT-PCR-DNA sequencing (22, 75) | |
| HCV | Interferon | Genetic variants of HCV predict response to interferon therapy | No previous methods | RT-PCR-CFLP (33), RT-LiPA (62), RT-PCR-RFLP (9, 10) RT-PCR-DNA sequencing (9, 10, 33, 62, 83) |
NIRCA, nonisotopic RNase cleavage assay; HDP, heteroduplex; RT, reverse transcription.
DESCRIPTIONS OF REPORTED GENETIC SUSCEPTIBILITY TESTING METHODS
Most genotypic methods include an initial step where the “target” nucleic acid is amplified. This is usually accomplished by PCR. Other less frequently used methods which amplify either target nucleic acid or nucleic acid contained in probes annealed to target nucleic acid include self-sustaining sequence replication, strand displacement amplification, ligase chain reaction, and Qβ replicase amplification. The signal generated from probes annealed to target nucleic acid can also be amplified; an application of this method, branched DNA (bDNA) assay, is covered in subsequent discussion. The reader is referred to a review by Podzorski and Persing (49) which provides detailed descriptions of the nucleic acid amplification techniques. The product of the PCR, referred to as an amplimer or amplicon, can be confirmed as the desired target nucleic acid (i.e., part or all of a resistance-associated genetic material) by electrophoretic mobility determinations, probe hybridization assays (Southern blotting of electrophoretic gels, slot, dot blot, enzyme-linked immunosorbent assay, or liquid hybridization formats), restriction fragment length polymorphism (RFLP) analysis, or DNA sequencing formats. Amplicons can also be assessed for specific mutations associated with antibiotic resistance by direct DNA sequencing methods and RFLP, single-strand conformation polymorphism (SSCP), dideoxy fingerprinting (ddF), Cleavase fragment polymorphism (CFLP), RNase cleavage, heteroduplex, line probe, molecular beacon, or microchip oligonucleotide array assays.
Detection of antimicrobial resistance genes. (i) PCR amplification of target DNA and amplicon confirmation by gel electrophoresis, probe hybridization techniques, or DNA sequencing.
Standard PCRs with amplicon sizing by gel electrophoresis are especially useful for identifying genes which encode antimicrobial resistance (18, 77, 78). These assays are highly specific, especially if there are no other nucleic sequences harbored by the organism which share significant homology with the target genetic material and large quantities of target nucleic acid are amplified. The latter condition exists when organisms are first propagated by culture and then isolated colonies are used for the PCR. Amplifying such large quantities of target nucleic acid reduces the sensitivity requirements for such assays, making it less likely that contamination with extraneous nucleic acid will be a significant problem. Therefore, provided that negative controls are used, specialized contained specimen processing areas and/or amplicon “sterilization” may be unnecessary. Amplicons can also be sterilized, that is, chemically or enzymatically modified, such that they cannot serve as a template for subsequent PCR assays which use the same oligonucleotide primers.
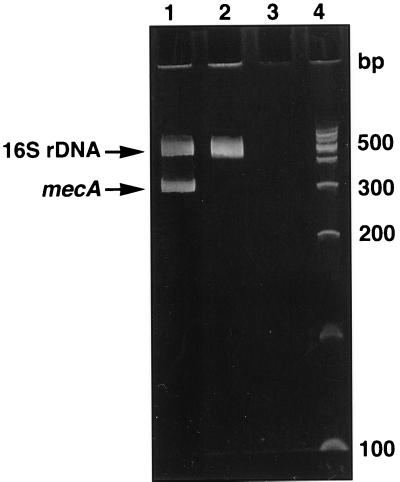
Figure 1 shows an example of a multiplex PCR method used in our laboratory to detect the mecA gene from isolated staphylococcal colonies which have been propagated by standard culture methods (18). Two distinct PCR amplicons are produced simultaneously in the same reaction tube, hence the term multiplex PCR. In one reaction, a portion of the mecA gene is amplified. For the other PCR, a nucleic acid sequence of the 16S rRNA gene unique to staphylococci is amplified. The second PCR assay confirms that the bacterium amplified is a staphylococcus. The PCR amplicons are electrophorized through an agarose gel, and their sizes are determined by comparing them to standard DNA fragments. If no bands are produced, either the organism tested is not a Staphylococcus species or the PCR did not occur. A positive result should produce amplicons of the appropriate size for mecA and 16S ribosomal DNA (16S rDNA) sequences based on the oligonucleotide primers used for PCR. Additional confirmatory steps not shown in Figure 1 can be performed. The mecA amplicon can by hybridized to labeled robes (signal hybridization), digested with restriction endonucleases (RFLP analyses), or the DNA sequence can be determined. Clinical evaluations for mecA by PCR and Southern blot hybridization (35, 36) are listed in Table 1 and are not covered in further detail. Also indicated in Table 1 are clinical studies that have used a chemiluminescent (28) or dot blot hybridization format (4, 35) to detect the mecA from cultured staphylococci. The reader is referred to comprehensive discussions on signal hybridization formats by Podzorski and Persing (49) and Tenover and colleagues (70). RFLP analyses and DNA sequencing for other organism-antimicrobic combinations are discussed subsequently in greater detail.
FIG. 1.
Multiplex PCR and gel electrophoresis for identifying the mecA gene in Staphylococcus spp. Two distinct PCRs were performed: one amplified a portion of 16S rDNA unique to Staphylococcus spp., and the second amplified a segment of the mecA gene. Lane 1, strain of methicillin-resistance S. aureus; electrophoretic bands of the appropriate size appear for the 16S rDNA and mecA. Lane 2, methicillin-susceptible S. aureus; an electrophoretic band for 16S rDNA is present but there is no band for mecA. Lane 3, negative control (reagents only), lane 4, DNA fragment standards. bp, base pair.
Multiplex PCR assays, like that described for the mecA gene, are relatively easy to perform and can be completed in 6 to 8 h. The approximate cost for reagents and supplies for each sample is less than $5. An initial capital outlay for a thermocycler (∼$5,000 to $10,000) and gel electrophoresis equipment (∼$3,000) is required. The multiplex mecA assay is more sensitive than current culture-based methods for detecting methicillin resistance related to the mecA gene, especially for coagulase-negative staphylococci (4, 18, 28, 29, 35, 36, 78). However, the turnaround time for results may be similar if this molecular assay is performed from isolated colonies of bacteria. Recently, Ubakata and colleagues (72) and Kitagawa and colleagues (26) have performed mecA assays by using PCR directly with broth from blood culture bottles. Direct detection of mecA-associated resistance in blood cultures should appreciably decrease turnaround time compared to that of culture-based methods. Substances that inhibit PCR, which are present in blood culture medium, notably sodium polyanethol sulfonate (SPS), may affect the sensitivity of these assays and methods to remove SPS should be employed (17).
(ii) bDNA assay.
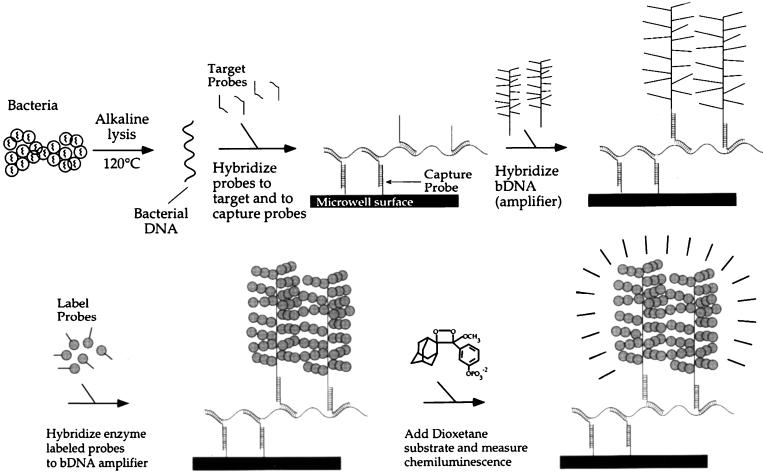
The bDNA assay is a non-PCR-based signal amplification method that is commercially produced by Chiron Diagnostics (Norwood, Mass.). It has been used to determine blood levels (viral loads) of HIV (25) and HCV (12). Our laboratory has adapted this assay to detect the mecA gene directly from blood cultures containing staphylococci (84). The bDNA method depends on the amplification of a signal molecule and not the target nucleic acid (Fig. 2). Signal generation is proportional to the amount of mecA gene sequences in the sample, which in turn is proportional to the number of staphylococcal cells analyzed. We have found the mecA bDNA assay to be as accurate as conventional PCR assays for detecting the mecA gene (29, 84). The procedure is partly automated but requires approximately 6 to 8 h to complete. Performing the test directly from blood culture broth should permit a decrease in turnaround time for methicillin susceptibility results for staphylococci. At this time, the bDNA assay for mecA detection is not commercially available and therefore reagent and instrument costs remain unknown.
FIG. 2.
bDNA method for identifying the mecA gene in Staphylococcus spp. A generic representation of the bDNA method, which has been used for detection of the mecA gene, is shown. DNA is released from staphylococcal cells and hybridized to both capture probes and target probes. bDNA molecules (amplifiers) are then hybridized to target probes. Enzyme-labeled probes are subsequently hybridized to the bDNA amplifier. A chemiluminsescent substrate, dioxetone, is added and emitted light is measured. (Adapted with permission from Chiron Diagnostics, Norwood, Mass.)
(iii) PCR-RFLP analysis.
In RFLP analysis, the amplified DNA is digested (fragmented) with restriction enzymes. These endonucleases will only cleave DNA molecules at specific sites, i.e., unique short sequences of nucleic acid. Therefore, if the sequence of the target DNA is known, RFLP analysis can be used to confirm the target DNA in its amplified form.
Recently, we have reported the use of RFLP analysis to differentiate among genes encoding vancomycin resistance (46). Because of conserved DNA sequences which exist among vancomycin resistance genes, more than one vancomycin resistance gene may be amplified with the same PCR oligonucleotide primers. RFLP analysis then can be used to differentiate the vancomycin resistance gene amplicons, providing that sites for a restriction endonuclease are present in one but not another gene.
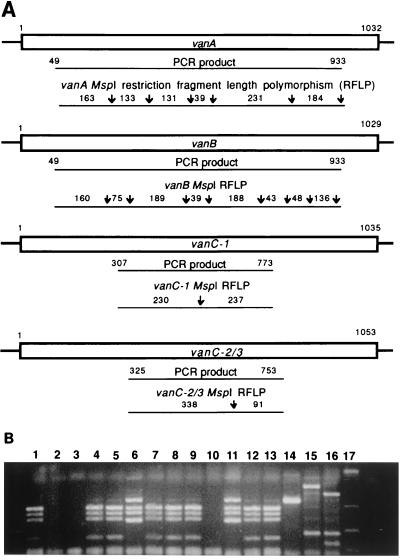
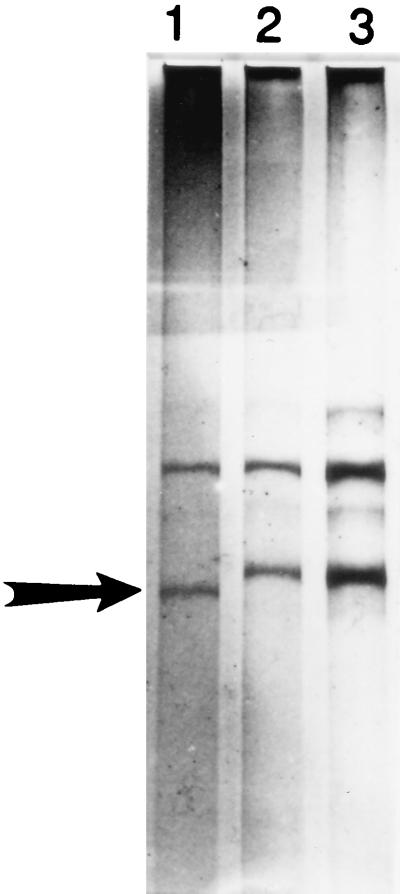
Figure 3A shows restriction fragments produced with the enzyme MspI for the vancomycin resistance genes vanA, vanB, vanC-1, and vanC-2/3. Figure 3B shows the electrophoretic gel RFLP patterns obtained with MspI and various control strain and clinical isolates of Enterococcus spp. containing vancomycin resistance genes. In essence, the RFLP procedure results in “bar codes” which specify distinct genes associated with vancomycin resistance. We have found this technique to be particularly useful for determining vancomycin resistance associated with the vanC gene family as this form of vancomycin resistance is not always detectable by conventional culture-based susceptibility methods (27). However, caution must be taken when identifying vanB and vanC-2 by RFLP analysis, as we have noted significant DNA sequence variation among these genes (47). Therefore, restriction sites may be gained or lost or PCR primers may be ineffective.
FIG. 3.
PCR-RFLP analysis for identifying genes associated with vancomycin resistance in Enterococcus spp. (A) Six primers are used in a multiplex reaction to generate four PCR products. Restriction fragment lengths predicted for vancomycin resistance genes vanA, vanB, vanC-1, and vanC-2/3 with the restriction enzyme (endonuclease) MspI are shown. (B) RFLP electrophoretic patterns obtained for various control strains and clinical isolates of Enterococcus sp. containing vancomycin resistance-associated genes. Lanes 2, 3, and 10, no restriction fragments produced; lanes 6 and 11, vanA pattern, lanes 1,4, 5, 7–9, 12, and 13 vanB patterns: lane 14, vanC-1 pattern; lane 15, vanC-2 pattern; lane 16, restriction fragment pattern unrelated to vanA, vanB, and vanC; lane 17, DNA fragment standards. (Adapted with permission from Patel et al. [46].)
A potential advantage of the RFLP analysis is that PCR amplicons may be digested by a restriction enzyme to the extent that full-length amplicons are no longer available for subsequent PCR assays. Therefore, the amplicons are sterilized. The PCR-RFLP procedure can be performed in 6 to 8 h. The approximate cost for reagents and supplies for each test sample is less than $5. The only additional reagent expense beyond that required for simple PCR-gel electrophoresis assays is the cost of restriction enzymes. Like PCR-gel electrophoresis assays, a thermocycler and electrophoresis equipment are required.
Detection of mutations associated with antimicrobial resistance. (i) PCR-RFLP analyses.
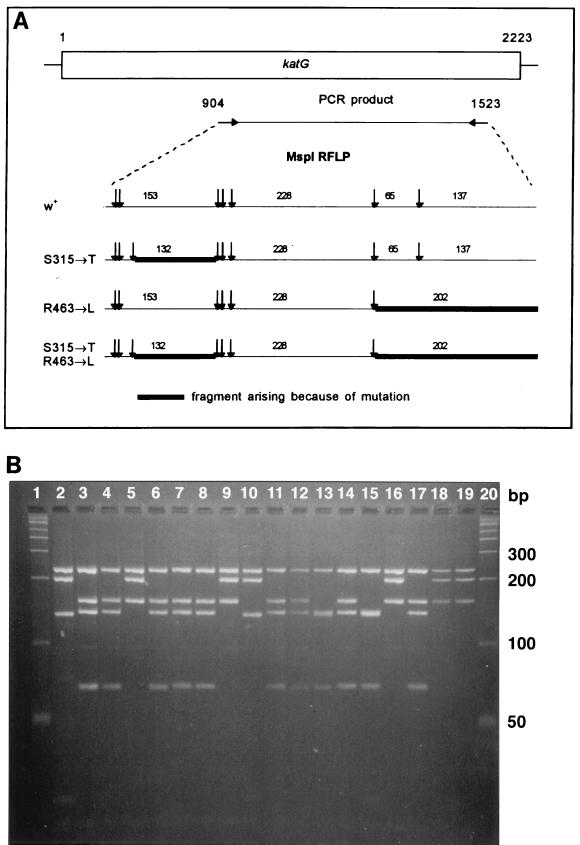
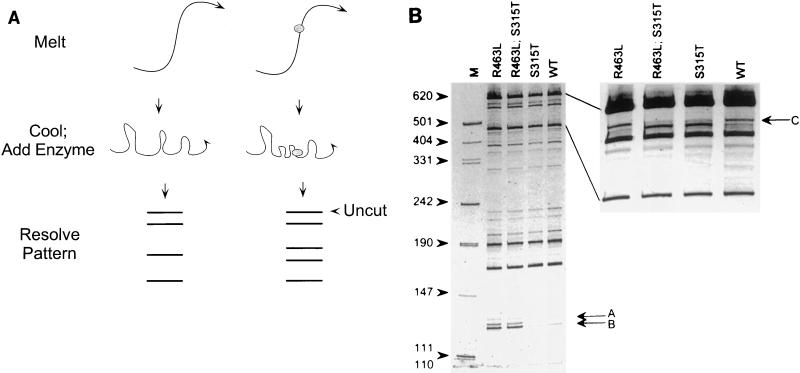
RFLP analysis is also useful for identifying mutations associated with antimicrobial resistance. We and others have shown that point mutations in the catalase-peroxidase (katG) gene of M. tuberculosis are associated with isoniazid resistance (5, 7, 19, 21, 34, 37, 39, 52, 69, 74, 76). Two of these mutations, which occur with significant frequency, are located in codons 315 (serine → threonine) and 463 (arginine → leucine) and can be identified by using the restriction endonuclease MspI (Fig. 4) (19, 34, 39, 74). For each of these mutations, different restriction fragments are produced by MspI digestion of katG amplicons compared with those of the wild-type katG amplicon. These fragments result from the gain of a MspI restriction site at the 315 codon or the loss of an MspI restriction site at the 463 codon in the respective mutant alleles compared with the wild-type allele. Therefore, specific DNA sequence interrogation in limited regions of katG is possible by virtue of RFLP analysis.
FIG. 4.
PCR-RFLP analysis for identifying isoniazid resistance-associated mutations in M. tuberculosis. (A) Restriction fragment length predicted for two mutations (codons 315 [serine → threonine] and 463 [arginine → leucine]) in the katG gene of M. tuberculosis with the restriction enzyme MspI. These mutations are associated with high- and low-level isoniazid resistance, respectively. W+, wild type. (Adapted with permission from Uhl et al. [74].) (B) RFLP electrophoresis patterns obtained for M. tuberculosis strains. Lanes 1 and 20, DNA fragment standards; lanes 2 and 10, DNA from strains with both the R463L and S315T mutations; lanes 5, 9, 16, 18, and 19, DNA from strains with the R463L mutation only; lanes 13 and 15, DNA from strains with the S315T mutation only. The remaining lanes have neither of these mutations. bp, base pair. (Adapted with permission from J. Uhl et al. [74].)
We have recently demonstrated that this technique is useful for detecting M. tuberculosis complex and isoniazid resistance-associated mutations directly from sputum specimens in 1 working day (73). RFLP analyses have also been demonstrated to be useful for identifying mutations associated with streptomycin resistance which occur in genes encoding 16S rRNA (rrs) and ribosomal protein S12 (rspL) in M. tuberculosis (39), for classifying genes encoding TEM- (2) and SHV-type (45) extended-spectrum β-lactamases in gram-negative bacteria, for identifying mutations in the protease gene of HIV (75), and for determining the genotype and subtype of HCV (9, 10).
Nucleic acid polymorphisms may be degenerate. That is, as part of a codon, they may encode for the same amino acid and so different base pair changes may result in the same amino acid change. In this case, RFLP analyses using the same restriction enzyme may not detect different base pair changes yet the end result is a resistance genotype. Therefore, negative results for RFLP analyses should be backed up with other susceptibility test methods.
(ii) PCR-SSCP analyses.
The technique of PCR-SSCP was first used to detect single nucleotide substitutions in human genes associated with hereditary disorders such as cystic fibrosis (11) and phenylketonuria (13, 30). Several investigators have reported on the utility of the PCR-SSCP method for identifying mutations in genes encoding SHV-type extended-spectrum β-lactamases in the family Enterobacteriaceae (15, 38) and in genes of M. tuberculosis associated with resistance to isoniazid (21, 52, 66, 69, 76), rifampin (16, 55, 66, 67, 68, 79), ethambutol (59), and the fluoroquinolones (65).
For this method, mobility shifts in high-resolution nondenaturing polyacrylamide gels are discernible for single-stranded mutated DNA versus wild-type DNA. After amplification of the target nucleic acid by PCR, it is denatured to a single strand and then electrophoresed. Mutations are inferred by the appearance of bands at positions different to those observed with the wild-type strain (Fig. 5). We have determined that the size of the PCR product is a critical factor for resolving differences of the denatured single-stranded DNA. In our hands, the optimal PCR product is in the range of 310 to 320 bp (69). When we limited the PCR product to this size range, we noted that the sensitivity and specificity of the PCR-SSCP method compared to those for PCR-RFLP analysis for detection of the 463 codon katG mutation (arginine-leucine) in M. tuberculosis were 100% (69).
FIG. 5.
PCR-SSCP analysis to identify isoniazid resistance-associated mutations in M. tuberculosis. Representative PCR-SSCP results for three M. tuberculosis strains, one clinical strain with the R463L mutation (lane 1), one clinical strain with the wild-type codon 463 (R463) (lane 2), and the control strain H37Rv also containing the wild-type codon 463 (R463) (lane 3). The arrows indicates the band position corresponding to the R463L mutation. (Adapted with permission from Temesgen et al. [69].)
Sarkar and colleagues, using the PCR-SSCP method for mutation detection in the factor 1X gene in humans, also noted a lack of sensitivity, especially when greater lengths of target DNA were analyzed (53). They subsequently developed a hybrid technique between dideoxy (Sanger) sequencing and SSCP, termed ddF, which provided a more efficient detection of mutations independent of the amplified product. This technique identifies DNA sequence changes by producing a ladder of bands with one of the four standard dideoxy sequencing reactions and then resolving the products on a nondenaturing polyacrylamide gel. In our laboratory, Felmlee and colleagues used this ddF method to determine rifampin resistance-associated mutations in clinical isolates of M. tuberculosis (16). When ddF results were compared with those for SSCP analysis, they noted that ddF results were more easily interpreted and contained more sequence-dependent information that facilitated differentiation of functionally important versus silent mutations. The ddF method requires the equipment costs (purchase or lease) for an automated DNA sequencer (purchase ∼$100,000 to $120,000). The approximate cost of reagents and supplies per sample is less than $10 in addition to baseline expenditures for a thermocycler and electrophoresis equipment.
A drawback of the conventional SSCP method has been the use of radioactivity for identifying amplification products. This has been obviated by either silver-staining methods (30, 69) or labeling PCR products with fluorescein (68). Furthermore, Telenti and colleagues have demonstrated that fluorescein-labeled products can be evaluated on automated DNA sequencers, which should make SSCP analysis a more acceptable testing method for the clinical laboratory (68). Another modification of the SSCP method by Selvakumar and colleagues (55) permits better visualization of the shifts in migration of single strands. This is accomplished by biotinylation of one of the PCR primers. One biotinylated stand of the PCR product is produced which is separated from the unbiotinylated strand by using streptavidin magnetic beads.
A recent report by Victor and colleagues emphasizes the importance of the positioning of PCR oligonucleotide primers used to produce amplified products for SSCP analysis (76). In cases where multiple PCR products are analyzed by SSCP, overlap of amplified sequences is essential to assure that mutations are not missed which are near the end of the PCR products, i.e., in the regions where PCR primers anneal. In this case, the S315T mutation in katG sequences of M. tuberculosis strains was not detected due to primer placement. Although this example relates to SSCP analysis, it may have relevance to other PCR-based assays.
The cost for reagents and supplies for the manual SSCP method is less than $10 per sample, although expenses for a thermocycler and electrophoresis equipment must be considered. If a DNA sequencer is used for either the SSCP or ddF technique, savings in personnel time should be realized; however, capital outlay for this instrument may be substantial. Both the SSCP and ddF techniques, in our hands, require approximately 2 days for completion.
(iii) PCR-CFLP analysis.
A genetically engineered structure-specific endonuclease, Cleavase I, has been used to detect mutations in the katG (5) and rpoB (20) genes of M. tuberculosis and to determine HCV genotypes (33). The Cleavase I enzyme recognizes the folded structures which DNA strands form after denaturation and cleaves at the junction of the single-stranded and duplexed areas (Fig. 6). After the fragments generated by Cleavase are separated by denaturing gel electrophoresis, unique polymorphisms occur with different mutations.
FIG. 6.
PCR-CFLP for identifying isoniazid resistance-associated mutations in M. tuberculosis. (A) Schematic representation of steps of CFLP pattern generation. Labeled fragments of DNA are heated to separate complementary strands. When the samples are cooled, the single strands of DNA assume folded hairpin-like structures; subtle differences in the sequence of the fragments can cause formation of different structures. The Cleavase I enzyme cleaves at the 5′ side of these structures, at the junction between duplexed and single-stranded regions. Separation and detection of the resulting fragments create signature banding patterns that can be compared to detect differences between the test molecules. (Adapted with permission from Brow et al. [5].) (B) Identification and positioning of mutations associated with isoniazid resistance in M. tuberculosis. For the gel on the left, amplicons generated with a 5′ labeled primer and an unlabeled primer were heat denatured for 15 s, rapidly cooled to 60°C, and digested with 25 U of Cleavase I for 2 min. The results are shown, with M indicating DNA size markers. Variant bands distinguishing mutant from wild-type (WT) DNA are marked A and B. Shown on the right is a second gel electrophoresis which was performed to expand the 500- to 600-nt region. Bands C in the wild type and the R463L mutant are similar, while the same band migrates faster in variants containing the S315T mutation. (Adapted with permission from Brow et al. [5].)
So far, this method has had limited use. According to the manufacturer, it will be commercially available in the future in kit form (Cleavase); Third Wave Technologies, Inc., Madison, Wis.) which should be easily adaptable to most laboratories. The total cost beyond that for a thermocycler and electrophoresis equipment is yet undetermined but the turnaround time for results appears to be less than 1 working day.
(iv)PCR-RNA/RNA duplex RNase cleavage assay.
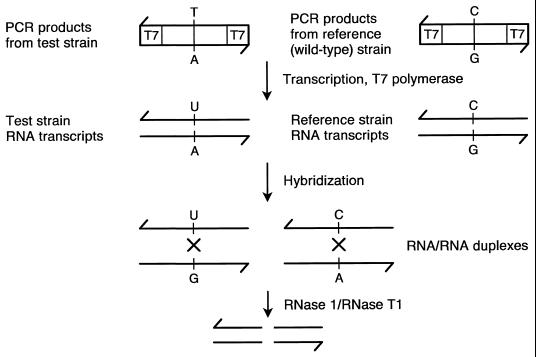
Nash and Inderlied have recently used a PCR-RNA/RNA duplex RNase cleavage assay to detect mutations in the 23S rRNA gene of Mycobacterium avium complex which are associated with macrolide resistance (40). The method is based on the ability of RNase 1 and RNase T1 to cleave mismatches (Fig. 7). Duplex RNA is transcribed from PCR products (test strain and wild-type strain) generated with primers containing opposing phage RNA polymerase promoters (T7/T7 or T7/Sp6). Following hybridization and RNase treatment, cleavage products of RNA/RNA duplexes occur due to mismatches. Mismatches are the result of mutation in the test strain compared with the wild-type strain.
FIG. 7.
PCR-nonisotopic RNase cleavage assay. Commercially available as the Mutation Screener assay (Ambion, Inc.) the PCR-nonisotopic RNase cleavage assay is based on the ability to selectively cleave unpaired bases in RNA/RNA duplexes. Duplex RNA is transcribed from PCR products generated with primers containing opposing phage RNA polymerase promoters (T7/T7 or T7/Sp6). Following hybridization and RNase treatment, the RNA/RNA cleavage products are analyzed by nondenaturing gel electrophoresis and detected by ethidium bromide staining. In the example shown, mismatches occur due to an opposing uracil (U) and guanine (G) and an opposing cystosine (C) and adenine (A) with the adenine representing a point mutation in the test strain. (Adapted with permission from M. Goldrick, Ambion, Inc.)
This method is commercialized and is available in kit form (Mutation Screener; Ambion, Inc., Austin, Tex.). It is relatively easy to perform but requires a longer time for completion than a PCR alone. Nash and Inderlied note that in their hands the assay can be completed in about 1 working day and that the cost (including reagent, supplies and personnel time) is less than $50 if one or more strains are tested simultaneously. Of course, added to this are expenditures for a thermocycler and electrophoresis equipment.
(v) PCR-universal heteroduplex generator analysis.
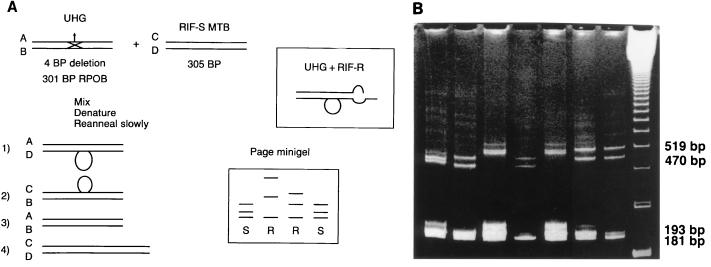
Williams and colleagues have used PCR-universal heteroduplex generator analysis to detect mutations in the rpoB genes of M. tuberculosis (80, 81, 82). DNA from a universal heteroduplex generator (Fig. 8) and the test strain are denatured simultaneously in the same reaction mixture and then allowed to reanneal. The heteroduplex generator in the hypothetical example in Fig. 8a has a 4-bp deletion. When separate strands of DNA from the heteroduplex generator and the test strain reanneal, four separate hybridizations can occur. Two of these hybridizations (heteroduplexes) will result in double-stranded DNA with a “bubble” occurring where no base-pair matches can hybridize. These hybrids will migrate as a single band when electrophoresed in a high-resolution gel. Two other bands will occur due to hybridization reactions (homoduplexes) which result in the formation of the double-stranded DNA of the heteroduplex generator and the test strain. If mutations are present in the test strain, the positions of the homoduplexes may vary compared to those of homoduplexes formed when no mutations are present. This assay requires the same technical expertise and time to complete (about 1 working day) as RFLP assays. Once a heteroduplex generator is produced, the cost for reagents and supplies and instruments is similar to that for RFLP assays. A thermocycler and gel electrophoresis equipment are required, and the approximate cost for reagents and supplies for each test sample is less than $5.
FIG. 8.
PCR-heteroduplex analysis to identify mutations associated with rifampin resistance in M. tuberculosis. (A) Hypothetical example of the universal heteroduplex generator technique. Different bands are produced in the polyacrylamide gel electrophoresis minigel depending on whether the test strain is susceptible or resistant to rifampin. UHG, universal heteroduplex generator; RIF-S, rifampin-susceptible test strain; RIF-R, rifampin-resistant test strain; BP, base pair; ROPβ, RNA polymerase β subunit gene. (Adapted with permission from D. Williams.) (B) In this example, a heteroduplex generator is used which contains four 3-bp deletions and three 2-bp substitutions. The presence of rifampin-susceptible M. tuberculosis is indicated by a four-band pattern. Two homoduplexes migrate in the gel at the equivalence of 181 (the universal heteroduplex generator double-stranded DNA) and 193 bp (the test sample PCR- generated double-stranded DNA). Two heteroduplexes migrate at the equivalence of 470 and 519 bp (each a hybrid of complementary strands of one strand of DNA from the heterodulex generator and on strand from the test sample DNA). The first five lanes (from left to right) contain rifampin-resistant strains, the next two lanes contain rifampin-susceptible strains, and the last lane contains molecular weight standards. (Adapted with permission from D. Williams.)
(vi) PCR-LiPA.
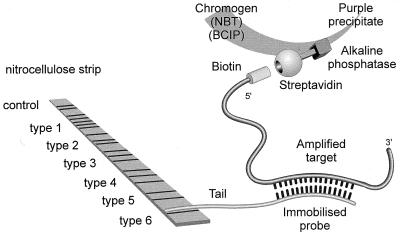
The PCR-line probe assay (LiPA) (Fig. 9) was first developed by Stuyer and colleagues to discriminate types and subtypes of HCV (62). A reverse transcriptase procedure followed by PCR is used to amplify HCV genetic material. The LiPA assay, which is based on the reverse hybridization principle, is then performed. Biotinylated PCR fragments are hybridized to a selection of highly specific immobilized probes. The biotin group in the hybridization complex is then revealed by incubation with a streptavidin-alkaline phosphatase complex and the appropriate chromogen compounds.
FIG. 9.
PCR-LiPA. Biotinylated PCR fragments are hybridized to a selection of highly specific probes immobilized on a nitrocellulose strip. The biotin group in the hybridization complex is revealed by incubation with a streptavidin-alkaline phosphatase complex and the chromogen compounds 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium (NBT). (Figure provided by F. Shapiro, Innogenetics.)
A second-generation LiPA assay was recently demonstrated to be highly sensitive and specific for detecting signature sequence motifs in HCV (63). The assay has also recently been used to detect mutations in the reverse transcriptase gene of HIV (64) and rifampin resistance-associated mutations in M. tuberculosis (51).
According to the manufacturer (Innogenetic, Ghent, Belgium), the assay can be performed in about 1 working day and should be adaptable to most clinical laboratories. The cost per test sample is approximately $80. Furthermore, a baseline expense for a thermocycler is required.
(vii) PCR-molecular beacon sequence analysis.
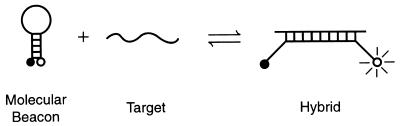
Piatek and colleagues (48) have recently used PCR-molecular beacon sequence analysis to detect mutations in the rpoB gene of M. tuberculosis associated with rifampin resistance. Molecular beacons are hairpin-shaped oligonucleotide probes that report the presence of specific nucleic acids in homogeneous solutions (71) (Fig. 10). After these oligonucleotides anneal to their targets, they undergo a conformational change that restores the fluorescence of an internally quenched fluorophore.
FIG. 10.
Principles of molecular beacons. The molecular beacon in its hairpin form shown on the left is nonfluorescent because the stem hybrid keeps the fluorophore close to the quencher. When the probe sequence in the loop hybridizes to its target, forming a rigid double helix, a conformational reorganization occurs that separates the quencher from the fluorophore, restoring fluorescence. (Adapted with permission from Tyagi et al. [71].)
This method is not yet commercially available. The cost per test sample is unknown; however, a thermocycler is required. Detection of color changes can be done manually, which make the assay particularly appealing for use in developing countries.
(viii) DNA oligonucleotide arrays on silicon microchips.
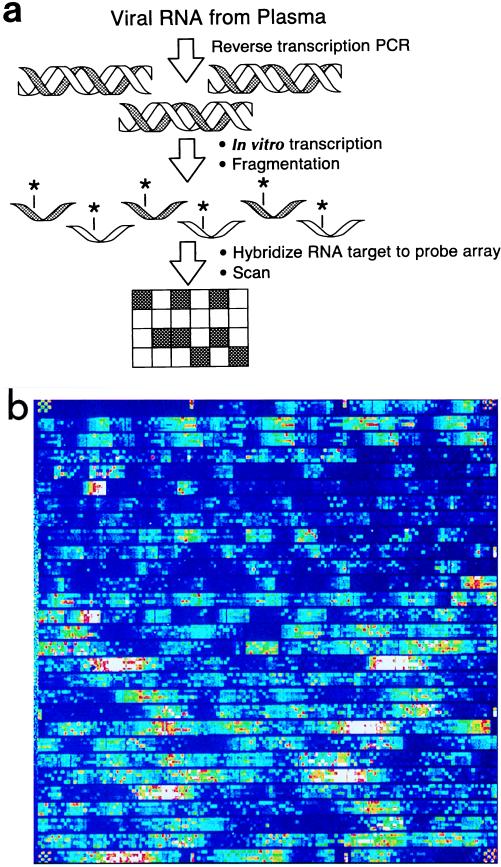
A promising new technology involves the synthesis of DNA probes directly onto silicon microchips (Fig. 11) (6, 32). At present, more than 50,000 oligonucleotide probes can be synthesized directly onto these chips. In one method developed for HIV antiviral-associated resistance mutations, RNA is first reverse transcribed to cDNA. cDNA is subsequently transcribed into RNA which is also biotin labeled. These labeled RNA molecules are then hybridized to complementary probes which have been synthesized directly onto silica microchips.
FIG. 11.
Silicon microchip assay for detection of mutations in the HIV genome associated with resistance to reverse transcriptase or protease inhibitors. (a) A schematic of the commercially available GeneChip HIV PRT Assay (Affymetrix, Santa Clara, Calif.) is shown. Viral RNA is reversed transcribed to cDNA which is then transcribed into RNA, labeled, and fragmented. These labeled RNA fragments are then hybridized to complementary probes synthesized directly onto a microchip. The hybridized array is then scanned to detect positive hybridization signals in the form of emitted light. (Adapted with permission from L. Constantine, Affymetrix.) (b) Example of DNA microchip array image. (Adapted with permission from L. Constantine, Affymetrix.)
Such matrix hybridization formats have great potential for rapid comprehensive detection of resistance genes or mutations associated with antimicrobial resistance. This is especially true if antimicrobial resistance in a specific organism can be due to multiple genes and/or multiple mutations. Examples of this include isoniazid resistance for M. tuberculosis and resistance to reverse transcriptase or protease inhibitors for HIV, as illustrated in Fig. 11. This technology also has the potential for significant automation and therefore should be easily adaptable to most clinical microbiology laboratories. The cost for reagents and instrumentation in addition to a thermocycler is around $150,000.
(ix) Automated DNA sequencing.
DNA sequencing remains the “gold standard” for identifying the products of amplification reactions. DNA sequencing of PCR products has become a much cheaper and faster method by virtue of automation (1, 3). Instruments are now available for semi-automated running and analyzing of sequence gels. Therefore, any resistance gene or resistance mutation can be determined relatively easily and economically by direct DNA sequencing. Musser and colleagues have published the results of a series of studies which used automated DNA sequencing as the primary method to determine antimicrobial resistance-associated mutations for a variety of drugs and M. tuberculosis (24, 37, 58, 59, 60, 61). In our institution, the cost for performing each test sample averages less than $10, but the cost may vary significantly from one institution to another. Initial expenditures for both a thermocycler and automated sequencer are required.
ACKNOWLEDGMENTS
Members of the Division of Clinical Microbiology at the Mayo Clinic (Rochester, Minn.) including David Persing, Glenn Roberts, Jon Rosenblatt, and Thomas Smith are thanked for their critical review of the manuscript. The following persons are recognized for their past or ongoing collaborative efforts with conventional and genetic antimicrobial susceptibility studies: David Persing, Glenn Roberts, Jon Resenblatt, Peggy Kohner, and James Uhl of the Division of Clinical Microbiology; Robin Patel, Zelalem Temesgen, and James Steckelberg of the Division of Infectious Diseases; Bruce Kline, Frank Rusnak, and Nancy Wengenack of the Department of Biochemistry and Molecular Biology; Mayo Clinic; Diana Williams, Louisiana State University, Baton Rouge, La; and Max Salfinger, New York State Department of Health, Albany, N.Y. Finally, Roberta Kondert (Mayo Clinic) is thanked for her efforts in preparing the manuscript.
REFERENCES
- 1.Anonymous. Methods of creating DNA molecules. In: Watson J D, Gilman M, Witkowski J, Moller Z, editors. Recombinant DNA. New York, N.Y: Scientific American Books; 1992. pp. 63–75. [Google Scholar]
- 2.Arlet G, Brami G, Décrè D, Flippo A, Gaillot O, Lagrange P H, Philippon A. Molecular characterization by PCR-restriction fragmented length polymorphism of TEM β-lactamases. FEMS Microbiol Let. 1995;134:203–208. doi: 10.1111/j.1574-6968.1995.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Albright L M. DNA sequencing. In: Ausubel F, Brent R, Kingston R, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, suppl. 23. Boston, Mass: John Wiley and Sons; 1998. pp. 7.0.1–7.7.7.23. [Google Scholar]
- 4.Bignardi G E, Woodford N, Chapman A, Johnson A P, Speller D C E. Detection of the mecA gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J Antimicrob Chemother. 1996;37:53–63. doi: 10.1093/jac/37.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Brow M A, Oldenburg M C, Lyamichev V, Heisler L M, Lyamichera N, Hall J G, Eagen N J, Olive D M, Smith L M, Fors L, Dahlberg J E. Differentiation of bacterial 16S rRNA genes and intergenic regions and Mycobacterium tuberculosis katG genes by structure-specific endonuclease cleavage. J Clin Microbiol. 1996;34:2139–3132. doi: 10.1128/jcm.34.12.3129-3137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellino A M. When the chips are down. Genome Res. 1997;7:943–946. doi: 10.1101/gr.7.10.943. [DOI] [PubMed] [Google Scholar]
- 7.Cockerill III F R, Uhl J R, Temesgen Z, Zhang Y, Stockman L, Roberts G D, Williams D L, Kline B C. Rapid identification of a point mutation of the Mycobacterium tuberculosis catalase-peroxidase (katG) gene associated with isoniazid resistance. J Infect Dis. 1995;171:240–245. doi: 10.1093/infdis/171.1.240. [DOI] [PubMed] [Google Scholar]
- 8.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Sivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 9.Constantine N, Abdel-Hamid M, Oldach D. Rapid genotyping of hepatitis C virus. N Engl J Med. 1995;333:800. doi: 10.1056/nejm199509213331213. [DOI] [PubMed] [Google Scholar]
- 10.Davidson F, Simmonds P, Ferguson J, Jarvis L, Dow B, Follett E, Seed C, Krusius T, Lin C, Medgyesi G, Kiyokawa H, Olim G, Duraisamy G, et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 11.Dean M, White W B, Amos J, Gerrard B, Stewart C, Khaw K-T, Leppert M. Multiple mutations in highly conserved residues are found in mildly affected cystic fibrosis patients. Cell. 1990;61:863–870. doi: 10.1016/0092-8674(90)90196-l. [DOI] [PubMed] [Google Scholar]
- 12.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dockhorn-Dworniczak B, Dworniczak B, Brammelkamp L, Bülles J, Horst J, Böcker W W. Non-isotopic detection of single-strand conformation polymorphism (PCR-SSCP): a rapid and sensitive technique in diagnosis of phenylketonuria. Nucleic Acids Res. 1991;19:2500. doi: 10.1093/nar/19.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragon E A, Spadoro J P, Madej R. Quality control of polymerase chain reaction. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: ASM Press; 1993. pp. 160–168. [Google Scholar]
- 15.Fatima-Hannachi M, Gascoyne-Binzi D M, Heritage J, Hawkey P M. Detection of mutations conferring extended-spectrum activity on SHV β-lactamases using polymerase chain reaction single strand conformational polymorphism (PCR-SSCP) J Antimicrob Chemother. 1996;37:797–802. doi: 10.1093/jac/37.4.797. [DOI] [PubMed] [Google Scholar]
- 16.Felmlee T A, Liu Q, Whelen A C, Williams D, Sommer S S, Persing D H. Genotypic detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand conformation polymorphism and dideoxy fingerprinting. J Clin Microbiol. 1995;33:1617–1623. doi: 10.1128/jcm.33.6.1617-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredricks D N, Relman D A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanthenolsulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geha D J, Uhl J R, Gustafero C A, Persing D H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas W H, Schilke K, Brand J, Amthor B, Weyer K, Fourie P B, Bretzel G, Sticht-Groh V, Bremer H J. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob Agents Chemother. 1997;41:1601–1603. doi: 10.1128/aac.41.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisler L, Lyamicheva N, Olive D M. Abstracts of the 96th General Meeting of the American Society for Microbiology. 1996. Detection of mutations related to rifampin resistance using Cleavase® fragment length polymorphism, abstr. U-95; p. 117. [Google Scholar]
- 21.Heym B, Alzari P M, Honoré N, Cole S T. Missense mutations in the catalse-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15:235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 22.Jacobsen G, Hängi M, Ott M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to human immunodeficiency virus type I proteinase inhibitors: mutations, kinetics, and frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby G, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapur V, Li L L, Iordanescu S, Hamrick M K, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoβ encoding RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitagawa Y, Yeda M, Ando N, Endo M, Ishibiki K, Kobayashi Y, Arai T, Kitajima M. Rapid diagnosis of methicillin-resistant Staphylococcus aureus bacteria by nested polymerase chain reaction. Ann Surg. 1996;224:665–671. doi: 10.1097/00000658-199611000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohner P C, Patel R, Uhl J R, Garin K M, Hopkins M K, Wegener L T, Cockerill F R., III Comparison of agar dilution, broth microdilution, E-test, disk diffusion, and automated Vitek methods for testing susceptibilities of Enterococcus spp. to vancomycin. J Clin Microbiol. 1997;35:3258–3263. doi: 10.1128/jcm.35.12.3258-3263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolbert C P, Connolly J E, Lee M J, Persing D H. Detection of the staphylococcal mecA gene by chemiluminescent DNA hybridization. J Clin Microbiol. 1995;33:2179–2182. doi: 10.1128/jcm.33.8.2179-2182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolbert C P, Varga-Delmore P, Arruda J, Zheng X, Lewis M E, Kolberg J, Persing D. A novel branched-DNA assay for detection of mecA gene in oxacillin-resistant and oxacillin-susceptible staphylococci. J Clin Microbiol. 1998;36:2640–2644. doi: 10.1128/jcm.36.9.2640-2644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labrune P, Melle D, Rey F, Berthelon M, Caillaud C, Rey J, Munnich M, Lyonnet S. Single-strand conformation polymorphism for detection of mutations and base substitutions in phenylketonuria. Am J Hum Genet. 1991;48:1115–1129. [PMC free article] [PubMed] [Google Scholar]
- 31.Larder B, Kemp S. Multiple mutations in HIV-1 reverse transcriptase confer high level resistance to zidovudine. 1989. Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 32.Lipshutz R J, Morris D, Chee M, Hubbell E, Kozal M J, Shah N, Shen N, Yang R, Fodor S P A. Using oligonucleotide arrays to assess genetic diversity. BioTechniques. 1995;19:442–447. [PubMed] [Google Scholar]
- 33.Marshall D J, Heisler L M, Lyamichev V, Murrine C, Olive D M, Ehrlich G D, Neri B P, de Arruda M. Determination of hepatitis C virus genotypes in the United States by cleavase fragment length polymorphism analysis. J Clin Microbiol. 1997;35:3156–3162. doi: 10.1128/jcm.35.12.3156-3162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marttila H J, Soini H, Huovinen P, Vijanen M K. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother. 1996;40:2187–2189. doi: 10.1128/aac.40.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo L, Wang Q. Rapid detection of methicillin-resistant staphylococci using polymerase chain reaction. Int J Infect Dis. 1997;2:15–20. [Google Scholar]
- 36.Mulder J G. Comparison of disk diffusion, the E test, and detection of mecA for determination of methicillin resistance in coagulase-negative staphylococci. Eur J Clin Microbiol Infect Dis. 1996;15:567–573. doi: 10.1007/BF01709365. [DOI] [PubMed] [Google Scholar]
- 37.Musser J M, Kapur V, Williams D L, Kreiswirth B N, van Soolingen D, Embden J D A. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 38.M’Zali F H, Heritage J, Gasoyne-Binzi D M, Snelling A M, Hawkey P M. PCR single-strand conformational polymorphism can be used to detect the gene encoding SHV-7 extended-spectrum β-lactamase and to identify different SHV genes within the same strain. Antimicrob Agents Chemother. 1998;41:123–125. doi: 10.1093/jac/41.1.123. [DOI] [PubMed] [Google Scholar]
- 39.Nachamkin I, Kang C, Weinstein M P. Detection of resistance to isoniazid, rifampin, and streptomycin in clinical isolates of Mycobacterium tuberculosis by molecular methods. Clin Infect Dis. 1997;24:894–900. doi: 10.1093/clinids/24.5.894. [DOI] [PubMed] [Google Scholar]
- 40.Nash K A, Inderlied C B. Rapid detection of mutations associated with macrolide resistance in Mycobacterium avium complex. Antimicrob Agents Chemother. 1996;40:1748–1750. doi: 10.1128/aac.40.7.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Committee for Clinical Laboratory Standards. Antimycobacterial susceptibility testing. Publication M24-P. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 42.National Committee for Clinical Laboratory Standards. Methods for antimicrobial susceptibility testing of anaerobic bacteria. 4th ed. Approved standard M11-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 43.National Committee for Clinical Laboratory Standards. Reference method for both dilution antifungal susceptibility testing of yeasts. Publication M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 44.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 8th informational supplement. Publication M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standard; 1998. [Google Scholar]
- 45.Nüesch-Inderbinen M T, Hächer H, Kayser F H. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E-test. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 46.Patel R, Uhl J, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel R, Uhl J R, Kohner P, Hopkins M K, Steckelberg J M, Kline B, Cockerill F R., III DNA sequence variation within vanA, vanB, vandC-1 and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob Agents Chemother. 1998;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piatek A S, Tyagi S, Pol A C, Telenti A, Miller L P, Kramer F R, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotech. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 49.Podzorski R P, Persing D H. Molecular detection and identification of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C: ASM Press; 1995. pp. 130–157. [Google Scholar]
- 50.Richman D D. Antiretroviral drug resistance: mechanisms, pathogenesis, clinical significance. Adv Exp Med Biol. 1996;394:383–395. doi: 10.1007/978-1-4757-9209-6_35. [DOI] [PubMed] [Google Scholar]
- 51.Rossau R, Traore H, De Beenouwer H, Mijs W, Jannes G, DeRijk P, Portaels F. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother. 1997;41:2093–2098. doi: 10.1128/aac.41.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouse D A, Zhongming L, Bai G-H, Morris S L. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995;39:2472–2477. doi: 10.1128/aac.39.11.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarkar G, Yoon H-S, Sommer S S. Dideoxy fingerprinting (ddF): a rapid and efficient screen for the presence of mutations. Genomics. 1992;13:441–443. doi: 10.1016/0888-7543(92)90266-u. [DOI] [PubMed] [Google Scholar]
- 54.Sasadeusz J J, Tufaro F, Safrin S, Schubert K, Hubinette M M, Cheung P K, Sachs S L. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J Virol. 1997;71:3872–3878. doi: 10.1128/jvi.71.5.3872-3878.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selvakumar N, Wilson S M, McNerney R, Narayanan P R. Single strand conformation polymorphism profiles with biotinylated PCR products to detect mutations in rpoβ gene of Mycobacterium tuberculosis. Curr Sci. 1997;73:774–777. [Google Scholar]
- 56.Shoek R, Gerard M, Sadzot-Delvaux C, Andrei G, Balzarini J, Reymen D, Ahadi N, De Bruyn J M, Pielte J, Rentier B. Meningoradiculoneuritis due to acyclovir-resistant varicella zoster virus in an acquired immune deficiency syndrome patient. J Med Virol. 1994;42:338–347. doi: 10.1002/jmv.1890420404. [DOI] [PubMed] [Google Scholar]
- 57.Smith J L, Cherrington J M, Jiles R E, Futter M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 58.Sreevatsan S, Pan X, Kreiswirth B N, Musser J M. Mutations associated by pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex orangisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sreevatsan S, Stockbauer K E, Pan X, Kreiswirth B N, Moghazeh S L, Jacobs W R, Jr, Telenti A, Musser J M. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sreevatsan S, Pan X, Stockbauer K E, Williams D L, Kreiswirth B N, Musser J M. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates for diverse geographic localities. Antimicrob Agents Chemother. 1996;40:1024–1026. doi: 10.1128/aac.40.4.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sreevatsan S, Pan X, Zhang Y, Deretic V, Musser J M. Analysis of the oxyR-aphC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997;41:600–606. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 63.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuyver L, Wysneur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takiff H E, Salazar L, Guerrero C, Philipp W, Huang W M, Kreiswirth B, Jacobs W R, Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 68.Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Temesgen Z, Satoh K, Uhl J R, Kline B C, Cockerill F R., III Use of polymerase chain reaction single-srtand conformation polymorphism (PCR-SSCP) analysis to detect a point mutation in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis. Mol Cell Probes. 1997;11:59–63. doi: 10.1006/mcpr.1996.0077. [DOI] [PubMed] [Google Scholar]
- 70.Tenover F C, Popovic T, Olsvik O. Genetic methods for detecting antibacterial resistance genes. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C: ASM Press; 1995. pp. 1368–1378. [Google Scholar]
- 71.Tyagi S, Bratu D P, Kramer F R. Multicolor beacons for allele discrimination. Nat Biotech. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 72.Ubakata K, Nakagami S, Nitta A, Yamane A, Kawkami S, Sugiura M, Masatoshi K. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992;30:1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uhl J R, Cockerill III F R, Kline B C, Stockman L, Robert G D, Williams D L, Fitzgerald M, Salfinger M. Abstracts of ASM Conference on Tuberculosis. Past, Present, and Future. Washington, D.C: ASM Press; 1997. Rapid simultaneous detection of Mycobacterium tuberculosis and isoniazid resistance directly from sputum. [Google Scholar]
- 74.Uhl J R, Sandhu G S, Kline B C, Cockerill F R., III . PCR-RFLP detection of point mutations in the catalse-peroxidase gene (katG) of Mycobacterium tuberculosis associated with isoniazid resistance. In: Persing D, editor. PCR protocols for emerging infectious disease. Washington, D.C: ASM Press; 1996. pp. 144–149. [Google Scholar]
- 75.Vasudevachari M B, Zhang Y M, Imamichi H, Imamichi T, Falloon J, Salzman N P. Emergence of protease inhibitor resistance mutations in human immunodeficiency virus type 1 isolates from patients and a rapid screening procedure for their detection. Antimicrob Agents Chemother. 1996;40:2535–2541. doi: 10.1128/aac.40.11.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Victor T C, Pretorius G S, Felix J V, Jordaan A M, van Helden P D, Eisenach K D. katG mutations in isoniazid-resistant strains of Mycobacterium tuberculosis are not infrequent. Antimicrob Agents Chemother. 1996;40:1572. doi: 10.1128/aac.40.6.1572. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wallet F, Roussel-Delvallez M, Courcol R J. Choice of a routine method for detecting methicillin resistance in staphylococci. J Antimicrob Chemother. 1996;37:901–909. doi: 10.1093/jac/37.5.901. [DOI] [PubMed] [Google Scholar]
- 78.Weller T M A, Crook D W, Crow M R, Ibrahim W, Pennington T H, Selkon J B. Methicillin susceptibility testing of staphylococci by E-test and comparison with agar dilution and mecA detection. J Antimicrob Chemother. 1997;39:251–253. doi: 10.1093/jac/39.2.251. [DOI] [PubMed] [Google Scholar]
- 79.Whelen A C, Felmlee T A, Hunt J M, Williams D L, Roberts G D, Stockman L, Persing D H. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR. J Clin Microbiol. 1995;33:556–561. doi: 10.1128/jcm.33.3.556-561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williams D L, Limbers C W, Spring L, Jayachandra S, Gillis T P. PCR-heteroduplex detection of rifampin-resistant Mycobacterium tuberculosis. In: Persing D, editor. PCR protocols for emerging infectious diseases. Washington, D.C: ASM Press; 1996. pp. 122–129. [Google Scholar]
- 81.Williams D L, Spring L, Gillis T P, Salfinger M, Persing D H. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin Infect Dis. 1998;26:446–450. doi: 10.1086/516313. [DOI] [PubMed] [Google Scholar]
- 82.Williams D L, Waguespack C, Gillis T P, Eisenach K, Crawford J, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zein N N, Rakela J, Krawitt E L, Reddy K R, Tominaga T, Persing D H. Hepatitis C virus genotypes in the United States: epidemiology, pathogenecity, and response to interferon therapy. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]
- 84.Zheng X, Kolbert C P, Delmore P, Arruda J, Lewis M, Kolberg J, Cockerill F R, Persing D H. Abstracts of the 37th Meeting of the International Conference on Antimicrobial Agents and Chemotherapy. Toronoto, Ontario, Canada. 1997. Direct mecA detection from blood culture bottles by branched DNA (bDNA) signal amplification, abstr. D-23; p. 87. [Google Scholar]