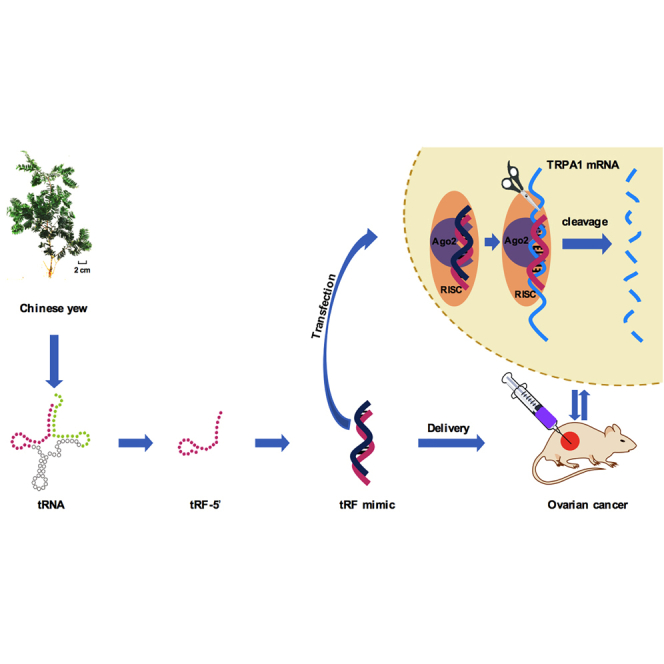
Abstract
Drug discovery from plants usually focuses on small molecules rather than such biological macromolecules as RNAs. Although plant transfer RNA (tRNA)-derived fragment (tRF) has been associated with the developmental and defense mechanisms in plants, its regulatory role in mammals remains unclear. By employing a novel reverse small interfering RNA (siRNA) screening strategy, we show that a tRF mimic (antisense derived from the 5′ end of tRNAHis(GUG) of Chinese yew) exhibits comparable anti-cancer activity with that of taxol on ovarian cancer A2780 cells, with a 16-fold lower dosage than that of taxol. A dual-luciferase reporter assay revealed that tRF-T11 directly targets the 3′ UTR of oncogene TRPA1 mRNA. Furthermore, an Argonaute-RNA immunoprecipitation (AGO-RIP) assay demonstrated that tRF-T11 can interact with AGO2 to suppress TRPA1 via an RNAi pathway. This study uncovers a new role of plant-derived tRFs in regulating endogenous genes. This holds great promise for exploiting novel RNA drugs derived from nature and sheds light on the discovery of unknown molecular targets of therapeutics.
Keywords: tRNA, tRF, ovarian cancer, Taxus chinensis, TRPA1
Graphical abstract
Introduction
The plant kingdom has been regarded as a significant source of medicinal products throughout the history of mankind.1 Herbal decoctions have been used for the prevention and treatment of diseases since ancient times and they remain in use nowadays.2 A wide variety of secondary metabolites, such as flavonoids, terpenes, and alkaloids, as obtained from this chemical treasure have been identified as active compounds. Despite the focus of most research on small molecule active principles, there has been increasing attention paid to biologics over the past few decades. Notably, crude plant extracts were found to be more effective than the single compound,3 which suggests that the whole plant extracts contain a cocktail of components where such substances as macromolecules may contribute significantly to the overall pharmacological effects of the herbal medicines.
Small RNAs (<200 nt) were discovered to play critical roles in a wide variety of biological processes for microorganisms, plants, animals, and humans, as well as in various human diseases through regulating those protein coding genes.4,5 Zhang et al.6 reported that an exogenous plant microRNA (miRNA), MIR168a, which is abundant in rice, is enriched in the sera of human subjects relying on rice as a staple food. MIR168a could bind to low-density lipoprotein (LDL) receptor adaptor protein 1 (LDLRAP1) mRNA to inhibit its expression in mice, thus suppressing the removal of LDL from the plasma. It was also found that a stable MIR2911 derived from Chinese medicine honeysuckle could be effective not only in producing inhibitory effects on various influenza A viruses but also in suppressing severe acute respiratory syndrome coronavirus-2 (SARS-COV-2) replication and accelerating the negative conversion of infected patients.7,8 In addition, miRNAs have been identified as playing some other therapeutic roles. For example, the plant miRNA miR159 detected from Western human sera could suppress the progression of breast cancer by targeting transcription factor 7.9 A small RNA HJT-sRNA-m7 derived from Chinese medicine Hongjingtian (Rhodiola crenulata) was found to be effective in treating pulmonary fibrotic disease by targeting α-SMA, fibronectin, and collagen type III α 1.10 Moreover, the mdo-miR7267-3p obtained from ginger could be encapsulated by ginger exosome-like nanoparticles to cross-kingdom regulate gut microbiota.11 These findings have indicated clearly that plants may be a desirable source of RNAs that can be used to regulate the expression of target genes for therapeutic purposes, especially for miRNAs. Notably, a recent report has suggested that the small RNA of corn, which has a larger size than miRNA, also exhibited anti-proliferative activity against HeLa cells,12 indicating that small RNA species other than miRNA can also exhibit pharmacological activities.
Known as the most abundant small RNA species, transfer RNAs (tRNAs) play a crucial role in protein translation and perform various regulatory functions required for several important biological processes, such as cell signaling, apoptosis, and stress response.13 It has been revealed that individual tRNAs purified from non-pathogenic Escherichia coli could significantly suppress the growth of colorectal cancer cells, indicating the gut microbiota's tRNAs as potential anti-cancer therapeutics for the first time.14 Most recently, tRNA-derived fragments (tRFs) have been identified as possessing such regulatory properties.15 The small RNAs of this class are not generated at random from tRNAs, and many of them are abundantly but specifically produced under certain conditions.16,17 The expressions of several tRFs are shown to have a positive association with cell proliferation in cancer cell lines. Furthermore, the endogenous tRFs derived from human breast cancer cells were found to be effective in suppressing breast cancer in vitro and in vivo by targeting the YBX1 gene,18 suggesting that tRFs could target the signaling pathways related to cell proliferation and thus enable an effective intervention with the treatment of cancers. These findings thus brought our attention to the potential suitability of plant-derived tRNAs and tRFs as RNA interference (RNAi)-based therapeutics.
As a species in the family Taxaceae, Chinese yew (Taxus chinensis (Pilger) Rehd. var. mairei) refers to an ornamental evergreen shrub or tree widely distributed in areas with high elevation across China. Chinese yew as a significant variety of medicinal plant has been used for the production of paclitaxel (taxol). In addition, it is the first anti-cancer small molecule,19 and has now been accepted as a standard first-line chemotherapeutic drug designed to treat multiple solid tumors, especially for ovarian cancer.20 However, the patients treated with taxol have been found to develop drug resistance.21 Therefore, it is necessary to develop a novel therapeutic strategy for cancer treatment. In this study, an investigation was conducted into the tRNA and tRF contained in Chinese yew for any potential positive effects on reducing the progression of ovarian cancer.
In this study, the tRNA sequence information on the Chinese yew was obtained by means of next-generation sequencing (NGS). After affinity purification and subsequent cytotoxicity determination, it was found that the individual tRNA contained in Chinese yew exhibited strong cytotoxicity on ovarian cancer cells. Through the design and screening of tRF, the tRF-T11 derived from the 5′ end of tRNAHis(GUG) with its antisense were discovered to produce comparable anti-cancer effects with taxol on ovarian cancer cell A2780 and its xenograft animal model. As further revealed by various mechanistic studies including dual-luciferase reporter assay and immunoprecipitation assay, tRF-T11 can interact with AGO2 to directly target TRPA1 and suppress its expression through the RNAi pathway in ovarian cancer cells.
Results
tRNA species contribute to the cytotoxicity of Taxus tRNA-enriched fraction
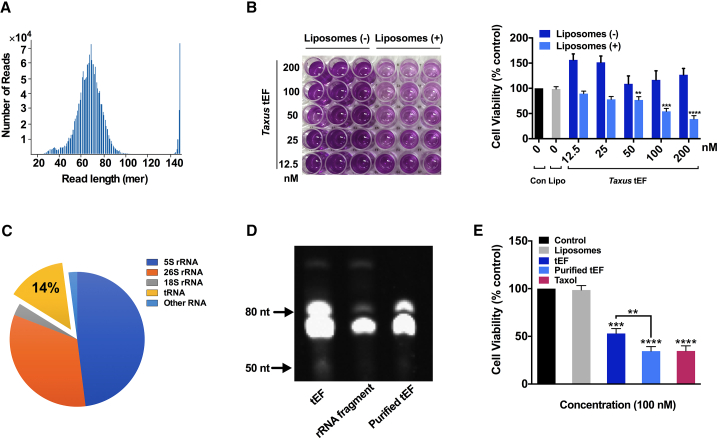
Total RNA was isolated from fresh branches of T. chinensis by a well-developed hexadecyl trimethyl ammonium bromide (CTAB) RNA extraction method (Figure S1A). Preparations obtained were of high integrity (RIN > 7.0) and purity (260/280 > 2.0, 260/230 > 2.0; Figure S1B). Subsequently, small RNA (<200 nt) was separated from large RNA with high purity (Figures S1C and S2). Separation of small RNA was carried out by using a 10% urea-polyacrylamide gel electrophoresis (urea-PAGE), in which the tRNA species were then gel fractionated and purified (Figure S3A) to remove 5S, 5.8S rRNA, and other RNA species (Figures S3B and S3C). NGS of the resultant tRNA-enriched fraction (tEF) generated over 1.7 million clean reads with a majority ranging from 60 to 80 mer in length (Figure 1A). Bioinformatics analysis identified and annotated over 20 Taxus tRNA sequences in the tEF using tRNAscan-SE 2.0 and NCBI Basic Local Alignment Search Tool (BLAST) program (Table S1). When transfected by liposomes, this tEF preparation has demonstrable cytotoxicity on ovarian cancer (OVCA) cell line A2780 with a half maximal inhibitory concentration (IC50) value of 128.8 nM, while no cytotoxicity was observed in controls without transfection (Figure 1B), implying that tRNAs in the tEF might act as effectors and contribute to the bioactivity. However, the content analysis of the reads in the tEF library revealed that rRNA fragments accounted for over 50% of the overall number of reads, while tRNA species only accounted for 14% (Figure 1C). Upon further purification to remove rRNA fragments using a Ribo-Zero rRNA removal kit (Illumina) (Figure 1D), the cytotoxic effects of retained tEF was observed to be significantly increased (Figure 1E), indicating the tRNA species are the principal factors responsible for the observed cytotoxic activities of Taxus tEF.
Figure 1.
Taxus tRNA species are major contributor of cytotoxicity of tEF
(A) Distribution of read length in the tEF as determined by NGS with a range from approximately 30 to 90 mer peaking at about 70 mer. (B) Taxus tEF with liposomal transfection exhibited significant cytotoxicity on A2780 cells in dose-dependent manner. Controls (Con) were without tEF and liposomes. Liposomes (Lipo) were without tEF. All data were normalized to Con. Representative MTT assay images are shown in the left panel. (C) NGS reads content analysis showed that only 14% of the tEFs belong to tRNA sequence. (D) Urea-PAGE analysis of the tEF before and after rRNA fragment purification. tEF = before purification, rRNA fragment = rRNA fragment removed from tEF, and purified tEF = after purification. (E) Comparison of the cytotoxic activities of tEF before and after rRNA fragment purification. Taxol was used as a positive control. Data are shown as the means ± SDs of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Individual tRNAs purified from Chinese yew are cytotoxic to cancer cells
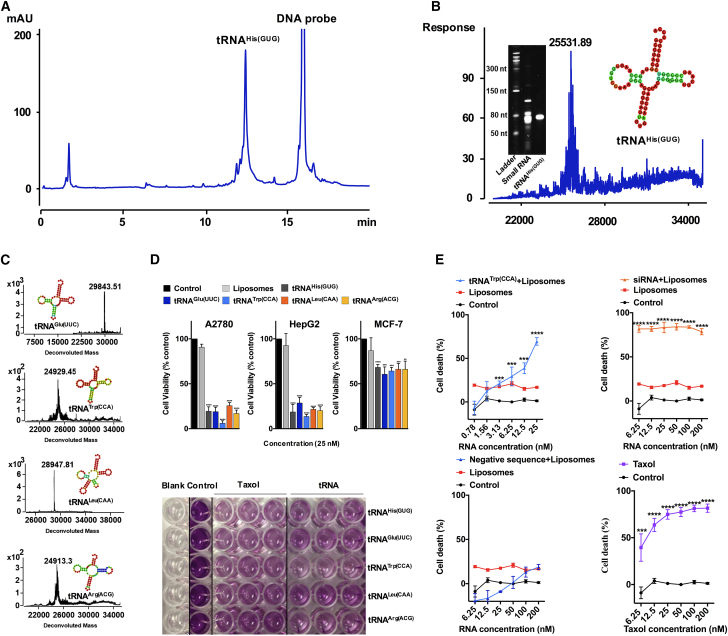
To further confirm the bioactivity of tRNA species, the five most abundant individual tRNAs (>500 reads per million) were captured based on a solid-phase DNA probe, followed by ultra-high performance liquid chromatography (UHPLC) to separate individual tRNA and its cognate DNA probe. UHPLC analysis demonstrated that the purified tRNAHis(GUG) displayed a single peak under UV 260 nm (Figure 2A) and a single gel band was clearly observed in the urea-PAGE analysis, confirming a high purity of the obtained tRNA (Figure 2B). Deconvolution mass spectrum indicated that the molecular weights of purified tRNAs are higher than those calculated based on unmodified sequence (Figures 2B and 2C). Interestingly, the top five abundant tRNAs at 25 nM were found to markedly reduced cell viability of A2780 cells (OVCA) and HepG2 cells (liver cancer) in vitro, but were much less effective on MCF-7 cells (breast cancer) (Figure 2D). Among the five tRNAs, tRNATrp(CCA) appeared to have the strongest cytotoxicity on A2780 cells, with an IC50 value of 14 nM, relatively comparable with that of taxol at 7 nM (Figure 2E; Table S2).
Figure 2.
Individual tRNAs exhibit anti-proliferation effects on cancer cells
(A) Representative UHPLC chromatogram of tRNAHis(GUG) with probe. (B) Deconvolution MS and urea-PAGE analysis (left inset) of purified tRNAHis(GUG) (cloverleaf-like secondary structure shown as right inset). (C) Deconvolution MS results of tRNAGlu(UUC), tRNATrp(CCA), tRNALeu(CAA), and tRNAArg(ACG). (D) Effects of the five most abundant tRNAs from Chinese yew on cell viability of A2780, HepG2, and MCF-7 cells at 25 nM. Representative MTT assay images of tRNAs on A2780 cells are presented in the lower panel. (E) Comparison of the dose-dependent cytotoxic effects of tRNATrp(CCA), siRNA, negative sequence, and taxol on A2780 cells. Data are shown as the means ± SDs of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Taxus tRF exhibits cytotoxicity in OVCA cells
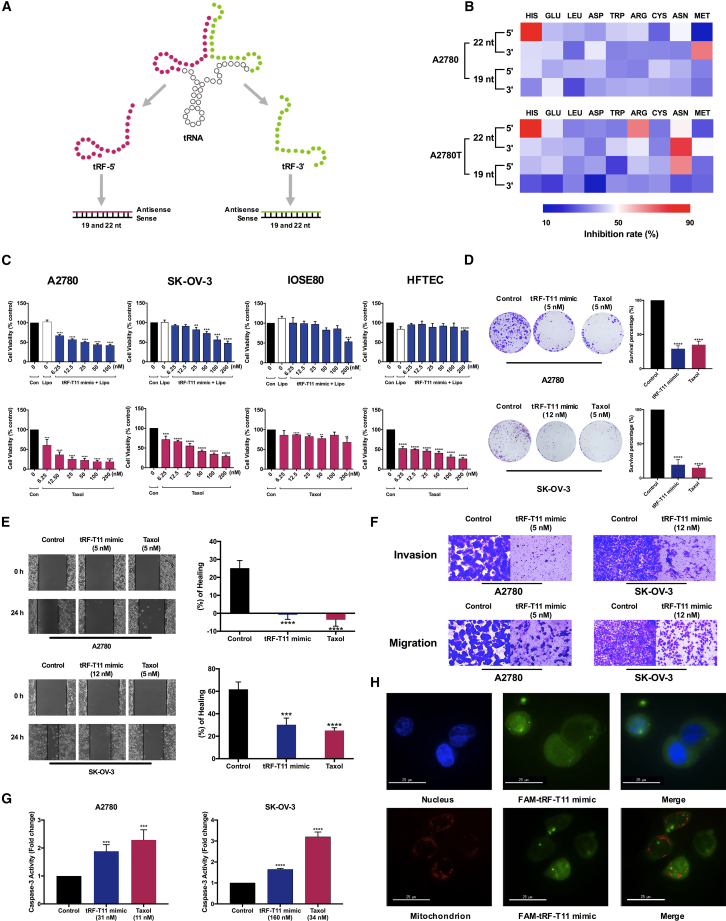
A recently reported tumor-suppressive role of endogenous tRFs on human breast cancer cells has led to a hypothesis that tRF derived from the tRNAs above may exhibit antitumor activity.18 Since tRFs from the 5′ terminal and 3′ ends (tRF-5′ and tRF-3′) in 19 or 22 nt are the most abundant tRF species,22,23 a total of 36 tRFs derived from the nine most abundant tRNAs of Chinese yew (Table S3), including tRF-5′ and tRF-3′ with lengths of, respectively, 19 and 22 nt, were selected as the antisense strands of tRF mimics in siRNA form (Figure 3A). High-throughput cytotoxic screening (Figure 3B) identified tRF-T11 mimic (a double-strand RNA with a 22-mer tRF-5′ derived from tRNAHis(GUG)) as the most active on both A2780 and its taxol-resistant strain A2780T cells. Since tRF-T11 as the antisense exhibited a worse cytotoxicity than tRF-T11 mimic, while its sense did not show significant inhibitory effects (Table 1), the double-strand tRF-T11 mimic was selected for further investigation of its biological and pharmacological activities. In A2780 and SK-OV-3 cells, the IC50 value of tRF-T11 mimic (31 nM and 158.7 nM) is comparable with that of taxol (10.8 nM and 33.8 nM), while it showed no significant effects on normal ovarian surface epithelial cells IOSE80 (Figure 3C). Importantly, the IC50 value of tRF-T11 mimic on A2780T cells remained unchanged, which is 80-fold lower than that of taxol (Figure S4). Since most high-grade serous adenocarcinomas originate from fallopian tube rather than ovary, which has become the most common type of OVCA with the highest mortality,24 the side effects of tRF-T11 mimic were also tested on the fallopian tube cell line HFTEC. The results also demonstrated that no significant effects of tRF-T11 mimic were observed, while a strong cytotoxicity of taxol appeared with an IC50 value of 11 nM (Figure 3C). Clonogenic assay demonstrated that tRF-T11 mimic can also significantly suppress the colony formation of A2780 and SK-OV-3 cell lines (Figure 3D). It also showed significant inhibitory effects on metastasis in both cell lines treated with tRF-T11 mimic by wound healing assay (Figure 3E). Furthermore, tRF-T11 mimic was found to markedly reduce the migration and invasion of OVCA cells (Figure 3F). In addition, the caspase-3 activity in A2780 and SK-OV-3 cells treated with tRF-T11 mimic was determined since caspase-3 is a key biomarker of apoptosis in cells.25 The results showed that tRF-T11 mimic could induce an approximately 1.6-fold increase of caspase-3 activity in the two ovarian cells after 18 h of treatment (Figure 3G). The above results demonstrated that tRF-T11 mimic exhibited significant anti-cancer activities against OVCA cells, especially for A2780, thus this cell line was selected for further investigations. Fluorescent dye (FAM)-labeled tRF-T11 mimic was mainly observable in the cytoplasm of A2780 cells and partly in the nucleus, but not in mitochondria (Figure 3H), suggesting that the cytoplasm or nucleus is the site of action of this tRF mimic, such as suppression of the expression of endogenous targets.26
Figure 3.
tRF-T11 mimic inhibits human OVCA cell proliferation
(A) Schematic diagram describing the design of tRF mimic from the 5′ and-3′ end of tRNA of 19 and 22 nt length. (B) Heatmap of the 36 synthesized tRF mimics according to their cytotoxicity against A2780 and A2780T cells. tRF-T11 mimic (top left corner) was identified to be the most effective. (C) Comparison of the dose-dependent effects of tRF-T11 mimic and taxol against A2780, SK-OV-3, IOSE80, and HFTEC cells. (D) Clonogenic assay of tRF-T11 mimic on A2780 and SK-OV-3 cells. (E) Wound healing assay of tRF-T11 mimic on A2780 and SK-OV-3 cells. (F) Transwell assay of tRF-T11 mimic on A2780 and SK-OV-3 cells. (G) Determination of caspase-3 activity in A2780 and SK-OV-3 cells treated with tRF-T11 mimic. (H) Fluorescent images showing the presence of FAM-labeled tRF-T11 mimic mainly in the cytoplasm and partly in the nucleus, but not in the mitochondria of A2780 cells. Scale bar, 25 μm. Data are presented as the means ± SDs of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Table 1.
IC50 values (nM) of RNA samples and taxol on A2780, SK-OV-3, A2780T, IOSE80, and HFTEC cells
| A2780 | SK-OV-3 | A2780T | IOSE80 | HFTEC | |
| tRF-T11 | 114.3 | 293.0 | 217.8 | ||
| Sense of tRF-T11 mimic | |||||
| tRF-T11 mimic | 31.0 | 158.7 | 32.3 | ||
| siRNA | 0.25 | 7.8 | 16.3 | ||
| Scramble control | |||||
| Taxol | 10.8 | 33.8 | 2600.0 | 11.0 |
Empty cells indicate that the values cannot be calculated.
tRF-T11 targets and inhibits TRPA1 expressions in vitro
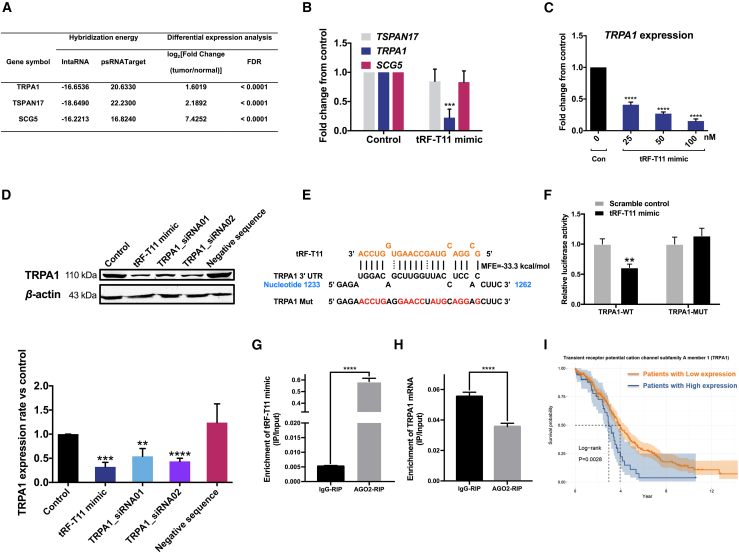
The functionality of exogenous tRFs may overlap that of miRNAs, a characteristic that has been widely documented for endogenous tRFs in both plants and mammalian cells.27,28 To further elucidate the mechanism of action that may underlie the functionality of tRFs, potential target genes of tRF-T11 were identified by in silico prediction using two independent bioinformatic tools, IntaRNAv229 and psRNATarget,30 followed by a differential expression analysis based on the gene expression profiles in normal and tumor ovarian tissue to prioritize the candidate targets. As a result, the top three differentially expressed genes, namely transient receptor potential cation channel subfamily A member 1 (TRPA1), tetraspanin 17 (TSPAN17), and secretogranin V (SCG5), were identified as the most likely candidates based on their minimum required energy to hybridize with tRF-T11 at their 3′ UTRs of mRNA (Figure 4A). Among these three genes, only TRPA1 was significantly downregulated in A2780 cells treated with tRF-T11 mimic (Figure 4B) in a dose-dependent manner (Figure 4C). Remarkably, tRF-T11 mimic was observed to be as effective as TRPA1-siRNA1 in suppressing TRPA1 expression (Figure 4D), suggesting that tRF-T11 may act as a translational suppressor of the TRPA1 gene by acting like an siRNA that silences a targeted gene. The minimum free energy of tRF-T11 binding TRPA1 was calculated to be −33.3 kcal/mol (Figure 4E). Dual-luciferase reporter assay was employed to determine whether tRF-T11 directly targets TRPA1. The TRPA1-wild type (WT) and TRPA1-mutant (MUT) luciferase reporter gene vectors and tRF-T11 mimic or its scrambled mimic were co-transfected into HEK-293T cells. The results showed that tRF-T11 mimic significantly reduced the luciferase activity of the TRPA1-WT reporter vector, while no significant change of the luciferase activity of the TRPA1-MUT reporter gene vector was observed (Figure 4F). This clearly demonstrated that tRF-T11 directly targets the predicted binding sites of 3′ UTR of TRPA1 mRNA to inhibit its expressions. Meanwhile, in the Argonaute-RNA immunoprecipitation (AGO-RIP) assay, tRF-T11 mimic was significantly enriched by almost 113 times in the AGO2-RIP complex (Figure 4G), which indicated the interaction between tRF-T11 mimic and AGO2 protein. It was also observed that the transfected tRF-T11 mimic significantly reduced the expression levels of TRPA1 mRNA in the AGO2-RIP complex of A2780 cells (Figure 4H). These data suggest that tRF-T11 interacts with AGO2 to directly target TRPA1 and inhibit its expression via the RNAi pathway in A2780 cells. Furthermore, by querying The Cancer Genome Atlas (TCGA) data, it was revealed that the survival probability of OVCA patients with low expression of TRPA1 is significantly higher than that of those with high expression of TRPA1 (Figure 4I), indicating that TRPA1 might be a new potential target for OVCA treatment, and tRF-T11 mimic might be effective to this treatment via targeting TRPA1.
Figure 4.
tRF-T11 targets TRPA1 and inhibits its expressions on both mRNA and protein levels
(A) Hybridization energies and differential expression analysis of predicted target genes identified by the IntaRNA and psRNATarget tools. (B) Suppression of TRPA1 expression (mRNA levels measured by qPCR) in A2780 cells treated with tRF-T11 mimic. Tetraspanin 17 (TSPAN17) and secretogranin V (SCG5) were not affected. (C) tRF-T11 mimic suppressed TRPA1 expression in a dose-dependent manner. (D) Comparison of the effects of tRF-T11 mimic and TRPA1-siRNA on TRPA1 expression in A2780 cells at 25 nM. Representative western blot bands are presented in the left panel. (E) Construction of the luciferase report plasmid carrying the luciferase-coding sequence attached with the WT or MUT 3′ UTR TRPA1 based on the putative binding sites of tRF-T11. (F) The relative luciferase activities were detected by transfecting pmiR-RB-Report h-TRPA1-WT or pmiR-RB-Report h-TRPA1-MUT and tRF-T11 mimic or its scramble mimic into HEK-293T cells. (G) tRF-T11 mimic was significantly enriched in AGO2-RIP complex of A2780 cells. (H) Suppression of TRPA1 expression (mRNA levels measured by qPCR) in AGO2-RIP complex of A2780 cells treated with tRF-T11 mimic. (I) Low expression of TRPA1 is associated with survival probability of OVCA patients. Data are shown as the means ± SDs of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Preparation of tRF-T11 mimic-containing nanoparticles
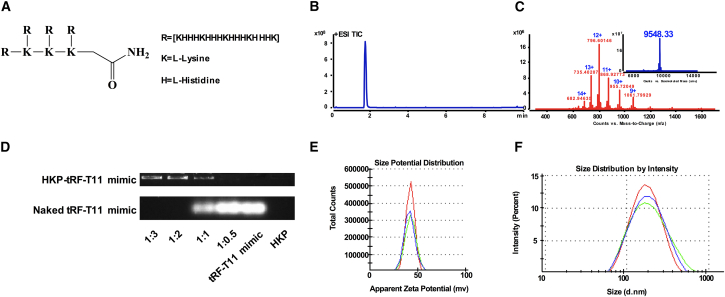
To encapsulate tRF-T11 mimic for in vivo study, a histidine-lysine polymer (HKP; Figure 5A) was employed as delivery agent since it is a nanoparticle system for siRNA drug delivery in vivo.31 The total ion chromatogram displayed a single peak of HKP (Figure 5B) and the deconvolution mass spectrum indicated its accurate molecular weight of 9,548.33 Da (Figure 5C). We used agarose gel electrophoresis analysis to investigate nanoparticle pharmaceutics formed under different mass ratios of tRF-T11 mimic and HKP. In Figure 5D, the upper panel indicates the encapsulated tRF-T11 mimic, while the lower panel shows naked RNA. The results show that when HKP was used at a mass ratio of 2 or above relative to the mimic, all tRF-T11 mimics were encapsulated. Furthermore, to evaluate the quality of nanoparticle pharmaceutics, we determined their particle size and zeta potential value in RNase-free water. The results indicated good dispersion (41.2 mV; Figure 5E) and appropriate particle size (164.2 nm; Figure 5F) and were obtained when the mass ratio was at 3. Thus, tRF-T11 mimic-containing nanoparticles were prepared at this mass ratio for in vivo experimentation.
Figure 5.
Optimization and preparation of tRF-T11 mimic-containing nanoparticles
(A) Chemical structure of HKP. (B) Total ion chromatogram showing a single peak of HKP. (C) Multiple charge distribution and deconvolution mass spectrum (inset) of HKP. (D) Agarose gel electrophoresis analysis of encapsulated tRF-T11 mimic by HKP under different mass ratios. When the mass ratio is above 1:2 (RNA:HKP), all RNAs were encapsulated. (E) The zeta potential and particle sizes (F) of the tRF-T11 nanoparticle in RNase-free water at mass ratio of 1:3. Data are shown as the means ± SDs of three independent experiments.
tRF-T11 mimic suppresses ovarian tumor growth in vivo
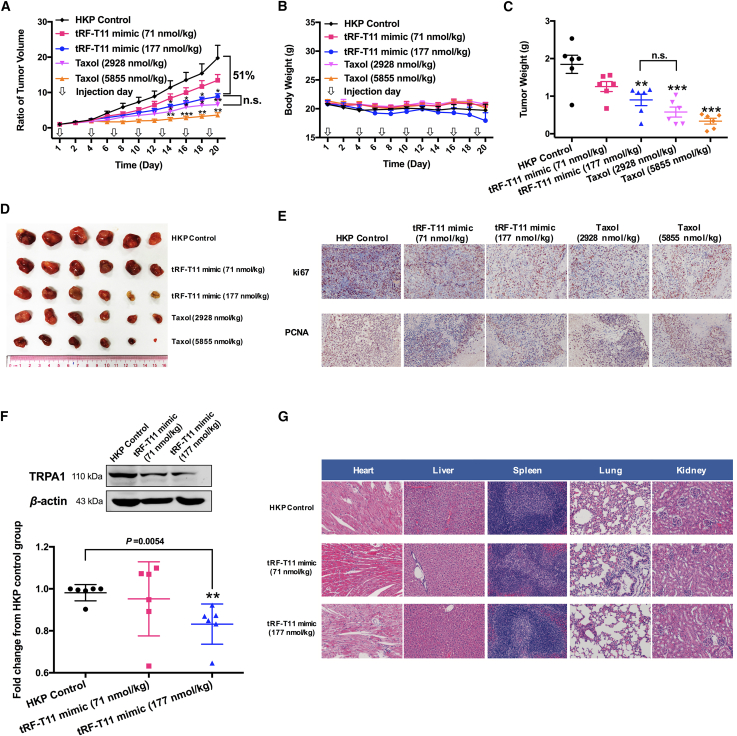
An A2780 cell xenograft tumor model was developed to determine whether tRF-T11 mimic could suppress OVCA growth in vivo. Female BALB/c nude mice were intratumorally injected once every 3 days with encapsulated tRF-T11 mimic (71 and 177 nmol/kg), taxol (2,928 and 5,855 nmol/kg), or blank HKP when the tumors reached 50 mm3. Treatment groups demonstrated a reduced tumor growth rate when compared with the untreated (HKP) control group (Figure 6A), while the body weight remained largely comparable (Figure 6B). The high-dose tRF-T11 mimic-treated group (177 nmol/kg) and the low-dose taxol-treated group produced results that were not statistically different, reaching an approximately 50% reduction in tumor size, in terms of both volume and weight after 20 days (Figures 6C and 6D). Surprisingly, the dose of tRF-T11 mimic (177 nmol/kg) is 16-fold lower than that of taxol (2,928 nmol/kg) to achieve comparable tumor-suppressive effects in vivo (Figures 6A and 6C). Also, tRF-T11 mimic significantly reduced the tumor size (Figure 6D) and showed no significant adverse impacts on the body weight of nude mice at the end of the experiment (Figure 6B). Furthermore, the expression levels of ki67 and PCNA in tumors of mice treated with tRF-T11 mimic or taxol significantly reduced compared with those in control group, suggesting that tRF-T11 mimic exhibited inhibitory effects on OVCA growth (Figure 6E). Western blot assay was further used to determine TRPA1 expressions in tRF-T11 mimic-treated group, and the results showed that mice treated with the high-dose tRF-T11 mimic had significantly reduced protein expression levels of TRPA1 in the tumors (Figure 6F) compared with those of control group, which is consistent with the results in vitro. In addition, morphological images and pathologic analysis showed that no significant histological differences were observed in the major organs between the tRF-T11 mimic-treated group and control group (Figures 6G and S5). This suggests that tRF-T11 mimic treatment is potentially safe. Thus, we concluded that tRF-T11 could suppress human OVCA growth via targeting and inhibiting the expressions of TRPA1 in vitro and in vivo.
Figure 6.
tRF-T11 mimic decreases OVCA tumor growth and suppresses TRPA1 expression in vivo
(A) Growth rate of tumors treated with encapsulated tRF-T11 mimic, taxol, or HKP alone as control. Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. (B) Body weights did not change significantly among all groups. (C) Weight of tumor removed from the mice after day 20. (D) Each tumor in all groups (n = 6 per group). (E) Representative images of ki67 and PCNA immunohistochemistry of tumor samples at 200× magnification. (F) Expression of TRPA1 in tumor tissues (n = 6 per group). Representative western blot images are presented in the top panel. Data are shown as the means ± SDs of three independent experiments. (G) Representative images of hematoxylin and eosin-stained major organs in A2780 xenografts nude mice from tRF-T11 mimic-treated group and control group. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. n.s., non-significant.
Low expression of TRPA1 is associated with higher survival probability in multiple types of cancer patients
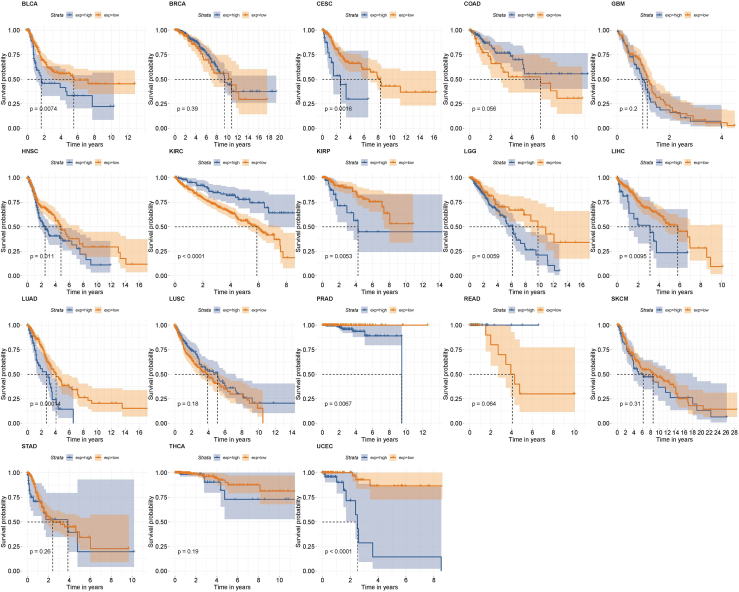
In addition to the potential role of TRPA1 in OVCA treatment as demonstrated in this study, there is also evidence that it is involved in breast cancer.32 Analysis of the TCGA database revealed that low expression of TRPA1 is associated with increased survival probability among patients with BLCA, CESC, HNSC, KIRP, LGG, LIHC, LUAD, PRAD, and UCEC, but not with BRCA, COAD, GBM, KIRC, LUSC, READ, SKCM, STAD, and THCA (Figure 7). The possible linkages of TRPA1 and other cancers should be further investigated in our future work.
Figure 7.
Survival analysis of TRPA1 in 18 kinds of cancer using TCGA data
For the first time, the authors revealed that a tRF mimic (antisense derived from Chinese yew) interacts with AGO2 to directly target TRPA1 via the RNAi pathway, resulting in suppressing OVCA growth. This uncovered a new role of plant-derived tRFs and broadened our knowledge of therapeutics from plants.
Discussion
Plant tRFs are closely associated with the developmental and defense mechanisms in plants.33 The miR164 and miR171 obtained from Chinese yew have been demonstrated to target and cleave biosynthetic genes of taxol, taxane 13α hydroxylase, and taxane 2α-O-benzoyl transferase.34 In this study, a bioinformatics approach was used to demonstrate the potential of tRFs derived from Chinese yew to target some genes for the biosynthesis of taxol with low MFE values (Table S4). It is suggested that these tRFs might play a role in regulating the biosynthetic process of taxol in Taxus plants.
In addition, this study emphasized the potential regulatory roles played by plant-derived tRNAs and tRFs in human cells. At the beginning of this study, tRF-T11 mimic was demonstrated to exhibit comparable inhibitory effects on cancer cells to taxol, both in vitro and in vivo. However, tRF-T11 mimic is clearly advantageous as it requires as little as 6% of the taxol dose (in molar terms and administered by intratumorally injection) for producing tumor-suppressing effects in a xenograft mouse model using A2780 OVCA cells. In further in-depth bioinformatics analysis, it was revealed that tRF-T11 could target the 3′ UTR of TRPA1 mRNA to inhibit its expression. Besides, dual-luciferase reporter assay experiments were conducted to confirm its binding sites on TRPA1 mRNA. Finally, AGO-RIP assay was performed to demonstrate that tRF-T11 interacts with AGO2 to inhibit TRPA1 via the RNAi pathway in A2780 cells. In the area of cross-kingdom regulation of exogenous small RNA, Prof. Chen-Yu Zhang's laboratory was the first to reveal that plant-derived miRNAs could function as cross-kingdom regulators to influence the expression level of homo-derived protein coding genes.6 Although we, and many other researchers, are focusing on plant-derived small RNA, our strategies are different: First, the identified tRF-T11 mimic is an artificially designed double-stranded oligonucleotide whose antisense is derived from the tRNA of Chinese yew. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis showed that tRF-T11 is not present in four kinds of cancer cells (Figure S6). This is different from the single-stranded plant miRNA, which is detectable in the human body.6, 7, 8, 9 Second, the administration mode of tRF-T11 mimic is intratumorally injection, which is also different from oral administration of plant miRNA for investigations on cross-kingdom regulation.
TRPA1 is referred to as a non-selective Ca2+ channel widely expressed in both neurons such as those in the sensory dorsal root and trigeminal ganglia, and such non-neuronal cells as epithelial and hair cells. It is known to play a certain role in sensory and homeostatic processes. In addition, it may play a critical role in inflammation and neuropathic pain.35 More recently, TRPA1 has been demonstrated as critical to the survival of inner cells in the breast and lung cancer spheroids with reactive oxygen species (ROS) accumulation. In addition, TRPA1 increased the resistance to ROS-producing chemotherapies, and the silencing of TRPA1 was effective in suppressing tumor progression in a xenograft animal model. Therefore, it is suspected that TRPA1 expression can contribute to preventing the occurrence of oxidative stress-induced cell death.32 In the present study, we carried out bioinformatics, dual-luciferase reporter assay, and AGO-RIP experiments to demonstrate that the molecular function of tRF-T11 is to interact with AGO2 to directly target TRPA1 via the RNAi pathway to suppress OVCA growth. Thus, Takahashi's findings could demonstrate the downstream mechanisms of tRF-T11 targeting TRPA1. More importantly, corroboration was obtained from our observation that the higher levels of TRPA1 expression in tumor tissues were associated with the lower chance of patient survival in a cohort consisting of 379 OVCA tissues and 88 normal ovarian tissues derived from UCSC Xena (Figure 4I). Thus, it is speculated that tRF-T11 reduced tumor growth mainly by suppressing the expression of TRPA1. Although taxol exhibits tumor-suppressing effects by stabilizing microtubules,36 there is evidence suggesting that the action of taxol is partially mediated by the activation of TRPA1 and TRPV4. Consequently, ROS accumulate to induce chemoresistance.37 Combined with the observation that tRF-T11 mimic is equally potent in drug-resistant A2780T cells and in non-resistant strains, it suggests a potential strategy of treatment through a combination of tRF-T11 mimic and taxol to improve the outcome of suppressing such tumors as OVCA. In addition, it is worth noting that the current study is far from sufficient to fully understand the potential of chemical modifications of Taxus tRFs. Since our previous studies have revealed that chemical modifications of tRNA are associated with its bioactivity,38,39 further studies will be conducted in the future to explore the structure-activity relationship and provide valuable information about the development of RNA mimics.
Since the first report on the capability of plant-derived miRNAs to regulate gene expression in mammalian systems,6 the findings obtained from many other studies have been strongly suggestive of a distinct potential for the small RNAs in plants to be developed into pharmacological agents.7, 8, 9, 10, 11 This study has extended this concept to include smaller tRNA and even smaller tRF, both of which are abundant in the small RNA of the plant kingdom. As the oligonucleotides in the structures of tRFs could be modified easily and efficiently through the phosphoramidite methodology,40,41 the possibility of a vast library of sequences, natural or modified, will promote the development of novel RNAi-based therapeutics with a potential that cannot be over-estimated. In October 2018, patisiran was granted approval from the US Food and Drug Administration (FDA) for the treatment of polyneuropathy caused by hereditary transthyretin-mediated amyloidosis (hATTR) in adult patients. It is not only the first drug officially approved for use in this condition but also the first siRNA drug of its kind approved by the FDA.42 As siRNA technology develops rapidly, siRNA can be readily designed and made available on the basis of a well-validated mRNA target,43 thus providing a new class of therapeutics against the targets previously unknown to be druggable.44 Thus, it is essential to identify a molecular target related to either disease etiology or progression for siRNA drug development. However, the identification of a critical molecular target in the pathogenesis and pathology of a disease involves a lengthy and difficult process.45, 46, 47 Therefore, a reversed strategy was adopted in this study, which first sought the pharmacologically active tRFs that are specifically produced from the 5′ and-3′ ends of plant tRNAs.48 Then, bioinformatics studies were conducted to predict and validate their mRNA targets. This approach proved successful in identifying tRF-T11 and its target TRPA1. It is suspected that a further optimization based on the target region on the TRPA1 mRNA can contribute to identifying more effective siRNA drug candidates.
In summary, we have developed an innovative strategy designed to draw the first conclusion that tRNAs are pharmacologically active as the most abundant class of small RNAs derived from medicinal plants. Also, the first evidence was provided to suggest that plant-derived tRFs are potent druggable RNA molecules, which significantly widens our knowledge about the active ingredients of medicinal plants and provides additional clues to exploring the active components contained in medicinal plants. Since our study revealed a first RNA sequence with antitumor activity derived from the Taxus plant, tRF-T11 represents the first natural RNA silencing human TRPA1. Overall, the novel strategy proposed in this paper can not only identify the pharmacologically active tRFs obtained from medicinal plants but also contribute to enhancing the efficiency and chance of identifying new drug targets, which holds massive potential for developing novel RNA drugs from nature and sheds light on how to find unknown molecular targets for therapeutics.
Materials and methods
Chemicals and reagents
Methanol was of mass spectrometry (MS) grade from Merck (Darmstadt, Germany). Formic acid (MS grade), hexafluoro-2-propanol (HFIP; ≥99.0%), trimethylamine (TEA; ≥99.5%), and guanidinium thiocyanate were purchased from Sigma (USA). MicroRNA marker and low-range ssRNA ladder were purchased from New England Biolabs (USA). Diethylpyrocarbonate (DEPC)-treated water, polyacrylamide containing a ratio of acrylamide/bis (19:1, w/w) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Thermo (USA). RPMI 1640 medium, MEM medium, DMEM, fetal bovine serum (FBS), penicillin, and streptomycin were purchased from Gibco (New Zealand). Taxol (purity ≥ 98%) was purchased from AdooQ bioscience (Nanjing, China). Deionized water was prepared by a Millipore Milli-Q Plus system (Millipore, USA). All reagents used were of analytical grade.
Plant specimens and animals
Plants of T. chinensis (Pilger) Rehd. var. mairei (Lemee et Levl.) Cheng et L.K. Fu used in this study were collected from Sanming City, Fujian Province, China. Branches of T. chinensis were freshly cut, frozen, and stored in liquid nitrogen until use. Animal care and use protocols were approved by the Ethics Committee of the College of Pharmaceutical Sciences (2018-052), Southwest University. The 6–8-week-old BALB/c nude mice were purchased from Shanghai SLAC Laboratory Animal Co., China) and fed under specific pathogen-free conditions at Southwest University.
Cell culture and antibodies
IOSE80 human ovarian surface epithelial cell line (Guandao Biological Engineering, China), A2780 human ovarian carcinoma cell line (KeyGen Biotech, China), and its taxol-resistant strain were cultured in RPMI 1640 medium. HepG2 human hepatocellular carcinoma cell line (ATCC) was cultured in MEM medium. SK-OV-3 human ovarian adenocarcinoma cell line (ATCC, USA), HFTEC fallopian tube cell line, and MCF-7 human breast cancer cell line (ATCC) were cultured in DMEM. All the cell lines were cultured in medium that contained 10% FBS, 1% penicillin/streptomycin (P/S) in a humidified 5% CO2 atmosphere at 37°C. For western blot assay, primary antibodies were purchased from the indicated sources, anti-TRPA1 antibody (1:500 dilution, Novus, USA); anti-β-actin antibody (1:1,000 dilution, Santa Cruz, USA). A fluorescent secondary antibody against rabbit was purchased from Li-cor (USA). For immunoprecipitation assay, anti-AGO2 antibody was purchased from Active Motif (USA), and anti-rat IgG antibody was purchased from Santa Cruz. For immunohistochemistry assay, primary antibodies were purchased from the indicated sources, anti-ki67 antibody (1:1,000 dilution, Abcam, USA) or anti-PCNA antibody (1:1,000 dilution, Novus, USA). A GTVison secondary antibody (GK600710A, HRP-conjugated anti-rabbit/mice antibody) was purchased from Gene Tech (Shanghai, China).
RNA extraction of Chinese yew
Plant tissues were ground into a fine powder in liquid nitrogen and were then homogenized (IKA, Germany) in hexadecyl trimethyl ammonium bromide solution. After incubation for 2 min at 65°C, the tissue lysate was cooled immediately on ice for 10 min, followed by centrifugation at 12,000 ×g for 15 min at 4°C. The supernatant was first extracted with an equal volume of an organic solvent containing phenol:chloroform:isopentanol (50:48:1). After centrifugation as above, a second extraction of the supernatant was performed with an equal volume of a mixture containing chloroform:isopentanol (24:1). Third, the supernatant was mixed with an equal volume of 6 M guanidinium thiocyanate solution (Tokyo Chemical Industry, Japan) and ethanol (Anaqua Chemicals Supply, USA) to a final concentration of 55%. The mixture was passed through a silicon-membrane column and washed with 80% ethanol. Finally, total RNA was eluted with RNase-free water, and small RNA species were enriched by using a mirVana miRNA isolation kit (Invitrogen, USA) following the manufacturer's instructions.
Preparation of tEF
Small RNA of Chinese yew was separated by 6% urea-PAGE. After SYBR Gold nucleic acid gel staining (Thermo, USA), polyacrylamide gels were examined using a UV lamp, and the region of gel fractions containing the tEF were excised by using a clean and sharp scalpel. The gel fractions were sliced into small pieces and tEF was recovered from the gel by electroelution in 3 kDa molecular weight cut-off dialysis tubing (Spectrum, USA) at 100 V for 1.5 h in 1× TAE buffer. The eluents in the dialysis tubing were recovered and tEF was finally desalted and concentrated by using the mirVana miRNA isolation kit.
tEF library construction and NGS
Sequencing libraries were generated by using a TruSeq small RNA Library Preparation Kit (Illumina, USA), followed by a round of adaptor ligation, reverse transcription and PCR. PCR products were then purified and quantified on an Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). Sequencing of the library preparations were performed at the Novogene Bioinformatics Institute (Beijing, China) on an Illumina HiSeq platform using the 150-bp paired-end (PE150) strategy to generate over 15 million raw paired reads. A total of 1,729,438 clean reads were obtained by removing low-quality regions and adaptor sequences. The tRNA genes were identified using the tRNAscan-SE 2.0 program (http://lowelab.ucsc.edu/tRNAscan-SE/) and annotated by searching the Nucleotide Collection (nr/nt) database using the BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Purification of individual tRNAs
Synthetic biotin-labeled single-strand DNA probes (BGI, China; Table S5) complementary to the target tRNA were mixed with small RNA of Chinese yew and denatured at 95°C for 5 min, followed by incubation at a temperature 5°C lower than the melting temperature (Tm) for 90 min49 Streptavidin magnetic beads (Beaverbio, China) were then added to the mixture and incubated for 30 min at the annealing temperatures. Then, the biotinylated DNA/tRNA-coated beads were isolated with a magnet and washed at 40°C. Subsequently, the magnetic beads were resuspended in RNase-free water, and the immobilized tRNA was released by incubation at 65°C for 5 min. The solutions were subjected to ultra-high-performance liquid ion-pair chromatography for further separation. The individual tRNA peak was collected and freeze dried to remove organic solvents and essential salts.
UHPLC-MS analysis
Briefly, a UHPLC Agilent 1290 system (Agilent, USA) equipped with an ACQUITY UPLC OST C18 column (2.1 × 100 mm internal diameter, 1.7 μm; Waters, USA) and a diode array detector. The column was maintained at 60°C and a gradient elution was performed with (A) 100 mM HFIP + 15 mM TEAA and (B) 50% MeOH in (A) as follows: 0–1.5 min, 2% (B); 1.5–8.3 min, 2%–28% (B); and 8.3–16.5 min, 28%–34% (B). The flow rate was set at 0.2 mL/min. MS data were acquired on an Agilent 6545 Q-TOF mass spectrometer with a gas temperature of 320°C, spray voltage of 3.5 kV, and sheath gas flow and temperature of 12 L/min and 350°C, respectively. Samples were analyzed in negative polarity over an m/z range of 500 to 3,200. Fractions from chromatographic peaks of individual tRNAs were collected and freeze dried.
RNA transfection and cytotoxicity assay
RNA samples dissolved in RNase-free water were stored at −20°C before use. Transfection of RNA samples was performed according to the manufacturer's instructions for Lipofectamine RNAiMAX Transfection Reagents (Thermo, USA). Briefly, cells (5 × 103 in 100 μL of culture medium) were seeded onto 96-well plates. After 20 h, cells were treated with varying concentrations of RNA sample solutions with Lipofectamine RNAiMAX Transfection Reagent in Opti-MEM medium (Thermo) according to the manufacturer's instructions. Cells without any treatment were used as control, and cells treated with liposomes were used as treatment control. Cell viability was determined after 48 h. MTT solutions (50 μL per well, 1 mg/mL, Thermo) were added to each well and incubated for 4 h at 37°C, followed by adding DMSO (200 μL). The absorbance values were colorimetrically determined at 570 nm using a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, USA). IC50 values were calculated by GraphPad Prism 5.0 (GraphPad, USA). An siRNA (forward, 5′-GGAGCGAUUUAGCCAAGAATT-3′: reverse, 5′-UUCUUGGCUAAAUCGCUCCTT-3′) was used as positive control. Negative sequence (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′; reverse, 5′-ACGUGACACGUUCGGAGAATT-3′) was used as negative control. Each experiment was carried out three times. The results are expressed as the means ± SD.
Clonogenic assay
The clonogenic assay was performed according to Franken et al. with minor modification.50 Briefly, cells were plated at a density of 500 cells/well with culture medium in 24-well plates. After 20 h, the medium was changed to medium containing RNA samples (5 nM for A2780 and 12 nM for SK-OV-3 cells) with liposomal transfection or taxol at 5 nM. Blank Opti-MEM was used as the control group. After 48 h of treatment, all media were changed to normal culture medium and the cells were cultured for another 14 days. The culture media were changed every 4 days. After fixation for 20 min with a 4% paraformaldehyde fixation solution (Beyotime, China), the cells were stained with crystal violet (Beyotime, China) for 10 min and photographed.
Wound healing assay
Cells (5 × 105 in 500 μL of culture medium) were grown in 24-well plates and for 20 h until confluent. A scratch was made by using a sterile 200-μL pipette tip and the medium was changed to medium containing RNA samples (5 nM for A2780 and 12 nM for SK-OV-3 cells) with liposomal transfection or taxol at 5 nM. The cells were viewed with a 10× objective and photographed using a phase contrast microscope (Leica microsystems, Germany) at 0 and 24 h. ImageJ software was used to quantify the area of wound created. Wound healing rate was calculated using the formula:
| Wound healing rate = [(Wound area at 0 h – Wound area at 24 h)/Wound area at 0 h] × 100. |
Transwell assay
Cell migration and invasion assays were carried out using a 24-well transwell chamber (8 μm pore size, Corning Costar, USA). For invasion assay, the membranes were first coated with Matrigel (Corning Costar) and dried at 37°C for 3 h. Cells were collected after transfection by tRF-T11 mimic for 24 h (5 nM for A2780 and 12 nM for SK-OV-3 cells). A2780 (1.5 × 105 cells) and SK-OV-3 (3 × 105 cells) resuspended in 200 μL of serum-free 1640 or DMEM were seeded in the upper chamber, while 600 μL of medium containing 20% FBS were added into the lower chamber. After incubation for 24 h in a humidified 5% CO2 atmosphere at 37°C, cells were fixed by 4% formaldehyde solution and methanol, each for 10 min, followed by staining with crystal violet for 15 min. The un-invaded cells were then wiped out using a cotton-tipped swab, and the invaded cells in the lower chamber were photographed using an Olympus IX51 (Olympus Optical, USA) and counted in five individual fields. The migration assays were exactly same as the invasion assay except that no Matrigel was used.
Caspase-3 activity determination
Caspase-3 activity was determined using a Caspase-3 colorimetic assay kit (Sigma, USA) according to the manufacturer's instructions. Briefly, A2780 and SK-OV-3 cells (2 × 106) were seeded onto the dish. After 20 h, cells were treated with tRF-T11 mimic (31 nM for A2780 and 160 nM for SK-OV-3 cells) or taxol (11 nM for A2780 and 34 nM for SK-OV-3 cells). After 18 h, cells were harvested by centrifugation at 600 × g for 5 min. Cell lysates were mixed with colorimetric peptide substrates Ac-DEVD-pNA for incubation overnight. The absorbance values were colorimetrically determined at 405 nm using a SpectraMax 190 microplate reader.
Acquisition of fluorescence images of transfected FAM-labeled tRF-T11 mimic in A2780 cells
A2780 cells (1 × 105) were seeded onto glass cell culture dishes (Nest Biotechnology, China). After 20 h, the medium was replaced by a transfection medium containing 50 nM FAM-labeled tRF-T11 mimic (GenePharma, China) for 6 h. On completion of the transfection, cells were rinsed three times with RPMI 1640 medium. Cells were incubated with 1 mL of Hoechst 33258 (Sigma, USA) for 10 min at 37°C for nuclear staining, or with 1 mL of MitoBeacon Red (GeneCopoeia, USA) for 30 min at 37°C for mitochondrial staining, followed by washing with RPMI 1640 medium. Fluorescence images were acquired with an API DeltaVision Live-cell Imaging System (Applied Precision, GE Healthcare, USA) and were captured at 60× magnification.
tRF-T11 target prediction and prioritization
The identification of functional target genes of tRF-T11 followed previously described small RNA or tRF target prediction methods and restriction criteria.51 Specifically, the 3′ UTR sequences of all mRNAs retrieved from GENCODE (v23) and the sequence of tRF-T11 derived from Taxus were first loaded into IntaRNAv2 and psRNATarget to predict the potential targets. The top 10 overlapping targets predicted were then prioritized and filtered by crosschecking their expression in tumor and normal ovarian tissues. Accordingly, the gene expression profiles were obtained for 379 OVCA tissues and 88 normal ovarian tissues from UCSC Xena, which contains curated comprehensive batch-effect-adjusted pancancer genomic data.52 Subsequently, EBSeq was employed to filter the target genes differentially expressed between tumor and normal ovarian tissues.53 The predicted targets satisfying two criteria, fold change (tumor/normal) >2 and false discovery rate (FDR) <0.1.
Quantitative real-time PCR
Total RNA was extracted from A2780 cells treatment with the tRF-T11 mimic for 48 h using TRIzol (Invitrogen, USA) on a silicon-membrane column. RNA was then reverse transcribed using the GoScript Reverse Transcription System (Promega, USA). Quantitative real-time PCR was performed on a high-productivity real-time quantitative PCR ViiA 7 system (Life Technologies, USA) using GoTaq qPCR Master Mix (Promega, USA) according to the manufacturers' instructions. The PCR primers were obtained commercially (BGI, China; Table S6). All data are presented as the mean fold change values of duplicates and normalized to β-actin expression using the 2−ΔΔCT method. Each experiment was carried out in triplicate and data are expressed as the means ± SD.
Western blot analysis
After treatment with tRF-T11 mimic or other RNA samples for 48 h, A2780 cells were lysed and protein extraction was carried out using RIPA buffer (Cell Signaling Technology, USA) supplemented with protease and phosphatase inhibitor cocktails (Roche, Switzerland). Protein concentrations were then determined by a BCA protein assay (Thermo, USA). Denatured proteins were separated by 8% SDS-PAGE and transferred to nitrocellulose membranes, followed by blocking with 5% BSA (Thermo, USA) for 2 h. Membranes containing proteins were incubated with primary antibody against TRPA1 or β-actin overnight, followed by washing in TBST and incubation with a fluorescent secondary antibody for detection. Band intensities were measured using ImageJ and were normalized to those of β-actin. The sequences of TRPA1_siRNA01, TRPA1_siRNA02 and the negative sequence (GenePharma) are shown in Table S7. Each experiment was carried out in triplicate and data are expressed as the means ± SD.
Dual-luciferase reporter assay
The 3′ UTR of the TRPA1 sequence containing the WT or MUT tRF-T11 binding sites was inserted into the Xhol and Notl restriction sites of pmiR-RB-REPORT vector (Ribobio, China). The luciferase reporter vectors were co-transfected with tRF-T11 mimic or non-target control into HEK293T cells using Lipofectamine RNAiMAX Transfection Reagents (Thermo). After transfection for 48 h, the Firefly and Renilla luciferase activities were detected with the Dual-Glo Luciferase Assay System (Promega) using the GLOMAX 96 spectrophotometer (Promega) according to the manufacturer's instructions. Scramble control (forward, 5′-CGCGACGUCUGAGGUAGACGAC-3′; reverse, 5′-GUCGUCUACCUCAGACGUCGCG-3′) was used as negative control. Each experiment was carried out triplicate and data are expressed as the means ± SD.
AGO-RIP assay
Briefly, tRF-T11 mimics were transfected into A2780 cells (2 × 107) at the concentration of 20 nM. After 48 h, the cell pellets were collected and lysed in RIPA buffer according to the manufacturers' instructions. After centrifugation at 20,000 × g for 15 min at 4°C, the supernatant was collected and incubated with Dynabeads M-280 Sheep Anti-Mouse immunoglobulin (Ig) G (Thermo) coated with rat anti-AGO2 antibody (Active Motif, USA) for 3 h at 4°C, while rabbit anti-rat IgG antibody was used as a negative control (Santa Cruz). The beads were washed by NET buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM NaF, and 0.05% NP-40) and were enriched with a magnetic stand. Total RNA was then extracted by TRIzol on a silicon-membrane column. TRPA1 mRNA and tRF-T11 mimic in the purified total RNA were detected by quantitative real-time PCR. Normalization of RIP results was carried out by quantifying in parallel the relative levels of U6 snRNA in the immunoprecipitation sample.
Preparation of HKP nanoparticles of tRF-T11 mimic
HKP (a gift from Dr. Yang Lu, Sirnaomics, USA) dissolved in RNase-free water (240 μg/mL) was first prepared overnight at 4°C. Equal volumes of tRF-T11 mimic solutions at a concentration of 80 μg/mL were then mixed and incubated at room temperature for 30 min. The particle sizes and zeta potential of the resulting nanoparticles were then evaluated by a Malvern Zetasizer Nano ZS90 (Malvern Instruments, UK).
In vivo models for tumor therapy
A2780 cells (4.0 × 106) were injected subcutaneously under the armpits of 6–8-week-old BALB/c nude mice. When the tumors reached 50 mm3, tRF-T11 mimic (GeneDesign, Osaka, Japan) encapsulated in HKP nanoparticles was administered by intratumoral injection (71 nmol/kg, 177 nmol/kg) once every 3 days. Taxol (2,928 nmol/kg or 5,855 nmol/kg) and HKP were used as positive and negative controls, respectively. The animals were sacrificed on day 20. Tumor diameters were measured at the points of maximum length and maximum width with digital calipers. Tumor volumes were calculated by the following formula: volume = (width)2 × length/2. Data were statistically analyzed using GraphPad Prism 5.0.
Histological analysis
The tissues were fixed in 4% PFA and embedded in paraffin. For immunohistochemistry, slides were deparaffinized in xylene and gradually rehydrated in a graded concentration of ethanol (100%, 95%, and 70%). Subsequently, antigen retrieval was carried out by incubating the slides with 10 mM sodium citrate (pH = 6.0) for 30 min at 95°C, followed by incubating with 3% H2O2 for 15 min and washing in PBS three times (3 min for each). The slides were then blocked by 5% BSA and then incubated with anti-ki67 antibody or anti-PCNA antibody at 4°C overnight, followed by washing in PBS and incubating with GTVison secondary antibody. Finally, antibody complexes were detected by 3,3′-diaminobenzidine (DAB) and counterstained with Mayer's hematoxylin. Paraffin sections of 3–5 μm were cut and were stained with hematoxylin and eosin to determine the side effects of tRF-T11 mimic treatment.
Statistical analysis
Data are presented as means ± SD. Statistical analyses were performed using GraphPad Prism using multiple two-tailed Student's t tests or two-way ANOVA followed by post hoc analysis. For analysis of tumor growth rate in animal studies, data are presented as means ± SEM. Throughout all figures, significance was tested at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We are grateful to Dr. Y. Lu (Sirnaomics, USA) for technical advice and the gift of HKP for RNA delivery in vivo. Funding: This work was financially funded by The Science and Technology Development Fund, Macau SAR (File no. 0023/2019/AKP, 015/2017/AFJ), the CUHK Direct Grant 4053364, and the funding from Innovation and Technology Commission of Hong Kong Government to the State Key Laboratory of Agrobiotechnology.
Author contributions
Z.-H.J., K.-Y.C., and T.-M.Y. conceived the study. K.-Y.C., T.-M.Y., and T.-F.C. designed the experiments. K.-Y.C., T.-M.Y., J.-Z.Z., J.G., and J.L. conducted the experiments. K.-Y.C., J.-Z.Z., and C.L. analyzed the data. K.-Y.C., T.-M.Y., E.L.L., B.-X.Z., and Z.-H.J. drafted the manuscript. All authors have reviewed and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.037.
Supplemental information
References
- 1.Yuan H., Ma Q., Ye L., Piao G. The traditional medicine and modern medicine from natural products. Molecules. 2016;21:559. doi: 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li F.S., Weng J.K. Demystifying traditional herbal medicine with modern approach. Nat. Plants. 2017;3:17109. doi: 10.1038/nplants.2017.109. [DOI] [PubMed] [Google Scholar]
- 3.Yarnell E. Synergy in herbal medicines: part 1. J. Restor. Med. 2015;4:60–73. [Google Scholar]
- 4.Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Standart N., Jackson R.J. MicroRNAs repress translation of m(7) Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L., Hou D.X., Chen X., Li D.H., Zhu L.Y., Zhang Y.J., Li J., Bian Z., Liang X.Y., Cai X., et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z., Li X.H., Liu J.X., Dong L., Chen Q., Liu J.L., Kong H.H., Zhang Q.Y., Qi X., Hou D.X., et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L.K., Zhou Z., Jiang X.M., Zheng Y., Chen X., Fu Z., Xiao G., Zhang C.Y., Zhang L.K., Yi Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6:54. doi: 10.1038/s41421-020-00197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin A.R., Fong M.Y., Somlo G., Wu J., Swiderski P., Wu X.W., Wang S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J.C., Liang Z., Xu J.T., Zhao Y., Li X.Y., Zhang Y.L., Zhao D.D., Chen R.X., Liu Y., Joshi T., et al. Plant-derived phosphocholine facilitates cellular uptake of anti-pulmonary fibrotic HJT-sRNA-m7. Sci. China Life Sci. 2019;62:309–320. doi: 10.1007/s11427-017-9026-7. [DOI] [PubMed] [Google Scholar]
- 11.Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J.Y., Deng Z.B., Luo C., et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe. 2018;24:637–652. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shekhawat M., Jahagirdar D., Yadav S., Sharma N.K. Induction of apoptosis in HeLa by corn small RNAs. Nutr. Cancer. 2019;71:348–358. doi: 10.1080/01635581.2018.1526307. [DOI] [PubMed] [Google Scholar]
- 13.Barciszewska M.Z., Perrigue P.M., Barciszewski J. tRNA--the golden standard in molecular biology. Mol. Biosyst. 2016;12:12–17. doi: 10.1039/c5mb00557d. [DOI] [PubMed] [Google Scholar]
- 14.Cao K.Y., Pan Y., Yan T.M., Jiang Z.H. Purification, characterization and cytotoxic activities of individual tRNAs from Escherichia coli. Int. J. Biol. Macromol. 2020;142:355–365. doi: 10.1016/j.ijbiomac.2019.09.106. [DOI] [PubMed] [Google Scholar]
- 15.Anderson P., Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeung M.L., Bennasser Y., Watashi K., Le S.Y., Houzet L., Jeang K.T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saikia M., Jobava R., Parisien M., Putnam A., Krokowski D., Gao X.H., Guan B.J., Yuan Y.Y., Jankowsky E., Feng Z.Y., et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodarzi H., Liu X.H., Nguyen H.C.B., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wani M.C., Taylor H.L., Wall M.E., Coggon P., McPhail A.T. Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 1971;93:2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- 20.Arbuck S.G., Christian M.C., Fisherman J.S., Cazenave L.A., Sarosy G., Suffness M., Adams J., Canetta R., Cole K.E., Friedman M.A. Clinical development of taxol. J. Natl. Cancer Inst. Monogr. 1993;15:11–24. [PubMed] [Google Scholar]
- 21.Sangrajrang S., Fellous A. Taxol resistance. Chemotherapy. 2000;46:327–334. doi: 10.1159/000007306. [DOI] [PubMed] [Google Scholar]
- 22.Martinez G., Choudury S.G., Slotkin R.K. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017;45:5142–5152. doi: 10.1093/nar/gkx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loss-Morais G., Waterhouse P.M., Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct. 2013;8:6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daly M.B., Dresher C.W., Yates M.S., Jeter J.M., Karlan B.Y., Alverts D.S., Lu K.H. Salpingectomy as a means to reduce ovarian cancer risk. Cancer Prev. Res. 2015;8:342–348. doi: 10.1158/1940-6207.CAPR-14-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Qare A.W., Abou-Donia M.B. Biomarkers of apoptosis: release of cytochrome c, activation of caspase-3, induction of 8-hydroxy-2'-deoxyguanosine, increased 3-nitrotyrosine, and alteration of p53 gene. J. Toxicol. Environ. Health B Crit. Rev. 2001;4:313–332. doi: 10.1080/109374001301419737. [DOI] [PubMed] [Google Scholar]
- 26.Keam S.P., Hutvagner G. tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel) 2015;5:1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1404. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng J., Ptashkin R.N., Chen Y., Cheng Z., Liu G., Phan T., Deng X., Zhou J., Lee I., Lee Y.S., et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 2015;23:1622–1629. doi: 10.1038/mt.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann M., Wright P.R., Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai X., Zhuang Z., Zhao P.X. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kesharwani P., Gajbhiye V., Jain N.K. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials. 2012;33:7138–7150. doi: 10.1016/j.biomaterials.2012.06.068. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi N., Chen H.Y., Harris I.S., Stover D.G., Selfors L.M., Bronson R.T., Deraedt T., Cichowski K., Welm A.L., Mori Y., et al. Cancer cells co-opt the neuronal redox-sensing channel TRPA1 to promote oxidative-stress tolerance. Cancer Cell. 2018;33:985–1003. doi: 10.1016/j.ccell.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren B., Wang X.T., Duan J.B., Ma J.X. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science. 2019;365:919–922. doi: 10.1126/science.aav8907. [DOI] [PubMed] [Google Scholar]
- 34.Hao D.C., Yang L., Xiao P.G., Liu M. Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiol. Plantarum. 2012;146:388–403. doi: 10.1111/j.1399-3054.2012.01668.x. [DOI] [PubMed] [Google Scholar]
- 35.Nilius B., Appendino G., Owsianik G. The transient receptor potential channel TRPA1: from gene to pathophysiology. Pflugers Arch-eur. J. Physiol. 2012;464:425–458. doi: 10.1007/s00424-012-1158-z. [DOI] [PubMed] [Google Scholar]
- 36.Yang C.H., Horwitz S.B. Taxol®: the first microtubule stabilizing agent. Int. J. Mol. Sci. 2017;18:1733. doi: 10.3390/ijms18081733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Materazzi S., Fusi C., Benemei S., Pedretti P., Patacchini R., Nilius B., Prenen J., Creminon C., Geppetti P., Nassini R. TRPA1 and TRPV4 mediate paclitaxel-induced peripheral neuropathy in mice via a glutathione-sensitive mechanism. Pflugers Arch-eur. J. Physiol. 2012;463:561–569. doi: 10.1007/s00424-011-1071-x. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y., Yan T.M., Wang J.R., Jiang Z.H. The nature of the modification at position 37 of tRNAPhe correlates with acquired taxol resistance. Nucleic Acids Res. 2021;49:38–52. doi: 10.1093/nar/gkaa1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan T.M., Pan Y., Yu M.L., Hu K., Cao K.Y., Jiang Z.H. Full-range profiling of tRNA modifications using LC–MS/MS at single-base resolution through a site-specific cleavage strategy. Anal. Chem. 2021;93:1423–1432. doi: 10.1021/acs.analchem.0c03307. [DOI] [PubMed] [Google Scholar]
- 40.Michelson A.M., Todd A.R. Nucleotides part XXXII. Synthesis of a dithymidine dinucleotide containing a 3′:5′-internucleotidic linkage. J. Chem. Soc. (Resumed) 1955:2632–2638. [Google Scholar]
- 41.Beaucage S.L., Iyer R.P. Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron. 1992;48:2223–2311. [Google Scholar]
- 42.Mauricio E.A., Gonzalez-Duarte A., Conceicao I., Brannagan T.H., Quan D., Mezei M., Schmidt H., Wixner J., Dinh Q., Berber E., et al. Autonomic manifestations of hereditary transthyretin-mediated amyloidosis: longterm safety and efficacy from the patisiran global open-label extension study. Muscle Nerve. 2019;60:S49–S50. [Google Scholar]
- 43.Anonymous Delivering the promise of RNA therapeutics. Nat. Med. 2019;25:1321. doi: 10.1038/s41591-019-0580-6. [DOI] [PubMed] [Google Scholar]
- 44.Wang T., Shigdar S., Shamaileh H., Gantier M.P., Yin W., Xiang D.X., Wang L., Zhou S.F., Hou Y.C., Wang P., et al. Challenges and opportunities for siRNA-based cancer treatment. Cancer Lett. 2017;387:77–83. doi: 10.1016/j.canlet.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 45.Hajduk P.J., Meadows R.P., Fesik S.W. Discovering high-affinity ligands for proteins. Science. 1997;278:497–499. doi: 10.1126/science.278.5337.497. [DOI] [PubMed] [Google Scholar]
- 46.LaVan D.A., Lynn D.M., Langer R. Moving smaller in drug discovery and delivery. Nat. Rev. Drug Discov. 2002;1:77–84. doi: 10.1038/nrd707. [DOI] [PubMed] [Google Scholar]
- 47.Bumcrot D., Manoharan M., Koteliansky V., Sah D.W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu L., Ow D.W., Dong Z. Transfer RNA-derived small RNAs in plants. Sci. China Life Sci. 2018;61:155–161. doi: 10.1007/s11427-017-9167-5. [DOI] [PubMed] [Google Scholar]
- 49.Tsurui H., Kumazawa Y., Sanokawa R., Watanabe Y., Kuroda T., Wada A., Watanabe K., Shirai T. Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem. 1994;221:166–172. doi: 10.1006/abio.1994.1393. [DOI] [PubMed] [Google Scholar]
- 50.Franken N.A., Rodermond H.M., Stap J., Haveman J., Bree C.V. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 51.Babski J., Maier L.K., Heyer R., Jaschinski K., Prasse D., Jager D., Randau L., Schmitz R.A., Marchfelder A., Soppa J. Small regulatory RNAs in archaea. RNA Biol. 2014;11:484–493. doi: 10.4161/rna.28452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldman M., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2019:326470. doi: 10.1101/326470. [DOI] [Google Scholar]
- 53.Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M.G., Haag J.D., Gould M.N., Stewart R.M., Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.