Abstract
The sigF gene encodes an alternate sigma factor found in Mycobacterium tuberculosis and related pathogenic mycobacteria. Determination of conditions of sigF expression is an important step in understanding the conditional gene regulation which may govern such processes as virulence and dormancy in mycobacteria. We constructed an in-frame translational lacZ-kan fusion within the sigF gene to determine the conditions of sigF expression. This reporter construct was expressed from a multicopy plasmid in a strain of BCG harboring an integrated luciferase reporter gene under the control of the mycobacteriophage L5 gp71 promoter. Antibiotic exposure, in particular, ethambutol, rifampin, streptomycin, and cycloserine treatment, increased the level of SigF reporter specific expression in a dose-dependent fashion. The level of SigF reporter specific expression increased over 100-fold in late-stationary-phase growth compared to that in exponential growth. During the exponential phase, SigF specific expression could be induced by a number of other stresses. Anaerobic metabolism induced SigF by greater than 150-fold, particularly in the presence of metronidazole. Cold shock increased the level of SigF specific expression, while heat shock decreased it. Oxidative stress was also an important inducer of SigF specific expression; a greater induction was seen with cumene hydroperoxide than with hydrogen peroxide. Comparisons of bacterial viability as determined by the luciferase assay or by plating serial dilutions revealed that luciferase gp71-dependent activity was an unreliable predictor of the numbers of CFU during stationary-phase growth and anaerobic metabolism. The induction of sigF following antibiotic exposure suggests that this bacterial transcription factor may control genes which are important for mycobacterial persistence in the host during chemotherapy.
Mycobacterium tuberculosis is an ancient, highly successful human pathogen. Skeletal evidence of tuberculosis infection can be found as far back as 4000 B.C. in the tombs of Egyptian mummies (19). Despite the development over 50 years ago of antibiotics that are active against this organism, tuberculosis latently infects approximately one-third of the world’s population and remains the leading infectious cause of death worldwide (36). The ability of M. tuberculosis to adapt to a wide range of host conditions, including survival within macrophages, and its capacity to enter a dormant state are critical to its success as a human pathogen.
Transcriptional regulators such as sigma factors are likely to play a large role in the bacterial adaptive responses needed for pathogenesis. Several alternate sigma factors have been correlated with virulence in other species. For example, AlgU controls the shift to mucoidy in Pseudomonas aeruginosa, a proven virulence mechanism in cystic fibrosis patients (13). In mycobacteria, mutations in sigA, a highly conserved essential gene, lead to attenuated virulence (10, 16), whereas the extracytoplasmic sigma factor SigE is involved in the stress response to heat shock, acidic pH, detergent stress, and oxidative stress (44). An alternate sigma factor, SigF, is found in slowly growing mycobacteria and has been associated with the stationary phase (12). The M. tuberculosis sigF gene is homologous to Bacillus subtilis sigF and sigB, which are sporulation and stress response sigma factor genes, respectively (11). It also shows homology to Staphylococcus aureus sigB, a sigma factor gene that participates in methicillin resistance (45), and to Listeria monocytogenes sigB, which plays a role in virulence (43). RNase protection assays have shown that M. tuberculosis sigF transcription is stimulated by entry into the stationary phase and certain stresses such as cold shock (12). However, RNase protection analysis of sigF transcription has been limited by the technical difficulties of reproducibly isolating high-quality mRNA from slowly growing mycobacteria. In the present study, we developed a double reporter assay in order to conduct a comprehensive analysis of the conditions under which SigF is expressed. Antibiotic stress, anaerobic conditions, stationary phase, oxidative stress, nutrient depletion, and cold shock were all shown to be important conditional expression stimuli.
MATERIALS AND METHODS
Plasmids and strains.
The plasmids and bacterial strains used in this study are described in Table 1. Recombinant plasmids were transformed into Escherichia coli by standard protocols (4). Transformations into Mycobacterium bovis BCG and Mycobacterium smegmatis were performed as described previously (11, 23) with a Bio-Rad apparatus (Bio-Rad Laboratories, Hercules, Calif.) and by allowing recovery for 24 or 3 h, respectively, in Middlebrook 7H9 liquid medium (Difco Laboratories, Detroit, Mich.) supplemented with albumin-dextrose complex (ADC) and Tween 80 (23), prior to plating on Middlebrook 7H10 solid selective medium (Difco Laboratories). Isolation and purification of plasmids were performed with the Qiagen system (Qiagen, Inc., Chatsworth, Calif.). The integrating luciferase plasmid pGS16 (24) was a generous gift from Gary J. Sarkis and Graham F. Hatfull.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristica | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−recA1 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 recA1 deoR Δ(lacZYA-argF)U169 (φ80 lacZ ΔM15) | 6 |
| M. bovis BCG | Pasteur strain | ATCC 35734 |
| M. smegmatis | mc26 1-2C | 47 |
| Plasmids | ||
| pUC19 | High-copy-number E. coli plasmid, Apr | 46 |
| pNBV1 | E. coli-mycobacterial shuttle plasmid, Hyr | 22 |
| pYZ99 | pUC18 containing sigF on a 2.8-kb BamHI fragment from M. tuberculosis H37Rv, Apr | 12 |
| pLZK82 | pUC19 containing lacZ-Kmr on a 4.4-kb BamHI fragment | 5 |
| pMH5 | Integration-proficient pUC119 derivative containing a 4.9-kb insert of the phage L5 genome, Kmr | 27 |
| pIJV1 | pUC19- and pMH5-derived integrating E. coli-mycobacterial shuttle plasmid | 15 |
| pCK2819-P | pUC19 containing sigF on a 2.8-kb PstI fragment | This study |
| pCK2819-EB | pUC19 containing sigF with an additional BamHI site adjacent to EcoRI on a 2.8-kb PstI fragment | This study |
| pCK0221 | pUC19 containing frameshifted sigF::lacZ on a 7.6-kb SapI-XbaI fragment, Kmr Apr | This study |
| pCK2295 | pUC19 containing in-frame sigF::lacZ on a 7.6-kb SapI-XbaI SapI-XbaI fragment, Kmr Apr | This study |
| pCK3127 | pNBV1 containing in-frame sigF::lacZ on a 7.6-kb SapI-XbaI fragment, Kmr Hyr | This study |
| pCK1107 | pNBV1 containing frameshifted sigF::lacZ on a 7.6-kb SapI-XbaI fragment, Kmr Hyr | This study |
| pCK3215 | pIJV1 containing in-frame sigF::lacZ on a 7.6-kb SapI-XbaI fragment, Kmr, integrating | This study |
| pGS16 | Integrating E. coli-mycobacterial shuttle plasmid containing FFlux Kmr Apr | 24 |
Abbreviations: Ap, ampicillin; Hy, hygromycin; Km, kanamycin.
Luciferase assays.
Luciferase assays were performed on a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, Calif.). Standardized curves of M. smegmatis and M. bovis BCG luciferase activity were constructed for both the logarithmic phase and the stationary phase with known quantities of bacteria, as determined by plating serial dilutions. Both fresh and frozen cultures were tested. The validity of the experimental results obtained by the luciferase assay was also periodically tested by obtaining colony counts. To perform measurements, each sample was vortexed for 60 s, and then 50 μl of culture was placed in a cuvette. A total of 100 μl of 1 mM d-luciferin (Analytical Luminescence Laboratory) in 0.1 M sodium citrate buffer (pH 5.0) was injected automatically, and readings were taken for 20 s. Each measurement was performed in triplicate, and the average value was recorded. Measurements were accepted only if the variance of all three readings was less than 15%. Readings from the luciferase assay were converted to colony counts by using a formula derived from the standardized curve.
Culture and stress conditions.
BCG strains containing pCK3127 or pCK3215 were grown to the early logarithmic phase in Middlebrook 7H9 liquid medium supplemented with ADC, Tween 80, and kanamycin (10 μg/ml) (23). In some instances, particularly under prolonged cultivation conditions, selective medium was used. The selective medium included cycloheximide (50 μg/ml), ampicillin (50 μg/ml), and polymyxin B (200 U/ml); these agents did not induce sigF expression in the exponential phase. The medium for BCG containing the reporter pGS16 also contained hygromycin (50 μg/ml). M. bovis BCG cultures were incubated in a rotary shaker at 37°C. For growth curve experiments, aliquots were removed daily for testing without adding additional medium or supplements to the original culture. Each stress experiment was performed with a fresh early-logarithmic-phase culture. For cold shock and heat shock experiments, samples were placed at the designated temperature without manipulation of the medium. For antibiotic stress and oxidative stress experiments, the compound of interest was added directly to the medium, and the cultures were returned to the 37°C rotary shaker for the duration of the experiment. Hydrogen peroxide was tested at concentrations ranging from 0.1 to 5 mM, cumene hydroperoxide from 0.1 to 1 mM, isoniazid from 0.01 to 0.1 μg/ml, rifampin from 0.05 to 0.4 μg/ml, ethambutol from 0.5 to 4 μg/ml, streptomycin was tested at from 0.5 to 4 μg/ml, and cycloserine from 1 to 50 μg/ml. To perform nutrient depletion experiments, early-logarithmic-phase cultures were sedimented, the supernatant was removed, and bacteria were resuspended in the test medium. Extensive washing procedures were not performed since cold shock was known to induce SigF. Colony counts were performed by plating serial dilutions of a fresh culture in duplicate onto solid 7H10 medium supplemented with ADC, cycloheximide (50 μg/ml), ampicillin (50 μg/ml), polymyxin B (200 U/ml), and kanamycin (10 μg/ml). The plates were incubated at 37°C in 5% CO2. The average colony count for the two plates in each series with between 50 and 300 colonies at 4 weeks was recorded.
A defined complete minimal medium was made with 100 ml of base salt solution, 0.2 ml of trace element solution, and 1.0 ml of each carbon source without the addition of ADC or Tween 80. The base salt solution is 4 g of NaCl, 0.2 g of MgSO4 · 7 H2O, 2 g of KH2PO4, and 2 g of (NH4)2HPO4 per liter of distilled H2O brought to pH 7.2 with NaOH. The trace element solution is 40 mg of ZnCl2, 200 mg of FeCl3 · 6H2O, 10 mg of CuCl2 · 2H2O, 10 mg of MnCl2 · 4 H2O, 10 mg of Na2B4O7 · 10H2O, and 10 mg of (NH4)6Mo7O24 · 4H2O per liter of distilled H2O. Carbon sources were made as 20% solutions in distilled deionized water and were then filter sterilized. For each nutrient depletion state, the designated substance was omitted from the medium.
Medium osmolarity was read directly with a 5100C vapor pressure osmometer (Wescor, Inc., Logan, Utah). Prior to the readings the osmometer was calibrated with 100- and 290-mOsm standards (Wescor, Inc.). For each specimen, 8 μl of sample was placed on a paper disc, and the average of three readings was taken.
Anaerobic experiments were performed by the method of slow stirring of Wayne and Hayes (41). Sealed screw-top tubes had a headspace/culture ratio of 0.5 and were stirred continuously at approximately 120 rpm during incubation at 37°C. Multiple sample tubes were incubated simultaneously so that tubes were opened a single time for measurements. After each experiment, the samples were vortexed for 60 to 90 s in a 5-ml Falcon tube (Becton Dickinson, Franklin Lakes, N.J.) with 0.5 ml of 3-mm sterile glass beads to eliminate clumping, and then the samples were aliquoted, placed into separate 1-ml Eppendorf tubes, and immediately frozen at −80°C for later luciferase and β-galactosidase assays. Measurements of optical density and numbers of CFU by plating on solid 7H10 medium for colony counts were performed immediately with fresh samples.
β-Galactosidase assays.
β-Galactosidase assays were performed with whole bacteria as rate assays that monitor the accumulation of the fluorescent cleavage product of methylumbelliferyl-β-d-galactopyranoside (MUG) on a Bowman Series 2 fluorescence spectrometer (SLM/Aminco, Urbana, Ill.) (18). Samples were initially tested both prior to and after freezing at −80°C, without an observable difference. Measurements were made at an excitation wavelength of 360 nm, an emission wavelength of 440 nm, a band width of 4 nm, and a sensitivity of 450 V. The slope of the fluorescence-time curve over 3 min was recorded and was normalized to a standard curve performed with known amounts of β-galactosidase enzyme. For each sample, 440 μl of buffer Z (28), 50 μl of culture that had been vortexed for 60 s, and 10 μl of 50 mM MUG in glycerol were added to a cuvette and were allowed to equilibrate for 60 s prior to measurement. Each measurement was performed in triplicate, and the average slope was recorded. Any curve without a smooth slope was rejected, and the sample was retested after additional sample vortexing. The lower limit of detection for this β-galactosidase assay was 0.01 U/ml of bacterial culture. Logarithmic-phase BCG harboring the integrated sigF reporter construct (pCK3215) produced 0.028 U of β-galactosidase activity per ml of culture, while an equivalent culture of BCG harboring the frameshifted negative control construct (pCK1107) produced an undetectable level of β-galactosidase activity (<0.01 U/ml of culture).
RESULTS
Construction and characterization of sigF reporter plasmids pCK3127 and pCK3215.
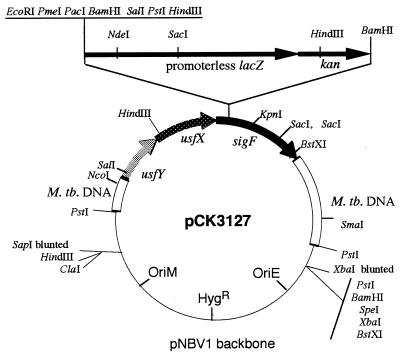
sigF-lacZ translational gene fusions, propagated as both a mycobacterial multicopy plasmid and a mycobacterial integrative single-copy construct, were made through a series of cloning steps. To generate pCK2819-P, the ends of the 2.8-kb BamHI fragment from pYZ99 (12) were blunted and ligated to PstI linkers, and the modified fragment was cloned into pUC19-PX (an altered pUC19 in which the EcoRI-HindIII polylinker site is replaced by a single PstI site and the NdeI site at position 183 is changed to XbaI). pCK2819-EB was created by changing the EcoRI site within the sigF gene (occurring at codons 35 and 36) in pCK2819-P to an EcoRI-BamHI site by linker addition. A 4.3-kb BamHI fragment bearing a lacZ-kan cassette from pLZK82 (5) was inserted into this new BamHI site in pCK2819-EB to yield pCK0221. Next, a PmeI- and PacI-containing synthetic linker (top strand, 5′-AA TTG TTT AAA CGC TTA ATT-3′; bottom strand, 5′-AAT AAT TAA GCG TTT AAA C-3′) was cloned into the unique EcoRI site of pCK0221 to yield pCK2295, a pUC-based plasmid with the lacZ gene translationally fused in the correct reading frame to the first 105 bp of the sigF-coding sequence and an additional 1.2 kb of M. tuberculosis DNA 5′ to the sigF gene. The sigF-lacZ-kan fusion block was excised as a 7.6-kb XbaI-SapI fragment, blunted, and cloned into the unique EcoRV site of the mycobacterial-E. coli shuttle plasmid pNBV1 (22) to give the multicopy, nonintegrating construct pCK3127 (Fig. 1) and into the SmaI site of pIJV1 (15) (a derivative of pMH5 [27] containing the mycobacterial L5 int-attP loci but lacking a kan resistance gene) to yield a mycobacterial integrative vector, pCK3215. A similar 7.6-kb XbaI-SapI fragment from pCK0221 was transferred to pNBV1 by the same strategy to yield pCK1107, a multicopy, nonintegrating construct analogous to pCK3127 but with the sigF and lacZ genes out of frame with respect to one another.
FIG. 1.
Plasmid map of pCK3127, an E. coli-mycobacterial shuttle plasmid containing an in-frame sigF::lacZ gene fusion. M. tb., M. tuberculosis.
Electrocompetent BCG cells were transformed with pCK1107 (multicopy, out-of-frame sigF-lacZ), pCK3215 (single copy, in-frame sigF-lacZ), and pCK3127 (multicopy, in-frame sigF-lacZ) and plated on 7H10-ADC agar supplemented with hygromycin and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal). As may be seen in Fig. 2, 5-week-old colonies with the out-of-frame plasmid pCK1107 failed to produce any β-galactosidase activity, as assessed by the accumulation of the nondiffusible, blue X-Gal hydrolysis product. In contrast, single-copy and multicopy in-frame sigF-lacZ fusion plasmids gave low-level and high-level blue signals, respectively, in a target pattern with SigF-LacZ expression restricted to the colony centers. This qualitative assessment suggested that mycobacterial sigF expression was conditionally regulated and that the stationary phase and/or starvation (conditions which prevail within the center of an expanding colony) were stimuli for its induction.
FIG. 2.
BCG harboring sigF::lacZ translational fusion constructs give “target colony” phenotypes with sigF expression centrally. (Top) BCG harboring a control plasmid with the sigF::lacZ fusion out of frame (pCK1107). (Middle) Integrated single-copy sigF::lacZ fusion gene (pCK3215). (Bottom) Multicopy plasmid-borne sigF::lacZ fusion (pCK3217). Colonies were grown on Middlebrook 7H10 agar supplemented with ADC, glycerol, cycloheximide, kanamycin, and X-Gal. After 5 weeks of incubation the colonies (each about 5 mm in diameter) were photographed.
Luciferase assay studies.
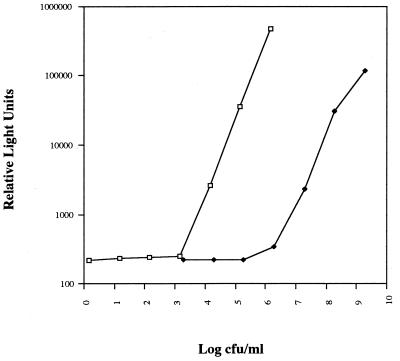
Comparisons of relative light units per milliliter of culture determined by the luciferase assay with the numbers of CFU determined by plating serial dilutions showed a linear relationship down to 103 CFU/ml for logarithmic-phase cultures. These results are similar to the results reported by Jacobs et al. (24). However, in stationary-phase cultures the sensitivity of the luciferase assay was only 105 CFU/ml (Fig. 3). In this construct, the lux gene is driven by Pgp71. The slopes of the lines for logarithmic-phase versus stationary-phase cultures were similar, and no difference was observed for M. bovis BCG strains containing the multicopy plasmid pCK3127. The sensitivity of the assay and the slope of the curve were the same for both fresh and frozen cultures and for M. bovis BCG versus M. smegmatis. Luciferase assays for cultures that had been incubated anaerobically were unreliable, giving numbers below the level of detection of the assay, despite reproducible colony counts of >105 CFU/ml. In view of the growth phase dependence of Pgp71::lux expression, we used standard CFU assays in the growth curve and anaerobic experiments for the denominator in our determinations of sigF specific expression. Luciferase assays with cultures that had been exposed to rifampin had luciferase activity/CFU ratios that were significantly higher than those prior to drug treatment, suggesting that the bacteria maintained luciferase enzymatic function beyond the time of cell death.
FIG. 3.
Correlation between mycobacteriophage L5 Pgp71-dependent luciferase expression and numbers of CFU for M. smegmatis harboring pGS16 with exponential- versus stationary-phase cultures. Serial dilutions of a logarithmic-phase culture (□) at an optical density at 600 nm of 0.05 were compared to the numbers of CFU. The optical density at 600 nm of the stationary-phase culture (⧫) was 1.0.
Stress experiments.
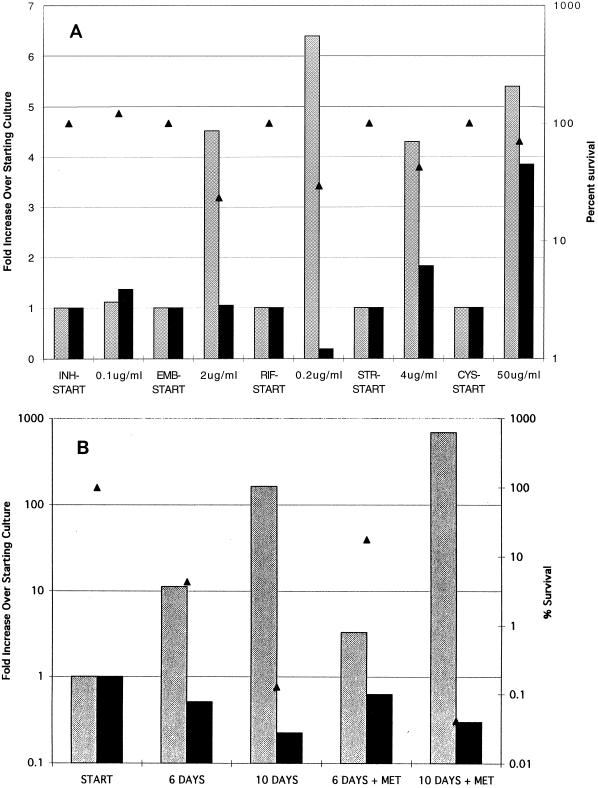
In order to investigate further the conditions under which SigF is expressed, a variety of stress conditions were evaluated. A summary of the results is presented in Table 2. To investigate whether SigF is involved in adaptation to antibiotics, a variety of agents clinically important in the treatment of human tuberculosis were tested, including four primary antituberculous antibiotics. Concentrations of 0.5, 1, 2, and 5 times the MIC of each drug were tested (7, 9, 30). A 4-h time point was selected in order to provide stress conditions but not significant killing of the organism. Rifampin, ethambutol, streptomycin, and d-cycloserine were shown to significantly increase the level of SigF specific expression, whereas little change was seen with isoniazid (Fig. 4A). A dose-response relationship was observed for those antibiotics that increased the level of SigF specific expression (data not shown). In most cases, the greatest induction was seen at the MIC for the organism rather than at the highest dose tested.
TABLE 2.
SigF activity summary
| Stress | Fold increase in activitya |
|---|---|
| Antibiotic (for 4 h) | |
| Rifampinb | 6 |
| Ethambutol | 4 |
| Cycloserineb | 5 |
| Streptomycinb | 4 |
| Isoniazid | 0 |
| Anaerobic | |
| Anaerobic for 10 days | 164 |
| Anaerobic plus metronidazole Flagyl for 10 days | 681 |
| Oxidative (for 4 h) | |
| Hydrogen peroxideb | 15 |
| Cumene hydroperoxideb | 53 |
| Temp | |
| Cold (27°C for 24 h) | 5 |
| Heat (42°C for 4 h) | −1.7 |
| Nutrient depletion (for 72 h) | |
| No glucoseb | 2 |
| No glycerolb | 8 |
| No magnesiumb | 2 |
| No potassiumb | 6 |
| No NaClb | 14 |
| No ammoniab | 14 |
| Growth phase | |
| Logarithmic | 1 |
| Stationary | 160 |
Ratio of β-galactosidase activity per CFU between stressed and unstressed cultures.
Denotes experiments performed with relative light units converted to CFU per milliliter.
FIG. 4.
SigF specific expression in response to antibiotic and anaerobic stress. Data are shown for units of β-galactosidase normalized to numbers of CFU (gray bars); β-galactosidase units per milliliter of culture (black bars), and percent survival (triangles). (A) Antibiotic stress measurements were performed at 4 h. INH, isoniazid; EMB, ethambutol; RIF, rifampin; STR, streptomycin; CYS, cycloserine. (B) Anerobic stress experiments were performed with and without metronidazole (MET) at 12 μg/ml.
We used the model of Wayne and Hayes with defined culture-to-headspace ratios and our reporter strain of BCG to evaluate the effect of oxygen depletion on sigF expression (41). In the model of Wayne and Hayes, mycobacterial cultures are subjected to progressively lower oxygen tensions when sealed tubes with limited headspaces are used and stirring is regulated to produce a state of nonreplicating persistence. As may be seen in Fig. 4B, SigF specific expression increased over 10-fold at 6 days and over 150-fold at 10 days following oxygen depletion. When metronidazole, an anaerobic antibiotic that has no activity against mycobacteria in the normal aerobic environment, is added to the culture, SigF-specific expression is induced even further. Although the colony counts were somewhat decreased at 10 days, the β-galactosidase signal did not decrease proportionately, indicating that the level of sigF transcription is increased per cell.
We also tested heat shock and cold shock to determine whether SigF expression was changed. A variety of temperatures and time points were assayed. Cold shock at 27°C for 24 h increased the level of SigF specific expression, whereas heat shock at 42°C for 4 h decreased it. Increasing lengths of exposure augmented the effect seen. Hydrogen peroxide and cumene hydroperoxide stress conditions were tested with several different doses near the MIC for the organism. Cumene hydroperoxide is an alkylperoxide produced by metabolism of unsaturated fatty acids and nucleic acids (3). Once again, a 4-h time point for measurements was chosen. SigF specific expression was induced 15-fold with hydrogen peroxide and 53-fold with cumene hydroperoxide.
Knowing that the level of SigF is dramatically increased during the late stationary phase, we also systematically tested nutrient depletion states. Early-logarithmic-phase cultures were transferred from 7H9 to experimental minimal media. Seventeen different culture media were tested, with each medium missing one ingredient. Tests were performed at both the 24- and 72-h time points. The results at 24 h were similar to the results at 72 h but were smaller in magnitude. SigF-specific expression was induced most strongly by sodium, nitrogen, and glycerol depletion and to a lesser extent by potassium and glucose depletion. Minimal change in SigF specific expression was seen with the absence of trace elements including iron, copper, boron, manganese, zinc, and molybdenum (data not shown). Metal chelating agents were not used in this experiment, so it is possible that the trace elements were not sufficiently depleted to observe a change. However, the impressive induction seen with other conditions makes this possibility less likely.
To investigate whether hypoosmolarity was responsible for the induction seen with nutrient depletion, we tested the osmolarity of 7H9, complete minimal medium, and the macronutrient depleted media. Rich medium (7H9) had an osmolarity of 182 mOsm, complete minimal medium had an osmolarity of 251 mOsm, and NaCl-depleted minimal medium had an osmolarity of 133 mOsm. All other nutrient-depleted media had osmolarities of greater than 200 mOsm. On the basis of these data it is unlikely that hypoosmolarity is responsible for the sigF induction seen with any of the nutrient depletion states except perhaps sodium chloride depletion.
Growth curves.
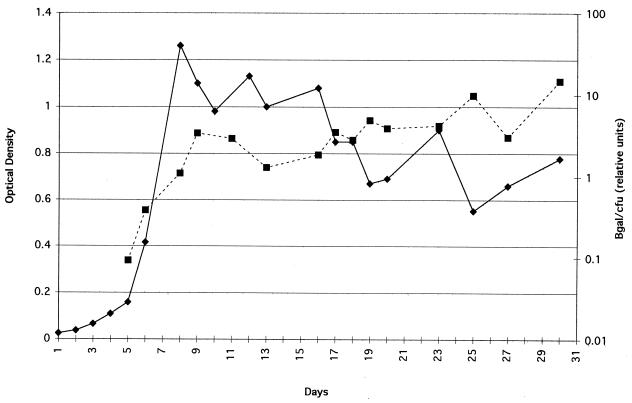
To investigate whether SigF is produced in similar amounts throughout the growth cycle, a longitudinal study of SigF specific expression during all phases of growth was performed. As shown in Fig. 5, SigF specific expression is minimal during the logarithmic phase but increases approximately 160-fold in the early stationary phase. These results are congruent with the results obtained by an RNase protection assay in which the sigF RNA level increased approximately 9.8-fold in the early stationary phase (12). Qualitatively identical curves were produced for bacteria containing the multicopy sigF reporter plasmid pCK3127 and the single-copy integrated reporter plasmid pCK3215, except that the single-copy vector produced about 10-fold less β-galactosidase activity than the multicopy form did.
FIG. 5.
Growth and SigF specific expression curves of M. bovis BCG containing pCK3215 in liquid culture. Aliquots were removed daily to measure the optical density; the numbers of CFU and amount of β-galactosidase (βgal) activity. Solid lines represent the optical density at 600 nm, and dashed lines represent relative units of β-galactosidase activity per CFU. Relative units of β-galactosidase activity/CFU = actual units of β-galactosidase activity/CFU × 1010.
DISCUSSION
We have shown that the mycobacterial transcription factor SigF is induced under a variety of stress conditions, most notably antibiotic stress, the nonreplicating persistence state induced by low oxygen tensions, nutrient depletion, oxidative stress, and stationary-phase growth. While an earlier report that used RNase protection assays showed that the sigF gene is transcriptionally upregulated under certain conditions (12), the present study significantly extends the previous work by showing that increased sigF transcription correlates with increased expression at the protein level, by monitoring sigF expression throughout the growth cycle, and by a comprehensive assessment of numerous stress conditions including antibiotic stress. The results presented in this paper give the SigF specific levels of expression of reporter activity per CFU; this interpretation of the data assumes that dead bacteria do not make or retain appreciable β-galactosidase activity. In some instances we also indexed the sigF::lacZ expression levels to that of a different promoter-reporter pair, namely, Pgp71::lux; however, we found that the levels of Pgp71::lux expression per cell differ dramatically between stationary- and exponential-phase growth (Fig. 3). Comparisons of promoters such as this are limited by the difficulty in identifying a promoter which is truly constitutive throughout all phases of culture growth and under all stress conditions.
The finding that sigF is induced by both stationary-phase growth and anaerobic metabolism as well as antibiotic exposure further reinforces a building body of evidence that tuberculosis latency and antibiotic resistance are closely related phenomena (34). In pyogenic bacteria such as S. aureus and Salmonella enteriditis, bacterial phenotypic adaptation to stationary-phase growth states places the organism in a state of relative antibiotic resistance known as tolerance (20). Antibiotic susceptibility testing of tolerant isolates reveals minimum bactericidal concentrations that are significantly higher than the MICs of the same drug when the drug is tested during logarithmic-phase growth. Tolerance has been shown to be clinically important in deep-seated infections such as S. aureus endocarditis, where studies have shown that patients infected with β-lactam-tolerant organisms have more complications than patients infected with fully sensitive strains of the same organism (35). A similar phenomenon has long been appreciated for M. tuberculosis, in which complete in vitro antibiotic killing occurs in a few days, but treatment of an in vivo infection requires a minimum of 6 to 12 months of therapy (2). Mitchison has explained this phenomenon using a “special populations” hypothesis for the action of antimycobacterial drugs, noting that organisms in a continuous growth phase are the easiest to kill, but populations of organisms that are in the specialized states of acid inhibition and intermittent growth require a specific drug that targets sterilization (29).
Two types of tolerance exist: genotypic and phenotypic. Bacterial strains with genotypic tolerance harbor mutations which reduce microbial lysis upon exposure to antibiotics. For example, Streptococcus pneumoniae R36A has a defect in the autolysin N-acetylmuramic acid-l-alanine amidase (40), and overproduction of certain extracellular proteases increases tolerance in B. subtilis (26). Phenotypic tolerance results when bacteria that are normally antibiotic sensitive encounter an environmental factor or enter a growth phase that decreases the level of antibiotic-mediated killing. Factors that have been described include low pH (21), the presence of serum (39), a high calcium or magnesium concentration (17), and stationary-phase growth (25). In mycobacteria, Wayne and colleagues have described a type of phenotypic tolerance in which cultures in the nonreplicating persistent state are relatively resistant to isoniazid and rifampin but sensitive to metronidazole (41, 42). In contrast, cultures in the usual aerobic logarithmic-phase growth conditions are resistant to metronidazole and sensitive to isoniazid and rifampin. The finding that SigF-specific expression is strongly induced by conditions in the model of Wayne and Hayes and is further increased by the addition of metronidazole suggests that SigF may control genes that affect nonreplicating persistence and modulate antibiotic resistance.
We have shown that sigF induction occurs with exposure to rifampin, streptomycin, ethambutol, cycloserine, and metronidazole but not with exposure to isoniazid. While it is possible that the upregulation of sigF is a stress response that is unrelated to the relative resistance of persistent M. tuberculosis to antibiotics, our observation that the stationary phase and antimicrobial exposure both lead to the induction of sigF suggests that SigF-dependent genes may confer a protective effect against certain antibiotics. Although our understanding of the mode of action of antituberculous drugs has improved in recent years (1, 8, 9, 30, 37), it is difficult to develop a unifying mechanism whereby a single transcription factor might confer resistance to multiple drugs, although not isoniazid. SigF-mediated entry into a stationary growth state may result in physiologic alterations that render certain antibiotic targets nonessential, resulting in phenotypic drug tolerance. Alternatively, increased production of a sigma factor may upregulate specific genes that overcome target inhibition produced by low levels of the drug. This mechanism is supported by the demonstration that increased alrA transcription results in d-cycloserine resistance (8). Induction of a factor that alters global membrane permeability is a third possibility, but it seems less likely since not all antibiotics cause an induction of sigF.
Finally, as part of these experiments, we have further characterized the conditions under which the luciferase reporter phage system (24) is useful as a surrogate marker of bacterial colony counts. In this system the luciferase gene is driven by the constitutively expressed gene 71 promoters of the temperate mycobacteriophage L5 (38). The gene 71 product maintains lysogeny by functioning as a repressor in an analogous function to the cI gene of bacteriophage lambda (14). The gene 71 promoters P1, P2, and P3 are strongly recognized by mycobacterial RNA polymerase, resulting in protein expression throughout the lysogenic and early lytic cycles. The promoters are presumably turned off during the late lytic phase due to a generalized inhibition of the host RNA polymerase once the gp71 protein is degraded (31). We found that luciferase production in the gp71 system was markedly decreased when it was tested in the stationary phase as well as in the nonreplicating persistent state induced by low oxygen tensions. It is likely that the gp71 promoters are recognized by a vegetative growth sigma factor that is less abundant or less functional under these stationary-phase conditions. Care should be exercised when this promoter-reporter system is used under conditions other than logarithmic-phase growth.
Tuberculosis latency and drug susceptibility are closely linked phenomena that are a driving force behind the modern-day tuberculosis epidemic. Recent global surveillance efforts in 35 countries by the World Health Organization and the International Union Against Tuberculosis and Lung Disease have shown a worldwide median prevalence of resistance to at least one drug of 12.6% (33). Because this study did not include some areas known to have high rates of tuberculosis, unregulated access to drugs, and relatively poor tuberculosis control efforts, the true prevalence is likely to be even higher. In addition, the development of new antituberculosis drugs has been slow and costly (32). An understanding of the mycobacterial adaptive mechanisms such as that mediated by sigF and other alternative sigma factors may offer novel approaches to the development new therapeutic agents with activity against this ancient and highly successful pathogen.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI-36973 and AI-37856 and by the American Lung Association of Maryland.
We thank Graham F. Hatfull and Gary J. Sarkis for supplying pGS16, Joseph S. Handler and Vincent W. Yang for use of the luminometer, and Floyd R. Bryant for use of the fluorometer. We also thank James Gomez for supplying pIJV1 and for helpful suggestions.
REFERENCES
- 1.Alcaide F, Pfyffer G E, Telenti A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–2273. doi: 10.1128/aac.41.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Thoracic Society. Treatment of tuberculosis and tuberculosis infection in adults and children. Am J Respir Crit Care Med. 1994;149:1359–1374. doi: 10.1164/ajrccm.149.5.8173779. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 5.Barcak G J, Chandler M S, Redfield R J, Tomb J. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 6.Bethesda Research Laboratories. Bethesda Research Laboratory Focus. Vol. 8. Gaithersburg, Md: Bethesda Research Laboratories; 1986. BRL pUC host: E. coli DH5αTM competent cells; p. 9. [Google Scholar]
- 7.Blanchard J S. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu Rev Biochem. 1996;65:215–239. doi: 10.1146/annurev.bi.65.070196.001243. [DOI] [PubMed] [Google Scholar]
- 8.Caceres N E, Harris N B, Wellehan J F, Feng Z, Kapur V, Barletta R G. Overexpression of the d-alanine racemase gene confers resistance to d-cycloserine in Mycobacterium smegmatis. J Bacteriol. 1997;179:5046–5055. doi: 10.1128/jb.179.16.5046-5055.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole S T. Mycobacterium tuberculosis: drug-resistance mechanisms. Trends Microbiol. 1994;2:411–415. doi: 10.1016/0966-842x(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 10.Collins D, Kawakami R, de Lisle G, Pascopella L, Bloom B, Jacobs W., Jr Mutation of the principal ς factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 1995;92:8036–8040. doi: 10.1073/pnas.92.17.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMaio J, Zhang Y, Ko C, Bishai W R. The Mycobacterium tuberculosis sigF gene is part of a gene cluster organized like the Bacillus subtilis sigF and sigB operons. Tubercle Lung Dis. 1997;78:3–12. doi: 10.1016/s0962-8479(97)90010-1. [DOI] [PubMed] [Google Scholar]
- 12.DeMaio J, Zhang Y, Ko C, Young D, Bishai W. A stationary phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deretic V, Schurr M, Boucher J, Martin D. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnelly-Wu M K, Jacobs W R, Jr, Hatfull G F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 15.Gomez, J. E., and W. R. Bishai. 1998. Unpublished data.
- 16.Gomez M, Doukhan L, Nair G, Smith I. sigA is an essential gene in Mycobacterium smegmatis. Mol Microbiol. 1998;29:617–628. doi: 10.1046/j.1365-2958.1998.00960.x. [DOI] [PubMed] [Google Scholar]
- 17.Goodell E W, Fazio M, Tomasz A. Effect of benzylpenicillin on the synthesis and structure of the cell envelope of N. gonorrhoeae. Antimicrob Agents Chemother. 1978;13:514–526. doi: 10.1128/aac.13.3.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grange J M. Fluorimetric assay of mycobacterial group-specific hydrolase enzymes. J Clin Pathol. 1978;31:378–381. doi: 10.1136/jcp.31.4.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas F, Haas S S. The origins of Mycobacterium tuberculosis and the notion of its contagiousness. In: Rom W N, Garay S M, editors. Tuberculosis. New York, N.Y: Little, Brown & Co.; 1996. pp. 3–19. [Google Scholar]
- 20.Handwerger S, Tomasz A. Antibiotic tolerance among clinical isolates of bacteria. Annu Rev Pharmacol Toxicol. 1985;25:349–380. doi: 10.1146/annurev.pa.25.040185.002025. [DOI] [PubMed] [Google Scholar]
- 21.Horne D, Tomasz A. pH dependent penicillin tolerance of group B streptococci. Antimicrob Agents Chemother. 1981;20:128–135. doi: 10.1128/aac.20.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard N S, Gomez J E, Ko C, Bishai W R. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene. 1995;166:181–182. doi: 10.1016/0378-1119(95)00597-x. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs W, Jr, Kalpana G, Cirillo J, Pascopella L, Snapper S, Udani R, Jones W, Barletta R, Bloom B. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs W R, Jr, Barletta R G, Udani R, Chan J, Kalkut G, Sosne G, Keiser T, Sarkis G J, Hatfull G F, Bloom B R. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–822. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 25.Jawetz E, Gunnison J B, Speck R S, Coleman V R. Studies on antibiotic synergism and antagonism. Arch Intern Med. 1951;87:349–359. doi: 10.1001/archinte.1951.03810030022002. [DOI] [PubMed] [Google Scholar]
- 26.Jolliffe L K, Doyle R J, Streips U N. Extracellular proteases increase tolerance of Bacillus subtilis to nafcillin. Antimicrob Agents Chemother. 1982;22:83–89. doi: 10.1128/aac.22.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Laboratory; 1972. [Google Scholar]
- 29.Mitchison D A. The Garrod Lecture: understanding the chemotherapy of tuberculosis—current problems. J Antimicrob Chemother. 1992;29:477–493. doi: 10.1093/jac/29.5.477. [DOI] [PubMed] [Google Scholar]
- 30.Musser J. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nesbit C E, Levin M E, Donnelly-Wu M K, Hatfull G F. Transcriptional regulation of repressor synthesis in mycobacteriophage L5. Mol Microbiol. 1995;17:1045–1056. doi: 10.1111/j.1365-2958.1995.mmi_17061045.x. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien R J, Vernon A A. New tuberculosis drug development: how can we do better? Am J Respir Crit Care Med. 1998;157:1705–1707. doi: 10.1164/ajrccm.157.6.1576. [DOI] [PubMed] [Google Scholar]
- 33.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S B, Kim S J, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994–1997. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 34.Parrish N, Dick J, Bishai W R. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 35.Rajashekaraiah K R, Rice T, Rao V S, Marsh D, Ramakrishna B, Kalick C A. Clinical significance of tolerant strains of Staphylococcus aureus in patients with endocarditis. Ann Intern Med. 1980;93:796–782. doi: 10.7326/0003-4819-93-6-796. [DOI] [PubMed] [Google Scholar]
- 36.Raviglione M, Snider D E, Kochi A. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 37.Samarawickrema N A, Brown D M, Upcroft J A, Thammapalerd N, Upcroft P. Involvement of superoxide dismutase and pyruvate:ferredoxin oxidoreductase in mechanisms of metronidazole resistance in Entamoeba histolitica. J Antimicrob Chemother. 1997;40:833–840. doi: 10.1093/jac/40.6.833. [DOI] [PubMed] [Google Scholar]
- 38.Sarkis G J, Jacobs W R, Jr, Hatfull G F. L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol Microbiol. 1995;15:1055–1067. doi: 10.1111/j.1365-2958.1995.tb02281.x. [DOI] [PubMed] [Google Scholar]
- 39.Storch G A, Krogstad D J, Parquette A. Antibiotic induced lysis of enterococci. J Clin Invest. 1981;68:639–645. doi: 10.1172/JCI110298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 41.Wayne L G, Hayes L G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wayne L G, Sramek H A. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiedmann M, Arvik T J, Hurley R J, Boor K J. General stress transcription factor ςB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Q-L, Kong D, Lam K, Husson R N. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J Bacteriol. 1997;179:2922–2929. doi: 10.1128/jb.179.9.2922-2929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S, deLencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Lathigra R, Garbe T, Catty D, Young D. Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol Microbiol. 1991;5:381–391. doi: 10.1111/j.1365-2958.1991.tb02120.x. [DOI] [PubMed] [Google Scholar]