Abstract
We investigated the relationship between susceptibility to β-lactam antibiotics and variation in the major outer membrane protein P2 (OmpP2; also called porin) of persistent nonencapsulated Haemophilus influenzae isolated from cystic fibrosis patients. Nine OmpP2 variants were selected from two distinct H. influenzae strains from two patients extensively treated with β-lactam antibiotics. The variants differed in their susceptibilities to at least two β-lactam antibiotics. By detergent extraction and column chromatography, OmpP2 was purified from two variants that were derived from strain 70 and that differed notably in their susceptibilities to β-lactam antibiotics. The proteins were reconstituted into black lipid membranes for measurement of porin function. OmpP2 from the more resistant isolate (isolate 70b) had a smaller channel conductance than OmpP2 of the more susceptible isolate (isolate 70f). DNA sequencing of ompP2 of these isolates revealed single nonsynonymous base differences; there were changes in the amino acid sequence corresponding to surface-exposed loops 4, 5, 6, and 8. Changes in loops 4, 5, and 6 were previously shown to result in antigenic differences. Beside these mutations, variants of strain 70 showed additional mutations in loop 1 and nonexposed loop 3. Taken together, our results suggest that in variants of strain 70, nonsynonymous point mutations accumulated both in the sequences of ompP2 coding for antigen-variable loops and in other loops, notably, loops 1 and 3. The latter changes are suggested to affect the permeability of the porin channel.
Haemophilus influenzae colonizes the human nasopharynx and is an important pathogenic bacterium in humans. Encapsulated strains (type b) cause systemic disease, including meningitis, epiglottitis, and cellulitis, in nonvaccinated individuals. Nonencapsulated H. influenzae is a frequent cause of respiratory tract infections including otitis media, sinusitis, pneumonia, and persistent infections in chronic bronchitis and cystic fibrosis (CF) patients (24).
Compared to H. influenzae type b, nonencapsulated H. influenzae shows much greater strain-to-strain diversity in the amino acid sequence of its major outer membrane proteins (Omps) including OmpP2 (48). OmpP2 has a molecular weight ranging from 35,000 to 42,000 (15, 48). The diversity of the amino acid sequence is found predominantly in surface-exposed loops 2, 4, 5, 6, and 8 (12).
During persistent infections in chronic bronchitis patients, the amino acid composition of OmpP2 of nonencapsulated H. influenzae was shown to vary due to the accumulation of point mutations (13). These changes were mapped in ompP2 to regions coding for loops 5 and 6 of the protein (2, 13, 19, 44). These changes in amino acid composition resulted in antigenic drift (13, 14, 18). More extensive amino acid variations have been observed when nonencapsulated H. influenzae strains persisted in the lower respiratory tracts of CF patients (29).
OmpP2 of H. influenzae type b is a member of the porin superfamily (22). Porins form nonspecific outer membrane diffusion channels for small water-soluble solutes including antibiotics (35). Several porin structures have been determined at the atomic level and have served to establish a consensus folding pattern of porins (7, 36, 40). This folding pattern consists of a β barrel made up of 16 or 18 membrane-spanning antiparallel β strands that are joined by short connecting turns on the periplasmic side of the membrane and loops of various lengths on the extracellular surface. Also, part of the consensus folding pattern is the folding of loop 3 to form a constriction in the barrel (45, 54). Topological models of OmpP2 of H. influenzae place loop 3 at the channel constriction (2, 44). Recently, Srikumar et al. (43) demonstrated that deletions in loop 4 resulted in an increased channel conductance and have proposed that loop 4 may serve to narrow the channel entrance. Mutations in the gene encoding the OmpF porin of Escherichia coli were detected as small changes in amino acid composition and in susceptibility to various antibiotics, particularly to β-lactams (3).
Because CF patients are extensively treated with antibiotics and especially β-lactam antibiotics (31), we wished to determine whether (i) differences in susceptibility to β-lactam antibiotics were associated with variations in OmpP2 of H. influenzae isolates from CF patients and (ii) variations in OmpP2 correlated with changes in pore properties. The H. influenzae OmpP2 variants differed in their susceptibilities to various β-lactam antibiotics and to chloramphenicol. OmpP2 proteins were purified from two variants and were incorporated into black lipid membranes to determine their channel properties (9). DNA sequencing of the genes encoding OmpP2 identified the differences between variant OmpP2 proteins.
MATERIALS AND METHODS
H. influenzae strains from CF patients.
A total of 247 nonencapsulated H. influenzae isolates were obtained from sputum samples from 39 CF patients (age range, 0.2 to 36 years; median age, 13 years) participating in a 2-year follow-up study (29). These isolates were characterized phenotypically by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis of their cell envelope proteins (Omp analysis) (29) and genotypically by randomly amplified polymorphic DNA (RAPD) analysis (51). Strains were found to differ in both their Omp and their RAPD patterns (29). Variants differed only in their Omp patterns but not in their RAPD patterns. These Omp variants were isolated from several patients over time (29). The antibiotics administered and the duration of antibiotic treatment of the patients were recorded. H. influenzae ATCC 49766 was used as a standard for β-lactam antibiotic sensitivity tests (1).
Strains were routinely plated on chocolate agar plates or were grown in brain heart infusion broth supplemented with 10 μg of hemin (X factor) per ml and 10 μg of NAD (V factor) per ml at 37°C in 5% CO2 for 18 h.
Antibiotic susceptibility testing.
All 247 H. influenzae isolates were tested for β-lactamase activity by using nitrocefin as the chromogenic cephalosporin (37). For the β-lactamase-negative isolates, susceptibility to ampicillin, amoxicillin-clavulanic acid, cefuroxime, cefotaxime, cefaclor, and imipenem was determined by disk diffusion on standard Haemophilus test medium (HTM) plates (diameter, 9 cm; Becton Dickinson, Erembodegem, Belgium) as described by Doern et al. (10). Sensidisks (also from Becton Dickinson) were used according to the method described by the National Committee for Clinical Laboratory Standards (approved standard M2-A4) (34). Selected isolates were further tested for susceptibility to penicillin, cephalothin, and chloramphenicol.
The broth microdilution test was used to assess the MICs of the three β-lactam antibiotics ampicillin, penicillin, and cephalothin and the MIC of chloramphenicol. Bacteria were grown overnight for 18 h (normally growing bacteria) or 36 h (slowly growing bacteria). A bacterial suspension of 2 × 108 to 8 × 108 CFU/ml was prepared in phosphate-buffered saline. According to the standard method of the National Committee for Clinical Laboratory Standards (33), 10 μl of the suspension was transferred to 10 ml of HTM broth to prepare the inoculum for the test. To each well of sterile microtiter plates (Hospidex, Nieuwkoop, The Netherlands) 50 μl of serial twofold dilutions of ampicillin, penicillin, cephalothin, or chloramphenicol in HTM was added. The concentrations of ampicillin ranged from 0.03 to 16 μg/ml, those of penicillin ranged from 0.006 to 3 μg/ml, those of cephalothin ranged from 0.125 to 64 μg/ml, and those of chloramphenicol ranged from 0.06 to 32 μg/ml. Control wells of HTM without antibiotics were included in each experiment. To each well 1 × 108 to 4 × 108 CFU/ml was added. The plates were incubated at 37°C in a 5% CO2 atmosphere for 18 h. When the medium in the control wells was not turbid, the plates were incubated for another 18 h. All MIC determinations were done in triplicate. The MIC was defined as the lowest antibiotic concentration that inhibited the growth of H. influenzae in the microtiter plates. H. influenzae ATCC 49247 (1) and QC2174 (10) were included in each test series for reference.
Sequence analysis of ompP2.
Fragments of DNA containing approximately 300 bp of ompP2 were obtained by PCR with chromosomal DNA and three ompP2-specific primer sets as described by Duim et al. (12). The PCR products were sequenced by direct sequencing of double-stranded DNA (Applied Biosystems) with dyed primers. All fragments were sequenced in both directions and in duplicate from at least two independently obtained PCR products. The analysis was performed in an automated fluorescent DNA sequencer (model 370A; Applied Biosystems). The sequences were analyzed with the computer programs included in the program package PC/GENE (IntelliGenetics, Inc., 1991). The DNA sequences were aligned with the program CLUSTAL by the method developed by Higgins and Sharp (20).
OmpP2 purification.
OmpP2 of H. influenzae was isolated by the method described by Munson et al. (32). Bacterial strains were cultured in brain heart infusion broth supplemented with 10 μg of both X and V factors per ml. The bacteria were harvested by centrifugation and were suspended in 10 mM HEPES (pH 7.4). The bacterial cell envelope fraction was isolated from the bacterial cells by ultrasonic treatment, followed by centrifugation as described by van Alphen et al. (49). The cytoplasmic membrane proteins were solubilized by treating the cell envelopes with 2% (wt/vol) sodium lauryl sarcosinate in 10 mM HEPES (pH 7.4). The insoluble fraction was collected by centrifugation, resuspended in 50 mM Tris-HCl (pH 8) containing 2% sodium deoxycholate, 200 mM sodium chloride, and 5 mM EDTA, centrifuged again, and washed once in the same buffer. OmpP2 was then solubilized from the pellet in 0.4% (wt/vol) Zwittergent Z-3,14 buffered with 25 mM imidazole at pH 6.5. The insoluble fraction was removed by centrifugation. The crude OmpP2 was collected from the supernatant by precipitation with ethanol at −20°C, followed by centrifugation at 1,700 × g for 30 min. The proteins were solubilized in 50 mM Tris-HCl (pH 8) containing 0.05% Zwittergent Z-3,14. OmpP2 was further purified by high-pressure liquid chromatography with a MonoQ HR 5/5 column (42). The salt concentration of the elution buffer (0.05% Zwittergent Z-3,14 in 50 mM Tris-HCl [pH 8]) was raised to 0.35 M with a 2-ml gradient. When no more protein was eluted with 0.35 M NaCl as detected by UV absorption, the salt concentration was raised in a gradient fashion to 1.0 M. Highly purified OmpP2 eluted with between 0.4 and 0.7 M NaCl. It was collected by precipitation with ethanol at −20°C, followed by centrifugation at 1,700 × g for 30 min. Purified protein was solubilized in 50 mM Tris-HCl (pH 8)–0.1% Zwittergent Z-3,14. The amount of protein was estimated with the dye-binding reagent of Bio-Rad; the purity of OmpP2 was assessed by SDS–11% PAGE (26), followed by Coomassie blue staining for proteins or by SDS–16% PAGE (41) followed by silver staining (46) for lipopolysaccharide. Purified H. influenzae lipopolysaccharide was prepared as described previously (50) and was used as a standard.
Pore function measurement.
Channel conductance was measured by the method described by Dahan et al. (9). Two Teflon chambers were separated by a thin Teflon foil with a small circular aperture. Lipid bilayers were formed across this aperture with a solution of 2.5% glyceryl monooleate (Sigma) dissolved in n-decane. The porin sample was added to the aqueous phase either before or after membrane formation. The conductance across the membrane was measured by determination of a fixed transmembrane potential.
Penicillin-binding assay.
The penicillin-binding assay method described by Mendelman et al. (27, 28) was used to test the binding of [3H]penicillin to exponentially growing or stationary-phase cells. Stationary-phase bacteria were obtained by cultivating bacteria on chocolate agar plates overnight, followed by resuspension in 5 ml of 50 mM Tris-HCl–1 mM MgCl2 (pH 7.8) to an optical density at 600 nm of 0.6. The bacteria were centrifuged and concentrated 10-fold in the same buffer. Logarithmic-phase bacteria were obtained by growing the cells in brain heart infusion broth supplemented with 10 μg of both X and V factors per ml until an optical density at 600 nm of 0.7 was achieved. The bacteria were spun down and resuspended in 0.5 ml of the Tris-HCl–MgCl2 buffer. For each bacterial suspension, 95 μl was incubated with 5 μl of [3H]penicillin (22 ng/ml, 44.5 μCi/μg; Amersham) for 30 min at 37°C. Penicillin-binding proteins were analyzed by SDS-PAGE, followed by autoradiography.
Nucleotide sequence accession numbers.
The nucleotide sequence data can be retrieved from the EMBL/Genbank/DDBJ Nucleotide Sequence Databases under the accession nos. AF052541 to AF052553.
RESULTS
Antibiotic susceptibilities of H. influenzae isolates.
Isolates (total = 247) were obtained from sputum samples from 39 CF patients, and 16 isolates produced β-lactamase. By the disk diffusion method for susceptibility testing, 47 (20.3%) of the 231 non-β-lactamase producing isolates showed intermediate or high levels of resistance to one or more of the following clinically useful β-lactam antibiotics: ampicillin, amoxicillin-clavulanic acid, cefuroxime, cefotaxime, cefaclor, and imipenem. On the basis of the results of Omp and RAPD analyses, the 47 isolates included 4 strains and 13 OmpP2 variants. Thirty additional isolates of these four strains were obtained at intervals of at least 1 month following initial isolation. OmpP2 variants that differed in their susceptibilities to more than one of the β-lactam antibiotics were isolated from only 3 of the 39 patients (patients 27, 28, and 30). Patient 27 was treated with β-lactam antibiotics for 12 weeks and with non-β-lactam antibiotics for 54 weeks. Patient 28 was treated with β-lactam antibiotics, mainly amoxicillin, for 2 years, and patient 30 was treated with β-lactam antibiotics for 3 months. The H. influenzae isolates from these patients included OmpP2 variants of strains 67, 69, 70, and 77. Isolates from each of these series had different OmpP2 proteins.
To determine whether differences in antibiotic susceptibility coincided with OmpP2 variation, H. influenzae isolates from patients 27, 28, and 30 were selected for further susceptibility testing and OmpP2 analysis. In addition, two related OmpP2 variants (82e and 82g) from patient 32, who was not treated with antibiotics during the study period, were included as controls. H. influenzae 70a, 70b, 70c, 70e, and 70f isolated from sputum specimens from patient 28 and H. influenzae isolates 77a, 77b, 77c, and 77f isolated from samples from patient 30 differed in their susceptibilities to at least two of the tested β-lactam antibiotics and to chloramphenicol. Two genotypically as well as phenotypically different isolates from sputum samples from patient 27 (isolates 67d and 69a) differed in their susceptibilities to ampicillin, penicillin, and cephalothin. Control OmpP2 variants 82e and 82g from CF patient 32 were similar with respect to their antibiotic susceptibilities.
The antibiotic susceptibilities of all 13 isolates were further analyzed by the more precise MIC microdilution method and are summarized in Table 1. Differences in the MICs of at least two of the three β-lactam antibiotics (ampicillin, penicillin, and cephalothin) were observed between OmpP2 variants isolated from patients 28 and 30 and between the distinct strains from patient 27. Also, differences in susceptibility to chloramphenicol were found for the H. influenzae isolates obtained from patients 28 and 30. The MICs of all tested antibiotics were similar for the OmpP2 variants of the strain from patient 32, who was not treated with antibiotics. These results show that for isolates from patients extensively treated with antibiotics, OmpP2 variation was associated with fourfold differences in the MICs of a variety of antibiotics including β-lactam and non-β-lactam antibiotics.
TABLE 1.
MICs of ampicillin, penicillin, cephalothin, and chloramphenicol for 13 H. influenzae isolates from four CF patients and antibiotic therapy of the patients
| Patient no. | Isolated H. influenzae straina | MIC (μg/ml)
|
Antibiotic treatment of patientb | |||
|---|---|---|---|---|---|---|
| Ampicillin | Penicillin | Cephalothin | Chloramphenicol | |||
| 27 | 67d | 8 | 3 | 4 | 0.5 | β-Lactams, 12 wk; non-β-lactams, 54 wk |
| 69a | 0.5 | 0.2 | 0.5 | 0.5 | ||
| 28 | 70a | 8 | 3 | 2 | 0.06 | β-Lactams, 104 wk; non-β-lactams, 11 wk |
| 70b | 8 | 3 | 2 | 0.5 | ||
| 70c | 4 | 3 | NDc | ND | ||
| 70e | 8 | 3 | 2 | 0.25 | ||
| 70f | 1 | 0.75 | 1 | 0.125 | ||
| 30 | 77b | 2 | 1.5 | 10 | 1 | β-Lactams, 13 wk; non-β-lactams, 5 wk |
| 77a | 4 | 1.5 | 2 | 2 | ||
| 77c | 4 | 1.5 | 4 | 1 | ||
| 77f | 1 | 0.75 | 1 | 0.5 | ||
| 32 | 82e | 0.125 | 0.1 | 0.25 | 0.125 | None |
| 82g | 0.25 | 0.2 | 0.25 | 0.25 | ||
The numbers indicate distinct strains that vary in their Omp and RAPD patterns; letters indicate the variants (identical RAPD patterns and different Omp compositions).
Number of weeks that the patients were treated with β-lactam antibiotics or with non-β-lactam antibiotics during the period that the strains were isolated.
ND, not done.
Pore properties of purified OmpP2.
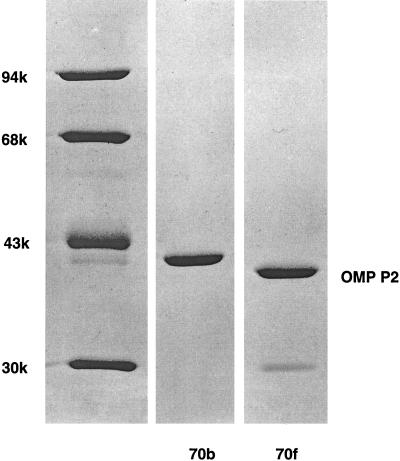
Since the greatest intrastrain differences in antibiotic susceptibility occurred between isolates 70b and 70f, we purified the OmpP2 proteins from these two isolates (Fig. 1) and subjected them to pore function analysis. The OmpP2 purification procedure described by Munson et al. (32) was followed by chromatography on a MonoQ column. A protein of 40 kDa was obtained from both strain 70b and strain 70f (Fig. 1). Densitometric scanning of silver-stained gels revealed that lipopolysaccharide contamination was 5 and 7% for the two protein samples, respectively.
FIG. 1.
Analysis of porins from variants strains H. influenzae 70b and 70f. A Coomassie brilliant blue-stained SDS-polyacrylamide gel is shown. The left contains a low-molecular-weight marker (in thousands, as indicated); lane 70b, 2 μg of protein of strain 70b; lane 70f, 1.5 μg of protein of strain 70f.
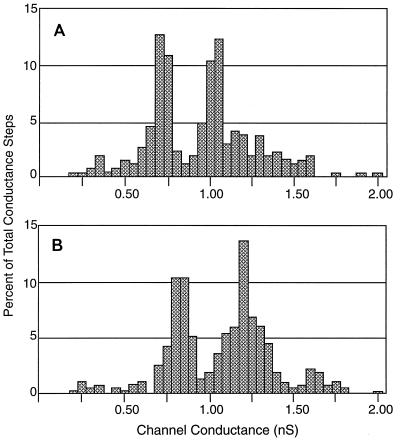
The channel properties of the OmpP2 preparations from variants 70b and 70f were characterized by determining single-channel conductances. By using approximately 15 ng of the porin and a transmembrane potential of 10 mV, stepwise increases in transmembrane conductance were recorded; this is characteristic for porins spontaneously inserting into the bilayer. The amplitudes of the conductance steps for the porins of variants 70b and 70f were measured, and the data are displayed as histograms (Fig. 2). Analysis of the conductance of purified OmpP2 of variant 70b revealed two channel populations (Fig. 2A): one with a conductance range of 0.65 to 0.75 nS and an average conductance of 0.71 nS and the other with a conductance range of 0.95 to 1.05 nS and an average conductance of 1.01 nS. Analysis of the purified OmpP2 of strain variant 70f showed two channel populations as well (Fig. 2B). However, these two populations had conductances significantly higher than those of their 70b counterparts. One population of channels showed a conductance range of 0.75 to 0.90 nS and an average conductance of 0.83 nS, and the other showed a conductance range of 1.05 to 1.35 nS and an average conductance of 1.20 nS.
FIG. 2.
Comparison of channel conductance as measured in planar bilayers for porins from H. influenzae OmpP2 strain variants 70b (A) and 70f (B). Conductance steps were recorded at a transmembrane potential of 10 mV and with 1 M KCl as the electrolyte. The bilayer was formed with monoolein dissolved in n-decane. The two porins were diluted with 50 mM Tris-HCl (pH 8.0) to 15 ng/μl. Approximately 1 μl of the diluted sample was added to the Teflon chamber. The total number of conductance steps analyzed was 257 for strain variant 70b and 349 for strain variant 70f.
ompP2 DNA and protein sequences.
By DNA sequence analysis of ompP2, differences in the sizes of the OmpP2 proteins of the five H. influenzae strains (different RAPD patterns and different Omp patterns) and their OmpP2 variants (identical RAPD patterns and different Omp patterns) were identified. Isolates showed interstrain diversity in regions of the gene coding for loops 2, 4, 5, 6, 7, and 8 (Fig. 3). The sequence changes that we identified resulted in substitutions, insertions, or deletions of amino acids. Sequence changes in loops 5 and 6 were apparently not related to antibiotic treatment, since the loops of the two variants of strain 82, isolated from a patient who was not treated with antibiotics, showed similar changes. Variations in loops 5 and 6 are known (6, 9) to be characteristic of antigenic variants of OmpP2. The membrane-spanning regions showed only a few conservative changes (data not shown). The ompP2 DNA sequences of the H. influenzae strains isolated from patient 27 (strains 67d and 69a) showed as much diversity as those of the H. influenzae strains isolated from different CF patients.
FIG. 3.
Alignment of the deduced amino acid composition of the variable regions of OmpP2 corresponding to surface-exposed loops according to the topology model of OmpP2 of nonencapsulated H. influenzae (3). The OmpP2 sequences of five strains and eight variants of these strains isolated from four CF patients are shown. The amino acid sequences of strain variants 67d, 70a, 77b, and 82e are used as the consensus sequences. The amino acid positions of the loops of strain 67d are indicated above the sequence. Dots indicate identical amino acids, and dashes indicate deletions. Amino acids are given as one-letter codes. Abbreviations: P, patient; S, strain numbers for OmpP2 variants; R, antibiotic-resistant variant; S, antibiotic-susceptible variant.
The five OmpP2 variants of strain 70 also showed amino acid substitutions in the regions corresponding to surface-exposed loops 4, 5, 6, 7, and 8. This variability was apparently unrelated to the antibiotic susceptibility pattern. In addition to these amino acid changes, the susceptible OmpP2 variant 70f showed the substitution Val30→Ile within loop 1 and the substitution Arg46→His at the border between loop 1 and β strand 2; resistant variants 70a, 70b, 70c, and 70e had an arginine in the same position. Variant 70c, which showed only a slight increase in susceptibility, also showed variation in loop 1: Val32→Asp. Among the OmpP2 variants of strain 70, changes in loop 3 were only partly associated with differences in the susceptibilities of these variants to the antibiotics tested. Multiple changes in loop 3 were detected only in variants 70f and 70c. None of the variants of strains 77 or 82 showed variation in this part of the sequence, suggesting that these alterations were not involved in antigenic drift but may influence antibiotic susceptibility.
Composition of penicillin-binding proteins.
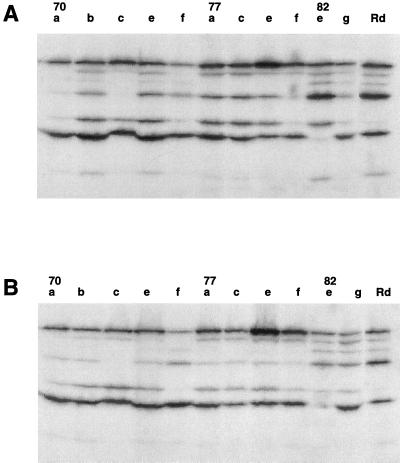
Despite numerous attempts, we did not succeed in transforming variant 70f with ompP2 of variant 70b. This may have been due to the poor growth of the strains and the fact that the difference in MIC was only 6 μg/ml. To exclude the possibility that other factors influenced antibiotic susceptibility, we tested the binding of radiolabeled penicillin to penicillin-binding proteins. H. influenzae 70, including variants of strains 70, 77, and 82, were incubated with 3H-labeled penicillin and analyzed by SDS-PAGE, followed by autoradiography. The results are shown in Fig. 4. H. influenzae Rd was included as control since it was previously shown to express all penicillin-binding proteins, three of which bind ampicillin with high affinities (27, 28).
FIG. 4.
Autoradiogram of the binding of [3H]penicillin to the penicillin-binding proteins of strains Rd, 70, 77, and 82, including OmpP2 variants (indicated by the lowercase letters), after the bacteria in the exponential (A) and stationary (B) phases of growth were harvested. The proteins were analyzed by SDS–11% PAGE before autoradiography.
Although the penicillin-binding protein binding patterns of OmpP2 variants 70a, 70b, 70c, 70e, and 70f were not identical, differences between the patterns were not correlated with differences in penicillin or ampicillin susceptibility. Although strain 70f was more susceptible to ampicillin than strain 70b, several penicillin-binding proteins of strain 70f showed reduced penicillin binding activity. A similar variation in the penicillin-binding pattern was observed for OmpP2 variants of strain 77. Similar penicillin-binding protein profiles were observed for exponentially growing and stationary-phase bacterial cultures, indicating that changes in the growth phase of the bacteria did not affect penicillin binding.
DISCUSSION
Of all β-lactamase-negative H. influenzae isolates examined in this study, one-fifth had reduced susceptibility to one or more β-lactam antibiotics. This frequency is within the range of resistance previously reported by others (11, 20, 22, 32, 38, 39). While OmpP2 variants were isolated from 23 of 40 patients (58%), only a minority of the variants showed reduced susceptibility to various broad-spectrum antibiotics (29). Furthermore, these resistant variants were isolated from only a small percentage (8%) of the CF patients. We previously reported (30) that among H. influenzae isolates from CF patients, there was no strong correlation between OmpP2 variation and antibiotic susceptibility. The isolated OmpP2 variants that differed in broad-spectrum antibiotic susceptibility were infrequently observed. As documented in this report, they were isolated only from patients extensively treated with antibiotics (Table 1). The disk diffusion test and the microdilution antibiotic susceptibility tests may not be sufficiently sensitive to detect differences in susceptibility that are attributable to differences in the diffusion rate of antibiotics through porins. However, E. coli mutants with increased permeability through the OmpF porin were identified by a similar agar diffusion method (3). Furthermore, Burns et al. (5) used the agar dilution test for MIC determinations and demonstrated reduced amounts of OmpP2 in H. influenzae isolates and a corresponding reduced susceptibility to chloramphenicol and β-lactam antibiotics. They also showed that these mutants had a lower outer membrane permeability for β-lactam antibiotics. In the microdilution test, differences in growth rate between resistant and susceptible strains could not have affected the susceptibility test readings since the test results were read after 24 h and again after another 18 h.
Our results suggest that the differences in the broad-spectrum antibiotic susceptibilities of H. influenzae strains from patients extensively treated with antibiotics can be correlated with changes in OmpP2 sequence and in the channel properties of these porins (Fig. 2 and 3). We cannot exclude the possibility that other determinants of antibiotic susceptibility may be changed in these variants as well. Changes in specific targets for the antibiotics are very unlikely, since the variants showed altered susceptibilities to a variety of unrelated antibiotics. The penicillin-binding proteins of the resistant and susceptible isolates obtained from patient 30 indeed varied in their reactivities with ampicillin, the drug administered to the patient (Fig. 4). However, since the susceptible strain showed less binding than the more resistant one, the resistance is not caused by reduced antibiotic binding to the penicillin-binding proteins. In addition, similar variations in binding to penicillin-binding proteins was observed among strains with identical susceptibilities.
The surface-exposed loops 2, 4, 5, 6, 7, and 8 of the ompP2 sequences of unrelated H. influenzae strains isolated from persistently infected CF patients differed predominantly (Fig. 3). Loops 2, 4, 5, and 6 of OmpP2 also showed strong diversity among H. influenzae isolates from sputum samples from patients with chronic bronchitis, and loop 8 showed diversity among isolates from patients with otitis media (12). The most extensive diversity, including amino acid insertions, was found in loop 6. Since the strong diversity in loop 6 has now been demonstrated for H. influenzae isolates that persist in both CF and chronic bronchitis patients, the variation in loop 6 may therefore be associated with the persistence of H. influenzae in the inflamed lower respiratory tract. Such variation may be a result of strong selective pressure, which is important for escape from antibody-mediated defense mechanisms of the host (13, 14, 17, 53). The sequences of the ompP2 genes of H. influenzae variants isolated from CF patients differed (Fig. 3) by single point mutations and often by one or more codon insertions, mainly in loops 4, 5, and 6. In isolates from CF patients, loop 6 was the most variable part of OmpP2 and was the longest loop, a result that matched the information for H. influenzae variants from chronic bronchitis patients. According to the secondary-structure models of Srikumar et al. (44) and Bell et al. (2), the amino acid substitutions may be positioned at the tip of the loop. Variation in loops 5 and 6 resembled strongly the antigenic variations observed among OmpP2 variants of H. influenzae isolates from chronic bronchitis patients (13, 14). In comparison with isolates from chronic bronchitis patients, the OmpP2 variants from CF patients had many more amino acid changes in loops 5 and 6 and also in several other loops (loops 1, 3, 4, 7, and 8) of OmpP2. Amino acid changes in OmpP2 loops 1, 3, 7, and 8 observed among variants isolated from sputum samples from CF patients were rarely found among H. influenzae variants from chronic bronchitis patients (13, 14).
Various considerations support the idea that some of these changes may influence the porin channel and consequently the susceptibility to β-lactam antibiotics (4, 6, 35). H. influenzae variants of strain 70 isolated from CF patient 28, who had been extensively treated with β-lactam antibiotics, showed substitutions in loop 3. According to the topological models of OmpP2 in the outer membrane (2, 44), the third surface-exposed loop folds into the channel and plays an essential role in determining cross-sectional diameter (43, 45, 52). Changes in loop 1 occurred only among the OmpP2 variants that differed in their antibiotic susceptibilities to the three tested β-lactam antibiotics as well as to chloramphenicol. Because Benson et al. (3) showed that changes in the amino acid sequences of porins were correlated with differences in uptake through the outer membrane, the variations in loop 1 may have affected pore function. In support of this hypothesis, channel conductance measurements showed significantly lower conductance values for the OmpP2 porin from the more resistant variant (variant 70b) than for the porin from the more susceptible variant (variant 70f) (Fig. 3). We propose that the smaller channel conductance of H. influenzae variant 70b OmpP2 could explain its lower susceptibility to β-lactam antibiotics and chloramphenicol compared to that of variant 70f.
Channel conductance measurements of OmpP2 purified from variants 70b and 70f revealed two channel populations (Fig. 3). An effect of lipopolysaccharide can be excluded since the amounts of lipopolysaccharide were low and lipopolysaccharide was not required for the pores to be functional (8). Contamination of the OmpP2 porin preparations (Fig. 1) with another pore-forming protein is an unlikely explanation for this phenomenon, because both channel populations of the porin from variant 70b showed a 14 to 16% smaller conductance than those of the corresponding populations of porin from variant 70f. It would be a very unlikely coincidence for two different proteins to change so that their conductances are altered to the same degree. In addition, the amount of protein used for the conductance measurements is low, and therefore, the concentration of the contaminants is almost negligible. Other explanations for the bimodal distribution merit consideration: the two populations of channel conductances may represent different porin conformational states, or the two channel populations may have been arisen from different subunit associations.
We conclude that the majority of variations observed in loops 4, 5, and 6 from OmpP2 proteins from strains from of CF patients likely cause antigenic differences and are not related to differences in pore function (of OmpP2). The results of channel conductance measurements, the amino acid analysis of OmpP2 variants, and the observed differences in antibiotic susceptibilities of the variants (Table 1) suggest that in some isolates OmpP2 variation might be responsible for the observed alteration in broad-spectrum antibiotic susceptibility.
Sequence analysis of the ompP2 (OmpP2) genes of H. influenzae isolates from CF patients showed that nearly all amino acid changes in the protein were, as in isolates from chronic bronchitis patients (13), the result of nonsynonymous point mutations. This observation indicates that variants are appearing under strong selective pressure. Changes in loops 5 and 6 have been associated with antigenic drift as a consequence of immune selection (13, 14). The changes in loops 1 and 3 may have been selected by the antibiotic therapy of the CF patient. The sequence variation in H. influenzae during persistent infections in CF patients was more extensive than that in H. influenzae during persistent infections in chronic bronchitis patients. This increased variability is most likely the consequence of the observed extensive persistence of the infection in CF patients (29), since persistence is sufficient for the appearance of variants (53). The very viscous mucus produced as a consequence of the chloride channel defect in CF patients (16) may provide an environment where H. influenzae is sheltered from both immune defense mechanisms and the bactericidal effect of antibiotics.
ACKNOWLEDGMENTS
Annette G. Regelink and Peter van Ulsen were funded by the National Asthma Foundation (grants 92.28 and 96.50). David Dahan was the recipient of a fellowship from the Fonds pour la Formation de Chercheurs et l’Aide à la Recherche (FCAR), Quebec, Quebec, Canada. James W. Coulton is the recipient of grant support (MT-14133) from the Medical Research Council of Canada.
REFERENCES
- 1.Barry A L, Jorgensen J H, Hardy D J, Allen S D, Baker C N. Haemophilus influenzae ATCC 49766, an alternative quality control strain for monitoring broth microdilution susceptibility tests with selected β-lactams. J Clin Microbiol. 1992;30:2033–2037. doi: 10.1128/jcm.30.8.2033-2037.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell J, Grass S, Jeanteur D, Munson R S., Jr Diversity of the P2 protein among nontypeable Haemophilus influenzae isolates. Infect Immun. 1994;62:2639–2643. doi: 10.1128/iai.62.6.2639-2643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson S A, Occi J L T, Sampson B A. Mutations that alter the pore function of the OmpF porin of Escherichia coli K12. J Mol Biol. 1988;203:961–970. doi: 10.1016/0022-2836(88)90121-0. [DOI] [PubMed] [Google Scholar]
- 4.Burns J L, Smith A L. A major outer-membrane protein functions as a porin in Haemophilus influenzae. J Gen Microbiol. 1987;133:1273–1277. doi: 10.1099/00221287-133-5-1273. [DOI] [PubMed] [Google Scholar]
- 5.Burns J L, Mendelman P M, Levy J, Stull T L, Smith A L. A permeability barrier as a mechanism of chloramphenicol resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1985;27:46–54. doi: 10.1128/aac.27.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulton J W, Mason P, Dorrance D. The permeability barrier of Haemophilus influenzae type b against β-lactam antibiotics. J Antimicrob Chemother. 1983;12:435–449. doi: 10.1093/jac/12.5.435. [DOI] [PubMed] [Google Scholar]
- 7.Cowan S W. Bacterial porins: lessons from three high-resolution structures. Curr Opin Struct Biol. 1993;3:501–507. [Google Scholar]
- 8.Dahan D, Srikumar R, Laprade R, Coulton J W. Purification and refolding of recombinant Haemophilus influenzae type b porin produced in Bacillus subtilis. FEBS Lett. 1996;392:304–308. doi: 10.1016/0014-5793(96)00841-1. [DOI] [PubMed] [Google Scholar]
- 9.Dahan D, Vachon V, Laprade R, Coulton J W. Voltage gating of porins from Haemophilus influenzae type b. Biochim Biophys Acta. 1994;1189:204–211. doi: 10.1016/0005-2736(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 10.Doern G V, Gerlach E H, Jorgensen J H, Murray P R, Thornsberry C, Washington J A., Jr Quality control limits for disk diffusion and microdilution susceptibility tests with Haemophilus test medium. Diagn Microbiol Infect Dis. 1991;14:485–493. doi: 10.1016/0732-8893(91)90004-y. [DOI] [PubMed] [Google Scholar]
- 11.Doern G V, Jorgensen J H, Thornsberry C, Preston D A, Tubert T, Redding J S, Maher L A. National collaborative study of the prevalence of antimicrobial resistance among clinical isolates of Haemophilus influenzae. Antimicrob Agents Chemother. 1988;32:180–185. doi: 10.1128/aac.32.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duim B, Dankert J, Jansen H M, van Alphen L. Genetic analysis of the diversity in outer membrane protein P2 of non-encapsulated Haemophilus influenzae. Microb Pathog. 1993;14:451–462. doi: 10.1006/mpat.1993.1044. [DOI] [PubMed] [Google Scholar]
- 13.Duim B, van Alphen L, Eijk P, Jansen H M, Dankert J. Antigenic drift of non-encapsulated Haemophilus influenzae major outer membrane protein P2 in patients with chronic bronchitis is caused by point mutations. Mol Microbiol. 1994;11:1181–1189. doi: 10.1111/j.1365-2958.1994.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 14.Duim B, Vogel L, Puijk W, Jansen H M, Meloen R H, Dankert J, van Alphen L. Fine mapping of outer membrane protein P2 antigenic sites varying during persistent infection of nonencapsulated Haemophilus influenzae. Infect Immun. 1996;64:4673–4679. doi: 10.1128/iai.64.11.4673-4679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forbes J K, Bruce K D, Ball A, Pennington T H. Variation in length and sequence of porin (OmpP2) alleles of non-capsulate Haemophilus influenzae. Mol Microbiol. 1992;6:2107–2112. doi: 10.1111/j.1365-2958.1992.tb01384.x. [DOI] [PubMed] [Google Scholar]
- 16.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groeneveld K, van Alphen L, Eijk P P, Jansen H M, Zanen H C. Changes in outer membrane proteins of nontypeable Haemophilus influenzae in patients with chronic obstructive pulmonary disease. J Infect Dis. 1988;158:360–365. doi: 10.1093/infdis/158.2.360. [DOI] [PubMed] [Google Scholar]
- 18.Groeneveld K, van Alphen L, Voorter C, Eijk P P, Jansen H M, Zanen H C. Antigenic drift of Haemophilus influenzae in patients with chronic obstructive pulmonary disease. Infect Immun. 1989;57:3038–3044. doi: 10.1128/iai.57.10.3038-3044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haase E M, Yi K, Morse G D, Murphy T F. Mapping of bactericidal epitopes on the P2 porin protein of nontypeable Haemophilus influenzae. Infect Immun. 1994;62:3712–3722. doi: 10.1128/iai.62.9.3712-3722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins D G, Sharp P M. CLUSTAL: a package for performing of multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 21.Hussey G, Hitchcock J, Hanslo D, Coetzee G, van Schalkwyk E, Pitout J, Schaaf H. Serotypes and antimicrobial susceptibility of Haemophilus influenzae. J Antimicrob Chemother. 1994;34:1031–1036. doi: 10.1093/jac/34.6.1031. [DOI] [PubMed] [Google Scholar]
- 22.Janteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 23.Kayser F H, Morenzoni G, Santanam P. The second European collaborative study on the frequency of antimicrobial resistance in Haemophilus influenzae. Eur J Clin Microbiol Infect Dis. 1990;9:810–817. doi: 10.1007/BF01967379. [DOI] [PubMed] [Google Scholar]
- 24.Kilian M. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol. 1976;93:9–62. doi: 10.1099/00221287-93-1-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee E-H, Collatz E, Trias J, Gutmann L. Diffusion of β-lactam antibiotics into proteoliposomes reconstituted with outer membranes of isogenic imipenem-susceptible and -resistant strains of Enterobacter cloacae. J Gen Microbiol. 1992;138:2347–2351. doi: 10.1099/00221287-138-11-2347. [DOI] [PubMed] [Google Scholar]
- 26.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the ‘major outer membrane protein’ of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 27.Mendelman P M, Chaffin D O, Stull T L, Rubens C E, Mack K D, Smith A L. Characterization of non-β-lactamase-mediated ampicillin resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1984;26:235–244. doi: 10.1128/aac.26.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendelman P M, Chaffin D O, Musser J M, de Groot R, Serfass D A, Selander R K. Genotypic and phenotypic diversity among ampicillin-resistant, non-β-lactamase-producing, nontypeable Haemophilus influenzae isolates. Infect Immun. 1987;55:2585–2589. doi: 10.1128/iai.55.11.2585-2589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Möller L V M, Regelink A G, Grasselier H, Dankert J, van Alphen L. Multiple Haemophilus influenzae strains and strain variants coincide in the lower respiratory tract of patients with cystic fibrosis. J Infect Dis. 1995;172:1388–1392. doi: 10.1093/infdis/172.5.1388. [DOI] [PubMed] [Google Scholar]
- 30.Möller, L. V. M., A. G. Regelink, H. Grasselier, L. van Alphen, and J. Dankert. 1998. Antimicrobial susceptibility of persistent Haemophilus influenzae in the respiratory tracts of patients with cystic fibrosis. 42:319–324. [DOI] [PMC free article] [PubMed]
- 31.Möller, L. V. M., I. Rupp, L. van Alphen, J. E. Dankert-Roelse, R. W. Griffioen, P. Braat, H. Grasselier, M. H. F. Wilkinson, V. Fidler, and J. Dankert. Diversity, variability and persistence of Haemophilus influenzae in patients with cystic fibrosis: a study on the relationship with clinical parameters. Submitted for publication.
- 32.Munson R S, Jr, Shenep J L, Barenkamp S J, Granoff D M. Purification and comparison of outer membrane protein P2 from Haemophilus influenzae type b isolates. J Clin Invest. 1983;72:677–684. doi: 10.1172/JCI111017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 35.Nikaido H. Role of permeability barriers in resistance to β-lactam antibiotics. Pharmacol Ther. 1985;27:197–231. doi: 10.1016/0163-7258(85)90069-5. [DOI] [PubMed] [Google Scholar]
- 36.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 37.O’Callighan C H, Morris A, Kirby S M, Shingler A H. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agent Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powell M, McVey D, Kassim M H, Chen H Y, Williams J D. Antimicrobial susceptibility of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella (Branhamella) catarrhalis isolated in the UK from sputa. J Antimicrob Chemother. 1991;28:249–259. doi: 10.1093/jac/28.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Powell M, Fah Y S, Seymour A, Yuan M, Williams J D. Antimicrobial resistance in Haemophilus influenzae from England and Scotland in 1991. J Antimicrob Chemother. 1992;29:547–554. doi: 10.1093/jac/29.5.547. [DOI] [PubMed] [Google Scholar]
- 40.Schultz G E. Porins: general to specific, native to engineered passive pores. Curr Opin Struct Biol. 1996;6:485–490. doi: 10.1016/s0959-440x(96)80113-8. [DOI] [PubMed] [Google Scholar]
- 41.Shägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 42.Srikumar R, Chin A C, Vachon V, Richardson C D, Ratcliffe M J H, Saarinen L, Käyhty H, Mäkelä P H, Coulton J W. Monoclonal antibodies specific to porin of Haemophilus influenzae type b: localization of their cognate epitopes and tests of their biological activities. Mol Microbiol. 1992;6:665–676. doi: 10.1111/j.1365-2958.1992.tb01514.x. [DOI] [PubMed] [Google Scholar]
- 43.Srikumar R, Dahan D, Arhin F F, Tawa P, Diederichs K, Coulton J W. Porins of Haemophilus influenzae type b mutated in loop 3 and loop 4. J Biol Chem. 1997;272:13614–13621. doi: 10.1074/jbc.272.21.13614. [DOI] [PubMed] [Google Scholar]
- 44.Srikumar R, Dahan D, Gras M F, Ratcliffe M J H, van Alphen L, Coulton J W. Antigenic sites on porin of Haemophilus influenzae type b: mapping with synthetic peptides and evaluation of structure predictions. J Bacteriol. 1992;174:4007–4016. doi: 10.1128/jb.174.12.4007-4016.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struyvé M, Visser J, Adriaanse H, Benz R, Tommassen J. Topology of PhoE porin: the ‘eyelet’ region. Mol Microbiol. 1993;7:131–140. doi: 10.1111/j.1365-2958.1993.tb01104.x. [DOI] [PubMed] [Google Scholar]
- 46.Tsai C-M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 47.Vachon V, Lyew D J, Coulton J W. Transmembrane permeability channels across the outer membrane of Haemophilus influenzae type b. J Bacteriol. 1985;162:918–924. doi: 10.1128/jb.162.3.918-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Alphen L, Riemens T, Poolman J, Zanen H C. Characteristics of major outer membrane proteins of Haemophilus influenzae. J Bacteriol. 1983;155:878–885. doi: 10.1128/jb.155.2.878-885.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Alphen L, Riemens T, Zanen H C. Antibody response against outer membrane components of Haemophilus influenzae type b strains in patients with meningitis. FEMS Lett. 1983;18:189–195. [Google Scholar]
- 50.Van Alphen L, Klein M, Geelen-van den Broek L, Riemens T, Eijk P, Kamerlingh J P. Biochemical characterization and world-wide distribution of serologically distinct lipopolysaccharides of Haemophilus influenzae type b. J Infect Dis. 1990;162:659–663. doi: 10.1093/infdis/162.3.659. [DOI] [PubMed] [Google Scholar]
- 51.Van Belkum A, Duim B, Regelink A, Möller L, Quint W, van Alphen L. Genomic DNA fingerprinting of clinical Haemophilus influenzae isolates by polymerase chain reaction (PCR) amplification: comparison with major outer-membrane protein and restriction fragment length polymorphism analysis. J Med Microbiol. 1994;41:63–68. doi: 10.1099/00222615-41-1-63. [DOI] [PubMed] [Google Scholar]
- 52.van der Ley P, de Graaff P, Tommassen J. A comparative study on the phoE genes of three enterobacterial species. Implications for structure-function relationship in a pore forming protein of the outer membrane. Eur J Biochem. 1987;164:469–475. doi: 10.1111/j.1432-1033.1987.tb11080.x. [DOI] [PubMed] [Google Scholar]
- 53.Vogel L, Duim B, Geluk F, Eijk P P, Jansen H M, Dankert J, van Alphen L. Immune selection for antigenic drift of major outer membrane protein P2 of Haemophilus influenzae during persistence in subcutaneous cages in rabbits. Infect Immun. 1996;64:980–986. doi: 10.1128/iai.64.3.980-986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;251:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]