Abstract
Genomic data are important for understanding the origin and evolution of traits. Under the context of rapidly developing of sequencing technologies and more widely available genome sequences, researchers are able to study evolutionary mechanisms of traits via comparative genomic methods. Compared with other vertebrates, bird genomes are relatively small and exhibit conserved synteny with few repetitive elements, which makes them suitable for evolutionary studies. Increasing genomic progress has been reported on the evolution of powered flight, body size variation, beak morphology, plumage colouration, high-elevation colonization, migration, and vocalization. By summarizing previous studies, we demonstrate the genetic bases of trait evolution, highlighting the roles of small-scale sequence variation, genomic structural variation, and changes in gene interaction networks. We suggest that future studies should focus on improving the quality of reference genomes, exploring the evolution of regulatory elements and networks, and combining genomic data with morphological, ecological, behavioural, and developmental biology data.
Keywords: Traits, bird, evolution, comparative genomics, genomic data, phylogenomic
1. INTRODUCTION
In recent decades, the development of genome sequencing technology has greatly enhanced biological research. Traditional phylogenetic reconstructions based on single or few genes have advanced into the phylogenomic era [1, 2]. On this basis, comparative genomics has made considerable progress [3, 4]. Combined with traditional phenotypic-based evolutionary biology, comparison between genomes with resolved evolutionary relationships can help to answer some key questions, such as the origin of and changes in traits, at the molecular level.
Birds, as the most diverse lineages of extant tetrapod vertebrates [5], include more than ten thousand species distributed on all continents and are characterized by complex and diverse morphology and habits [6]. Specifically, the genomes of birds are compact and structurally stable [7, 8], making them irreplaceable models for studying the dynamic changes in the genome during the evolutionary process. Since the first bird genome was published in 2004 (domestic chicken (Gallus gallus)) [9], more than 500 genomes of birds have been released (data from www.ncbi.nlm.nih.gov) with 267 genomes contributed by phase II of the Bird 10,000 Genomes (B10K) project [10, 11]. The ongoing B10K project aims to sequence all extant bird species to analyse the evolutionary history and biodiversity patterns of birds from a comprehensive genomic perspective [12], further reflecting the significance of genomic data for bird trait evolutionary studies.
Profiting from the accumulation of genomic data in recent years, much progress has been made in the fields of comparative genomics about avian trait evolution at different evolutionary scales and degrees of postnatal effects. In this review, we first introduce the features of the bird genome and then briefly review the progress of seven relatively thoroughly studied avian traits in comparative genomics. Furthermore, we summarize the genetic mechanisms of trait evolution, including small-scale sequence variation, genomic structural variation, and changes in gene interaction networks. Finally, we propose future research directions and summarize required approaches.
2. CHARACTERISTICS OF AVIAN GENOME
Compared with other animal groups, the genome of birds is generally small, ranging from 1 Gb to 2.1 Gb [13]. It is hypothesized that smaller genomes are conducive to reducing metabolic costs, which is generally an adaptation to flight [14-17]. Evidence supporting this hypothesis is that birds and bats, representing the only extant vertebrate groups that evolved powered flight, both have smaller genomes [13, 16]. Additionally, the genome size of birds has been shown to be related to the metabolic consumption of flight movement. For example, hummingbirds have the highest metabolism and the smallest genome, while ratite birds have the largest genome but the least flight ability [13]. Although studies have shown that genome shrinkage may have occurred before the origin of powered flight [15], there is numeric evidence indicating that the bird genome has decreased during the evolution of flight [17, 18]. In addition, the range of variation in genome size within bird groups is relatively narrow (for example, the genome size of mammals ranges from 1.6 G to 6.3 G) [13]. Some hypotheses suggest that the smaller genome size variation within birds is due to less DNA acquisition and loss during bird evolution [19, 20]. In contrast, a recent study has shown that DNA acquisition by transposon amplification and DNA loss by deletions are widespread during the evolution of birds, and the two processes are in dynamic balance, maintaining the relatively constant size of the bird genome [13].
Limited by its size, the composition of the bird genome is characterized by fewer repetitive elements [15, 21], a low proportion of non-coding regions, and a large amount of gene loss [21]. Although it is difficult to accurately measure the proportion of repetitive sequences of bird genomes due to the limitations of short read sequencing [22, 23], the proportion of repetitive sequences in birds (5%-31%) is estimated to be far less than that in mammals (35%-52%) [11, 21, 24]. Protein-coding genes in birds are usually shorter, and the proportion of non-coding regions is lower (only half of that in mammals) [15, 21], suggesting that the genomes of birds are more condensed. This denser gene distribution may be mainly caused by shorter introns and intergenic regions [21]. In addition, there are large segmental deletions in the avian genome, leading to the loss of many coding genes [21].
Chromosomal evolution is slow in birds, which often leads to the conservation of gene order [25]. High synteny and gene order conservation can even be observed between passerine birds and chickens, which diverged approximately 88 million years ago [2], making it possible to map genome sequences of a bird species to another distantly related species [26, 27]. It is assumed that non-allelic homologous recombination (NAHR) occurring in repetitive regions may cause chromosomal rearrangement [24, 28]. The proportion of repeats in the avian genome is relatively small, as a result, the probability of occurrence of NAHR tends to be low [25]. However, a genomic comparison between chicken and zebra finch shows that evolutionary breakpoint regions (where contiguous syntenic chromosomal segments show different ancestry [29], a sign of chromosome rearrangement) are not related to recombination hotspots [30]. This dispute may come from the limitation of assembly quality in repeated regions [31]. Compared with the total number of bird species, the number of chromosome-level bird genomes is relatively low. More high-quality assemblies are needed to effectively analyse the structural changes in avian genomes during evolution [31]. To date, the B10K Reference Genome Project has submitted 37 high-quality bird genomes to NCBI (Accession: PRJNA489244).
In summary, avian genomes are characterized by small genome size, compact gene organization, and stable structure, which make them suitable for studying the genetic mechanisms of trait evolution by comparative genomics.
3. RESEARCH PROGRESS ON THE GENETIC BASES OF BIRD TRAIT EVOLUTION
In recent years, much progress has been made in understanding the evolution of bird traits. Studies of avian-specific traits cover a wide range and represent substantial progress. To systematically introduce the progress of comparative genomics in this field, only several well-studied traits were reviewed. These traits are related specifically to birds and have sustained and in-depth research results throughout the history of evolutionary biology. However, it is worth noting that different traits have different evolutionary scales and are affected by environmental changes to varying degrees. The traits introduced here are organized by the scale of evolution and the degree of genetic impact, including powered flight, body size variation, beak morphology, plumage colouration, high-elevation colonization, migration, and vocalization. Powered flight and body size variation are key characteristics of birds, the evolution of which runs through the whole bird lineage. Studies of these large-scale morphological characteristics help to promote our understanding of genomic changes during macroevolution. Beak morphology, plumage colouration, and high-elevation colonization usually have lineage specificity and are more likely than other traits to be determined by genetic factors. These traits are usually related to selection pressure and reflect the adaptative characteristics of species, knowledge of which has important application potential in agriculture and animal husbandry. Compared with the abovementioned traits, migration and vocalization are behavioural traits and are much more plastic. Research on the evolution of bird behaviour provides a feasible model for studying the evolution of complex behaviours and cognitive activities, as well as their related genomic bases.
3.1. Powered Flight
Among vertebrates, birds and bats independently evolved the ability of powered flight [32], which improves their competence in searching for suitable habitats, avoiding predator attacks, and foraging [33]. For flight adaptation, a series of changes have taken place in the morphology and anatomy of these taxa, including feathers, body shape, miniaturization of size, lengthening of forelimbs, the shape of wings, hollow and healing bones, etc. [32-34]. Some of these traits may show evolutionary trends before the origin of flight [35], but their roles in flight adaptation are beyond doubt.
By combining morphological evolution and phylogeny, Puttick et al. (2014) found that body size is evolving towards miniaturization, leading to an increase in relative forelimb length and an improvement in flight ability. In addition, birds have evolved unique hollow air-filled bones to reduce their body weight [36]. The bone-associated genes of birds and bats were identified, with many genes related to bone metabolism, bone fusion, and muscle development, being positively selected in the two lineages [33]. The statistical character of the genome is also associated with flight. The negative correlation between flight ability (measured by flight muscle size and heart index) and genome size indicates that smaller genomes are associated with stronger flyers [18].
Phylogenetic studies have shown that there are many independent flight capacity loss events in birds, which occur not only in Palaeognathae [37, 38], but also in rails and ducks [39]. These birds share many convergent phenotypic characteristics, including reduced metabolic rates, forelimb shortening, pectoral muscle reduction, keel degeneration, and body size enlargement [40, 41]. By comparing the phylogeny, development, epigenetics, and genomes of 11 paleognathous birds, Sackton et al. (2019) found that the loss of flight ability is closely related to the convergent evolution of regulatory elements in development pathways. Since flight is a process of high energy consumption, the loss of flight ability also affects energy metabolism. For example, convergent amino acid substitutions of ATGL and ACOT7 in flight-degenerate birds lead to a transformation of the main energy source of lipids to carbohydrates [42].
In conclusion, powered flight creates higher requirements for energy metabolism, and probably limits the genome size of birds [18]; genes related to body size, bone development, and metabolism are under selection during the dramatic morphological changes associated with the gain and loss of flight ability [33, 36-38, 40-42]. However, it is believed that flying ability may have multiple parallel origins in dinosaurs [43]. From the perspective of genome evolution, explaining why birds apply current body architecture, whether bird genomes are small because of the constraint of flight, and whether flight ability comes from a single event or multiple steps will enhance our understanding of powered flight evolution.
3.2. Body Size Variation
The body size of birds is affected by many factors. At the macro-evolutionary scale, flight adaptation spurs decreases in body size [32], while the degradation of flight ability induces increases in body size. Temperature may also affect bird body size. According to Bergman's law [44], endotherms living in cold areas tend to be larger to reduce energy dissipation. Statistical studies have shown that most birds (more than 72%) conform to this rule [45]. In addition, body size is also associated with the rate of species differentiation and the intensity of sexual selection; in large-bodied birds, high degrees of polygyny and rapid molecular evolution rates are positively correlated with the rate of diversification [46].
As mentioned above, the active flight process consumes large amounts of energy. The size of the bird genome is limited to reduce the resource consumption caused by replication and other processes [14-17]. A typical inference is that the genomes of hummingbirds (i.e., the smallest birds), which perform energy-intensive hovering flight, should be smaller than those of other bird species [47]. After multiple species comparison, Gregory et al. (2009) confirmed this hypothesis and found that most genome reduction occurred before the diversification of hummingbirds and that there were four unrelated events during which the genome size of hummingbirds increased, which may have been caused by biogeography or demographic history. Through a comparative genomic study, Ji and DeWoody (2016) not only proved a positive correlation between body mass and genome size but also emphasized the role of indels in genome size variation.
Studies based on genome data [48] and microsatellites [49] both confirmed a negative correlation between body size and genomic diversity. The variation in genomic diversity may result from the difference in mutation rate. Gillman et al. (2012) compared the cytochrome b gene across species and found that species with smaller body sizes have a higher mutation rate [50]. This trend is supported by another study based on the motif mismatch patterns of microsatellites, displaying a negative correlation between body size and mutation rate [51].
Because body size is affected by both genetic and non- genetic factors, such as development, physiological metabolism, and environment (e.g., food resources), it is difficult to find genes that specifically control body size. A comparative genomic study of 48 avian species that represent most bird orders detected many genes under positive selection, including AHSG and P2RX7, which are associated with bone development, probably explaining the body size variation among these species [21]. Through an intercross population study, Zhou et al. (2018) identified a regulatory mutation that influences the expression of IGF2BP1, which may lead to an increase in body size and feed efficiency [52].
In summary, body size variation is a relatively complicated trait that is influenced by multiple factors. Some statistical characteristics of the genome are associated with body size variation, including genome size [17], length of indels [17], genome diversity [48, 49], and mutation rate [50, 51]. On the other hand, body mass is also related to organism-level traits and life-history traits, such as metabolic rate and lifespan [53]. To date, genes specifically related to body size are rare in avian genomic research, which may be the result of either a lack of study or the polygenic nature of this trait.
3.3. Beak Morphology
The beak is an important specialized structure of the craniofacial bone of birds, playing an important role in survival and reproductive behaviours such as probing for food, preening, attacking, courtship, and feeding young. The morphology of the beak is highly diversified, as a result of the adaptation to different habits and varied environments. Beak morphology often varies in different dimensions, including length, height, width, and curvature, which has been reported to be controlled by different molecular mechanisms, suggesting that avian beaks are a polygenic trait [54-56]. Previous studies have shown that the expression of BMP4 [57], CALM1 [58], TGFBR2 [59], CTNNB1 [59], DKK3 [59], and some other genes are related to the size and shape of beaks. Since a few years ago, comparative genomic methods have been used to explore the genetic underpinnings of evolutionary variation in beak morphology under strong natural selection. As a result, it is possible to identify related genes or regions of the genome showing high differentiation between populations with different beak morphs and significantly correlating with beak morphology.
Darwin’s finches constitute a classical model in studies of beak evolution. Rands et al. (2013) published the genome assembly of the large ground finch (Geospiza magnirostris) and determined that two genes (IGF2R and POU1F1) related to beak morphology are under positive selection [60]. Through whole-genome re-sequencing analysis of all Darwin’s finches and related species, Lamichhaney et al. (2015) found that a region containing ALX1 is related to the beak diversity of Darwin’s finches, and the transcription factor encoded by this gene affects the process of craniofacial development [61]. Subsequently, a genome scan of medium ground finches (G. fortis), which experienced drought stress from 2004 to 2005, revealed that HMGA2 affects the fitness of individuals by determining beak size, leading to rapid adaptive radiation [62]. In addition to single gene variation, studies based on genomic comparisons proved that the size and shape of the beak are regulated by a set of genes controlling the development process [63].
Studies have shown that the variation in bill length of the great tit (Parus major) in England and Holland is highly related to COL4A5, and the evolution of longer beaks may be an adaptation to artificial feeding [56]. Genome scanning and detection of outlier regions across the entire geographic range of the great tit found that the differentiation of beak-related genes (COL4A5 and BMPR1A) exists in both UK and Scotland populations because of recent natural selection [64].
The ground tit (Pseudopodoces humilis) lives at the highest altitude compared with its relatives in the family Paridae [65, 66]. To adapt to a high plateau terrestrial lifestyle, the beak of the ground tit is long and curved and obviously differs from that of its relatives [54, 55]. A comparative transcriptome study found that the evolution of tit beak morphs is probably controlled by multiple genes that are differentially expressed in embryonic upper beaks [55]. Among these genes, FGF13 and ITGB3 affect the diversification and distribution of osteoblasts and osteoclasts, thus playing an important role in the process of beak development [55]. By comparing the genome of P. humilis with those of other tits, seven candidate genes related to bone development and morphology were found, of which COL27A1 is affected by selective sweeps with a high fixation rate in non-synonymous mutations, indicating that this may be the major gene in controlling beak lengthening in tits [54].
In addition to studies focusing on microevolution, a macroevolutionary study that combines phenotypic data with genomic data revealed that both coding genes and non- coding regions related to BMP and Wnt signalling pathways play a role in the evolution of bird beaks and that regulatory elements may be the major drivers of morphological differentiation on the macroevolutionary scale [67]. Moreover, regulatory changes can also occur at the expression level. Schubert et al. (2012) found that microRNAs differentially expressed in cranial neural crest cells affect facial development and beak morphological variation [68].
These studies demonstrate that 1) genes related to beak development are usually under natural selection and can be identified by a whole genome scan or selective pressure detection; 2) beak morphology is polygenic [69, 70]; 3) candidate genes are usually scattered throughout the genome [69]; 4) the genomic bases of beak variation are species-specific, with a few shared major effect genes and signalling pathways [71] and 5) the roles of non-coding and regulatory elements are not negligible. However, it is necessary to point out that candidate genes detected by in silico studies require subsequent experimental validation. In addition, current results are highly discrete, and how to integrate those results into a more uniform evolutionary model requires further investigation.
3.4. Plumage Colouration
Plumage colouration, as one of the most important avian morphological traits, is influenced by both natural selection and sexual selection [72]. The molecular mechanisms of plumage colouration include the synthesis and distribution of melanins, carotenoids, porphyrins, pterins, polyenes, and structural colours [73]. Limited by technologies, early studies focused on a few target genes, such as MC1R [74-76] and FST [77]. However, these studies are based on coding sequences and whole-body colour variations. The advances of high-throughput sequencing technology enable us to study plumage colour variation through genomic or transcriptomic data [72].
Mutations of key genes in plumage colouration pathways may lead to significant differences. For example, the difference between green and blue colour types in budgerigar (Melopsittacus undulatus) is caused by a single amino acid substitution (R644W) of the polyketide synthase gene (MuPKS), which results in the loss of MuPKS activity and the failure to deposit yellow pigments [78]. The structural variation in supergenes [79] can also cause phenotypic differences. Studies have shown that structural variation in a supergene leads to three different male phenotypes of the ruff (Philomachus pugnax), with obvious differences in behaviour, feather colour and size [80, 81].
By linking genome data with plumage colour variation between species, it is possible to identify genomic regions underlying plumage colour variation between species, which usually contain pigment synthesis genes [72]. Many differentiated regions in the genomes of munia (Lonchura) species contain genes related to colour formation (ASIP, EDN3, IGSF11, KITLG, MC1R, and SOX10), proving that recombination of ancestral genetic variation leads to phenotypic variation [82]. Extreme colour differences among subpopulations in the limestone wren-babbler (Napothera crispifrons) may be caused by a few genomic loci associated with the expression of pigmentation, including RAB3IP and SLC16A3 [83].
In addition, many genetic variations causing plumage colour differences are located in non-coding regions, which indicates that differences in gene expression levels play an important role in the pattern formation of colour patches [72]. For example, Corvus corone and C. corni have obvious differences in plumage colouration [84]. A previous study showed that the coding region of MC1R is not related to the phenotypic differentiation of these two species [85]. After re-sequencing and transcriptome analysis, Poelstra et al. (2014) found that the pigment genes associated with grey and black feather follicles are differentially expressed in these two species, indicating that the feather colour difference may be caused by the difference in gene expression regulation [86]. Similarly, feather colour variation between dark-eyed juncos (Junco hyemalis) is mainly related to the differential expression of ASIP, MFSD12, KCNJ13, and HAND2 [87]. By comparing the genomes of nine sympatric capuchino seedeater (Sporophila) species, it was found that the melanogenesis pathway and its putative regulatory regions are under divergent selection, which contributes to species differentiation [88]. Moreover, a backcrossing-based genomic and transcriptomic study proved that differential expression of BCO2 caused by cis-regulatory element variation is related to sexual dichromatism in canaries [89, 90]. In addition to differences in expression quantity and space-time variation, the splicing process after transcription may also lead to different final expression products. Genomic and transcriptomic analysis proved that differences in alternative splicing results in differences in feather colour between two Chrysolophus species [91].
Collectively, among sister species or subspecies, whole- body colour differences are relatively rare, while most colour variations have a patch-specific style, indicating that changes in expression may play a major role [72]. On the other hand, major changes in key genes or at the genomic level may drive colour differentiation on a larger evolutionary scale. Nevertheless, plumage colour differentiation is only a part of the process of species differentiation; it may be difficult to distinguish genes specifically responsible for plumage colour variation at the macroevolution scale. Another puzzling question is the relationship between plumage colouration and speciation. As one of the main targets of sexual selection, differentiation in colour may drive reproductive isolation, which subsequently leads to speciation. Evidently, the species boundary between hybridizing species is sustained by a few differential islands related to colouration in the genome, with a shallowly differentiated background in crows and warblers [86, 92]. However, the role of differentiation islands has been questioned recently. A growing number of studies have proven that genomic islands may also be generated from neutral demographic processes [93] or recombination [94, 95], challenging traditional ideas and methods. Therefore, the causal relationship between plumage colour differentiation and speciation is an important issue in the studies.
3.5. High-elevation Colonization
High mountains and plateaus are characterized by extreme environmental factors such as low oxygen levels, low temperatures and strong ultraviolet radiation [65], forcing species to evolve unique adaptive characteristics. In addition, studying the adaptation of different species to similar extreme environments will improve the understanding of genetic convergence and the influence of evolutionary history [96]. Compared with other vertebrates, birds have higher metabolic requirements due to their flight ability; thus, they have to face unique challenges in high-elevation colonization. As a result, high-elevation colonization by birds is one of the hot spots in comparative genomics, and much progress has been made in this area in recent decades.
Through comparative genomic analysis for species endemic to high elevations, genes and pathways under natural selection can be discovered intuitively. By comparing the genome of the ground tit, the great tit, the yellow-cheeked tit (Machlolophus spilonotus), and the Mongolian ground jay (Podoces hendersoni), Qu et al. (2013) found a series of genomic changes in adaptation to a high-altitude environment, including elevated genes related to the hypoxia response and energy metabolism. In Tibetan chickens, some genes involved in the calcium signalling pathway are under selection [97]; nevertheless, these genes are different from those in ground tits, suggesting that similar adaptive traits may have different genetic bases among species. Both Tibetan chickens and buff-throated partridges (Tetraophasis szechenyii) have an enhanced DNA damage repair system [98, 99], which may be an adaptation to high ultraviolet radiation. In addition, the ground tit and the buff-throated partridge show a loss of immune- and olfactory-related gene families, possibly due to the lower risk of infection in the relatively homogenized environment of the plateau where they are distributed [65, 99].
Comparisons of different species pairs at high and low altitudes help to identify convergent changes in the process of high-elevation adaptation. Studies have shown that the haemoglobin-O2 affinity of high-altitude Passeriformes is stronger than that of low-altitude relatives, which may be caused by similar functional effects that are attributed to different amino acid substitutions [96] or recurrent non-synchronous substitutions produced by mutation hotspots in the genome [100]. Comparative transcriptomic studies have revealed that high-altitude species also share similar changes in gene expression in lung, cardiac muscle, kidney, liver, and flight muscle [101, 102]. From the perspective of energy metabolism, colonization history affects adaptation strategies; species with a short colonization history to a high plateau tend to have improved glucose utilization, while species with longer colonization history tend to have improved fatty acid biosynthesis and oxidation [102].
Mitochondria are directly related to energy metabolism, and their variation may be associated with the genetic mechanism of high-elevation adaptation. Compared with low-altitude geese, bar-headed geese (Anser indicus) are adapted to high-altitude flight, with a significant difference in the activity of cytochrome c oxidase as a result of functional changes caused by non-synonymous substitution [103]. Gu et al. (2016) compared the mitochondrial genomes of alpine pheasants with those of low-altitude Phasianidae species and found that ATP6 and ND6 are associated with high-altitude adaptation [104].
These studies illustrate that high-elevation colonization in birds is affected by both natural selection and evolutionary history. The extreme environment in highlands forms intense selection pressure on living organisms. Consequently, a wide range of genes are under selection, including genes involved in the hypoxia response, energy metabolism, DNA repair, and the circulatory system [65, 96-99, 103]; in contrast, genes involved in olfactory perception and the immune system tend to degenerate in this environment due to relaxed selection [65, 99]. Evolutionary history influences high-elevation adaptation in two ways. First, adaptation can occur in a lineage-specific style. During the process of colonization, species with divergent genetic backgrounds undergo different changes in similar genes or pathways [96, 102]. Second, colonization time affects the extent to which organisms can adapt to the environment. Differences in the time under selection pressure lead to divergent adaptive evolution in metabolic strategies [102]. In conclusion, high-elevation colonization is the result of both selection pressure and the impact of evolutionary history of the species. How to tease apart the roles of these two factors will be a key question in future studies.
3.6. Migration
Migration, an individual’s adaptation to seasonal changes in the environment, is one of the most wonderful spectacles in nature and is undertaken by a variety of animals [105]. Migration requires morphological, physiological, and behavioural adaptations, some of which have been shown to be highly heritable, yet the underlying genetic basis of this process remains unclear [106]. With the innovation of tracking technology, especially the continuous miniaturization of trackers (e.g., geolocators) [107], the migration routes and migration strategies of a wider range of avian groups have been discovered. Moreover, the development of sequencing technology, such as the reduced-representation genome sequencing [108], has advanced the study of the genetic basis underlying bird migration [109].
Migration behaviour differentiation is often associated with genetic differentiation [109]. Biogeographic models show that different migration behaviours can promote the evolution of new species, and new migration directions lead to genetic and phenotypic differentiation [110, 111]. Differences in migration routes and overwintering places and temporal breeding segregation may be associated with sympatric speciation [112]. A study using a microarray compared the differentially expressed genes of two willow warbler subspecies (Phylloscopus trochilus trochilus and P. t. acredula) with different migratory strategies; the results showed that the differentially expressed genes (e.g., ADCYAP1) were linked to neuronal firing and neuronal synapse formation [106]. Swainson’s thrushes (Catharus ustulatus) include two subspecies with different migration routes [113]. By integrating light-level geolocators and next-generation sequencing, Delmore et al. (2016) identified a region on chromosome 4 strongly associated with migratory orientation, which includes genes involved with the circadian clock, nervous system, and cell signalling. Another comprehensive study also illustrated that ADCY8, which is associated with long-term memory, may contributes to the populational divergence by migratory routes and distance in peregrine falcons (Falco peregrinus) [114]. Golden-winged warblers (Vermivora chrysoptera) and blue-winged warblers (V. cyanoptera) have different plumage colouration and wintering areas [115]. An analysis of whole-genome re-sequencing data identified a single gene (VPS13A) associated with distinct differences in their wintering areas [115]. It is speculated that VPS13A may be associated with removing reactive oxygen species during migration [115].
In addition to complete migration, partial migration is widely present in birds. Partial migration refers to migrants and residents coexisting within a population, which provides opportunities to understand the underlying genetic mechanisms of migration, especially in populations with different breeding areas [116]. A comparative transcriptomic study of partially migratory populations of European blackbirds (Turdus merula) identified 4 differentially expressed genes between migrants and residents, which are related to hyperphagia, moulting and enhanced DNA replication and transcription [117]. In addition, other candidate genes that may be related to migratory behaviour have been identified by genomic studies of different species. For instance, DRD4 and CRY1 are both found in blackcaps (Sylvia atricapilla) and European blackbirds [117, 118].
Seasonal migration is based on a series of morphological, sensory, and physiological adaptations [119]. Current studies have identified several candidate genes associated with migration. However, the function of these genes has not been verified. In addition, birds have different migratory strategies and mechanisms, and the genetic basis of migration may also be lineage specific. Current research on migratory birds mainly concentrates on a single species, lacking integration of different species and migratory strategies. Moreover, migration studies are easily influenced by the perspective of the northern hemisphere [120], while the origin of bird migration remains controversial. Combining migration tracking methods with omics methods as well as other types of data will help us to further understand the migration of birds.
3.7. Vocalization
Vocalization is one of the main ways for birds to communicate, playing an important role in individual identification, group cooperation, sexual selection, and vigilance [121]. Different species have different abilities to learn and remember songs. Current studies have found that the vocal learning ability evolved independently in the vocal learners, including songbirds, parrots, and hummingbirds [122]. These species have specialized vocal motor learning brain regions that are absent in the vocal non-learning species [122-124]. Vocal learning includes two processes, called sensory learning and sensorimotor learning, that are controlled by specific neural pathways, including primary and secondary auditory cortical regions, vocal motor areas, and a cortical basal ganglia-thalamic loop [125, 126]. Genomic mechanisms underlying vocalization in birds (especially songbirds) can also promote our understanding of the evolution of cognitive ability [121, 126].
In early studies, genes related to sound stimulation and learning were mainly identified by comparisons of expression level [127-129]. The results showed that ZENK (synonyms of four mammalian orthologous genes, ZIF-268, EGR1, NGFI-A, and KROX24) [129] is regulated in auditory brain areas in response to hearing sounds and vocal areas in response to singing, while FOXP2 is downregulated in Area X during undirected singing [130].
With the accumulation of genomic data, the molecular mechanisms of vocalization evolution are further revealed. Studies have shown that vocalization of birds is affected by hormones and mediated by androgen receptors (AR) and estrogen receptor alpha (ER α) and that the distribution of ER α in vocal control area is species-specific [131]. After species-level comparisons, Frankl-Vilches and Gahr (2018) found that the difference in promoter regions affected the expression of these genes.
The results of transcriptomic comparisons between the zebra finch (Taeniopygia guttata), the double-barred finch (T. bichenovii), and their interspecific first-generation hybrids showed that region-specific transcriptional regulation contributes to the production of species-specific songs [132]. Through an integrated analysis of the brain region gene expression databases, Pfenning et al. (2014) [133] identified hundreds of genes with convergent expression behavior among the vocal learners in birds and humans. Another genome-scale gene expression study illustrated that about 10% of all genes in the zebra finch genome are regulated by singing behavior, with specific genes in specific song nuclei [134].
At the macroevolutionary scale, the diversity of bird songs is also related to the species radiation. Combining the source of singing ability (clade with postnatal songs and clade with innate songs) with phylogeny, Mason et al. (2017) showed that macroevolution and sound evolution are interrelated and that postnatal vocalization evolves faster than innate vocalization [135].
Most of these studies are based on the spatio-temporal expression of target genes [127-129] or transcriptome divergence [132]. In addition, comparative studies of DNA sequences also reveal changes in regulatory elements that affect the expression of target genes [131]. It can be inferred that vocalization in some birds is affected by genetic background, development of neuron system, and vocal learning. The contribution of these factors varies among different species [135]. As a consequence, it may be difficult to eliminate the influence of developmental or postnatal factors and study the effects of genomic variation on vocalization.
3.8. Summary
To summarize, comparative genomic studies of various bird traits are based on two logics. Through the comparison of genomic data at a large evolutionary scale, changes in key traits in the evolution of bird lineages are revealed. On the other hand, comparisons between sister species or among populations illustrate small-scale evolutionary changes under specific selection pressure. The choice of data and methods are dependent on the evolutionary scale and the degree of effect of postnatal factors. Integrating the above research progress and results from other vertebrate lineage studies, we next summarize the potential mechanisms of trait evolution.
4. POTENTIAL MECHANISMS OF TRAIT EVOLUTION
In this review, trait refers to a relatively broad concept, ranging from a major change in morphology (e.g., flight ability) to a change in the catalytic activity of an enzyme with significant effects (e.g., synthesis of melanin). The specific scale of traits depends on the scientific problems studied. Biological phenomena come from multi-level complex networks, where genotypic space maps to phenotypic space through a series of complex relationships and finally determines the fitness landscapes [136]. Consequently, evolution, in terms of changes from ancestry, may occur at any level.
4.1. Small-scale Sequence Variation
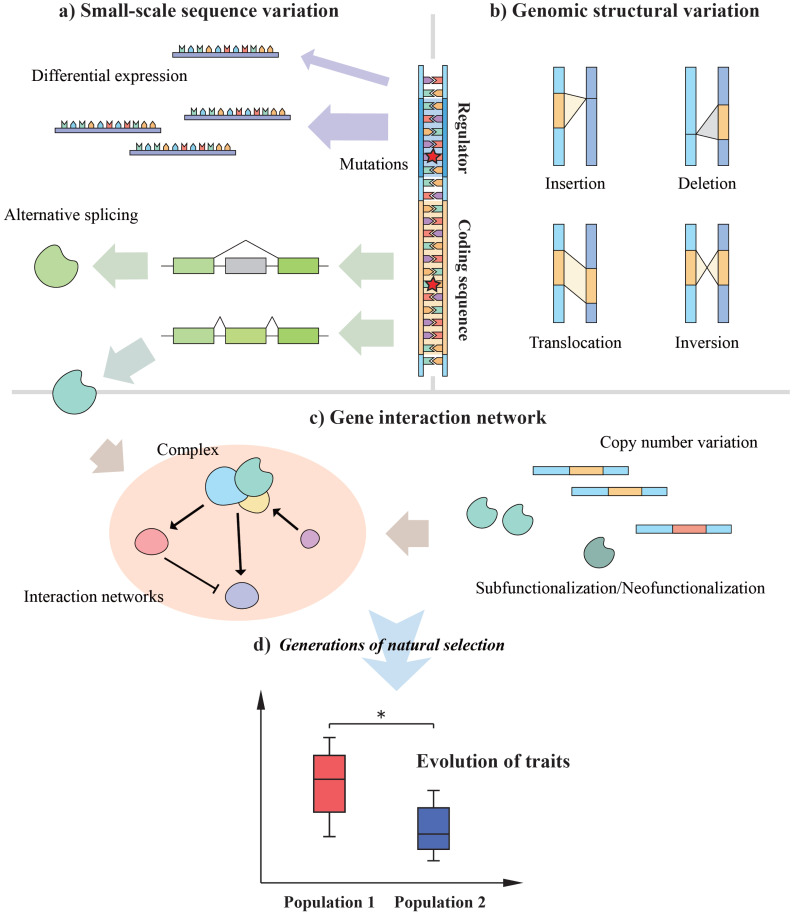
Small-scale variation usually refers to nucleotide substitutions or insertions/deletions with an affected range of less than 1 kilobase [137]. Occurring in the key positions of the genome with specific functions, these mutations may lead to significant changes in traits (Fig. 1a). Based on the molecular functions of mutated regions, they can be subdivided into coding region variations and regulatory region variations.
Fig. (1)(a-d).
Mechanisms of trait evolution, in terms of changes from ancestry. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
If the variation in a coding region is a non-synonymous substitution, it will lead to amino acid substitution, which may greatly affect the properties and functions of protein products. By way of illustration, mutations in MC1R [74-76] and MuPKS [78] lead to feather colour variation. From the whole genome perspective, genes with adaptive mutations, such as COL4A5 [56, 64], BMPR1A [64], and COL27A1 [54], leave positive selection imprints and differentiate significantly among different phylogenetic groups. A whole-genome scan for positive selection may help to identify those genes. For example, an association between variations in XRCC5, PRIMPOL, MDM2, SIRT1 and owls’ adaptation to ultraviolet radiation was identified through genomic scanning [138]. Another possibility is that the variation does not directly cause a change in expression products, but affects the expression time and position, isoform type or expression quantity from the regulation level. This variation may come from cis-acting elements close to key genes [131, 139, 140] or from alternative splicing [91]. A growing number of studies have proven that variation in traits may have regulatory change bases; for example, the evolution of sexual dimorphism may arise from sex differences in patterns of splicing [141], and FOXP2 upregulation during times of song plasticity is essential for song learning [128].
In addition, transcriptome sequencing studies have directly proved a correlation between gene expression differences and trait differences [55, 86, 87, 101, 102, 132]. Comparative transcriptome analysis is helpful to quickly and intuitively identify the genes related to the evolution of traits. However, the results may be influenced by random factors and individual differences [142]. To further elucidate the molecular mechanism of differential expression and its evolutionary significance, comparative genomics studies are required.
4.2. Genomic Structural Variation
It is necessary to keep in mind that genes do not exist in the form of isolated units in the genome. Functionally related genes may be close to each other and share the same regulatory system to form “supergenes” [79]. In addition, the order of genes on chromosomes may also affect phenotypes through temporal differences in expression (e.g., Hox gene family [143]). Therefore, variations in genomic structure, including translocation, inversion, insertion, and deletion of large fragments, can also have a significant impact on traits (Fig. 1b).
The insertion and deletion of large fragments affect the existence or total number of specific genes. The condensed and contracted nature of bird genomes indicates that a large number of genes are lost during the process of evolution; this loss may be lethal or severely reduce fitness in mammals [144]. For instance, SLC2A4 in the solute carrier family encodes an insulin-sensitive glucose transporter with essential physiological functions in glucose homeostasis and is completely lost in the avian lineage as a result of chromosome rearrangements [144]. Moreover, a deletion affecting PITX1, together with ectopic expression of TBX5, contributes to the parallel evolution of foot feathering in chickens and pigeons [145, 146]. Inversion and translocation affect the regulatory network of gene expression by disrupting the location of related functional elements [147]. A study of the differentiation between carrion crows and hooded crows proved that an inversion in chromosome 18 leads to the formation of a genetically differentiated island, which contains genes (RGS9, PRCKA, CACNG4, and CACNG1) related to feather colour formation, causing the morphological differentiation of the two species [86].
Compared with small-scale changes, genomic structural variation involves a wider range of genes and more complex regulatory mechanisms [148], which can explain the large-scale variations in traits and the common changes in multiple associated traits. However, research on genome structure variation usually requires highly contiguous assemblies [149, 150]. At present, to include more species in evolutionary studies, some comparative genomic studies adopt the strategy of re-sequencing. This method maps reads obtained from sequencing to a few specific reference genomes, which leads to a loss of information on structural variation [151, 152]. However, with the development of technology, the cost of chromosome-level de novo sequencing will be further reduced in the future, and large-scale genome structure variation studies will become possible [151].
4.3. Gene Interaction Network
In addition to the evolution of traits caused by single gene mutations and structural variation, changes arising from gene interaction networks can also affect the phenotype of organisms (Fig. 1c). Studies of convergent evolution have proven that repeated adaptive changes may occur not in specific genes, but in the same metabolic pathways [101, 102, 153]. As described above, to improve the oxygen carrying capacity of haemoglobin during the process of adaptation to hypoxic stress at high-elevations, different birds have variations in different genes or mutation sites [96, 153], but these variants occur in similar pathways and are located in the same gene interaction network [101]. In addition, many traits are polygenic at the molecular level, such as the resistance to gapeworm in the house sparrow (Passer domesticus) [154].
Changes in a few key nodes in gene networks may dramatically affect traits. For example, mutations occurring in genes with developmental input-output functions may cause major evolutionary innovations at the morphological level [155]. In the context of avian evolution, changes in regulatory elements of key developmental genes may play an important role in the process of flight loss among paleognathous birds [40]. Apart from mutation events, gene loss events may affect functionally related downstream genes, leading to significant phenotypic changes. However, a study focusing on the loss of urate oxidase (UOX) activity has illustrated that this hypothesis stands for hominoids, while birds and reptiles reserve the UOX locus and some downstream genes, indicating that those genes either have evolved new functions or are kept for alternative functions [156].
Changes in the topological structure of the gene interaction network may also affect the phenotype of organisms. The expression of interacting genes usually stays proportional through the dosage effect [157], but this balance can be disturbed by duplication or loss events that are ubiquitous through genome evolution. According to the ‘2R’ hypothesis, whole genome duplication (WGD) occurred twice during the origin of tetrapods, supplying additional genetic materials for evolutionary innovation and laying a foundation for the blooming of the lineage [158]. However, due to the limitation of genome size, no WGD event has been reported specific to avian lineages to date; in contrast, duplications happen on a smaller scale with only a few genes involved [159]. When a specific gene is duplicated, two genes with similar functions are often differentiated, which changes the original gene network. A case of subfunctionalization was reported in fish, where duplicated MITF genes degenerated to different exons [160]. Moreover, subfunctionalization may also occur at the expression level. After an expansion in birds, different β-keratin paralogues recruited distinct regulators to diverge in the time and tissue of expression, generating structural differences in feathers, beaks, scales, and claws [161]. On the other hand, when duplicated genes are lost, paralogous genes in the genome may provide functional compensation [162], minimizing the functional change caused by gene loss events, as mentioned for the evolution of glucose transporters in birds [144].
5. PERSPECTIVES
Thanks to the development of comparative genomics, researchers have gained a deeper understanding of the molecular mechanisms underlying bird trait evolution. However, new questions and challenges emerge with this achievement. We suggest that future research should consider three aspects: 1) improving data quality, 2) analysing non-coding regions and networks, and 3) combining evolutionary studies with developmental biology.
Data quality is one of the most important factors to determine the reliability of comparative genomic analysis. Different assembly strategies will generate significant variation in both genome completeness and annotation [163]. In addition, complex repeats and haplotype heterozygosity may cause assembly errors [31]. Low-quality reference genomes may lead to questionable conclusions in comparative genomic studies. For example, the presence or absence of specific genes in a genome plays a key role in the analysis of gene family expansion and contraction. Through a comparison between previous genome assemblies and more reliable versions generated by the Vertebrate Genomes Project (VGP), Ko et al. (2021) found that false duplicated sequences accounted for 4 to 16% of previous genome assemblies [164]. Another study applying a similar comparison strategy showed that among previous reference genomes, 3 to 11% of genome sequences were lost and 25% to 60% of genes were completely or partially lost [165], which are false losses caused by sequencing and assembly errors. These two studies pointed out that previous genome assemblies have a high rate of both false duplication [164] and missing sequences [165]. Additionally, as mentioned above, the limitation of assembly capacity in repeat regions constrains studies aiming to resolve the role of NAHR in genomic structural evolution. The application of new technologies, including third-generation sequencing [166] and Hi-C [167], may provide feasible solutions [31].
The second direction for future development is in noncoding regions and networks. Early evolutionary studies have focused on the changes in protein-coding genes during the evolution of life, but an increasing number of studies have shown that noncoding regions and other regulatory elements (e.g., microRNAs, lncRNAs, etc.) play an important role in the phenotypic variation in organisms [40]. In addition, duplication and loss events cause great changes to the network and thus influence the evolutionary routine of traits at the morphological level [144, 161]. In genomic studies, changes in non-coding regions and networks are identified through genome comparisons. The association between these changes and phenotypic variation are subsequently proven through statistical methods. However, traditional methods may lead to false positive results [94, 95], and functional verification is necessary for validation.
Finally, the genome determines the initial state of an individual’s life, while spatio-temporal expression of genetic information determines the development of life. It has been proven that developmental interactions may obfuscate mapping from genotype to phenotype [168]; therefore, the combination of genomic information and developmental biology will be an important task in evolutionary biology research and will involve the cross integration of multiple disciplines. A growing number of studies in the field of evolutionary developmental biology will bring new insights into the evolution of bird traits. Emerging single-cell sequencing technology [169] helps to resolve the heterogeneity of gene expression, which is of great significance to understand the dynamic process of how static genetic information is expressed.
CONCLUSION
In summary, much progress has been made in studies of avian-specific trait evolution. The development of comparative genomics further promotes our understanding of the identity and diversity of organisms from the phenotypic level to the molecular level. The improvement of data quality, the expansion of research content, and the elevation from static data to dynamic data will further push forward the development of this subject.
ACKNOWLEDGEMENTS
We thank Yanzhu Ji for the help to improve this manuscript. We also thank editor for the manuscript invitation and the anonymous reviewer for the valuable comments and suggestions.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was funded by the State Key Program of NSFC (3213000355, 31630069 [F.L.], NSFC32020103005 [Y.Q.]), the Strategic Priority Research Program, Chinese Academy of Sciences (XDA19050202 [F.L.]), and the National Science and Technology Major Project (2018ZX10101004 [G.S.]).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Young A.D., Gillung J.P. Phylogenomics - principles, opportunities and pitfalls of big-data phylogenetics. Syst. Entomol. 2020;45(2):225–247. doi: 10.1111/syen.12406. [DOI] [Google Scholar]
- 2.Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T., Suh A., Weber C.C., da Fonseca R.R., Li J., Zhang F., Li H., Zhou L., Narula N., Liu L., Ganapathy G., Boussau B., Bayzid M.S., Zavidovych V., Subramanian S., Gabaldón T., Capella-Gutiérrez S., Huerta-Cepas J., Rekepalli B., Munch K., Schierup M., Lindow B., Warren W.C., Ray D., Green R.E., Bruford M.W., Zhan X., Dixon A., Li S., Li N., Huang Y., Derryberry E.P., Bertelsen M.F., Sheldon F.H., Brumfield R.T., Mello C.V., Lovell P.V., Wirthlin M., Schneider M.P.C., Prosdocimi F., Samaniego J.A., Vargas Velazquez A.M., Alfaro-Núñez A., Campos P.F., Petersen B., Sicheritz-Ponten T., Pas A., Bailey T., Scofield P., Bunce M., Lambert D.M., Zhou Q., Perelman P., Driskell A.C., Shapiro B., Xiong Z., Zeng Y., Liu S., Li Z., Liu B., Wu K., Xiao J., Yinqi X., Zheng Q., Zhang Y., Yang H., Wang J., Smeds L., Rheindt F.E., Braun M., Fjeldsa J., Orlando L., Barker F.K., Jønsson K.A., Johnson W., Koepfli K-P., O’Brien S., Haussler D., Ryder O.A., Rahbek C., Willerslev E., Graves G.R., Glenn T.C., McCormack J., Burt D., Ellegren H., Alström P., Edwards S.V., Stamatakis A., Mindell D.P., Cracraft J., Braun E.L., Warnow T., Jun W., Gilbert M.T.P., Zhang G. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson A.H., Bowers J.E., Burow M.D., Draye X., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Wright R.J. Comparative genomics of plant chromosomes. Plant Cell. 2000;12(9):1523–1540. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardison R.C. Comparative genomics. PLoS Biol. 2003;1(2):E58. doi: 10.1371/journal.pbio.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prum R.O., Berv J.S., Dornburg A., Field D.J., Townsend J.P., Lemmon E.M., Lemmon A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526(7574):569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 6.Gill F., Donsker D. IOC world bird list (v8. 1). doi: 10.14344/ioc.ml. 2018 8. [DOI] [Google Scholar]
- 7.Toews D.P.L., Campagna L., Taylor S.A., Balakrishnan C.N., Baldassarre D.T., Deane-Coe P.E., Harvey M.G., Hooper D.M., Irwin D.E., Judy C.D., Mason N.A., McCormack J.E., McCracken K.G., Oliveros C.H., Safran R.J., Scordato E.S.C., Stryjewski K.F., Tigano A., Uy J.A.C., Winger B.M. Genomic approaches to understanding population divergence and speciation in birds. Auk. 2016;133(1):13–30. doi: 10.1642/AUK-15-51.1. [DOI] [Google Scholar]
- 8.Kapusta A., Suh A. Evolution of bird genomes-a transposon’s- eye view. Ann. N. Y. Acad. Sci. 2017;1389(1):164–185. doi: 10.1111/nyas.13295. [DOI] [PubMed] [Google Scholar]
- 9.International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 10.Jarvis E.D. Perspectives from the avian phylogenomics project: questions that can be answered with sequencing all genomes of a vertebrate Class. Annu. Rev. Anim. Biosci. 2016;4:45–59. doi: 10.1146/annurev-animal-021815-111216. [DOI] [PubMed] [Google Scholar]
- 11.Feng S., Stiller J., Deng Y., Armstrong J., Fang Q., Reeve A.H., Xie D., Chen G., Guo C., Faircloth B.C., Petersen B., Wang Z., Zhou Q., Diekhans M., Chen W., Andreu-Sánchez S., Margaryan A., Howard J.T., Parent C., Pacheco G., Sinding M.S., Puetz L., Cavill E., Ribeiro A.M., Eckhart L., Fjeldså J., Hosner P.A., Brumfield R.T., Christidis L., Bertelsen M.F., Sicheritz-Ponten T., Tietze D.T., Robertson B.C., Song G., Borgia G., Claramunt S., Lovette I.J., Cowen S.J., Njoroge P., Dumbacher J.P., Ryder O.A., Fuchs J., Bunce M., Burt D.W., Cracraft J., Meng G., Hackett S.J., Ryan P.G., Jønsson K.A., Jamieson I.G., da Fonseca R.R., Braun E.L., Houde P., Mirarab S., Suh A., Hansson B., Ponnikas S., Sigeman H., Stervander M., Frandsen P.B., van der Zwan H., van der Sluis R., Visser C., Balakrishnan C.N., Clark A.G., Fitzpatrick J.W., Bowman R., Chen N., Cloutier A., Sackton T.B., Edwards S.V., Foote D.J., Shakya S.B., Sheldon F.H., Vignal A., Soares A.E.R., Shapiro B., González-Solís J., Ferrer-Obiol J., Rozas J., Riutort M., Tigano A., Friesen V., Dalén L., Urrutia A.O., Székely T., Liu Y., Campana M.G., Corvelo A., Fleischer R.C., Rutherford K.M., Gemmell N.J., Dussex N., Mouritsen H., Thiele N., Delmore K., Liedvogel M., Franke A., Hoeppner M.P., Krone O., Fudickar A.M., Milá B., Ketterson E.D., Fidler A.E., Friis G., Parody-Merino A.M., Battley P.F., Cox M.P., Lima N.C.B., Prosdocimi F., Parchman T.L., Schlinger B.A., Loiselle B.A., Blake J.G., Lim H.C., Day L.B., Fuxjager M.J., Baldwin M.W., Braun M.J., Wirthlin M., Dikow R.B., Ryder T.B., Camenisch G., Keller L.F., DaCosta J.M., Hauber M.E., Louder M.I.M., Witt C.C., McGuire J.A., Mudge J., Megna L.C., Carling M.D., Wang B., Taylor S.A., Del-Rio G., Aleixo A., Vasconcelos A.T.R., Mello C.V., Weir J.T., Haussler D., Li Q., Yang H., Wang J., Lei F., Rahbek C., Gilbert M.T.P., Graves G.R., Jarvis E.D., Paten B., Zhang G. Dense sampling of bird diversity increases power of comparative genomics. Nature. 2020;587(7833):252–257. doi: 10.1038/s41586-020-2873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G., Rahbek C., Graves G.R., Lei F., Jarvis E.D., Gilbert M.T. Genomics: Bird sequencing project takes off. Nature. 2015;522(7554):34. doi: 10.1038/522034d. [DOI] [PubMed] [Google Scholar]
- 13.Kapusta A., Suh A., Feschotte C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA. 2017;114(8):E1460–E1469. doi: 10.1073/pnas.1616702114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes A.L., Hughes M.K. Small genomes for better flyers. Nature. 1995;377(6548):391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- 15.Organ C.L., Shedlock A.M., Meade A., Pagel M., Edwards S.V. Origin of avian genome size and structure in non-avian dinosaurs. Nature. 2007;446(7132):180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- 16.Organ C.L., Shedlock A.M. Palaeogenomics of pterosaurs and the evolution of small genome size in flying vertebrates. Biol. Lett. 2009;5(1):47–50. doi: 10.1098/rsbl.2008.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji Y., DeWoody J.A. Relationships among powered flight, metabolic rate, body mass, genome size, and the retrotransposon complement of volant birds. Evol. Biol. 2016;44(2):261–272. doi: 10.1007/s11692-016-9405-4. [DOI] [Google Scholar]
- 18.Wright N.A., Gregory T.R., Witt C.C. Metabolic ‘engines’ of flight drive genome size reduction in birds. Proceedings of the Royal Society B: Biological Sciences. 1779;2014(281):20132780. doi: 10.1098/rspb.2013.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shedlock A.M. Phylogenomic investigation of CR1 LINE diversity in reptiles. Syst. Biol. 2006;55(6):902–911. doi: 10.1080/10635150601091924. [DOI] [PubMed] [Google Scholar]
- 20.Janes D.E., Organ C.L., Fujita M.K., Shedlock A.M., Edwards S.V. Genome evolution in Reptilia, the sister group of mammals. Annu. Rev. Genomics Hum. Genet. 2010;11:239–264. doi: 10.1146/annurev-genom-082509-141646. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G., Li C., Li Q., Li B., Larkin D.M., Lee C., Storz J.F., Antunes A., Greenwold M.J., Meredith R.W., Ödeen A., Cui J., Zhou Q., Xu L., Pan H., Wang Z., Jin L., Zhang P., Hu H., Yang W., Hu J., Xiao J., Yang Z., Liu Y., Xie Q., Yu H., Lian J., Wen P., Zhang F., Li H., Zeng Y., Xiong Z., Liu S., Zhou L., Huang Z., An N., Wang J., Zheng Q., Xiong Y., Wang G., Wang B., Wang J., Fan Y., da Fonseca R.R., Alfaro-Núñez A., Schubert M., Orlando L., Mourier T., Howard J.T., Ganapathy G., Pfenning A., Whitney O., Rivas M.V., Hara E., Smith J., Farré M., Narayan J., Slavov G., Romanov M.N., Borges R., Machado J.P., Khan I., Springer M.S., Gatesy J., Hoffmann F.G., Opazo J.C., Håstad O., Sawyer R.H., Kim H., Kim K.W., Kim H.J., Cho S., Li N., Huang Y., Bruford M.W., Zhan X., Dixon A., Bertelsen M.F., Derryberry E., Warren W., Wilson R.K., Li S., Ray D.A., Green R.E., O’Brien S.J., Griffin D., Johnson W.E., Haussler D., Ryder O.A., Willerslev E., Graves G.R., Alström P., Fjeldså J., Mindell D.P., Edwards S.V., Braun E.L., Rahbek C., Burt D.W., Houde P., Zhang Y., Yang H., Wang J., Jarvis E.D., Gilbert M.T., Wang J., Wang J., Avian Genome Consortium Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346(6215):1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissensteiner M.H., Suh A. Repetitive DNA: the dark matter of avian genomics, in Avian genomics in ecology and evolution. Springer. 2019:93–150. doi: 10.1007/978-3-030-16477-5_5. [DOI] [Google Scholar]
- 23.Peona V., Blom M.P.K., Xu L., Burri R., Sullivan S., Bunikis I., Liachko I., Haryoko T., Jønsson K.A., Zhou Q., Irestedt M., Suh A. Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol. Ecol. Resour. 2021;21(1):263–286. doi: 10.1111/1755-0998.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damas J., O’Connor R.E., Griffin D.K., Larkin D.M. Avian genomics in ecology and evolution. 2019. Avian chromosomal evolution. pp. 69–92. [DOI] [Google Scholar]
- 25.Ellegren H. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol. Evol. 2010;25(5):283–291. doi: 10.1016/j.tree.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D., Song G., Gao B., Cheng Y., Qu Y., Wu S., Shao S., Wu Y., Alström P., Lei F. Genomic differentiation and patterns of gene flow between two long-tailed tit species (Aegithalos). Mol. Ecol. 2017;26(23):6654–6665. doi: 10.1111/mec.14383. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D., Tang L., Cheng Y., Hao Y., Xiong Y., Song G., Qu Y., Rheindt F.E., Alstrom P., Jia C., Lei F. ‘Ghost introgression’ as a cause of deep mitochondrial divergence in a bird species complex. Mol. Biol. Evol. 2019;36(11):2375–2386. doi: 10.1093/molbev/msz170. [DOI] [PubMed] [Google Scholar]
- 28.Lynch M., Walsh B. The origins of genome architecture. MA: Sinauer Associates Sunderland; 2007. p. Vol. 98. [Google Scholar]
- 29.Sankoff D. The where and wherefore of evolutionary breakpoints. J. Biol. 2009;8(7):66. doi: 10.1186/jbiol162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanov M.N., Farré M., Lithgow P.E., Fowler K.E., Skinner B.M., O’Connor R., Fonseka G., Backström N., Matsuda Y., Nishida C., Houde P., Jarvis E.D., Ellegren H., Burt D.W., Larkin D.M., Griffin D.K. Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genomics. 2014;15:1060. doi: 10.1186/1471-2164-15-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhie A., McCarthy S.A., Fedrigo O., Damas J., Formenti G., Koren S., Uliano-Silva M., Chow W., Fungtammasan A., Kim J., Lee C., Ko B.J., Chaisson M., Gedman G.L., Cantin L.J., Thibaud-Nissen F., Haggerty L., Bista I., Smith M., Haase B., Mountcastle J., Winkler S., Paez S., Howard J., Vernes S.C., Lama T.M., Grutzner F., Warren W.C., Balakrishnan C.N., Burt D., George J.M., Biegler M.T., Iorns D., Digby A., Eason D., Robertson B., Edwards T., Wilkinson M., Turner G., Meyer A., Kautt A.F., Franchini P., Detrich H.W., III, Svardal H., Wagner M., Naylor G.J.P., Pippel M., Malinsky M., Mooney M., Simbirsky M., Hannigan B.T., Pesout T., Houck M., Misuraca A., Kingan S.B., Hall R., Kronenberg Z., Sović I., Dunn C., Ning Z., Hastie A., Lee J., Selvaraj S., Green R.E., Putnam N.H., Gut I., Ghurye J., Garrison E., Sims Y., Collins J., Pelan S., Torrance J., Tracey A., Wood J., Dagnew R.E., Guan D., London S.E., Clayton D.F., Mello C.V., Friedrich S.R., Lovell P.V., Osipova E., Al-Ajli F.O., Secomandi S., Kim H., Theofanopoulou C., Hiller M., Zhou Y., Harris R.S., Makova K.D., Medvedev P., Hoffman J., Masterson P., Clark K., Martin F., Howe K., Flicek P., Walenz B.P., Kwak W., Clawson H., Diekhans M., Nassar L., Paten B., Kraus R.H.S., Crawford A.J., Gilbert M.T.P., Zhang G., Venkatesh B., Murphy R.W., Koepfli K-P., Shapiro B., Johnson W.E., Di Palma F., Marques-Bonet T., Teeling E.C., Warnow T., Graves J.M., Ryder O.A., Haussler D., O’Brien S.J., Korlach J., Lewin H.A., Howe K., Myers E.W., Durbin R., Phillippy A.M., Jarvis E.D. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021;592(7856):737–746. doi: 10.1038/s41586-021-03451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puttick M.N., Thomas G.H., Benton M.J. High rates of evolution preceded the origin of birds. Evolution. 2014;68(5):1497–1510. doi: 10.1111/evo.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machado J.P., Johnson W.E., Gilbert M.T., Zhang G., Jarvis E.D., O’Brien S.J., Antunes A. Bone-associated gene evolution and the origin of flight in birds. BMC Genomics. 2016;17(1):371. doi: 10.1186/s12864-016-2681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altshuler D.L., Bahlman J.W., Dakin R., Gaede A.H., Goller B., Lentink D., Segre P.S., Skandalis D.A. The biophysics of bird flight: functional relationships integrate aerodynamics, morphology, kinematics, muscles, and sensors. Can. J. Zool. 2015;93(12):961–975. doi: 10.1139/cjz-2015-0103. [DOI] [Google Scholar]
- 35.Padian K., Chiappe L.M. The origin and early evolution of birds. Biol. Rev. Camb. Philos. Soc. 1998;73(1):1–42. doi: 10.1017/S0006323197005100. [DOI] [Google Scholar]
- 36.Gutzwiller S.C., Su A., O’Connor P.M. Postcranial pneumaticity and bone structure in two clades of neognath birds. Anat. Rec. (Hoboken) 2013;296(6):867–876. doi: 10.1002/ar.22691. [DOI] [PubMed] [Google Scholar]
- 37.Yonezawa T., Segawa T., Mori H., Campos P.F., Hongoh Y., Endo H., Akiyoshi A., Kohno N., Nishida S., Wu J., Jin H., Adachi J., Kishino H., Kurokawa K., Nogi Y., Tanabe H., Mukoyama H., Yoshida K., Rasoamiaramanana A., Yamagishi S., Hayashi Y., Yoshida A., Koike H., Akishinonomiya F., Willerslev E., Hasegawa M. Phylogenomics and morphology of extinct paleognaths reveal the origin and evolution of the ratites. Curr. Biol. 2017;27(1):68–77. doi: 10.1016/j.cub.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 38.Baker A.J., Haddrath O., McPherson J.D., Cloutier A. Genomic support for a moa-tinamou clade and adaptive morphological convergence in flightless ratites. Mol. Biol. Evol. 2014;31(7):1686–1696. doi: 10.1093/molbev/msu153. [DOI] [PubMed] [Google Scholar]
- 39.McNab B.K. Energy conservation and the evolution of flightlessness in birds. Am. Nat. 1994;144(4):628–642. doi: 10.1086/285697. [DOI] [Google Scholar]
- 40.Sackton T.B., Grayson P., Cloutier A., Hu Z., Liu J.S., Wheeler N.E., Gardner P.P., Clarke J.A., Baker A.J., Clamp M., Edwards S.V. Convergent regulatory evolution and loss of flight in paleognathous birds. Science. 2019;364(6435):74–78. doi: 10.1126/science.aat7244. [DOI] [PubMed] [Google Scholar]
- 41.Nesbitt S.J., Clarke J.A. The anatomy and taxonomy of the exquisitely preserved Green River Formation (early Eocene) lithornithids (Aves) and the relationships of Lithornithidae. Bull. Am. Mus. Nat. Hist. 2016;406(406):1–91. doi: 10.1206/0003-0090-406.1.1. [DOI] [Google Scholar]
- 42.Pan S., Lin Y., Liu Q., Duan J., Lin Z., Wang Y., Wang X., Lam S.M., Zou Z., Shui G., Zhang Y., Zhang Z., Zhan X. Convergent genomic signatures of flight loss in birds suggest a switch of main fuel. Nat. Commun. 2019;10(1):2756. doi: 10.1038/s41467-019-10682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M., O’Connor J.K., Xu X., Zhou Z. A new Jurassic scansoriopterygid and the loss of membranous wings in theropod dinosaurs. Nature. 2019;569(7755):256–259. doi: 10.1038/s41586-019-1137-z. [DOI] [PubMed] [Google Scholar]
- 44.Bergmann C. About the relationships between heat conservation and body size of animals. Gött. Stud. 1847;1:595–708. [Google Scholar]
- 45.Meiri S., Dayan T. On the validity of Bergmann’s rule. J. Biogeogr. 2003;30(3):331–351. doi: 10.1046/j.1365-2699.2003.00837.x. [DOI] [Google Scholar]
- 46.Iglesias-Carrasco M., Jennions M.D., Ho S.Y.W., Duchêne D.A. Sexual selection, body mass and molecular evolution interact to predict diversification in birds. Proc. Biol. Sci. 2019;286(1899):20190172. doi: 10.1098/rspb.2019.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory T.R., Andrews C.B., McGuire J.A., Witt C.C. The smallest avian genomes are found in hummingbirds. Proc. Biol. Sci. 2009;276(1674):3753–3757. doi: 10.1098/rspb.2009.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brüniche-Olsen A., Kellner K.F., DeWoody J.A. Island area, body size and demographic history shape genomic diversity in Darwin’s finches and related tanagers. Mol. Ecol. 2019;28(22):4914–4925. doi: 10.1111/mec.15266. [DOI] [PubMed] [Google Scholar]
- 49.Eo S.H., Doyle J.M., DeWoody J.A. Genetic diversity in birds is associated with body mass and habitat type. J. Zool. 2011;283(3):220–226. doi: 10.1111/j.1469-7998.2010.00773.x. [DOI] [Google Scholar]
- 50.Gillman L.N., McCowan L.S.C., Wright S.D. The tempo of genetic evolution in birds: body mass and climate effects. J. Biogeogr. 2012;39(9):1567–1572. doi: 10.1111/j.1365-2699.2012.02730.x. [DOI] [Google Scholar]
- 51.Fan H., Guo W. A genome-wide investigation of microsatellite mismatches and the association with body mass among bird species. PeerJ. 2018;6:e4495. doi: 10.7717/peerj.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z., Li M., Cheng H., Fan W., Yuan Z., Gao Q., Xu Y., Guo Z., Zhang Y., Hu J., Liu H., Liu D., Chen W., Zheng Z., Jiang Y., Wen Z., Liu Y., Chen H., Xie M., Zhang Q., Huang W., Wang W., Hou S., Jiang Y. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018;9(1):2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Healy K., Guillerme T., Finlay S., Kane A., Kelly S.B., McClean D., Kelly D.J., Donohue I., Jackson A.L., Cooper N. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. Biol. Sci. 2014;281(1784):20140298. doi: 10.1098/rspb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Y., Miller M.J., Zhang D., Song G., Jia C., Qu Y., Lei F. Comparative genomics reveals evolution of a beak morphology locus in a high-altitude songbird. Mol. Biol. Evol. 2020;37(10):2983–2988. doi: 10.1093/molbev/msaa157. [DOI] [PubMed] [Google Scholar]
- 55.Cheng Y., Gao B., Wang H., Han N., Shao S., Wu S., Song G., Zhang Y.E., Zhu X., Lu X., Qu Y., Lei F. Evolution of beak morphology in the Ground Tit revealed by comparative transcriptomics. Front. Zool. 2017;14:58. doi: 10.1186/s12983-017-0245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bosse M., Spurgin L.G., Laine V.N., Cole E.F., Firth J.A., Gienapp P., Gosler A.G., McMahon K., Poissant J., Verhagen I., Groenen M.A.M., van Oers K., Sheldon B.C., Visser M.E., Slate J. Recent natural selection causes adaptive evolution of an avian polygenic trait. Science. 2017;358(6361):365–368. doi: 10.1126/science.aal3298. [DOI] [PubMed] [Google Scholar]
- 57.Abzhanov A., Protas M., Grant B.R., Grant P.R., Tabin C.J. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305(5689):1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 58.Abzhanov A., Kuo W.P., Hartmann C., Grant B.R., Grant P.R., Tabin C.J. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442(7102):563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 59.Mallarino R., Grant P.R., Grant B.R., Herrel A., Kuo W.P., Abzhanov A. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proc. Natl. Acad. Sci. USA. 2011;108(10):4057–4062. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rands C.M., Darling A., Fujita M., Kong L., Webster M.T., Clabaut C., Emes R.D., Heger A., Meader S., Hawkins M.B., Eisen M.B., Teiling C., Affourtit J., Boese B., Grant P.R., Grant B.R., Eisen J.A., Abzhanov A., Ponting C.P. Insights into the evolution of Darwin’s finches from comparative analysis of the Geospiza magnirostris genome sequence. BMC Genomics. 2013;14:95. doi: 10.1186/1471-2164-14-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamichhaney S., Berglund J., Almén M.S., Maqbool K., Grabherr M., Martinez-Barrio A., Promerová M., Rubin C.J., Wang C., Zamani N., Grant B.R., Grant P.R., Webster M.T., Andersson L. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 62.Lamichhaney S., Han F., Berglund J., Wang C., Almén M.S., Webster M.T., Grant B.R., Grant P.R., Andersson L. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science. 2016;352(6284):470–474. doi: 10.1126/science.aad8786. [DOI] [PubMed] [Google Scholar]
- 63.Lawson L.P., Petren K. The adaptive genomic landscape of beak morphology in Darwin’s finches. Mol. Ecol. 2017;26(19):4978–4989. doi: 10.1111/mec.14166. [DOI] [PubMed] [Google Scholar]
- 64.Spurgin L.G., Bosse M., Adriaensen F., Albayrak T., Barboutis C., Belda E., Bushuev A., Cecere J.G., Charmantier A., Cichon M., Dingemanse N.J., Doligez B., Eeva T., Erikstad K.E., Fedorov V., Griggio M., Heylen D., Hille S., Hinde C.A., Ivankina E., Kempenaers B., Kerimov A., Krist M., Kvist L., Laine V.N., Mänd R., Matthysen E., Nager R., Nikolov B.P., Norte A.C., Orell M., Ouyang J., Petrova-Dinkova G., Richner H., Rubolini D., Slagsvold T., Tilgar V., Török J., Tschirren B., Vágási C.I., Yuta T., Groenen M.A.M., Visser M.E., van Oers K., Sheldon B.C., Slate J. The great tit HapMap project: a continental-scale analysis of genomic variation in a songbird. Preprint. 2020 doi: 10.1111/1755-0998.13969. [DOI] [PubMed] [Google Scholar]
- 65.Qu Y., Zhao H., Han N., Zhou G., Song G., Gao B., Tian S., Zhang J., Zhang R., Meng X., Zhang Y., Zhang Y., Zhu X., Wang W., Lambert D., Ericson P.G., Subramanian S., Yeung C., Zhu H., Jiang Z., Li R., Lei F. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 66.Jiang Z., Gao B., Lei F., Qu Y. Population genomics reveals that refugial isolation and habitat change lead to incipient speciation in the Ground tit. Zool. Scr. 2019;48(3):277–288. doi: 10.1111/zsc.12340. [DOI] [Google Scholar]
- 67.Yusuf L., Heatley M.C., Palmer J.P.G., Barton H.J., Cooney C.R., Gossmann T.I. Noncoding regions underpin avian bill shape diversification at macroevolutionary scales. Genome Res. 2020;30(4):553–565. doi: 10.1101/gr.255752.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Powder K.E., Ku Y.C., Brugmann S.A., Veile R.A., Renaud N.A., Helms J.A., Lovett M., Lovett M. A cross-species analysis of microRNAs in the developing avian face. PLoS One. 2012;7(4):e35111. doi: 10.1371/journal.pone.0035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaves J.A., Cooper E.A., Hendry A.P., Podos J., De León L.F., Raeymaekers J.A., MacMillan W.O., Uy J.A. Genomic variation at the tips of the adaptive radiation of Darwin’s finches. Mol. Ecol. 2016;25(21):5282–5295. doi: 10.1111/mec.13743. [DOI] [PubMed] [Google Scholar]
- 70.Lundregan S.L., Hagen I.J., Gohli J., Niskanen A.K., Kemppainen P., Ringsby T.H., Kvalnes T., Pärn H., Rønning B., Holand H., Ranke P.S., Båtnes A.S., Selvik L.K., Lien S., Saether B.E., Husby A., Jensen H. Inferences of genetic architecture of bill morphology in house sparrow using a high-density SNP array point to a polygenic basis. Mol. Ecol. 2018;27(17):3498–3514. doi: 10.1111/mec.14811. [DOI] [PubMed] [Google Scholar]
- 71.Mundy N.I. Population genomics fits the bill: genetics of adaptive beak variation in Darwin’s finches. Mol. Ecol. 2016;25(21):5265–5266. doi: 10.1111/mec.13868. [DOI] [PubMed] [Google Scholar]
- 72.Funk E.R., Taylor S.A. High-throughput sequencing is revealing genetic associations with avian plumage color. Auk. 2019;136(4):ukz048. doi: 10.1093/auk/ukz048. [DOI] [Google Scholar]
- 73.McGRAW K.J. Pterins, porphyrins, and psittacofulvins. Bird coloration: mechanisms and measurements. 2006;1:354. [Google Scholar]
- 74.Theron E., Hawkins K., Bermingham E., Ricklefs R.E., Mundy N.I. The molecular basis of an avian plumage polymorphism in the wild: a melanocortin-1-receptor point mutation is perfectly associated with the melanic plumage morph of the bananaquit, Coereba flaveola. Curr. Biol. 2001;11(8):550–557. doi: 10.1016/S0960-9822(01)00158-0. [DOI] [PubMed] [Google Scholar]
- 75.Mundy N.I., Badcock N.S., Hart T., Scribner K., Janssen K., Nadeau N.J. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science. 2004;303(5665):1870–1873. doi: 10.1126/science.1093834. [DOI] [PubMed] [Google Scholar]
- 76.Baião P.C., Schreiber E., Parker P.G. The genetic basis of the plumage polymorphism in red-footed boobies (Sula sula): a melanocortin-1 receptor (MC1R) analysis. J. Hered. 2007;98(4):287–292. doi: 10.1093/jhered/esm030. [DOI] [PubMed] [Google Scholar]
- 77.Lehtonen P.K., Laaksonen T., Artemyev A.V., Belskii E., Berg P.R., Both C., Buggiotti L., Bureš S., Burgess M.D., Bushuev A.V., Krams I., Moreno J., Mägi M., Nord A., Potti J., Ravussin P.A., Sirkiä P.M., Sætre G.P., Winkel W., Primmer C.R. Candidate genes for colour and vision exhibit signals of selection across the pied flycatcher (Ficedula hypoleuca) breeding range. Heredity. 2012;108(4):431–440. doi: 10.1038/hdy.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cooke T.F., Fischer C.R., Wu P., Jiang T.X., Xie K.T., Kuo J., Doctorov E., Zehnder A., Khosla C., Chuong C.M., Bustamante C.D. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars. Cell. 2017;171(2):427–439.e21. doi: 10.1016/j.cell.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson M.J., Jiggins C.D. Supergenes and their role in evolution. Heredity. 2014;113(1):1–8. doi: 10.1038/hdy.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Küpper C., Stocks M., Risse J.E., Dos Remedios N., Farrell L.L., McRae S.B., Morgan T.C., Karlionova N., Pinchuk P., Verkuil Y.I., Kitaysky A.S., Wingfield J.C., Piersma T., Zeng K., Slate J., Blaxter M., Lank D.B., Burke T. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 2016;48(1):79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lamichhaney S., Fan G., Widemo F., Gunnarsson U., Thalmann D.S., Hoeppner M.P., Kerje S., Gustafson U., Shi C., Zhang H., Chen W., Liang X., Huang L., Wang J., Liang E., Wu Q., Lee S.M., Xu X., Höglund J., Liu X., Andersson L. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Genet. 2016;48(1):84–88. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- 82.Stryjewski K.F., Sorenson M.D. Mosaic genome evolution in a recent and rapid avian radiation. Nat. Ecol. Evol. 2017;1(12):1912–1922. doi: 10.1038/s41559-017-0364-7. [DOI] [PubMed] [Google Scholar]
- 83.Gwee C.Y., Lee Q.L., Mahood S.P., Hung Le M., Tizard R., Eiamampai K., Round P.D., Rheindt F.E. The interplay of colour and bioacoustic traits in the differentiation of a Southeast Asian songbird complex. Mol. Ecol. 2020;30(1):297–309. doi: 10.1111/mec.15718. [DOI] [PubMed] [Google Scholar]
- 84.Randler C. Assortative mating of carrion Corvus corone and hooded crows C. cornixin the hybrid zone in eastern Germany. Ardea. 2007;95(1):143–149. doi: 10.5253/078.095.0116. [DOI] [Google Scholar]
- 85.Haas F., Pointer M.A., Saino N., Brodin A., Mundy N.I., Hansson B. An analysis of population genetic differentiation and genotype-phenotype association across the hybrid zone of carrion and hooded crows using microsatellites and MC1R. Mol. Ecol. 2009;18(2):294–305. doi: 10.1111/j.1365-294X.2008.04017.x. [DOI] [PubMed] [Google Scholar]
- 86.Poelstra J.W., Vijay N., Bossu C.M., Lantz H., Ryll B., Müller I., Baglione V., Unneberg P., Wikelski M., Grabherr M.G., Wolf J.B. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344(6190):1410–1414. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- 87.Abolins-Abols M., Kornobis E., Ribeca P., Wakamatsu K., Peterson M.P., Ketterson E.D., Milá B. Differential gene regulation underlies variation in melanic plumage coloration in the dark-eyed junco (Junco hyemalis). Mol. Ecol. 2018;27(22):4501–4515. doi: 10.1111/mec.14878. [DOI] [PubMed] [Google Scholar]
- 88.Campagna L., Repenning M., Silveira L.F., Fontana C.S., Tubaro P.L., Lovette I.J. Repeated divergent selection on pigmentation genes in a rapid finch radiation. Sci. Adv. 2017;3(5):e1602404. doi: 10.1126/sciadv.1602404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gazda M.A., Araújo P.M., Lopes R.J., Toomey M.B., Andrade P., Afonso S., Marques C., Nunes L., Pereira P., Trigo S., Hill G.E., Corbo J.C., Carneiro M. A genetic mechanism for sexual dichromatism in birds. Science. 2020;368(6496):1270–1274. doi: 10.1126/science.aba0803. [DOI] [PubMed] [Google Scholar]
- 90.Chen N. A gene for color differences between sexes. Science. 2020;368(6496):1185–1186. doi: 10.1126/science.abc2242. [DOI] [PubMed] [Google Scholar]
- 91.Gao G., Xu M., Bai C., Yang Y., Li G., Xu J., Wei Z., Min J., Su G., Zhou X., Guo J., Hao Y., Zhang G., Yang X., Xu X., Widelitz R.B., Chuong C.M., Zhang C., Yin J., Zuo Y. Comparative genomics and transcriptomics of Chrysolophus provide insights into the evolution of complex plumage coloration. Gigascience. 2018;7(10):giy113. doi: 10.1093/gigascience/giy113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Toews D.P., Taylor S.A., Vallender R., Brelsford A., Butcher B.G., Messer P.W., Lovette I.J. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr. Biol. 2016;26(17):2313–2318. doi: 10.1016/j.cub.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 93.Hofer T., Foll M., Excoffier L. Evolutionary forces shaping genomic islands of population differentiation in humans. BMC Genomics. 2012;13(1):107. doi: 10.1186/1471-2164-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Booker T.R., Yeaman S., Whitlock M.C. Variation in recombination rate affects detection of outliers in genome scans under neutrality. Mol. Ecol. 2020;29(22):4274–4279. doi: 10.1111/mec.15501. [DOI] [PubMed] [Google Scholar]
- 95.Stevison L.S., McGaugh S.E. It’s time to stop sweeping recombination rate under the genome scan rug. Mol. Ecol. 2020;29(22):4249–4253. doi: 10.1111/mec.15690. [DOI] [PubMed] [Google Scholar]
- 96.Zhu X., Guan Y., Signore A.V., Natarajan C., DuBay S.G., Cheng Y., Han N., Song G., Qu Y., Moriyama H., Hoffmann F.G., Fago A., Lei F., Storz J.F. Divergent and parallel routes of biochemical adaptation in high-altitude passerine birds from the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. USA. 2018;115(8):1865–1870. doi: 10.1073/pnas.1720487115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang M.S., Li Y., Peng M.S., Zhong L., Wang Z.J., Li Q.Y., Tu X.L., Dong Y., Zhu C.L., Wang L., Yang M.M., Wu S.F., Miao Y.W., Liu J.P., Irwin D.M., Wang W., Wu D.D., Zhang Y.P. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 2015;32(7):1880–1889. doi: 10.1093/molbev/msv071. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Q., Gou W., Wang X., Zhang Y., Ma J., Zhang H., Zhang Y., Zhang H. Genome resequencing identifies unique adaptations of Tibetan chickens to hypoxia and high-dose ultraviolet radiation in high-altitude environments. Genome Biol. Evol. 2016;8(3):765–776. doi: 10.1093/gbe/evw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou C., James J.G., Xu Y., Tu H., He X., Wen Q., Price M., Yang N., Wu Y., Ran J., Meng Y., Yue B. Genome-wide analysis sheds light on the high-altitude adaptation of the buff-throated partridge (Tetraophasis szechenyii). Mol. Genet. Genomics. 2020;295(1):31–46. doi: 10.1007/s00438-019-01601-8. [DOI] [PubMed] [Google Scholar]
- 100.Galen S.C., Natarajan C., Moriyama H., Weber R.E., Fago A., Benham P.M., Chavez A.N., Cheviron Z.A., Storz J.F., Witt C.C. Contribution of a mutational hot spot to hemoglobin adaptation in high-altitude Andean house wrens. Proc. Natl. Acad. Sci. USA. 2015;112(45):13958–13963. doi: 10.1073/pnas.1507300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hao Y., Xiong Y., Cheng Y., Song G., Jia C., Qu Y., Lei F. Comparative transcriptomics of 3 high-altitude passerine birds and their low-altitude relatives. Proc. Natl. Acad. Sci. USA. 2019;116(24):11851–11856. doi: 10.1073/pnas.1819657116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiong Y., Hao Y., Cheng Y., Fan L., Song G., Li D., Qu Y., Lei F. Comparative transcriptomic and metabolomic analysis reveals pectoralis highland adaptation across altitudinal songbirds. Preprint. 2020 doi: 10.1111/1749-4877.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scott G.R., Schulte P.M., Egginton S., Scott A.L., Richards J.G., Milsom W.K. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol. Biol. Evol. 2011;28(1):351–363. doi: 10.1093/molbev/msq205. [DOI] [PubMed] [Google Scholar]
- 104.Gu P., Liu W., Yao Y.F., Ni Q.Y., Zhang M.W., Li D.Y., Xu H.L. Evidence of adaptive evolution of alpine pheasants to high-altitude environment from mitogenomic perspective. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 2016;27(1):455–462. doi: 10.3109/19401736.2014.900667. [DOI] [PubMed] [Google Scholar]
- 105.Winger B.M., Auteri G.G., Pegan T.M., Weeks B.C. A long winter for the Red Queen: rethinking the evolution of seasonal migration. Biol. Rev. Camb. Philos. Soc. 2019;94(3):737–752. doi: 10.1111/brv.12476. [DOI] [PubMed] [Google Scholar]
- 106.Boss J., Liedvogel M., Lundberg M., Olsson P., Reischke N., Naurin S., Åkesson S., Hasselquist D., Wright A., Grahn M., Bensch S. Gene expression in the brain of a migratory songbird during breeding and migration. Mov. Ecol. 2016;4(1):4. doi: 10.1186/s40462-016-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bridge E.S., Kelly J.F., Contina A., Gabrielson R.M., MacCurdy R.B., Winkler D.W. Advances in tracking small migratory birds: a technical review of light-level geolocation. J. Field Ornithol. 2013;84(2):121–137. doi: 10.1111/jofo.12011. [DOI] [Google Scholar]
- 108.Davey J.W., Hohenlohe P.A., Etter P.D., Boone J.Q., Catchen J.M., Blaxter M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011;12(7):499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 109.Turbek S.P., Scordato E.S.C., Safran R.J. The role of seasonal migration in population divergence and reproductive isolation. Trends Ecol. Evol. 2018;33(3):164–175. doi: 10.1016/j.tree.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 110.Irwin D.E. Speciation: new migratory direction provides route toward divergence. Curr. Biol. 2009;19(24):R1111–R1113. doi: 10.1016/j.cub.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 111.Delmore K.E., Hübner S., Kane N.C., Schuster R., Andrew R.L., Câmara F., Guigó R., Irwin D.E. Genomic analysis of a migratory divide reveals candidate genes for migration and implicates selective sweeps in generating islands of differentiation. Mol. Ecol. 2015;24(8):1873–1888. doi: 10.1111/mec.13150. [DOI] [PubMed] [Google Scholar]
- 112.Bearhop S., Fiedler W., Furness R.W., Votier S.C., Waldron S., Newton J., Bowen G.J., Berthold P., Farnsworth K. Assortative mating as a mechanism for rapid evolution of a migratory divide. Science. 2005;310(5747):502–504. doi: 10.1126/science.1115661. [DOI] [PubMed] [Google Scholar]
- 113.Delmore K.E., Toews D.P., Germain R.R., Owens G.L., Irwin D.E. The genetics of seasonal migration and plumage color. Curr. Biol. 2016;26(16):2167–2173. doi: 10.1016/j.cub.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 114.Gu Z., Pan S., Lin Z., Hu L., Dai X., Chang J., Xue Y., Su H., Long J., Sun M., Ganusevich S., Sokolov V., Sokolov A., Pokrovsky I., Ji F., Bruford M.W., Dixon A., Zhan X. Climate- driven flyway changes and memory-based long-distance migration. Nature. 2021;591(7849):259–264. doi: 10.1038/s41586-021-03265-0. [DOI] [PubMed] [Google Scholar]
- 115.Toews D.P.L., Taylor S.A., Streby H.M., Kramer G.R., Lovette I.J. Selection on VPS13A linked to migration in a songbird. Proc. Natl. Acad. Sci. USA. 2019;116(37):18272–18274. doi: 10.1073/pnas.1909186116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hegemann A., Fudickar A.M., Nilsson J.A. A physiological perspective on the ecology and evolution of partial migration. J. Ornithol. 2019;160(3):893–905. doi: 10.1007/s10336-019-01648-9. [DOI] [Google Scholar]
- 117.Franchini P., Irisarri I., Fudickar A., Schmidt A., Meyer A., Wikelski M., Partecke J. Animal tracking meets migration genomics: transcriptomic analysis of a partially migratory bird species. Mol. Ecol. 2017;26(12):3204–3216. doi: 10.1111/mec.14108. [DOI] [PubMed] [Google Scholar]
- 118.Ruegg K., Anderson E.C., Boone J., Pouls J., Smith T.B. A role for migration-linked genes and genomic islands in divergence of a songbird. Mol. Ecol. 2014;23(19):4757–4769. doi: 10.1111/mec.12842. [DOI] [PubMed] [Google Scholar]
- 119.Liedvogel M., Akesson S., Bensch S. The genetics of migration on the move. Trends Ecol. Evol. 2011;26(11):561–569. doi: 10.1016/j.tree.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 120.Pulido F. The genetics and evolution of avian migration. Bioscience. 2007;57(2):165–174. doi: 10.1641/B570211. [DOI] [Google Scholar]
- 121.Clayton D.F. The genomics of memory and learning in songbirds. Annu. Rev. Genomics Hum. Genet. 2013;14:45–65. doi: 10.1146/annurev-genom-090711-163809. [DOI] [PubMed] [Google Scholar]
- 122.Matsunaga E., Okanoya K. Evolution and diversity in avian vocal system: an Evo-Devo model from the morphological and behavioral perspectives. Dev. Growth Differ. 2009;51(3):355–367. doi: 10.1111/j.1440-169X.2009.01091.x. [DOI] [PubMed] [Google Scholar]
- 123.Nottebohm F., Kelley D.B., Paton J.A. Connections of vocal control nuclei in the canary telencephalon. J. Comp. Neurol. 1982;207(4):344–357. doi: 10.1002/cne.902070406. [DOI] [PubMed] [Google Scholar]
- 124.Nottebohm F., Stokes T.M., Leonard C.M. Central control of song in the canary, Serinus canarius. J. Comp. Neurol. 1976;165(4):457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 125.Woolley S.C., Sakata J.T. Mechanisms of species diversity in birdsong learning. PLoS Biol. 2019;17(12):e3000555. doi: 10.1371/journal.pbio.3000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brainard M.S., Doupe A.J. What songbirds teach us about learning. Nature. 2002;417(6886):351–358. doi: 10.1038/417351a. [DOI] [PubMed] [Google Scholar]
- 127.Haesler S., Wada K., Nshdejan A., Morrisey E.E., Lints T., Jarvis E.D., Scharff C. FoxP2 expression in avian vocal learners and non-learners. J. Neurosci. 2004;24(13):3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scharff C., Haesler S. An evolutionary perspective on FoxP2: strictly for the birds? Curr. Opin. Neurobiol. 2005;15(6):694–703. doi: 10.1016/j.conb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 129.Mello C.V., Vicario D.S., Clayton D.F. Song presentation induces gene expression in the songbird forebrain. Proc. Natl. Acad. Sci. USA. 1992;89(15):6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Teramitsu I., White S.A. FoxP2 regulation during undirected singing in adult songbirds. J. Neurosci. 2006;26(28):7390–7394. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Frankl-Vilches C., Gahr M. Androgen and estrogen sensitivity of bird song: a comparative view on gene regulatory levels. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2018;204(1):113–126. doi: 10.1007/s00359-017-1236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang H., Sawai A., Toji N., Sugioka R., Shibata Y., Suzuki Y., Ji Y., Hayase S., Akama S., Sese J., Wada K. Transcriptional regulatory divergence underpinning species-specific learned vocalization in songbirds. PLoS Biol. 2019;17(11):e3000476. doi: 10.1371/journal.pbio.3000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pfenning A.R., Hara E., Whitney O., Rivas M.V., Wang R., Roulhac P.L., Howard J.T., Wirthlin M., Lovell P.V., Ganapathy G., Mouncastle J., Moseley M.A., Thompson J.W., Soderblom E.J., Iriki A., Kato M., Gilbert M.T.P., Zhang G., Bakken T., Bongaarts A., Bernard A., Lein E., Mello C.V., Hartemink A.J., Jarvis E.D. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346(6215):1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Whitney O., Pfenning A.R., Howard J.T., Blatti C.A., Liu F., Ward J.M., Wang R., Audet J-N., Kellis M., Mukherjee S., Sinha S., Hartemink A.J., West A.E., Jarvis E.D. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346(6215):1256780. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mason N.A., Burns K.J., Tobias J.A., Claramunt S., Seddon N., Derryberry E.P. Song evolution, speciation, and vocal learning in passerine birds. Evolution. 2017;71(3):786–796. doi: 10.1111/evo.13159. [DOI] [PubMed] [Google Scholar]
- 136.Stadler P.F., Stephens C.R. Landscapes and effective fitness. Comments Theor. Biol. 2003;8(4-5):389–431. doi: 10.1080/08948550302439. [DOI] [Google Scholar]
- 137.Pollex R.L., Hegele R.A. Copy number variation in the human genome and its implications for cardiovascular disease. Circulation. 2007;115(24):3130–3138. doi: 10.1161/CIRCULATIONAHA.106.677591. [DOI] [PubMed] [Google Scholar]
- 138.Zhou C., Jin J., Peng C., Wen Q., Wang G., Wei W., Jiang X., Price M., Cui K., Meng Y., Song Z., Li J., Zhang X., Fan Z., Yue B. Comparative genomics sheds light on the predatory lifestyle of accipitrids and owls. Sci. Rep. 2019;9(1):2249. doi: 10.1038/s41598-019-38680-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Toomey M.B., Marques C.I., Andrade P., Araújo P.M., Sabatino S., Gazda M.A., Afonso S., Lopes R.J., Corbo J.C., Carneiro M. A non-coding region near Follistatin controls head colour polymorphism in the Gouldian finch. Proc. Biol. Sci. 2018;285(1888):285. doi: 10.1098/rspb.2018.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim K.W., Jackson B.C., Zhang H., Toews D.P.L., Taylor S.A., Greig E.I., Lovette I.J., Liu M.M., Davison A., Griffith S.C., Zeng K., Burke T. Genetics and evidence for balancing selection of a sex-linked colour polymorphism in a songbird. Nat. Commun. 2019;10(1):1852. doi: 10.1038/s41467-019-09806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rogers T.F., Palmer D.H., Wright A.E. Sex-specific selection drives the evolution of alternative splicing in birds. Mol. Biol. Evol. 2020;38(2):519–530. doi: 10.1093/molbev/msaa242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bruning O., Rodenburg W., Wackers P.F., van Oostrom C., Jonker M.J., Dekker R.J., Rauwerda H., Ensink W.A., de Vries A., Breit T.M. Confounding factors in the transcriptome analysis of an in-vivo exposure experiment. PLoS One. 2016;11(1):e0145252. doi: 10.1371/journal.pone.0145252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 144.Xiong Y., Lei F. SLC2A12 of SLC2 gene family in bird provides functional compensation for the loss of SLC2A4 gene in other vertebrates. Mol. Biol. Evol. 2021;38(4):1276–1291. doi: 10.1093/molbev/msaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Domyan E.T., Kronenberg Z., Infante C.R., Vickrey A.I., Stringham S.A., Bruders R., Guernsey M.W., Park S., Payne J., Beckstead R.B., Kardon G., Menke D.B., Yandell M., Shapiro M.D. Molecular shifts in limb identity underlie development of feathered feet in two domestic avian species. eLife. 2016;5:e12115. doi: 10.7554/eLife.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bortoluzzi C., Megens H.J., Bosse M., Derks M.F.L., Dibbits B., Laport K., Weigend S., Groenen M.A.M., Crooijmans R.P.M.A. Parallel genetic origin of foot feathering in birds. Mol. Biol. Evol. 2020;37(9):2465–2476. doi: 10.1093/molbev/msaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McBroome J., Liang D., Corbett-Detig R. Fine-scale position effects shape the distribution of inversion breakpoints in Drosophila melanogaster. Genome Biol. Evol. 2020;12(8):1378–1391. doi: 10.1093/gbe/evaa103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chiang C., Scott A.J., Davis J.R., Tsang E.K., Li X., Kim Y., Hadzic T., Damani F.N., Ganel L., Montgomery S.B., Battle A., Conrad D.F., Hall I.M., Hall I.M., GTEx Consortium The impact of structural variation on human gene expression. Nat. Genet. 2017;49(5):692–699. doi: 10.1038/ng.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Peona V., Weissensteiner M.H., Suh A. How complete are “complete” genome assemblies?-An avian perspective. Mol. Ecol. Resour. 2018;18(6):1188–1195. doi: 10.1111/1755-0998.12933. [DOI] [PubMed] [Google Scholar]
- 150.Weissensteiner M.H., Bunikis I., Catalán A., Francoijs K.J., Knief U., Heim W., Peona V., Pophaly S.D., Sedlazeck F.J., Suh A., Warmuth V.M., Wolf J.B.W. Discovery and population genomics of structural variation in a songbird genus. Nat. Commun. 2020;11(1):3403. doi: 10.1038/s41467-020-17195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ho S.S., Urban A.E., Mills R.E. Structural variation in the sequencing era. Nat. Rev. Genet. 2020;21(3):171–189. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huddleston J., Eichler E.E. An incomplete understanding of human genetic variation. Genetics. 2016;202(4):1251–1254. doi: 10.1534/genetics.115.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.ZHu X., Guan Y., Qu Y., David G., Song G., Lei F. Elevational divergence in the great tit complex revealed by major hemoglobin genes. Curr. Zool. 2018;64(4):455–464. doi: 10.1093/cz/zox042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lundregan S.L., Niskanen A.K., Muff S., Holand H., Kvalnes T., Ringsby T.H., Husby A., Jensen H. Resistance to gapeworm parasite has both additive and dominant genetic components in house sparrows, with evolutionary consequences for ability to respond to parasite challenge. Mol. Ecol. 2020;29(20):3812–3829. doi: 10.1111/mec.15491. [DOI] [PubMed] [Google Scholar]
- 155.Garcia H.G., Berrocal A., Kim Y.J., Martini G., Zhao J. Lighting up the central dogma for predictive developmental biology. Gradients and Tissue Patterning. 2020;137:1–35. doi: 10.1016/bs.ctdb.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 156.Keebaugh A.C., Thomas J.W. The evolutionary fate of the genes encoding the purine catabolic enzymes in hominoids, birds, and reptiles. Mol. Biol. Evol. 2010;27(6):1359–1369. doi: 10.1093/molbev/msq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Birchler J.A., Veitia R.A. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dehal P., Boore J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3(10):e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Velová H., Gutowska-Ding M.W., Burt D.W., Vinkler M. Toll- like receptor evolution in birds: gene duplication, pseudogenization, and diversifying selection. Mol. Biol. Evol. 2018;35(9):2170–2184. doi: 10.1093/molbev/msy119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Altschmied J., Delfgaauw J., Wilde B., Duschl J., Bouneau L., Volff J.N., Schartl M. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics. 2002;161(1):259–267. doi: 10.1093/genetics/161.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bhattacharjee M.J., Yu C.P., Lin J.J., Ng C.S., Wang T.Y., Lin H.H., Li W.H. Regulatory divergence among beta-keratin genes during bird evolution. Mol. Biol. Evol. 2016;33(11):2769–2780. doi: 10.1093/molbev/msw165. [DOI] [PubMed] [Google Scholar]
- 162.Diss G., Ascencio D., DeLuna A., Landry C.R. Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J. Exp. Zoolog. B Mol. Dev. Evol. 2014;322(7):488–499. doi: 10.1002/jez.b.22555. [DOI] [PubMed] [Google Scholar]
- 163.Tsuchiya M.T.N., Dikow R.B., Koepfli K.P., Frandsen P.B., Rockwood L.L., Maldonado J.E. Whole genome sequencing of procyonids reveals distinct demographic histories in kinkajou (Potos flavus) and northern raccoon (Procyon lotor). Genome Biol. Evol. 2020;13(1):evaa255. doi: 10.1093/gbe/evaa255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ko B.J., Lee C., Kim J., Rhie A., Yoo D., Howe K., Wood J., Cho S., Brown S., Formenti G., Jarvis E.D., Kim H. Widespread false gene gains caused by duplication errors in genome assemblies. bioRxiv. 2021 doi: 10.1101/2021.04.09.438957. [DOI] [PMC free article] [PubMed]
- 165.Kim J., Lee C., Ko B.J., Yoo D., Won S., Phillippy A., Fedrigo O., Zhang G., Howe K., Wood J., Durbin R., Formenti G., Brown S., Cantin L., Mello C.V., Cho S., Rhie A., Kim H., Jarvis E.D. False gene and chromosome losses affected by assembly and sequence errors. bioRxiv. 2021 doi: 10.1101/2021.04.09.438906. [DOI]
- 166.Xiao T., Zhou W. The third generation sequencing: the advanced approach to genetic diseases. Transl. Pediatr. 2020;9(2):163–173. doi: 10.21037/tp.2020.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Belton J.M., McCord R.P., Gibcus J.H., Naumova N., Zhan Y., Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Conith A.J., Hope S.A., Chhouk B.H., Craig Albertson R. Weak genetic signal for phenotypic integration implicates developmental processes as major regulators of trait covariation. Mol. Ecol. 2021;30(2):464–480. doi: 10.1111/mec.15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nawy T. Single-cell sequencing. Nat. Methods. 2014;11(1):18. doi: 10.1038/nmeth.2771. [DOI] [PubMed] [Google Scholar]