Abstract
With the deepening of our understanding in life science, molecular biology, nanotechnology, optics, electrochemistry and other areas, an increasing number of biosensor design strategies have emerged in recent years, capable of providing potential practical applications for point-of-care (POC) diagnosis in various human diseases. Compared to conventional biosensors, the latest POC biosensor research aims at improving sensor precision, cost-effectiveness and time-consumption, as well as the development of versatile detection strategies to achieve multiplexed analyte detection in a single device and enable rapid diagnosis and high-throughput screening. In this review, various intriguing strategies in the recognition and transduction of POC (from 2018 to 2021) are described in light of recent advances in CRISPR technology, electrochemical biosensing, and optical- or spectra-based biosensing. From the perspective of promoting emerging bioanalytical tools into practical POC detecting and diagnostic applications, we have summarized key advances made in this field in recent years and presented our own perspectives on future POC development and challenges.
POC diagnostics are driven by the rapid advances in CRISPR, electrochemical and optical biosensors. Related emerging strategies are described and discussed from the perspective of facilitating the practical application of biosensors in POC testing.
1. Introduction
The ongoing global outbreak of COVID-19 which started in 2019/2020 has not only panicked billions of people, but also raised public attention about the importance of early detection of infectious diseases.1–3 As a result, many research institutions and companies are striving to develop effective methods to rapidly detect SARS-CoV-2 RNA, antibodies, antigens and viruses in order to help identify and isolate infected patients.1,4,5 Regardless of whether the test is conducted using devices made of colloidal gold nanoparticles (AuNPs) or the polymerase chain reaction (PCR), they are all considered as point-of-care (POC) detection, which is defined as a method that uses fast analysis and accurate diagnostics to produce a clinically relevant output near the patient's bedside, thereby allowing healthcare workers or patients to make clinical decisions by themselves.6–10 Over the past few decades, biosensing technology has made significant progress by taking advantage of a large number of novel materials that serve as particularly useful platforms for a wide range of biosensing applications.11–19 For example, gold nanomaterials were fabricated on Si/Au/Si3N4 chips to detect circulating tumor DNA, proteins and other kinds of biological targets.20 In addition, responsive optical nanomaterials, as one of the most active research fields in nano-optics, could achieve POC detection by allowing biomarkers to be detected with color or spectral changes.16 These newly developed methods outperform conventional assays in terms of sensitivity, selectivity, practicality and most importantly, the potential for POC applications to be realized in less developed and resource-limited areas.21,22
Ideally, a POC system should minimize user operations to eliminate errors, be easy to use, possess the required sensitivity, selectivity and accuracy needed to detect certain clinical biomarkers, be able to generate diagnostic results directly from self-testing, and be inexpensive enough for most families.23,24 The World Health Organization Special Programme for Research and Training in Tropical Diseases (WHO/TDR) has put forward a set of criteria for ideal diagnostic tools which was termed ‘ASSURED’ (Affordable, Sensitive, Specific, User friendly, Rapid and Robust, Equipment free and Deliverable to end users).25,26 These criteria have become the gold standard for the development of biosensors, especially those suitable in POC tests for resource-limited areas. Current mainstream biomolecular quantitative techniques, including mass spectrometry (MS), ELISA, western blot and northern blot, however, are prohibitively expensive, complex, and time-consuming, making them unsuitable for the immediate care needed to assess various human diseases today.8,27–29 In response, POC approaches, including those based on field-effect transistors, fluorescence-based biosensors, surface plasmon resonance (SPR) sensors, and electrochemical biosensors, have emerged as a result of rapid scientific endeavors in the fields of electrochemistry, chemical biology, chemistry and engineering.18,30,31
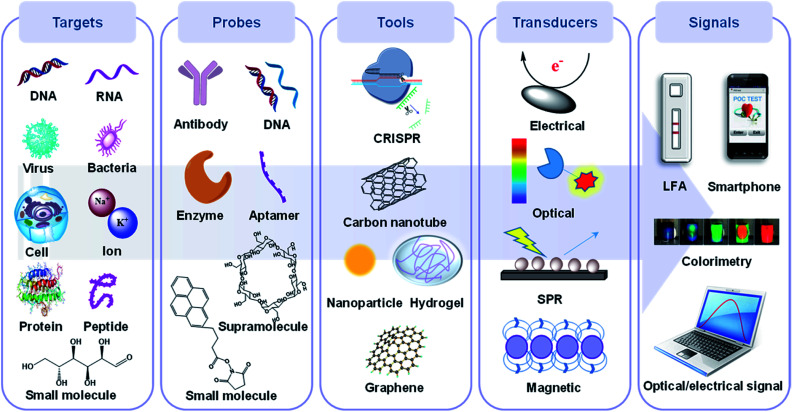
As shown in Fig. 1, a POC device is generally made up of five parts: a target, probe, sensing tool, transducer, and signal readout device, with the first three basic elements being enhanced by recent incredible developments in biomolecular immobilization, signal transduction and device integration.32,33 Briefly, biotargets are first recognized by a probe in a selective manner. Next, this recognition process is translated by a specific transducer into in vitro signals, which are then displayed on a digital platform. Overall, the selectivity of a POC device depends on the first step while the sensitivity is mainly decided by the subsequent steps.34
Fig. 1. The components of a POC device including a potential target, probe, sensing tool and transducer, signal acquisition and processing, and their corresponding elements. Biological targets mainly consist of nucleic acids, whole cells, bacteria/virus particles, proteins and small molecules, while probes usually contain synthetic or natural antibodies, oligonucleotides, proteins and other supra/small molecules. Sensing tools primarily refer to the sensing materials and platforms such as nanomaterials, hydrogels and CRISPR. Transducers convert the probe recognition into signals, which are then displayed on readable devices.
With the rapid development of chemistry, biology, materials science, microelectronics and their interdisciplines in recent years, POC technologies for different diseases are changing with each passing day. More and more methods are being transformed into practical products, such as the lateral flow assay (LFA) for detection of pregnancy and portable devices for detection of diabetes. Among them, DNA/RNA, antibodies, supramolecules, etc. are routinely used as probes to improve the specificity at the detection stage, while colorimetric methods are commonly employed as signal transduction methods to make POC detection more intuitive. In addition to these mature techniques that have been employed in POC detection products, there are also some newly developed strategies which have the potential to be adapted in new generations of POC diagnostics. Herein, we review the progress made in this vibrant research area, with a focus on key advances of potential chemistry-based recognition and transduction technologies for POC that have been developed over the last four years (2018 to 2021), including revolutionary clustered regularly interspaced short palindromic repeats (CRISPR) for the sensing stage and electrochemical/optical-based technologies for the transducing stage of POC strategies. We further classify both the CRISPR- and electrochemical-based technologies by the type of targets, and optical-based biosensors by their mechanisms with enhanced visualization, such as surface enhanced Raman scattering (SERS), SPR, fluorescence, colorimetry and their hybrids. The highlighted work herein focuses primarily on sensing tools, signal transducers and corresponding readout platforms. We will center on the CRISPR-based POC systems in the section on sensing tools because there have been considerable recent efforts aimed at development in this area compared to conventional sensing tools. Whereafter, cutting-edge techniques for POC systems in both electrochemical and optical areas are highlighted as they are the most extensively used transduction methods which have generated majority of innovations and are closest to real POC applications. The objectives of this minireview are: (1) to highlight a number of intriguing POC applications based on the above-mentioned technologies and their recent advancements; (2) to give an overview of their current status towards POC development on the basis of selected innovative research areas; (3) to discuss the drawbacks of the above-mentioned methods; (4) to provide a summary and set forth some perspectives regarding existing challenges in this research field.
2. CRISPR-based POC detection
The 2020 Nobel Prize in chemistry was awarded to Charpentier and Doudna for their work on the discovery of CRISPR and its applications in gene-editing. Since the seminal discovery of CRISPR, the related biotechnologies have significantly improved our capabilities in the rapid and sensitive diagnosis of various human diseases. CRISPR has the potential to achieve two (i.e. recognition and transduction) out of the five above-mentioned components in an ultimate POC device, as shown in Fig. 1. Although there have been many comprehensive and professional review papers published on CRISPR-based diagnostics in the last few years,35–37 none of them has focused specifically on the design principles for different kinds of targets in POC diagnostics. Furthermore, as a rapidly changing field, new CRISPR-based bioanalytical strategies are being constantly reported in the scientific literature on a weekly/monthly basis. We therefore chose to dedicate this section to highlight the potential and most recent advances in CRISPR-based bioanalytical tools covering the detection of diverse biological targets from 2020 to 2021. In this section, we first discussed briefly the different CRISPR-Cas systems and cleavage activities commonly used for POC detection. Next, we summarized their latest advances in detecting nucleic acids, small molecules, metal ions and proteins by using on-target and collateral cleavage activities in a POC manner, as well as the role of CRISPR in enabling integrated electrochemical and optical POC testing. Finally, the challenges and the great promise of CRISPR-based biosensing developments are highlighted.
2.1. Construction and cleavage activity of different CRISPR-Cas systems for POC sensing
The CRISPR-Cas system, present in most bacteria and archaea, is made up of a CRISPR-associated (Cas) nuclease and guide RNA (gRNA), which work together to confer immunity against foreign viruses by cleaving nucleic acids.38–40 In the emerging POC applications, Cas9 (type II), Cas12 (type V) and Cas13 (type VI) of the Class-2 CRISPR-Cas systems are the most widely used, by the simple reason that only a single, RNA-guided, multidomain Cas protein is required to assemble the CRISPR effector complex.41,42 As shown in Table 1, although all three types of CRISPR systems are classified as Class-2, they differ in their targeting nucleic acids; the Cas9 and Cas12 systems target the DNA specifically, while the Cas13 system targets the RNA. The gRNA is made up of two parts, with the first being a programmable CRISPR RNA (crRNA) that identifies and hybridizes with the target nucleic acid sequence, and the second being a trans-activating RNA (tracrRNA) that binds to crRNA and guides the nuclease to cleave the DNA. In the type II-Cas9 system, both crRNA and tracrRNA are required, while type V and type VI systems require only one single crRNA to achieve the above-mentioned functions.38,43,44 The first step of target recognition involves identification of the specific protospacer adjacent motif (PAM) and protospacer flanking sites (PFS) by Cas9/Cas12 and Cas13, respectively, followed by evaluation of sequence complementarity in the target. This two-step target recognition process ensures a much greater selectivity than conventional nucleic acid probes which mainly rely on sequence complementarity. Upon successful target recognition, all Class-2 CRISPR-Cas systems make use of either the on-target (cis-cleavage) or collateral cleavage (trans-cleavage) of the corresponding nuclease to carry out subsequent signal transduction events.45
Key features of Class-2 CRISPR-Cas systems and the representative POC sensing platforms summarized in this review.
| Type II-Cas9 | Type V-Cas12 | Type VI-Cas13 | |
|---|---|---|---|
| Subtypes | Deactivated Cas9 (dCas9) | Cas12a (Cpf1) | Cas13a (C2c2) |
| Cas9 nickase (nCas9) | Cas12b (C2c1) | Cas13b | |
| Cas12f (Cas14) | |||
| Target (activator) | dsDNA | dsDNA | RNA |
| ssDNA | |||
| gRNA | crRNA + tracrRNA | crRNA | crRNA |
| Platforms | NASBACC48 | DETECTR54 | SHERLOCK58 |
| CRISDA49 | HOLMES43 | HUDSON-SHERLOCK44 | |
| CRISPR-Chip50 | CRISPR-hydrogel69 | CARMEN61 |
The cis-cleavage can be achieved by Cas9, Cas12 and Cas13, while Cas12 and Cas13 are also capable of collateral cleavage.41 From a POC test perspective, this type of collateral cleavage activity has the potential to facilitate rapid and accurate detection of biomarkers by generating readily amplifiable output signals. Through the modification of gRNA sequences, CRISPR-Cas systems can direct Cas proteins to cleave any sequence of interest. Together, these two key characteristics (i.e. programmable target recognition and signal amplification) make CRISPR-Cas systems highly versatile tools for the POC detection of nucleic acids, small molecules, proteins, metal ions and so forth.
2.2. Cis- and trans-cleavage activity for POC detection of nucleic acids
The development of PCR and DNA sequencing methods have made it possible to diagnose many infectious and genetic diseases rapidly and accurately, and to monitor their therapeutic effects.46 These methods, however, have trade-offs between costs, sensitivity, selectivity and efficiency. Therefore, there is an urgent need for strategies capable of performing highly sensitive and selective single-base detection of nucleic acids. Fortunately, CRISPR-Cas systems have brought new solutions to these issues due to their high programmability and accurate gene-editing capabilities.40 In this section, we highlight the most recent CRISPR-based POC technologies for detection of nucleic acids according to the different classifications of CRISPR-Cas systems shown in Table 1.
In 2016, Doudna and Collins independently proposed and demonstrated new ways to detect RNA by using CRISPR-Cas systems.47,48 Doudna's study reported that C2c2 (Cas13a) enzyme possesses two distinct RNase activities, i.e. cis- and trans-cleavage activities, which function in concert to provide powerful RNA detection and degradation capability. In the other study reported by Collins and co-workers, isothermal RNA amplification was used with a toehold switch RNA sensor to detect Zika virus in clinical samples. When coupled with a CRISPR-Cas9 module, the reported sensor could discriminate virus strains down to a single-base level. Since then, several innovative sensing platforms based on Cas9 have been established, such as CRISDA in 2018 and CRISPR-chip in 2019.49,50 The CRISDA is capable of amplifying and sensing double-stranded DNA (dsDNA) with attomolar sensitivity and single-base specificity based on a strand displacement amplification method;49 however, the amplification process will undoubtedly increase the detection complexity and time (>3 h) for the entire detection cycle. To achieve the ultimate goal of developing easy-to-operate POC devices, amplification-free strategies should be highlighted. The CRISPR-chip achieved the detection of unamplified target genes by combining CRISPR-Cas9 with high targeting capability and a graphene-based field effect transistor (gFET) with high sensitivity.50
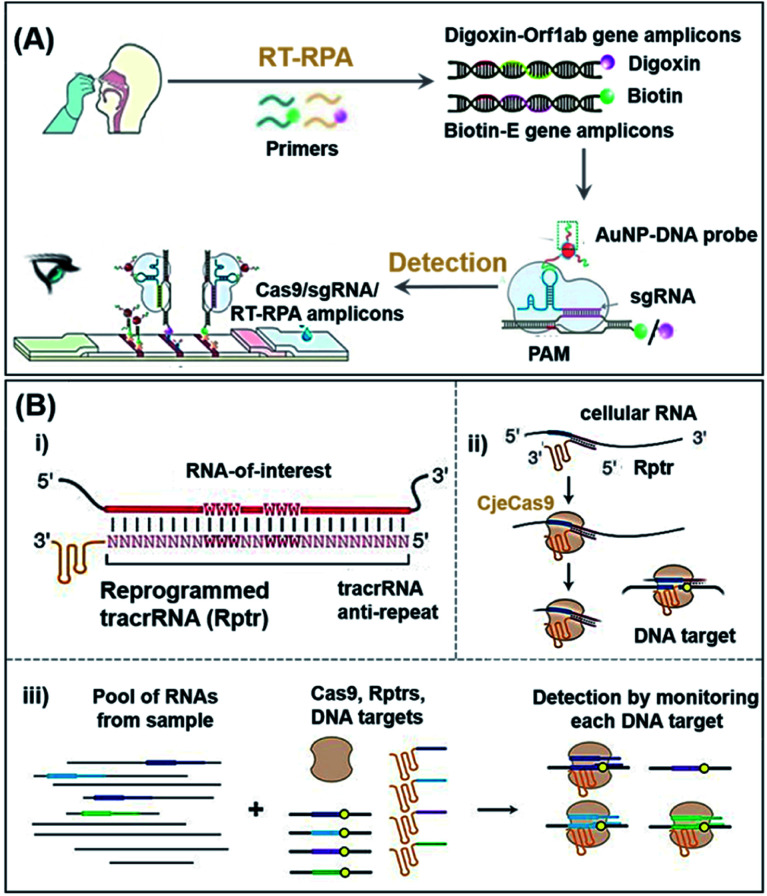
COVID-19 is a pandemic caused by SARS-CoV-2, with over 226.5 million confirmed cases and 4.6 million related deaths as of 15 September 2021.51 Xiong et al. proposed a diagnostic method for simultaneous dual-gene detection of SARS-CoV-2 based on CRISPR/Cas9-mediated triple-line lateral flow assay (LFA).52 As shown in Fig. 2A, the swab samples were labelled with digoxin and biotin on open reading frame 1ab (Orf1ab) and envelope (E) genes of the virus, respectively, by using multiplex reverse transcription-recombinase polymerase amplification (RT-RPA). Upon identification by Cas9 linked with a AuNP–DNA probe, the complex could be detected by the digoxin antibody and streptavidin immobilized inside the test lines of LFA. With the ability to simultaneously detect two genes in a single LFA, this platform achieved a sensitivity of 100 RNA copies per test (25 μL) with 100% and 97.14% predictive agreement of negative and positive results within a quick turnaround time (<1 h) for completing SARS-CoV-2 detection, although an amplification process was adapted. Although the authors have switched to the LFA detection process, the complex amplification process still needs to be addressed if non-expert detection is to be achieved.
Fig. 2. The potential POC sensing platform for nucleic acids enabled by CRISPR-Cas9. (A) Schematic reaction mechanism of simultaneous dual-gene amplifying of SARS-CoV-2 by RT-RPA and sensing by LFA. Reproduced from ref. 52 with permission. Copyright 2021 Wiley-VCH. Using RT-RPA, the E gene and Orf1ab gene regions of extracted samples are amplified and labeled with biotin and digoxin, respectively. In a typical test, the aforementioned complexes are dropped onto the sample pad and allowed to flow to the conjugate pad, where the AuNP–DNA probes hybridize to the scaffold sequence. Finally, due to the accumulation of AuNPs, the colorful bands could be observed with the naked-eye. (B) Overview of a multiplexable LEOPARD platform for parallel RNA detection. (i) Design process of Rptr; (ii) NcrRNA generation process; (iii) scheme of the multiplexed detection process for RNAs of interest. Rptrs convert sensed RNAs into ncrRNAs, which would direct Cas9 to match DNA. Cas9 binding or cleavage of a DNA sequence then indicate the presence of the sensed RNA in the sample. Reproduced from ref. 53 with permission. Copyright 2021 American Association for the Advancement of Science.
Typically, hybridization of a tracrRNA to crRNAs propels the operation of CRISPR-Cas9 systems. Jiao et al. recently discovered, however, that a tracrRNA could also hybridize to cellular RNAs, resulting in the formation of “noncanonical” crRNAs (ncrRNA) capable of further guiding the subsequent DNA targeting by Cas9.53 As shown in Fig. 2B, with the ability of linking any RNA of interest to DNA targeting by the reprogrammed tracrRNA, this new discovery has enabled the development of a multiplexable diagnostic platform termed LEOPARD for parallel RNA detection (Fig. 2B(iii)). In a proof-of-concept demonstration, the authors showed that LEOPARD should have the potential to be further developed into a new-generation CRISPR-Cas9 platform for POC detection with scalable multiplex properties in a single reaction.
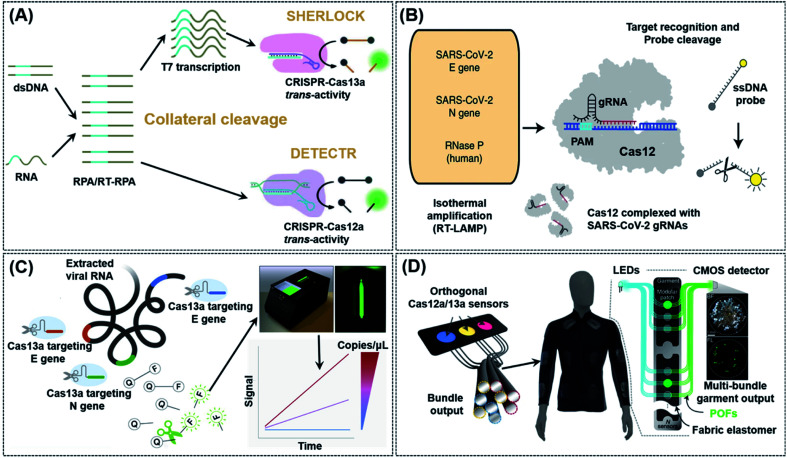
As indicated in Table 1, both Type V (Cas12a) and VI CRISPR (Cas13a) can achieve cis- and trans-cleavage, and simplify POC biosensor applications by using non-specific nucleic acid cleavage as a transducer to generate an electrochemical or optical signal. Based on these characteristics, Doudna's group developed a diagnostic system called DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR), which can be used to quickly and easily detect small amounts of DNA in a clinical sample in real-time by using Cas12a.54 The amplification of DNA/RNA by RPA/RT-RPA makes it easier for recognition by Cas12a, which subsequently and arbitrarily cut nearby ssDNA, causing the reporter molecule to fluoresce and achieving efficient detection of nucleic acids even in mixed samples (Fig. 3A). Chiu et al. reported a DETECTR-based detection method that identified SARS-CoV-2 by using RNA extracted from swab samples of COVID-19 patients.55 Upon isothermal amplification of the DNA reversely transcribed from virus RNA, the E (envelope) and N (nucleoprotein) genes of SARS-CoV-2 were detected by means of CRISPR-Cas12 trans-cleavage to release reporting molecules, which confirmed the presence of the virus (Fig. 3B). Compared to RT-PCR tests, this method has a positive and negative predictive agreement of 95% and 100%, respectively. Furthermore, a RT-PCR test usually takes more than 24 h to detect SARS-CoV-2 and it requires specialized equipment and laboratories, while the DETECTR only requires two fixed operating temperatures and can provide a visual readout after ∼45 min, similar to a home pregnancy test. The authors have listed the minimum equipment needed to run the protocol which includes Eppendorf tubes with reagents, heat blocks or a water bath (37 °C and 62 °C), nuclease-free water, pipettes and tips, and lateral flow strips. In addition, the complicated RNA extraction process and heating requirement in the amplification process are challenges that must be resolved before this technology may be used in households. Divalent cations have been found to accelerate the pre-crRNA process in the Cas12a activation step and DNA cleavage process.56 Based on this theory, Ma et al. first screened several divalent cations and found that manganese ions had the greatest promotion effect. Following which, the authors used a manganese-enhanced Cas12a (MeCas12a) to boost the signal by 13-fold for tested crRNAs, thereby allowing the detection of single-digit copies of RNA fragments without the need for specialized equipment.57
Fig. 3. CRISPR-Cas12/Cas13 based biosensing strategies used in nucleic acid detection. (A) Schematic illustration of typical collateral cleavage-based CRISPR-Cas systems, SHERLOCK and DETECTR. The Cas protein's trans-cleavage activity is activated when the crRNA binds to the nucleic acid target, resulting in the cleavage of multiple reporter molecules to achieve detection. (B) Schematic diagram of the DETECTR workflow for SARS-CoV-2. Conventional RNA extraction was used as an input to DETECTR, which was visualized by a fluorescent reader or lateral flow strip. This assay employs RT-LAMP to perform simultaneous reverse transcription and isothermal amplification of RNA extracted from swabs in universal transport medium (UTM), followed by Cas12 detection of predefined coronavirus sequences and cleavage of a reporter molecule to confirm virus detection. Reproduced from ref. 55 with permission. Copyright 2020 Springer Nature. (C) Amplification-free quantitative detection method for combining crRNAs targeting multiple regions of the viral RNA enhances sensitivity towards SARS-CoV-2 with a portable, mobile phone-based device. By combining multiple crRNAs to increase Cas13a activation, this assay can analyze the change in fluorescence over time with a mobile phone camera in a compact device that includes low-cost laser illumination and collection optics rather than solely endpoint fluorescence. Ref. 62 with permission. Copyright 2021 Elsevier Inc. (D) Design of a CRISPR-based wearable device. Distributed continuous sensing of garment activity can be achieved through multi-bundle imaging. The various optical fibers carrying the output emission signals from different sensors can be routed into a single bundle for centralized imaging analysis. Ref. 64 with permission. Copyright 2021 Springer Nature.
Based on the discovery that Cas12a can be used to detect RNA with the trans-cleavage activity, Zhang's group also developed and optimized another sensing strategy, termed specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) in 2017 by using Cas13.58 In their study, the authors demonstrated that SHERLOCK was able to identify RNA sequences of Zika, Dengue, and even some cancer-related mutations in a diverse range of human samples. There are, however, several limitations to this technology, including its inability to quantify targets and heavy reliance on fluorescence detectors for signal readout. In an effort to overcome these drawbacks, the same group developed SHERLOCK version 2 (SHERLOCKv2) with increased sensitivity and the ability to perform quantitative and multiplexed detection by using a lateral flow readout.59 SHERLOCKv2 involves the introduction of a variety of Cas13 enzymes from different species of bacteria, such as LwaCas13a and PsmCas13b. SHERLOCKv2 worked analogously as DETECTR, except that a specific fluorescent reporter gene was required. The improved specificity of SHERLOCKv2 enabled it to detect multiple sequences simultaneously. Similar to DETECTR, the isothermal nuclear amplification technology was also used in SHERLOCKv2 to improve detection sensitivity. Further amplification of detection signals was achieved by introducing CRISPR Type-III Csm6 enzyme as activating species into the system.
Inspired by the success of SHERLOCK/SHERLOCKv2, other CRISPR-Cas13 based strategies have also been developed to detect SARS-CoV-2 and Ebola viruses.35,60 An effective viral detection method requires the simultaneous detection of multiple strains of a virus in order to anticipate rapid virus mutations. Most conventional detection methods, however, do not possess such capability. Moreover, they are not easily scalable, making them unsuitable in case of a pandemic like COVID-19. To solve these issues, Ackerman et al. developed combinatorial arrayed reactions for multiplexed evaluation of nucleic acids named CARMEN-Cas13.61 This detection method successfully integrated nucleic acid detection using CRISPR-Cas13 with the microarray technology. CARMEN-Cas13 was also reusable and cost-effective, enabling the authors to simultaneously detect and differentiate 169 human-related viruses from ≥10 published genome sequences. Notwithstanding, most of the above-mentioned methods based on Cas13a either rely on sophisticated instruments for signal readouts or require a separate signal amplification step, and from the POC point-of-view, these issues will need to be properly addressed in future. Recently, Fozouni et al. proposed an amplification-free method to quantitatively detect SARS-CoV-2 from nasal-swabbed RNA simply by reading signals off from a compact microscope installed on a mobile phone (Fig. 3C).62 By employing multiple crRNAs, this method possessed a high sensitivity capable of detecting ∼100 copies of RNA per μL of sample within 30 min, and could also sensitively detect pre-extracted target RNA within 5 min. This assay method is potentially close to being a new-generation POC detection method for RNA targets, such as those from SARS-CoV-2. Wearable sensors also have been considered as a new development opportunity for POC systems due to the rapid development of mobile technology and wearable electronic materials.34,63 Further effort will be required to fully transform this work to a widely available point-of-care device. The central issue is an extraction-free protocol for sample preparation. In a recent study from Collins et al., the authors integrated CRISPR-Cas systems into a freeze-dried, cell-free (FDCD) wearable platform (Fig. 3D).64 The sensing mechanism was based on SHERLOCK but possessed the additional property of being able to detect both DNA and RNA by using Cas12a and Cas13a, respectively. By means of multi-bundle imaging, the fabric-based wearable POC device was able to achieve continuous detection of three common resistance markers from the bacteria Staphylococcus aureus. Face masks designed on the basis of this system were able to conveniently detect SARS-CoV-2 with a limit of detection (LoD) of 500 copies (17 aM) in 1.5 h. This design has many aspects to learn from, such as no requirement of manual swabbed samples; nevertheless the excessively long detection time still needs to be resolved in the future.
2.3. Trans-cleavage activity for POC detection of proteins, small molecules and metal ions
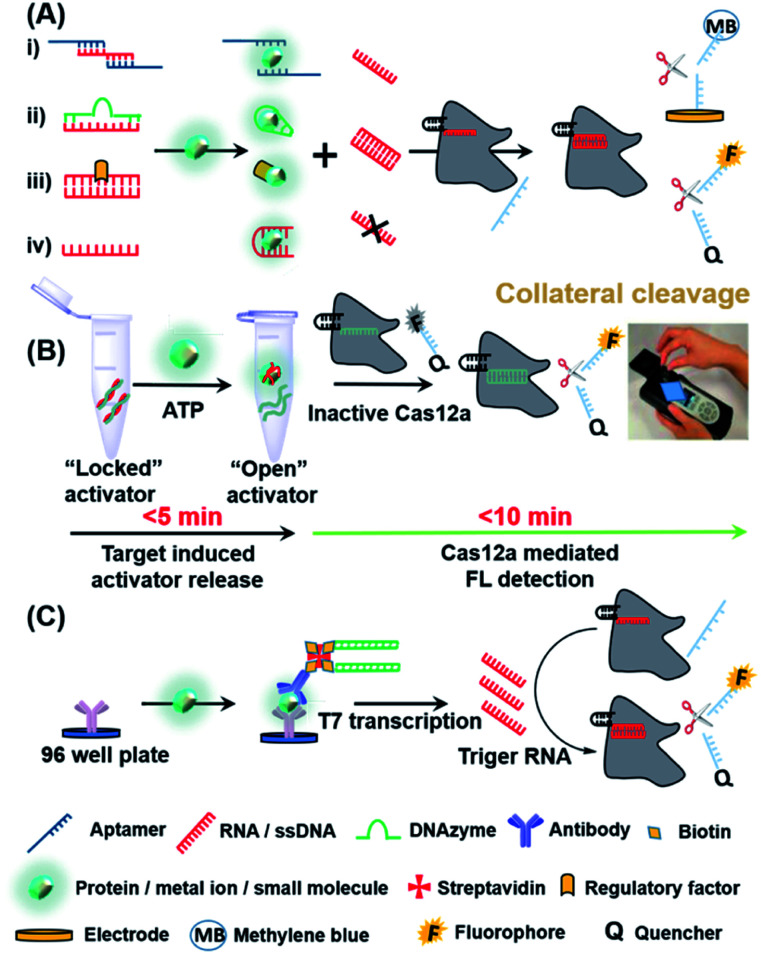
Besides nucleic acids, other diagnostic biomarkers such as proteins and small molecules, are also important for POC testing. Due to its powerful programmability and gene-recognition capabilities, especially with the collateral cleavage properties, CRISPR-Cas has been used to indirectly detect a wide range of non-nucleic acid targets. We summarize below the main mechanisms (Fig. 4A) for CRISPR-based POC detection of proteins, small molecules and metal ions, all of which are based on the concept of molecular translation. The basic principle is to release or bind the trigger of a CRISPR-Cas system by a target molecule, thus resulting in either the activation or restriction, respectively, of collateral cleavage activity in the Cas protein and ultimately providing fluorescence (FL) or electrical readout signals. Fig. 4A(i)–(iv) represent the sensing process by using designed aptamers (for small molecules/proteins/peptides), DNAzymes (for metal ions), regulatory proteins (for small molecules) and oligonucleotides (for proteins), respectively.
Fig. 4. CRISPR-based strategies used in the detection of non-nucleic acid targets. (A) Molecular translation mechanisms for the CRISPR-based POC detection for non-nucleic acid targets. (B) Detection strategy and workflow of the fDNAs-regulated CRISPR-Cas12a sensor for adenosine triphosphate (ATP) using a hand-hold portable device. Target binding to the fDNA in the presence of ATP can induce the DNA activator to dehybridize from the fDNA and become an “open activator” that binds to and activates the Cas12a. Reproduced from ref. 65 with permission. Copyright 2020 American Chemical Society. (C) Scheme of CLISA for femtomolar high throughput detection of proteins assisted by CRISPR-Cas13a. The capture antibody binds to the antigen of interest first, followed by a biotinylated detection antibody that is streptavidin-linked to a biotin-dsDNA template. Following transcription, the amplified RNAs activate trans-cleavage, allowing for detection. Reproduced from ref. 68 with permission. Copyright 2020 American Chemical Society.
Xiong et al. recently reported a novel CRISPR-Cas12a sensor regulated by functional DNA (fDNA) molecules such as aptamers and DNAzymes to specifically detect non-nucleic acid targets including small molecules and metal ions.65 As shown in Fig. 4B, the recognition of ATP resulted in the unlocking of the activator (a strand of fDNA) from the aptamer–activator complex. Cas12a subsequently recognized the released DNA activator and performed a trans-cleavage. The activated non-specific ssDNA nuclease further cleaved the quencher-ssDNA-fluorescence reporter (FQ-reporter) in two sides and eventually fluorescent signals were produced. By using a portable fluorescence reader, the authors showed that not only ATP but also Na+ (mechanism shown in Fig. 4A(ii)) in serum could be rapidly detected (<15 min) at room temperature, indicating that the device is suitable for POC detection. According to the authors, this sensor can in principle be further extended for rapid and highly sensitive detection of proteins, viruses and even cells. Similar to small molecules and metal ions, proteins can also be detected by CRISPR systems. Dai et al. developed an universal electrochemical biosensor by utilizing the CRISPR-Cas12a system (E-CRISPR).66 In this study, the target protein was first treated with a fixed concentration of an aptamer (which can bind to the protein target). Next, Cas12a–crRNA was designed to specifically recognize the aptamer, and finally electrical signals were applied to determine the remaining concentration of the target-specific aptamer indirectly based on the trans-cleavage of a nonspecific DNA reporter. By using this electrochemical detection, the authors were able to detect proteins with a LoD of 0.2 nM. Liang et al. developed a CaT-SMelor platform (CRISPR-Cas12a- and bacterial allosteric transcription factors-mediated small molecule detector) based on a similar mechanism shown in Fig. 4A(iii).67 In the presence of a small-molecule target, the competitive binding between the regulatory factor and the target could release the trigger dsDNA for Cas12a, thereby cleaving the FQ-reporter and allowing the concentration of the small molecule to be quantified. The powerful and ubiquitous antibody–antigen interaction is a useful tool for the development of universal biosensors. Chen et al. proposed an universal CRISPR-Cas13a signal amplification linked immunosorbent assay (CLISA) for the detection of proteins at femtomolar concentrations.68 As shown in Fig. 4C, the authors first used an antibody immobilized in a 96-well microplate to capture the corresponding protein antigen. Next, a detection antibody linked with a biotin-streptavidin–biotin-dsDNA complex was used to recognize the protein antigen. Upon signal amplification of the dsDNA by T7 RNA polymerase, the CLISA achieved LoD values of 2.29 fM and 0.81 fM for human interleukin-6 and human vascular endothelial growth factor, respectively.
The use of responsive hydrogels is vital in various biotechnology applications, ranging from scaffold designs for tissue engineering to biosensors in the detection of different biological analytes of interest. For instance, hydrogels that are responsive to DNA are ideal for interacting with synthetic or naturally occurring DNA. Existing DNA-responsive hydrogels, however, necessitate the use of high-concentration DNA triggers for actuation. In order to accommodate the insertion of the modified triggers, the hydrogel system needs to be redesigned, which could lead to a conflict with its structural requirements. In light of this, English et al. designed a programmable CRISPR-based DNA hydrogel sensor that could be tailored to function according to the user-defined nucleic acid sequences.69 During the process of target sensing, this technology utilized the trans-cleavage ability of Cas12a to cut off the links between DNA embedded in the hydrogel, thus resulting in hydrogel hydrolysis. By using such CRISPR-responsive smart hydrogels, the authors further demonstrated the successful release of other targets including small molecules, enzymes, nanoparticles and live cells that were pre-anchored in the DNA, thereby suggesting that these hydrogels could be used in the POC detection of such targets in future.
CRISPR has certainly injected new blood into the field of POC diagnostics. However, due to the fragile property of most Cas proteins, the storage and transportation of the CRISPR-Cas based POC products put forward strict requirements on its stability. Although reports have shown that the storage temperature of CRISPR-Cas systems can be increased to about 20 °C through embedding the systems into paper-based devices, manufacturing highly robust CRISPR-Cas based POC test kits and their marketing still face key challenges.
3. Electrochemistry-based POC detection
The field of electrochemical sensing is well-established, but in recent years has been accelerated and given a new life in POC diagnostics upon successful merging with nanotechnology, biotechnology and a number of other emerging strategies in target recognition and signal transduction. Electrochemical-based strategies are largely associated with the transducer part in the design of a POC device, and are responsible for the conversion of target sensing to electrochemical readouts. Electrochemical sensors can be divided into five main types according to their signal-generating modes, including voltammetric/amperometric, impedimetric, conductometric, potentiometric, and field effect transistor (FET) biosensors.70 In general, regardless of the type of biosensors, electrical signals are generated by a physical or chemical reaction between the probe and the target, and the quantitative/semi-quantitative/qualitative detection of the target is subsequently achieved on the basis of signal changes.
In this section, we introduce the most recent advances in the development of various electrochemical sensors for the detection of diverse targets that are suitable in a POC setting. As a kind of relatively mature transduction technology, electrochemical techniques have already been used in real POC applications,71,72 such as the blood-glucose meter.73 However, there are still certain challenges to overcome for the electrochemical techniques to be used in some urgently needed diagnostic field. Firstly, due to the sensitivity of electrochemical signals, the interference of irrelevant molecules in body fluids has a great influence on the accuracy of the results. However, the abundance of most biomarkers is lower than that of interfering molecules, which has led to a scarcity of biosensors for early-stage screening for many diseases and cancers. Secondly, there are still problems in the current development goals of POC to achieve multiplex detection. Previous reviews in this area mainly focused on sensor materials or mechanisms for classification and discussion,74–80 therefore, in this part we highlight a series of newly developed methods that successfully addressed challenges previously faced by conventional electrochemical biosensors, especially simple fabrication, ultra-sensitivity, detection in complex samples, and multiplexed target detection.70,81,82 We focus on the biotargets of nucleic acids, proteins, and bacteria/virus since they are most important biotargets for the early-stage diagnosis and multiplex screening of diseases. Besides, due to the cost effectiveness of various electric (signal) transduction systems,70,83 such as linear sweep voltammetry and differential pulse voltammetry, the entire sensing system could become readily applicable in resource-limited areas. In short, key recent developments in electrochemical biosensing systems have promoted their widespread applications in the global healthcare system in a positive way. This section is also divided into two parts, one for nucleic acids and the other for other types of biomarkers.
3.1. Electrochemical detection of nucleic acids
The very low concentrations of nucleic acids in clinical samples require the corresponding biosensors to possess both high sensitivity and specificity to distinguish specific sequences from related molecules.84 Kelley and co-workers have pioneered the use of electrochemical methods to detect nucleic acids.70,85–88 In one classical example, the authors developed the so-called neutralizer displacement assay (NDA) with record-breaking sensitivity across all of the major classes of nucleic acids.85,87 They also developed a minimally invasive disease diagnosis strategy that employed DNA clutch probes (DCPs) to detect mutations in circulating tumor DNA (ctDNA) with a LoD of 1 fg μL−1.88 By using Si3N4/Au/Si electrodes and synthetic molecules such as peptide nucleic acids (PNAs) as probes, such methods made use of potentiometric electrochemical signals to generate sensitive readouts. Although this design exhibits outstanding sensitivity and selectivity for mutations of ctDNA, a critical challenge for detection of circulating nucleic acids is to differentiate multiple mutated sites from highly expressed wild type sequence in blood samples. Afterwards, the authors proposed a combinatorial-probe-based strategy for high-throughput ctDNA electrochemical biosensing.20 This method was demonstrated to be an effective assay with potential for POC applications by directly quantifying multiple mutation-bearing nucleic acid sequences from human blood within 30 min.
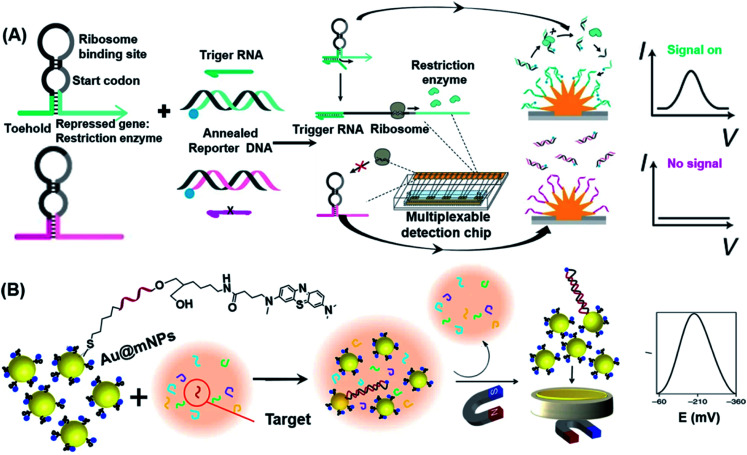
The development of gene circuits offers new ideas for designing novel biosensors. Since many of the gene circuits are located in living cells, the resulting biosensors are renewable, highly selective, easy to produce and cost-effective.89 In a cell-free system, gene-circuit sensors could also be used to concentrate the enzymes needed for gene transcription and translation.90 Previously, such sensors relied on optical reporter proteins that could only generate at most three reporter signals in one single reaction, hence limiting their capability to measure multiple distinct signals. To overcome this limitation, Mousavi et al. developed a platform based on electrochemical signals that allows multiplexed reporting for cell-free gene-circuit biosensors.91 As shown in Fig. 5A, the system includes nanostructured gold electrodes, a capture DNA fixed to the electrode surface, and an annealed reporter DNA that exists in the system in a free state. In the presence of the target RNA (trigger RNA), the toehold switch RNA was opened and translated, enabling restriction enzymes encoded by the RNA to be translated for recognition and cleavage of the reporter dsDNA. Due to the lower free energy of binding between ssDNA and capture DNA, the resulting binding was stable, therefore driving methylene blue (MB) close to the gold electrode to generate an electric current via the mechanism of electron transfer. This novel electrochemical interface was further demonstrated by its ability to detect a synthetic gene network actuated by a small molecule, and its multiplexed detection of colistin antibiotic resistance genes using the designed detection chip.
Fig. 5. Electrochemical-based POC strategies used in nucleic acid detection. (A) Scheme illustration of the synthetic gene detection method that combines cell-free transcription and translation systems with engineered gene circuits on nanostructured microelectrodes. In short, this approach employs toehold switch-based RNA sensors which will express one of ten restriction-enzyme-based reporters in the presence of trigger RNA. Afterwards, restriction enzymes cleave annealed reporter DNA which was followed by releasing reporter DNA labelled with the redox reporter (blue circle). After the capture of reporter DNA by probe DNA on nanostructured microelectrodes, it will generate an electrochemical signal. Reproduced from ref. 91 with permission. Copyright 2020 Springer Nature. (B) Schematic representation of steps involved in miRNA detection by using Au@mNPs. Au@MNPs are modified with methylene-blue-labelled probe DNA which is complementary to target miRNA. Unhybridized sequences are washed away after magnetic separation of Au@MNPs from the solution. Magnetically collected Au@MNPs (both hybridized and unhybridized) on the surface of a gold microelectrode provide the electrochemical signal. Reproduced from ref. 93 with permission. Copyright 2018 Springer Nature.
MicroRNAs (miRNAs) are a class of non-coding RNAs with a length of 18–24 nucleotides that are closely associated with the formation of tumors by acting as either tumor suppressor genes or oncogenes. They can degrade the mRNA of the target gene or inhibit its translation through complete or incomplete pairing with the target gene 3′UTR, thus participating in the regulation of life activities such as ontogenesis, apoptosis, proliferation and differentiation. Hence, they represent some of the most important biomarkers for cancer.92 Tavallaie et al. reported a strategy to sensitively detect miRNAs (as low as 10 aM to 1 nM in unprocessed blood samples) by modifying gold-coated magnetic nanoparticles, which serve as dispersible electrodes with miRNA-specific DNA (Au@mNPs, Fig. 5B).93 During their circulation in blood, target miRNAs captured by Au@mNPs could be magnetically isolated and converted into measurable electrical signals for direct miRNA detection from the blood. As such, this type of sensor might hold great promise in determining miRNA levels with just a simple finger prick in future.
One problem with designing biosensors for POC applications utilizing such nanostructures in the above-mentioned two examples is that the reproducibility of the materials might create deviations in detection results, and thus researchers may be more interested in exploring how to solve it in the future.
In addition to the above-mentioned biosensors, there are also other electrochemical strategies for the detection of nucleic acids, such as those that make use of immunomagnetic beads and selective DNA hybridization to detect methylation of tumor suppressor genes from not only raw serum and tumor cells, but also diseased tissues.83 Alafeef et al. developed a graphene–ssDNA–AuNP platform with digital electrochemical output for the real-time monitoring of SARS-CoV-2.84 This method achieved a sensitivity of 231 (copies per μL)−1 and limit of detection of 6.9 copies per μL without any further amplification. With the advancement of genetics and electrochemical science, we are quite optimistic that more novel electrochemistry-based methods for detecting nucleic acids will be reported in the future.
3.2. Electrochemical detection of non-nucleic acids
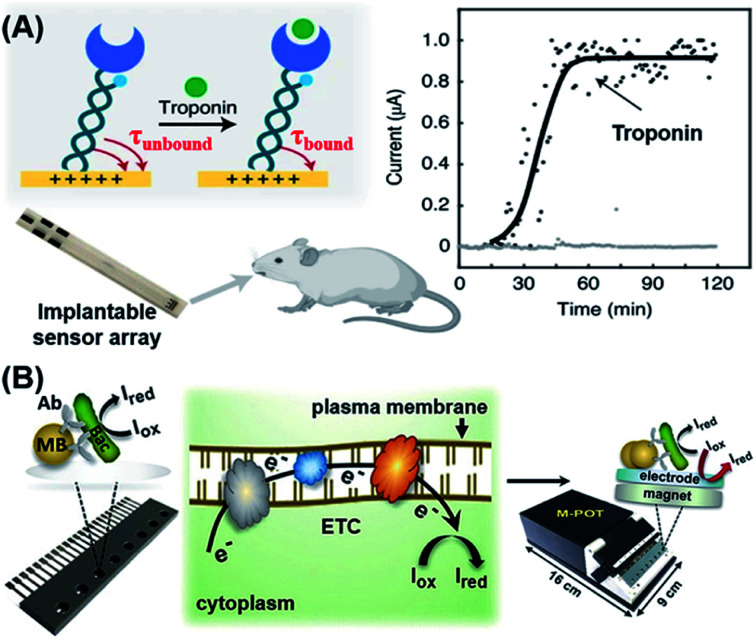
The electrochemical method is a feasible POC strategy for the detection of proteins, small molecules and bacteria.94–96 The most commonly used probes for detecting these molecules are aptamers, antibodies and enzymes. Based on their previous research,97 Kelley's group designed a peptide-mediated electrochemical steric hindrance hybridization assay,98 which can be utilized to detect antibodies against the gp41 protein of HIV-1 with a LOD of 10 nM. As the DNA-based signaling mechanism of this design is robust and given the versatility and modularity of PNA as a linker joining peptides (antigens) with DNA strands, this method may be readily adapted for detection of other antibodies having known peptide epitopes. Recently, Kelley's group reported a new reagent-free electrochemical sensing method based on the motion of a cleverly designed inverted molecular pendulum, with field-induced transport modulated by the existence of a bound analyte (Fig. 6A).99,100 By using the electron-transfer kinetics of an attached reporter molecule to measure the electric field-mediated transport of the sensor, the authors were able to detect cardiac troponin and SARS-CoV-2 spike protein with a LoD of 1 pg mL−1 (∼40 fM). Moreover, the authors also demonstrated that this mechanism could be used to detect biomolecules not only in blood, tears, urine, saliva and sweat, but also in living animals (i.e. in situ detection) by characterizing the molecular pendulum motion with time-resolved electrochemical curves.
Fig. 6. Electrochemical-based POC biosensors for the detection of proteins and bacteria. (A) Reagentless electrochemical biosensor of protein detection by monitoring the kinetics of transport of a molecular pendulum complex constructed with dsDNA, a specific antibody and a redox reporter. This approach relies on the motion of an inverted molecular pendulum that exhibits field-induced transport. The motion is regulated by the presence of bound analytes which is quantified by the sensor's electric field-mediated transport. Reproduced from ref. 99 with permission. Copyright 2021 Springer Nature. (B) Workflow of an immuno-affinity amperometric method for the detection of bacterial infection which is composed of three steps: (1) Bacterial capture and enrichment from complex matrices; (2) chemical reduction and (3) amperometric read-out. Reproduced from ref. 106 with permission. Copyright 2018 Wiley-VCH.
Bacterial infection, especially in blood circulation, poses a serious threat to human health. The misuse and overuse of antimicrobial drugs have led to an increase in bacterial resistance, thus increasing the risk of bacterial infection and transmission. Therefore, there is an urgent need for POC detection of bacterial infection.101 To detect bacterial infection in the bloodstream, advances in genotyping techniques have in recent years greatly accelerated the identification of pathogens. Most existing diagnostic methods, however, still heavily rely on time-consuming antimicrobial culture methods.102–105 Zhu et al. recently developed a sensitive immuno-affinity amperometric method that could detect bacterial infections within several hours.106 In their sensor design, bacterial pathogens were firstly captured and enriched from complex matrices by using super-paramagnetic beads (MB) that had been functionalized with the corresponding antibodies (Fig. 6B). Subsequently, suitable metabolic activity indicators were introduced which underwent chemical reduction (Iox to Ired) in the presence of bead-captured bacteria in the respiratory electron transport chain (ETC) on the plasma membrane of metabolically active cells. All of these reactions took place in a microchip containing multiple potentiostats that could generate electrochemical signals to be directly read by a readout device. This method serves as a useful reference for rapid and high-throughput detection of bacterial species, live-cell quantification and antibiotic sensitivity testing in clinical settings.
The various detection strategies described above provide an overview of some recent developments of robust electrochemical biosensors for convenient detection of most types of biomolecules, and can serve as meaningful references for the future construction of on-site diagnostic systems. In addition, various creative combinations of sensing, recognition and transduction elements described herein could also help to open up new opportunities for the development of future POC detection systems. It is now possible to achieve multiplexed electrochemical signaling simultaneously by multiple probes with different activation. Hence, next generation electrochemical biosensors can be designed to identify more electrochemical redox species with differentiable potentials. Furthermore, the stability and activity of integrated probes used in electrochemical platforms should be further improved as most existing methods could cause significant declines in biomolecular activity after repeated uses.
4. Optical-based POC detection
Optical signals are the most important and intuitive detection signals. The concentration of biomolecules in a test sample can be detected qualitatively, semi-quantitatively and quantitatively by measuring changes in optical signals with superior visibility. Optical signals are produced by means of reflection, diffraction or absorption of external light or internal fluorescence emission, quenching, chemiluminescence, etc.16,107–111 Unlike other systems, optical biosensors do not require complex instruments, and in some cases can even achieve non-invasive and naked-eye detection, making them more cost-effective than other systems.112 For instance, optical signals can be digitalized into readable images by means of a digital camera, smartphone camera, or photodetector.113,114 The signals can then be processed and analyzed by using appropriate imaging software that translates pixel density into quantitative results for a target analyte in the detection zone. Therefore, with the development of portable spectrometers, optical detection is considered to be the most suitable method for POC detection and the most convenient method for naked-eye detection. The application of in vitro optical detection, however, still faces significant challenges due to limitations associated with optical signal strengths, accuracy of detection instruments and poor signal-to-noise ratios (SNR) of optical readers.34 To develop optical biosensors into suitable POC devices, researchers have made improvements in probe sensitivity, signal transduction methods, preparation of sensing materials, signal enhancement and noise reduction. In this section, we review key recent progress of optical POC diagnostics by different ways optical signals are produced.
4.1. Optical POC sensors based on the structural color
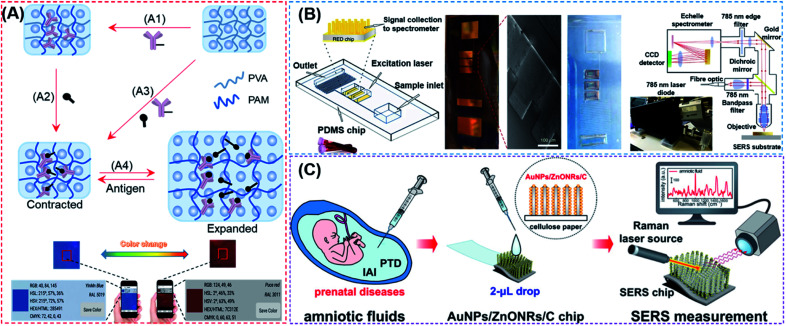
The structural color, also known as the physical color, is generated by a specific structure resulting from light diffraction and reflection of incident light.115 Structures that produce structural colors are typically assembled from photonic crystals (PCs), which are made up of spatially arranged periodic dielectric materials that uniquely change the propagations of light, resulting in high efficiency reflection at specific wavelengths.116 Optical cables, sensors, and quantum computers can all benefit from the PC structure.117,118 Hydrogels are considered as important sensing materials since they are easy to modify chemically, cross-linked with biomolecules, and can respond to acoustic, optical, electromagnetic, pH, and other types of stimuli.119,120 Because of their numerous advantages over other existing bioanalytical approaches, biosensors paired with PC technology and hydrogels hold much promise in this area. Since the first demonstration by Asher and co-workers in which PCs were combined with intelligent hydrogels that regulate photonic band gaps by expanding or shrinking,121 a variety of biosensors based on PC hydrogels (PCHs) have been reported, with the potential to be developed into POC devices for the detection of many types of biomarkers, such as glucoses, lectin protein, kinases, circulating tumor cells, hazardous ions, mycotoxins, Candida albicans and nucleic acids.113,122–131 Such innovative studies have clearly demonstrated the effectiveness of structural color-based strategies in the development of novel POC devices. Current PCH biosensors, however, are all designed to immobilize only the probe molecules into hydrogels. These PCH-based biosensing strategies require large changes in the volume or refractive index of the hydrogels due to intermolecular bonding, thus severely limiting the types of analytes that may be detected and making them ill-suited for more widespread use in POC diagnostics. We first reported a competition-based concept for the design of PCH biosensors by using antibody–antigen interactions (Fig. 7A).132 By introducing antigen–antibody pairs within the hydrogel simultaneously using non-covalent bonding, the resulting competition-based PCH biosensor was versatile, independent of the type of target molecules, and produced color changes that can be quantitatively detected by visual observation and by using a smartphone. More importantly, the use of this strategy in the detection of kinase–phosphatase activities and phosphorylation of peptides demonstrated its broad development potential in the detection of post-translational modification (PTM) of proteins in biological fluids.
Fig. 7. (A) Overall fabrication and workflow of competition-based universal photonic crystal hydrogel biosensors by using antibody–antigen interactions. The photonic crystal hydrogel sensors are modified with the complex of antibody–antigen which both were covalently linked on the scaffold hydrogel. After the detection of free targets, the breakage between immobilized antibody and antigen will cause significant expansion and a color change of the hydrogel from the contracted state which can be detected by the naked eye and smart phone. This universal design enabled the successful quantitative detection of small molecules, peptides and proteins with the naked eye and smart phone. Reproduced from ref. 132 with permission. Copyright 2020 American Chemical Society. (B) Rapid POC microengineered device technology for TBI biodiagnostics. The left figure shows the construction of this device. The middle figure shows the optical image and SEM image of the copper master used for fabrication of the PDMS, and a photograph of the microfluidic device with an input of 1–2 drops of whole human blood. Briefly, the whole blood will be separated by an optofluidic lab-on-a-chip to plasma. The optical information enhanced by the microfabricated SERS chip is sent to a PC to display the collected spectra using a designed portable spectrometer. Reproduced from ref. 135 with permission. Copyright 2020 Springer Nature. (C) AuNPs/ZnONRs/C chip for SERS analysis of amniotic fluids to detect prenatal diseases. The procedures are as follows: collection of amniotic fluids; dropping 2 μL of amniotic fluid onto the AuNPs/ZnONRs/C chip and real-time Raman measurement. Reproduced from ref. 136 with permission. Copyright 2018 American Chemical Society.
PCH has the potential to be used in wearable devices due to the flexibility of hydrogels. The lack of malleable electrical transducers, however, makes it difficult to perform quantifiable measurements when using such wearable PCH biosensors. To solve this issue, Snapp et al. detailed a colorimetric sensor with optoelectrical quantification based on an integrated system of PCs over a crumpled graphene photo-transducer.118 Although their research mainly focused on strain detection, the authors indicated that the novel combination of a photonic sensing element and a deformable transducer in such devices could offer a fresh perspective on the design of wearable and visual biosensors. Most studies described above could serve as useful references for the future development of more sophisticated PCH sensors capable of POC detection of various biological analytes. Since the structural color has the advantages of being photobleaching-free and having excellent signal stability, this type of biosensor could provide a new choice for the development of POC systems as long as the photonic crystal sensor can be reasonably embedded into sensitive and portable matrices in future.
4.2. Optical POC sensors based on SERS
In 1974, Fleischmann et al. discovered that the inelastic scattering from pyridine was considerably enhanced upon close contact with a silver electrode.133 Since then, SERS spectroscopy has revealed itself to be a promising analytical tool for ultra-sensitive detection, owing to its high precision, narrow Raman spectral widths, short analysis times, and easy surface modification with harnessed materials. The design of multifunctional SERS sensors is also possible.134 Although some researchers believe that the enhanced local electromagnetic field caused by SPR is the mechanism of SERS, it is still difficult to be accurately understood and unanimously accepted at present. We will therefore discuss the recent development in POC biosensors by using both SERS and SPR detection mechanisms.
Conventional SERS-based biosensors require a relatively long turnaround time from sample collection in order to obtain actionable results. It is also difficult to construct a field-deployable diagnostic platform and a companion diagnostic device. On this account, Rickard et al. reported a sensing strategy by combining SERS with an optofluidic device for rapid and label-free detection of biomarkers for traumatic brain injury (TBI) in biofluids with a picomolar LoD.135 As shown in Fig. 7B, the authors constructed the SERS-active substrate by using electrohydrodynamically fabricated submicrometre pillars, which were subsequently integrated into an optofluidic chip, followed by the use of a plasmon-active nanometric gold layer to coat the pillars. With the resulting device, N-acetylasparate, a potential biomarker released from the central nervous system after TBI, could be detected immediately from finger-prick blood samples, demonstrating that this strategy is ideal for constructing SERS devices and may be used for future on-site and real-time target detection. Notably, the authors designed a miniaturized Raman system for multiplexed and high-throughput analysis of biomarkers.
The sensitivity and precision of SERS-based devices can be enhanced by changing the nanostructure of the SERS substrate to increase the enhancement factor. Kim et al. reported a chip functionalized with SERS by decorating gold nanoparticles (AuNPs) on vertically grown ZnO nanorod (ZnO NR) arrays on cellulose paper (Fig. 7C).136 The developed nanostructure was demonstrated to increase the Raman signals by 1.25 × 107 with excellent reproducibility. Upon further integration of the designed sensor with a multivariate statistics-derived machine-learning-trained bio-classification approach, the authors were able to detect trace concentrations of human amniotic fluids in patients with subclinical intra-amniotic infection (IAI) and preterm delivery (PTD). By using nitrocellulose paper as a substrate (which is also often used in other SERS devices),137 the resulting SERS-based biosensors could be used as disposable and low-cost POC devices. The above-mentioned tactics are more convenient than other conventional SERS approaches in terms of detecting chips and detection equipment, but they still require the operation of specialized persons.
4.3. Optical POC sensors based on surface plasmon resonance
Plasmonics is the phenomenon that occurs when free electrons oscillate on a metal surface under light stimulation. Resonance can be achieved when incident light has the same frequency as that of the surface electrons oscillating against the attraction force to their positive nuclei, which is known as SPR.138,139 SPR has been employed in various biosensing and bioimaging methods to effectively enhance the luminance of fluorescent tags by increasing the ratio of fluorescence signal to background noise. Such enhancement and confinement of light at the interface between two media makes plasmonic platforms excellent for fabricating POC diagnostic devices.138,140,141
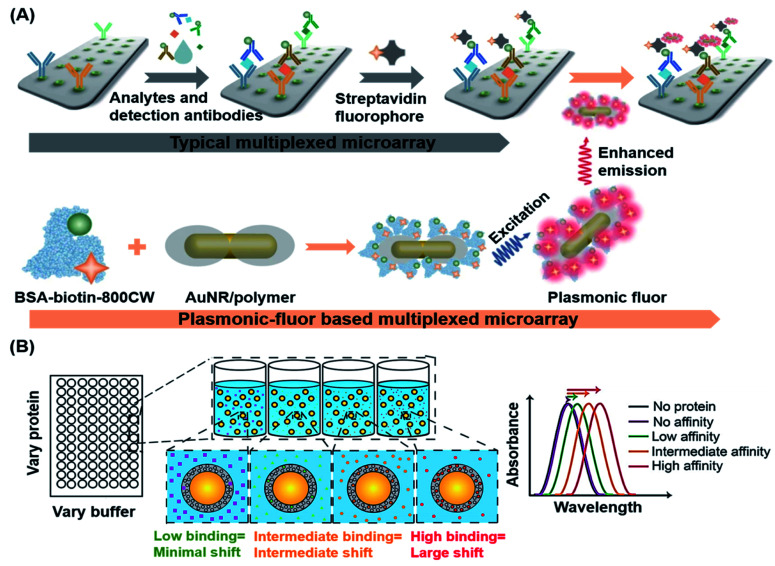
Singamaneni et al. recently designed and synthesized ultra-bright plasmonic-fluorophores as enhanced fluorescence reporters in plasmon-enhanced fluorescence immunoassays.142 The plasmonic-fluorophores consisted of a bovine serum albumin covalently conjugated with 210 IRDye 800CW fluorophores, a polymer-coated gold nanorod (AuNR) acting as a plasmonic antenna that allowed highly concentrated light to be irradiated into the fluorophores around the nanoparticles, thereby increasing the emission rate of fluorophores (Fig. 8A). Such plasmonic-fluorophores displayed fluorescence intensities that were approximately 6700-fold higher than those of sensors containing single fluorophores. When compared to control immunoassays, the detection limit of plasmonic-fluorophore-linked immunoassays was improved by 4750 fold.
Fig. 8. (A) Schematic illustration of the plasmonic-fluorophore structure and its working principle as a biolabel to enhance the FL and S/N ratio of immunoassays. The plasmonic-fluorophore consists of a plasmonically-active core (AuNR), a polymer shell as a spacer layer, light emitters, and biotin served as a universal biorecognition element. In the detection process, the analytes are recognized by the detection antibodies on the microarray and in the sample mixture to generate a sandwich structure, followed by binding with streptavidin and plasmonic fluorophores to achieve detection with enhanced optical signals. Reproduced from ref. 142 with permission. Copyright 2020 Springer Nature. (B) Schematic depiction of the LSPR-based biosensor for label-free detection of tear biomarkers based on different affinity interactions. Based on the sensing strategy that the dependence of the refractive index changes on the amount of protein bound is critical, little to no shift in LSPR wavelength is expected for low affinity contacts or low protein concentrations. Larger red shifts in the LSPR wavelength are expected as protein concentration or affinity increases. Reproduced from ref. 143 with permission. Copyright 2018 American Chemical Society.
Apart from using SPR for fluorescence enhancement, localized SPR (LSPR) of noble-metal nanomaterials could also be used to affect changes in the refractive index, making LSPR an effective transduction strategy for POC diagnostics. On the basis of this characteristic, Peppas's group developed a label-free method for detection of biomarkers present in human tears by using silica gold nanoshells coated with poly(N-isopropylacrylamide-co-methacrylic acid) nanogels (AuNS@PNM), which served as the corresponding protein receptors.143 As shown in Fig. 8B, different changes in the refractive index of AuNS@PNM were detected upon binding of different proteins having different charges (including lysozyme and lactoferrin which are known biomarkers of chronic dry eye), and the resulting biosensor displayed large, concentration-dependent red shifts in the LSPR wavelength and was readily measured with a portable spectrometer.
4.4. Optical POC sensors based on luminescence and colorimetry assays
Luminescence, including fluorescence, chemiluminescence and electroluminescence, has become an attractive method, and a potentially useful alternative, for disease detection and diagnosis, largely due to its rapid, non-invasive, real-time, low-cost, and high-resolution properties.34,144,145 Upon direct or indirect interaction with the biological analyte of interest, photophysical properties of the recognition system would present signals that can be readily detected by using a spectrometer or by the naked eye.146 Amongst various luminescence-based methods, fluorescence resonance energy transfer (FRET)-based sensing systems have been widely employed for fluorescence-based POC testing.108 Although some CRISPR- and SERS-based POC methods described earlier also used fluorescence as the signal transducer, this section is listed separately for better organization and more convenient interpretation since there are still many emerging POC methods in the field of fluorescence and chemiluminescence.
Lu et al. reported a facile “lock–key” strategy using imidazolium functionalized polydiacetylenes (iPDAs) to rapidly and specifically identify lysophosphatidic acid (LPA) for the early diagnosis of ovarian cancer.147 Based on the synergistic electrostatic and hydrophobic interactions between LPA and iPDAs, LPA “key” was able to specifically insert into the iPDAs “lock”, leading to conformation transition of the iPDA accompanied with a blue-to-red color change within 5 min. Such an approach was integrated in a lateral flow test strip to successfully distinguish the ovarian cancer patients' blood from healthy people's blood samples with 100% accuracy. Although other fluorescent biosensors may take a slightly longer turnaround time, the overall detection time of fluorescent sensors meet the ASSURED criteria. Therefore, most optical POC sensors based on luminescence focused on the improvement of their sensitivity.
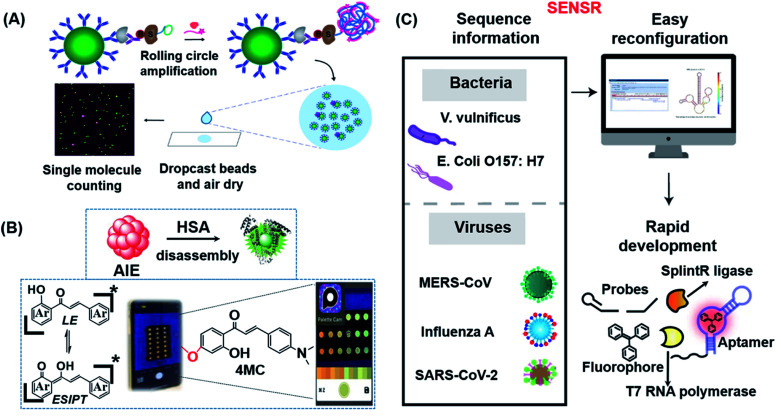
In 2018, Walt's group pioneered single-molecule immunoassays (Simoa) based on a fluorogenic enzymatic reaction in femtoliter arrays, which were subsequently commercialized by Quanterix.148 While Simoa is the current gold standard for the ultra-sensitive detection of proteins, its sensitivity is hampered by low sampling efficiencies. To address this limitation, the same group recently developed a significantly simplified and more sensitive digital ELISA platform, known as dropcast single-molecule assays (dSimoa), by bead-drop casting onto a glass slide which was subsequently air-dried to form a monolayer film with signals that can be read with a digital signal readout device (Fig. 9A).149 Compared to the original Simoa, this platform eliminated the need for bead loading into microwells or droplets for signal compartmentalization by localizing beads carrying the target molecules, thus allowing more beads to be analyzed and thereby increasing detection sensitivity by 25 fold.
Fig. 9. (A) Schematic illustration of dSimoa. From left to right shown in the scheme, the main structure was composed of antibody-coated paramagnetic beads, the target protein, the biotinylated detector antibody, the labeled streptavidin–DNA conjugate, DNA polymerase and the probe. RCA is conducted on the beads after the development of a single immunocomplex on antibody–magnetic beads–streptavidin–DNA sandwiches to construct a lengthy concatemer connected to each immunocomplex. The concentrated beads are then dropcast onto a microscope slide and fluorescence imaging of the dropcast film is used to count single target molecules. Reproduced from ref. 149 with permission. Copyright 2020 American Chemical Society. (B) Schematic showing the design of the paper-based ratiometric fluorescence analytical device and the chemical structure of 4MC. In this assay, the concentration of HSA was determined by comparing the ratios of emission from probes in aggregated and monomeric states, which results in a visible red-to-green color change on a simple, portable paper-based analytical device developed by integrating the identification probe into the detecting pad. Reproduced from ref. 150 with permission. Copyright 2020 Wiley-VCH. (C) Illustration of the broad adaptability of SENSR and its easy reconfiguration and rapid development for various pathogens. The assay relies on a sustained isothermal reaction cascade producing an RNA aptamer that binds to a fluorogenic dye. T7 RNA polymerase transcribes the RNA aptamer from a promoter DNA probe and a reporter DNA probe that hybridize with the target single-stranded RNA sequence via the SplintR ligase. After the one-pot isothermal reaction, the complex shows strong fluorescence in the reaction tube. Two pathogenic microbes and three viruses were targeted by redesigning the probe sequences. Reproduced from ref. 151 with permission. Copyright 2020 Springer Nature.
The majority of the probes developed in the field of fluorescent biosensors are activatable probes (i.e. off–on types) which have the unique properties of being “turned-on” in response to certain stimuli with measurable fluorescence signals that are proportional to the analyte concentration. Although a number of activatable probes have shown great potential as biosensing probes, light source attenuation and photobleaching of fluorescent molecules are common factors that can easily make the detection results inaccurate. To overcome such limitations, Liu's group successfully developed a dual-state emissive chalcone probe (4MC) based on aggregation-induced emission (AIE) for the POC monitoring of human serum albumin (HSA) in older and chronically unwell patients (Fig. 9B).150 The concentration of HSA was determined by the specific HSA-induced disassembly of 4MC nanoaggregates into 4MC–HSA complexes, which resulted in a distinct ratiometric emission color change (from red to green; Fig. 9B). Probe 4MC was subsequently integrated into paper-based analytical devices for naked-eye detection of HSA in whole blood.
An insufficient LoD is still the most concerned issue for most in vitro POC sensors. Lee's group developed a one-pot and ligation-dependent isothermal reaction cascade, named SENSR, that could be used to rapidly and sensitively detect RNAs (Fig. 9C).151 The reported SENSR was made up of two simple enzymatic reactions: a ligation reaction by SplintR ligase followed by transcription by T7 RNA polymerase. The resulting transcript formed an RNA aptamer that bound to a fluorogenic dye, generating a planar RNA-stabilized fluorogenic dye structure that only fluoresced upon detection of the RNA targets present in the test sample. One important characteristic of this method is that the LoD could be as low as 0.1 aM (∼6 copies of RNA in a 100 μL reaction). As shown in Fig. 9C, this strategy can be easily reconfigured for the detection of various pathogens in clinical samples.
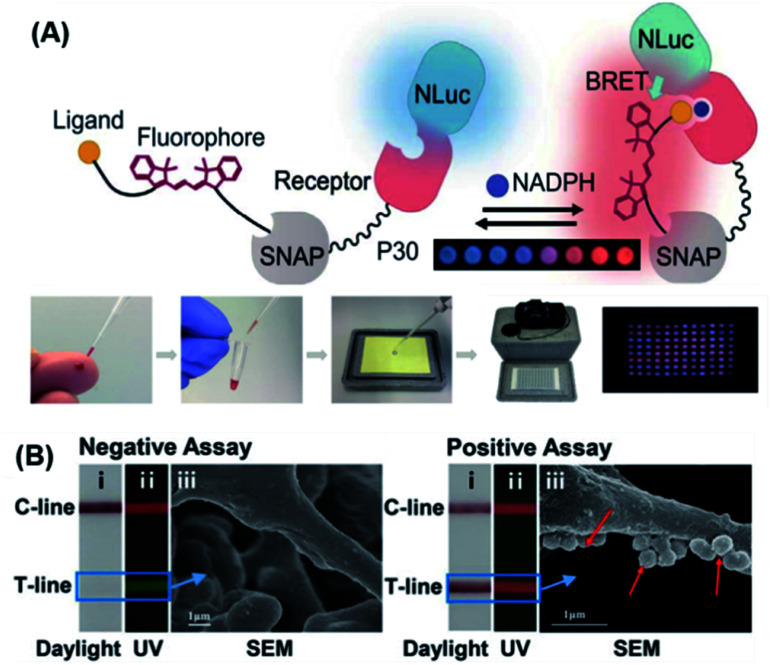
Compared to other luminescence-based methods, chemiluminescence has higher sensitivity and a better SNR. Furthermore, chemiluminescence readout instruments are simpler than other optical instruments, making them more likely to be used in POC detection.109 Potential chemiluminescence-based POC systems simplify readouts, increase sensitivity, and render the sensor to be independent of external light sources.152 Johnsson's group designed semi-synthetic bioluminescent sensor proteins through the genetic fusion of antibody fragments to NanoLuc luciferase and SNAP-tag. Binding of the drug displaces the tethered fluorescent competitor and disrupts bioluminescent resonance energy transfer (BRET) in the ratiometric sensor. The color shifts from red to blue in a drug-concentration-dependent manner, which permits direct POC quantification using a digital camera.153 Inspired by this work, they reported a semi-synthetic bioluminescent sensor for the direct quantification of phosphorylated nicotinamide adenine dinucleotide (NADPH) by using paper-based assays.154 As shown in Fig. 10A, the biosensor was made up of a NanoLuc luciferase-labeled NADPH sensor protein and a SNAP-tag synthetically labeled with a competitive fluorescent tether. In the presence of NADPH, the fluorescent tether bound to the receptor protein and resulted in BRET, thereby shifting the color (from blue to red) and enabling direct NADPH quantification by using a digital camera. This approach in principle allows any metabolite that is oxidized by NADPH to be used in quantitative POC assays. As a proof-of-concept, the authors demonstrated that 96 phenylalanine-spiked blood samples could be successfully quantified with just one camera shot. More recently, a similar biosensor was also developed for NAD+ detection.155
Fig. 10. (A) The design principle of a BRET sensor for POC testing of NADPH and the results of parallel analysis of 96 phenylalanine samples using naked eye. For this method, the metabolite is oxidized by nicotinamide adenine dinucleotide phosphate, and the sensor changes color when the reduced cofactor is present, allowing for metabolite measurement using a digital camera. Reproduced from ref. 154 with permission. Copyright 2018 American Association for the Advancement of Science. (B) Ratiometric fluorescent lateral flow immunoassay (RFLFIA) strip under daylight (i), UV light (ii), and SEM images (iii) after treatment with negative and positive assays for acute myocardial infarction (50 ng mL−1). In this design, the RFLFIA strip works according to the ratiometric change of the fluorescence signal, arising from blending of fluorescence emitted by two composite nanostructures conjugated to capture and probe antibodies and the inner filter effect of gold nanoparticles. Finally, detection is achieved by using a custom smartphone-based analytical device. Reproduced from ref. 159 with permission. Copyright 2021 Wiley-VCH.
Portable lateral flow immunoassay (LFIA) test strips are widely used in POC testing. They are inexpensive, rapid (10–20 min), selective in their detection of target analytes, and can be read by the naked eye in various signal readout devices.156,157 Although there are a number of LFIA methods available, most of them cannot qualitatively distinguish targets at low concentrations with weakly colored test-lines because of analyte-independent parameters such as photobleaching, light scattering, fluctuation of excitation light, etc. Ratiometric fluorescence technology is regarded as a solution since it relies on the ratio of the intensity of two or more emission bands.158 On this basis, Wang et al. developed a new type of ratiometric fluorescence-based LFIA (RFLFIA) strip for naked-eye detection of heart-type fatty acid-binding protein (H-FABP) by using a smartphone.159 The RFLFIA strip was constructed with a suitable reporter and capture probes made from two kinds of quantum dots conjugated with two different types of H-FABP antibodies. The sensing mechanism of RFLFIA involved a target-induced inner filter effect between the two quantum dots, ultimately resulting in a fluorescence color change (from green to red), which was accompanied by a change in the morphology (SEM image) of the strip membrane (Fig. 10B).
The abovementioned methods have greatly improved the detection accuracy and capability for multiplex detection. Due to the optical bleaching of fluorescent molecules and the complexity of chemical preparation routes, however, some important problems still need to be solved, including the stable preservation of fluorescent molecules, further simplification of the adopted chemistries, etc. for such methods to achieve practical and robust POC applications.
5. Summary and outlook
In recent years, new strategies aimed at ultimately achieving POC detection have been successfully developed to detect a wide range of biomolecules, from nucleic acids to human cancer cells. With the growing understanding of various human diseases and the continued advancement in different scientific fields (chemistry, life science, materials, etc.), cutting-edge research is now focusing not only on method development to detect the biological analyte of interest, but also more importantly on devising versatile approaches that can be used to detect a class of targets containing multiple kinds of biomolecules. The growth of these biosensor strategies has facilitated the quantitative analysis of targets from real clinical samples and strongly promoted emerging medical industries to become more involved in the further development of clinical POC systems. In this minireview, we have discussed the recent work on POC detection/diagnostic systems based on different design principles, including CRISPR which truly represents the latest advances in biotechnology, electrochemistry and optics. The detectable target molecules that can be covered range from nucleic acids, including DNA, RNA, and microRNA, to proteins, small molecules, bacteria, and human cells. We have also described in detail various device configurations in terms of sensing, probes, transducers, and others, which may serve as important references for interested researchers working in related fields.
Future POC systems will be able to use a single chip to achieve multiplexed sensing or even high-throughput screening of different biomolecules and simultaneously diagnose different human diseases according to highly reliable and readily distinguishable readout signals, thereby enabling POC sensors to be truly time-efficient, cost-effective, fast-responding and readily deployable in resource-poor areas. For example, electrochemical probes can be designed to include multiple signaling probes on a single chip but with different activation potentials so that the concentration of different targets can be measured. In addition, the precision of the POC system can be improved by not only concentrating the targets, but also enhancing sensing signals or reducing background signals. Inspired by PCR, the amplification strategy may be applied to other biomolecules in additional to nucleic acids. Furthermore, the fusion of different technologies may likely result in new and more effective approaches, as has been successfully demonstrated by the numerous developments of CRISPR-based methods coupled with electrochemistry, PCs and SPR. In the coming years, we may also expect some innovative machine-learning research for POC detection as well, by the use of technologies based on artificial intelligence (AI). Eventually, all of these technologies are expected to be integrated into wearable devices that have real-time monitoring capabilities. In our opinion, we believe that the development of POC systems will have a bright future as long as the issue of mass production is resolved.
Author contributions
J. Q. and S. Q. Y. presented the outline of this review. J. Q. wrote the draft and the graphic design. W. W., L. G. and S. Q. Y. revised and edited the draft. S. Q. Y. conceived and supervised this project. All authors participated in the discussion of the draft and carefully revised the draft before the final submission.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the GSK-EDB Trust Fund (R-143-000-688-592) and the Synthetic Biology Research & Development Programme (SBP) of National Research Foundation (SBP-P4 and SBP-P8) of Singapore.
Notes and references
- Yao L. Zhu W. Shi J. Xu T. Qu G. Zhou W. Yu X. F. Zhang X. Jiang G. Chem. Soc. Rev. 2021;50:3656–3676. doi: 10.1039/D0CS00595A. [DOI] [PubMed] [Google Scholar]
- Udugama B. Kadhiresan P. Kozlowski H. N. Malekjahani A. Osborne M. Li V. Y. C. Chen H. Mubareka S. Gubbay J. B. Chan W. C. W. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Gupta A. Madhavan M. V. Sehgal K. Nair N. Mahajan S. Sehrawat T. S. Bikdeli B. Ahluwalia N. Ausiello J. C. Wan E. Y. Freedberg D. E. Kirtane A. J. Parikh S. A. Maurer M. S. Nordvig A. S. Accili D. Bathon J. M. Mohan S. Bauer K. A. Leon M. B. Krumholz H. M. Uriel N. Mehra M. R. Elkind M. S. V. Stone G. W. Schwartz A. Ho D. D. Bilezikian J. P. Landry D. W. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B. D. Machhi J. Herskovitz J. Oleynikov M. D. Blomberg W. R. Bajwa N. Soni D. Das S. Hasan M. Patel M. Senan A. M. Gorantla S. McMillan J. Edagwa B. Eisenberg R. Gurumurthy C. B. Reid S. P. M. Punyadeera C. Chang L. Gendelman H. E. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W. Newbigging A. M. Le C. Pang B. Peng H. Cao Y. Wu J. Abbas G. Song J. Wang D. B. Cui M. Tao J. Tyrrell D. L. Zhang X. E. Zhang H. Le X. C. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Quesada-Gonzalez D. Merkoci A. Chem. Soc. Rev. 2018;47:4697–4709. doi: 10.1039/C7CS00837F. [DOI] [PubMed] [Google Scholar]
- Roberts J. D. Wells G. A. Le May M. R. Labinaz M. Glover C. Froeschl M. Dick A. Marquis J.-F. O'Brien E. Goncalves S. Druce I. Stewart A. Gollob M. H. So D. Y. F. Lancet. 2012;379:1705–1711. doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- Wu L. Qu X. Chem. Soc. Rev. 2015;44:2963–2997. doi: 10.1039/C4CS00370E. [DOI] [PubMed] [Google Scholar]
- Zhou W. Gao X. Liu D. Chen X. Chem. Rev. 2015;115:10575–10636. doi: 10.1021/acs.chemrev.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia S. Conoci S. ACS Sens. 2017;2:876–891. doi: 10.1021/acssensors.7b00299. [DOI] [PubMed] [Google Scholar]
- Markwalter C. F. Kantor A. G. Moore C. P. Richardson K. A. Wright D. W. Chem. Rev. 2018;119:1456–1518. doi: 10.1021/acs.chemrev.8b00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. C. Jana T. Kesavamoorthy R. Shi L. Virji M. A. Finegold D. N. Asher S. A. J. Am. Chem. Soc. 2004;126:2971–2977. doi: 10.1021/ja038187s. [DOI] [PubMed] [Google Scholar]
- Suni I. I. TrAC, Trends Anal. Chem. 2008;27:604–611. doi: 10.1016/j.trac.2008.03.012. [DOI] [Google Scholar]
- Furst A. Landefeld S. Hill M. G. Barton J. K. J. Am. Chem. Soc. 2013;135:19099–19102. doi: 10.1021/ja410902j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman A. Kalra P. Bansal V. Bruno J. G. Sharma T. K. Sens. Actuators, B. 2017;246:535–553. doi: 10.1016/j.snb.2017.02.060. [DOI] [Google Scholar]
- Li Z. Yin Y. Adv. Mater. 2019;31:e1807061. doi: 10.1002/adma.201807061. [DOI] [PubMed] [Google Scholar]
- Nekoueian K. Amiri M. Sillanpaa M. Marken F. Boukherroub R. Szunerits S. Chem. Soc. Rev. 2019;48:4281–4316. doi: 10.1039/C8CS00445E. [DOI] [PubMed] [Google Scholar]
- Yang M. Liu Y. Jiang X. Chem. Soc. Rev. 2019;48:850–884. doi: 10.1039/C8CS00303C. [DOI] [PubMed] [Google Scholar]
- Darwish N. T. Sekaran S. D. Khor S. M. Sens. Actuators, B. 2018;255:3316–3331. doi: 10.1016/j.snb.2017.09.159. [DOI] [Google Scholar]
- Das J. Ivanov I. Safaei T. S. Sargent E. H. Kelley S. O. Angew. Chem., Int. Ed. 2018;57:3711–3716. doi: 10.1002/anie.201800455. [DOI] [PubMed] [Google Scholar]
- Sun J. Xianyu Y. Jiang X. Chem. Soc. Rev. 2014;43:6239–6253. doi: 10.1039/C4CS00125G. [DOI] [PubMed] [Google Scholar]
- Inan H. Poyraz M. Inci F. Lifson M. A. Baday M. Cunningham B. T. Demirci U. Chem. Soc. Rev. 2017;46:366–388. doi: 10.1039/C6CS00206D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekjahani A. Sindhwani S. Syed A. M. Chan W. C. W. Acc. Chem. Res. 2019;52:2406–2414. doi: 10.1021/acs.accounts.9b00200. [DOI] [PubMed] [Google Scholar]
- Wu Y. Tilley R. D. Gooding J. J. J. Am. Chem. Soc. 2019;141:1162–1170. doi: 10.1021/jacs.8b09397. [DOI] [PubMed] [Google Scholar]
- Mabey D. Peeling R. W. Ustianowski A. Perkins M. D. Nat. Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- Land K. J. Boeras D. I. Chen X. S. Ramsay A. R. Peeling R. W. Nat. Microbiol. 2019;4:46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti D. van't Erve M. Stefania R. Ruggiero M. R. Tapparo M. Geninatti Crich S. Aime S. Angew. Chem., Int. Ed. 2014;53:3488–3491. doi: 10.1002/anie.201310959. [DOI] [PubMed] [Google Scholar]
- Nayak S. Blumenfeld N. R. Laksanasopin T. Sia S. K. Anal. Chem. 2017;89:102–123. doi: 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasseri B. Soleimani N. Rabiee N. Kalbasi A. Karimi M. Hamblin M. R. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour C. R. Parker L. A. Hutter E. M. Weckhuysen B. M. Nat. Rev. Chem. 2021;5:370–387. doi: 10.1038/s41570-021-00275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh N. Jolliffe K. A. New E. J. Angew. Chem., Int. Ed. 2020;59:20290–20301. doi: 10.1002/anie.202007673. [DOI] [PubMed] [Google Scholar]
- Li P. Lee G. H. Kim S. Y. Kwon S. Y. Kim H. R. Park S. ACS Nano. 2021;15:1960–2004. doi: 10.1021/acsnano.0c06688. [DOI] [PubMed] [Google Scholar]
- Christodouleas D. C. Kaur B. Chorti P. ACS Cent. Sci. 2018;4:1600–1616. doi: 10.1021/acscentsci.8b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S. Trung T. Q. Lee N. E. Chem. Soc. Rev. 2020;49:1812–1866. doi: 10.1039/C9CS00319C. [DOI] [PubMed] [Google Scholar]
- Kaminski M. M. Abudayyeh O. O. Gootenberg J. S. Zhang F. Collins J. J. Nat. Biomed. Eng. 2021;5:643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- Dai Y. Wu Y. Liu G. Gooding J. J. Angew. Chem., Int. Ed. 2020;59:20754–20766. doi: 10.1002/anie.202005398. [DOI] [PubMed] [Google Scholar]
- Aman R. Mahas A. Mahfouz M. ACS Synth. Biol. 2020;9:1226–1233. doi: 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- Chen J. S. Doudna J. A. Nat. Rev. Chem. 2017;1:1–15. doi: 10.1038/s41570-016-0001. [DOI] [Google Scholar]
- Barrangou R. Fremaux C. Deveau H. Richards M. Boyaval P. Moineau S. Romero D. A. Horvath P. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Shivram H. Cress B. F. Knott G. J. Doudna J. A. Nat. Chem. Biol. 2021;17:10–19. doi: 10.1038/s41589-020-00700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G. J. Doudna J. A. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S. Abudayyeh O. O. Makarova K. S. Wolf Y. I. Gootenberg J. S. Semenova E. Minakhin L. Joung J. Konermann S. Severinov K. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Y. Cheng Q. X. Wang J. M. Li X. Y. Zhang Z. L. Gao S. Cao R. B. Zhao G. P. Wang J. Cell Discovery. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold C. Freije C. A. Gootenberg J. S. Abudayyeh O. O. Metsky H. C. Durbin A. F. Kellner M. J. Tan A. L. Paul L. M. Parham L. A. Garcia K. F. Barnes K. G. Chak B. Mondini A. Nogueira M. L. Isern S. Michael S. F. Lorenzana I. Yozwiak N. L. MacInnis B. L. Bosch I. Gehrke L. Zhang F. Sabeti P. C. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar-Oliver A. Gersbach C. A. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Yin F. Song L. Mao X. Li F. Fan C. Zuo X. Xia Q. Chem. Rev. 2021;121:10469–10558. doi: 10.1021/acs.chemrev.1c00241. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A. O'Connell M. R. Knight S. C. Burstein D. Cate J. H. Tjian R. Doudna J. A. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K. Green A. A. Takahashi M. K. Braff D. Lambert G. Lee J. W. Ferrante T. Ma D. Donghia N. Fan M. Daringer N. M. Bosch I. Dudley D. M. O'Connor D. H. Gehrke L. Collins J. J. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Zhou W. Hu L. Ying L. Zhao Z. Chu P. K. Yu X. F. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajian R. Balderston S. Tran T. deBoer T. Etienne J. Sandhu M. Wauford N. A. Chung J. Y. Nokes J. Athaiya M. Paredes J. Peytavi R. Goldsmith B. Murthy N. Conboy I. M. Aran K. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roser M., Ritchie H., Ortiz-Ospina E. and Hasell J., Published online at OurWorldInData.org, 2021, https://ourworldindata.org/coronavirus
- Xiong E. Jiang L. Tian T. Hu M. Yue H. Huang M. Lin W. Jiang Y. Zhu D. Zhou X. Angew. Chem., Int. Ed. 2021;60:5307–5315. doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- Jiao C. Sharma S. Dugar G. Peeck N. L. Bischler T. Wimmer F. Yu Y. Barquist L. Schoen C. Kurzai O. Sharma C. M. Beisel C. L. Science. 2021;372:941–948. doi: 10.1126/science.abe7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S. Ma E. Harrington L. B. Da Costa M. Tian X. Palefsky J. M. Doudna J. A. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P. Deng X. Yu G. Fasching C. L. Servellita V. Singh J. Miao X. Streithorst J. A. Granados A. Sotomayor-Gonzalez A. Zorn K. Gopez A. Hsu E. Gu W. Miller S. Pan C. Y. Guevara H. Wadford D. A. Chen J. S. Chiu C. Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D. Ren K. Qiu X. Zheng J. Guo M. Guan X. Liu H. Li N. Zhang B. Yang D. Ma C. Wang S. Wu D. Ma Y. Fan S. Wang J. Gao N. Huang Z. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- Ma P. Meng Q. Sun B. Zhao B. Dang L. Zhong M. Liu S. Xu H. Mei H. Liu J. Adv. Sci. 2020;7:2001300. doi: 10.1002/advs.202001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S. Abudayyeh O. O. Lee J. W. Essletzbichler P. Dy A. J. Joung J. Verdine V. Donghia N. Daringer N. M. Freije C. A. Myhrvold C. Bhattacharyya R. P. Livny J. Regev A. Koonin E. V. Hung D. T. Sabeti P. C. Collins J. J. Zhang F. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S. Abudayyeh O. O. Kellner M. J. Joung J. Collins J. J. Zhang F. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K. G. Lachenauer A. E. Nitido A. Siddiqui S. Gross R. Beitzel B. Siddle K. J. Freije C. A. Dighero-Kemp B. Mehta S. B. Carter A. Uwanibe J. Ajogbasile F. Olumade T. Odia I. Sandi J. D. Momoh M. Metsky H. C. Boehm C. K. Lin A. E. Kemball M. Park D. J. Branco L. Boisen M. Sullivan B. Amare M. F. Tiamiyu A. B. Parker Z. F. Iroezindu M. Grant D. S. Modjarrad K. Myhrvold C. Garry R. F. Palacios G. Hensley L. E. Schaffner S. F. Happi C. T. Colubri A. Sabeti P. C. Nat. Commun. 2020;11:4131. doi: 10.1038/s41467-020-17994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman C. M. Myhrvold C. Thakku S. G. Freije C. A. Metsky H. C. Yang D. K. Ye S. H. Boehm C. K. Kosoko-Thoroddsen T. F. Kehe J. Nguyen T. G. Carter A. Kulesa A. Barnes J. R. Dugan V. G. Hung D. T. Blainey P. C. Sabeti P. C. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P. Son S. Diaz de Leon Derby M. Knott G. J. Gray C. N. D'Ambrosio M. V. Zhao C. Switz N. A. Kumar G. R. Stephens S. I. Boehm D. Tsou C. L. Shu J. Bhuiya A. Armstrong M. Harris A. R. Chen P. Y. Osterloh J. M. Meyer-Franke A. Joehnk B. Walcott K. Sil A. Langelier C. Pollard K. S. Crawford E. D. Puschnik A. S. Phelps M. Kistler A. DeRisi J. L. Doudna J. A. Fletcher D. A. Ott M. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J. Torrente-Rodríguez R. M. Wang M. Gao W. Adv. Funct. Mater. 2019;30:1906713. doi: 10.1002/adfm.201906713. [DOI] [Google Scholar]
- Nguyen P. Q. Soenksen L. R. Donghia N. M. Angenent-Mari N. M. de Puig H. Huang A. Lee R. Slomovic S. Galbersanini T. Lansberry G. Sallum H. M. Zhao E. M. Niemi J. B. Collins J. J. Nat. Biotechnol. 2021;39:1366–1374. doi: 10.1038/s41587-021-00950-3. [DOI] [PubMed] [Google Scholar]
- Xiong Y. Zhang J. Yang Z. Mou Q. Ma Y. Xiong Y. Lu Y. J. Am. Chem. Soc. 2020;142:207–213. doi: 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y. Somoza R. A. Wang L. Welter J. F. Li Y. Caplan A. I. Liu C. C. Angew. Chem., Int. Ed. 2019;58:17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M. Li Z. Wang W. Liu J. Liu L. Zhu G. Karthik L. Wang M. Wang K. F. Wang Z. Yu J. Shuai Y. Yu J. Zhang L. Yang Z. Li C. Zhang Q. Shi T. Zhou L. Xie F. Dai H. Liu X. Zhang J. Liu G. Zhuo Y. Zhang B. Liu C. Li S. Xia X. Tong Y. Liu Y. Alterovitz G. Tan G. Y. Zhang L. X. Nat. Commun. 2019;10:3672. doi: 10.1038/s41467-019-11648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. Tian T. Xiong E. Wang P. Zhou X. Anal. Chem. 2020;92:573–577. doi: 10.1021/acs.analchem.9b04403. [DOI] [PubMed] [Google Scholar]
- English M. A. Soenksen L. R. Gayet R. V. de Puig H. Angenent-Mari N. M. Mao A. S. Nguyen P. Q. Collins J. J. Science. 2019;365:780–785. doi: 10.1126/science.aaw5122. [DOI] [PubMed] [Google Scholar]
- Labib M. Sargent E. H. Kelley S. O. Chem. Rev. 2016;116:9001–9090. doi: 10.1021/acs.chemrev.6b00220. [DOI] [PubMed] [Google Scholar]
- daSilva E. T. S. G. Souto D. E. P. Barragan J. T. C. Giarola J. d. F. de Moraes A. C. M. Kubota L. T. ChemElectroChem. 2017;4:778–794. doi: 10.1002/celc.201600758. [DOI] [Google Scholar]
- Labib M. Sargent E. H. Kelley S. O. Chem. Rev. 2016;116:9001–9090. doi: 10.1021/acs.chemrev.6b00220. [DOI] [PubMed] [Google Scholar]
- Teymourian H. Barfidokht A. Wang J. Chem. Soc. Rev. 2020;49:7671–7709. doi: 10.1039/D0CS00304B. [DOI] [PubMed] [Google Scholar]
- Wongkaew N. Simsek M. Griesche C. Baeumner A. J. Chem. Rev. 2019;119:120–194. doi: 10.1021/acs.chemrev.8b00172. [DOI] [PubMed] [Google Scholar]
- Furst A. L. Francis M. B. Chem. Rev. 2019;119:700–726. doi: 10.1021/acs.chemrev.8b00381. [DOI] [PubMed] [Google Scholar]
- Dhanjai Sinha A. Kalambate P. K. Mugo S. M. Kamau P. Chen J. Jain R. TrAC, Trends Anal. Chem. 2019;118:488–501. doi: 10.1016/j.trac.2019.06.014. [DOI] [Google Scholar]
- Blair E. O. Corrigan D. K. Biosens. Bioelectron. 2019;134:57–67. doi: 10.1016/j.bios.2019.03.055. [DOI] [PubMed] [Google Scholar]
- Ji C. Zhou Y. Leblanc R. M. Peng Z. ACS Sens. 2020;5:2724–2741. doi: 10.1021/acssensors.0c01556. [DOI] [PubMed] [Google Scholar]
- Nekoueian K. Amiri M. Sillanpaa M. Marken F. Boukherroub R. Szunerits S. Chem. Soc. Rev. 2019;48:4281–4316. doi: 10.1039/C8CS00445E. [DOI] [PubMed] [Google Scholar]
- Dai Y. Liu C. C. Angew. Chem., Int. Ed. 2019;58:12355–12368. doi: 10.1002/anie.201901879. [DOI] [PubMed] [Google Scholar]
- Kim J. Campbell A. S. de Avila B. E. Wang J. Nat. Biotechnol. 2019;37:389–406. doi: 10.1038/s41587-019-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst A. L. Muren N. B. Hill M. G. Barton J. K. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14985–14989. doi: 10.1073/pnas.1417351111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povedano E. Valverde A. Montiel V. R. Pedrero M. Yanez-Sedeno P. Barderas R. San Segundo-Acosta P. Pelaez-Garcia A. Mendiola M. Hardisson D. Campuzano S. Pingarron J. M. Angew. Chem., Int. Ed. 2018;57:8194–8198. doi: 10.1002/anie.201804339. [DOI] [PubMed] [Google Scholar]
- Alafeef M. Dighe K. Moitra P. Pan D. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J. Cederquist K. B. Zaragoza A. A. Lee P. E. Sargent E. H. Kelley S. O. Nat. Chem. 2012;4:642–648. doi: 10.1038/nchem.1367. [DOI] [PubMed] [Google Scholar]
- Wan Y. Zhou Y. G. Poudineh M. Safaei T. S. Mohamadi R. M. Sargent E. H. Kelley S. O. Angew. Chem., Int. Ed. 2014;53:13145–13149. doi: 10.1002/anie.201407982. [DOI] [PubMed] [Google Scholar]
- Das J. Ivanov I. Montermini L. Rak J. Sargent E. H. Kelley S. O. Nat. Chem. 2015;7:569–575. doi: 10.1038/nchem.2270. [DOI] [PubMed] [Google Scholar]
- Das J. Ivanov I. Sargent E. H. Kelley S. O. J. Am. Chem. Soc. 2016;138:11009–11016. doi: 10.1021/jacs.6b05679. [DOI] [PubMed] [Google Scholar]
- Wan X. Volpetti F. Petrova E. French C. Maerkl S. J. Wang B. Nat. Chem. Biol. 2019;15:540–548. doi: 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- Yang Y. Song Y. Bo X. Min J. Pak O. S. Zhu L. Wang M. Tu J. Kogan A. Zhang H. Nat. Biotechnol. 2020;38:217–224. doi: 10.1038/s41587-019-0321-x. [DOI] [PubMed] [Google Scholar]
- Sadat Mousavi P. Smith S. J. Chen J. B. Karlikow M. Tinafar A. Robinson C. Liu W. Ma D. Green A. A. Kelley S. O. Pardee K. Nat. Chem. 2020;12:48–55. doi: 10.1038/s41557-019-0366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A. Croce C. M. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Tavallaie R. McCarroll J. Le Grand M. Ariotti N. Schuhmann W. Bakker E. Tilley R. D. Hibbert D. B. Kavallaris M. Gooding J. J. Nat. Nanotechnol. 2018;13:1066–1071. doi: 10.1038/s41565-018-0232-x. [DOI] [PubMed] [Google Scholar]
- Chinnadayyala S. R. Park J. Le H. T. N. Santhosh M. Kadam A. N. Cho S. Biosens. Bioelectron. 2019;126:68–81. doi: 10.1016/j.bios.2018.10.038. [DOI] [PubMed] [Google Scholar]
- Hou Y. H. Wang J. J. Jiang Y. Z. Lv C. Xia L. Hong S. L. Lin M. Lin Y. Zhang Z. L. Pang D. W. Biosens. Bioelectron. 2018;99:186–192. doi: 10.1016/j.bios.2017.07.035. [DOI] [PubMed] [Google Scholar]
- Li B.-R. Hsieh Y.-J. Chen Y.-X. Chung Y.-T. Pan C.-Y. Chen Y.-T. J. Am. Chem. Soc. 2013;135:16034–16037. doi: 10.1021/ja408485m. [DOI] [PubMed] [Google Scholar]
- Mahshid S. S. Camire S. Ricci F. Vallee-Belisle A. J. Am. Chem. Soc. 2015;137:15596–15599. doi: 10.1021/jacs.5b04942. [DOI] [PubMed] [Google Scholar]
- Mahshid S. S. Mahshid S. Vallee-Belisle A. Kelley S. O. Anal. Chem. 2019;91:4943–4947. doi: 10.1021/acs.analchem.9b00648. [DOI] [PubMed] [Google Scholar]
- Das J. Gomis S. Chen J. B. Yousefi H. Ahmed S. Mahmud A. Zhou W. Sargent E. H. Kelley S. O. Nat. Chem. 2021;13:428–434. doi: 10.1038/s41557-021-00644-y. [DOI] [PubMed] [Google Scholar]
- Yousefi H. Mahmud A. Chang D. Das J. Gomis S. Chen J. B. Wang H. Been T. Yip L. Coomes E. Li Z. Mubareka S. McGeer A. Christie N. Gray-Owen S. Cochrane A. Rini J. M. Sargent E. H. Kelley S. O. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- Furst A. L. Hoepker A. C. Francis M. B. ACS Cent. Sci. 2017;3:110–116. doi: 10.1021/acscentsci.6b00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P. R. Masur H. Crit. Care Med. 2012;40:3277–3282. doi: 10.1097/CCM.0b013e318270e771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opota O. Croxatto A. Prod'hom G. Greub G. Clin. Microbiol. Infect. 2015;21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Reddy Jr B. Hassan U. Seymour C. Angus D. C. Isbell T. S. White K. Weir W. Yeh L. Vincent A. Bashir R. Nat. Biomed. Eng. 2018;2:640–648. doi: 10.1038/s41551-018-0288-9. [DOI] [PubMed] [Google Scholar]
- Hassan U. Ghonge T. Reddy Jr B. Patel M. Rappleye M. Taneja I. Tanna A. Healey R. Manusry N. Price Z. Jensen T. Berger J. Hasnain A. Flaugher E. Liu S. Davis B. Kumar J. White K. Bashir R. Nat. Commun. 2017;8:15949. doi: 10.1038/ncomms15949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. Jovic M. Lesch A. Tissieres Lovey L. Prudent M. Pick H. Girault H. H. Angew. Chem., Int. Ed. 2018;57:14942–14946. doi: 10.1002/anie.201808666. [DOI] [PubMed] [Google Scholar]
- Gauglitz G. Anal. Bioanal. Chem. 2020;412:3317–3349. doi: 10.1007/s00216-020-02581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. Huang C. Emery B. P. Sedgwick A. C. Bull S. D. He X. P. Tian H. Yoon J. Sessler J. L. James T. D. Chem. Soc. Rev. 2020;49:5110–5139. doi: 10.1039/C9CS00318E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W. Ye H. Yuan Z. Liu X. Chen X. Yang W. TrAC, Trends Anal. Chem. 2020;123:115767. doi: 10.1016/j.trac.2019.115767. [DOI] [Google Scholar]
- Liu B. Monshat H. Gu Z. Lu M. Zhao X. Analyst. 2018;143:2448–2458. doi: 10.1039/C8AN00144H. [DOI] [PubMed] [Google Scholar]
- Yang J. Wang K. Xu H. Yan W. Jin Q. Cui D. Talanta. 2019;202:96–110. doi: 10.1016/j.talanta.2019.04.054. [DOI] [PubMed] [Google Scholar]
- Kal-Koshvandi A. T. TrAC, Trends Anal. Chem. 2020;128:115920. doi: 10.1016/j.trac.2020.115920. [DOI] [Google Scholar]
- Elsherif M. Hassan M. U. Yetisen A. K. Butt H. ACS Nano. 2018;12:5452–5462. doi: 10.1021/acsnano.8b00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. Chen Y. Tang Y. Cheng G. Yu X. He H. Cao G. Lu H. Liu Z. Zheng S. Y. ACS Sens. 2019;4:3298–3307. doi: 10.1021/acssensors.9b01927. [DOI] [PubMed] [Google Scholar]
- Hou J. Li M. Song Y. Angew. Chem., Int. Ed. 2018;57:2544–2553. doi: 10.1002/anie.201704752. [DOI] [PubMed] [Google Scholar]
- Fenzl C. Hirsch T. Wolfbeis O. S. Angew. Chem., Int. Ed. 2014;53:3318–3335. doi: 10.1002/anie.201307828. [DOI] [PubMed] [Google Scholar]
- He M. Gales J. P. Ducrot E. Gong Z. Yi G. R. Sacanna S. Pine D. J. Nature. 2020;585:524–529. doi: 10.1038/s41586-020-2718-6. [DOI] [PubMed] [Google Scholar]
- Snapp P. Kang P. Leem J. Nam S. Adv. Funct. Mater. 2019;29:1902216. doi: 10.1002/adfm.201902216. [DOI] [Google Scholar]
- Jung I. Y. Kim J. S. Choi B. R. Lee K. Lee H. Adv. Healthcare Mater. 2017;6:1601475. doi: 10.1002/adhm.201601475. [DOI] [PubMed] [Google Scholar]
- Culver H. R. Clegg J. R. Peppas N. A. Acc. Chem. Res. 2017;50:170–178. doi: 10.1021/acs.accounts.6b00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz J. H. Asher S. A. Nature. 1997;389:829–832. doi: 10.1038/39834. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Zhao X. Gu Z. Adv. Funct. Mater. 2010;20:2970–2988. doi: 10.1002/adfm.201000098. [DOI] [Google Scholar]
- Inan H. Poyraz M. Inci F. Lifson M. A. Baday M. Cunningham B. T. Demirci U. Chem. Soc. Rev. 2017;46:366–388. doi: 10.1039/C6CS00206D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z. Sasmal A. Liu X. Asher S. A. ACS Sens. 2017;2:1474–1481. doi: 10.1021/acssensors.7b00426. [DOI] [PubMed] [Google Scholar]
- MacConaghy K. I. Geary C. I. Kaar J. L. Stoykovich M. P. J. Am. Chem. Soc. 2014;136:6896–6899. doi: 10.1021/ja5031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan C. Wang H. Han Q. Ma X. Zhang D. Xu Y. Chen B. Li M. Zhao Y. ACS Appl. Mater. Interfaces. 2018;10:21206–21212. doi: 10.1021/acsami.8b06882. [DOI] [PubMed] [Google Scholar]
- Qin J. Dong B. Li X. Han J. Gao R. Su G. Cao L. Wang W. J. Mater. Chem. C. 2017;5:8482–8488. doi: 10.1039/C7TC02140B. [DOI] [Google Scholar]
- Qin J. Dong B. Cao L. Wang W. J. Mater. Chem. C. 2018;6:4234–4242. doi: 10.1039/C8TC00242H. [DOI] [Google Scholar]
- Deng G. Xu K. Sun Y. Chen Y. Zheng T. Li J. Anal. Chem. 2013;85:2833–2840. doi: 10.1021/ac3033728. [DOI] [PubMed] [Google Scholar]
- Cai Z. Kwak D. H. Punihaole D. Hong Z. Velankar S. S. Liu X. Asher S. A. Angew. Chem., Int. Ed. 2015;54:13036–13040. doi: 10.1002/anie.201506205. [DOI] [PubMed] [Google Scholar]
- Li M. He F. Liao Q. Liu J. Xu L. Jiang L. Song Y. Wang S. Zhu D. Angew. Chem., Int. Ed. 2008;47:7258–7262. doi: 10.1002/anie.200801998. [DOI] [PubMed] [Google Scholar]
- Qin J. Li X. Cao L. Du S. Wang W. Yao S. Q. J. Am. Chem. Soc. 2020;142:417–423. doi: 10.1021/jacs.9b11116. [DOI] [PubMed] [Google Scholar]
- Fleischmann M. Hendra P. J. McQuillan A. J. Chem. Phys. Lett. 1974;26:163–166. doi: 10.1016/0009-2614(74)85388-1. [DOI] [Google Scholar]
- Granger J. H. Schlotter N. E. Crawford A. C. Porter M. D. Chem. Soc. Rev. 2016;45:3865–3882. doi: 10.1039/C5CS00828J. [DOI] [PubMed] [Google Scholar]
- Rickard J. J. S. Di-Pietro V. Smith D. J. Davies D. J. Belli A. Oppenheimer P. G. Nat. Biomed. Eng. 2020;4:610–623. doi: 10.1038/s41551-019-0510-4. [DOI] [PubMed] [Google Scholar]
- Kim W. Lee S. H. Kim J. H. Ahn Y. J. Kim Y. H. Yu J. S. Choi S. ACS Nano. 2018;12:7100–7108. doi: 10.1021/acsnano.8b02917. [DOI] [PubMed] [Google Scholar]
- Tran V. Walkenfort B. Konig M. Salehi M. Schlucker S. Angew. Chem., Int. Ed. 2019;58:442–446. doi: 10.1002/anie.201810917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Salazar J. R. Oliveira Jr O. N. Chem. Rev. 2018;118:10617–10625. doi: 10.1021/acs.chemrev.8b00359. [DOI] [PubMed] [Google Scholar]
- Kravets V. G. Kabashin A. V. Barnes W. L. Grigorenko A. N. Chem. Rev. 2018;118:5912–5951. doi: 10.1021/acs.chemrev.8b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T. Liang W. Li Y. Sun Y. Xiang Y. Zhang Y. Dai Z. Duo Y. Wu L. Qi K. Shivananju B. N. Zhang L. Cui X. Zhang H. Bao Q. Nat. Commun. 2019;10:28. doi: 10.1038/s41467-018-07947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Guardado A. Barkam S. Peppler M. Biswas A. Dennis W. Das S. Seal S. Chanda D. Nano Lett. 2019;19:449–454. doi: 10.1021/acs.nanolett.8b04253. [DOI] [PubMed] [Google Scholar]
- Luan J. Seth A. Gupta R. Wang Z. Rathi P. Cao S. Gholami Derami H. Tang R. Xu B. Achilefu S. Morrissey J. J. Singamaneni S. Nat. Biomed. Eng. 2020;4:518–530. doi: 10.1038/s41551-020-0547-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver H. R. Wechsler M. E. Peppas N. A. ACS Nano. 2018;12:9342–9354. doi: 10.1021/acsnano.8b04348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iravani B. Arshamian A. Ohla K. Wilson D. A. Lundstrom J. N. Nat. Commun. 2020;11:648. doi: 10.1038/s41467-020-14520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. Jangili P. Son S. Ji M. S. Won M. Kim J. S. Adv. Mater. 2020;32:2001945. doi: 10.1002/adma.202001945. [DOI] [PubMed] [Google Scholar]
- Mou X. Z. Chen X. Y. Wang J. Zhang Z. Yang Y. Shou Z. X. Tu Y. X. Du X. Wu C. Zhao Y. Qiu L. Jiang P. Chen C. Huang D. S. Li Y. Q. ACS Appl. Mater. Interfaces. 2019;11:23093–23101. doi: 10.1021/acsami.9b09279. [DOI] [PubMed] [Google Scholar]
- Wang Y. Pei H. Jia Y. Liu J. Li Z. Ai K. Lu Z. Lu L. J. Am. Chem. Soc. 2017;139:11616–11621. doi: 10.1021/jacs.7b06885. [DOI] [PubMed] [Google Scholar]
- Wang X. Cohen L. Wang J. Walt D. R. J. Am. Chem. Soc. 2018;140:18132–18139. doi: 10.1021/jacs.8b11185. [DOI] [PubMed] [Google Scholar]
- Wu C. Garden P. M. Walt D. R. J. Am. Chem. Soc. 2020;142:12314–12323. doi: 10.1021/jacs.0c04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. Lv T. Zhu K. Li Y. Wang L. Gooding J. J. Liu G. Liu B. Angew. Chem., Int. Ed. 2020;59:3131–3136. doi: 10.1002/anie.201915046. [DOI] [PubMed] [Google Scholar]
- Woo C. H. Jang S. Shin G. Jung G. Y. Lee J. W. Nat. Biomed. Eng. 2020;4:1168–1179. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Konforti M. Green O. Hupfeld M. Fieseler L. Heinrich N. Ihssen J. Vorberg R. Wick L. Spitz U. Shabat D. Angew. Chem., Int. Ed. 2019;58:10361–10367. doi: 10.1002/anie.201904719. [DOI] [PubMed] [Google Scholar]
- Xue L. Yu Q. Griss R. Schena A. Johnsson K. Angew. Chem., Int. Ed. 2017;56:7112–7116. doi: 10.1002/anie.201702403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q. Xue L. Hiblot J. Griss R. Fabritz S. Roux C. Binz P. A. Haas D. Okun J. G. Johnsson K. Science. 2018;361:1122–1126. doi: 10.1126/science.aat7992. [DOI] [PubMed] [Google Scholar]
- Yu Q. Pourmandi N. Xue L. Gondrand C. Fabritz S. Bardy D. Patiny L. Katsyuba E. Auwerx J. Johnsson K. Nat. Metab. 2019;1:1219–1225. doi: 10.1038/s42255-019-0151-7. [DOI] [PubMed] [Google Scholar]
- Liu Y. Zhan L. Qin Z. Sackrison J. Bischof J. C. ACS Nano. 2021;15:3593–3611. doi: 10.1021/acsnano.0c10035. [DOI] [PubMed] [Google Scholar]
- Sciutto G. Zangheri M. Anfossi L. Guardigli M. Prati S. Mirasoli M. Di Nardo F. Baggiani C. Mazzeo R. Roda A. Angew. Chem., Int. Ed. 2018;57:7385–7389. doi: 10.1002/anie.201713298. [DOI] [PubMed] [Google Scholar]
- Liu Z. Zhou X. Miao Y. Hu Y. Kwon N. Wu X. Yoon J. Angew. Chem., Int. Ed. 2017;56:5812–5816. doi: 10.1002/anie.201702114. [DOI] [PubMed] [Google Scholar]
- Wang J. Jiang C. Jin J. Huang L. Yu W. Su B. Hu J. Angew. Chem., Int. Ed. 2021;60:13042–13049. doi: 10.1002/anie.202103458. [DOI] [PubMed] [Google Scholar]