Abstract
Recent times have experienced more than ever the impact of viral infections in humans. Viral infections are known to cause diseases not only in humans, but also in plants and animals. Here, we have compiled the literature review of aptamers selected and used for detection and inhibition of viral infections in all three categories: humans, animals, and plants. This review gives an indepth introduction to aptamers, different types of aptamer selection (SELEX) methodologies, the benefits of using aptamers over commonly used antibody-based strategies, and the structural and functional mechanism of aptasensors for viral detection and therapy. The review is organized based on the different characterization and read-out tools used to detect virus-aptasensor interactions with a detailed index of existing virus-targeting aptamers. Along with addressing recent developments, we also discuss a way forward with aptamers for DNA nanotechnology-based detection and treatment of viral diseases. Overall, this review will serve as a comprehensive resource for aptamer-based strategies in viral diagnostics and treatment.
Keywords: viruses, sensing, inhibition, aptamers, aptasensors, DNA nanostructures
Graphical Abstract

Viral infections are a major cause of disease in humans, plants, and animals. Aptamers (shown in an assembly line on a conveyor belt) are being developed as molecular tools for creating diagnostics and therapeutics for viral infections.
INTRODUCTION
Detection and treatment of viral infections is an ever-necessary aspect of biomedical science with viruses such as human immunodeficiency virus-1 (HIV-1) and hepatitis C virus (HCV) killing millions of people every year1-3. While several diagnostic methods have been developed for viral infections, global pandemics such as the current COVID-19 scenario demonstrates the need for multiple alternate strategies for rapid detection of viral infections. There is also a need for more efficient diagnostic tools with focus on aspects such as rapid detection, accuracy, affordability, and portability of the assay so that detection strategies are useful in low-resource settings. For treating viral infections, new methods that address molecular mechanisms of viral infection could be potent in creating therapeutics for a range of viral diseases with similar infection routes. Recently, several materials have been developed toward more effective viral diagnostics and treatment4-6. For example, graphene-based materials have been used for detection of viruses7 and nanoporous carbon-based materials and thin film based coatings have been used in developing protective equipment against viruses8. Nucleic acid engineering has also been a rapidly developing area in biosensing and drug delivery. Among these, aptamer-based methods have attracted more attention due to their applicability in a wide range of disciplines such as pathogen and toxin diagnostics9, therapeutics10,11, water quality control12, and imaging13.
Aptamers are single stranded oligonucleotide (ssDNA or ssRNA) ligands comprised of 10 to 100 nucleotides that can exhibit high affinity and specificity to their selected target. Originally discovered by Gold and Szostak14,15, they have now been developed by several research groups and considered as the nucleic acid equivalent of the antibody. Their low immunogenicity, small size, batch to batch reproducibility, ability to be chemically modified, and low cost of production have positioned aptamers to replace antibodies in many circumstances16-19. More specifically, aptamers are structurally more flexible and are magnitudes smaller than antibodies, facilitating their ability to recognize regions of the antigen that are otherwise inaccessible to antibodies. Their smaller size also aids in higher cell entry for in situ diagnostics, imaging, and disease treatment. Aptamers can be synthesized in large scale using phosphoramidite chemistry at a low cost. Further, aptamers can recognize a wide range of targets, tolerate various storage and usage conditions at different temperatures and can return to their original conformation after denaturation and annealing, allowing repeated use20,21. The in vitro selection process of specific aptamers is less time consuming and cheaper compared to antibody production where generation and screening of monoclonal antibodies is laborious and requires highly specialized facilities. To date, aptamers have been selected with nM to μM KD, targeting ions, small molecules as well as biological molecules and cells in both buffer and physiological conditions, including blood plasma and serum, and employed in environmental sensing, bioimaging, disease diagnosis and treatment. Despite these advantages of aptamers over antibodies, it has to be pointed out that compared to antibodies, aptamers are easier to degrade in vivo and thus need to be modified either chemically or enzymatically for enhanced biostability. Also, aptamers are quickly excreted by renal filtration from the bloodstream and thus need to be attached to other higher-molecular-weight molecules such as cholesterol or polyethylene glycol for prolonged bioavailability when used as diagnosis/imaging reagents or as part of drug complexes. In Table 1 we briefly summarize the pros and cons of using aptamers for virus detection and inhibition when compared with antibody-based methods22-24.
Table 1.
Comparison of the properties of antibodies and aptamers in virus diagnostics and therapeutics.
| Properties | Aptamer | Antibody |
|---|---|---|
| Material | Oligonucleotides (DNA or RNA) | Proteins |
| Target | Wide range of targets such as elements, ions, peptides, proteins, cells, and viruses | Proteins and peptides |
| Size | ~ 20 kDa | ~ 150 kDa |
| Immunogenicity | Low | High |
| Development period | 3-6 weeks | Months |
| Manufacturing | Chemical synthesis | Biological manufacturing |
| Storage | Room temperature | Cold temperature |
| Shelf-life | Unlimited | Limited |
| Binding affinity | Nanomolar to picomolar range | Nanomolar to picomolar range |
| Stability | Stable in various environmental conditions | Special conditions are required for handling and storage |
| Clinical application | Immature | Mature |
| Specificity | High | High |
| Chemical modifications | Easy and controllable | Limited and uncontrollable |
Aptamer structures consists of a diverse set of secondary motifs such as stem-loop, hairpin, pseudoknot, kissing loop, three-way junction, G-quadruplex, and internal bulge structures25. Aptamer-target binding occurs through a variety of non-covalent interactions such as hydrogen bonding, electrostatic, Van der Waals and hydrophobic interaction. The binding affinity of aptamers to their targets is also mediated by the environment, including buffer and ion composition and concentration, pH, and temperature26. The overall three-dimensional shape and conformation of the target molecule defines the strength of the binding affinity of the selected aptamer27 (Figure 1). For example, aptamers can differentiate between closely associated molecules and even between different chirality and recognize a specific epitope of a target molecule28. Previous studies have established that aptamers can bind to a variety of targets such as bacteria and viruses29, proteins30, prions31, and other small molecules15. Specifically, various aptamers have been developed to recognize key structural or metabolic determinants associated with bacterial and viral pathogens23. This specificity towards clinically-relevant biomarkers makes aptamers useful in biosensing. Recent studies have developed a range of aptamers for different viruses including vaccinia virus, dengue virus, severe acute respiratory syndrome (SARS), hepatitis C virus (HCV), human immunodeficiency virus (HIV), apple stem pitting virus, norovirus, rabies virus, bovine viral diarrhoea virus, hepatitis B, Ebola and influenza32,33. For these interactions to be used in sensing, aptamers are coupled with a variety of detection formats such as fluorescence, radioisotope, electrochemical, optical, colorimetric, and enzyme-linked assays. In addition to detection capabilities, aptamers can block proteins or molecules from binding their target and inhibit viral infection by stopping further replication, thus preventing the infection from spreading. This aptamer recognition can also act as a targeting tool for drug delivery (for example, using an aptamer that is specific to a tumor cell receptor). In this review, we discuss the structure, function, and development of aptamers, as well as their use in viral detection, inhibition, and therapy. We also discuss an outlook into promising developments in DNA nanotechnology that has the potential to use aptamers for sensing and therapeutic applications.
Figure 1.
Aptamer configurations and targeting. (a) Secondary structures of the aptamers. Mechanism of aptamer binding through molecular recognition and folding for (b) biosensing and (c) drug delivery.
APTAMER SELECTION BY SELEX
Aptamers can be selected specifically for single atoms/ions, molecules, viruses, bacteria, eukaryotic cells, and tissues by using different types of systematic evolution of ligands by exponential enrichment (SELEX) strategies and modifications to the incubation conditions (e.g., pH, temperature, buffer, etc.) (Figure 2)34,35. Aptamer selection relies on the distinctive secondary and tertiary structures that assist in target binding. The SELEX method starts with a combinatorial library that consists of two constant primer regions flanking a randomized segment of 20-50 nucleotides. Iterative rounds of incubation, separation and amplification enrich oligonucleotides that bind the intended target from the initial library. Incubation of the libraries with the target can be carried out in several ways. Usually, the target of interest is immobilized on a surface that can be washed or separated from the bulk liquid (centrifugation or magnetic separation of particles)36. The bound sequences that remain after binding and washing are then released chemically and/or thermally. This provides a phenotype-genotype linkage, where the desired binding sequences can then be amplified using polymerase chain reaction (PCR) (for DNA-based libraries) or reverse transcription PCR (for RNA-based libraries). This process is repeatedly performed for 8 to 15 rounds to get a desired pool of target-specific aptamers. Increasingly stringent conditions during library binding are utilized to obtain high-affinity aptamers, and negative (against the solid support) and counter (against unintended targets) selections are performed to increase the specificity of the evolved pool. At the end of the SELEX procedure, the identity of the enriched aptamer pool is determined by cloning and sequencing37,38. The SELEX technique is a potential method for assessing aptamers against a variety of target molecules and is crucial for developing novel aptamer-based detection procedures.
Figure 2. Outline of SELEX.
(a) A degenerate nucleic-acid sequence library is incubated with the target molecule under defined solution conditions. (b) Target-bound nucleic acids are partitioned. (c–e) Species with lower binding affinity are removed and the bound species are eluted, allowing preferential amplification of higher affinity species. This enriched pool is then used as the starting point in subsequent cycles. Typically, 10 to 20 cycles are carried out before aptamer characterization. In early rounds, species with no affinity are competed out of the pool. In later rounds, molecules with affinity compete for binding sites on the target. Such competition results in enhancement of the pool binding-affinity in a manner similar to Darwinian evolution. Recent technical developments described in the text are listed alongside each step in brackets. CE, capillary electrophoresis; SELEX, systematic evolution of ligands by exponential enrichment; SPR, surface plasmon resonance. Image reproduced with permission from ref. 75. Copyright 2006 Springer Nature.
Cell SELEX.
Since its inception in 1990, there have been many advancements to the SELEX technology. Cell-SELEX is a modified form of SELEX used to develop aptamers against a whole cell39. Cell-SELEX provides a wide flexibility to target unknown cell particles and detection of various cell types (bacteria, viruses, and tumor cells), primarily targeting the extracellular proteins present on the outer membrane of the cells or the characteristic structures specific to the cells. Cell-based SELEX processes have an additional step such as centrifugation or washing depending on the nature of the cells (adhesive or suspension). The bound sequences are collected and incubated with a negative control cell where the unbound nucleotides are then collected and used for negative selection round40. Once the aptamers are developed, they can be used for diverse applications such as drug delivery, cell- specific therapy, and cell surface diagnostics41,42. Since the aptamers developed by Cell-SELEX may target molecules that have not been characterized as cell-specific surface molecules, they might be novel biomarkers. As a result, Cell-SELEX may be used to explore new biomarkers for a specific cell. Further, Cell-SELEX may be used to manufacture aptamers against a particular target protein present on living cells such as transmembrane proteins (receptor kinases, G protein-coupled receptors, and ion channels)43,44. For example, Tang et al. used Cell-SELEX to produce aptamers by against adenocarcinoma epithelial cells infected with the vaccina virus (A549)45.
Complex-target SELEX.
This process involves the use of genetically modified cells. The whole cell is used as a target and these cells include genetically modified cells which over-express a target recombinant protein on the cell surface. Using a parallel selection process, multiple aptamers are selected for multiple targets in a single experiment and sequential target selection (X-SELEX) selects aptamers that bind to the multiple forms of a single target. The main advantage of this methodology is the ability to target and specifically differentiate microbial strains without knowing the details of the membrane structure or molecules present in any particular microorganism46. Pan et al. isolated aptamers for Rous sarcoma viruses (RSV), an enveloped avian retrovirus using this method47. Aptamers were specific to RSV surface glycoproteins that are necessary for binding and entry into host cells. Inhibition of viral infection was identified after 12 rounds of selection by chosen pools.
Genomic SELEX.
The conventional DNA library uses a chemically synthesized library whereas the genomic library uses the genomic DNA library48. Genomic SELEX is prepared via random priming, PCR amplification and in vitro transcription which creates an initial library49. This library is then transcribed into RNA and used for the selection process. At first, a counter selection method is performed at the immobilization matrix level. Due to the reversal of the transcription in several rounds, it can cause severe effects on highly structured RNA and is more acute in the case of genomic SELEX. Nucleic acid-binding proteins with diverse specificities and affinities are the most prevalent baits used in Genomic SELEX. Some examples of such nucleic acid binding proteins (associated to regulatory non-coding RNAs) are those involved in transcriptional and post-transcriptional silencing, chromatin remodeling, and components of machinery that participate in transport, RNA processing, and translation50-53. The utility of such proteins as targets enhances the potency of Genomic SELEX for analyzing RNA-protein interactions.
Microfluidic SELEX (M-SELEX).
This process generates DNA aptamers by employing conventional SELEX within a microfluidic system54. M-SELEX increases the stringency of the selection by utilizing a minimum amount of target molecules. It is a cost-effective method that consumes a small number of reagents, can be automated and multiplexed, and reduces the time required for the selection of new aptamers. For example, M-SELEX was used to isolate aptamers against hepatitis C virus (HCV) RNA polymerase55. A wide variety of M-SELEX methods have now emerged from employing microfluidic incubation, amplification, and separation techniques.
Magnetic bead-based microfluidic SELEX.
As one of the most widely used methods, this process uses a micro or nanosized magnetic bead as solid support for target binding. Here, a magnetically activated chip-based separation takes place in a continuous flow56-58. The screening of aptamers in magnetic bead-based M-SELEX takes place by incubating target-immobilized magnetic bead with a random nucleic acid library. Afterward, unbound nucleic acids are separated from target-bound aptamers by performing a stringent washing in the microfluidic channel. After separation, the external magnet is removed, and the attached, selected aptamers are collected for further PCR amplification. Small molecules, proteins, and cell surfaces have been used as effective targets for magnetic bead-based selection approaches56. For example, Soh's team designed a high-efficiency continuous-flow magnetic activated chip-based separation (CMACS) system that combines microfluidics technology with magnetic bead-assisted SELEX. The highly localized magnetophoretic forces and magnetic field gradients present in this system allows separation of the target protein with high purity. In addition, nonspecific binding was reduced by using carboxylic acid-coated beads on negatively charged oligonucleotides, further increasing the efficiency of the selection process59.
Capillary electrophoretic (CE) SELEX.
This method was the first microfluidic technique to yield a highly rapid SELEX process60-63. The difference in the electrophoretic mobility of the components in a mixture is used as the separation tool in capillary electrophoresis. The change in the size and charge of the target-aptamer complex decreases their mobility compared to unbound DNA or RNA sequences with high negative charge density. Several aptamers targeting HIV-1 reverse transcriptase, Lactobacillus acidophilus, and adenosine have been isolated by CE SELEX64-66.
Sol-gel method.
This is another prominent microfluidic-based SELEX that overcomes the uncertainty behind the effects of target immobilization on its conformation as well as blockage of binding sites67. Here, the sol consists of silica derivatives and the addition of chemical additives solidifies the sol into a nanoporous framework which enables the trapping of the target molecule within the gel. The nanoporous gel provides an aqueous domain to conserve the biological activity and stability of the entrapped target. Park et al. developed the first nanoporous sol-gel protein microfluidic array for entrapping target molecules and enabled the selection of RNA aptamers against multiple target molecules68. Jiyoung et al. used a sol-gel M-SELEX for high throughput characterization and selection of RNA aptamers. This microchip used a sol-gel network to immobilize the targeted protein and a localized heat source to selectively extract the RNA aptamers from the targeted protein69.
AFM-SELEX.
In this method, the aptamer is immobilized on a cantilever via biotin-streptavidin and target molecules are immobilized on a gold chip70. If the target-aptamer affinity is very strong, the biotin-streptavidin interaction of the aptamer to cantilever breaks and the aptamer remains on the gold surface. The DNA is then recovered by heat elution followed by PCR for amplification70. Such a method has been used to select a DNA aptamer against thrombin with very strong affinity (KD = 200 pM)71.
Toggle-SELEX.
Toggle-SELEX is a selection method for aptamers that can bind to a particular target present in different organisms. For instance, RNA aptamers that can bind to both porcine and human thrombin are selected by “toggling” the target between the human and porcine thrombin during alternate rounds of selection. In the first round, the library is incubated with both porcine and human thrombin. Aptamers that are bound to the protein are then recovered and amplified72. Using this process, Derbyshire et al isolated aptamers capable of targeting several aminoglycosides73. While toggle-SELEX is useful to develop cross-reactive aptamers, sometimes affinity may be compromised during selection process74.
APTASENSOR BASED DETECTION OF VIRAL PATHOGENS
As evidenced by the ongoing COVID-19 pandemic, diagnosis of viral pathogens at early stages of infection is crucial for the prevention and early treatment of viral infections. Current gold standard methods for detecting viral infections include nucleic acid testing (NAT)76 and antigen-antibody based ELISA tests77, and other common methods include viral plaque assay78, flow cytometry79, and hemagglutination assay80. NAT methods are amplification-based enzymatic assays that detect viral genetic material (DNA or RNA) typically using PCR. Although NAT is sensitive, it requires labor-intensive, laboratory-based sample preparation protocols for viral lysis, extraction of genetic materials, purification of the isolated materials, thermal cycling for enzymatic amplification of viral nucleic acid sequences, and interpretation of complex results by skilled personnel. Immune assays test for viral antigens or antibodies and are in general rapid, but less sensitive. There have been tremendous efforts in the development of alternate methods for faster and low-cost methods for detecting cellular and disease biomarkers81,82. In this section, we discuss aptamer-based sensors, (called aptasensors) that are surface-based assays (where the aptamer is immobilized on a surface) or solution-based assays (where they are mixed with analytes in solution). In such assays, aptamer-analyte binding transduces a detectable signal which can be readout by electrochemical, optical, or enzyme-linked methods.
Electrochemical aptasensors
Electrochemical aptasensors rely on the immobilization of an aptamer ligand on a conductive surface. Gold or carbon-based electrodes are commonly used for this purpose, as their surfaces are hydrophobic, inert, and easily functionalized to provide robust attachment of many aptamer ligands. Different chemical strategies have been developed to immobilize aptamers on electrode surfaces. These include chemisorption (attachment of the aptamer to the surface by noncovalent interactions such as electrostatic, hydrogen bonding and π-π stacking), biotinylation of aptamers to bind avidin-modified surfaces, covalent linkage mechanisms such as click chemistry (azide-modified aptamer to the alkyne-modified surfaces) and chemical crosslinking (coupling of the amine-modified aptamers to carboxyl-modified surface)83. Electrochemical aptasensors use electrochemically active species to provide a readout, where an electrode-immobilized aptamer serves as a transducer. Some examples of electrochemically active species include organic molecules such as ferrocene, methylene blue (MB) and thionine, which are redox-active molecules that interact with DNA via intercalation and electrostatic interactions. Their presence facilitates the transfer of electrons from the aptamer-target binding site to the electrode surface and provides a more sensitive electrochemical output83,84. Electrochemical changes that result from the formation of aptamer-analyte complex can be detected in the form of current, potential and impedance, typically measured by amperometric, potentiometric, and conductometric techniques. These include electrochemical impedance spectroscopy, cyclic voltammetry, differential pulse voltammetry, square wave voltammetry, field effect transistor, linear sweep voltammetry, and potentiometry85. Electrochemical aptasensors present advantages such as repeatability, accuracy, high sensitivity, low cost, easy miniaturization and robustness86. These detection methods can also be transformed into a chip-based platform for use as portable devices at point-of-care28.
Voltammetry and electrical impedance aptasensor
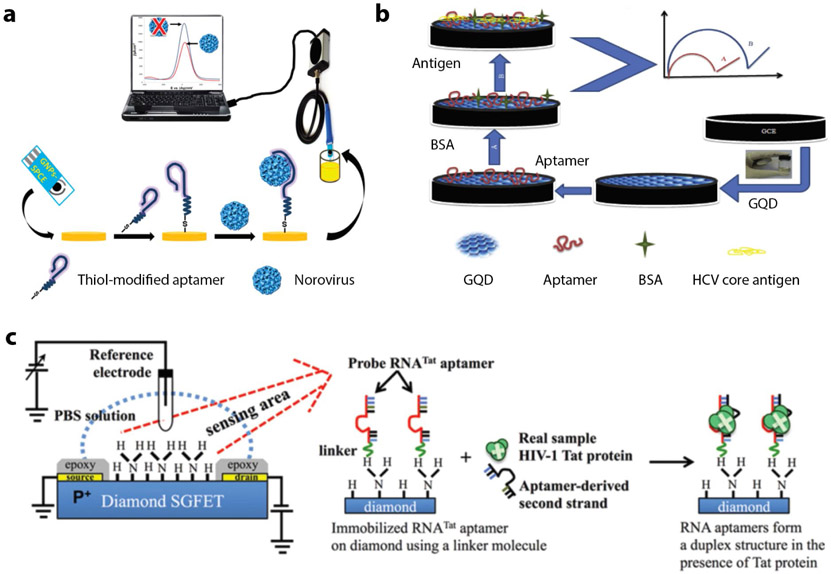
Giamberardino et al. developed an ultrasensitive electrochemical norovirus detection system using aptamers evolved to bind both murine norovirus (MNV) and human norovirus (HuNoV) with picomolar affinity87 (Figure 3a). Aptamer AG3 that selectively binds MNV and the HuNoV strain GII.3 was modified with a thiol-group at the 5’ end and subsequently immobilized on gold nanoparticle-modified carbon electrodes. Using square wave voltammetry readout technique and a ferricyanide/ruthenium hexamine redox reporter system, the norovirus aptasensor exhibited a limit of detection (LoD) of 10 aM, or 180 virus particles for MNV. Lum et al. developed an impedimetric aptasensor for the detection of the avian influenza virus (AIV) H5N1. A biotin-labelled, H5N1-specific DNA aptamer was immobilized on streptavidin-modified gold interdigitated microelectrodes that were embedded in a microfluidics chip88. The virus was allowed to interact with the aptamer-coated electrodes for 30 mins before measuring impedance. The difference in the impedance of the virus:aptamer-electrode complex and the aptamer-electrode alone indicates presence of the virus, with an LoD of 0.0128 hemagglutinating units (HAU). Detection methods for influenza A89 and vaccinia virus86 were also constructed using such an electrochemical aptasensor approach. A similar technique was used in detecting HCV90 using an aptamer evolved to bind the HCV core antigen that was chemisorbed on graphene quantum dot (GQD)-coated electrodes (Figure 3b). Electrochemical impedance spectroscopy was used to monitor changes in electrical signals upon aptamer interaction with HCV antigen. The purported mechanism is that the aptamer: HCV antigen complex increases the ability of the redox species in solution to reach the electrode. This “signal-on” mechanism provided an LoD of 3.3 pg/ml.
Figure 3. Electrochemical aptasensors.
(a) A thiolated norovirus-specific DNA aptamer self-assembled onto a gold nanoparticle-modified screen-printed carbon electrode. Binding of the virus to the immobilized aptamer causes a decrease in the redox current, measured via square wave voltammetry. Reproduced from ref.87. (b) Use of glassy carbon electrode (GCE) with graphene quantum dots for HCV core antigen detection. Reproduced with permission from ref. 90. Copyright 2017 Elsevier. (c) Schematic structure of diamond-FET-based RNA aptamer for HIV-1 Tat protein detection based on changes in the surface charge. Reproduced with permission from ref. 92. Copyright 2013 Elsevier.
Field-effect transistor-based aptasensors
A field-effect transistor (FET) is a voltage-controlled three-electrode (source, gate and drain) system that acts as a transducer and converts signals generated by the detected target to an electrical readout. Aptamers are immobilized on a FET electrode surface that measures the charge distribution when the target binds to the aptamer. Various biomolecules such as proteins91, viruses92 and nucleic acids93 have been detected by FET-integrated aptasensors. Ruslinda et al. demonstrated diamond FET for the detection of HIV-1 using RNA aptamers as the sensor element92 (Figure 3c). The group used the RNATAT aptamer to detect HIV1 trans-activator transcription (Tat) protein that contributes to several pathological symptoms of HIV-1 infection and plays a critical role in virus replication.
Piezoelectric aptasensors
Certain materials possess the ability to generate electric charge in response to applied mechanical stress, known as the piezoelectric effect. Quartz crystal microbalance (QCM) is the most common piezoelectric sensor used for the detection of biomarkers94. QCM-based strategies have also used aptamers immobilized on gold-coated quartz as sensing elements. Binding of the target to the aptamers increases the mass on the surface of the crystals which generates a detectable signal due to the decrease in the resonance frequency of the crystal. Wang et.al. developed a ssDNA-crosslinked polymeric hydrogel to form a network of water-insoluble polymer chains in a QCM aptasensor for the rapid detection of AIV H5N195. An aptamer specific to AIV H5N1 surface protein was hybridized to the ssDNA, thus crosslinking the hydrogel in a shrunken state. The aptamer-hydrogel complex was fixed on the gold surface of the QCM sensor using a self-assembled monolayer method. When the surface protein binds to the aptamer, the aptamer is released from the hydrogel complex, causing the hydrogel to swell, with the changes transduced to a detectable decreased frequency. The assay time for this method was 30 min with a detection limit of 0.0128 HAU95.
Enzyme-linked electrochemical aptasensors
Enzyme-based biosensors have also been used in combination with aptamers to detect viral pathogens. For example, an electrochemical aptasensor was coupled to a glucose oxidase (GO) enzyme-based readout for the detection of H5N1 (Figure 4a)96. They used a complex consisting of gold nanoparticles, glucose oxidase, concanavalin A (AuNPs-GOx-ConA) and the capturing aptamers were embedded on magnetic beads. The complex triggered an enzymatic catalysis that in turn increased the ion concentration and decreased the impedance, with the changes measured by electrical impedance spectroscopy. The LoD for the technique was 8x10−4 HAU in a 200 μl reaction. Another study involving enzymatic electrochemical detection of H5N1 utilized AuNP-modified electrodes which were coated with the capturing aptamers (Figure 4b)97. The functionalization of the electrodes with 3-mercaptopropionic acid and the presence of the anti-H5N1 antibodies modified with alkaline phosphatase generates an electrochemical signal with an LoD of 100 fM.
Figure 4. Enzyme-linked electrochemical aptasensors.
(a) Enzyme catalysis in ultra-low ion strength media to develop an ion strength increase-based impedance biosensor for H5N1 virus. Reproduced from ref.96. (b) Schematic diagram of H5N1 viral protein detection using the enzymatic reaction of the substrate 4-amino phenyl phosphate with the surface formed aptamer/H5N1/antiH5N1-alkaline phosphatase on gold nanoparticle-modified screen-printed carbon electrode. Reproduced with permission from ref. 97. Copyright 2015 Elsevier. (c) Working principle of the norovirus nanozyme aptasensor. Reproduced with permission from ref. 100. Copyright 2019 American Chemical Society.
Nanozymes, artificial nanomaterials that imitate properties of natural enzymes, have also been used in aptasensors. The chemical makeup of nanozymes include simple metal and metal oxide nanoparticles, metal nanoclusters, quantum dots, nanotubes, nanowires, or metal organic framework98. Compared to conventional enzymes, nanozymes exhibit improved catalytic activity, lower cost of production, greater robustness and modification capabilities, and long-term storage and shelf-life99. Such a nanozyme based aptasensor was used to detect Norovirus (HuNoV)100. This aptasensor employs the murine norovirus (MNV) specific aptamer (AG3) as the molecular recognition element, with a detection limit of 200 virus particles/ml in 10 min (Figure 4c).
Optical detection-based aptasensors
The aptasensor field is replete with optical sensors that rely on fluorescence, luminescence, colorimetry and plasmon or refractive index-related changes for analyte detection. Optical biosensing is divided into two modes: label-free, where target binding is analyzed by a coupled material that transduces an optical signal, and label-based, where colorimetric, luminescent, or fluorescent labels on either the aptamer or the target or both generate an optical signal. Optical biosensors use various molecular recognition elements such as nucleic acids, enzymes, antibodies, whole cells, and tissues42,101.
Surface plasmon resonance aptasensors
Surface plasmon resonance (SPR) aptasensors are based on the change in the refractive index of a metal, typically gold, surface due to the resonant oscillation of free electrons. In the SPR technique102, polarized light of a specific wavelength and incident angle passes through a prism and is reflected off the gold surface. SPR aptasensors are label-free and have recently been developed to be miniaturized, portable, and automated103. For example, Tombelli et al. developed an SPR aptasensor for HIV-1 detection with an LoD of 0.25 ppm (ref.104). Here, a biotinylated RNA aptamer that recognizes the Tat protein was immobilized on an avidin-modified gold surface. Bai et al. developed a portable and fast SPR-based aptasensor for AIV H5N1 (Figure 5a).105 Using streptavidin-biotin interaction, a DNA aptamer that targets the glycosylated hemagglutinin (HA) viral protein was immobilized on gold surface. The resulting sensor was shown to detect in vitro isolated AIV H5N1 as well as AIV H5N1 from poultry swabs with an LoD of 0.128 HAU. This sensor allowed rapid detection within minutes, and with poultry swab preparation takes a total time of only 1.5 h.
Figure 5. Optical aptasensors.
(a) SPR based aptasensor: upon binding the target (virus) the surface plasmon angle of the reflected light changes resulting in a difference in plasmon resonance. (b) LSPR based biosensor for real-time detection: a large array of nanoantennas is incorporated into a microfluidic chamber system that guides analyte solutions precisely over the sensitive area. Optical readout is realized with a spectrometer and spectra are continuously recorded upon chemical reactions; the inset illustrates the investigated biochemical reaction, which is immobilization, backfilling, and hybridization of short DNA sequences. Reproduced from ref.107. (c) Schematic illustration of the preparation of aptamer-Ag@SiO2 sensor and the determination of rHA protein of H5N1. Reproduced with permission from ref. 108. Copyright 2015 Elsevier. (d) Schematic of selective virus sizing and counting by fluorescent nanoparticle tracking. Reproduced from ref.109. (e) SERS imaging-based assay using a 3D nano-popcorn plasmonic aptasensor: (i) Detection of DNA using Cy3-labeled aptamer probes (left) or recognition of A/H1N1 virus (right), (ii) resulting in increased Raman signal (left) or decreased Raman signal intensity (right), respectively. Reproduced with permission from ref. 117. Copyright 2020 Elsevier. (f) A sandwich-like aptasensor for influenza virus detection: 1) primary aptamer is immobilized onto Ag nanoparticles, 2) virus is captured with primary aptamers, 3) secondary aptamers interact with virus, providing the SERS signal. Reproduced from ref.118, (g) Schematic representation of the construction of a chemiluminescence aptasensor based on magnetic separation and immunoassay. Reproduced from ref.124, (h) Working principle for the single universal aptamer detection of different kinds of influenza viruses under two different reaction conditions. Reproduced with permission from ref. 127. Copyright 2016 Elsevier. (i) Molecular beacon aptamer strategy for analyzing the viral protein (Tat). (j) Protein-binding aptamer assisted detection of the H1N1 influenza A virus based on fluorescence polarization. Reproduced with permission from ref. 132. Copyright 2013 Royal Society of Chemistry.
Localized SPR (LSPR) is an optical phenomenon that occurs in metallic nanostructures (nanospheres, nanodiscs, and nanorods). Normally these localized metal nanostructures are designed within microfluidic channels to detect ssDNA in low ng/mL range giving cheaper alternative for biomolecule sensing. Incident light at the specific plasmonic wavelength irradiates the metallic structure, which induces collective electron charge oscillations. This ultimately leads to a shift in absorbance in the ultraviolet-visible region, which can be used to detect target binding106. Klinghammer et al. developed an aptasensor based on this mechanism using arrays of gold nanorods (Figure 5b)107. They used different complementary DNA (cDNA) strand hybridization kinetics to monitor the optical nanostructure resonance of densely packed gold nanorods upon binding of biomolecules. Red shift of 2 nm and 5 nm were detected upon binding of 25 and 100 bp respectively.
Another process of metal-enhanced fluorescence (MEF) occurs when there is an increased emission around specific metallic materials. When fluorophores are close to the surface of metal nanoparticles, the LSPR of the metal nanoparticles coupled with the excited fluorophores improves the detection sensitivity significantly. The signals can be detected by surface plasmon field-enhanced fluorescence. Pang et al. developed an aptasensor using this principle for detecting H5N1 virus (Figure 5c)108. A guanine-rich DNA aptamer that recognizes recombinant hemagglutinin (rHA) was immobilized on core-shell AgSiO2 nanoparticles and thiazole orange was added as a fluorescence amplification reporter. Target binding causes the aptamer to fold into a G-quadruplex structure. The thiazole orange previously in solution then binds to the G-quadruplex and its interaction with the core-shell nanoparticle causes an increase in fluorescence. Using this assay, H5N1 rHA could be detected in 30 mins in both buffer and when spiked in serum. Another study used aptamer-based fluorescent nanoparticles for the detection of the respiratory syncytial virus (RSV) (Figure 5d)109. This method could detect viral particles that are 80-100 nm.
Surface-enhanced Raman scattering (SERS) aptasensors
Raman spectroscopy is a promising optical technique due to its simple and cheap instrumentation and requires minimal sample preparation110. The SERS enhancement is due to (i) an enhanced electromagnetic field on a rough metallic surface which amplifies the incident light and therefore the Raman modes being detected, and (ii) direct enhancement of the Raman signal by the resonant surface. Together, these mechanisms can provide signal enhancements of 1010 to 1011 potentially allowing for single-molecule detection111,112. SERS-based biosensing can be categorized as direct or indirect. In the direct technique, detection is based on the Raman spectrum of the analyte itself without any reporter molecules (label-free techniques). This technique has been used for detecting hepatitis B virus (HBV)113, adenovirus, rhinovirus and HIV114. The indirect SERS-based technique involves the use of molecular recognition elements such as antibodies, aptamers or other specific binding molecules placed close to the “hot spots” (interparticle gap in a nanocluster) on the surface. These methods rely on the change in Raman signals from the recognition element (e.g., aptamers) or reporter molecules and not the analyte itself. The reporter molecules used in SERS-based methods are water-soluble, and easily conjugated or intercalated to oligonucleotides110,115,116. For example, Chen et al. a developed a SERS-based aptasensor using a Cy3-labelled DNA aptamer for the detection of influenza A (H1N1) virus with high sensitivity (Figure 5e)117. The biosensor consisted of a three-dimensional (3D) nano-popcorn plasmonic substrate fabricated by depositing gold layers on a polyethylene naphthalate (PEN) polymer substrate. On binding the virus, the Cy3-labelled DNA aptamer is released from the nano-popcorn substrate surface, causing a decrease in the Raman peak intensity. In another example, Kukushkin et al developed an aptasensor for the detection of influenza virus including H1, H3, H5 haemaglutinin subtypes (Figure 5f)118. The aptamer RHA0385 showed strain specificity to both recombinant hemagglutinins and whole cell viruses. The aptasensor consisted of a primary aptamer attached to the metal particles of the SERS substrate, and secondary aptamer labelled with Raman-active molecules. The influenza virus was captured and bound to the labelled secondary aptamer. The LoD for this aptasensor was 104 virus particles per sample or 10−4 HAU per sample.
Chemiluminescent aptasensors
The energy transition between the molecular orbitals emits light which is termed as luminescence. When this phenomenon occurs in the presence of a chemical reaction, it is termed as chemiluminescence119,120. Chemiluminescent methods have often been implemented as signal amplifiers to provide detection limits of 10−12 to 10−21 mol24,121,122. Ahn et al. developed an aptasensor to detect the nucleocapsid protein of SARS coronavirus (SARS-CoV)123. This aptasensor utilizes an aptamer-target-antibody sandwich method, where the nucleocapsid proteins are recognized by surface-immobilized aptamers. This assay provided detection at a limit of 2 pg/ml. Xi et al. developed a chemiluminescent aptasensor for the detection of the hepatitis B surface antigen (Figure 5g)124. This aptasensor used aptamers immobilized on Fe3O4-SiO2 magnetic nanoparticles that allowed for detection of the target biomarker with a linear range of 1-200 ng/ml. The detection limit for the aptasensor was 100 pg/ml and worked well even in the presence of interfering substances present in blood.
Fluorescent aptasensors
Fluorescent biosensors rely on changes in fluorescence polarization, wavelength, or intensity as a means of detection. These changes are produced upon the interaction of the target analyte and fluorescently-labelled aptamers125. Percze et al. developed a fluorescence polarization assay for the detection of respiratory syncytial virus (RSV), a viral pathogen affecting young infants126. Wang et al. constructed an integrated microfluidic device for fluorescence-based multi-virus detection of influenza A H1N1, H3N2 and influenza B virus (Figure 5h)127. This aptasensor consisted of aptamer-modified magnetic beads to detect RSV and another fluorescent-labelled aptamer was used for counter selection against human rhinovirus (HRV).
Some fluorescence-based methods use Förster resonance energy transfer (FRET). FRET involves two fluorophores of particular electromagnetic properties, where the emission spectrum of a donor molecule overlaps with the excitation spectrum of an acceptor molecule. Distance changes between the fluorophore pairs can therefore be monitored based on the wavelength and intensity of the emitted light, providing a detectable output for target recognition128,129. FRET-based systems can be incorporated into aptasensors in a variety of ways. Frequently, an aptamer is modified with both a donor and acceptor molecule such that when there is no target present, the donor and acceptor are too far for FRET to occur. Then, upon target binding, there is a conformational change in the aptamer that brings the FRET pair closer together, causing a change in FRET signal. In another method, the target molecule and the aptamer are both modified with either part of the donor/acceptor pair. The binding of the aptamer to the target brings the pair in proximity to undergo FRET110. Yamamoto and Kumar developed a quencher based method to detect HIV-1 Tat protein (Figure 5i)130. For this aptasensor, an RNA aptamer specific to Tat contained a hairpin structure with fluorophore and quencher modifications at 5’-end and 3’-end respectively. Upon Tat protein binding, the hairpin structure is opened, moving the fluorophore/quencher pair away from each other and causing fluorescence.
Quantum dots (QD) are spherical, inorganic, fluorescent nanocrystals which are extensively used as fluorescent probes. Compared to traditional organic dyes, QDs exhibit greater stability, reduced susceptibility to photobleaching, and greater and more precise spectral properties for multi-signal detection131. Ghanbari et al. used RNA aptamers conjugated to QDs to detect HCV NS3 protein88. HCV NS3 was produced in vitro, immobilized on a glass slide, then probed with the aptamer-QD complex to detect HCV at a limit of 5 ng/ml. Another study involved aptamers modified with QDs for detecting H1N1 with a detection level of 3.45 nM (Figure 5j)132. This study involved a protein-binding bifunctional aptamer and DNA-functionalized QD probe. The aptamer is complementary to part of the target DNA sequence of H1N1 Influenza virus and the rest of the sequence is a recognition sequence for streptavidin. In the absence of the target molecule, the QDs and the aptamer-DNA-streptavidin complex are free in solution and do not hybridize. In the presence of the target, the target hybridizes with aptamer-DNA and the aptamer-DNA binds to the DNA-QD forming a complex and providing a readout.
Colorimetric aptasensor
Colorimetric detection measures changes in color shifts that can be inspected either by the naked eye or a spectrophotometer24. While several traditional methods are available for analyzing DNA and RNA biomarkers, they still face a number of drawbacks133. For instance, radioactive and fluorescence probe based Southern blots are criticized for their toxicity and massive cost134,135 while PCR-based methods require precise instruments that can be large and expensive, and require skilled personnel136. Colorimetric detection has various advantages such as eliminating the use of radioisotopes, reduced costs related to required equipment and on-site and real-time quantification and detection137. The traditional colorimetric aptasensor works by first incubating the aptamer with virus, then adding catalytic-active substances that attach to the trapped virus. To change the color of the sample, appropriate chromogenic reagents are introduced24. For example, Chen et al. employed magnetic bead-modified aptamers specific to H3N2 to detect influenza A virus138. The sensor consisted of AuNPs modified with concanavalin A and glucose oxidase (ConA-GOx-AuNP). The complex attached to the virus through concanavalin A-glycan interaction, with glucose oxidase transforming a chemical signal into a color change, with an LoD of 11.16 g/ml138. Liu et al139 developed a colorimetric assay using graphene/AuNP hybrids to detect hepatitis C virus140. The ssDNA aptamer reduces the catalytic activity of graphene/AuNPs by preventing the contact between active interface and peroxidase substrates. Interaction of the virus with the aptamer restores the catalytic activity, with color change produced by the substrate 3,3′,5,5′-tetramethylbenzidine (TMB).
Non-electrochemical surface immobilized antibody-coupled aptasensors
Aptasensors can be coupled with antibodies to be used as direct or sandwich type immunoassays. Enzyme-linked immunosorbent assay (ELISA) is one of the most used diagnostic methods for the detection of proteins and antigens. When aptamers are used in substitution for the antibodies, the method is termed as enzyme-linked oligosorbent assay (ELOSA), enzyme-linked oligonucleotide assay (ELONA) or enzyme-linked aptasorbent assay (ELASA)141. The direct method of ELONA constitutes a plate coated with the target and a biotinylated aptamer that binds to the target (Figure 6a, top). Horseradish peroxidase (HRP)-conjugated streptavidin then binds to the biotinylated aptamers causing chemiluminescnce in the presence of the enzyme substrate TMB. In the sandwich based ELONA, the primary aptamer is immobilized on the surface of the plate and recognizes the target molecule142 (Figure 6a, bottom).
Figure 6. Non-electrochemical aptasensors.
(a) Mechanism for direct and indirect ELONA where the virus is immobilized on the surface or the aptamer is immobilized on surface, respectively. (b) Mechanism for aptamer based lateral flow assay: LFA strip includes positive control line with antibody binding to the target virus and a test line with streptavidin immobilized aptamer. Upon binding of the target virus, the AuNP-Ab complex shows the right signal; in absence of virus no line is visible in test region.
An enzyme linked aptamer assay (ELAA) based study was conducted for detecting the influenza A strain H5N1 using an aptamer that targets the HA protein142. This sandwich-based aptasensor comprises of amino groups conjugated at the 3’ terminus of aptamers that were immobilized in the wells. The target HA protein or semi-purified influenza virus was then added to the wells followed by addition of streptavidin-horseradish peroxidase. The LoD was 0.1 μg/well and the assay also discriminated between H1N1 and H3N2 subtypes of the virus. Other analogous ELAAs methods have been used for the detection of the human norovirus143, Zika virus144, and HCV145.
Lateral flow assays (LFA) are a type of immunochromatographic assay commonly used for the detection and quantification of an analyte146. These assays employ layered pads that generate a series of capillary beds to allow for directional fluid movement. They are frequently used to detect a wide range of targets, including viral targets for HIV and HBV147,148. LFA strips consist of different overlapped layers of sample pads, conjugate pad, nitrocellulose analytical membrane and absorbance pad mounted on a sticking backing sheet. The two most common LFAs are the competitive and the sandwich or complementary oligonucleotide assay. In sandwich assay, aptamers tagged with enzymes/AuNP/fluorescent dyes form a complex with the analyte at the conjugate pad. The complex flows to the test zone via the capillary pull of the strip and is captured by the antibody or another aptamer which forms a sandwich between the two aptamers or antibody (Figure 6b). This complex results in a visible color change in the test zone, typically a red line. The excess labelled aptamer moves to the control zone which is captured by another oligonucleotide complementary to the aptamer or protein that binds the antibody and causes another color change (the red line) to validate the assay (as a positive control). Le et al. developed a dual recognition element lateral flow assay (DRELFA) method to detect strain-specific influenza viruses in a multiplexed fashion149. Compared to the current antibody based conventional LFA, this aptasensor can discriminate between different strains of influenza virus. Further, by combining nucleic acid aptamers with antibodies, this device can overcome limitations such as antibody cross reactivity and slow aptamer kinetics. The aptasensor comprises a sample pad conjugated with biotinylated aptamer and an AuNP-labelled monoclonal antibody to form the dual-recognition complex; specifically, this sensor was constructed using a strain-specific biotinylated RNA aptamer and an AuNP-labelled monoclonal antibody that can detect specific strains of H3N2 influenza virus. In the presence of the virus, the test line shows a color change due to the formation of the biotin-streptavidin-aptamer and AuNP-conjugated antibody complex. With an LoD of 2x106 virus particles, this assay was able to differentiate between subtypes of the three different strains of influenza virus and showed no cross reactivity when compared to conventional LFA.
APTAMERS IN THERAPEUTICS
Several viral infections now have vaccinations and post-infection treatments that inhibit viral infection150-152. In some cases, viruses have still been shown to invade the immune system post treatment. Therefore, to inhibit the infection, there is an immediate need of antiviral biological molecules that can interrupt the viral life cycle and thus inhibit further infection. There are several ways by which viral infection can be inhibited, such as by interrupting the viral entry into the host cell and interfering with the viral replication machinery and thus preventing release of viruses to infect other cells153. However, the emergence of viral drug-resistant strains and the cytotoxicity of present inhibitory methods to host cells have affected treatment using anti-viral therapeutics154. Thus, searching for new and effective therapeutic tools is a prime need.
Aptamers have shown promising progress in the field of therapeutics and several aptamers are in pre-clinical stages155-157. Therapeutic aptamers bind directly to the viral targets and inhibit the downstream signaling of the replication cycle. Aptamers work in one of the following ways: they prevent structural changes in the target molecule, inhibit dimerization through associated molecules or can phosphorylate the proteins involved in downstream signaling158. There are other advantages of using aptamers as antiviral therapeutics. First, aptamers bind to the target tightly via surface-contact and disrupt protein-protein or protein-nucleic acid interactions159. Second, aptamers can form thermodynamically stable secondary structures by folding itself, regulated by Watson and Crick base pairing23,75. Third, aptamers for a particular target can be identified within a month by SELEX process and can be adaptable for several chemical modifications160. These advantages have brought aptamers into focus and has been extensively studied in the field of drug delivery.
One example of aptamer developed for treatment against viral infections is B40, an RNA aptamer that inhibits the HIV-1 envelope glycoprotein (gp120) from binding the C─C chemokine receptor-5, a T cell co-receptor 161-164. In another example, Cell surface SELEX was used to construct a ssDNA aptamer ZE2 that interacts exclusively with E2, a surface glycoprotein of HCV to impede the initial attachment of HCV with the host cells165. In vitro studies show that HCV particles are trapped by aptamer ZE2, making the aptamer potentially useful for anti-HCV therapy165. The antigen hemagglutinin (HA), expressed on surface of influenza viruses is a prime target molecule for aptamers. As compared to the conventional anti-HA monoclonal antibody, the aptamer P30-10-16 interacts with HA with a greater affinity (15-fold)166. Kwon et al. synthesized an RNA aptamer that binds to glycosylated receptor of the HA and neutralizes the receptor binding site of HA, thus restricting the attachment of the virus to the host cell167. Yuan et al. generated ssDNA aptamers against amino acid residues present in PA subunit of N-terminus of the polymerase of the influenza A virus. The PAN-2 aptamer they synthesized has an IC50 value of 10 nM and offered cross protection against influenza viruses (H1N1, H5N1, H7N7, and H7N9)168. There are only a few studies on aptamers against viruses such as human papillomavirus (HPV)169-172, hepatitis B virus (HBV)173-175, dengue viruses (DENVs)176, severe acute respiratory syndrome coronavirus (SARS-CoV)177, and rabies178,179. Some of these include the HBs-A22 RNA aptamer that targets the HBV surface, S15 tar ssDNA aptamer that targets the envelope protein of DENV-2, and GE54 tar ssDNA aptamer that targets the glycoprotein expressed by rabies virus (RABV)178,179. Valencia-Resendiz et al. reported that RNA aptamers for HPV16 L1 virus inhibits infection at early stages by interacting with the viral particles180. Yadavalli et al. isolated a DNA aptamer specific to gD protein of HSV-1 and demonstrated superior binding affinity and inhibition of viral reproduction and entry in in vitro, in vivo and ex vivo studies181. The different mechanisms through which aptamers inhibit viral infection are discussed in this section.
Aptamer suppression of viral attachment to host cells
Aptamers can obstruct the entry of viral particles into host cells by binding to viral surface proteins, thus affecting the ability of the virus to interact with related receptors on the host cell. Nucleolin, a eukaryotic cellular protein, is one such protein that aids in the attachment and entry of different viruses182. Balinsky et al. showed that the interaction of nucleolin with DENV capsid protein aids the formation of infectious virus particles183. The nucleolin-DENV interaction was hindered by the addition of RNA aptamer AS1411 that binds nucleolin. Similarly, HCV entry into the host requires the interaction of viral surface E2 proteins with host membrane cell receptors. An aptamer developed against the HCV E2 glycoprotein showed inhibition of viral entry into host cells184. The selected aptamer could significantly block the binding of HCV (90%) to CD81 host receptor, thereby inhibiting the infection of human hepatocytes. The first identified anti-HIV aptamer forms a tetramolecular parallel G-quadruplex (d(TTGGGGTT)) structure185,186. This aptamer is anionic and strongly interacts with the cationic V3 loop of the HIV envelope glycoprotein, gp120. This interaction inhibits host/virus surface adsorption and cell fusion, inhibiting HIV entry into the host cell185,186. However, HIV entry can still occur through other means, such as by interacting with other cellular receptors including heparan sulfate proteoglycans and nucleolin present on cell surfaces187. Application of AS1411, the nucleolin-binding RNA aptamer, at low nanomolar concentrations also showed antiviral activity by interfering with HIV attachment via nucleolin-based pathway24,188. Jeon et al. designed a DNA aptamer A22 against HA regions of influenza virus that blocked the binding of the virus to target cell receptors. Animal studies showed that A22-treated mice lose weight at a slower rate than the control group and infiltration of mononuclear cells in the alveoli was also decreased in the A22-treated group189. Choi et al. selected an aptamer C7-35M against H9N2 avian influenza virus that blocks viral infection in a dose-dependent manner 190. Similarly, Gopinath et al. constructed an aptamer against HSV-1 which impedes viral entry.191 The IC50 value was measured to be 0.8 μM and it can specifically distinguish HSV-1 from HSV-2.
Aptamers inhibiting virus replication cycle
Aptamers have been employed for viral inhibition and show promise as therapeutic agents by inhibiting the replication of the viral genome24. For example, the RNA aptamer B.2 was developed as a therapeutic against HCV192,193. Specifically, the aptamer forms a stem-loop structure and can bind and inhibit the HCV 5B polymerase, a non-structural RNA-dependent RNA polymerase that catalyzes RNA replication. The aptamer and the template RNA have different binding domains; B.2 non-competitively binds the RNA polymerase and therefore weakens the polymerase ability to bind the RNA template194. In vitro studies by Bellecave et al found two aptamers, 27v and 127v, which could inhibit the same polymerase but through competition for the polymerase binding site on the template RNA195. The aptamers function at different stages of the replication cycle of viral RNA: 27v blocks both initiation and elongation whereas 127v blocks initiation and post-initiation events of the viral RNA replication. Similarly, an RNA aptamer developed against the HIV nucleocapsid protein (critical for replication, encapsidation of viral genomes and assembly of viral particles196) hindered viral packaging by competing for psi RNA binding to the nucleocapsid protein197. Another truncated RNA ligand, RNA tat, was developed using SELEX and reduced HIV-1 replication by 70% (ref.198).
Aptamers have also been developed against human cytomegalovirus (HCMV), a member of the betaherpesvirinae subfamily and affects immunocompromised individuals. It is a major cause of many birth defects, and its impact is increased due to the rise in HIV-infected patients and immunosuppressive treatment. Similarly, RNA aptamers were generated to target the gp120 glycoprotein involved in viral replication and were able to suppress HIV-1 replication by preventing viral-mediated T-cell reduction199,200.
Inhibition of viral enzymes by activity of polymerase
The nonstructural protein-5B (NS5B), an RNA-polymerase in HCV replication, is a promising target for aptamer therapy to suppress HCV infection. Biroccio et al. selected an RNA aptamer B.2 to target the functional part of NS5B, a GTP binding site. In vitro studies for polymerase activity demonstrated aptamer concentration-dependent inhibition of polymerase activity201. Bellecave et al. developed a DNA aptamer 27v that inhibits the activity of NS5B by competing with the viral RNA template for the polymerase-binding site. 27v aptamer-treated hepatoma cell line (human) infected with the HCV JFH1 strain showed reduced amount of viral RNA195. The interaction between pregenomic RNA with viral protein R is an essential part of HBV replication. Feng et al. discovered an RNA aptamer S9 with a strong affinity for viral polymerases of HBV, showing 80-85% suppression of HBV replication in a human cell line infected with HBV174. Similarly, DeStefano and Nair showed that a DNA aptamer targeted towards the reverse transcriptase of HXB2 strain of HIV suppressed viral replication in vitro202. The aptamer prevents the viral replication by competing with natural template for the enzyme’s binding site.
Inhibition of other enzyme activity associated with viral replication
In addition to polymerases, other enzymes are also involved in viral replication in an indirect manner such as nonstructural protein 3 (NS3) which has two domains, one with protease activity and one with helicase activity. For HCV replication, both these domains are essential. A strong affinity is found in the helicase domain for the poly(U) motif present in 3’ UTR of viral genome. Umehara et al. developed bivalent aptamers with sequences linked by a poly(U) linker. G925-S50 and NEO-35-s41 aptamers developed by the group was shown to reduce NS3 activity by inhibiting both helicase activity (IC50 0.2 μM/15 nM) and protease activity (IC50 0.2 μM/20 nM). Fukuda et al. used RNA aptamer ΔNEO-III-14U which has a poly(U) sequence and impedes helicase and polymerase activity of NS3, evaluated by in vivo and in vitro tests203.
Nonstructural protein 5A (NS5A) is another protein required for HCV virion assembly and replication. Yu et al., isolated aptamers NS5A-4 and NS5A-5 which showed inhibition of viral infection in Huh7.5 cells. The treated group showed one-fold reduction in viral RNA level compared to the control group as evaluated by RT-PCR. Gao et al. developed aptamers NS2-1, NS2-2, and NS2-3 to target the nonstructural protein 2 (NS2) of HCV and showed successful inhibition of viral replication204. Jang et al. designed an RNA aptamer ES15 to bind nonstructural protein 10 (nsP10) which has NTPase/helicase activity. The aptamer repressed viral enzyme activity by up to 85% in in vitro studies with an IC50 of 1.2 nM (ref205).
Blocking of nucleic acid sequences associated with viral replication
Certain genomic sequences interact with proteins associated with viral replication, translation, transcription initiation and assembly. Designing aptamers that can target these genomic sequences are promising for antiviral treatment strategies. The mRNA of HCV has an internal ribosome entry site (IRES) which is associated with viral replication and is a potential target for antiviral therapy. IRES which has four domains (I-IV) in its 5’UTR, plays a role in viral replication initiation and mRNA translation. Konno et al designed an RNA aptamer AP30 consisting of sequences 5’-GAGUAC-3’ and 5’-UGGAUC-3’, to target the first domain (SL-D1 and SL-E1 loops) of IRES and showed in vitro suppression of viral replication.206,207 An RNA aptamer (2-02) with sequence 5’-UAUGGCU-3’ targeting the domain II of IRES has been synthesized by Kikuchi et al208. In a separate study, the same group developed aptamer 3-07, which targets the third domain of the IRES. This mechanism was substantially more potent at suppressing viral infection by blocking in vitro IRES-dependent translation compared to the aptamer against the second domain of IRES. Combined blocking of II and III-IV domains of IRES has also received considerable attention as the IIIe and IIId regions are essential for translation in HCV209. Two aptamers (0207 and 0702) which are the combined form of 3-07 and 2-02 reported to have 10-fold higher binding affinity and the IC50 value lower the translational activity by 10-fold. Romero-Lopez et al developed a construct (HH363-24) that targets the IIId domain and cleaves the 3’ end of HCV genome sequence210. The HH363-24 construct impedes both the replication and translation of virus. Other research has focused on aptamers that interact with the long terminal repeats of HIV-1 (ref.211).
Aptamers for delivering therapeutics to viral infected cells
Aptamers can also be used to target drugs to specific diseased regions. For example, Liu et al. constructed an RNA aptamer conjugated with fluorescein isothiocyanate (HBs-A22) to target HbsAg surface antigen present in cells infected with HBV,173. In another example, Zhou et al created a chimeric construct which contains a siRNA molecule and anti-gp120 aptamer and targeted the mRNA for the tat/rev protein in HIV-1 in the Chinese Hamster Ovary (CHO) cell line241. Neff et al. performed a similar study using HIV-1 NL4-3 infected humanized mice (RAG-Hu). 2’-fluoro modifications further improved the biostability of this siRNA-aptamer chimera in mouse serum212. The aptamer-based treatment showed inhibition of viral activity as confirmed by 75-90% decrease in mRNA transcript level for tat/rev protein in T lymphocytes of mice. Zhu et al. used a different strategy by converting anti-CD4 RNA aptamer to a DNA aptamer and conjugating an siRNA molecule for targeting mRNA of HIV-1 protease213. The therapeutic activity was evaluated by qRT-PCR which quantified the reduction in expression for the mRNA protease in pcDNA-HIV-PR plasmid transfected CD4+ T cells.
Other approaches and aptamer-based therapeutics in clinical trials
Different aptamer-siRNA chimeras have been developed against HIV-1 infection such as anti-CD4 aptamer/anti-gag-siRNA and anti-CD4 aptamer/anti-CCR5-siRNA214. Bruno et al developed aptamers against Dengue fever and west Nile virus infections215. An RNA aptamer (CL9) containing the cytosolic receptor RIG-I was generated by Hwang et al. and was shown to activate innate antiviral immune response216. The increased antiviral response and production of IFNβ is the effect of cytosolic receptor RIG-I which helps in recognizing the pattern for foreign molecular agent in the cells infected with the virus. In vitro studies showed that CL9 inhibits cells from invasion. Table 2 lists the aptamer candidates that are currently evaluated in clinical trials for viral therapeutics.
Table 2.
Aptamers in clinical trials.
| Target | Oligonucleotide | Functional activity | Clinical trial |
|---|---|---|---|
| HIV-1 Tat | RNA198 | Reducing TAT-mediated HIV replication. | Phase I |
| HIV-2 Tat | RNA217 | Reducing Tat-2 transactivation 20 and HIV-1 replication. | Phase I |
| HIV-1 Rev response element | RNA218,219 | HIV replication in vitro and in vivo. | Phase I |
| Hepatitis C NS3 | RNA220,221 | Reduction in NS3 activity in vitro; Reduction in NS3 protease activity; Reduction in MBP-NS3 protease activity in vivo. | Phase I |
DNA APTAMERS FOR SARS-COV-2 DETECTION AND INHIBITION
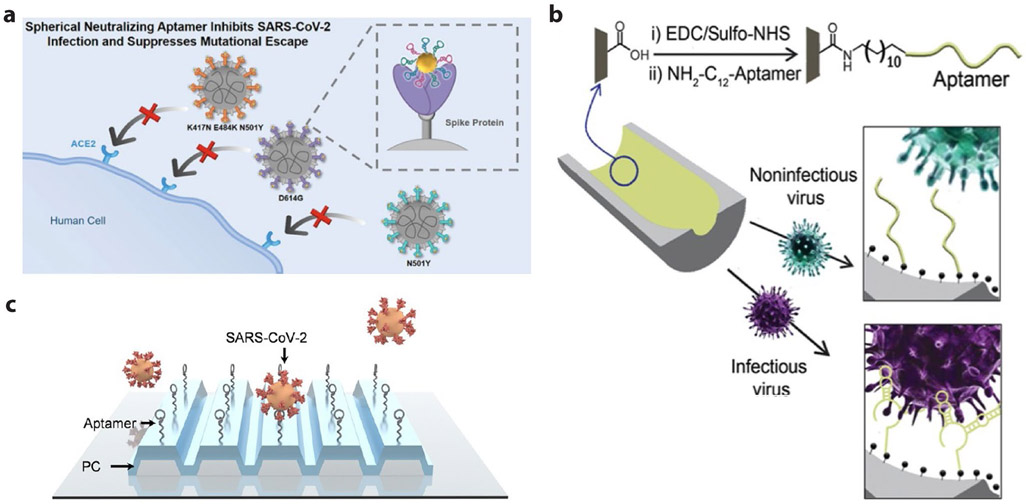
Since the outbreak of the COVID-19 pandemic, DNA aptamers targeting either SARS-CoV-2 nucleocapsid (N)222 or spike (S)223-229 protein have been selected and employed in the development of viral sensors or inhibitors. Yang and colleagues obtained three effective aptamers, named as CoV2-RBD-1C223, CoV-2-RBD-4C223, and CoV2-6C3224, which bind to spike receptor binding domain (RBD) region with nM KD. Among them, circular CoV2-6C3 dimer shows increased stability in human plasma than linear monovalent aptamer. In addition, it displays a high antiviral potency with an IC50 of 0.42 nM and reduces the amount of viral genome in the infected cells by 87.1% compared to the viral culture that was not treated with the aptamer224. The same group recently constructed a spherical cocktail of neutralizing aptamer-gold nanoparticle (SNAP) decorated by all three aptamers (Figure 7a). Exploiting the synergetic blocking strategy from the multivalent aptamer and steric hindrance effect of the gold scaffold, the cocktail SNAP neutralizes both wildtype strain and three variants (commonly called D614G, Alpha, and Beta) with a further improved IC50 at fM level230. Potency of the cocktail SNAP is about 2 to 3-fold better than the performance of other reported neutralizing aptamers in a monovalent225 or a circular divalent form224. In a separate study by Pun and colleagues, a DNA aptamer, named SNAP1, was selected showing <80 nM KD, and binding to the spike N-terminal domain as revealed by high-resolution cryo-EM imaging228. The aptamer detects UV-inactivated SARS-CoV-2 with an LoD of 5x105 copies/mL when used in lateral flow assay or ELISA, suggesting SNAP1 is a valuable ligand capable of COVID-19 diagnostics in point of care settings. Additionally, a dimeric DNA aptamer form, denoted as DSA1N5 by Li and colleagues, was derived from two previously obtained aptamers, MSA1 and MSA5, in the same group227. DSA1N5 recognizes the spike protein of wildtype, alpha, or delta SARS-CoV-2 strains with a KD of 120, 290, or 480 pM (ref.229). After being immobilized onto gold electrodes to produce a sensor rapidly generating electrochemical signals, the aptamer can detect 1x103 virus particles per mL in 1:1 diluted saliva of both wildtype and alpha/delta variants within 10 mins. The study provides the first aptamer for rapid test of SARS-CoV-2 delta variant.
Figure 7. DNA aptamers selected for SARS-CoV-2 viral detection and inhibition.
(a) Schematic of SNAP to block the interaction between the RBD of SARS-CoV-2 and host ACE2 with synergetic strategy of multivalent multisite binding and steric hindrance. Reproduced from ref.230. (b) Scheme of Infectious virus detection using aptamer-functionalized nanopore sensors. Reproduced from ref.231 (c).Working principle of label-free optical detection for intact SARS-CoV-2 using surface immobilized DNA aptamers and PRISM system. Reproduced with permission from ref. 232. Copyright 2022 American Chemical Society.
In addition to aforementioned aptamers that were selected by targeting specific SARS-CoV-2 surface antigens (N or S protein), Lu and colleagues have evolved an aptamer, called SARS2-AR10, that was selected against intact virions by performing a counterselection using UV-inactivated virus particles231. As a result, the aptamer can distinguish active/infectious SARS-CoV-2 virus from the noninfectious form. SARS2-AR10 aptamer was integrated with a solid-state nanopore system, which renders strong confinement to virus, to selectively detect intact SARS-CoV-2 containing samples with an LoD of 1x104 copies/mL (Figure 7b). The same aptamer was recently immobilized onto a customized photonic crystal surface for digital detection of intact SARS-CoV-2 virions with an LoD of 1x104 copies/mL (ref.232), using a label-free imaging technique, called photonic resonator interferometric scattering microscopy (PRISM233) (Figure 7c). These sensors offer a tool to minimize or eliminate the chance of false positive results resulting from PCR based detection of SARS-CoV-2 RNA genome residue rather than the infectious virions.
DNA APTAMERS IN PLANT VIRAL INFECTION CONTROL
Viral plant infections pose a growing concern in the agricultural field which can result in low quality grains, fruits, vegetables and flowers and lead to huge economic loss. The most common viruses include Tobacco mosaic virus (TMV causes infection in Tobaco and Solanaceae plant), Tomato spotted wilt virus (TSWV causes infection Nicotiana, groundnut), African cassava mosaic virus (ACMV infects genus Begomovirus such as Datura, Cassava, and Nicotiana), Tomato yellow leaf curl virus (TYLCV infects Solanaceae plant family such as tomato), Cucumber mosaic virus (CMV causes infection in Cucumberaceae family of plant such as carrot, pepper, bean, spinach), Potato virus Y (PVY infects weed plants, pepper, Solanaum eseculentum, Solanaum tuberosum,Nicotiana), Plum pox virus (PPV infects peaches, almonds, plums, apricots, nectarines), Potexvirus like Potato virus X (PVX infects Potato), Cauliflower mosaic virus (CaMV infects species of Resedaceae and Brassicaceae ), and Brome mosaic virus (BMV infects species of poaceae family like barley). The pathogen's information has crucial role in the proper diagnosis and therapy of any disease and it is essential for protecting the crops from infection and saving the farmers from commercial loss234.
Approaches to prevent or inhibit viral infection in plants are based on gene silencing, metabolic regulation, hormones, proteolysis and immunological receptor signaling235,236. The few modern methods that can control viral infection in plants are rotation of crop, pathogen-free plant production through tissue culture and through integrated vector management. Traditional breeding techniques can take a long time and genetic modification with naturally occurring resistance genes (R genes) can produce virus resistant strains. Alternatively, the use of symbiotic fungal interaction with arbuscular mycorrhizae can improve the natural defense mechanism plants have against plant viruses237,238. Recent studies have demonstrated the use of peptide and DNA aptamers to control viral plant infection. These aptamers bind specifically to virus coat proteins such as capsid protein, nucleoprotein and movement proteins which thereby prevents infection. Specifically, peptide aptamers are highly target-specific and can function in both intracellular and extracellular environments and interfere with viral gene expression or replication238,239.
Lopez-Ochoa et al. reported a set of peptide aptamers that bind the N terminus of the Rep protein from the Tomato golden mosaic virus (TGMV)240. Peptide aptamers A22 and A64 expressed in transgenic tomato lines have been used for treating viral diseases, specifically by interacting with viral Rep proteins that are involved in the replication, transcription, and infection241. The two aptamers bind to different regions in the N-terminus of Rep proteins of geminiviruses such as tomato yellow leaf curl or tomato mottle virus and interfere with the replication activity of the virus. A22 recognizes the first 35 amino acids of Rep whereas A64 primarily interacts with residues 64 through 97 including a highly conserved motif geminivirus Rep sequence (GRS). Another study detected the apple stem pitting virus (ASPV) using a label-free SPR approach242. DNA aptamers were developed against two viral coat proteins HS-MT32 and HS-PSA-H. Here, thiol-modified aptamers were immobilized on the gold surface of the SPR chip. To avoid non-specific adsorption, the surface was modified with random oligonucleotides of same length as that of aptamers. The sensitivity of SPR to the viral fragments was verified using SEM imaging showing aptamer-modified chips binding to the viral fragments. 2.2 × 107 virus fragments per cm2 were visible on the aptamer modified surface. Using cell-SELEX, Ye et al. produced three DNA aptamers targeting GCRV-infected cells, that could be utilized for developing quick detection technologies and antiviral therapeutics for GCRV infection243. Application of aptamers technology offers broad-spectrum in viral infection resistance. Integrating conventional breeding techniques with peptide aptamers might be a potential route to tackle new variants and virus species. RNA interference (RNAi), which is a homology-dependent, plays a role in preventing infections that results transgenic resistance towards plant infections244.
ROLE OF DNA NANOTECHNOLOGY IN VIRAL DIAGNOSTICS AND THERAPY
Synthetic scaffolds such as polymers, nanofibers, nanoparticles, and liposomes have emerged as advanced platforms for infectious disease detection and treatment245-248. However, these materials do not allow control over surface probe or ligand density for biosensing applications249. To address these drawbacks, aptamer-labelled DNA nanostructures have been explored as biosensing and diagnostic platforms250-254. DNA is a spatially controllable and versatile material that can be used for the bottom-up construction of nanostructures255. These DNA nanostructures have been robustly self-assembled into 2D and 3D geometries of specific shapes and sizes. Moreover, the chemical nature and predictability of DNA base-pairing allow for the precise decoration of DNA nanostructures with ligands at sub-nanometer resolution. Potential ligands include proteins, nanoparticles, oligonucleotides, fluorophores, and other biomolecules256-260. Further, the biocompatibility, biostability and nontoxicity of DNA nanostructures have made them useful in biosensing and drug delivery applications.
On a separate note, flexibility of nucleic acid backbone may prevent aptamers from reaching to optimal folding poses for better binding affinities. DNA nanostructures can provide an ideal platform with excellent addressability to fix flexible aptamers into a specific configuration. Tan and colleagues recently stabilized an anti-lysozyme aptamer by fixing the termini of the aptamer with a length-optimized triplex structure on a DNA tetrahedron nanostructure261. As a result, the target binding affinity of the aptamer increased by ~10 fold. Additionally, the aptasensor built on the DNA tetrahedron nanostructure achieved a 180-fold better LoD. We expect this emerging aptamer-DNA nanostructure hybridization strategy to have the potential to greatly improve the performance of existing aptasensors.
DNA nanostructures for viral detection
Recently, DNA nanostructures have garnered tremendous interest in biosensing due to their high surface-to-volume ratio, which provides greater space for responsive elements and therefore greater changes in signal generation251. This biosensing ability has been explored for the early detection of pathogens in human samples262 with enhanced target specificity and avidity263. In this section, aptasensors utilizing a DNA nanostructure scaffold are discussed, categorizing the techniques based on the same output methods discussed previously.
DNA-antibody nanostructure as electrochemical immunosensors
DNA nanostructures can be employed as electrochemical sensors in which an antibody-labelled DNA nanostructure is attached to a gold electrode (examples in Ref.264). For instance, a DNA-antibody nanostructure has been used as an electrochemical immunosensor for the rapid detection of Streptococcus pneumoniae265. In this study, a hollow structured DNA tetrahedron was assembled and functionalized on the surface of gold electrodes and the surface was further passivated with bovine serum albumin (BSA) (Figure 8a). Later, pneumococcal surface protein A (PspA) antibody was tagged on to the top vertex of the DNA tetrahedron via carboxyl group conjugation. Electrochemical detection occurred by the introduction of ferrocene carboxylic acid-conjugated antibodies (FeC-Ab) onto the electrode surface. This electro-active tag reacts with PspA and produces a peak current corresponding to the target concentration, measured using square wave voltammetry technique. This method allowed detection of PspA peptide and S. pneumoniae lysate from synthetic and real human samples from the axilla, nasal cavity, and mouth.
Figure 8. DNA nanostructure based viral detection.
(a) Schematic illustration of construction of DNA-tetrahedra-based electrochemical immunosensor, which senses the pathogen using redox-labelled antibody attached to the top vertex of the tetrahedron. (b) Star-shaped DNA architecture, carrying five molecular beacon-like motifs and five FRET pairs. Binding of DENV reconfigures the structure, resulting in a FRET signal due to the change in distance between the dye pairs. (c) Fluorescently labelled DNA nanobarcodes for detecting a mixture of viral pathogens. (d) Detection of HIV DNA by fluorescent labelled DNA sensors combined with enzyme-based rolling cycle amplification. (e) DNA nanoswitches reconfigure from a linear “off” state to a looped “on” state on detecting viral RNA.
Fluorescent DNA nanostructures
DNA nanostructure-based biosensors may also utilize FRET as its detectable output, hybridizing specific nanostructure strands with aptamers that are labelled with fluorophore or fluorophore-quencher pairs266. Similar to the mechanism employed for regular FRET-based aptasensing, the placement of the FRET pair is rationally designed so that the fluorescence changes when the target is present. Kwon et. al developed a star-shaped DNA nanoarchitecture for the detection and inhibition of dengue virus (DENV)267,268. This two-dimensional nanoarchitecture contains five fluorophore-quencher pairs which act as molecular beacons as well as ten DENV envelope protein domain-III (ED3)-binding aptamers (Figure 8b). The resulting structure precisely matches the pentagonal arrangement of ED3 clusters on the DENV surface. The molecular beacons are placed such that each edge of the inner pentagonal scaffold consists of a fluorescein (FAM, fluorophore)-modified ssDNA and a BHQ-1 (quencher)-modified ssDNA hybridized to a hairpin structure that allows for FRET-based sensing capabilities. In the absence of DENV, the hairpin structure is maintained, and fluorescence is quenched due to the closed proximity of FAM and BHQ-1. In the presence of DENV, the ED3 aptamers bind the protein targets on the DENV surface and cause a structural expansion of the entire DNA star architecture. To accommodate for this change, the hairpin structure dehybridizes and the FAM and BHQ-1 molecules move apart and allow fluorescence to occur. This platform detected DENV in human blood serum and plasma with high sensitivity of 1 × 102 p.f.u./ml and 1 × 103 p.f.u./ml respectively.
In another study, DNA dendrimer-based fluorescently-labelled barcodes were used for the multiplexed detection of pathogen DNA269. This structure contains two kinds of fluorescence dyes (Alexa Fluor 488 and BODIPY 630/650) and a probe that is complementary to the target pathogen DNA used for the specific detection (Figure 8c). The dendrimer DNA structure is formed from a Y-shaped DNA attached to a detection probe and a fluorescent dye. Multiplexed detection is carried out by introducing different nanobarcodes containing specific target probes into a sample containing DNA from four targets Bacillus anthracis, Francisella tularensis, Ebola virus and SARS coronavirus. Binding of the target with the probe yields a particular pseudocolour corresponding to the specific target, generated from the merged images of 4 possible combinations. The detected pathogen is then identified by decoding with a preassigned barcode library.
Zheng and co-workers used a DNA nanomachine on gold nanoparticles (AuNPs) for the selective and ultrasensitive detection of HIV nucleic acids270. This strategy combines rolling circle amplification (RCA) and catalytic recycling for a DNA-walker cascade amplification on AuNP surface (Figure 8d). For the RCA reaction, HIV specific nucleic acid was taken as a primer DNA which has hybridization sequence to the 3’ and 5’ ends of a DNA padlock, which can synthesize long ssDNA from short circular padlock DNA using DNA/RNA primer via RCA. Here, addition of phi29 DNA polymerase helped in driving the RCA reaction. The by-product of RCA reaction consists of catalysts created by the extended strand containing a recognition sequence that is nicked by the nicking enzyme Nb. BtsI and can be used as triggers for initiating the DNA nanomachine. This catalyst initiates the opening of hairpins to yield signals via duplex formation on the AuNP surface and causes DNA walker cascade amplification. This results in the liberation of FAM-labelled DNA payload and induces fluorescence signals in the presence of HIV targets. The specificity of the DNA nanomachine was confirmed with four different DNA sequences, with an LoD of 1.46 fM.
Atomic force microscopy-based readout
Atomic force microscopy (AFM) has also been utilized in DNA nanostructure-based sensors. This scanning-probe microscopy technique relies on mechanical interactions with a molecular surface to provide a visual image. Biological targets are identified through visual identification of induced structural changes. In one such example, a self-assembled rectangular DNA origami nanochip was constructed through a bottom-up process for the rapid detection of Human Papillomavirus (HPV), an important target for gynaecologic diagnosis271. Two staples of the DNA origami nanochip were modified to contain single stranded extensions that act as DNA probes complementary to the HPV target. Binding of the viral DNA to the single stranded probes causes the formation of a doubled stranded DNA helix on the origami surface that was visualized using AFM.
Gel-based viral detection using DNA devices
Dynamic DNA devices that reconfigure on recognizing biomarkers have also been used in viral detection. Halvorsen and colleagues developed DNA nanoswitches to detect biomarkers such as DNA272, microRNAs273 and ribonucleases (Figure 8e)274. The DNA nanoswitch is a long duplex constructed using the single stranded M13 scaffold and short complementary backbone oligonucleotides. Two of the backbone oligonucleotides were modified to contain single stranded extensions that are partly complementary to a target nucleic acid. On binding the target, the nanoswitch reconfigures from the linear “off” state to a looped “on” state, and the two states were resolved on an agarose gel. The “on” state of the DNA nanoswitch is universal for any type of target. Recently, the group used this strategy to detect Zika virus275, showing a detection sensitivity of ~105 copies/μl without any amplification, and an LoD of ~ 100 copies/μl when the strategy was coupled with an enzyme based isothermal amplification step. The assay was able to discriminate Zika and Dengue viruses, as well as different strains of the Zika virus. They also demonstrated the utility of the assay to detect SARS-CoV-2 virus responsible for the current COVID19 pandemic. This DNA nanoswitch based assay also offers multiplexed detection of several RNA targets, as well as multiplexed barcoded detection of different types of biomarkers such as proteins, DNA, RNA, and antibodies in a single reaction276.
DNA nanostructures for viral therapy
DNA nanostructures also offers advantageous properties for drug delivery such as high solubility, non-toxicity, and biodegradability. Moreover, DNA nanostructures possess high cellular uptake properties without the utilization of transfection agents, which make them a suitable tool for health care applications277. Many studies have therefore employed aptamer-labelled, drug-conjugated DNA nanostructures as targeted drug delivery vehicles. Mela et al. constructed an aptamer-functionalized DNA origami rectangular frame structure loaded with the antibacterial peptide lysozyme to destroy gram-positive (Bacillus subtilis) and gram-negative (Escherichia coli) bacteria278. This DNA origami structure contained fourteen bacteria-selective aptamers on the origami’s four edges, and five wells where the lysozyme is immobilized through a biotin/streptavidin interaction (Figure 9a). The delivery system was efficient against both types of bacteria.
Figure 9. DNA nanostructures for viral therapy.
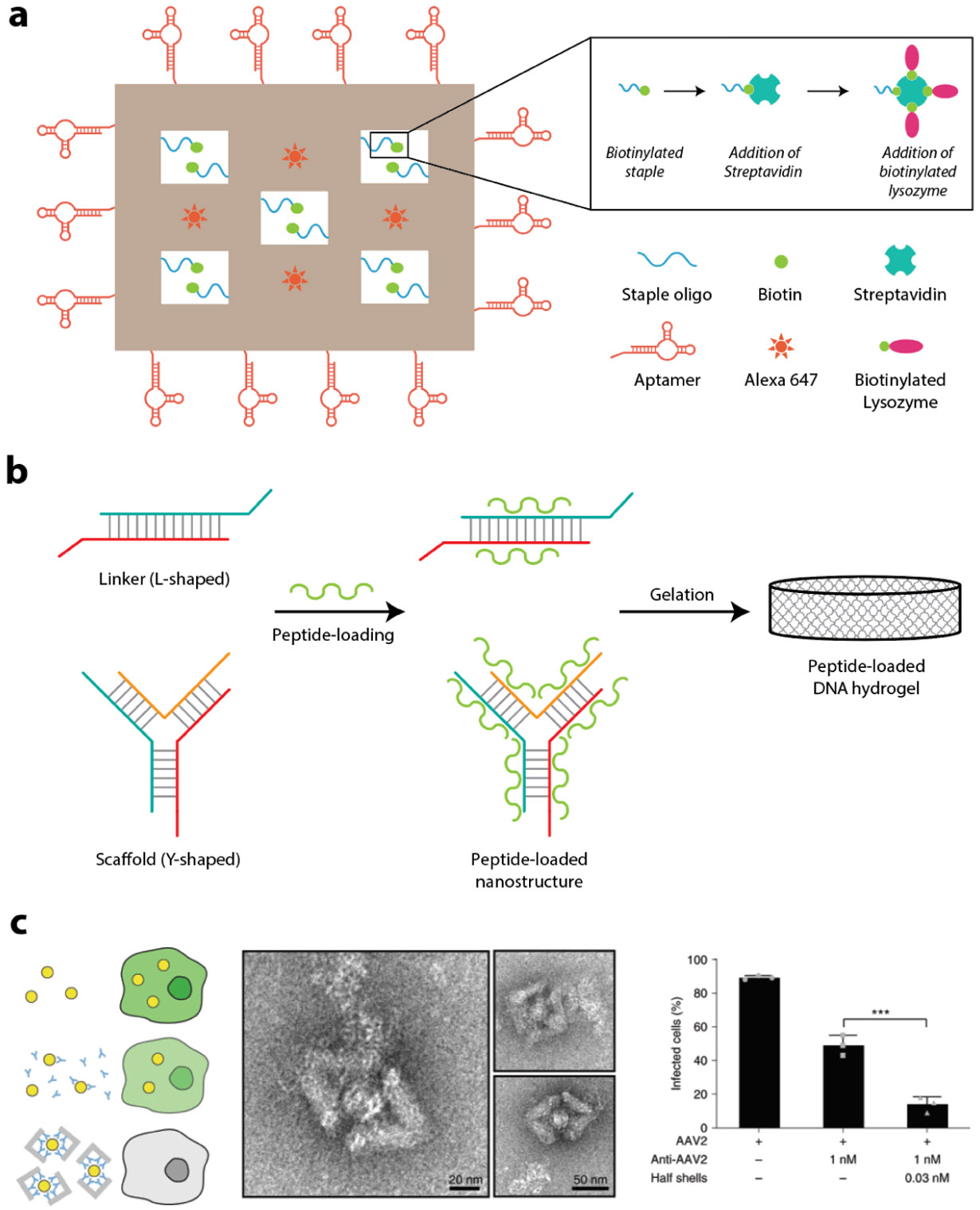
(a) Scheme of aptamer-functionalized DNA origami nanostructure loaded with antibacterial peptide. The aptamers are decorated around the origami to target four bacteria in parallel and treat the disease. (b) Schematic representation of synthesis of peptide-loaded DNA hydrogel via electrostatic crosslinking process for controlled drug delivery. (c) Left: scheme of inhibiting AAV2 infection by DNA origami half shells; Middle: TEM images of AAV2 virus particles captured by DNA origami half shells; Right: Quantification of infected cells for conditions with AAV2 virus only, AAV2 plus antibody at 1 nM (IC50 concentration), and AAV2 plus DNA origami half shells decorated with antibodies inside shells. Reproduced with permission from ref. 259. Copyright 2021 Springer Nature.
In another study, an antibacterial DNA nanostructure-based hydrogel was developed for the treatment of a cutaneous wound279. Hydrogel formation occurs based on electrostatic interactions between polyanionic DNA nanostructures and cationic antimicrobial peptides (AMPs) (Figure 9b). Antimicrobial peptide L12 was released from the hydrogel in response to pathogenic S. aureus infections. This strategy showed controlled L12 release and superior antimicrobial activity towards methicillin-resistant S. aureus infections. Moreover, ex vivo antimicrobial assay of L12 loaded DNA hydrogels against S. aureus infected porcine skin showed bio-responsive delivery systems, nuclease sensitive degradation, and significant potency against S. aureus and MRSA infections with 24 hours of application. Further, the anti-inflammatory effect of the hydrogel was demonstrated in vivo in mice, revealing faster wound healing rates within 10 days of treatment, an approach that can potentially be translated for treating viral infections in humans. In addition to 2D DNA platforms, Dietz and colleagues recently created a programmable icosahedral canvas platform for constructing unique DNA origami icosahedral/cage structures whose cavities are large enough to wrap entire virions for effective virus inhibition259. Antibodies against adeno-associated virus serotype 2 (AAV2) were attached to the inside of the shells to pilot the in vitro test of antiviral efficacy, which shows that the cages not only decreased the number of infected host cells, but also substantially lowered their viral loads when compared to free antibodies (Figure 9c). Decorated with viral antigens on the outer surface of the same shells, the authors created DNA nanodevices that were used for the detection of viral infections as well as the antigen-triggered release of molecular payload280. DNA origami platform can provide excellent spatial addressability to enable precise display of multiple virus antigen-targeting aptamers to mirror the spatial arrangement of the target viral surface antigens. Such pattern-matching interactions should be able to further improve viral detection and inhibition performance compared to monovalent aptamers267.
SUMMARY AND OUTLOOK
In this review, we have summarized the selection, characterization and use of existing nucleic acid aptamers for diagnosis and treatment of viral infections. Aptamers can be selected against whole cell virus or for viral components such as surface proteins or against blood markers that are upregulated upon viral infection. In response to emerging viruses like SARS-CoV-2, new aptamers can be readily and economically produced using SELEX to target whole virions or epitopes of novel viruses and the mutants. They can then be quickly plugged into existing virus detection platforms that use electrochemistry, fluorescence, optical, or AFM imaging for detection signal readouts. Compared to antibodies, aptamers in general have lower target binding affinities that may compromise the sensitivity of an aptamer-based sensor. However, as nucleic acids, aptamers can be easily docked within designer DNA nanostructures via DNA base pairing to achieve optimal binding poses, multivalency, and/or pattern-matching interactions with targeted viral antigens for greatly enhanced binding affinity and avidity259,261,267. The resulting higher binding affinity can facilitate the development of rapid, inexpensive, and sensitive strategies for virus sensing early after infection, which is critical for curbing the spread of highly contagious infectious diseases like COVID-19281-283. Additionally, aptamers can be strategically evolved to distinguish infectious virus particles from noninfectious forms231, which provides a unique and novel solution to address the problem for people being able to know when they are no longer infectious and can come out of quarantine, as nucleic acid tests are known to generate false positive results from the presence of nucleic acid molecules from degraded viruses284,285. These emerging platforms and technologies are well worth further investigations for the development of better viral sensors to mitigate future epidemics and pandemics.
For applications in therapeutics, aptamers have unique advantages such as their higher penetration in tissues, easy chemical synthesis, high specificity, and ease of conjugation to therapeutic RNAs, proteins, peptides, small drug molecules and nanoparticles. Yet aptamer-based structures are still underdeveloped in the context of viral therapeutics with many areas for improvement and development. For example, due to the small size, aptamers have a shorter duration of renal filtration. Biostability and bioavailability of an aptamer can also be challenged by nuclease action. Recently developed approaches such as backbone or nucleotide modification are promising strategies for reducing aptamer degradation in physiological conditions20,286. Furthermore, aptamers can be easily conjugated with bulk molecules or with DNA nanostructures, which can not only encapsulate the aptamers, but trigger the delivery of drugs upon locking/unlocking mechanism using aptamer-target binding287. Thus, molecular platforms built using designer DNA nanostructures have the potential to create the next generation of aptamer-based therapeutics. In terms of using DNA nanostructures for such biological applications, stability against nucleases, robust functionality in a variety of biofluids such as serum and whole blood, and easy readout are some of the aspects that could be addressed in future research. Some of these challenges are already being addressed, including the development of a variety of strategies to enhance nuclease resistance of DNA nanostructures288. However, DNA nanostructures also have several advantages. The methods have been shown to be scalable both in terms of the size289 and amount290 of DNA nanostructures that can be produced, with minimal cost associated with DNA synthesis and assembly291, and new reports have shown clinical utility of DNA nanostructure based sensors292. Developments in aptamers combined with advances in DNA nanotechnology can serve as potential alternatives to traditional methods in viral detection and treatment.
Supplementary Material
ACKNOWLEDGEMENTS
B.C. thanks support from DBT, India (BT/PR21542/NNT/28/1561/2018) and DBT, Ramalingaswami Fellowship (BT//RLF/Re-entry/13/2013). M.E.K thanks support from NIH Postdoctoral Fellowship (F32GM137477). J.D. thanks support from Natural Science Foundation of Shaanxi Province (Nos. 2017JQ2035, 2018JQ2023) and Fundamental Research Funds for the Central Universities (No. 300102129101). X.W. thanks support from NIH NIAAA (AA029348), NIH NIAID (AI159454), NIH NIDCR (DE030852), and NSF RAPID (20-27778).
Footnotes
Supporting Information. A supplementary table and references of virus-targeting aptamers for detection and/or inhibition of virial infections.
REFERENCE
- (1).Assaad F; Borecka I, Nine-year study of WHO virus reports on fatal viral infections. Bull World Health Organ 1977, 55 (4), 445–453. [PMC free article] [PubMed] [Google Scholar]
- (2).Tobin NH; Campbell AJP; Zerr DM; Melvin AJ, Life-Threatening Viral Diseases and Their Treatment. In Pediatric Critical Care, 2011, 1324–1335. [Google Scholar]
- (3).Keaney D; Whelan S; Finn K; Lucey B, Misdiagnosis of SARS-CoV-2: A Critical Review of the Influence of Sampling and Clinical Detection Methods. Med. Sci 2021, 9 (2), 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ménard-Moyon C; Bianco A; Kalantar-Zadeh K, Two-Dimensional Material-Based Biosensors for Virus Detection. ACS Sensors 2020, 5 (12), 3739–3769. [DOI] [PubMed] [Google Scholar]
- (5).Kevadiya BD; Machhi J; Herskovitz J; Oleynikov MD; Blomberg WR; Bajwa N; Soni D; Das S; Hasan M; Patel M; Senan AM; Gorantla S; McMillan J; Edagwa B; Eisenberg R; Gurumurthy CB; Reid SPM; Punyadeera C; Chang L; Gendelman HE, Diagnostics for SARS-CoV-2 infections. Nat. Mater 2021, 20 (5), 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Myhrvold C; Freije CA; Gootenberg JS; Abudayyeh OO; Metsky HC; Durbin AF; Kellner MJ; Tan AL; Paul LM; Parham LA; Garcia KF; Barnes KG; Chak B; Mondini A; Nogueira ML; Isern S; Michael SF; Lorenzana I; Yozwiak NL; MacInnis BL; Bosch I; Gehrke L; Zhang F; Sabeti PC, Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360 (6387), 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vermisoglou E; Panacek D; Jayaramulu K; Pykal M; Frebort I; Kolar M; Hajduch M; Zboril R; Otyepka M, Human virus detection with graphene-based materials. Biosens. Bioelectron 2020, 166, 112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tang Z; Kong N; Zhang X; Liu Y; Hu P; Mou S; Liljestrom P; Shi J; Tan W; Kim JS; Cao Y; Langer R; Leong KW; Farokhzad OC; Tao W, A materials-science perspective on tackling COVID-19. Nat. Rev. Mater 2020, 5 (11), 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Davydova A; Vorobjeva M; Pyshnyi D; Altman S; Vlassov V; Venyaminova A, Aptamers against pathogenic microorganisms. Crit Rev Microbiol 2016, 42 (6), 847–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Keefe AD; Pai S; Ellington A, Aptamers as therapeutics. Nat. Rev. Drug Discov 2010, 9 (7), 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nimjee SM; White RR; Becker RC; Sullenger BA, Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol 2017, 57, 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Soukarie D; Ecochard V; Salome L, DNA-based nanobiosensors for monitoring of water quality. Int. J. Hyg. Environ. Health 2020, 226, 113485. [DOI] [PubMed] [Google Scholar]
- (13).Gomes SDR; Azéma L; Allard M; Toulmé J-J, Aptamers as imaging agents. Expert Opin. Med. Diagn 2010, 4 (6), 511–518. [DOI] [PubMed] [Google Scholar]
- (14).Tuerk C; Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249 (4968), 505–510. [DOI] [PubMed] [Google Scholar]
- (15).Ellington AD; Szostak JW, In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346 (6287), 818–822. [DOI] [PubMed] [Google Scholar]
- (16).Jayasena SD, Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem 1999, 45 (9), 1628–1650. [PubMed] [Google Scholar]
- (17).Carlson B, Aptamers : the new frontier in drug development? Biotechnol. Healthc 2007, 4 (2), 31–36. [PMC free article] [PubMed] [Google Scholar]
- (18).Song KM; Lee S; Ban C, Aptamers and their biological applications. Sensors 2012, 12 (1), 612–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Beier R; Boschke E; Labudde D, New strategies for evaluation and analysis of SELEX experiments. Biomed. Res. Int 2014, 2014, 849743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Afrasiabi S; Pourhajibagher M; Raoofian R; Tabarzad M; Bahador A, Therapeutic applications of nucleic acid aptamers in microbial infections. J. Biomed. Sci 2020, 27 (1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Groff K; Brown J; Clippinger AJ, Modern affinity reagents: Recombinant antibodies and aptamers. Biotechnol. Adv 2015, 33 (8), 1787–1798. [DOI] [PubMed] [Google Scholar]
- (22).Kim T-H; Lee S-W, Aptamers for Anti-Viral Therapeutics and Diagnostics. Int. J. Mol. Sci 2021, 22 (8), 4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nimjee SM; Rusconi CP; Sullenger BA, Aptamers: an emerging class of therapeutics. Annu. Rev. Med 2005, 56, 555–583. [DOI] [PubMed] [Google Scholar]
- (24).Zou X; Wu J; Gu J; Shen L; Mao L, Application of Aptamers in Virus Detection and Antiviral Therapy. Front Microbiol. 2019, 10, 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Micura R; Höbartner C, Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes. Chem. Soc. Rev 2020, 49 (20), 7331–7353. [DOI] [PubMed] [Google Scholar]
- (26).Hasegawa H; Savory N; Abe K; Ikebukuro K, Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21 (4), 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zhang N; Chen Z; Liu D; Jiang H; Zhang Z-K; Lu A; Zhang B-T; Yu Y; Zhang G, Structural Biology for the Molecular Insight between Aptamers and Target Proteins. Int. J. Mol. Sci 2021, 22 (8), 4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Conrad RC; Baskerville S; Ellington AD, In vitro selection methodologies to probe RNA function and structure. Mol. Divers 1995, 1 (1), 69–78. [DOI] [PubMed] [Google Scholar]
- (29).Hamula CL; Zhang H; Guan LL; Li XF; Le XC, Selection of aptamers against live bacterial cells. Anal. Chem 2008, 80 (20), 7812–7819. [DOI] [PubMed] [Google Scholar]
- (30).Li Y; Lee J-S, Recent developments in affinity-based selection of aptamers for binding disease-related protein targets. Chemical Papers 2019, 73 (11), 2637–2653. [Google Scholar]
- (31).Takemura K; Wang P; Vorberg I; Surewicz W; Priola SA; Kanthasamy A; Pottathil R; Chen SG; Sreevatsan S, DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Exp. Biol. Med 2006, 231 (2), 204–214. [DOI] [PubMed] [Google Scholar]
- (32).Parekh P; Tang Z; Turner PC; Moyer RW; Tan W, Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem 2010, 82 (20), 8642–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pan Q; Luo F; Liu M; Zhang XL, Oligonucleotide aptamers: promising and powerful diagnostic and therapeutic tools for infectious diseases. J. Infect 2018, 77 (2), 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Klug SJ; Famulok M, All you wanted to know about SELEX. Mol. Biol. Rep 1994, 20 (2), 97–107. [DOI] [PubMed] [Google Scholar]
- (35).Amiri S; Navaee A; Salimi A; Ahmadi R, Zeptomolar detection of Hg 2+ based on label-free electrochemical aptasensor: One step closer to the dream of single atom detection. Electrochem. Commun 2017, 78, 21–25. [Google Scholar]
- (36).Hong KL; Sooter LJ, Single-Stranded DNA Aptamers against Pathogens and Toxins: Identification and Biosensing Applications. Biomed. Res. Int 2015, 2015, 419318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ilgu M; Fazlioglu R; Ozturk M; Ozsurekci Y; Nilsen-Hamilton M, Aptamers for diagnostics with applications for infectious diseases. In Recent Advances in Analytical Chemistry, IntechOpen: 2019. [Google Scholar]
- (38).Li Y; Lee J-S, Recent developments in affinity-based selection of aptamers for binding disease-related protein targets. Chemical Papers 2019, 1–17. [Google Scholar]
- (39).Ohuchi S., Cell-SELEX Technology. BioResearch Open Access 2012, 1 (6), 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ulrich H; Wrenger C, Disease-specific biomarker discovery by aptamers. Cytometry A 2009, 75 (9), 727–733. [DOI] [PubMed] [Google Scholar]
- (41).Kumar V; Sharma A, Recent Updates for Isolation of Aptamers for Various Biothreat Agents Using Different Strategies and Their Role in Detection Applications. In Aptamers, Springer: 2019; pp 19–36. [Google Scholar]
- (42).Teng J; Yuan F; Ye Y; Zheng L; Yao L; Xue F; Chen W; Li B, Aptamer-Based Technologies in Foodborne Pathogen Detection. Front Microbiol. 2016, 7, 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hopkins AL; Groom CR, The druggable genome. Nat. Rev. Drug. Discov 2002, 1 (9), 727–730. [DOI] [PubMed] [Google Scholar]
- (44).Rask-Andersen M; Almén MS; Schiöth HB, Trends in the exploitation of novel drug targets. Nat. Rev. Drug. Discov 2011, 10 (8), 579–590. [DOI] [PubMed] [Google Scholar]
- (45).Tan W; Moyer RW; Turner P; Parekh P; Tang Z, Generating Aptamers for Recognition of Virus-Infected Cells. Clin. Chem 2009, 55 (4), 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Torres-Chavolla E; Alocilja EC, Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron 2009, 24 (11), 3175–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Pan W; Craven RC; Qiu Q; Wilson CB; Wills JW; Golovine S; Wang JF, Isolation of virus-neutralizing RNAs from a large pool of random sequences. Proc. Natl. Acad. Sci. U.S.A 1995, 92 (25), 11509–11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Zimmermann B; Bilusic I; Lorenz C; Schroeder R, Genomic SELEX: A discovery tool for genomic aptamers. Methods 2010, 52 (2), 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ostrander EA; Jong PM; Rine J; Duyk G, Construction of small-insert genomic DNA libraries highly enriched for microsatellite repeat sequences. Proc. Natl. Acad. Sci. U. S. A 1992, 89 (8), 3419–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Khalil AM; Guttman M; Huarte M; Garber M; Raj A; Rivea Morales D; Thomas K; Presser A; Bernstein BE; van Oudenaarden A; Regev A; Lander ES; Rinn JL, Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (28), 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zaratiegui M; Irvine DV; Martienssen RA, Noncoding RNAs and Gene Silencing. Cell 2007, 128 (4), 763–776. [DOI] [PubMed] [Google Scholar]
- (52).Halic M; Beckmann R, The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Bio 2005, 15 (1), 116–125. [DOI] [PubMed] [Google Scholar]
- (53).Eddy SR, Noncoding RNA genes. Curr. Opin. Genet & Dev 1999, 9 (6), 695–699. [DOI] [PubMed] [Google Scholar]
- (54).Hybarger G; Bynum J; Williams RF; Valdes JJ; Chambers JP, A microfluidic SELEX prototype. Anal. Bioanal. Chem 2006, 384 (1), 191–198. [DOI] [PubMed] [Google Scholar]
- (55).Cho S; Lee SH; Chung WJ; Kim YK; Lee YS; Kim BG, Microbead-based affinity chromatography chip using RNA aptamer modified with photocleavable linker. Electrophoresis 2004, 25 (21-22), 3730–3739. [DOI] [PubMed] [Google Scholar]
- (56).Lou X; Qian J; Xiao Y; Viel L; Gerdon AE; Lagally ET; Atzberger P; Tarasow TM; Heeger AJ; Soh HT, Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (9), 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Qian J; Lou X; Zhang Y; Xiao Y; Soh HT, Generation of highly specific aptamers via micromagnetic selection. Anal. Chem 2009, 81 (13), 5490–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Wang M; Wang Q; Li X; Lu L; Du S; Zhang H, Selection and identification of diethylstilbestrol-specific aptamers based on magnetic-bead SELEX. Microchem. J 2020, 159, 105354. [Google Scholar]
- (59).Xu Y; Yang X; Wang E, Review: Aptamers in microfluidic chips. Anal. Chim. Acta 2010, 683 (1), 12–20. [DOI] [PubMed] [Google Scholar]
- (60).Yang J; Bowser MT, Capillary electrophoresis-SELEX selection of catalytic DNA aptamers for a small-molecule porphyrin target. Anal. Chem 2013, 85 (3), 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Mendonsa SD; Bowser MT, In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc 2004, 126 (1), 20–21. [DOI] [PubMed] [Google Scholar]
- (62).Mendonsa SD; Bowser MT, In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal. Chem 2004, 76 (18), 5387–5392. [DOI] [PubMed] [Google Scholar]
- (63).Mosing RK; Bowser MT, Isolating Aptamers Using Capillary Electrophoresis–SELEX (CE–SELEX). In Nucleic Acid and Peptide Aptamers, 2009; 33–43. [DOI] [PubMed] [Google Scholar]
- (64).Pavski V; Le XC, Detection of Human Immunodeficiency Virus Type 1 Reverse Transcriptase Using Aptamers as Probes in Affinity Capillary Electrophoresis. Anal. Chem 2001, 73 (24), 6070–6076. [DOI] [PubMed] [Google Scholar]
- (65).Meng C; Zhao X; Qu F; Mei F; Gu L, Interaction evaluation of bacteria and protoplasts with single-stranded deoxyribonucleic acid library based on capillary electrophoresis. J. Chromatogr. A 2014, 1358, 269–276. [DOI] [PubMed] [Google Scholar]
- (66).André C; Xicluna A; Guillaume Y-C, Aptamer-oligonucleotide binding studied by capillary electrophoresis: Cation effect and separation efficiency. Electrophoresis 2005, 26 (17), 3247–3255. [DOI] [PubMed] [Google Scholar]
- (67).Bae H; Ren S; Kang J; Kim M; Jiang Y; Jin MM; Min IM; Kim S, Sol-Gel SELEX Circumventing Chemical Conjugation of Low Molecular Weight Metabolites Discovers Aptamers Selective to Xanthine. Nucleic Acid Ther. 2013, 23 (6), 443–449. [DOI] [PubMed] [Google Scholar]
- (68).Park S.-m.; Ahn J-Y; Jo M; Lee D.-k.; Lis JT; Craighead HG; Kim S, Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab on a Chip 2009, 9 (9), 1206–1212. [DOI] [PubMed] [Google Scholar]
- (69).Park S.-m.; Ahn J-Y; Jo M; Lee D.-k.; Lis JT; Craighead HG; Kim S, Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab Chip 2009, 9 (9). [DOI] [PubMed] [Google Scholar]
- (70).Miyachi Y; Shimizu N; Ogino C; Kondo A, Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010, 38 (4), e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Takenaka M; Okumura Y; Amino T; Miyachi Y; Ogino C; Kondo A, DNA-duplex linker for AFM-SELEX of DNA aptamer against human serum albumin. Bioorg. Med. Chem. Lett 2017, 27 (4), 954–957. [DOI] [PubMed] [Google Scholar]
- (72).White R; Rusconi C; Scardino E; Wolberg A; Lawson J; Hoffman M; Sullenger B, Generation of species cross-reactive aptamers using "toggle" SELEX. Mol. Ther 2001, 4 (6), 567–573. [DOI] [PubMed] [Google Scholar]
- (73).Derbyshire N; White SJ; Bunka DHJ; Song L; Stead S; Tarbin J; Sharman M; Zhou D; Stockley PG, Toggled RNA Aptamers Against Aminoglycosides Allowing Facile Detection of Antibiotics Using Gold Nanoparticle Assays. Anal. Chem 2012, 84 (15), 6595–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Malhotra S., Aptamer Selection: Choosing the Appropriate SELEX. Indian J. Pure Appl. Biosci 2021, 9 (2), 240–253. [Google Scholar]
- (75).Bunka DH; Stockley PG, Aptamers come of age - at last. Nat. Rev. Microbiol 2006, 4 (8), 588–596. [DOI] [PubMed] [Google Scholar]
- (76).Jackson BR; Busch MP; Stramer SL; AuBuchon JP, The cost-effectiveness of NAT for HIV, hCv, and HBV in whole-blood donations. Transfusion 2003, 43 (6), 721–729. [DOI] [PubMed] [Google Scholar]
- (77).Vainionpää R; Leinikki P, Diagnostic Techniques: Serological and Molecular Approaches. In Encyclopedia of Virology, 2008; pp 29–37. [Google Scholar]
- (78).Baer A; Kehn-Hall K, Viral concentration determination through plaque assays: using traditional and novel overlay systems. J. Vis. Exp 2014, (93), e52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Yang S-Y; Lien K-Y; Huang K-J; Lei H-Y; Lee G-B, Micro flow cytometry utilizing a magnetic bead-based immunoassay for rapid virus detection. Biosens. Bioelectron 2008, 24 (4), 855–862. [DOI] [PubMed] [Google Scholar]
- (80).Killian ML, Hemagglutination Assay for Influenza Virus. In Animal Influenza Virus, 2014; pp 3–9. [DOI] [PubMed] [Google Scholar]
- (81).Alahi MEE; Mukhopadhyay SC, Detection Methodologies for Pathogen and Toxins: A Review. Sensors 2017, 17 (8), 1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Rajapaksha P; Elbourne A; Gangadoo S; Brown R; Cozzolino D; Chapman J, A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144 (2), 396–411. [DOI] [PubMed] [Google Scholar]
- (83).Vasilescu A; Marty J-L, Electrochemical aptasensors for the assessment of food quality and safety. TrAC Trends Analyt. Chem 2016, 79, 60–70. [Google Scholar]
- (84).Bhatt R; Bagri LP; Saini R; Bajpai AK, Sensors: Advanced Aptasensors Design. 2018. [Google Scholar]
- (85).Mishra GK; Sharma V; Mishra RK, Electrochemical Aptasensors for Food and Environmental Safeguarding: A Review. Biosensors 2018, 8 (2), 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Labib M; Zamay AS; Muharemagic D; Chechik AV; Bell JC; Berezovski MV, Aptamer-based viability impedimetric sensor for viruses. Anal. Chem 2012, 84 (4), 1813–1816. [DOI] [PubMed] [Google Scholar]
- (87).Giamberardino A; Labib M; Hassan EM; Tetro JA; Springthorpe S; Sattar SA; Berezovski MV; DeRosa MC, Ultrasensitive norovirus detection using DNA aptasensor technology. PloS one 2013, 8 (11), e79087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Lum J; Wang R; Hargis B; Tung S; Bottje W; Lu H; Li Y, An impedance aptasensor with microfluidic chips for specific detection of H5N1 avian influenza virus. Sensors 2015, 15 (8), 18565–18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Kirkegaard J; Rozlosnik N, Screen-Printed All-Polymer Aptasensor for Impedance Based Detection of Influenza A Virus. Methods. Mol. Biol 2017, 1572, 55–70. [DOI] [PubMed] [Google Scholar]
- (90).Ghanbari K; Roushani M; Azadbakht A, Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem 2017, 534, 64–69. [DOI] [PubMed] [Google Scholar]
- (91).Maehashi K; Katsura T; Kerman K; Takamura Y; Matsumoto K; Tamiya E, Label-free protein biosensor based on aptamer-modified carbon nanotube field-effect transistors. Anal. Chem 2007, 79 (2), 782–787. [DOI] [PubMed] [Google Scholar]
- (92).Ruslinda AR; Tanabe K; Ibori S; Wang X; Kawarada H, Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 Tat protein. Biosens. Bioelectron 2013, 40 (1), 277–282. [DOI] [PubMed] [Google Scholar]
- (93).Veigas B; Fortunato E; Baptista PV, Field effect sensors for nucleic Acid detection: recent advances and future perspectives. Sensors 2015, 15 (5), 10380–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Lim HJ; Saha T; Tey BT; Tan WS; Ooi CW, Quartz crystal microbalance-based biosensors as rapid diagnostic devices for infectious diseases. Biosens. Bioelectron 2020, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Wang R; Li Y, Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron 2013, 42, 148–155. [DOI] [PubMed] [Google Scholar]
- (96).Fu Y; Callaway Z; Lum J; Wang R; Lin J; Li Y, Exploiting enzyme catalysis in ultra-low ion strength media for impedance biosensing of avian influenza virus using a bare interdigitated electrode. Anal. Chem 2014, 86 (4), 1965–1971. [DOI] [PubMed] [Google Scholar]
- (97).Diba FS; Kim S; Lee HJ, Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron 2015, 72, 355–361. [DOI] [PubMed] [Google Scholar]
- (98).Nath I; Chakraborty J; Verpoort F, Metal organic frameworks mimicking natural enzymes: a structural and functional analogy. Chem. Soc. Rev 2016, 45 (15), 4127–4170. [DOI] [PubMed] [Google Scholar]
- (99).Jiang D; Ni D; Rosenkrans ZT; Huang P; Yan X; Cai W, Nanozyme: new horizons for responsive biomedical applications. Chem. Soc. Rev 2019, 48 (14), 3683–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Weerathunge P; Ramanathan R; Torok VA; Hodgson K; Xu Y; Goodacre R; Behera BK; Bansal V, Ultrasensitive colorimetric detection of murine norovirus using NanoZyme aptasensor. Anal. Chem 2019, 91 (5), 3270–3276. [DOI] [PubMed] [Google Scholar]
- (101).Salouti M; Derakhshan FK, Biosensors and Nanobiosensors in Environmental Applications. In Biogenic Nano-Particles and their Use in Agro-ecosystems, Springer: 2020; pp 515–591. [Google Scholar]
- (102).Prabowo BA; Purwidyantri A; Liu KC, Surface Plasmon Resonance Optical Sensor: A Review on Light Source Technology. Biosensors 2018, 8 (3), 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Zeni L; Perri C; Cennamo N; Arcadio F; D’Agostino G; Salmona M; Beeg M; Gobbi M, A portable optical-fibre-based surface plasmon resonance biosensor for the detection of therapeutic antibodies in human serum. Sci. Rep 2020, 10 (1), 11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Tombelli S; Minunni M; Luzi E; Mascini M, Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 2005, 67 (2), 135–141. [DOI] [PubMed] [Google Scholar]
- (105).Bai H; Wang R; Hargis B; Lu H; Li Y, A SPR aptasensor for detection of avian influenza virus H5N1. Sensors 2012, 12 (9), 12506–12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Shimanoe K; Endo S; Matsuyama T; Wada K; Okamoto K, Localized surface plasmon resonance in deep ultraviolet region below 200 nm using a nanohemisphere on mirror structure. Sci. Rep 2021, 11 (1), 5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Klinghammer S; Uhlig T; Patrovsky F; Bohm M; Schutt J; Putz N; Baraban L; Eng LM; Cuniberti G, Plasmonic Biosensor Based on Vertical Arrays of Gold Nanoantennas. ACS Sens. 2018, 3 (7), 1392–1400. [DOI] [PubMed] [Google Scholar]
- (108).Pang Y; Rong Z; Wang J; Xiao R; Wang S, A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF). Biosens. Bioelectron 2015, 66, 527–532. [DOI] [PubMed] [Google Scholar]
- (109).Szakacs Z; Meszaros T; de Jonge MI; Gyurcsanyi RE, Selective counting and sizing of single virus particles using fluorescent aptamer-based nanoparticle tracking analysis. Nanoscale 2018, 10 (29), 13942–13948. [DOI] [PubMed] [Google Scholar]
- (110).Fu Z; Lu YC; Lai JJ, Recent Advances in Biosensors for Nucleic Acid and Exosome Detection. Chonnam. Med. J 2019, 55 (2), 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Blackie EJ; Le Ru EC; Etchegoin PG, Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. J. Am. Chem. Soc 2009, 131 (40), 14466–14472. [DOI] [PubMed] [Google Scholar]
- (112).Le Ru EC; Blackie EJ; Meyer M; Etchegoin PG, Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study. J. Phys. Chem. C 2007, 111 (37), 13794–13803. [Google Scholar]
- (113).Lu Y; Lin Y; Zheng Z; Tang X; Lin J; Liu X; Liu M; Chen G; Qiu S; Zhou T; Lin Y; Feng S, Label free hepatitis B detection based on serum derivative surface enhanced Raman spectroscopy combined with multivariate analysis. Biomed. Opt. Express 2018, 9 (10), 4755–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Driskell JD; Shanmukh S; Liu Y-J; Hennigan S; Jones L; Zhao Y-P; Dluhy RA; Krause DC; Tripp RA, Infectious Agent Detection With SERS-Active Silver Nanorod Arrays Prepared by Oblique Angle Deposition. IEEE Sens. J 2008, 8 (6), 863–870. [Google Scholar]
- (115).Ambartsumyan O; Gribanyov D; Kukushkin V; Kopylov A; Zavyalova E, SERS-Based Biosensors for Virus Determination with Oligonucleotides as Recognition Elements. Int. J. Mol. Sci 2020, 21 (9), 3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Wang Y; Schlucker S, Rational design and synthesis of SERS labels. Analyst 2013, 138 (8), 2224–2238. [DOI] [PubMed] [Google Scholar]
- (117).Chen H; Park SG; Choi N; Moon JI; Dang H; Das A; Lee S; Kim DG; Chen L; Choo J, SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron 2020, 167, 112496. [DOI] [PubMed] [Google Scholar]
- (118).Kukushkin VI; Ivanov NM; Novoseltseva AA; Gambaryan AS; Yaminsky IV; Kopylov AM; Zavyalova EG, Highly sensitive detection of influenza virus with SERS aptasensor. PLoS One 2019, 14 (4), e0216247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Giuffrida MC; Cigliana G; Spoto G, Ultrasensitive detection of lysozyme in droplet-based microfluidic devices. Biosens. Bioelectron 2018, 104, 8–14. [DOI] [PubMed] [Google Scholar]
- (120).Jolly P; Damborsky P; Madaboosi N; Soares RR; Chu V; Conde JP; Katrlik J; Estrela P, DNA aptamer-based sandwich microfluidic assays for dual quantification and multi-glycan profiling of cancer biomarkers. Biosens. Bioelectron 2016, 79, 313–319. [DOI] [PubMed] [Google Scholar]
- (121).Mazaafrianto DN; Maeki M; Ishida A; Tani H; Tokeshi M, Recent Microdevice-Based Aptamer Sensors. Micromachines 2018, 9 (5), 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Yi-Xian W; Zun-Zhong Y; Cheng-Yan S; Yi-Bin Y, Application of aptamer based biosensors for detection of pathogenic microorganisms. Chinese J. Anal. Chem 2012, 40 (4), 634–642. [Google Scholar]
- (123).Ahn DG; Jeon IJ; Kim JD; Song MS; Han SR; Lee SW; Jung H; Oh JW, RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst 2009, 134 (9), 1896–1901. [DOI] [PubMed] [Google Scholar]
- (124).Xi Z; Huang R; Li Z; He N; Wang T; Su E; Deng Y, Selection of HBsAg-Specific DNA Aptamers Based on Carboxylated Magnetic Nanoparticles and Their Application in the Rapid and Simple Detection of Hepatitis B Virus Infection. ACS Appl. Mater. Interfaces 2015, 7 (21), 11215–11223. [DOI] [PubMed] [Google Scholar]
- (125).Lakowicz JR, Principles of Fluorescence Spectroscopy. Springer Science+Business Media, LLC: 1999. [Google Scholar]
- (126).Percze K; Szakacs Z; Scholz E; Andras J; Szeitner Z; Kieboom CH; Ferwerda G; Jonge MI; Gyurcsanyi RE; Meszaros T, Aptamers for respiratory syncytial virus detection. Sci. Rep 2017, 7, 42794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Wang C-H; Chang C-P; Lee G-B, Integrated microfluidic device using a single universal aptamer to detect multiple types of influenza viruses. Biosens. Bioelectron 2016, 86, 247–254. [DOI] [PubMed] [Google Scholar]
- (128).Forster TH, 10th Spiers Memorial Lecture. Transfer mechanisms of electronic excitation. Discuss. Faraday Soc 1959, 27, 7–17. [Google Scholar]
- (129).Zhang X; Hu Y; Yang X; Tang Y; Han S; Kang A; Deng H; Chi Y; Zhu D; Lu Y, FOrster resonance energy transfer (FRET)-based biosensors for biological applications. Biosens. Bioelectron 2019, 138, 111314. [DOI] [PubMed] [Google Scholar]
- (130).Yamamoto R; Kumar PK, Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes to Cells 2000, 5 (5), 389–396. [DOI] [PubMed] [Google Scholar]
- (131).Hu J; Wang Z.-y.; Li C.-c.; Zhang C.-y., Advances in single quantum dot-based nanosensors. Chem. Commun 2017, 53 (100), 13284–13295. [DOI] [PubMed] [Google Scholar]
- (132).Zhang J; Tian J; He Y; Chen S; Jiang Y; Zhao Y; Zhao S, Protein-binding aptamer assisted signal amplification for the detection of influenza A (H1N1) DNA sequences based on quantum dot fluorescence polarization analysis. Analyst 2013, 138 (17), 4722–4727. [DOI] [PubMed] [Google Scholar]
- (133).Li; Rothberg LJ, Label-Free Colorimetric Detection of Specific Sequences in Genomic DNA Amplified by the Polymerase Chain Reaction. J. Am. Chem. Soc 2004, 126 (35), 10958–10961. [DOI] [PubMed] [Google Scholar]
- (134).Taleat Z; Mathwig K; Sudhölter EJR; Rassaei L, Detection strategies for methylated and hypermethylated DNA. TrAC Trends Anal. Chem 2015, 66, 80–89. [Google Scholar]
- (135).Chen C-A; Wang C-C; Jong Y-J; Wu S-M, Label-Free Fluorescent Copper Nanoclusters for Genotyping of Deletion and Duplication of Duchenne Muscular Dystrophy. Anal. Chem 2015, 87 (12), 6228–6232. [DOI] [PubMed] [Google Scholar]
- (136).Mao X; Liu S; Yang C; Liu F; Wang K; Chen G, Colorimetric detection of hepatitis B virus (HBV) DNA based on DNA-templated copper nanoclusters. Anal. Chim. Acta 2016, 909, 101–108. [DOI] [PubMed] [Google Scholar]
- (137).Liu J; Lu Y, Adenosine-Dependent Assembly of Aptazyme-Functionalized Gold Nanoparticles and Its Application as a Colorimetric Biosensor. Anal. Chem 2004, 76 (6), 1627–1632. [DOI] [PubMed] [Google Scholar]
- (138).Chen C; Zou Z; Chen L; Ji X; He Z, Functionalized magnetic microparticle-based colorimetric platform for influenza A virus detection. Nanotechnology 2016, 27 (43), 435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Liu M; Zhao H; Chen S; Yu H; Quan X, Interface engineering catalytic graphene for smart colorimetric biosensing. Acs Nano 2012, 6 (4), 3142–3151. [DOI] [PubMed] [Google Scholar]
- (140).Wang H; Cui L-F; Yang Y; Sanchez Casalongue H; Robinson JT; Liang Y; Cui Y; Dai H, Mn3O4-Graphene Hybrid as a High-Capacity Anode Material for Lithium Ion Batteries. J. Am. Chem. Soc 2010, 132 (40), 13978–13980. [DOI] [PubMed] [Google Scholar]
- (141).Rasoulinejad S; Gargari SLM, Aptamer-nanobody based ELASA for specific detection of Acinetobacter baumannii isolates. J. Biotechnol 2016, 231, 46–54. [DOI] [PubMed] [Google Scholar]
- (142).Shiratori I; Akitomi J; Boltz DA; Horii K; Furuichi M; Waga I, Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun 2014, 443 (1), 37–41. [DOI] [PubMed] [Google Scholar]
- (143).Escudero-Abarca BI; Suh SH; Moore MD; Dwivedi HP; Jaykus L-A, Selection, characterization and application of nucleic acid aptamers for the capture and detection of human norovirus strains. PloS One 2014, 9 (9), e106805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Lee KH; Zeng H, Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem 2017, 89 (23), 12743–12748. [DOI] [PubMed] [Google Scholar]
- (145).Park JH; Jee MH; Kwon OS; Keum SJ; Jang SK, Infectivity of hepatitis C virus correlates with the amount of envelope protein E2: development of a new aptamer-based assay system suitable for measuring the infectious titer of HCV. Virology 2013, 439 (1), 13–22. [DOI] [PubMed] [Google Scholar]
- (146).Eilers A; Witt S; Walter J, Aptamer-Modified Nanoparticles in Medical Applications. Adv. Biochem. Eng. Biotechnol 2020, 174, 161–193. [DOI] [PubMed] [Google Scholar]
- (147).Heiat M; Ranjbar R; Alavian SM, Classical and modern approaches used for viral hepatitis diagnosis. Hepat. Mon 2014, 14 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Dhiman A; Kalra P; Bansal V; Bruno JG; Sharma TK, Aptamer-based point-of-care diagnostic platforms. Sens. Actuators B Chem 2017, 246, 535–553. [Google Scholar]
- (149).Le TT; Chang P; Benton DJ; McCauley JW; Iqbal M; Cass AEG, Dual Recognition Element Lateral Flow Assay Toward Multiplex Strain Specific Influenza Virus Detection. Anal. Chem 2017, 89 (12), 6781–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Mescalchin A; Restle T, Oligomeric nucleic acids as antivirals. Molecules 2011, 16 (2), 1271–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Alter G; Heckerman D; Schneidewind A; Fadda L; Kadie CM; Carlson JM; Oniangue-Ndza C; Martin M; Li B; Khakoo SI; Carrington M; Allen TM; Altfeld M, HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 2011, 476 (7358), 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Ding Q; Cao X; Lu J; Huang B; Liu YJ; Kato N; Shu HB; Zhong J, Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol 2013, 59 (1), 52–58. [DOI] [PubMed] [Google Scholar]
- (153).Haasnoot J; Berkhout B, Nucleic acids-based therapeutics in the battle against pathogenic viruses. Handb. Exp. Pharmacol 2009, (189), 243–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Arts EJ; Hazuda DJ, HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med 2012, 2 (4), a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Gopinath SC, Antiviral aptamers. Arch. Virol 2007, 152 (12), 2137–2157. [DOI] [PubMed] [Google Scholar]
- (156).Jones M; Nunez M, Liver toxicity of antiretroviral drugs. Semin. Liver Dis 2012, 32 (2), 167–176. [DOI] [PubMed] [Google Scholar]
- (157).Jordheim LP; Durantel D; Zoulim F; Dumontet C, Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov 2013, 12 (6), 447–464. [DOI] [PubMed] [Google Scholar]
- (158).Ni X; Castanares M; Mukherjee A; Lupold SE, Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem 2011, 18 (27), 4206–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Zhou J; Bobbin ML; Burnett JC; Rossi JJ, Current progress of RNA aptamer-based therapeutics. Front Genet. 2012, 3, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Wang RE; Wu H; Niu Y; Cai J, Improving the Stability of Aptamers by Chemical Modification. Curr. Med. Chem 2011, 18 (27), 4126–4138. [DOI] [PubMed] [Google Scholar]
- (161).Sayer N; Ibrahim J; Turner K; Tahiri-Alaoui A; James W, Structural characterization of a 2′F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun 2002, 293 (3), 924–931. [DOI] [PubMed] [Google Scholar]
- (162).Dey AK; Griffiths C; Lea SM; James W, Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11 (6), 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Cohen C; Forzan M; Sproat B; Pantophlet R; McGowan I; Burton D; James W, An aptamer that neutralizes R5 strains of HIV-1 binds to core residues of gp120 in the CCR5 binding site. Virology 2008, 381 (1), 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Zhou J; Swiderski P; Li H; Zhang J; Neff CP; Akkina R; Rossi JJ, Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009, 37 (9), 3094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Bereswill S; Chen F; Hu Y; Li D; Chen H; Zhang X-L, CS-SELEX Generates High-Affinity ssDNA Aptamers as Molecular Probes for Hepatitis C Virus Envelope Glycoprotein E2. PLoS One 2009, 4 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Gopinath SCB; Misono TS; Kawasaki K; Mizuno T; Imai M; Odagiri T; Kumar PKR, An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol 2006, 87 (3), 479–487. [DOI] [PubMed] [Google Scholar]
- (167).Park M-S; Kwon H-M; Lee KH; Han BW; Han MR; Kim DH; Kim D-E, An RNA Aptamer That Specifically Binds to the Glycosylated Hemagglutinin of Avian Influenza Virus and Suppresses Viral Infection in Cells. PLoS One 2014, 9 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Yuan S; Zhang N; Singh K; Shuai H; Chu H; Zhou J; Chow BKC; Zheng B-J, Cross-Protection of Influenza A Virus Infection by a DNA Aptamer Targeting the PA Endonuclease Domain. Antimicrob. Agents Chemother 2015, 59 (7), 4082–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Nicol C; Bunka DHJ; Blair GE; Stonehouse NJ, Effects of single nucleotide changes on the binding and activity of RNA aptamers to human papillomavirus 16 E7 oncoprotein. Biochem. Biophys. Res. Commun 2011, 405 (3), 417–421. [DOI] [PubMed] [Google Scholar]
- (170).Toscano-Garibay JD; Benítez-Hess ML; Alvarez-Salas LM, Isolation and Characterization of an RNA Aptamer for the HPV-16 E7 Oncoprotein. Arch. Med. Res 2011, 42 (2), 88–96. [DOI] [PubMed] [Google Scholar]
- (171).Burk RD; Nicol C; Cesur Ö; Forrest S; Belyaeva TA; Bunka DHJ; Blair GE; Stonehouse NJ, An RNA Aptamer Provides a Novel Approach for the Induction of Apoptosis by Targeting the HPV16 E7 Oncoprotein. PLoS One 2013, 8 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Belyaeva T; Nicol C; Cesur Ö; Travé G; Blair G; Stonehouse N, An RNA Aptamer Targets the PDZ-Binding Motif of the HPV16 E6 Oncoprotein. Cancers 2014, 6 (3), 1553–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Liu J; Yang Y; Hu B; Ma Z.-y.; Huang H.-p.; Yu Y; Liu S.-p.; Lu M.-j.; Yang D.-l., Development of HBsAg-binding aptamers that bind HepG2.2.15 cells via HBV surface antigen. Virol. Sin 2010, 25 (1), 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Feng H; Beck J; Nassal M; Hu KH, A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS One 2011, 6 (11), e27862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Zhang Z; Zhang J; Pei X; Zhang Q; Lu B; Zhang X; Liu J, An aptamer targets HBV core protein and suppresses HBV replication in HepG2.2.15 cells. Int. J. Mol. Med 2014, 34 (5), 1423–1429. [DOI] [PubMed] [Google Scholar]
- (176).Chen HL; Hsiao WH; Lee HC; Wu SC; Cheng JW, Selection and Characterization of DNA Aptamers Targeting All Four Serotypes of Dengue Viruses. PLoS One 2015, 10 (6), e0131240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Shum KT; Tanner JA, Differential Inhibitory Activities and Stabilisation of DNA Aptamers against the SARS Coronavirus Helicase. Chembiochem 2008, 9 (18), 3037–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Liang H-R; Hu G-Q; Li L; Gao Y-W; Yang S-T; Xia X-Z, Aptamers targeting rabies virus-infected cells inhibit street rabies virus in vivo. Int. Immunopharmacol 2014, 21 (2), 432–438. [DOI] [PubMed] [Google Scholar]
- (179).Liang H-R; Hu G-Q; Xue X-H; Li L; Zheng X-X; Gao Y-W; Yang S-T; Xia X-Z, Selection of an aptamer against rabies virus: A new class of molecules with antiviral activity. Virus Res. 2014, 184, 7–13. [DOI] [PubMed] [Google Scholar]
- (180).Valencia-Reséndiz DG; Palomino-Vizcaino G; Tapia-Vieyra JV; Benítez-Hess ML; Leija-Montoya AG; Alvarez-Salas LM, Inhibition of Human Papillomavirus Type 16 Infection Using an RNA Aptamer. Nucleic Acid Ther. 2018, 28 (2), 97–105. [DOI] [PubMed] [Google Scholar]
- (181).Yadavalli T; Agelidis A; Jaishankar D; Mangano K; Thakkar N; Penmetcha K; Shukla D, Targeting Herpes Simplex Virus-1 gD by a DNA Aptamer Can Be an Effective New Strategy to Curb Viral Infection. Mol. Ther. Nucleic Acids 2017, 9, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Hovanessian AG, Midkine, a cytokine that inhibits HIV infection by binding to the cell surface expressed nucleolin. Cell Res. 2006, 16 (2), 174–181. [DOI] [PubMed] [Google Scholar]
- (183).Balinsky CA; Schmeisser H; Ganesan S; Singh K; Pierson TC; Zoon KC, Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol 2013, 87 (24), 13094–13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Chen F; Hu Y; Li D; Chen H; Zhang X-L, CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for hepatitis C virus envelope glycoprotein E2. PloS One 2009, 4 (12), e8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Buckheit RW Jr.; Roberson JL; Lackman-Smith C; Wyatt JR; Vickers TA; Ecker DJ, Potent and specific inhibition of HIV envelope-mediated cell fusion and virus binding by G quartet-forming oligonucleotide (ISIS 5320). AIDS Res. Hum. Retroviruses 1994, 10 (11), 1497–1506. [DOI] [PubMed] [Google Scholar]
- (186).Stoddart CA; Rabin L; Hincenbergs M; Moreno M; Linquist-Stepps V; Leeds JM; Truong LA; Wyatt JR; Ecker DJ; McCune JM, Inhibition of human immunodeficiency virus type 1 infection in SCID-hu Thy/Liv mice by the G-quartet-forming oligonucleotide, ISIS 5320. Antimicrob. Agents Chemother 1998, 42 (8), 2113–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Nisole S; Said EA; Mische C; Prevost M-C; Krust B; Bouvet P; Bianco A; Briand J-P; Hovanessian AG, The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J. Biol. Chem 2002, 277 (23), 20877–20886. [DOI] [PubMed] [Google Scholar]
- (188).Perrone R; Butovskaya E; Lago S; Garzino-Demo A; Pannecouque C; Palu G; Richter SN, The G-quadruplex-forming aptamer AS1411 potently inhibits HIV-1 attachment to the host cell. Int. J. Antimicrob. Agents 2016, 47 (4), 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Jeon SH; Kayhan B; Ben-Yedidia T; Arnon R, A DNA Aptamer Prevents Influenza Infection by Blocking the Receptor Binding Region of the Viral Hemagglutinin. J. Biol. Chem 2004, 279 (46), 48410–48419. [DOI] [PubMed] [Google Scholar]
- (190).Choi SK; Lee C; Lee KS; Choe S-Y; Mo IP; Seong RH; Hong S; Jeon SH, DNA aptamers against the receptor binding region of hemagglutinin prevent avian influenza viral infection. Mol. Cells 2011, 32 (6), 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Gopinath SCB; Hayashi K; Kumar PKR, Aptamer That Binds to the gD Protein of Herpes Simplex Virus 1 and Efficiently Inhibits Viral Entry. J. Virol 2012, 86 (12), 6732–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Luo G; Hamatake RK; Mathis DM; Racela J; Rigat KL; Lemm J; Colonno RJ, De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol 2000, 74 (2), 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Cheney IW; Naim S; Lai VC; Dempsey S; Bellows D; Walker MP; Shim JH; Horscroft N; Hong Z; Zhong W, Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 2002, 297 (2), 298–306. [DOI] [PubMed] [Google Scholar]
- (194).Biroccio A; Hamm J; Incitti I; De Francesco R; Tomei L, Selection of RNA aptamers that are specific and high-affinity ligands of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol 2002, 76 (8), 3688–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Bellecave P; Cazenave C; Rumi J; Staedel C; Cosnefroy O; Andreola ML; Ventura M; Tarrago-Litvak L; Astier-Gin T, Inhibition of hepatitis C virus (HCV) RNA polymerase by DNA aptamers: mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob. Agents Chemother 2008, 52 (6), 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Nagy PD; Pogany J, The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol 2011, 10 (2), 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Kim SJ; Kim MY; Lee JH; You JC; Jeong S, Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun 2002, 291 (4), 925–931. [DOI] [PubMed] [Google Scholar]
- (198).Yamamoto R; Katahira M; Nishikawa S; Baba T; Taira K; Kumar PKR, A novel RNA motif that binds efficiently and specifically to the Tat protein of HIV and inhibits the transactivation by Tat of transcription in vitro and in vivo. Genes to Cells 2000, 5 (5), 371–388. [DOI] [PubMed] [Google Scholar]
- (199).Dey AK; Khati M; Tang M; Wyatt R; Lea SM; James W, An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J. Virol 2005, 79 (21), 13806–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Khati M; Schüman M; Ibrahim J; Sattentau Q; Gordon S; James W, Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′ F-RNA aptamers. J. Virol 2003, 77 (23), 12692–12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Biroccio A; Hamm J. r.; Incitti I; De Francesco R; Tomei L, Selection of RNA Aptamers That Are Specific and High-Affinity Ligands of the Hepatitis C Virus RNA-Dependent RNA Polymerase. J. Virol 2002, 76 (8), 3688–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).DeStefano JJ; Nair GR, Novel Aptamer Inhibitors of Human Immunodeficiency Virus Reverse Transcriptase. Oligonucleotides 2008, 18 (2), 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Fukuda K; Umehara T; Sekiya S; Kunio K; Hasegawa T; Nishikawa S, An RNA ligand inhibits hepatitis C virus NS3 protease and helicase activities. Biochem. Biophys. Res. Commun 2004, 325 (3), 670–675. [DOI] [PubMed] [Google Scholar]
- (204).Yu X; Gao Y; Xue B; Wang X; Yang D; Qin Y; Yu R; Liu N; Xu L; Fang X; Zhu H, Inhibition of hepatitis C virus infection by NS5A-specific aptamer. Antiviral Res. 2014, 106, 116–124. [DOI] [PubMed] [Google Scholar]
- (205).Jang KJ; Lee N-R; Yeo W-S; Jeong Y-J; Kim D-E, Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun 2008, 366 (3), 738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Konno K; Fujita S; Iizuka M; Nishikawa S; Hasegawa T; Fukuda K, Isolation and characterization of RNA aptamers specific for the HCV minus-IRES domain I. Nucleic Acids Symp. Ser 2008, 52 (1), 493–494. [DOI] [PubMed] [Google Scholar]
- (207).Konno K; Iizuka M; Fujita S; Nishikawa S; Hasegawa T; Fukuda K, An Rna Aptamer Containing Two Binding Sites against the Hcv Minus-Ires Domain I. Nucleosides, Nucleotides Nucleic Acids 2011, 30 (3), 185–202. [DOI] [PubMed] [Google Scholar]
- (208).Kikuchi K, RNA Aptamers Targeted to Domain II of Hepatitis C Virus IRES That Bind to Its Apical Loop Region. J. Biochem 2003, 133 (3), 263–270. [DOI] [PubMed] [Google Scholar]
- (209).Kikuchi K; Umehara T; Nishikawa F; Fukuda K; Hasegawa T; Nishikawa S, Increased inhibitory ability of conjugated RNA aptamers against the HCV IRES. Biochem. Biophys. Res. Commun 2009, 386 (1), 118–123. [DOI] [PubMed] [Google Scholar]
- (210).Romero-López C; Berzal-Herranz B; Gómez J; Berzal-Herranz A, An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antiviral Res. 2012, 94 (2), 131–138. [DOI] [PubMed] [Google Scholar]
- (211).Srisawat C; Engelke DR, Selection of RNA aptamers that bind HIV-1 LTR DNA duplexes: strand invaders. Nucleic Acids Res. 2010, 38 (22), 8306–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Neff CP; Zhou J; Remling L; Kuruvilla J; Zhang J; Li H; Smith DD; Swiderski P; Rossi JJ; Akkina R, An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med 2011, 3 (66), 66ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Zhu Q; Shibata T; Kabashima T; Kai M, Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur. J. Med. Chem 2012, 56, 396–399. [DOI] [PubMed] [Google Scholar]
- (214).Wheeler LA; Trifonova R; Vrbanac V; Basar E; McKernan S; Xu Z; Seung E; Deruaz M; Dudek T; Einarsson JI; Yang L; Allen TM; Luster AD; Tager AM; Dykxhoorn DM; Lieberman J, Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Invest 2011, 121 (6), 2401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Bruno JG; Carrillo MP; Richarte AM; Phillips T; Andrews C; Lee JS, Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes 2012, 5, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Hwang S-Y; Sun H-Y; Lee K-H; Oh B-H; Cha YJ; Kim BH; Yoo J-Y, 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 2012, 40 (6), 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Browning CM; Cagnon L; Good PD; Rossi J; Engelke DR; Markovitz DM, Potent Inhibition of Human Immunodeficiency Virus Type 1 (HIV-1) Gene Expression and Virus Production by an HIV-2 Tat Activation-Response RNA Decoy. J. Virol 1999, 73 (6), 5191–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Lee TC; Sullenger BA; Gallardo HF; Ungers GE; Gilboa E, Overexpression of RRE-derived sequences inhibits HIV-1 replication in CEM cells. New. Biol 1992, 4 (1), 66–74. [PubMed] [Google Scholar]
- (219).Kohn DB; Bauer G; Rice CR; Rothschild JC; Carbonaro DA; Valdez P; Hao Q; Zhou C; Bahner I; Kearns K; Brody K; Fox S; Haden E; Wilson K; Salata C; Dolan C; Wetter C; Aguilar-Cordova E; Church J, A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 1999, 94 (1), 368–371. [PubMed] [Google Scholar]
- (220).Kumar PKR; Machida K; Urvil PT; Kakiuchi N; Vishnuvardhan D; Shimotohno K; Taira K; Nishikawa S, Isolation of RNA Aptamers Specific to the NS3 Protein of Hepatitis C Virus from a Pool of Completely Random RNA. Virology 1997, 237 (2), 270–282. [DOI] [PubMed] [Google Scholar]
- (221).Fukuda K; Vishnuvardhan D; Sekiya S; Hwang J; Kakiuchi N; Taira K; Shimotohno K; Kumar PKR; Nishikawa S, Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur J. Biochem 2000, 267 (12), 3685–3694. [DOI] [PubMed] [Google Scholar]
- (222).Chen Z; Wu Q; Chen J; Ni X; Dai J, A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein. Virol. Sin 2020, 35 (3), 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Song Y; Song J; Wei X; Huang M; Sun M; Zhu L; Lin B; Shen H; Zhu Z; Yang C, Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem 2020, 92 (14), 9895–9900. [DOI] [PubMed] [Google Scholar]
- (224).Sun M; Liu S; Wei X; Wan S; Huang M; Song T; Lu Y; Weng X; Lin Z; Chen H; Song Y; Yang C, Aptamer Blocking Strategy Inhibits SarS-CoV-2 Virus Infection. Angew. Chem. Int. Ed. Engl 2021, 60 (18), 10266–10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Liu X; Wang Y. l.; Wu J; Qi J; Zeng Z; Wan Q; Chen Z; Manandhar P; Cavener VS; Boyle NR; Fu X; Salazar E; Kuchipudi SV; Kapur V; Zhang X; Umetani M; Sen M; Willson RC; Chen S. h.; Zu Y, Neutralizing Aptamers Block S/RBD-ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. Int. Ed. Engl 2021, 60 (18), 10273–10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Schmitz A; Weber A; Bayin M; Breuers S; Fieberg V; Famulok M; Mayer G, A SARS-CoV-2 Spike Binding DNA Aptamer that Inhibits Pseudovirus Infection by an RBD-Independent Mechanism. Angew. Chem. Int. Ed. Engl 2021, 60 (18), 10279–10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Li J; Zhang Z; Gu J; Stacey HD; Ang JC; Capretta A; Filipe CDM; Mossman KL; Balion C; Salena Bruno J.; Yamamura D; Soleymani L; Miller MS; Brennan John D.; Li Y; Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021, 49 (13), 7267–7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Kacherovsky N; Yang LF; Dang HV; Cheng EL; Cardle II; Walls AC; McCallum M; Sellers DL; DiMaio F; Salipante SJ; Corti D; Veesler D; Pun SH, Discovery and Characterization of Spike N-Terminal Domain-Binding Aptamers for Rapid SARS-CoV-2 Detection. Angew. Chem. Int. Ed. Engl 2021, 60 (39), 21211–21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Zhang Z; Pandey R; Li J; Gu J; White D; Stacey HD; Ang JC; Steinberg CJ; Capretta A; Filipe CDM; Mossman K; Balion C; Miller MS; Salena BJ; Yamamura D; Soleymani L; Brennan JD; Li Y, High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva. Angew. Chem. Int. Ed. Engl 2021, 60 (45), 24266–24274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Sun M; Liu S; Song T; Chen F; Zhang J; Huang J.-a.; Wan S; Lu Y; Chen H; Tan W; Song Y; Yang C, Spherical Neutralizing Aptamer Inhibits SARS-CoV-2 Infection and Suppresses Mutational Escape. J. Am. Chem. Soc 2021, 143 (51), 21541–21548. [DOI] [PubMed] [Google Scholar]
- (231).Peinetti AS; Lake RJ; Cong W; Cooper L; Wu Y; Ma Y; Pawel GT; Toimil-Molares ME; Trautmann C; Rong L; Marinas B; Azzaroni O; Lu Y, Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci. Adv 2021, 7 (39), eabh2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Li N; Wang X; Tibbs J; Che C; Peinetti AS; Zhao B; Liu L; Barya P; Cooper L; Rong L; Wang X; Lu Y; Cunningham BT, Label-Free Digital Detection of Intact Virions by Enhanced Scattering Microscopy. J. Am. Chem. Soc 2021, 144 (4), 1498–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Li N; Canady TD; Huang Q; Wang X; Fried GA; Cunningham BT, Photonic resonator interferometric scattering microscopy. Nat. Commun 2021, 12 (1), 1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Mendoza-Figueroa JS; Soriano-García M; Valle-Castillo LB; Méndez-Lozano J, Peptides and Peptidomics: A Tool with Potential in Control of Plant Viral Diseases. Adv. Microbiol 2014, 4 (9), 539–548. [Google Scholar]
- (235).Komorowska B; Hasiów-Jaroszewska B; Minicka J, Application of nucleic acid aptamers for detection of Apple stem pitting virus isolates. Mol. Cell. Probes 2017, 36, 62–65. [DOI] [PubMed] [Google Scholar]
- (236).Muthamilarasan M; Prasad M, Plant innate immunity: An updated insight into defense mechanism. J. Biosci 2013, 38 (2), 433–449. [DOI] [PubMed] [Google Scholar]
- (237).Maffei G; Miozzi L; Fiorilli V; Novero M; Lanfranco L; Accotto GP, The arbuscular mycorrhizal symbiosis attenuates symptom severity and reduces virus concentration in tomato infected by Tomato yellow leaf curl Sardinia virus (TYLCSV). Mycorrhiza 2014, 24 (3), 179–186. [DOI] [PubMed] [Google Scholar]
- (238).Yadav PK; Kumar S; Yadav S; Kumar S, Role of Aptamers in Plant Defense Mechanism Against Viral Diseases. In Aptamers, Springer: 2019; 169–174. [Google Scholar]
- (239).Mendoza-Figueroa J; Kvarnheden A; Méndez-Lozano J; Rodríguez-Negrete E-A; de los Monteros RA-E; Soriano-García M, A peptide derived from enzymatic digestion of globulins from amaranth shows strong affinity binding to the replication origin of Tomato yellow leaf curl virus reducing viral replication in Nicotiana benthamiana. Pestic. Biochem. Phys 2018, 145, 56–65. [DOI] [PubMed] [Google Scholar]
- (240).Lopez-Ochoa L; Ramirez-Prado J; Hanley-Bowdoin L, Peptide aptamers that bind to a geminivirus replication protein interfere with viral replication in plant cells. J. Virol 2006, 80 (12), 5841–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Reyes MI; Nash TE; Dallas MM; Ascencio-Ibanez JT; Hanley-Bowdoin L, Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to tomato yellow leaf curl virus and tomato mottle virus infection in tomato. J. Virol 2013, 87 (17), 9691–9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (242).Lautner G; Balogh Z; Bardoczy V; Meszaros T; Gyurcsanyi RE, Aptamer-based biochips for label-free detection of plant virus coat proteins by SPR imaging. Analyst 2010, 135 (5), 918–926. [DOI] [PubMed] [Google Scholar]
- (243).Yu Q; Liu M; Wu S; Xiao H; Qin X; Li P, Generation and characterization of aptamers against grass carp reovirus infection for the development of rapid detection assay. J. Fish Dis 2020, 44 (1), 33–44. [DOI] [PubMed] [Google Scholar]
- (244).Shepherd DN; Martin DP; Thomson JA, Transgenic strategies for developing crops resistant to geminiviruses. Plant Sci. 2009, 176 (1), 1–11. [Google Scholar]
- (245).Sanjay ST; Dou M; Sun J; Li X, A paper/polymer hybrid microfluidic microplate for rapid quantitative detection of multiple disease biomarkers. Sci. Rep 2016, 6, 30474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Paul KB; Kumar S; Tripaty S; Singh V; Vanjari SRK; Singh SG, Highly-sensitive label-free differential pulse voltammetric immunosensor for diagnosis of infectious diseases based on electrospun copper doped ZnO nanofiber biosensing platform. Procedia Technol. 2017, 27, 219–220. [Google Scholar]
- (247).Oyen WJ; Boerman OC; Storm G; Bloois L. v.; Koenders EB; Claessens RA; Perenboom RM; Crommelin DJ; van der Meer JW; Corstens FH; Detecting infection and inflammation with technetium-99m-labeled Stealth liposomes. 1996. [PubMed] [Google Scholar]
- (248).Qasim M; Lim D-J; Park H; Na D, Nanotechnology for diagnosis and treatment of infectious diseases. J. Nanosci. Nanotechnol 2014, 14 (10), 7374–7387. [DOI] [PubMed] [Google Scholar]
- (249).Psimadas D; Georgoulias P; Valotassiou V; Loudos G, Molecular nanomedicine towards cancer: (1)(1)(1)In-labeled nanoparticles. J. Pharm. Sci 2012, 101 (7), 2271–2280. [DOI] [PubMed] [Google Scholar]
- (250).Mathur D; Medintz IL, The Growing Development of DNA Nanostructures for Potential Healthcare-Related Applications. Adv. Healthc. Mater 2019, 8 (9), e1801546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Chandrasekaran AR, DNA nanobiosensors: an outlook on signal readout strategies. J. Nanomater 2017, 2017, 2820619. [Google Scholar]
- (252).Chi Q; Yang Z; Xu K; Wang C; Liang H, DNA nanostructure as an efficient drug delivery platform for immunotherapy. Front. Pharmacol 2019, 10, 1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (253).Sakai Y; Islam MS; Adamiak M; Shiu SC-C; Tanner JA; Heddle JG, DNA Aptamers for the Functionalisation of DNA Origami Nanostructures. Genes 2018, 9 (12), 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Walia S; Chandrasekaran AR; Chakraborty B; Bhatia D, Aptamer-Programmed DNA Nanodevices for Advanced, Targeted Cancer Theranostics. ACS Appl. Bio Mater 2021, 4 (7), 5392–5404. [DOI] [PubMed] [Google Scholar]
- (255).Zhang G; Surwade SP; Zhou F; Liu H, DNA nanostructure meets nanofabrication. Chem. Soc. Rev 2013, 42 (7), 2488–2496. [DOI] [PubMed] [Google Scholar]
- (256).Saccà B; Niemeyer CM, Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev 2011, 40 (12), 5910–5921. [DOI] [PubMed] [Google Scholar]
- (257).Julin S; Nummelin S; Kostiainen MA; Linko V, DNA nanostructure-directed assembly of metal nanoparticle superlattices. J. Nanopart. Res 2018, 20 (5), 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (258).Zhang X; Yadavalli VK, Functional self-assembled DNA nanostructures for molecular recognition. Nanoscale 2012, 4 (7), 2439–2446. [DOI] [PubMed] [Google Scholar]
- (259).Sigl C; Willner EM; Engelen W; Kretzmann JA; Sachenbacher K; Liedl A; Kolbe F; Wilsch F; Aghvami SA; Protzer U; Hagan MF; Fraden S; Dietz H, Programmable icosahedral shell system for virus trapping. Nat. Mater 2021, 20 (9), 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (260).Chauhan N; Wang X, Nanocages for virus inhibition. Nat. Mater 2021, 20 (9), 1176–1177. [DOI] [PubMed] [Google Scholar]
- (261).Zhao L; Qi X; Yan X; Huang Y; Liang X; Zhang L; Wang S; Tan W, Engineering Aptamer with Enhanced Affinity by Triple Helix-Based Terminal Fixation. J. Am. Chem. Soc 2019, 141 (44), 17493–17497. [DOI] [PubMed] [Google Scholar]
- (262).Lin M; Song P; Zhou G; Zuo X; Aldalbahi A; Lou X; Shi J; Fan C, Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc 2016, 11 (7), 1244–1263. [DOI] [PubMed] [Google Scholar]
- (263).Deal BR; Ma R; Ma VP-Y; Su H; Kindt JT; Salaita K, Engineering DNA-Functionalized Nanostructures to Bind Nucleic Acid Targets Heteromultivalently with Enhanced Avidity. J. Am. Chem. Soc 2020, 142 (21), 9653–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (264).Xiao M; Lai W; Man T; Chang B; Li L; Chandrasekaran AR; Pei H, Rationally Engineered Nucleic Acid Architectures for Biosensing Applications. Chem. Rev 2019, 119 (22), 11631–11717. [DOI] [PubMed] [Google Scholar]
- (265).Wang J; Leong MC; Leong EZW; Kuan WS; Leong DT, Clinically relevant detection of Streptococcus pneumoniae with DNA-antibody nanostructures. Anal. Chem 2017, 89 (12), 6900–6906. [DOI] [PubMed] [Google Scholar]
- (266).Lertanantawong B; Krissanaprasit A; Chaibun T; Gothelf KV; Surareungchai W, Multiplexed DNA detection with DNA tweezers in a one-pot reaction. Mater. Sci. Ener. Technol 2019, 2 (3), 503–508. [Google Scholar]
- (267).Kwon PS; Ren S; Kwon SJ; Kizer ME; Kuo L; Xie M; Zhu D; Zhou F; Zhang F; Kim D; Fraser K; Kramer LD; Seeman NC; Dordick JS; Linhardt RJ; Chao J; Wang X, Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem 2020, 12 (1), 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Ren S; Fraser K; Kuo L; Chauhan N; Adrian AT; Zhang F; Linhardt RJ; Kwon PS; Wang X, Designer DNA nanostructures for viral inhibition. Nat. Protoc 2022, 17 (2), 282–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (269).Li YG; Cu YTH; Luo D, Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat. Biotechnol 2005, 23 (7), 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (270).Zheng J; Ji X; Du M; Tian S; He Z, Rational construction of a DNA nanomachine for HIV nucleic acid ultrasensitive sensing. Nanoscale 2018, 10 (36), 17206–17211. [DOI] [PubMed] [Google Scholar]
- (271).Li X; Li Y; Hong L, A Novel Self-Assembling DNA Nano Chip for Rapid Detection of Human Papillomavirus Genes. PloS One 2016, 11 (10), e0162975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Chandrasekaran AR; Zavala J; Halvorsen K, Programmable DNA Nanoswitches for Detection of Nucleic Acid Sequences. ACS Sens. 2016, 1 (2), 120–123. [Google Scholar]
- (273).Chandrasekaran AR; MacIsaac M; Dey P; Levchenko O; Zhou L; Andres M; Dey BK; Halvorsen K, Cellular microRNA detection with miRacles: microRNA-activated conditional looping of engineered switches. Sci. Adv 2019, 5 (3), eaau9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (274).Chandrasekaran AR; Trivedi R; Halvorsen K, Ribonuclease-Responsive DNA Nanoswitches. Cell Rep. Phys. Sci 2020, 1 (7), 100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Zhou L; Chandrasekaran AR; Punnoose JA; Bonenfant G; Charles S; Levchenko O; Badu P; Cavaliere C; Pager CT; Halvorsen K, Programmable low-cost DNA-based platform for viral RNA detection. Sci. Adv 2020, 6 (39), eabc6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Chandrasekaran AR; MacIsaac M; Vilcapoma J; Hansen CH; Yang D; Wong WP; Halvorsen K, DNA Nanoswitch Barcodes for Multiplexed Biomarker Profiling. Nano Lett. 2021, 21 (1), 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (277).Bastings MMC; Anastassacos FM; Ponnuswamy N; Leifer FG; Cuneo G; Lin C; Ingber DE; Ryu JH; Shih WM, Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018, 18 (6), 3557–3564. [DOI] [PubMed] [Google Scholar]
- (278).Mela I; Vallejo-Ramirez PP; Makarchuk S; Christie G; Bailey D; Henderson RM; Sugiyama H; Endo M; Kaminski CF, DNA Nanostructures for Targeted Antimicrobial Delivery. Angew. Chem. Int. Ed. Engl 2020, 59 (31), 12698–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Obuobi S; Tay HK-L; Tram NDT; Selvarajan V; Khara JS; Wang Y; Ee PLR, Facile and efficient encapsulation of antimicrobial peptides via crosslinked DNA nanostructures and their application in wound therapy. J. Control. Release 2019, 313, 120–130. [DOI] [PubMed] [Google Scholar]
- (280).Engelen W; Sigl C; Kadletz K; Willner EM; Dietz H, Antigen-Triggered Logic-Gating of DNA Nanodevices. J. Am. Chem. Soc 2021, 143 (51), 21630–21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).Read JM; Bridgen JRE; Cummings DAT; Ho A; Jewell CP, Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. Philos. Trans. R. Soc. Lond. B. Biol. Sci 2020, 376 (1829), 20200265.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).Li Q; Guan X; Wu P; Wang X; Zhou L; Tong Y; Ren R; Leung KSM; Lau EHY; Wong JY; Xing X; Xiang N; Wu Y; Li C; Chen Q; Li D; Liu T; Zhao J; Li M; Tu W; Chen C; Jin L; Yang R; Wang Q; Zhou S; Wang R; Liu H; Luo Y; Liu Y; Shao G; Li H; Tao Z; Yang Y; Deng Z; Liu B; Ma Z; Zhang Y; Shi G; Lam TTY; Wu JTK; Gao GF; Cowling BJ; Yang B; Leung GM; Feng Z, Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med 2020, 382, 1199–1207.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (283).Wu JT; Leung K; Leung GM, Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 2020, 395 (10225), 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (284).Findings from investigation and analysis of re-positive cases. https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030&act=view&list_no=367267&nPage=1.
- (285).Lu J; Peng J; Xiong Q; Liu Z; Lin H; Tan X; Kang M; Yuan R; Zeng L; Zhou P; Liang C; Yi L; du Plessis L; Song T; Ma W; Sun J; Pybus OG; Ke C, Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020, 59, 102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (286).Rothlisberger P; Hollenstein M, Aptamer chemistry. Adv. Drug. Deliv. Rev 2018, 134, 3–21. [DOI] [PubMed] [Google Scholar]
- (287).Douglas SM; Bachelet I; Church GM, A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335 (6070), 831–834. [DOI] [PubMed] [Google Scholar]
- (288).Chandrasekaran AR, Nuclease resistance of DNA nanostructures. Nat. Rev. Chem 2021, 5 (4), 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Tikhomirov G; Petersen P; Qian L, Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552 (7683), 67–71. [DOI] [PubMed] [Google Scholar]
- (290).Praetorius F; Kick B; Behler KL; Honemann MN; Weuster-Botz D; Dietz H, Biotechnological mass production of DNA origami. Nature 2017, 552 (7683), 84–87. [DOI] [PubMed] [Google Scholar]
- (291).Coleridge EL; Dunn KE, Assessing the cost-effectiveness of DNA origami nanostructures for targeted delivery of anti-cancer drugs to tumours. Biomed. Phys. Eng. Express 2020, 6 (6). [DOI] [PubMed] [Google Scholar]
- (292).Smith DM; Keller A, DNA Nanostructures in the Fight Against Infectious Diseases. Adv. Nanobiomed. Res 2021, 2000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.