Abstract
Introduction: The community mitigation measures taken because of the COVID-19 pandemic had side effects on the circulation of the most frequent respiratory viruses during 2020. In the case of respiratory syncytial virus (RSV), an important paediatric pathogen, a decrease in the number of cases and delayed outbreaks was previously described.
Aim and Methods: The genetic characteristics of the RSV circulating strains in paediatric patients in Buenos Aires, Argentina before and during the COVID-19 pandemic were studied. RSV (+) samples taken from hospitalised patients with respiratory tract infections (2018- 2021) were analysed through G gene sequencing and evolutionary analyses.
Results: No RSV hospitalised paediatric patients were registered in Buenos Aires during 2020; however, RSV reemerged in 2021 with a lower number of cases and a delayed outbreak, peaking in July-August. A total of 147 G gene sequences were analysed. RSV-B (N = 85) predominated during 2018 and 2021 whereas in 2019 RSV-A were more prevalent (N = 62). All RSV-A sequences were ON1-like strains, and all RSV-B were BA-like. Phylogenetic analyses showed that the same genetic lineages circulated before and after 2020, but RSVs from 2021 corresponded to new viral introductions rather than cryptic circulation of the previous genetic clusters in Buenos Aires during 2020.
Conclusions: Following the reopening of borders, the reemergence of RSV in Argentina brought new viral introductions from other countries. Therefore, it is important to continue a deep global molecular surveillance to characterise RSV strains in post-pandemic circulation with an impact in future vaccine implementation.
Keywords: Respiratory syncytial virus, COVID-19 pandemic, Molecular epidemiology, Genetic lineages
1. Introduction
Human respiratory syncytial virus (RSV) is one of the most common viral pathogens causing acute lower respiratory tract infections (ALRTI) in paediatric patients [1]. In Argentina, RSV has contributed to approximately 80% of paediatric viral ALRTI yearly [2].
RSV has been classified into two antigenic subgroups A and B. Within each subgroup there are numerous genotypes, which have historically been classified based on the glycoprotein (G) gene sequence (3). Outbreaks are commonly produced by both subgroups, and currently the most frequent genotypes worldwide are ON1 for RSV-A and BA for RSV-B, characterised by duplications of a 72-nt and a 60-nt in the G gene, respectively [2, 4, 5].
In December 2019, the world experienced the beginning of the COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The global impact at the public health level was catastrophic, with millions of hospitalisations and deaths. Argentina's first COVID-19 case was detected on 3 March 2020 when the country had already prepared for its arrival, including rapid detection of cases, patient's isolation, tracing and quarantine of contacts [6, 7]. Nevertheless, COVID-19 cases increased, and consequently mitigation measures were taken, including the closure of borders, educational institutions, public places, stores, and strict social isolation as part of a comprehensive lockdown. Up to 6 November 2021, a total of 5.3 M cases and 116 K deaths were reported in Argentina due to COVID-19 [8].
Community mitigation measures to fight against COVID-19 also affected the circulation of other respiratory viruses, such as RSV. The decrease or even the absence of cases of RSV during 2020, and the appearance of delayed annual RSV outbreaks in both 2020 and 2021 have been reported in other countries [9, 10, 11]. In this context, the question that arises in Argentina is: what happened with RSV during 2020 and what genetic characteristics do the reemerged RSV strains have after the mitigation measures were taken due to the COVID-19 pandemic?
1.1. Objective
Perform a molecular epidemiological analysis of the RSV circulating strains before and during the COVID-19 pandemic in Buenos Aires, Argentina.
2. Methodology
2.1. Samples, G gene sequencing and phylogenetic analysis
Viral RNA was extracted from nasopharyngeal RSV-positive samples collected from hospitalised paediatric patients (range 0–12 years-old) due to ALRTI at the Ricardo Gutierrez Children's Hospital in Buenos Aires, from 2018 to 2021. Samples were randomly selected throughout each annual outbreak. The full G gene was sequenced as previously described [4].
Sequence alignments were performed with MUSCLE [12]. Phylogenetic analyses were performed by Maximum Likelihood using IQ-TREE v.2.1 with ultrafast bootstrap and SH-aLRT (1000 replicates each) to assess the phylogenetic clades statistical support [13].
Genotyping analyses were performed according to Goya et al. 2019, using the ReSVidex online tool and checked by phylogenetic analyses [3,14].
When needed, reference sequences with a collection date from 2017 onwards from other countries were downloaded from the GISAID and GenBank databases considering the best ten BLAST hits sequences (Supplementary Table 1).
3. Results
3.1. RSV epidemiology
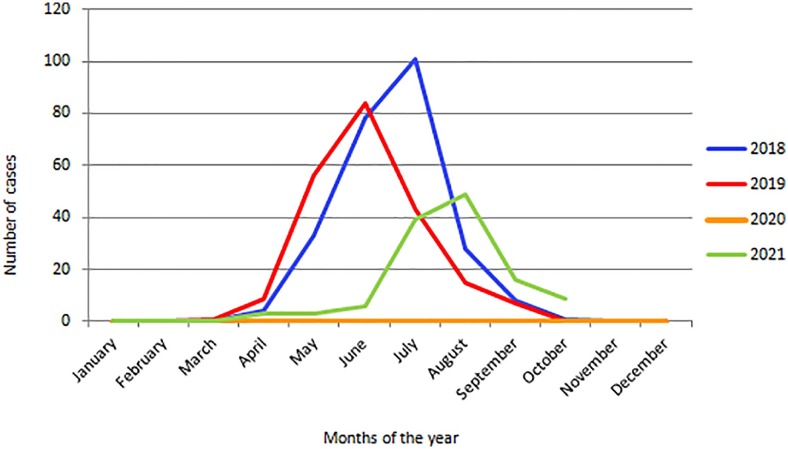
In 2018 and 2019, the outbreaks of RSV in hospitalised paediatric patients with ALRTI showed the same seasonal pattern as the one that was seen during the last decade (2). A total of 253 RSV-positive cases were detected in 2018 with peaks in July and 215 in 2019 with peaks in June (Fig. 1 ). Surprisingly, in 2020 no hospitalisations due to RSV were registered, coinciding with the implementation of community mitigation measures due to the COVID-19 pandemic (Table 1 ). In this context, an additional study was conducted in 82 paediatric outpatients with respiratory symptoms with a negative diagnosis for SARS-CoV-2 between July and November 2020 to detect possible cryptic RSV circulation in the mentioned population. No RSV-positive cases were found in the studied group.
Fig. 1.
Seasonality of the RSV outbreaks between 2018 and 2021 at the Ricardo Gutiérrez Children's Hospital, Buenos Aires, Argentina. Only hospitalised cases were considered.
Table 1.
RSV Epidemiology. The total number of hospitalised paediatric patients per year and age distribution in months are detailed.
| Year | Total No. of hospitalised patients due to RSV | Age group (months) | No. of cases by age (%) |
|---|---|---|---|
| 2018 | 253 | 0 to 6 | 103 (40.70%) |
| 6 to 12 | 86 (34%) | ||
| 12 to 24 | 36 (14.23%) | ||
| >24 | 28 (11.07%) | ||
| 2019 | 215 | 0 to 6 | 76 (35.35%) |
| 6 to 12 | 58 (26.98%) | ||
| 12 to 24 | 42 (19.53%) | ||
| >24 | 39 (18.14%) | ||
| 2021 | 116 | 0 to 6 | 41 (34.34%) |
| 6 to 12 | 37 (31.90%) | ||
| 12 to 24 | 13 (11.21%) | ||
| >24 | 25 (21.55%) |
In 2021, a total of 116 RSV-positive cases in hospitalised paediatric patients were registered. Nevertheless, the total number of hospitalisations during the reemergence of RSV in 2021 was lower in comparison to the 2018 and 2019 outbreaks. In addition, the 2021 outbreak was delayed, starting in April, peaking in July-August, and ending in November (Fig. 1). The patients’ age distribution analysis showed that each year the highest percentage of hospitalised patients corresponded to the 0–12 months range (>60% of cases) (Table 1).
3.2. RSV genotyping
A total of 57 G-gene sequences were obtained from 2018, 44 from 2019 to 46 from 2021. RSV-A predominated in 2019 whereas RSV-B were more frequent in 2018 and 2021. All the RSV-A sequences were ON1-like, comprising two genetic lineages GA2.3.5 and GA2.3.6b. In addition, the RSV-B sequences were BA-like associated to the genetic lineage GB5.0.5a (Table 2 ).
Table 2.
G-gene sequences analyses. The subgroup and genetic lineages were obtained from the genotyping analyses. The classification of genetic lineages was established according to Goya et al. (3).
| Year | No. of hospitalised cases due to RSV | Subgroup | Sequences per subgroup per year (%) | Genetic lineage | No. of G-gene sequences | Cases sequenced per total RSV-positive cases (%) |
|---|---|---|---|---|---|---|
| 2018 | 253 | A | 21.05 | GA2.3.5 | 10 | 22.53 |
| GA2.3.6b | 2 | |||||
| B | 78.95 | GB5.0.5a | 45 | |||
| 2019 | 215 | A | 84.10 | GA2.3.5 | 21 | 20.46 |
| GA2.3.6b | 16 | |||||
| B | 15.90 | GB5.0.5a | 7 | |||
| 2021 | 116 | A | 28.26 | GA2.3.5 | 5 | 39.65 |
| GA2.3.6b | 8 | |||||
| B | 71.74 | GB5.0.5a | 33 |
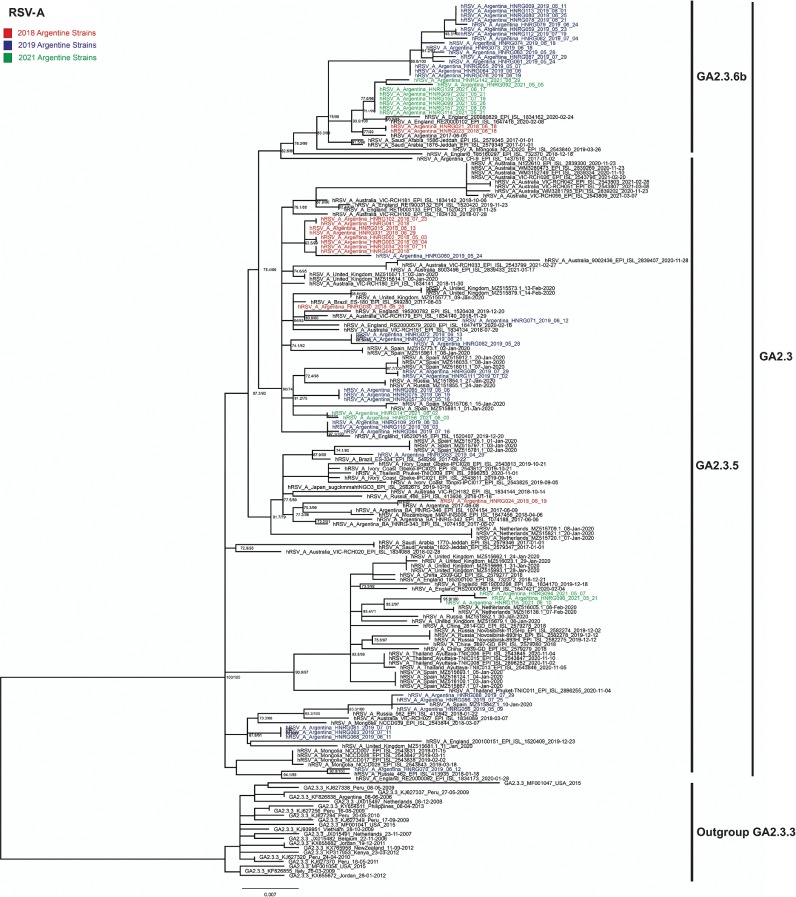
Phylogenetic analyses to assess the relationships amongst Argentine 2021-sequences with locally and globally sequences from 2017 to 2021 reported in public databases were performed. The results suggest that the Argentine sequences detected in 2021 corresponded to multiple new viral introductions to our country. The GA2.3.5 2021-sequences shown in Fig. 2 are distributed in two different phylogenetic clades, one shares a most recent common ancestor (MRCA) with sequences that circulated in Spain in 2020 and in Argentina in 2019. The other one shares an MRCA with sequences from the Netherlands in 2020. The GA2.3.6b 2021-sequences are associated in a well-supported monophyletic clade and share an MRCA with sequences from England 2020.
Fig. 2.
Maximum Likelihood phylogenetic analysis of RSV-A. Argentine sequences from 2018 to 2021 were analysed with sequences from the same period downloaded from GISAID and GenBank databases. Argentine sequences are highlighted in colours according to the year. The genotypes are indicated on the right. The model established by IQTREE was GTR + F + G4. Nodes supports: SH-aLRT support (%)/ultrafast bootstrap support (%) (1000 replicates each). Only supports over 70/70 are shown.
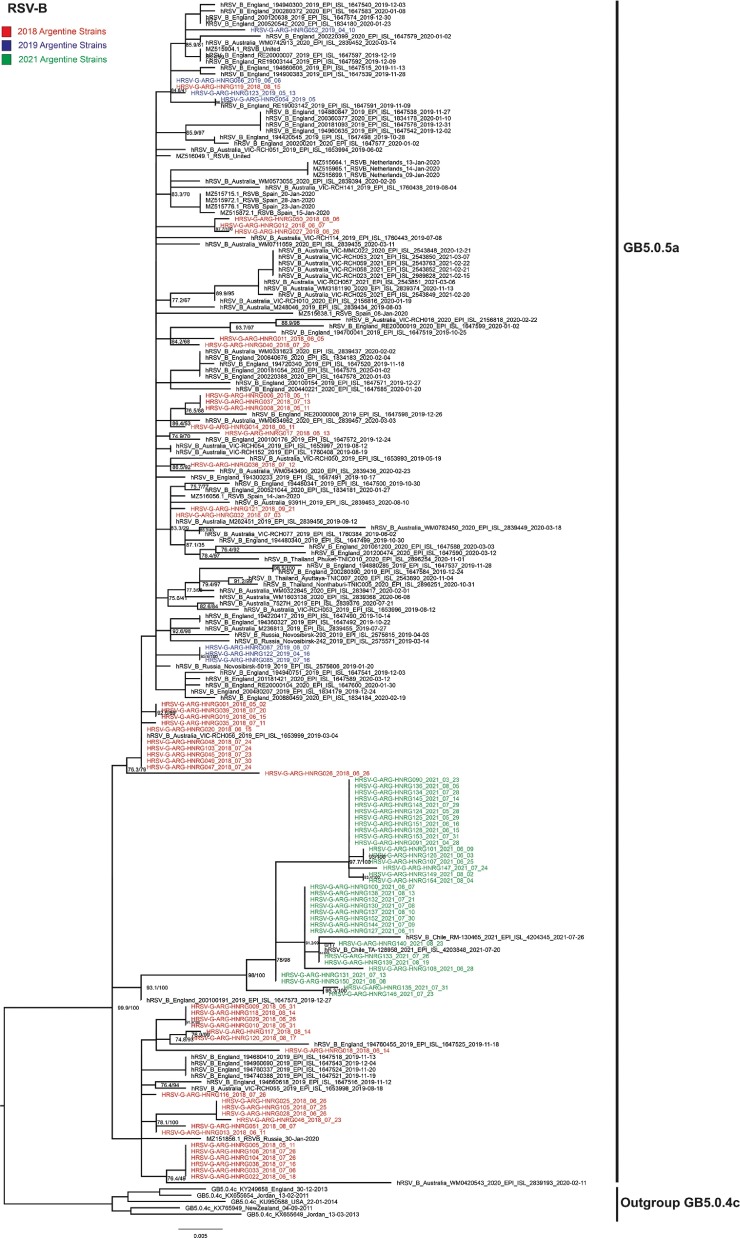
Regarding RSV-B, GB5.0.5a 2021-sequences shown in Fig. 3 are associated with a well-supported monophyletic clade with two sequences from Chile 2021, unrelated to previous Argentine sequences.
Fig. 3.
Maximum Likelihood phylogenetic analysis of RSV-B. Argentine sequences from 2018 to 2021 were analysed with sequences from the same period downloaded from GISAID and GenBank databases. Argentine sequences are highlighted in colours according to the year. The genotypes are indicated on the right. The model established by IQTREE was TIM3 + F + G4. Nodes supports: SH-aLRT support (%)/ultrafast bootstrap support (%) (1000 replicates each). Only supports over 70/70 are shown.
4. Discussion
Epidemic outbreaks of respiratory viruses depend on regional factors (demography, age distribution, susceptibility, etc.), but also on global factors (e.g., transmission due to long-distance air travel) [15]. Isolation, social distancing measures and border closures or limited international travel in response to the COVID-19 pandemic since March 2020 in Argentina seriously affected the circulation of RSV. Usual RSV outbreaks in Argentina were disrupted in 2020 when no RSV cases were detected. In fact, cases of bronchiolitis in children under two years of age decreased by 84.5% from 2019 to 2020 in Argentina, evidencing the effect of lockdown on respiratory infections [7].
Furthermore, 2021 underwent a delayed RSV outbreak with a lower total number of cases. Since January 2021, the lockdown in Argentina began to decrease, borders reopened for the Argentines who travelled abroad, educational institutions resumed later that year under the ‘bubbles’ approach (children attended in consistent non-overlapping groups) and there was a concomitant social relaxation. Nevertheless, the children were the last to leave the lockdown in Argentina. In this context and considering the importance of household transmission [16], it is plausible that RSV has reemerged in Argentina with such a delayed outbreak.
After a year of strict confinement, those 1-year-olds not exposed to RSV could have been more susceptible to severe infections the following year. However, this possible increase in hospitalised cases was not registered (Table 1) with the majority of those under 12 months. This reinforces the idea that the implementation of any prophylaxis (vaccine, monoclonal antibodies) should target children under 1 year.
The performed genotyping analyses suggest that the same three genetic lineages co-circulated during the three-year period, but the evolutionary evidence together with the epidemiological data support the idea that the lineages that spread in 2021 might be new introductions to our country. Moreover, to the extent that RSV sequence-databases grow and there is more global representativeness in geographic and temporal terms, it will be possible to determine from which countries these RSV strains could have been introduced.
Finally, it would be important to continue a deep global molecular surveillance to characterise RSV strains and other respiratory viruses in the pre- and post-pandemic era to analyse the effect of the mitigation measures with an impact in the implementation of future vaccines.
Funding
This work was supported by ANPCyT, Ministerio de Ciencia, Tecnología e Innovación de la Nación Argentina (Grant PICT03820/2019). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding was received for this study.
Ethical statement
The project was reviewed and approved by the Medical Ethics and Research Committees of the Ricardo Gutierrez Children's Hospital, Buenos Aires, Argentina (IRB No. 17.21). Parental informed consent was not obtained because patient information was anonymised before analysis.
Author contributions
Conceptualisation: Goya Stephanie, Mistchenko Alicia S., Viegas Mariana. Data curation: Acuña Dolores; Nabaes Jodar Mercedes S.
Formal analysis: Acuña Dolores, Goya Stephanie, Nabaes Jodar Mercedes S., Mischenko Alicia S., Mariana Viegas
Funding acquisition; Mistchenko Alicia S., Viegas Mariana
Methodology: Acuña Dolores, Nabaes Jodar Mercedes S., Goya Stephanie, Grandis Erica, Mischenko Alicia S., Mariana Viegas
Writing - original draft: Acuña Dolores, Goya Stephanie
Writing - review & editing: Acuña Dolores, Goya Stephanie, Nabaes Jodar Mercedes S., Mistchenko Alicia S., Viegas Mariana
Declaration of Competing Interests
The authors declare no competing interests.
Acknowledgement
We would like to thank the health care workers for their hard work especially during the pandemic and for continuing to treat all diseases other than COVID-19. We would like to especially thank the diagnostic team of the Virology Laboratory of the Ricardo Gutierrez Children's Hospital for their dedicated effort. We would also like to extend our sincere gratitude to Dr. Carolina Torres for her contributions in phylogenetic analyses and Laura Valinotto, Silvina Lusso and Mónica Natale for their cooperation throughout this work.We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which part of this research is based (Supplementary Table 1).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105126.
Appendix. Supplementary materials
References
- 1.Shi T., McAllister D.A., O'Brien K.L., Simoes E.A.F., Madhi S.A., Gessner B.D., et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017 doi: 10.1016/S0140-6736(17)30938-8. 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viegas M., Goya S., Mistchenko A.S. Sixteen years of evolution of human respiratory syncytial virus subgroup A in Buenos Aires, Argentina: GA2 the prevalent genotype through the years. Infection Genet. Evolution. 2016 doi: 10.1016/j.meegid.2016.04.034. https://doi: 10.1016/j.meegid.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 3.Goya S., Galiano M., Nauwelaers I., Trento A., Openshaw P.J., Mistchenko A.S., et al. Toward unified molecular surveillance of RSV: a proposal for genotype definition. Influenza Other Respiratory Viruses. 2020 doi: 10.1111/irv.12715. doi:10.1111/irv.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rojo G.L., Goya S., Orellana M., Sancilio A., Rodriguez Pérez A., Montali C., et al. Unravelling respiratory syncytial virus outbreaks in Buenos Aires, Argentina: molecular basis of the spatio-temporal transmission. Virology. 2017 doi: 10.1016/j.virol.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Tabor D.E., Fernandes F., Langedijk A.C., Wilkins D., Lebbink R.J., Tovchigrechko A., et al. Global molecular epidemiology of respiratory syncytial virus from the 2017-2018 INFORM-RSV study. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01828-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ministry of Health of the Argentine Nation, https://www.argentina.gob.ar/noticias/salud-confirma-el-primer-caso-de-coronavirus-en-el-pais, 2019.
- 7.Ministry of Health of the Argentine Nation, https://www.argentina.gob.ar/noticias/argentina-adopta-medidas-oportunas-para-minimizar-la-trasmision-de-covid-19, 2019.
- 8.Ministry of Health of the Argentine Nation, Integrated surveillance bulletin N574, https://bancos.salud.gob.ar/recurso/boletin-integrado-de-vigilancia-n574-se-44 2021.
- 9.Edwards K.M. The impact of social distancing for severe acute respiratory syndrome coronavirus 2 on respiratory syncytial virus and influenza burden. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciaa1543. 10.1093/cid/ciaa1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trenholme A., Webb R., Lawrence S., Arrol S., Taylor S., Ameratunga S. et al. (2021). COVID-19 and infant hospitalizations for seasonal respiratory virus infections, New Zealand, 2020. Emerging Infect. Dis. doi: 10.3201/eid2702.204041. [DOI] [PMC free article] [PubMed]
- 11.Di Mattia G., Nenna R., Mancino E., Rizzo V., Pierangeli A., Villani A., et al. During the COVID-19 pandemic where has respiratory syncytial virus gone? Pediatr. Pulmonol. 2021 doi: 10.1002/ppul.25582. doi: 10.1002/ppul.25582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004 doi: 10.1093/nar/gkh340. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015 doi: 10.1093/molbev/msu300. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cacciabue M., Goya S. (2020), https://sourceforge.net/projects/resvidex/.
- 15.Zheng Z., Pitzer V.E., Warren J.L., Weinberger D.M. Community factors associated with local epidemic timing of respiratory syncytial virus: a spatiotemporal modeling study. Sci. Adv. 2021 doi: 10.1126/sciadv.abd6421. doi: 10.1126/sciadv.abd6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agoti C.N., Phan M.V.T., Munywoki P.K., et al. Genomic analysis of respiratory syncytial virus infections in households and utility in inferring who infects the infant. Sci. Report. 2019 doi: 10.1038/s41598-019-46509-w. 10.1038/s41598-019-46509-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.