Abstract
Lipidomics is a relatively recent molecular research field, and explores lipids (fats) and their biology using advanced mass spectrometry technologies. Although this field has expanded significantly in biomedical and biotechnological disciplines, it is still in its infancy in molecular parasitology. Our goal here is to review and discuss technical aspects of MS-based lipidomics and its recent applications to parasitic worms, as well as challenges and future directions for worm lipid research. In a multi-omic paradigm, we expect that the exploration of lipidomic data for parasitic worms will yield important insights into lipid-associated biological pathways and processes, including the regulation of essential signalling pathways, parasite invasion, establishment, adaptation and development.
Keywords: Lipid (fat), Lipidome, Lipidomics, Mass spectrometry, Parasitic worm, Helminth
Graphical abstract
Highlights
-
•
Lipids are involved in critical biological functions in parasitic worms.
-
•
Lipidomics is an emerging research field in molecular helminthology.
-
•
This article covers technological advances and applications to parasitic worms.
-
•
It also discusses challenges and future directions for lipidomic research.
1. Introduction
Over the past two decades, substantial progress has been made in the field of molecular parasitology using genomic, transcriptomic and proteomic technologies (International Helminth Genomes, 2019; Jex et al., 2019; Ma et al., 2020b; McVeigh, 2020). Numerous studies of parasitic worms have focused on the identification and characterisation of genes and proteins (Ma et al., 2020a; McVeigh, 2020). Some of these studies have identified and/or annotated a large number of genes and proteins which are associated with the biosynthesis and transportation of lipids (e.g. International Helminth Genomes, 2019), and significant changes in transcription/expression profiles in the transition from free-living to parastitic stages (Laing et al., 2013; Wang et al., 2019b). Other studies have shown that worm-derived lipids are involved in the regulation of essential signalling pathways during worm development, host invasion and/or establishment of infection (Magalhaes et al., 2010; Ma et al., 2019c). This evidence indicates that multi-omic investigations of parasites could benefit from the inclusion of systematic lipidomic data sets.
Lipids have marked chemical diversity (Fahy et al., 2005). According to the International Lipid Classification and Nomenclature Committee, lipids can be classified into at least eight categories, namely fatty acyls (FA), glycerolipids (GL), glycerophospholipids (GP), sphingolipids (SP), sterol lipids (ST), prenol lipids (PR), saccharolipids (SL) and polyketides (PK) (Fahy et al., 2005). Based on their structural and physiochemical features (e.g. motifs, polarity, charge, shape and molecular mass), lipids within each of these eight categories can be divided into classes and then subdivided into individual molecular species. To date (December 2020), the world's largest public lipid-only database – LIPID MAPS Structure Database (LMSD) – contains 44,978 unique lipid structures (https://www.lipidmaps.org/).
In the field of helminthology, the study of lipids is not new. Previous biochemical studies have used proximate analysis, thin-layer and/or gas liquid chromatography to identify lipids in parasitic worms, such as Ascaris lumbricoides (see Greichus & Greichus, 1966), Strongyloides ratti (see Minematsu et al., 1990), Brugia malayi (see Smith et al., 1996) and Dictyocaulus viviparus (see Becker et al., 2017). However, due to the limited capacity of these techniques to resolve the composition of lipids, there has been only a vague appreciation of the lipid make-up in parasitic worms. Recently, though, advanced mass spectrometry (MS)-based lipidomics has provided a powerful analytical platform to comprehensively characterise the composition and abundance of lipids in organisms and/or tissues thereof (Rustam & Reid, 2018). The use of complementary technologies, such as fluorescence spectroscopy and high performance liquid chromatography (HPLC), will enable the dynamics and interactions of lipids to be explored, and lipid-associated signalling pathways to be elucidated (Han, 2016; Yang & Han, 2016).
Although lipidomics is an emerging and rapidly developing field in biology and biomedicine, it is only starting to enter the molecular helminthology arena. To date, a number of lipidomic studies has been conducted of parasitic worms of major socioeconomic importance, including Schistosoma mansoni, Haemonchus contortus and Ascaris suum. In this article, we (i) review current workflows used for MS-based lipidomics; (ii) summarise lipidomic studies of parasitic worms conducted to date; and (iii) discuss the challenges and future directions for lipidomics in the field of molecular helminthology.
2. Mass spectrometry-based analytical workflows for lipidomic investigations
Depending on the desired research outcomes, lipidomic studies can be “targeted” or “untargeted”: (i) targeted lipidomics usually characterises a pre-defined subset of lipids of interest, to address one or more particular biological questions, while (ii) untargeted lipidomics usually aims to identify and quantify, in an unbiased manner, all lipids present in a sample (e.g. organ, organ system, tissue, cell-type or fluid).
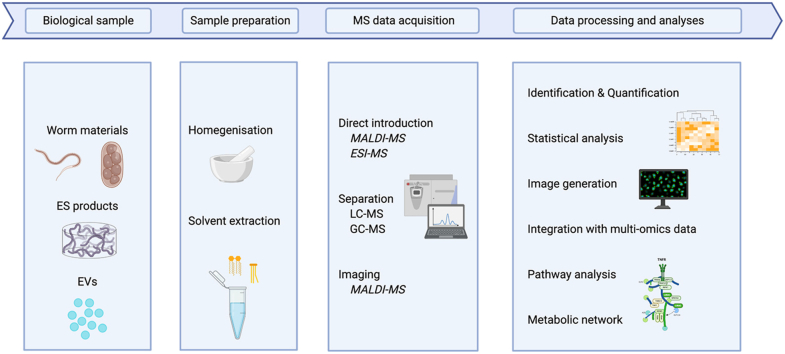
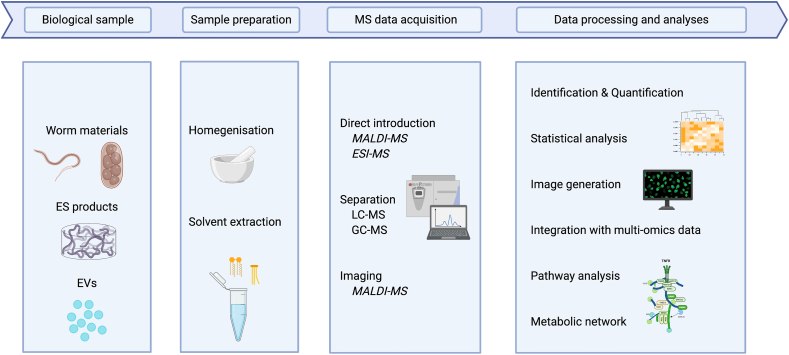
Most current protocols start with lipid extraction from biological samples using an optimised extraction procedure (based on the specific lipids of interest). Subsequently, mass spectrometry is applied either directly (i.e. direct infusion – “shotgun”) or with prior chromatographic fractionation (typically liquid chromatography, LC) to analyse the complexity of extracted lipids. Then, bioinformatics is employed to process the raw data, to identify the lipids and to quantify their absolute or relative abundances (Yang & Han, 2016; Rustam & Reid, 2018). With rapid advances in MS and bioinformatics, both direct infusion and LC-linked lipidomics allow the analysis of complex mixtures in a rapid and large-scale manner. For instance, it only takes ~1 h to quantify complex mixtures of > 1,000 unique lipid features via high-resolution, accurate-mass LC-tandem mass spectrometry (MS/MS) (Kiyonami et al., 2016). On the other hand, imaging-associated MS – such as high-resolution matrix-assisted laser desorption/ionisation mass spectrometry imaging (MALDI-MSI) (Wang et al., 2020a) – has been used successfully to establish the spatial distribution of lipids in vivo, within intact tissue samples, in an untargeted manner. A summary of typical workflows for MS-based lipidomic analyses of parasitic worms is given in Fig. 1. In the following, we summarise recent applications of such workflows to parasitic worms (cf. Table 1).
Fig. 1.
Typical workflows for mass spectrometry-based lipidomic analyses of parasitic worms. The steps of lipidomic analysis of worm samples include sample preparation, MS data acquisition, and data processing and analyses. Abbreviations: MS, mass spectrometry; ES, excretory-secretory; EVs, extracellular vesicles; MALDI, matrix-assisted laser desorption/ionisation; ESI, electrospray ionisation; LC, liquid chromatography; GC, gas chromatography
Table 1.
Summary of key lipidomic studies of parasitic worms
| Helminth species | Samples/developmental stages | Analytical platform/approach | Aim of study | Reference |
|---|---|---|---|---|
| Schistosoma mansoni | Male and female adults | MALDI MSI | Untargeted lipidomics | Ferreira et al. (2014b) |
| Eggs, miracidia and cercariae | MALDI MSI | Untargeted lipidomics | Ferreira et al. (2014a) | |
| Praziquantel treated male and female adults | MALDI MSI | Untargeted lipidomics | Ferreira et al. (2015) | |
| Tegumental surface membranes1 | HPLC-MS | Targeted lipidomics (glycerophospholipids) | Retra et al. (2015) | |
| Eggs, cercariae, adults as well as their excretory-secretory (ES) products | ESI(+) Q-TOF LC-MS/MS | Untargeted lipidomics | Giera et al. (2018) | |
| Eggs, cercariae, adults as well as their ES products | ESI(−) Q-Trap LC-MS/MS | Targeted lipidomics (eicosanoid and docosanoid) | Giera et al. (2018) | |
| Eggs, cercariae, adults as well as their ES products | GC–MS | Targeted lipidomics (fatty acid) | Giera et al. (2018) | |
| Eggs and miracidia | LC-MS | Targeted lipidomics (glycerophospholipids) | Bexkens et al. (2019) | |
| Cryosections of male and female adults | AP-SMALDI MSI | Untargeted lipidomics | Kadesch et al. (2019) | |
| Tegumental surface of male and female adults | AP-SMALDI MSI | Untargeted lipidomics | Kadesch et al. (2020) | |
| Heligmosomoides polygyrus | Extracellular vesicles (EVs) | UHPLC-MS/MS | Untargeted lipidomics | Simbari et al. (2016) |
| Ascaris suum | Female adult | UHPLC-MS/MS | Targeted lipidomics (steroids) | Ma et al. (2019a) |
| L3-egg, L3-lung, L4, male and female adults, as well as organ system from male and female adults | UHPLC-MS/MS | Untargeted lipidomics | Wang et al. (2020d) | |
| Haemonchus contortus | L3, L4, adults | UHPLC-MS/MS | Untargeted lipidomics | Wang et al. (2018) |
| L3 and L4 (dafachronic acid treated) | UHPLC-MS/MS | Untargeted lipidomics | Ma et al. (2020b) | |
| Organ system of female adults | UHPLC-MS/MS | Untargeted lipidomics | Wang et al. (2020c) | |
| Toxocara canis | Female adult | ESI(−) UHPLC-MS/MS | Targeted lipidomics (steroids) | Ma et al. (2019a) |
| Trichinella papuae | L1 (muscle-stage) | ESI(+/−) LC-MS/MS | Untargeted lipidomics | Mangmee et al. (2020) |
| Onchocerca volvulus | Male and female adults | Shotgun ESI Q-TOF MS/MS | Targeted lipidomics (glycerophospholipids) | Wewer et al. (2017) |
| Onchocerca ochengi | Male and female adults | Shotgun ESI Q-TOF MS/MS | Targeted lipidomics (glycerophospholipids) | Wewer et al. (2017) |
| Litomosoides sigmodontis | Male and female adults | Shotgun ESI Q-TOF MS/MS | Targeted lipidomics (glycerophospholipids) | Wewer et al. (2017) |
Abbreviations: L1, the first-stage larvae; L3-egg, the infective third-stage larvae, L3-lung, the third-stage larvae migrated to host lung tissue; L4, the fourth-stage larvae; female adult, the adult-stage female worm; male adult, the adult-stage male worm; MALDI, matrix-assisted laser desorption/ionisation; MSI, mass spectrometry imaging; HPLC, high performance liquid chromatography; MS, mass spectrometry; ESI, electrospray ionisation; Q-TOF, quad time of flight; Q-Trap, Quadropule ion trap; GC, gas chromatography; LC, liquid chromatography; SMALDI, scanning microprobe matrix-assisted laser desorption/ionisation; UHPLC, ultra-high performance liquid chromatography.
3. Lipidomic studies of parasitic worms
3.1. Trematodes
3.1.1. Schistosoma mansoni
This is one of the most prevalent schistosomes, causing hepato-intestinal schistosomiasis in humans. There are ∼200 million people affected by schistosomiasis, causing a neglected tropical disease (NTD) burden estimated at ∼ 1.9 million disability-adjusted life years (DALYs) (GBD 2016 DALYs and HALE Collaborators, 2017; McManus et al., 2018). Thus far, the lipidome of S. mansoni is amongst the best characterised for a parasitic helminth. Initial lipidomic studies of this worm (Van Hellemond et al., 2006) were focused mainly on specific biological questions relating to specific lipid synthesis and fatty acid modifications, and served as a foundation for subsequent, comprehensive analyses of lipids in S. mansoni.
Lipids of the tegumental surface have been explored; the tegument is a double membrane structure and the outer surface of flatworms. Thus, it is at the ‘parasite-host interface’ and plays roles including the active uptake of nutrients, the elimination of ‘waste-products’ from the worm as well as immune modulation and parasite survival (Van Hellemond et al., 2006). Using a MS-based approach, Brouwers et al. (1997) reported abundant glycerophosphocholine (PC) and glycerophosphoethanolamine (PE) species in the tegumental membranes, although less abundant glycerophospholipids, such as phosphatidylinositols (PI), phosphatidylserines (PS) and lysophospholipids, were not detectable. Subsequently, using an HPLC-MS method, the same research group conducted a comprehensive and selective analysis of lipids species of the major glycerophospholipid classes, namely PC, PE, PI, PS and lysophospholipids, in adult S. mansoni and its tegumental membranes (Retra et al., 2015). This work successfully identified glycerophospholipids, including octadecenoic acid-containing PC and eicosenoic acid (20:1)-containing LPE and LPS, which were enriched in the tegument. Another recent investigation (Bexkens et al., 2019) explored the biosynthesis and transport/incorporation of exogenous lipids in S. mansoni eggs and female adults. By tracking [14C]-labelled FAs via energy metabolism, this study showed that S. mansoni was not able to oxidise FAs and that host-derived FAs accumulated in the eggs within female worms, which are likely required for biosynthetic purposes.
In a more global context, Giera et al. (2018) employed three complementary MS-based lipidomic approaches to characterise the lipidomes of three developmental stages (i.e. egg, cercariae and adults) of S. mansoni as well as excretory/secretory (ES) products from each of these stages. Quadrupole ion trap (Q-Trap) LC-MS/MS was used for the analysis of eicosanoids and docosanoids, and gas chromatography (GC)-MS) and quadrupole time of flight (Q-TOF) LC-MS/MS platforms were employed to discern total FAs and other major lipid classes. This global lipidomic study allowed a first lipid database to be established to enable studies of the biology of lipids in schistosomes, but also identified lipids (including prostaglandin D2 and E2) proposed to have immunomodulatory roles and possible implications for developing interventions for inflammatory disorders in humans (Giera et al., 2018).
Schistosomes, as multicellular organisms, have differentiated tissues and highly specialised organs with diverse molecular compositions (Wendt et al., 2020). Advanced MS imaging (MSI) provides an ideal tool to identify the spatial distribution of lipids in intact parasitic worms. As proof of concept, Ferreira et al. (2014b) used MALDI-MSI to explore the distribution of lipids in the surface of adult S. mansoni (see Ferreira et al., 2014b). Using an image acquisition of 50 μm-raster width, lipids of multiple classes (e.g. diradylglycerols (DG), triradylglycerols (TG) and PC) were characterised in tegument and suckers. The complementary use of MALDI-TOF-MS plus infusion electrospray ionisation (ESI)-MS defined lipid profiles in adult worms and other developmental stages (i.e. eggs, and cercariae) that were distinct among different S. mansoni strains (Ferreira et al., 2014b). In a subsequent study, the established MALDI-MSI-based platform was utilised to identify molecular pathways in adult worms affected by praziquantel treatment (Ferreira et al., 2015).
Limited by the procedure of sample preparation and relatively low image resolution, the MSI method used initially (cf. Ferreira et al., 2014b) was unable to discern lipids in internal organs and the anterior and posterior parts of worms. However, Kadesch et al. (2019) recently overcame these limitations through the optimisation of both sample preparation and image acquisition (at lateral resolutions of 10 μm and 5 μm), successfully establishing a novel, untargeted protocol for the detailed analysis of lipids in cryosections of S. mansoni. Using this optimised MALDI-MSI workflow, the same research group (see Kadesch et al., 2019) comprehensively characterised the lipidomes of cryo-sectioned adult worms (separate males and females as well as pairs) of this species in a spatially-resolved manner (Kadesch et al., 2020). For the first time, these authors were able to characterise pronounced differences in lipid classes, abundance, FA composition and the degree of saturation between the tegument and parenchyma of S. mansoni.
MS-based lipidomics has also been used to explore the host-parasite interplay between S. mansoni and a mammalian host. Bexkens et al. (2020) characterised and compared the lipid compositions of host-derived, circulating extracellular vesicles (EVs) in S. mansoni-infected hamsters with those of uninfected hamsters. Intriguingly, although no parasite-derived proteins or lipids were detected in EVs in infected hamster blood, published evidence (Bexkens et al., 2020) indicates that worm infection can influence the protein and/or lipid composition of host-derived EVs.
3.2. Parasitic nematodes
3.2.1. Heligmosomoides polygyrus (or H. bakeri; see Behnke & Harris, 2010)
For parasitic nematodes, lipidomics was first applied to the intestinal nematode H. polygyrus (Simbari et al., 2016) to investigate the lipid composition of EVs. Using ultra-high performance liquid chromatography (UHPLC)-MS/MS, Simbari et al. (2016) showed an up to 62-fold enrichment of ether-linked glycerophospholipids and a lower abundance of cholesterol and sphingomyelin in EVs secreted by H. polygyrus as compared with host-derived EVs. Biophysical analyses – using a membrane fusion assay – and studies of synthetic vesicles (Simbari et al., 2016) have indicated that ether-linked glycerophospholipids provide the stability and rigidity in nematode EV membranes, compensating for low levels of cholesterol and sphingomyelin.
3.2.2. Onchocerca volvulus, O. ochengi and Litomosoides sigmodontis
In addition to the study of exosome stability, lipidomics has also been employed to identify parasite-specific molecules as potential biomarkers for filariasis (including onchocerciasis, or river blindness). Using a direct infusion ESI Q-TOF MS/MS approach, the composition of glycerophospholipids was investigated in three distinct species of filarioids – i.e. O. volvulus, O. ochengi and L. sigmodontis as well as in the host serum and fluid from within nodules (Wewer et al., 2017). Targeted shotgun lipidomic analysis revealed substantial amounts of ether-linked PC and PE in all three species. Although no parasite-specific lipids were detected in the serum from infected hosts, nematode-derived ether-linked PE species (i.e. PE(e38:4), PE(e40:8) and PE(e40:9)) were enriched in fluid taken from O. ochengi nodules from the host, suggesting that such accumulated ether-linked PE lipids might be used as biomarkers for the diagnosis of river blindness (Wewer et al., 2017).
3.2.3. Haemonchus contortus
This haematophagous (‘barberʼs pole’) worm infects hundreds of millions of small ruminants (e.g. sheep and goats) worldwide, causing worldwide economic losses estimated at $3.3 billion per annum (Evans & Chapple, 2002; Gasser & von Samson-Himmelstjerna, 2016). Genomic (Laing et al., 2013; Schwarz et al., 2013; Palevich et al., 2019; Doyle et al., 2018, 2020), transcriptomic (Salle et al., 2018; Ma et al., 2018b, Ma et al., 2019a, Ma et al., 2019b, Ma et al., 2019c; Stasiuk et al., 2019) and proteomic (Wang et al., 2019a, b, 2020b) investigations of H. contortus have not only detected a wide range of molecules (i.e. genes and proteins) involved in lipid metabolism and transportation, but also revealed marked alterations in transcription or expression of selected lipid-associated genes and proteins during the development, which could relate to dynamic changes in the lipidome of H. contortus.
Some studies have taken the first step to study the lipidomes of H. contortus and its stages, organ systems and tissues. Recently, we characterised the developmental lipidome of H. contortus using an untargeted approach (Wang et al., 2018). A total of > 500 lipid species representing 18 lipid classes within four lipid categories were identified and quantified in six key developmental stages/sexes. The relative quantification data showed a significant shift in lipid abundance during the transition from the free-living to the parasitic phase of the life-cycle, particularly for lipid species (e.g. TG, PC, PE and ether-linked lipids), with essential roles in cellular processes and functions (linked to nutrient acquisition and/or membrane remodelling), suggesting dynamic changes as the nematode adapts to varying environments without and within the host animal (Wang et al., 2018). Extending this work, we used a similar, untargeted approach to gain insight into lipid composition and abundance in the organ systems (i.e. alimentary and reproductive systems) of the adult female of H. contortus (see Wang et al., 2020c). We discovered very high levels of energy storage-related TG lipids in the gravid uteri of female worms, suggesting that they are essential for egg and larval survival and development.
Apart from studying lipid composition, there has been an increased focus on understanding the molecular and biochemical basis of dauer signalling pathway-regulated development of parasitic nematodes (including hypobiosis and larval activation) (Wang et al., 2009; Zhi et al., 2012; Albarqi et al., 2016; Ma et al., 2019b). Lipidomic investigations are likely to become critical to underpin this area. Recently, we used targeted lipidomics to measure endogenous delta(7)-dafachronic acid (a steroidal hormone that binds to the nuclear hormone receptor DAF-12 to regulate development), following the exsheathment and activation of free-living larvae of H. contortus (see Ma et al., 2019c). We observed a positive correlation between the concentration (0–1.25 μM) of delta(7)-dafachronic acid and larval development after 48 h of in vitro culture. Furthermore, untargeted lipidomics was conducted to measure the potential impact of the negative feedback circuit – a signalling mechanism to regulate nematode development – on lipid metabolism; the results indicated that the dafachronic acid-DAF-12 module promotes the degradation of glycerolipids (e.g. DG(32:1) and TG(42:2)) and the biosynthesis of glycerophospholipids (e.g. PC(35:4), PC(33:0) and PI(35:4)) (Ma et al., 2019c).
3.2.4. Ascaris suum and Toxocara canis
These are two important ascaridoid nematodes which cause ascariasis and toxocariasis, respectively, in animals including humans (Crompton, 2001; Ma et al., 2018a). Recently, lipidomics has served as a complementary means of verifying the bioinformatically-inferred dauer-like signalling pathways of A. suum and T. canis (see Ma et al., 2019a). UHPLC-MS/MS analysis was employed to detect endogenous steroidal hormones in these two ascaridoid species, and specifically identified two distinct isomers of endogenous dafachronic acid (i.e. delta(4)- and delta(7)- dafachronic acids) in adult worms. Interestingly, delta(7)-DA dominated in A. suum, whereas delta(4)-DA was enriched in T. canis (see Ma et al., 2019a).
Recently, we also investigated the lipidomes of key developmental stages/sexes as well as organ systems of A. suum using high throughput LC-MS/MS (Wang et al., 2020d). Approximately 600 lipid species belonging to 18 lipid classes were identified. Similar to results for H. contortus (see Wang et al., 2018, 2020c), there was a substantial difference in the composition and abundance of lipids with essential roles in cellular processes and functions (e.g. energy storage regulation and/or membrane structure) among different developmental stages and organ systems, likely reflecting the differing demands for lipids for parasite development, reproduction and adaptation to constantly changing environments. Notably, the semi-quantitative method (i.e. normalised molar amounts in relation to the dry weight of worm material) established in this study provides an efficient and cost-effective means of undertaking comparative lipidomic analyses of the model species Caenorhabditis elegans and other parasitic nematodes of interest.
3.2.5. Trichinella papuae
The lipidome of first-stage larvae (L1s) of T. papuae has also been characterised by LC-MS/MS to identify potential therapeutic targets (Mangmee et al., 2020). In total, 255 glycerophospholipids, 62 glycerolipids, 33 sphingolipids and 25 FAs with unique molecular structures were identified in a single experiment using this approach. The comparison of T. papuae and the human lipidomes showed that more than half of lipid species identified in T. papuae were absent from the human lipidome, indicating distinctive lipid profiles in the parasite and the host.
4. Challenges and future perspectives
Although lipidomic investigations have been conducted only on a limited number of helminth species, some progress has been made to enhance our knowledge of lipid-associated signalling pathways, lipid synthesis and fatty acid modifications. However, the more insights we gain from these studies, the more we will appreciate the complexity of the lipidomes and lipid functions in parasitic worms. In parasitology, lipidomics is in its infancy, and there is whole new ‘fatty world’ to be discovered in worms.
4.1. Need for worm-specific lipid standards
For MS-based lipidomic explorations, confident identification and accurate quantification of lipids rely on comparisons with authentic internal standards; ideally, using representative lipids from the same lipid class that exhibit similar ionisation and fragmentation behaviours under the same analytical conditions (Sumner et al., 2007; Rustam & Reid, 2018). Unfortunately, no specific standards are commercially available for worms, including the free-living nematode C. elegans (see Witting & Schmitt-Kopplin, 2016). Since worms synthesise odd chain FAs (Watts & Ristow, 2017), and their lipidomes contain larger proportions of odd chain FA-containing lipids compared with mammals (Wang et al., 2018), many mammal-specific reference standards (which contains odd chain fatty acids; e.g. PC(17:0/17:0) are not suitable for lipidomic studies of worms. As we have shown recently (Wang et al., 2020d), lipid standards comprising isotope-labelled analogues can offer a solution for quantitative analyses of worm lipids, but there is still a need for specific standards, such as dafachronic acids, to serve as proxy for the precise characterisation of worm specific lipids of biological importance (cf. Ma et al., 2019a; Ma et al., 2019c). Moreover, it will be critical that chemical structural information on worm specific lipids be entered into databases and be accessible for accurate and comprehensive characterisation/annotation using informatics (e.g. via LipidSearch).
4.2. Integrative ‘omics
With the development of advanced nucleic acid sequencing, MS-based ‘omics (e.g. proteomics), bioinformatics and functional genomic technologies, huge data sets and information have become available for parasitic worms over the last two decades. This context creates unprecedented opportunities to integrate such data sets with lipidomic data, which will be critical for systems biological investigations. Recent studies illustrate the usefulness of a multi-omics approach. For example, Ma et al. (2019b) reconstructed the dauer signalling pathway model for H. contortus by integrating genomic, transcriptomic and proteomic data sets. Subsequently, these workers (Ma et al., 2019c) combined lipidomic with transcriptomic and proteomic data to explore the roles of dafachronic acid (a bile-acid like steroid hormone) in promoting larval development, in modulating dauer-like signal transduction and in lipid metabolism in H. contortus.
Encouragingly, a recent large-scale genome-lipid association mapping study using a mouse model demonstrated the potential of cross-linking available genetic information with global lipid data (Linke et al., 2020). If such large-scale mapping work could be extended to selected worm species for which chromosome-contiguous genomes have been defined, this would open up opportunities to elucidate the genetic regulation of lipid metabolism.
4.3. MSI-based lipidomics of parasitic nematodes
As described in Section 2, the MS-based imaging technique provides a ‘non-invasive’ tool to identify and localise lipids in intact worms. Despite this first success for flatworm research (Ferreira et al., 2014b; Kadesch et al., 2020), there is a dearth of MS-based imaging studies in parasitic nematodes, although some initial work has been conducted on C. elegans. Three preliminary studies using MALDI-MS and TOF-secondary ion MS-based platforms (Geier et al., 2012; Patti et al., 2014; Menger et al., 2015) have been assessed and have showed potential for applications to nematodes. However, compared with what has been achieved in schistosomes, the spatial resolution of lipids was not sufficient for routine use in C. elegans. Witting & Schmitt-Kopplin (2016) recommended that improvements in sample preparation and instrumentation (relating to spatial resolution) will be needed to make this work well. In the near future, we hope that such tools will allow the identification and quantitation of lipid species in situ in whole worms, and that analyses of the temporal and spatial changes in lipid profiles will ‘shine a light’ on the biology of lipids in parasitic nematodes.
4.4. Single cell lipidomics
To date, no study has comprehensively explored the lipidomes of single cells from worms. Compared with transcriptomics and proteomics, single cell lipidomics is still in its infancy (reviewed by Couvillion et al., 2019). The analytical sensitivity of modern MS instrumentation is as low as the concentration of amol/mg dry protein of tissue, which is within the range of that in single cells (i.e. ranging from fmol to amol) (Hu & Zhang, 2018). In practice, the bottleneck in applying lipidomics to single cells is sample preparation (e.g. isolation and extraction of purified single cells) rather than MS detection. The recent, successful application of single cell RNA sequencing to the flatworm S. mansoni (see Wendt et al., 2020) and the free-living nematode C. elegans (see Packer et al., 2019) demonstrate the feasibility of capturing the most immediate and dynamic molecular alterations in individual cells of worms. We hope that similar developments will accelerate the study of lipidomes of other parasites.
5. Concluding remarks
Although lipidomics is in its infancy in parasitology, studies in other biomedical disciplines have demonstrated some key biological roles that lipids play in organisms, tissues and cells (e.g. Yang & Han, 2016). Given the technological advances in mass spectrometry made recently, there is major merit in including lipidomics in the helminthology “tool kit” to enable ‘systems biological’ investigations of parasitic worms using multi-omic approaches. As this new frontier develops and expands, we hope to see a deepening of the knowledge and understanding of the structures and functions of different lipid species/classes in socioeconomically important parasitic worms in a search for new and innovative approaches for the diagnosis, treatment and/or control of helminthiases.
Declaration of competing interests
The authors declare that they have no competing interests.
Acknowledgements
Research funding from the Australian Research Council and National Health and Medical Research Council is gratefully acknowledged. Funding bodies played no role in the design of the study or collection, analysis or interpretation of data, or in the writing of the manuscript.
Contributor Information
Tao Wang, Email: tao.wang1@unimelb.edu.au.
Robin B. Gasser, Email: robinbg@unimelb.edu.au.
References
- Albarqi M.M., Stoltzfus J.D., Pilgrim A.A., Nolan T.J., Wang Z., Kliewer S.A., Mangelsdorf D.J., Lok J.B. Regulation of life cycle checkpoints and developmental activation of infective larvae in Strongyloides stercoralis by dafachronic acid. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A.C., Willenberg I., Springer A., Schebb N.H., Steinberg P., Strube C. Fatty acid composition of free-living and parasitic stages of the bovine lungworm Dictyocaulus viviparus. Mol. Biochem. Parasitol. 2017;216:39–44. doi: 10.1016/j.molbiopara.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Behnke J., Harris P.D. Heligmosomoides bakeri: a new name for an old worm? Trends Parasitol. 2010;26:524–529. doi: 10.1016/j.pt.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Bexkens M.L., Mebius M.M., Houweling M., Brouwers J.F., Tielens A.G.M., van Hellemond J.J. Schistosoma mansoni does not and cannot oxidise fatty acids, but these are used for biosynthetic purposes instead. Int. J. Parasitol. 2019;49:647–656. doi: 10.1016/j.ijpara.2019.03.005. [DOI] [PubMed] [Google Scholar]
- Bexkens M.L., van Gestel R.A., van Breukelen B., Urbanus R.T., Brouwers J.F., Nieuwland R., Tielens A.G.M., van Hellemond J.J. Schistosoma mansoni infection affects the proteome and lipidome of circulating extracellular vesicles in the host. Mol. Biochem. Parasitol. 2020;238 doi: 10.1016/j.molbiopara.2020.111296. [DOI] [PubMed] [Google Scholar]
- Brouwers J.F., Smeenk I.M., van Golde L.M., Tielens A.G. The incorporation, modification and turnover of fatty acids in adult Schistosoma mansoni. Mol. Biochem. Parasitol. 1997;88:175–185. doi: 10.1016/s0166-6851(97)00091-1. [DOI] [PubMed] [Google Scholar]
- Couvillion S.P., Zhu Y., Nagy G., Adkins J.N., Ansong C., Renslow R.S., et al. New mass spectrometry technologies contributing towards comprehensive and high throughput omics analyses of single cells. Analyst. 2019;144:794–807. doi: 10.1039/c8an01574k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton D.W. Ascaris and ascariasis. Adv. Parasitol. 2001;48:285–375. doi: 10.1016/s0065-308x(01)48008-0. [DOI] [PubMed] [Google Scholar]
- Doyle S.R., Laing R., Bartley D.J., Britton C., Chaudhry U., Gilleard J.S., et al. A genome resequencing-based genetic map reveals the recombination landscape of an outbred parasitic nematode in the presence of polyploidy and polyandry. Genome Biol. Evol. 2018;10:396–409. doi: 10.1093/gbe/evx269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.R., Tracey A., Laing R., Holroyd N., Bartley D., Bazant W., et al. Extensive genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus. bioRxiv. 2020 doi: 10.1038/s42003-020-01377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Chapple N. The animal health market. Nat. Rev. Drug Discov. 2002;1:937–938. doi: 10.1038/nrd975. [DOI] [PubMed] [Google Scholar]
- Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Jr., Murphy R.C., et al. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Ferreira M.S., de Oliveira D.N., de Oliveira R.N., Allegretti S.M., Catharino R.R. Screening the life cycle of Schistosoma mansoni using high-resolution mass spectrometry. Anal. Chim. Acta. 2014;845:62–69. doi: 10.1016/j.aca.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Ferreira M.S., de Oliveira D.N., de Oliveira R.N., Allegretti S.M., Vercesi A.E., Catharino R.R. Mass spectrometry imaging: a new vision in differentiating Schistosoma mansoni strains. J. Mass Spectrom. 2014;49:86–92. doi: 10.1002/jms.3308. [DOI] [PubMed] [Google Scholar]
- Ferreira M.S., de Oliveira R.N., de Oliveira D.N., Esteves C.Z., Allegretti S.M., Catharino R.R. Revealing praziquantel molecular targets using mass spectrometry imaging: an expeditious approach applied to Schistosoma mansoni. Int. J. Parasitol. 2015;45:385–391. doi: 10.1016/j.ijpara.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., von Samson-Himmelstjerna G. Haemonchus contortus and haemonchosis – past, present and future trends. Adv. Parasitol. 2016;93 [Google Scholar]
- GBD 2016 DALYs and HALE Collaborators Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1260–1344. doi: 10.1016/S0140-6736(17)32130-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier F.M., Fearn S., Bundy J.G., McPhail D.S. In: Surf. Interface Anal. Napolitani E., Giubertoni D., Bersani M., Anderle M., Licciardello A., editors. 2012. ToF-SIMS analysis of biomolecules in the model organism Caenorhabditis elegans. pp. 234–236. [Google Scholar]
- Giera M., Kaisar M.M.M., Derks R.J.E., Steenvoorden E., Kruize Y.C.M., Hokke C.H., et al. The Schistosoma mansoni lipidome: leads for immunomodulation. Anal. Chim. Acta. 2018;1037:107–118. doi: 10.1016/j.aca.2017.11.058. [DOI] [PubMed] [Google Scholar]
- Greichus A., Greichus Y.A. Chemical composition and volatile fatty acid production of male Ascaris lumbricoides before and after starvation. Exp. Parasitol. 1966;19:85–90. doi: 10.1016/0014-4894(66)90056-7. [DOI] [PubMed] [Google Scholar]
- Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016;12:668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- Hu T., Zhang J.L. Mass-spectrometry-based lipidomics. J. Separ. Sci. 2018;41:351–372. doi: 10.1002/jssc.201700709. [DOI] [PubMed] [Google Scholar]
- International Helminth Genomes C. Comparative genomics of the major parasitic worms. Nat. Genet. 2019;51:163–174. doi: 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jex A.R., Gasser R.B., Schwarz E.M. Transcriptomic resources for parasitic nematodes of veterinary importance. Trends Parasitol. 2019;35:72–84. doi: 10.1016/j.pt.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Kadesch P., Quack T., Gerbig S., Grevelding C.G., Spengler B. Lipid topography in Schistosoma mansoni cryosections, revealed by microembedding and high-resolution atmospheric-pressure matrix-assisted laser desorption/ionization (MALDI) mass spectrometry Imaging. Anal. Chem. 2019;91:4520–4528. doi: 10.1021/acs.analchem.8b05440. [DOI] [PubMed] [Google Scholar]
- Kadesch P., Quack T., Gerbig S., Grevelding C.G., Spengler B. Tissue- and sex-specific lipidomic analysis of Schistosoma mansoni using high-resolution atmospheric pressure scanning microprobe matrix-assisted laser desorption/ionization mass spectrometry imaging. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyonami R., Peake D.A., Liu X., Huang Y. Thermo Fisher Scientific; 2016. Large-scale Lipid Profiling of a Human Serum Lipidome Using a High-Resolution, Accurate-Mass LC/MS/MS Approach. Application Note No. 647. [Google Scholar]
- Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke V., Overmyer K.A., Miller I.J., Brademan D.R., Hutchins P.D., Trujillo E.A., et al. A large-scale genome-lipid association map guides lipid identification. Nat. Metab. 2020;2:1149–1162. doi: 10.1038/s42255-020-00278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Holland C.V., Wang T., Hofmann A., Fan C.K., Maizels R.M., et al. Human toxocariasis. Lancet Infect. Dis. 2018;18:e14–24. doi: 10.1016/S1473-3099(17)30331-6. [DOI] [PubMed] [Google Scholar]
- Ma G., Wang T., Korhonen P.K., Ang C.S., Williamson N.A., Young N.D., et al. Molecular alterations during larval development of Haemonchus contortus in vitro are under tight post-transcriptional control. Int. J. Parasitol. 2018;48:763–772. doi: 10.1016/j.ijpara.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Ma G., Wang T., Korhonen P.K., Nie S., Reid G.E., Stroehlein A.J., et al. Comparative bioinformatic analysis suggests that specific dauer-like signalling pathway components regulate Toxocara canis development and migration in the mammalian host. Parasit. Vectors. 2019;12:32. doi: 10.1186/s13071-018-3265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Wang T., Korhonen P.K., Stroehlein A.J., Young N.D., Gasser R.B. Dauer signalling pathway model for Haemonchus contortus. Parasit. Vectors. 2019;12:187. doi: 10.1186/s13071-019-3419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Wang T., Korhonen P.K., Young N.D., Nie S., Ang C.S., et al. Dafachronic acid promotes larval development in Haemonchus contortus by modulating dauer signalling and lipid metabolism. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Gasser R.B., Wang T., Korhonen P.K., Young N.D. Toward integrative 'omics of the barberʼs pole worm and related parasitic nematodes. Infect. Genet. Evol. 2020;85:104500. doi: 10.1016/j.meegid.2020.104500. [DOI] [PubMed] [Google Scholar]
- Ma G., Wang T., Korhonen P.K., Hofmann A., Sternberg P.W., Young N.D., Gasser R.B. Elucidating the molecular and developmental biology of parasitic nematodes: moving to a multiomics paradigm. Adv. Parasitol. 2020;108:175–229. doi: 10.1016/bs.apar.2019.12.005. [DOI] [PubMed] [Google Scholar]
- Magalhaes K., Almeida P.E., Atella G., Maya-Monteiro C.M., Castro-Faria-Neto H., Pelajo-Machado M., et al. Schistosomal-derived lysophosphatidylcholine are involved in eosinophil activation and recruitment through Toll-like receptor-2-dependent mechanisms. J. Infect. Dis. 2010;202:1369–1379. doi: 10.1086/656477. [DOI] [PubMed] [Google Scholar]
- Mangmee S., Adisakwattana P., Tipthara P., Simanon N., Sonthayanon P., Reamtong O. Lipid profile of Trichinella papuae muscle-stage larvae. Sci. Rep. 2020;10:10125. doi: 10.1038/s41598-020-67297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus D.P., Dunne D.W., Sacko M., Utzinger J., Vennervald B.J., Zhou X.N. Schistosomiasis. Nat. Rev. Dis. Primers. 2018;4:13. doi: 10.1038/s41572-018-0013-8. [DOI] [PubMed] [Google Scholar]
- McVeigh P. Post-genomic progress in helminth parasitology. Parasitology. 2020;147:835–840. doi: 10.1017/S0031182020000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger R.F., Clendinen C.S., Searcy L.A., Edison A.S., Yost R.A. MALDI mass spectrometric imaging of the nematode Caenorhabditis elegans. Curr. Metabolomics. 2015;3:130–137. [Google Scholar]
- Minematsu T., Yamazaki S., Uji Y., Okabe H., Korenaga M., Tada I. Analysis of polyunsaturated fatty acid composition of Strongyloides ratti in relation to development. J. Helminthol. 1990;64:303–309. doi: 10.1017/s0022149x00012347. [DOI] [PubMed] [Google Scholar]
- Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365(6459) doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N., Maclean P.H., Baten A., Scott R.W., Leathwick D.M. The genome sequence of the anthelmintic-susceptible New Zealand Haemonchus contortus. Genome Biol. Evol. 2019;11:1965–1970. doi: 10.1093/gbe/evz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G.J., Tautenhahn R., Johannsen D., Kalisiak E., Ravussin E., Bruning J.C., et al. Meta-analysis of global metabolomic data identifies metabolites associated with life-span extension. Metabolomics. 2014;10:737–743. doi: 10.1007/s11306-013-0608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retra K., deWalick S., Schmitz M., Yazdanbakhsh M., Tielens A.G., Brouwers J.F., van Hellemond J.J. The tegumental surface membranes of Schistosoma mansoni are enriched in parasite-specific phospholipid species. Int. J. Parasitol. 2015;45:629–636. doi: 10.1016/j.ijpara.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Rustam Y.H., Reid G.E. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem. 2018;90:374–397. doi: 10.1021/acs.analchem.7b04836. [DOI] [PubMed] [Google Scholar]
- Salle G., Laing R., Cotton J.A., Maitland K., Martinelli A., Holroyd N., et al. Transcriptomic profiling of nematode parasites surviving vaccine exposure. Int. J. Parasitol. 2018;48:395–402. doi: 10.1016/j.ijpara.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E.M., Korhonen P.K., Campbell B.E., Young N.D., Jex A.R., Jabbar A., et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbari F., McCaskill J., Coakley G., Millar M., Maizels R.M., Fabrias G., et al. Plasmalogen enrichment in exosomes secreted by a nematode parasite versus those derived from its mouse host: implications for exosome stability and biology. J. Extracell. Vesicles. 2016;5:30741. doi: 10.3402/jev.v5.30741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V.P., Selkirk M.E., Gounaris K. Identification and composition of lipid classes in surface and somatic preparations of adult Brugia malayi. Mol. Biochem. Parasitol. 1996;78:105–116. doi: 10.1016/s0166-6851(96)02615-1. [DOI] [PubMed] [Google Scholar]
- Stasiuk S.J., MacNevin G., Workentine M.L., Gray D., Redman E., Bartley D., et al. Similarities and differences in the biotransformation and transcriptomic responses of Caenorhabditis elegans and Haemonchus contortus to five different benzimidazole drugs. Int. J. Parasitol. Drugs Drug Resist. 2019;11:13–29. doi: 10.1016/j.ijpddr.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A., et al. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hellemond J.J., Retra K., Brouwers J.F., van Balkom B.W., Yazdanbakhsh M., Shoemaker C.B., Tielens A.G. Functions of the tegument of schistosomes: clues from the proteome and lipidome. Int. J. Parasitol. 2006;36:691–699. doi: 10.1016/j.ijpara.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhou X.E., Motola D.L., Gao X., Suino-Powell K., Conneely A., et al. Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc. Natl. Acad. Sci. USA. 2009;106:9138–9143. doi: 10.1073/pnas.0904064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Nie S., Ma G., Korhonen P.K., Koehler A.V., Ang C.S., et al. The developmental lipidome of Haemonchus contortus. Int. J. Parasitol. 2018;48:887–895. doi: 10.1016/j.ijpara.2018.06.002. [DOI] [PubMed] [Google Scholar]
- Wang T., Ma G., Ang C., Korhonen P.K., Rong X., Nie S., et al. Somatic proteome of Haemonchus contortus. Int. J. Parasitol. 2019;49:311–320. doi: 10.1016/j.ijpara.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Wang T., Ma G., Ang C.S., Korhonen P.K., Koehler A.V., Young N.D., et al. High throughput LC-MS/MS-based proteomic analysis of excretory-secretory products from short-term in vitro culture of Haemonchus contortus. J. Proteomics. 2019;204:103375. doi: 10.1016/j.jprot.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Wang T., Cheng X., Xu H., Meng Y., Yin Z., Li X., Hang W. Perspective on advances in laser-based high-resolution mass spectrometry imaging. Anal. Chem. 2020;92:543–553. doi: 10.1021/acs.analchem.9b04067. [DOI] [PubMed] [Google Scholar]
- Wang T., Ma G., Ang C.S., Korhonen P.K., Stroehlein A.J., Young N.D., et al. The developmental phosphoproteome of Haemonchus contortus. J. Proteomics. 2020;213:103615. doi: 10.1016/j.jprot.2019.103615. [DOI] [PubMed] [Google Scholar]
- Wang T., Ma G., Nie S., Williamson N.A., Reid G.E., Gasser R.B. Lipid composition and abundance in the reproductive and alimentary tracts of female Haemonchus contortus. Parasit. Vectors. 2020;13:338. doi: 10.1186/s13071-020-04208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Nie S., Ma G., Vlaminck J., Geldhof P., Williamson N.A., et al. Quantitative lipidomic analysis of Ascaris suum. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.L., Ristow M. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics. 2017;207:413–446. doi: 10.1534/genetics.117.300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt G., Zhao L., Chen R., Liu C., OʼDonoghue A.J., Caffrey C.R., et al. A single-cell RNA-seq atlas of Schistosoma mansoni identifies a key regulator of blood feeding. Science. 2020;369:1644–1649. doi: 10.1126/science.abb7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wewer V., Makepeace B.L., Tanya V.N., Peisker H., Pfarr K., Hoerauf A., Dormann P. Lipid profiling of the filarial nematodes Onchocerca volvulus, Onchocerca ochengi and Litomosoides sigmodontis reveals the accumulation of nematode-specific ether phospholipids in the host. Int. J. Parasitol. 2017;47:903–912. doi: 10.1016/j.ijpara.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting M., Schmitt-Kopplin P. The Caenorhabditis elegans lipidome: a primer for lipid analysis in Caenorhabditis elegans. Arch. Biochem. Biophys. 2016;589:27–37. doi: 10.1016/j.abb.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Yang K., Han X. Lipidomics: techniques, applications, and outcomes related to biomedical sciences. Trends Biochem. Sci. 2016;41:954–969. doi: 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X., Zhou X.E., Melcher K., Motola D.L., Gelmedin V., Hawdon J., et al. Structural conservation of ligand binding reveals a bile acid-like signaling pathway in nematodes. J. Biol. Chem. 2012;287:4894–4903. doi: 10.1074/jbc.M111.315242. [DOI] [PMC free article] [PubMed] [Google Scholar]