Abstract
Plasmodium malariae and Plasmodium vivax are protozoan parasites that can cause malaria in humans. They are genetically indistinguishable from, respectively, Plasmodium brasilianum and Plasmodium simium, i.e. parasites infecting New World non-human primates in South America. In the tropical rainforests of the Brazilian Atlantic coast, it has long been hypothesized that P. brasilianum and P. simium in platyrrhine primates originated from P. malariae and P. vivax in humans. A recent hypothesis proposed the inclusion of Plasmodium falciparum into the transmission dynamics between humans and non-human primates in the Brazilian Atlantic tropical rainforest. Herein, we assess the occurrence of human malaria in simians and sylvatic anophelines using field-collected samples in the Capivari-Monos Environmental Protection Area from 2015 to 2017. We first tested simian blood and anopheline samples. Two simian (Aloutta) blood samples (18%, n = 11) showed Plasmodium cytb DNA sequences, one for P. vivax and another for P. malariae. From a total of 9,416 anopheline females, we found 17 pools positive for Plasmodium species with a 18S qPCR assay. Only three showed P. cytb DNA sequence, one for P. vivax and the others for rodent malaria species (similar to Plasmodium chabaudi and Plasmodium berghei). Based on these results, we tested 25 rodent liver samples for the presence of Plasmodium and obtained P. falciparum cytb DNA sequence in a rodent (Oligoryzomys sp.) liver. The findings of this study indicate complex malaria transmission dynamics composed by parallel spillover-spillback of human malaria parasites, i.e. P. malariae, P. vivax, and P. falciparum, in the Brazilian Atlantic forest.
Keywords: Anopheles, Malaria, Plasmodium, Plasmodium falciparum, Plasmodium malariae, Plasmodium vivax
Graphical abstract
Highlights
-
•
Human malaria parasites circulate in sylvatic cycles in the Brazilian Atlantic forest.
-
•
Plasmodium vivax and Plasmodium malariae identified in simian blood samples.
-
•
Plasmodium falciparum detected in a rodent liver sample.
-
•
Anopheline vectors found to carry human and rodent malaria parasites.
-
•
Local vector ecology and biology are key to the spillover-spillback of human malaria parasites.
1. Introduction
The five agents of human malaria are vector-borne protozoans of the genus Plasmodium, four are considered primarily human parasites, and one is a zoonotic parasite. The bites of Anopheles mosquito species transmit these malaria parasites to their vertebrate hosts, including primates. Plasmodium falciparum is responsible for most malaria deaths globally, reaching > 400 thousand cases worldwide in 2019, especially in sub-Saharan Africa (WHO, 2020). In contrast, the mortality associated with the other three species is significantly lower. Plasmodium vivax is the second most important because of high morbidity and high prevalence in endemic areas of Southeast Asia and Latin America (WHO, 2020). Plasmodium ovale and the pantropical distributed P. malariae, only account for a fraction of the clinical cases worldwide (Rutledge et al., 2017). These four Plasmodium species differ in aspects of their biology, likely because they evolved as human parasites from distinct phylogenetic lineages that are still under investigation (Escalante & Pacheco, 2019). The zoonotic malaria parasite, Plasmodium knowlesi, infects Macaca spp. in Southeast Asia and belongs to a phylogenetic clade that also includes P. vivax (Muehlenbein et al., 2015; Yusof et al., 2016; Yakob et al., 2018). This parasite is one of the most important causes of human malaria in Malaysia (Singh & Daneshvar, 2013). There is another causative species of zoonotic malaria, Plasmodium cynomolgi, which infects macaques, and has also been reported in humans (Grignard et al., 2019; Hartmeyer et al., 2019), but this species seems to be less frequent. Adding to these multiple parasites and the diversity of Anopheles species that can act as vectors worldwide, it is evident that the emergence of human malaria involved evolutionary processes and interspecies spillovers, which occurred in a variety of ecological settings that allow for malaria transmission (Escalante & Pacheco, 2019).
The African origin of P. falciparum has long been accepted (Garnham, 1966; Coatney, 1971; Escalante & Ayala, 1994). This parasite belongs to the subgenus Laverania, the African ape Plasmodium clade (Krief et al., 2010; Loy et al., 2017; Otto et al., 2018). The populations of P. falciparum followed the increase in the size of the human population and its global expansion out of Africa (Joy et al., 2003; Tanabe et al., 2010; Loy et al., 2017; Otto et al., 2018; Rodrigues et al., 2018). Part of this process was the colonization of the Americas, an event that likely occurred during the transatlantic slave trade 1533–1851 (Rodrigues et al., 2018).
The introduction of P. falciparum in the Americas required adapting to new-world Anopheles spp. (Molina-Cruz et al., 2016). These vector species have distinct ecological and genetic characteristics as they shared their most recent common ancestor with African vectors as long as 100 Mya ago (Foster et al., 2017). The main malarial vector in Brazil is Anopheles (Nyssorhynchus) darlingi, an anopheline species responsible for 99% of the reported malaria cases annually in the Amazon basin (Sallum et al., 2019). In the Atlantic tropical rainforest of Brazil, the local vector Anopheles (Kertezsia) cruzii can transmit human malaria (Carlos et al., 2019). However, this local vector has a broader host species preference (Multini et al., 2020), ranging from humans to domestic and sylvatic animals (Medeiros-Sousa et al., 2019). In addition, larvae of this species develop in the bromeliad phytotelma in shaded or partially shaded plants inside the forest (Marques et al., 2012).
Although currently under control, the Atlantic coast of Brazil saw past epidemics of malaria caused by P. falciparum and P. vivax in the 1950s until it was eliminated from urban areas (e.g. Rio de Janeiro) in the 1970s (Ferreira & Castro, 2016). Imported cases, P. vivax or P. falciparum from endemic malaria regions such as the Amazon, frequently occur in extra-Amazonian sites where they can infect local vectors (de Pina-Costa et al., 2014). When local vectors are infected, they can generate autochthonous malaria cases, often associated with tourism or occasional activities carried out inside the forest (de Alvarenga et al., 2016). The alternative hypothesis for these autochthonous malaria cases is transmission inside the forest, which means transmission from an infected non-human primate (zoonotic malaria) (Brasil et al., 2017). Autochtonous P. vivax cases in humans can be caused by the genetically indistinguishable form Plasmodium simium, circulating in non-human primates (Brasil et al., 2017; de Alencar et al., 2018; Abreu et al., 2019). Likewise, autochthonous P. malariae cases can also result from circulation and transmission of Plasmodium brasilianum from simians to humans (Coatney, 1971; Guimarães et al., 2012). Further support for the zoonotic malaria hypothesis is the biting behavior of An. cruzii in the canopy where non-human primates forage and on the ground level where humans walk inside forest (Medeiros-Sousa et al., 2021). The origin of zoonotic malaria caused by P. simium is a reverse zoonosis of the human parasite, i.e. spillover of P. vivax from human population to non-human primates during the colonization period in Brazil (de Oliveira et al., 2021). Now, the simian lineage (P. simium) is there circulating in nature and causing subpatent or patent infections in humans (Brasil et al., 2017). The spillback of the simian lineage is recognized as zoonotic malaria. Spillover-spillback mechanism could have hypothetically supported the persistence and adaptation of other invasive human malaria parasites arriving in the Americas, such as P. falciparum.
Plasmodium falciparum DNA and immunological responses were found in residents living in forested regions of São Paulo and Rio de Janeiro, suggesting its transmission among asymptomatic individuals (Maselli et al., 2014; Sallum et al., 2014; Miguel et al., 2019). A study by Laporta et al. (2015) investigated local foci of P. falciparum among local anophelines. Accordingly, 4.4% (21/480) of anophelines were found infected with P. falciparum, and most infected females (86%, 18/21) were An. cruzii (see Laporta et al., 2015). Finally, there is evidence of P. falciparum detected in Amazonian non-human primates in Brazil and Colombia (Araújo et al., 2013; Rondón et al., 2019).
The main working hypothesis tested here is that the most threatening human malaria parasite (P. falciparum) has a transmission cycle involving non-human primates in the Brazilian Atlantic Forest (Duarte et al., 2008; Laporta et al., 2015; Laporta, 2017; Assis et al., 2021). We tested this hypothesis using field-collected anopheline and simian blood samples in an environmental protection area (Medeiros-Sousa et al., 2019). We also tested these samples for the presence of other human malaria parasites (P. vivax and P. malariae) (Demari-Silva et al., 2020). As a complementary hypothesis, we tested for the presence of Plasmodium spp. in rodent liver samples considering that rodents can act as reservoirs of malaria parasites. We discuss these results in light of the Plasmodium spp. resilience post-elimination.
2. Materials and methods
2.1. Study area
The forest physiognomies of the Atlantic forest biome were reduced over the last century to 11–16% of their original domain that covered the entire South-to-North gradient of the Brazilian Atlantic coast (Ribeiro et al., 2009). The most extensive conserved region with these forest remnants is in the southeastern São Paulo State (Fig. 1A). Field investigations were carried out in the Capivari-Monos EPA (Environmental Protection Area) at ∼800 m above sea level (Fig. 1B) (Duarte et al., 2013; Medeiros-Sousa et al., 2019). The climate in the region is classified as a tropical monsoon climate (modified Köppen AM-type classification) with excessive annual precipitation (> 2,500 mm) and dry, mild winters (Rolim et al., 2007). Field collections were carried out in sites 1–4 where few humans are present, non-human primates are abundant, and the dominance of An. cruzii is documented (Duarte et al., 2013; Medeiros-Sousa et al., 2019) (Fig. 1C).
Fig. 1.
Study area. A Atlantic tropical rainforest remnants. B Southeastern Atlantic Forest. C Field collections were conducted in Capivari-Monos EPA (−46.7, −23.9): 1, Embura village; 2, Marsilac village; 3, Transition zone; 4, Cachoeira do Marsilac (Medeiros-Sousa et al., 2019). Abbreviation: SP, São Paulo metropolitan urban area with a population of ∼20 million people. Source: SOS Mata Atlântica/INPE, 2016
2.2. Study design
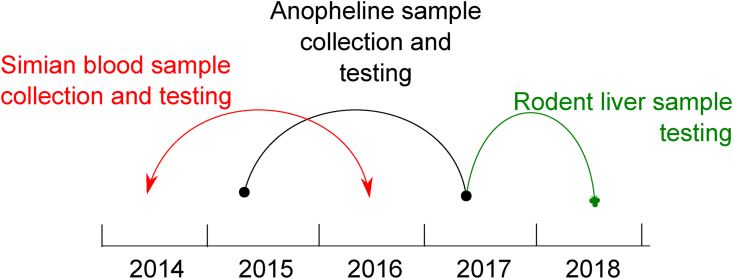
The study includes samples from three sources: (i) simian blood samples; (ii) anopheline samples; and (iii) rodent liver samples. With the first two sources, we tested for the presence of human malaria, and with the latter, for the presence of any Plasmodium in the rodent liver. We made prospective studies for collecting and testing simian blood samples from July 2014 to June 2016 and anopheline samples from March 2015 to May 2017. After the outcomes of both studies, we made a retrospective study with rodent liver samples (collected in 2012) from June 2017 to September 2018 (Fig. 2).
Fig. 2.
Timeline of the activities carried out during the investigation
Injured howler monkeys (Alouatta clamitans) by any cause (n = 11) were received from the study area and during the study period (Fig. 2). They were treated and sent back to nature by the municipal department for forestry and wildlife (DEPAVE/PMSP). Blood samples of 5 ml were collected in EDTA tubes for laboratory testing. The faunal survey conducted by DEPAVE/PMSP in the study area in 2012 obtained liver tissue samples from 25 rodent species. These samples were kept stored at −80 °C before the laboratory testing (Fig. 2).
2.3. Field collections and sample DNA extraction
Anopheline sampling was performed monthly during the study period (Fig. 2). In each of the four study sites (Fig. 1C), we employed: (i) Shannon traps from 18:00 h to 22:00 h (depending on the crepuscular time); (ii) CDC light traps with CO2 (dry ice) from 18:00 h to 6:00 h at ground level and in the canopy (10-m height); and (iii) backpack aspirator sampling on vegetation that could represent shelters for adults (20-min sampling).
Mosquito specimens were euthanized immediately before morphological identification using the keys of Forattini (2002). Non-engorged anopheline females were stored individually in isopropanol until DNA extraction. DNA was extracted in pools (a maximum of ten specimens/pool) using the Qiagen™ DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacturerʼs protocol.
Howler monkey blood samples collected as part of the DEPAVE/PMSP 2014 and 2016 surveys were stored in −20 °C (DEPAVE/PMSP, 2012). Rodent liver tissue samples were obtained from the mammal census performed by DEPAVE/PMSP in the study area in 2012 (SVMA/PMSP, 2011). DNA extraction from blood and liver samples followed the protocol provided by the Qiagen™ DNeasy Blood and Tissue kit.
2.4. Laboratory testing
Anophelines, simian blood samples and rodent liver samples were tested for the presence of Plasmodium DNA using the TaqMan qPCR assay (18S rRNA gene) following Bickersmith et al. (2015). Assays to detect genus Plasmodium, P. vivax and P. falciparum were performed separately using the TaqMan Universal Master Mix (Applied Biosystems, Foster City, CA, USA). The same methodology (Bickersmith et al., 2015) was used to detect P. malariae but with the primers and probes described by Rougemont et al. (2004). DNA extracted from known positive P. falciparum samples, P. brasilianum blood smear, and a patient blood with P. vivax were used as positive controls. An aliquot of ultrapure water was used as a negative control.
Positive samples for Plasmodium DNA using 18S qPCR were further analyzed. A nested PCR assay amplifying a ∼402 bp cytochrome b (cytb) fragment was performed with the cytb-1 primers (Siregar et al., 2015) followed by sequencing for molecular identification. PCR products were purified with ExoSAP-IT PCR Product Cleanup (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturerʼs instructions and forwarded with the respective primers to a gene sequencing company (Genomic Engenharia Molecular, São Paulo, SP, Brazil). An alternative protocol for amplifying a fragment of ∼424 bp mitochondrial Plasmodium small subunit rRNA (18S rRNA) gene, according to Siregar et al. (2015) was also employed. To generate more sequence data for phylogenetic analysis, we re-tested 15 anopheline samples confirmed positive for P. falciparum by Laporta et al. (2015) with the cytb nested PCR assay. Plasmodium 18S and cytb sequences generated in this study were deposited in the GenBank database under the accession numbers MF573323 (18S) and MF573300, MF573301, MT770753, MF476105, MT779799, MT779800, and MT779801 (cytb).
Because morphological identification of rodents at the species level was not possible, we applied the cytb protocol for species identification (Smith & Patton, 1993).
2.5. Phylogenetic analysis
The sequences generated in this study were compared with databases available using the BLAST tool (https://blast.ncbi.nlm.nih.gov) on GenBank (Altschul et al., 1997). A nucleotide alignment was generated using ClustalX v2.0.12 and Muscle as implemented in SeaView v4.3.5 (Gouy et al., 2010) with manual editing. The alignment was constructed with 32 cytb partial sequences (331 bp excluding gaps) belonging to the genus Plasmodium, including the newly generated sequences and sequences available on GenBank (Benson et al., 2013) for Plasmodium spp. that infect mammals (e.g. P. falciparum, P. vivax, P. simium, P. malariae, P. knowlesi and Plasmodium berghei amongst others). Sequences of species that infect birds, Plasmodium relictum and Plasmodium gallinaceum, were included as the outgroup. Sequences that showed a similarity > 95% using BLAST (Altschul et al., 1997) were included.
The phylogenetic tree was inferred based on this alignment using the Bayesian method implemented in MrBayes v3.2.6 with the default priors (Ronquist & Huelsenbeck, 2003). The general time reversible model with gamma-distributed substitution rates and a proportion of invariant sites (GTR + Γ + I) was estimated as the best-fit model of nucleotide substitution based on the lowest Bayesian Information Criterion (BIC) scores as estimated with MEGA v7.0.14 (Kumar et al., 2016). Bayesian support was inferred for the nodes in MrBayes by sampling every 1,000 generations from two independent chains lasting 2 × 106 Markov Chain Monte Carlo (MCMC) steps. Chains were assumed to have converged once the value of the potential scale reduction factor (PSRF) was between 1.00 and 1.02 and the average SD of the posterior probability was < 0.01 (Ronquist & Huelsenbeck, 2003). Then, 25% of the samples were discarded once convergence was reached as “burn-in”. GenBank accession numbers for all sequences used in the analysis are given in the phylogenetic tree.
3. Results
3.1. Simian blood samples
Out of 11 simian blood samples tested, we obtained two (18%) P. vivax-positive and two (18%) P. malariae-positive in 18S qPCR (Table 1). From the positive samples, we obtained one 18S sequence of P. malariae and one cytb sequence of P. vivax (Table 1).
Table 1.
Testing of simian blood samples (Alouatta guariba clamitans) for Plasmodium spp., Capivari-Monos EPA, Brazilian Atlantic rainforest, 2014–2016
| ID (DEPAVE) | 18S Plasmodium | cytb Plasmodium | GenBank ID |
|---|---|---|---|
| 10 (56067) | P. vivax | P. vivaxa | MF573301 |
| 16 (63739) | P. malariaea | – | MF573323 |
| 17 (64241) | P. malariae | – | – |
| 19 (65181) | – | nd | nd |
| 20 (65218) | – | nd | nd |
| 24 (62454) | – | nd | nd |
| 30 (68145) | – | nd | nd |
| 31 (68334) | – | nd | nd |
| 36 (70954) | – | nd | nd |
| 38 (73727) | P. vivax | – | – |
| 41 (75124) | – | nd | nd |
Note: P. vivax- and P. malariae-positive or negative (−) results for testing for parasite species presence in the sample.
Abbreviation: ID(DEPAVE), Divisão Técnica de Medicina Veterinária e Manejo da Fauna Silvestre, São Paulo, SP, Brazil; nd, not done.
Sequenced sample.
3.2. Anopheline samples
A total of 9,416 anopheline females were collected; of these, 9,328 specimens were An. cruzii (> 99%), 82 An. strodei, 3 Anopheles (Anopheles) pseudotibiamaculatus, 2 Anopheles (Nyssorhynchus) evansae, and 1 Anopheles (Nyssorhynchus) albitarsis. Seventeen pools returned Plasmodium-positive 18S qPCR (Table 2). Three Plasmodium cytb sequences were confirmed in An. cruzii (Table 2). The obtained P. vivax cytb sequence was 100% similar to the one found in howler-monkeys in the study region (Table 1). The obtained P. berghei and Plasmodium chabaudi cytb sequences (Table 2) motivated testing of rodent liver samples.
Table 2.
Testing of anopheline samples for Plasmodium spp., Capivari-Monos EPA, Brazilian Atlantic rainforest, 2015–2017
| ID (pool) trap | Site | 18S Plasmodium | cytb Plasmodium | GenBank ID |
|---|---|---|---|---|
| 84 (10 An. cruzii) Sh | 4 (forest) | P. vivax | – | – |
| 85 (10 An. cruzii) Sh | – | P. chabaudi | MT770753 | |
| 87 (10 An. cruzii) Sh | Plasmodium spp. | P. berghei | MF476105 | |
| 92 (10 An. cruzii) Sh | P. vivax | – | – | |
| 291 (10 An. cruzii) Sh | P. vivax | – | – | |
| 361 (10 An. cruzii) Sh | P. vivax | P. vivax | MF573300 | |
| 381 (10 An. cruzii) Sh | P. malariae | – | – | |
| 553 (10 An. cruzii) Sh | P. malariae | – | – | |
| 386 (10 An. cruzii) Sh | 3 (transition zone) | P. vivax | – | – |
| 598 (1 An. cruzii) CDCg | P. malariae | – | – | |
| 600 (1 An. cruzii) CDCg | P. falciparum | – | – | |
| 621 (1 An. strodei) Sh | P. vivax + P. falciparum | – | – | |
| 638 (1 An. strodei) Sh | P. vivax + P. malariae | – | – | |
| 678 (7 An. strodei) Sh | Plasmodium spp. | – | – | |
| 599 (10 An. cruzii) Asp | 2 (Marsilac village) | P. falciparum | – | – |
| 684 (5 An. cruzii) CDCc | Plasmodium spp. | – | – | |
| 01 (1 An. evansae) CDCg | 1 (anthropic area) | P. vivax | – | – |
| 09 (1 An. cruzii) CDCc | P. vivax | – | – |
Note: Plasmodium-positive or negative (−) results for testing for parasite species presence in the sample.
Abbreviations: Sh, Shannon traps; CDCg, CDC trap on the ground; CDCc, CDC trap in the canopy; Asp, backpack aspirator.
Out of 15 anopheline samples from Laporta et al. (2015), we obtained two P. falciparum cytb sequences, one in An. cruzii (GenBank: MT779800) and another in Anopheles (Nyssorhynchus) strodei (GenBank: MT779801).
3.3. Rodent liver samples
One rodent species (Oligoryzomys cf. flavescens) was positive for P. falciparum using 18S and cytb protocols out of 25 rodent specimens tested (Supplementary Table S1).
3.4. Phylogenetic analyses
The phylogenetic relationships between the cytb sequences generated in this study (n = 7) and those available on GenBank are illustrated in Fig. 3. Sequences MF573300 (from An. cruzii) and MF573301 (from A. clamitans) were placed in a clade containing sequences of P. vivax and P. simium (Fig. 3). Sequences MT770753 and MF476105 obtained from An. cruzii clustered in clades containing sequences of P. chabaudi and P. berghei, respectively. Sequences MT779800 (from An. cruzii), MT779801 (from An. strodei), and MT779799 (from O. flavescens) were placed together in a clade containing sequences of P. falciparum (Fig. 3). Although the alignment comprised only 331 bp, most of the nodes of interest had high posterior probability values supporting the identification of the parasites found in this study.
Fig. 3.
A Bayesian phylogenetic hypothesis of Plasmodium spp. parasites infecting mammals and mosquitoes from the Capivari-Monos EPA, Brazil. We constructed a phylogenetic tree based on partial sequences of the cytb gene (31 sequences; 331 bp excluding gaps). Posterior probability values are shown above or below the branches. Plasmodium relictum and Plasmodium gallinaceum were used as the outgroup. In addition to parasite and host names, lineage identification (if available) and GenBank accession numbers are provided in parentheses for all sequences used in the analysis. Sequences colored in red and blue show similarities with human malaria parasites, P. vivax and P. falciparum, respectively. Sequences colored in green represent rodent malaria parasites, similar to P. berghei and P. chabaudi. The reference sequences in the clades containing colored sequences are highlighted in bold
4. Discussion
Our results indicate that P. vivax, P. malariae and P. falciparum are transmitted among non-human primates, primarily by An. cruzii, in the Atlantic tropical rainforest of Brazil. In particular, consistent with other studies, we found P. vivax and P. malariae in anopheline vector species and non-human primates (Duarte et al, 2008, 2013; Abreu et al., 2019). However, the presence of P. falciparum in An. cruzii in the sylvatic transmission cycle remains puzzling.
Our previous findings of the presence of P. falciparum in a sylvatic transmission cycle detected in An. cruzii and An. strodei (see Laporta et al., 2015) add to the current finding of P. falciparum in a rodent liver (Fig. 3). The presence of P. falciparum DNA in a rodent liver sample may represent an unsuccessful infection stopped at the liver stage because rodents are likely dead-end hosts for human malaria pathogens (Laporta et al., 2013). Although P. falciparum is considered eliminated from the Atlantic Forest, there seems to be transmission undetected by the traditional vector-borne disease surveillance methods. The lack of detection can be explained by low prevalence and focal circulation in a “post-elimination” phase. Subpatent or undetectable malaria transmission appears to be common in South America, e.g. Molina Gómez et al. (2017); Manrique et al. (2019). Exploring the possibility that P. falciparum remains undetected in these areas of Brazil is a matter that requires active surveillance and further investigations.
We can speculate that P. falciparum-infected (likely asymptomatic) humans could infect local vectors. Consistent with this scenario, a recent cross-sectional study carried out on humans living on the border of the Atlantic tropical rainforest region of Rio de Janeiro identified P. falciparum (0.3%), P. vivax (0.6%) and P. malariae (1.9%) in humans with malaria (Miguel et al., 2019). These authors also reported positive serological testing for P. falciparum (3.5%), P. vivax (7.7%) and P. malariae (30.9%). All thick blood smears were negative, indicating that the individuals had submicroscopic, asymptomatic infections. Overall, people who entered the forest were more likely to exhibit reactive serology (Miguel et al., 2019). These data corroborate evidence found in Espírito Santo and São Paulo states (Curado et al., 2006; Duarte et al., 2006; Cerutti et al., 2007). Thus, our findings are consistent with a scenario where infected asymptomatic individuals may be entering the forest environment frequently. However, it is difficult to detect these infectious individuals without carrying out a longitudinal study.
Alternatively, our findings are also consistent with non-human primates being competent hosts for P. falciparum (see Duarte et al., 2008; Monteiro et al., 2020). Plasmodium falciparum DNA was detected in two fecal samples of red howler monkeys (Alouatta seniculus) from Colombia (Rondón et al., 2019) and one Alouatta guariba clamitans from the Atlantic tropical rainforest (Duarte et al., 2008). Still, testing this hypothesis is logistically complicated even when using fecal samples (Rondón et al., 2019) because of the likely low prevalence of P. falciparum parasites in non-human primates (Duarte et al., 2008), if any. Although explaining negative results is particularly difficult, a case could be made that the complexities of detecting parasites with low frequency may explain why other studies have failed to detect P. falciparum in Brazilian non-human primates (Abreu et al., 2019). Altogether, the available data indicate a continuous forest cycle involving P. falciparum-infected zoophilic mosquitoes and that such event occurs with low frequency. Whether it results from asymptomatic human patients or non-human primates cannot be determined in this study. Nevertheless, a small fraction of the mosquito population is responsible for this residual transmission, challenging investigations. Longitudinal, long-term studies will be necessary to uncover the mechanisms that can maintain this putative silenced transmission. Whatever scenario is sustaining transmission, a P. falciparum malaria case at the forest border does not fit the case definition of imported malaria based on travel history (de Pina-Costa et al., 2014; Lorenz et al., 2015).
Our results increase the body of evidence supporting that humans introduced malarial parasites to the native non-human primate species that are now maintaining a forest transmission cycle in the Brazilian Atlantic Forest. Transference of human parasites to animals has been reported for other parasitic diseases (e.g. Cryptosporidium hominis, P. ovale wallikeri, strongylid nematodes) (Estrada-Peña et al., 2014; Hasegawa et al., 2014; Mapua et al., 2018; Pafčo et al., 2019). Reverse zoonosis of human P. vivax into simian P. simium in the past Brazilian colonization (de Oliveira et al., 2021) is the result of spillover of the human parasite to New World monkeys from European or African people who arrived at the Americas during the colonization period. The spillback of the simian lineage (P. simium) is recognized as zoonotic malaria (Brasil et al., 2017), an alternative hypothesis for malaria infections in the Atlantic Forest opposite to the classical case definition based on imported malaria vectorized by the local vector. Likewise, human P. malariae may have been affected by the same process in South America. Further supporting evidence of a complex human-non-human primate cycle is a higher genetic diversity found in the Brazilian Amazon and Atlantic Forest populations of P. brasilianum vs P. malariae (Guimarães et al., 2012; Lalremruata et al., 2015). This pattern suggests that the P. brasilianum populations could be the source of at least part of the P. malariae cases. It is worth noting that P. malariae has a broader distribution than P. vivax. Indeed, P. malariae has been detected in approximately 31 species of New World monkeys (de Alvarenga et al., 2017; Erkenswick et al., 2017; Rondón et al., 2019) from Costa Rica to Brazil. This broad host and geographic ranges are unique among primate malaria parasites. Reverse zoonosis of P. malariae in South America may have occurred before the colonization period because spillback of the simian lineage (P. brasilianum) has been considered to explain zoonotic malaria by P. malariae in humans across the continent and not only in the Atlantic Forest (Rondón et al., 2019).
In the case of the parasites similar to African rodent malaria agents (P. chabaudi and P. berghei) obtained from the An. cruzii pools, we can only speculate that this could be an unknown rodent parasite in South America whose relationship with the African species needs to be explored. Interestingly, we observed 18% of blood meals being taken from rodents by An. cruzii (68 samples with rodent DNA/373 engorged females) in the study area (Evangelista et al., unpublished observations). Evidence of a rodent-specific Plasmodium species has been previously found, i.e. Plasmodium spp. in capybaras (dos Santos et al., 2009). More data are needed to understand what these rodent parasites are.
Another layer of complexity in malaria dynamics is the anopheline vector composition in the Atlantic Forest. Although An. cruzii is the dominant vector, we observed An. strodei and An. evansae, together with An. cruzii, infected with human malaria on the forest edges. This diversity in the vector composition was previously observed (Duarte et al., 2013; Laporta et al., 2015). While An. cruzii is sylvatic, the other vectors can survive man-made changes in the natural ecosystem and proliferate in an anthropic environment (Forattini & Massad, 1998). The role of local vectors as “bridge vectors” of malaria parasites in the human environment cannot be neglected.
Overall, malaria transmission dynamics in the Brazilian Atlantic rainforest is consistent with a mosaic of cycles involving human malaria parasites being transmitted among local non-human primates. Such dynamics is maintained by vectors feeding upon a broad range of vertebrate hosts, in this case, the dominant vector An. cruzii combined with local vectors. Thus, the vector host range seems crucial to explain the proposed spillover-spillback process.
5. Conclusions
We tested the hypothesis of the transmission cycle of human malaria parasites (P. vivax, P. malariae and P. falciparum) involving non-human primates and anophelines in the Brazilian Atlantic Forest. Although the role of long-lasting asymptomatic infections in humans cannot be ruled out, particularly in the case of the P. falciparum infections, these results yield additional evidence indicating that non-human primates could act as reservoirs for human malaria. The evidence is clearer for P. vivax and P. malariae. These parasites have been found in both non-human primates and the dominant vector (An. cruzii). Furthermore, outbreaks that have been reported (Brasil et al., 2017) are more likely the result of spillback. Assessing whether spillbacks constitute significant risk for the reintroduction of malaria into the human population, particularly in urban areas, is a matter that requires longitudinal studies and scaling up molecular surveillance on the forest edges, the human-non-human primate interface.
Funding
This research was funded by the São Paulo Research Foundation (FAPESP) grant numbers (awarded to) 2014/10919-4 (AMRCD), 2014/50444-5 (MTM), 2014/26229-7 (MAMS) and 2014/09774-1 (GZL). The CNPq fellowship is granted to ACL (grant n. 302375/2020-1), MAMS (grant n. 301877/2016-5) and GZL (grant n. 307432/2019-0).
Ethical approval
All procedures were authorized by the Federal Environment Institute (ICMBio; n. 47812-4). Laboratory testing from simian blood and rodent liver samples were approved by the Tropical Medicine Institute Ethics Committee on the use of animals in research (IMTUSP; n. 260108–012.028/2013 and n. 000421A).
CRediT author statement
Ana Maria Ribeiro de Castro Duarte: Conceptualization, Validation, Formal analysis, Investigation, Data curation, Writing–original draft, Visualization, Supervision, Funding acquisition. Licia Natal Fernandes: Investigation, Data curation. Fabiana Santos Silva: Investigation, Data curation. Igor Lucoves Sicchi: Investigation, Data curation. Luis Filipe Mucci: Conceptualization, Methodology, Investigation. Izilda Curado: Conceptualization, Methodology. Aristides Fernandes: Investigation, Data curation. Antônio Ralph Medeiros-Sousa: Conceptualization, Methodology, Investigation. Walter Ceretti-Junior: Investigation, Data curation. Mauro Toledo Marrelli: Supervision, Funding acquisition. Eduardo Evangelista: Investigation, Data curation. Renildo Teixeira: Investigation, Data curation. Juliana Laurito Summa: Investigation, Project administration. Marcello Shiavo Nardi: Investigation, Project administration. Margoth Ramos Garnica: Investigation, Data curation. Ana Carolina Loss: Formal analysis, Investigation, Data curation. Julyana Cerqueira Buery: Investigation, Data curation. Crispim Cerutti Jr.: Conceptualization, Resources, Writing–review & editing. M. Andreína Pacheco: Phylogenetic analysis, Writing–review & editing. Ananias Alberto Escalante: Writing–review & editing, Supervision. Maria Anice Mureb Sallum: Writing - review & editing, Supervision. Gabriel Zorello Laporta: Conceptualization, Formal analysis, Investigation, Data curation, Writing - original draft, Visualization, Supervision, Funding acquisition.
Data availability
The sequences generated in this study were deposited in the GenBank database under the accession numbers MF573323 (18S) and MF573300, MF573301, MT770753, MF476105, MT779799, MT779800 and MT779801 (cytb).
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge the contributions of the following members of the São Paulo State SUCEN (Amanda Lumi Kawanami, Iole Arumi Sei, João Carlos do Nascimento, Paulo Frugoli dos Santos, Luis Milton Bonafé, Antônio Waldomiro de Oliveira, Gabriel Marcelino Neto and Luiz Sposito Jr.), Joyce Montes (JMBio), Thaysa Carolina Cantanhêde Figueiredo and Leila Matajs (Insituto Pedro Matajs), for their assistance with fieldwork planning and/or field collections; Almir Robson Ferreira (IMTUSP) for helping with scientific illustrations; Heitor Franco de Andrade Junior (IMTUSP) for assistance with data interpretation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crpvbd.2021.100032.
Contributor Information
Ana Maria Ribeiro de Castro Duarte, Email: amrcd2@gmail.com.
Gabriel Zorello Laporta, Email: gabriel.laporta@fmabc.br.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplementary Table S1.Testing of rodent liver samples for Plasmodium spp., Capivari-Monos EPA, Brazilian Atlantic rainforest, 2017–2018.
References
- Abreu F.V.S. de, Santos E.D., Mello A.R.L., Gomes L.R., Alvarenga D.A.M. de, Gomes M.Q., et al. Howler monkeys are the reservoir of malarial parasites causing zoonotic infections in the Atlantic forest of Rio de Janeiro. Plos Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo M.S., Messias M.R., Figueiró M.R., Gil L.H.S., Probst C.M., Vidal N.M., et al. Natural Plasmodium infection in monkeys in the state of Rondônia (Brazilian western Amazon) Malar. J. 2013;12:180. doi: 10.1186/1475-2875-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis G.M.P., Alvarenga D.A.M., Costa Pereira M.O., Sánchez-Arcila J.C., de Pina Costa A., Souza Junior J.C., et al. Profiling humoral immune response against pre-erythrocytic and erythrocytic antigens of malaria parasites among Neotropical primates in the Brazilian Atlantic Forest. Front. Cell. Infect. Microbiol. 2021;11:678996. doi: 10.3389/fcimb.2021.678996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickersmith S.A., Lainhart W., Moreno M., Chu V.M., Vinetz J.M., Conn J.E. A sensitive, specific and reproducible real-time polymerase chain reaction method for detection of Plasmodium vivax and Plasmodium falciparum infection in field-collected anophelines. Mem. Inst. Oswaldo Cruz. 2015;110:573–576. doi: 10.1590/0074-02760150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P., Zalis M.G., de Pina-Costa A., Siqueira A.M., Júnior C.B., Silva S., et al. Outbreak of human malaria caused by Plasmodium simium in the Atlantic forest in Rio de Janeiro: A molecular epidemiological investigation. Lancet Glob. Health. 2017;5:e1038–e1046. doi: 10.1016/S2214-109X(17)30333-9. [DOI] [PubMed] [Google Scholar]
- Carlos B.C., Rona L.D.P., Christophides G.K., Souza-Neto J.A. A comprehensive analysis of malaria transmission in Brazil. Pathog. Glob. Health. 2019;113:1–13. doi: 10.1080/20477724.2019.1581463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti C.Jr., Boulos M., Coutinho A.F., Hatab M., do C.L.D., Falqueto A., Rezende H.R., et al. Epidemiologic aspects of the malaria transmission cycle in an area of very low incidence in Brazil. Malar. J. 2007;6:33. doi: 10.1186/1475-2875-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coatney G.R. The simian malarias: Zoonoses, anthroponoses, or both? Am. J. Trop. Med. Hyg. 1971;20:795–803. doi: 10.4269/ajtmh.1971.20.795. [DOI] [PubMed] [Google Scholar]
- Curado I., Dos Santos Malafronte R., de Castro Duarte A.M.R., Kirchgatter K., Branquinho M.S., Bianchi Galati E.A. Malaria epidemiology in low-endemicity areas of the Atlantic forest in the Vale do Ribeira, São Paulo, Brazil. Acta Trop. 2006;100:54–62. doi: 10.1016/j.actatropica.2006.09.010. [DOI] [PubMed] [Google Scholar]
- de Alencar F.E.C., Malafronte R.D.S., Cerutti Junior C., Natal Fernandes L., Buery J.C., Fux B., et al. Assessment of asymptomatic Plasmodium spp. infection by detection of parasite DNA in residents of an extra-Amazonian region of Brazil. Malar. J. 2018;17:113. doi: 10.1186/s12936-018-2263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alvarenga D.A.M., de Pina-Costa A., Brasil P., de Brito C.F.A., Daniel-Ribeiro C.T. Malaria attack in southeastern Brazil: A probable locally acquired new infection. Infect. Ecol. Epidemiol. 2016;6:32308. doi: 10.3402/iee.v6.32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alvarenga D.A.M., Pina-Costa A., Bianco C., Moreira S.B., Brasil P., Pissinatti A., et al. New potential Plasmodium brasilianum hosts: Tamarin and marmoset monkeys (family Callitrichidae) Malar. J. 2017;16:71. doi: 10.1186/s12936-017-1724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira T.C., Rodrigues P.T., Early A.M., Duarte A.M.R.C., Buery J.C., Bueno M.G., et al. Plasmodium simium: Population genomics reveals the origin of a reverse zoonosis. J. Infect. Dis. 2021 (in press) doi: 10.1093/infdis/jiab214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pina-Costa A., Brasil P., Di Santi S.M., de Araujo M.P., Suárez-Mutis M.C., Santelli A.C.F. e S., et al. Malaria in Brazil: What happens outside the Amazonian endemic region. Mem. Inst. Oswaldo Cruz. 2014;109:618–633. doi: 10.1590/0074-0276140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demari-Silva B., Laporta G.Z., Oliveira T., Sallum M. Plasmodium infection in Kerteszia cruzii (Diptera: Culicidae) in the Atlantic tropical rain forest, southeastern Brazil. Infect. Genet. Evol. 2020;78:104061. doi: 10.1016/j.meegid.2019.104061. [DOI] [PubMed] [Google Scholar]
- DEPAVE/PMSP . 2012. Sao Paulo city case study: Re-introducing of howler monkey in the city goal and description of initiative.https://www.prefeitura.sp.gov.br/cidade/secretarias/upload/chamadas/saopaulocity_case_study_1334931581.pdf [Google Scholar]
- Duarte de A.M.R.C., Malafronte R., Dos S., Cerutti C., Curado I., de Paiva B.R., Maeda A.Y., et al. Natural Plasmodium infections in Brazilian wild monkeys: Reservoirs for human infections? Acta Trop. 2008;107:179–185. doi: 10.1016/j.actatropica.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Duarte A.M.R.C., Pereira D.M., de Paula M.B., Fernandes A., Urbinatti P.R., Ribeiro A.F., et al. Natural infection in anopheline species and its implications for autochthonous malaria in the Atlantic Forest in Brazil. Parasit. Vectors. 2013;6:58. doi: 10.1186/1756-3305-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte de A.M.R.C., Porto M.A.L., Curado I., Malafronte R.S., Hoffmann E.H.E., de Oliveira S.G., et al. Widespread occurrence of antibodies against circumsporozoite protein and against blood forms of Plasmodium vivax, P. falciparum and P. malariae in Brazilian wild monkeys. J. Med. Primatol. 2006;35:87–96. doi: 10.1111/j.1600-0684.2006.00148.x. [DOI] [PubMed] [Google Scholar]
- dos Santos L.C., Curotto S.M.R., de Moraes W., Cubas Z.S., Costa-Nascimento M. de J., de Barros Filho I.R., et al. Detection of Plasmodium sp. in capybara. Vet. Parasitol. 2009;163:148–151. doi: 10.1016/j.vetpar.2009.03.042. [DOI] [PubMed] [Google Scholar]
- Erkenswick G.A., Watsa M., Pacheco M.A., Escalante A.A., Parker P.G. Chronic Plasmodium brasilianum infections in wild Peruvian tamarins. PloS One. 2017;12 doi: 10.1371/journal.pone.0184504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A.A., Ayala F.J. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl. Acad. Sci. USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A.A., Pacheco M.A. Malaria molecular epidemiology: An evolutionary genetics perspective. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec. AME-0010-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Peña A., Ostfeld R.S., Peterson A.T., Poulin R., de la Fuente J. Effects of environmental change on zoonotic disease risk: An ecological primer. Trends Parasitol. 2014;30:205–214. doi: 10.1016/j.pt.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Ferreira M.U., Castro M.C. Challenges for malaria elimination in Brazil. Malar. J. 2016;15:284. doi: 10.1186/s12936-016-1335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forattini O.P., Massad E. Culicidae vectors and anthropic changes in a southern Brazil natural ecosystem. Ecosyst. Health. 1998;4:9–19. doi: 10.1046/j.1526-0992.1998.00067.x. [DOI] [Google Scholar]
- Forattini O.P. USP; São Paulo: 2002. Medical Culicidology: Identification, Biology, Epidemiology. 1. ed. [Google Scholar]
- Foster P.G., de Oliveira T.M.P., Bergo E.S., Conn J.E., Sant’Ana D.C., Nagaki S.S., et al. Phylogeny of Anophelinae using mitochondrial protein coding genes. R. Soc. Open Sci. 2017;4:170758. doi: 10.1098/rsos.170758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham P.C. Comments on biology of human malaria. Mil. Med. 1966;131(Suppl.):961–962. [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grignard L., Shah S., Chua T.H., William T., Drakeley C.J., Fornace K.M. Natural human infections with Plasmodium cynomolgi and other malaria species in an Elimination Setting in Sabah, Malaysia. J. Infect. Dis. 2019;220:1946–1949. doi: 10.1093/infdis/jiz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães L.O., Bajay M.M., Wunderlich G., Bueno M.G., Röhe F., Catão-Dias J.L., et al. The genetic diversity of Plasmodium malariae and Plasmodium brasilianum from human, simian and mosquito hosts in Brazil. Acta Trop. 2012;124:27–32. doi: 10.1016/j.actatropica.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Hartmeyer G.N., Stensvold C.R., Fabricius T., Marmolin E.S., Hoegh S.V., Nielsen H.V., et al. Plasmodium cynomolgi as cause of malaria in tourist to Southeast Asia, 2018. Emerg. Infect. Dis. 2019;25:1936–1939. doi: 10.3201/eid2510.190448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H., Modrý D., Kitagawa M., Shutt K.A., Todd A., Kalousová B., et al. Humans and great apes cohabiting the forest ecosystem in Central African Republic harbour the same hookworms. PloS Negl. Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy D.A., Feng X., Mu J., Furuya T., Chotivanich K., Krettli A.U., et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- Krief S., Escalante A.A., Pacheco M.A., Mugisha L., André C., Halbwax M., et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalremruata A., Magris M., Vivas-Martínez S., Koehler M., Esen M., Kempaiah P., et al. Natural infection of Plasmodium brasilianum in humans: Man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine. 2015;2:1186–1192. doi: 10.1016/j.ebiom.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporta G.Z. Spotlight on Plasmodium falciparum evolutionary system in the southeastern Atlantic forest. Biota Neotrop. 2017;17 doi: 10.1590/1676-0611-bn-2016-0314. [DOI] [Google Scholar]
- Laporta G.Z., Burattini M.N., Levy D., Fukuya L.A., de Oliveira T.M.P., Maselli L.M.F., et al. Plasmodium falciparum in the southeastern Atlantic forest: A challenge to the bromeliad-malaria paradigm? Malar. J. 2015;14:181. doi: 10.1186/s12936-015-0680-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporta G.Z., Lopez de Prado P.I.K., Kraenkel R.A., Coutinho R.M., Sallum M.A.M. Biodiversity can help prevent malaria outbreaks in tropical forests. Plos Negl. Trop. Dis. 2013;7:e2139. doi: 10.1371/journal.pntd.0002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C., Virginio F., Aguiar B.S., Suesdek L., Chiaravalloti-Neto F. Spatial and temporal epidemiology of malaria in extra-Amazonian regions of Brazil. Malar. J. 2015;14 doi: 10.1186/s12936-015-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy D.E., Liu W., Li Y., Learn G.H., Plenderleith L.J., Sundararaman S.A., et al. Out of Africa: Origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int. J. Parasitol. 2017;47:87–97. doi: 10.1016/j.ijpara.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique P., Miranda-Alban J., Alarcon-Baldeon J., Ramirez R., Carrasco-Escobar G., Herrera H., et al. Microsatellite analysis reveals connectivity among geographically distant transmission zones of Plasmodium vivax in the Peruvian Amazon: A critical barrier to regional malaria elimination. PloS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapua M.I., Fuehrer H.-P., Petrželková K.J., Todd A., Noedl H., Qablan M.A., Modrý D. Plasmodium ovale wallikeri in western lowland gorillas and humans, Central African Republic. Emerg. Infect. Dis. 2018;24:1581–1583. doi: 10.3201/eid2408.180010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques T.C., Bourke B.P., Laporta G.Z., Sallum M.A.M. Mosquito (Diptera: Culicidae) assemblages associated with Nidularium and Vriesea bromeliads in Serra do Mar, Atlantic forest, Brazil. Parasit. Vectors. 2012;5:41. doi: 10.1186/1756-3305-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maselli L.M.F., Levy D., Laporta G.Z., Monteiro A.M., Fukuya L.A., Ferreira-da-Cruz M.F., et al. Detection of Plasmodium falciparum and Plasmodium vivax subclinical infection in non-endemic region: Implications for blood transfusion and malaria epidemiology. Malar. J. 2014;13:224. doi: 10.1186/1475-2875-13-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros-Sousa A.R., de Oliveira Christe R., de Castro Duarte A.M.R., Mucci L.F., Ceretti-Junior W., Marrelli M.T. Effects of anthropogenic landscape changes on the abundance and acrodendrophily of Anopheles (Kerteszia) cruzii, the main vector of malaria parasites in the Atlantic Forest in Brazil. Malar. J. 2019;18:110. doi: 10.1186/s12936-019-2744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros-Sousa A.R., Laporta G.Z., Coutinho R.M., Mucci L.F., Marrelli M.T. A mathematical model for zoonotic transmission of malaria in the Atlantic Forest: Exploring the effects of variations in vector abundance and acrodendrophily. Plos Negl. Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0008736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel R.B., Albuquerque H.G., Sanchez M.C.A., Coura J.R., Santos S. da S., Silva S. da, et al. Asymptomatic Plasmodium infection in a residual malaria transmission area in the Atlantic Forest region: Implications for elimination. Rev. Soc. Bras. Med. Trop. 2019;52 doi: 10.1590/0037-8682-0537-2018. [DOI] [PubMed] [Google Scholar]
- Molina Gómez K., Caicedo M.A., Gaitán A., Herrera-Varela M., Arce M.I., Vallejo A.F., et al. Characterizing the malaria rural-to-urban transmission interface: The importance of reactive case detection. Plos Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Cruz A., Zilversmit M.M., Neafsey D.E., Hartl D.L., Barillas-Mury C. Mosquito vectors and the globalization of Plasmodium falciparum malaria. Annu. Rev. Genet. 2016;50:447–465. doi: 10.1146/annurev-genet-120215-035211. [DOI] [PubMed] [Google Scholar]
- Monteiro E.F., Fernandez-Becerra C., Araujo M., Messias M.R., Ozaki L.S., Duarte A., et al. Naturally acquired humoral immunity against malaria parasites in non-human primates from the Brazilian Amazon, Cerrado and Atlantic Forest. Pathogens. 2020;9:525. doi: 10.3390/pathogens9070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlenbein M.P., Pacheco M.A., Taylor J.E., Prall S.P., Ambu L., Nathan S., et al. Accelerated diversification of nonhuman primate malarias in Southeast Asia: Adaptive radiation or geographic speciation? Mol. Biol. Evol. 2015;32:422–439. doi: 10.1093/molbev/msu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multini L.C., Wilke A.B.B., Marrelli M.T. Neotropical Anopheles (Kerteszia) mosquitoes associated with bromeliad-malaria transmission in a changing world. Acta Trop. 2020;205:105413. doi: 10.1016/j.actatropica.2020.105413. [DOI] [PubMed] [Google Scholar]
- Otto T.D., Gilabert A., Crellen T., Böhme U., Arnathau C., Sanders M., et al. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat. Microbiol. 2018;3:687–697. doi: 10.1038/s41564-018-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pafčo B., Kreisinger J., Čížková D., Pšenková-Profousová I., Shutt-Phillips K., Todd A., et al. Genetic diversity of primate strongylid nematodes: Do sympatric nonhuman primates and humans share their strongylid worms? Mol. Ecol. 2019;28:4786–4797. doi: 10.1111/mec.15257. [DOI] [PubMed] [Google Scholar]
- Ribeiro M.C., Metzger J.P., Martensen A.C., Ponzoni F.J., Hirota M.M. The Brazilian Atlantic forest: How much is left, and how is the remaining forest distributed? Implications for conservation. Biol. Conserv. 2009;142:1141–1153. doi: 10.1016/j.biocon.2009.02.021. [DOI] [Google Scholar]
- Rodrigues P.T., Valdivia H.O., de Oliveira T.C., Alves J.M.P., Duarte A.M.R.C., Cerutti-Junior C., et al. Human migration and the spread of malaria parasites to the New World. Sci. Rep. 2018;8:1993. doi: 10.1038/s41598-018-19554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolim G. de S., Camargo M.B.P. de, Lania D.G., Moraes J.F.L. de. Classificação climática de Köppen e de Thornthwaite e sua aplicabilidade na determinação de zonas agroclimáticas para o estado de são Paulo. Bragantia. 2007;66:711–720. doi: 10.1590/S0006-87052007000400022. [DOI] [Google Scholar]
- Rondón S., León C., Link A., González C. Prevalence of Plasmodium parasites in non-human primates and mosquitoes in areas with different degrees of fragmentation in Colombia. Malar. J. 2019;18:276. doi: 10.1186/s12936-019-2910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rougemont M., Van Saanen M., Sahli R., Hinrikson H.P., Bille J., Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J. Clin. Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge G.G., Böhme U., Sanders M., Reid A.J., Cotton J.A., Maiga-Ascofare O., et al. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature. 2017;542:101–104. doi: 10.1038/nature21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallum M.A.M., Conn J.E., Bergo E.S., Laporta G.Z., Chaves L.S.M., Bickersmith S.A., et al. Vector competence, vectorial capacity of Nyssorhynchus darlingi and the basic reproduction number of Plasmodium vivax in agricultural settlements in the Amazonian Region of Brazil. Malar. J. 2019;18:117. doi: 10.1186/s12936-019-2753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallum M.A.M., Daniel-Ribeiro C.T., Laporta G.Z., Ferreira-da-Cruz M. de F., Maselli L.M.F., Levy D., Bydlowski S.P. Finding connections in the unexpected detection of Plasmodium vivax and Plasmodium falciparum DNA in asymptomatic blood donors: A fact in the Atlantic forest. Malar. J. 2014;13:337. doi: 10.1186/1475-2875-13-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 2013;26:165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siregar J.E., Faust C.L., Murdiyarso L.S., Rosmanah L., Saepuloh U., Dobson A.P., Iskandriati D. Non-invasive surveillance for Plasmodium in reservoir macaque species. Malar. J. 2015;14:404. doi: 10.1186/s12936-015-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.F., Patton J.L. The diversification of South American murid rodents: Evidence from mitochondrial DNA sequence data for the akodontine tribe. Biol. J. Linn. Soc. 1993;50:149–177. doi: 10.1111/j.1095-8312.1993.tb00924.x. [DOI] [Google Scholar]
- SVMA/PMSP . 2011. Plano de manejo – APA Capivari Monos.https://www.prefeitura.sp.gov.br/cidade/secretarias/meio_ambiente/publicacoes_svma/index.php?p=26341 [Google Scholar]
- Tanabe K., Mita T., Jombart T., Eriksson A., Horibe S., Palacpac N., et al. Plasmodium falciparum accompanied the human expansion out of Africa. Curr. Biol. 2010;20:1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva: 2020. World malaria report 2020.https://www.who.int/publications/i/item/9789240015791 [Google Scholar]
- Yakob L., Lloyd A.L., Kao R.R., Ferguson H.M., Brock P.M., Drakeley C., Bonsall M.B. Plasmodium knowlesi invasion following spread by infected mosquitoes, macaques and humans. Parasitology. 2018;145:101–110. doi: 10.1017/S0031182016002456. [DOI] [PubMed] [Google Scholar]
- Yusof R., Ahmed M.A., Jelip J., Ngian H.U., Mustakim S., Hussin H.M., et al. Phylogeographic evidence for 2 genetically distinct zoonotic Plasmodium knowlesi parasites, Malaysia. Emerg. Infect. Dis. 2016;22:1371–1380. doi: 10.3201/eid2208.151885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1.Testing of rodent liver samples for Plasmodium spp., Capivari-Monos EPA, Brazilian Atlantic rainforest, 2017–2018.
Data Availability Statement
The sequences generated in this study were deposited in the GenBank database under the accession numbers MF573323 (18S) and MF573300, MF573301, MT770753, MF476105, MT779799, MT779800 and MT779801 (cytb).