Abstract
Monitoring for lamivudine (3TC) resistance is important both for the clinical management of human immunodeficiency virus type 1 (HIV-1)-infected patients treated with 3TC and for surveillance of transmission of 3TC-resistant HIV-1. We developed a novel non-culture-based assay for the rapid analysis of phenotypic resistance to 3TC of HIV-1 in plasma. The assay measures the susceptibility of HIV-1 reverse transcriptase (RT) activity to 3TC triphosphate (3TC-TP) in plasma. RT detection was done by the Amp-RT assay, an ultrasensitive PCR-based RT assay. Under our assay conditions, we found that 5 μM 3TC-TP inhibited RT activity from wild-type (WT), zidovudine-resistant, or nevirapine-resistant HIV-1 but not from HIV-1 carrying either the M184V mutation or multidrug (MD) resistance mutations (77L/116Y/151M or 62V/75I/77L/116Y/151M). Mixing experiments showed a detection threshold of 10% 3TC-resistant virus (M184V) in a background of WT HIV-1. To validate the assay for the detection of phenotypic resistance of HIV-1 to 3TC in plasma samples, HIV-1 RT in 30 plasma specimens collected from 15 patients before and during therapy with 3TC was tested for evidence of phenotypic resistance by the Amp-RT assay. The results were compared with those of genotypic analysis. The RT in 12 samples was found to be 3TC sensitive, while the RT in 18 samples had evidence of phenotypic resistance. All 12 samples with 3TC-sensitive RT had WT genotypes at codon 184 and were retrieved before treatment with 3TC. In contrast, all 18 specimens with 3TC-resistant RT were posttherapy samples. This assay provides a simple, rapid, and reliable method for the detection of phenotypic resistance of HIV-1 to 3TC in plasma.
Lamivudine [3TC; (−)-β-2′,3′-dideoxy-3′-thiacytidine] is one of several nucleoside analogs that are currently approved for the treatment of human immunodeficiency virus type 1 (HIV-1) infections (5). 3TC has potent anti-HIV-1 activity and minimal toxicity, and its triphosphate (3TC-TP) inhibits HIV-1 reverse transcriptase (RT) by acting as a competitive inhibitor of 2′-deoxycytidine-5′-triphosphate (dCTP) and as a chain terminator (1). 3TC is one of the most commonly used drugs in combination therapy as first-line treatment for HIV-1-infected patients (4, 5). 3TC administered in combination with zidovudine (AZT) and protease inhibitors slows the progression of HIV-1 disease and reduces levels of HIV-1 RNA to less than 500 copies per ml in 90% of patients for as long as 1 year (13).
The use of 3TC in both monotherapy or combination therapy, however, has resulted in the emergence of 3TC-resistant variants of HIV-1 (13, 21, 33, 40). This resistance is conferred by mutations at codon 184 of the HIV-1 RT gene, in which the wild-type (WT) methionine (M; ATG) residue is replaced with either a valine (V; GTG) or an isoleucine (I; ATA) residue (3, 31, 38). The presence of the M184V mutation has been associated with a >500-fold resistance to 3TC and with the loss of the antiretroviral and clinical benefits of 3TC (41).
It is therefore important to monitor HIV-1 for 3TC resistance in patients treated with 3TC. Phenotypic assays provide definitive information on resistance to 3TC and are well suited for assessments of the complex resistance patterns that may arise from combination therapy. However, most phenotypic assays developed to date are based on virus isolation and culture and are therefore labor intensive, costly, and unsuitable for rapid clinical monitoring or surveillance of drug resistance. In addition, these assays are fraught with biologic variabilities, including those related to viral isolation and tropism (23, 25). To circumvent the problem of virus isolation and tropism, recombinant virus assays in which an infectious virus is generated by recombination of patient-derived RT sequences with an RT-deleted HIV-1 backbone were developed (16, 22). However, these improved assays still require 2 to 3 weeks and may not be easily adapted to clinical laboratories.
In the absence of rapid phenotypic assays, several genotypic tests are being used to monitor for the presence of resistance mediated by the M184V mutation (21, 33, 37). However, clinical monitoring of 3TC resistance by genotypic testing may not detect resistance mediated by unrecognized mutations. In addition, genotypic testing cannot detect potential synergistic or antagonistic effects of complex mutation patterns arising from combination therapy with different RT inhibitors. The transient suppression of phenotypic resistance to AZT conferred by the M184V or the L74V mutation illustrates the effect that combinations of mutations may have in a given phenotype (26, 36, 38).
In this report, we describe the development and application of a rapid non-culture-based assay for the analysis of phenotypic resistance of HIV-1 to 3TC in plasma samples. The assay is based on the direct analysis of the susceptibility of plasma HIV-1 RT to inhibition by 3TC-TP. We describe the ability of the assay to successfully detect the phenotypic resistance of HIV-1 to 3TC in plasma samples from 3TC-treated persons. We also identify resistance to 3TC in HIV-1 RT carrying mutations associated with resistance to multiple nucleoside analogs (multidrug [MD] resistance).
MATERIALS AND METHODS
Principle of the phenotypic analysis of 3TC resistance.
The phenotypic assay is based on the analysis of the susceptibility of the RT activity of HIV-1 from plasma to inhibition by 3TC-TP. RT activity in plasma is detected by the Amp-RT assay, an ultrasensitive PCR-based RT assay (12, 14, 43). The susceptibility of the RT activity in plasma to 3TC-TP is determined on the basis of the level of inhibition produced by 3TC-TP and is measured by running quantitative Amp-RT reactions in the presence and absence of 3TC-TP.
RT analysis by the Amp-RT assay.
Amp-RT detects RT activity by using a known nonretroviral heteropolymeric RNA template and a complementary DNA oligonucleotide primer. The cDNA is detected by PCR amplification and probing with an internal oligonucleotide (12, 14, 43).
For detection of RT activity in culture supernatant, 10 μl of the supernatant was used directly in the Amp-RT assay. For testing of plasma, a volume of 100 μl was clarified by centrifugation at 10,000 × g for 5 min and was then ultracentrifuged at a fixed angle at 99,000 × g for 1 h at 4°C. The viral pellet was resuspended in 100 μl of RT buffer (50 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2), and aliquots of 2 to 10 μl were used for analysis by the Amp-RT assay.
RT levels were quantitated by enzyme-linked immunosorbent assay, with a standard curve generated with known units of RT activity from a reference HIV-1 stock (12). The Amp-RT signals were expressed as units of RT activity per milliliter and reflect the average of duplicate or triplicate results. Qualitative detection of the Amp-RT products was done by Southern blot hybridization as described previously (12).
Analysis of phenotypic resistance to 3TC by the Amp-RT assay.
For RT detection and analysis of phenotypic resistance to 3TC, samples (10 μl of culture supernatant or virus pellets from 2 to 10 μl of plasma) were applied to an RT buffer containing 10 ng of encephalomyocarditis virus RNA template, 10 U of RNasin, 100 ng of 5′-biotin-labeled EMCR2 antisense primer, 20 μM (each) dATP, dGTP, and dTTP, and 5 μM dCTP (12). To determine the susceptibility of the RT to 3TC-TP, an additional Amp-RT reaction was done in the presence of 3TC-TP. Reactions were incubated at 37°C for 2 h and then heated at 95°C for 5 min to destroy RT activity. PCR amplification of encephalomyocarditis virus cDNA was made as described previously (12) after the addition of each deoxynucleoside triphosphate at a concentration of 200 μM. Quantitative detection of the products obtained by the Amp-RT assay was done as described above.
Susceptibility of HIV-1 RT to 3TC-TP was determined from the level of inhibition of RT activity by 3TC-TP. We calculated the percentage of inhibition by using the ratio of the RT level obtained in the Amp-RT reactions containing 3TC-TP to the RT level seen in Amp-RT reactions done in the absence of 3TC-TP multiplying that ratio by 100. The drug concentrations resulting in 50 and 90% inhibition (IC50 and IC90, respectively) were also determined by testing RT in the presence of several 3TC-TP concentrations. These values were calculated by the method of Chou and Talalay (6).
Detection of 3TC resistance mutations.
Mutations at codon 184 of HIV-1 RT were detected by using the HIV-1 Line probe assay (LiPA), which detects both WT Met and mutant Val (37).
Study population.
A total of 30 EDTA-anticoagulated plasma samples from 15 HIV-1-infected patients from the Veteran Affairs Medical Center in Decatur, Ga., were studied. The samples were collected from patients before and during antiretroviral therapy with 3TC. The antiretroviral therapy histories for all patients are presented in Table 1. The Amp-RT-based phenotypic assay was done under code with respect to the date of serial bleeding and RT genotype. One plasma specimen from a blood donor who tested antibody negative for HIV-1, HIV-2, human T-cell leukemia virus type I (HTLV-I), and HTLV-II was used in the assay as a negative control.
TABLE 1.
Direct analysis of phenotypic resistance of HIV-1 RT activity to 3TC in plasma from 15 HIV-1-infected persons by determination of levels of RT inhibition by 3TC-TPa
| Sample | Week | Treatment | RT activity (U/ml)
|
% Inhibition of RT | Mutations
|
||
|---|---|---|---|---|---|---|---|
| No 3TC-TP | With 3TC-TP (5 μM) | Codon 184 | Other | ||||
| 1B | Preb | AZT-ddC | 1.7 × 10−9 | NDc | 100 | M184 | none |
| 1E | 12 | AZT-3TC | 2.1 × 10−6 | 5.1 × 10−7 | 75.6 | V/M184 | L41, Y215 |
| 1F | 18 | AZT-3TC | 1.3 × 10−7 | 3.3 × 10−8 | 75.9 | V184 | none |
| 2B | Pre | AZT-ddC | 2.2 × 10−6 | 6.3 × 10−9 | 99.7 | M184 | none |
| 2F | 12 | AZT-3TC | 1.5 × 10−6 | 4.7 × 10−7 | 68 | V184 | none |
| 3B | Pre | AZT | 1.8 × 10−6 | 1.7 × 10−8 | 99.4 | M184 | none |
| 3F | 12 | AZT-3TC | 1.6 × 10−8 | 1.2 × 10−9 | 92.3 | M/V184 | none |
| 3G | 21 | AZT-3TC | 5.1 × 10−9 | 1.0 × 10−8 | 0 | V184 | none |
| 4A | Pre | AZT | 1.6 × 10−7 | 1.2 × 10−10 | 99.9 | M184 | R70 |
| 4B | 12 | AZT-3TC | ND | ND | V184 | R70 | |
| 5A | Pre | AZT | 1.2 × 10−8 | ND | 100 | M184 | Y215 |
| 5B | 4 | AZT-3TC | ND | ND | M/V184 | Y215 | |
| 5C | 10 | AZT-3TC | ND | ND | V184 | Y215 | |
| 6A | Basal | Basal | 7.7 × 10−5 | 3.6 × 10−8 | 99.9 | M184 | none |
| 6B | 12 | AZT-3TC | 2.3 × 10−5 | 1.5 × 10−5 | 34.7 | V184 | none |
| 7A | Basal | Basal | 1.2 × 10−8 | ND | 100 | M/I184 | none |
| 7C | 28 | AZT-3TC | ND | ND | V184 | none | |
| 8A | Pre | AZT | 4.4 × 10−5 | 5.7 × 10−8 | 99.9 | M184 | L41, Y215 |
| 8B | 8 | AZT-3TC | 5.0 × 10−5 | 4.3 × 10−5 | 15 | V184 | L41, Y215 |
| 9A | Pre | d4Td | 2.9 × 10−6 | 6.3 × 10−8 | 97.8 | M184 | L41, Y215 |
| 9F | 12 | AZT-3TC | 2.1 × 10−6 | 1.1 × 10−6 | 47 | V184 | L41, Y215 |
| 10A | 4 | AZT-3TC | 8.9 × 10−5 | 9.1 × 10−6 | 87.8 | M184 | none |
| 10C | 12 | AZT-3TC | 1.9 × 10−7 | 1.9 × 10−7 | 0 | M/V184 | none |
| 10G | 52 | AZT-3TC | 2.6 × 10−5 | 3.5 × 10−5 | 0 | V184 | L41, Y215 |
| 11A | 12 | AZT-3TC | 1.9 × 10−7 | 1.3 × 10−7 | 29.1 | V/M184 | R70 |
| 11C | 36 | AZT-3TC | 1.7 × 10−7 | 7.0 × 10−8 | 58.5 | V184 | R70 |
| 12A | Pre | AZT | 2.5 × 10−5 | 9.2 × 10−7 | 96.3 | M184 | none |
| 12E | 28 | AZT-3TC | 1.7 × 10−6 | 5.1 × 10−7 | 70 | V184 | R70, Y215 |
| 13A | Pre | d4T | 1.5 × 10−6 | 6.2 × 10−8 | 95.9 | M184 | N69, R70 |
| 13F | 60 | d4T-3TC-INDe | 6.3 × 10−5 | 7.4 × 10−5 | 0 | V184 | L41, D/N69, R70 |
| 14A | 1 | AZT-3TC | 2.5 × 10−7 | 1.3 × 10−8 | 94.7 | M184 | L41, D69, R70, Y215 |
| 14F | 44 | IND-3TC | 2.9 × 10−9 | 5.1 × 10−9 | 0 | V184 | D69, R70, Y215 |
| 15A | Pre | AZT | 8.9 × 10−5 | 4.0 × 10−6 | 95.5 | M184 | D69 |
| 15D | 40 | AZT-3TC-IND | 1.6 × 10−10 | 2.4 × 10−10 | 0 | V184 | D69, Y215 |
Previous treatments were as follows: patient 9 was on AZT before the first sample was obtained, patient 13 was on AZT-ddC before the first sample was obtained, and patient 14 was on ddI for a period of time between the times that the first and second samples were obtained.
Pre, pretherapy.
ND, not detected.
D4T, stavudine.
IND, indinavir.
Viruses and 3TC-TP.
For assay development and validation, HIV-1 molecular infectious clones xxBRUpitt and M184Vpitt were used as WT and 3TC-resistant (M184V mutation) HIV-1 reference strains, respectively (31). Other reference viruses included M184V/Y181CEU, 181Y/CEU, and HIV-1RTMC/MT-2, which represented 3TC- and nevirapine-resistant (M184V/Y181C), nevirapine-resistant (Y181C), and AZT-resistant (D67N/K70R/T215F/K219Q) HIV-1 isolates, respectively (24). WT HIV-1SUM9 and multiple dideoxynucleoside-resistant isolates HIV-1SUM8 (Q151M mutation), HIV-1SUM12 (F77L/F116Y/Q151M), and HIV-1SUM13 (A62V/V75I/F77L/F116Y/Q151M) were kindly provided by H. Mitsuya (35).
3TC-TP was synthesized by the method of Ludwig and Eckstein (28). The crude 3TC-TP was purified by fast-performance liquid chromatography with HiLoad 26/10, a Q Sepharose Fast Flow Pharmacia column, and a gradient of TEAB (triazol ammonium bicarbonate) buffer (pH 7.0). The compound was characterized by UV, proton, and phosphorous nuclear magnetic resonance imaging, mass spectroscopy, and high-pressure chromatography. The concentration of 3TC-TP resulting in 50% inhibition of incorporation of [3H]dCTP into an (rI)n-dC12–18 template primer by recombinant p66/p51 HIV-1 RT (Biotechnology General, Rehovot, Israel) was 1.3 μM, as determined by the decrease in the formation of acid-insoluble product compared to the amount formed by the untreated control (10, 30).
RESULTS
Test conditions that differentiate WT and 3TC-resistant HIV-1 RTs.
To determine the optimal ratio of 3TC-TP and dCTP needed to inhibit WT but not 3TC-resistant RT, we tested RT activity from WT (xxBRUpitt) and 3TC-resistant (M184Vpitt) HIV-1 molecular clones in the presence of increasing concentrations of 3TC-TP (from 0.1 to 10 μM) and a fixed concentration of 5 μM dCTP. Since previous reports have shown that augmented chain termination by 3TC-TP is observed with decreasing concentrations of both 3TC-TP and dCTP, we used a low concentration of dCTP in the RT step (2). We selected the dCTP concentration of 5 μM because it was found to be the lowest concentration that did not compromise the performance of the Amp-RT assay (data not shown), which previously was done with excess dCTP (12).
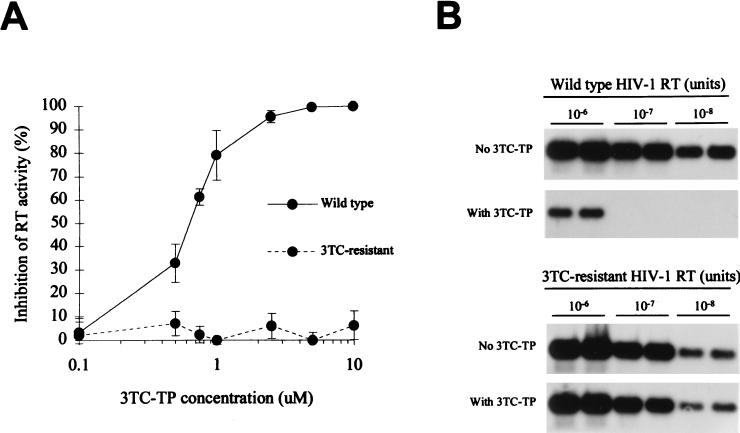
Figure 1A illustrates the inhibition seen with 10−7 U of RT activity, in which complete inhibition of RT activity from WT but not from 3TC-resistant virus was accomplished with 5 μM 3TC-TP. This concentration of 3TC-TP resulted in the inhibition of 10−7 and 10−8 U of RT activity from WT HIV-1, which are equivalent to 105 and 104 HIV-1 particles of the reference virus per ml, respectively (Fig. 1B) (12). With a higher input of RT (10−6 U of RT activity; the equivalent to 106 HIV-1 particles of the reference virus per ml), these conditions did not result in complete inhibition. The residual RT activity in the Amp-RT reaction containing 3TC-TP was found to be 0.4% of the Amp-RT signal from the control reaction that had no 3TC-TP. This reduction in RT signal is equivalent to a 2.35 log10 drop in virus load as detected by the Amp-RT assay. No significant inhibition of the 3TC-resistant HIV-1 was seen when it was tested with either a high or a low input of RT, demonstrating the ability of the assay to distinguish between WT and 3TC-resistant RTs within a wide range of RT levels (Fig. 1B). On the basis of these results, we adopted as a primary screening assay in all subsequent tests for the detection of 3TC resistance the Amp-RT assay conditions in which 5 μM 3TC-TP and 5 μm dCTP were used, unless otherwise indicated.
FIG. 1.
Detection of phenotypic resistance to 3TC by measuring levels of RT inhibition by the Amp-RT assay. (A) Inhibition of 10−7 U of RT activity from WT (xxBRUpitt) and 3TC-resistant (M184Vpitt) HIV-1 by different concentrations of 3TC-TP. (B) Inhibition of RT activity (from 10−6 to 10−8 U) from WT (xxBRUpitt) and 3TC-resistant (M184Vpitt) HIV-1 by 5 μM 3TC-TP.
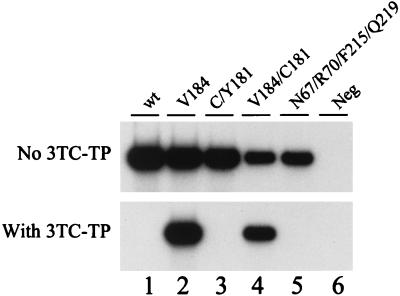
To demonstrate that the Amp-RT-based phenotypic assay is specific for the detection of 3TC resistance, we tested several HIV-1 reference clones with well-characterized phenotypic resistance to nucleoside and nonnucleoside RT inhibitors. Figure 2 shows that resistance to 3TC was seen only with RTs carrying the M184V mutation. As expected, HIV-1 RTs carrying AZT (D67N, K70R, T215F, K219Q) or nevirapine (Y181C) resistance mutations were all found to be susceptible to 3TC-TP. These results confirm that the assay was specific for viruses with phenotypic resistance to 3TC and indicate that the presence of other mutations associated with AZT and nevirapine resistance does not affect the inhibition of RT activity by 3TC-TP.
FIG. 2.
Analysis of the specificity of the Amp-RT-based phenotypic assay for detection of 3TC resistance by testing RTs from several HIV-1 reference clones in the presence and absence of 5 μM 3TC-TP. Lane 1, WT clone xxBRUpitt; lane 2, 3TC-resistant clone M184Vpitt; lane 3, nevirapine-resistant clone 181C/YEU; lane 4, 3TC- and nevirapine-resistant clone M184V/Y181CEU; lane 5, AZT-resistant clone HIV-1RTMC/MT-2; lane 6, water control (Neg, negative).
Detection threshold of 3TC resistance in mixtures of WT and 3TC-resistant viruses.
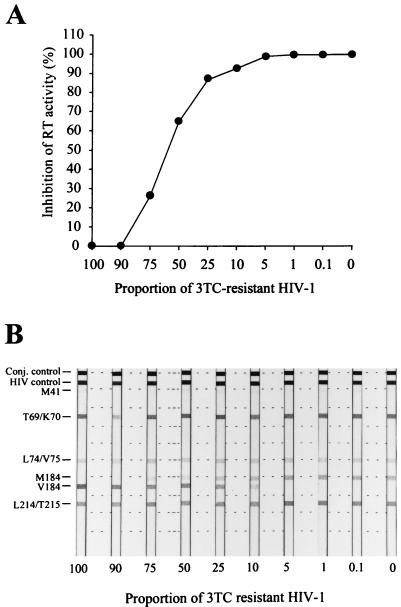
To determine the detection threshold of the Amp-RT-based assay, we tested mixtures of WT (xxBRUpitt) and 3TC-resistant (M184Vpitt) virus for evidence of 3TC resistance. The reference viruses were adjusted so that they had similar levels of RT activity before virus mixtures were prepared. We compared the level of inhibition of RT activity by 3TC-TP observed in each mixture with the proportion of 3TC-resistant virus used. The assay detection threshold was found to be 10% 3TC-resistant viruses in a background of WT HIV-1 (Fig. 3A). A good correlation between the proportion of virus carrying the M184V mutation and the level of inhibition was also observed. For instance, in mixtures containing 25 or 75% 3TC-resistant virus, the observed inhibition was 87 and 26%, respectively, which very likely represents the signals from the 3TC-resistant RT and therefore suggests that only WT RT activity was inhibited.
FIG. 3.
Evaluation of the detection threshold of 3TC resistance by the Amp-RT assay and comparison with the genotypic analysis at codon 184 by the HIV-1 LiPA. (A) Levels of RT inhibition by 3TC-TP (5 μM) in mixtures of WT clone xxBRUpitt and 3TC-resistant HIV-1 clone M184Vpitt. (B) Genotypic detection of the M184V mutation in the same mixtures by the HIV-1 LiPA. Conj. control, conjugate control.
We also used the same mixtures to compare the detection threshold of the Amp-RT-based phenotypic assay with the genotypic detection threshold for viruses carrying the M184V mutation by the HIV-1 LiPA (Fig. 3B). The detection threshold for 3TC-resistant virus by the LiPA was also 10%, indicating that both assays can reliably detect low levels of either genotypic or phenotypic resistance to 3TC. However, the signal intensities in mixtures containing 50% WT and 50% 3TC-resistant viruses were not similar in the LiPA (Fig. 3B). This may be due to different levels of HIV-1 RNA in both reference viruses resulting from adjustment of virus RT activity rather than adjustment of RNA levels or to different efficiencies in the hybridization of the WT- or 184V-specific probes.
MD resistance mutations confer phenotypic resistance to 3TC.
To determine if mutations other than 184V confer resistance to 3TC, we analyzed HIV-1 clones containing mutations associated with resistance to several dideoxynucleoside analogs. The IC50s and IC90s of 3TC for viruses containing one (Q151M; HIV-1SUM 8), three (F77L/F116Y/Q151M; HIV-1SUM 12), and all five (A62V/V75I/F77L/F116Y/Q151M; HIV-1SUM 13) mutations associated with MD resistance were determined by the Amp-RT assay. Control WT HIV-1 RTs were also tested.
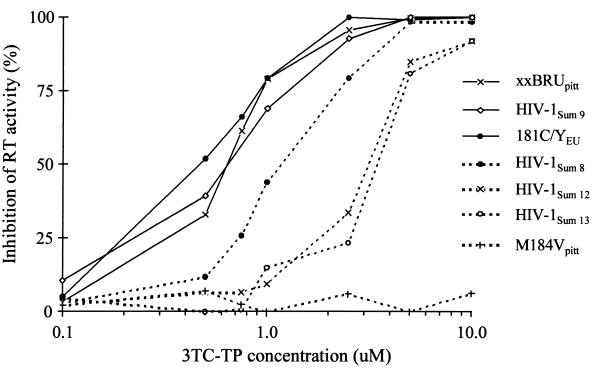
The RT from HIV-1 carrying the Q151M mutation had a slightly reduced susceptibility to 3TC, with IC50s and IC90s approximately twofold higher than those for WT reference viruses (Fig. 4 and Table 2). However, the presence of additional MD resistance mutations resulted in higher levels of resistance to 3TC, with an increase in IC50s of about six- and eightfold for virus with three or all five MD resistance mutations, respectively, compared to the IC50s and IC90s for WT viruses. These results suggest that these MD resistance mutations in HIV-1 RT confer phenotypic resistance to 3TC.
FIG. 4.
Determination by the Amp-RT assay of the IC50s and IC90s of 3TC-TP for HIV-1 RT carrying MD resistance mutations in the presence of increasing concentrations of 3TC-TP. HIV-1SUM9 and xxBRUpitt are WT; Y181CEU is nevirapine resistant, M184Vpitt is 3TC resistant, and HIV-1SUM8, HIV-1SUM12, and HIV-1SUM13 are MD-resistant HIV-1 clones.
TABLE 2.
IC50s, IC90s, and fold increases in resistance for various HIV-1 clones
| Virus | Mutation(s) | IC50 (μM) | IC90 (μM) |
|---|---|---|---|
| xxBRUpitt | 0.6 | 1.3 | |
| 181C/YEU | Y181C | 0.4 | 0.7 |
| HIV-1SUM9 | 0.4 | 0.9 | |
| HIV-1SUM8 | Q151M | 0.9 (2)a | 2.8 (2.8) |
| HIV-1SUM12 | F77L/F116Y/Q151M | 2.7 (6.4) | 6.8 (6.8) |
| HIV-1SUM13 | A62V/V75I/F77L/F116Y/Q151M | 3.3 (8) | 8.2 (8.2) |
| M184Vpitt | M184V | >10 (>20) | >10 (>10) |
Values in parentheses are fold resistance (fold increase in ICs relative to the mean IC50s and IC90s for wild-type HIV-1).
Analysis of phenotypic resistance to 3TC in plasma HIV-1 RT and correlation with mutations at codon 184.
We evaluated the performance of the Amp-RT-based phenotypic assay with plasma samples by testing 30 specimens collected from 15 HIV-1-infected patients before and during treatment with 3TC. The results are shown in Table 1. Plasma HIV-1 RT in all pretreatment samples (n = 12) had WT phenotype, with RT inhibition values of >95%. The observed inhibition of the RT activity in these samples ranged from 95.9 to 100% (mean, 98.7% ± 1.8%; median, 99.8%). Of these samples, 11 had WT viral genotypes at codon 184 and one had a mixture of viruses of the WT and mutant (M184I) genotypes (sample 7A). Interestingly, mutations at codon 69, which are associated with resistance to dideoxycytosine (ddC) (11, 32), were observed in viruses in samples from the two 3TC-naive individuals whose RT had lower susceptibilities to 3TC-TP (samples 13A and 15A; RT inhibition values, 95.9 and 95.5%, respectively).
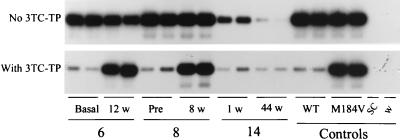
In contrast, values of RT inhibition of ≤95% were seen only for viruses in samples obtained from patients after 1 to 60 weeks of antiretroviral therapy with 3TC (n = 18). By LiPA, viruses in 12 of these samples had the M184V mutation, viruses in 4 samples had mixtures of WT and M184V genotypes, and viruses in 2 samples (samples 10A and 14A) had only WT genotypes. The mean inhibition in the samples with viruses with evidence of only the 184V mutation was 30.8% (median, 24.9%), reflecting the high frequency of 3TC-resistant viruses. The mean inhibition of RT activity in samples with mixtures of viruses with WT and resistant genotypes was 49.3% (median, 52.4%), indicating lower levels of resistance, which was expected since the viruses in these samples have higher proportions of WT RT. The lowest level of resistance to 3TC among viruses from posttherapy samples was seen in two specimens collected after 1 and 4 weeks of therapy (samples 14A and 10A; RT inhibition values, 94.7 and 87.8%, respectively). The viruses in both samples had the WT genotype at codon 184, and the virus in sample 10A had ddC and AZT resistance mutations (D69 and L41/R70/Y215, respectively). The inability to detect the M184V mutation among the viruses in these samples by LiPA may likely be due to difficulty in detecting the M184V mutation at the detection threshold of the LiPA. Both patients had evidence of the M184V viral genotype and had high levels of resistance to 3TC after 12 and 44 weeks of 3TC treatment (Table 1). Figure 5 illustrates representative results for plasma from three patients and shows the presence of phenotypic differences of RT activity from specimens collected before and during antiretroviral therapy with 3TC.
FIG. 5.
Detection of phenotypic resistance to 3TC of HIV-1 in plasma by the Amp-RT assay. Results of duplicate tests with plasma samples from three HIV-1-infected patients (patients 6, 8, and 14) obtained at different time points before therapy (basal and pre-therapy) and during antiretroviral therapy (1 to 44 weeks) with 3TC are shown. SC, HIV-1-, HIV-2-, HTLV-I-, and HTLV-II-seronegative control; w, water control.
DISCUSSION
The efficacy of antiretroviral therapy with 3TC is strongly limited by the emergence of 3TC-resistant HIV-1 variants. We have developed a rapid non-culture-based assay for the analysis of phenotypic resistance of HIV-1 RT to 3TC in plasma samples. The assay uses a small volume of plasma, and the HIV-1 RT phenotype for 3TC is determined on the basis of the level of RT inhibition by a single 3TC-TP concentration. Compared to standard culture-based phenotypic assays, this novel approach has several advantages. First, test results are obtained in 1 to 2 days, providing rapid information on resistance to 3TC that is of clinical relevance to treatment decisions and patient management. Second, testing is done directly with virion-associated RT from plasma, and therefore, unlike culture-based methods, the assay does not select for particular viral phenotypes (23, 27). Third, the assay has a low detection threshold for the presence of 3TC-resistant viruses and may be useful for the early detection of 3TC resistance.
Our data demonstrate that the assay can be used to successfully monitor resistance to 3TC mediated by mutations at codon 184. Decreased RT inhibition by 3TC-TP occurred in samples obtained from persons after treatment with 3TC and coincided with the emergence of resistant genotypes. In addition to providing phenotypic information on resistance to 3TC, the Amp-RT reaction done without 3TC-TP provides information on the level of virus in plasma on the basis of the RT level and therefore may be used to simultaneously monitor the virologic response to treatment with 3TC (12).
In this study, our primary objective was to validate the assay to monitor resistance mediated by mutations at codon 184, since resistance to 3TC has mainly been associated with mutations in this position of the HIV-1 RT gene (3, 31, 38). However, we also examined the effect that other resistance mutations may have on the susceptibility of RT to 3TC-TP. We provide evidence of resistance to 3TC in RTs carrying MD resistance mutations, thus expanding the pattern of MD resistance conferred by these mutations to 3TC. Our results obtained with an RT-based assay confirm the low level of resistance to 3TC observed in some culture-based assays (18, 19). The alteration of the RT’s substrate recognition caused by these mutations has been implicated in resistance to multiple dideoxynucleosides and may likely explain the observed resistance to 3TC-TP (29, 35).
The association between MD resistance mutations and phenotypic resistance to 3TC is of special importance. These mutations have been found in viruses from 8 to 16% of patients receiving combination therapy with AZT and dideoxyinosine (ddI) or AZT and ddC (20, 34) and may likely compromise future treatment with 3TC, despite the lower level of resistance of viruses with these mutations compared to that of viruses with the M184V mutation. Similar levels of resistance to ddI or ddC mediated by other mutations have been associated with drug failures (8). The finding of phenotypic resistance to 3TC mediated by MD resistance mutations supports the use of the present phenotypic assay for the monitoring of 3TC resistance conferred by non-M184V mutations.
Our assay was designed for the rapid evaluation of resistance to 3TC and included tests with an optimal concentration of 3TC-TP. Using this assay format, we found an interesting association between mutations at codon 69 and borderline susceptibility to 3TC-TP, suggesting that this mutation may confer some level of resistance to 3TC, a finding which is also consistent with recent observations (42). However, to confirm the role of mutations at codon 69 in the observed borderline susceptibility to 3TC, additional phenotypic testing by culture-based assays is required in these samples.
In addition to clinical monitoring of 3TC resistance, the present assay may also be used as a rapid method for surveillance of transmission of 3TC resistance among persons with newly diagnosed HIV-1 infections. Transmission of drug-resistant HIV-1 raises public health concerns because of its potential to compromise the efficacy of antiretroviral therapy both in the initial treatment of HIV-1-infected persons and in therapy for the prevention of perinatal transmission. While transmission of HIV strains with resistance to AZT and nevirapine has been documented (7, 9, 17, 39), little is known about the transmission of 3TC-resistant HIV-1. However, because of the wide use of 3TC, the risk of transmission of 3TC-resistant viruses is likely increasing. Therefore, the present assay may be used as a tool for the rapid detection of resistance to 3TC in 3TC-naive HIV-1-infected patients.
In conclusion, in contrast to culture-based methods, this RT-based phenotypic assay is a rapid and simple method for the direct analysis of phenotypic resistance of HIV-1 to 3TC mediated by mutations at codon 184 and at other positions such as those associated with MD resistance. The use of small volumes of plasma, the rapidity of testing, and the lack of selection bias all make the Amp-RT-based phenotypic assay a useful method for clinical monitoring and management of HIV-1 resistance to 3TC. This testing approach may also be expanded to analysis of resistance to nonnucleoside RT inhibitors and other nucleoside analogs such as ddC, ddI, and AZT (Q151M-mediated resistance) (15).
ACKNOWLEDGMENTS
We thank Hiroaki Mitsuya and John Mellors for providing molecular infectious clones. HIV-1RTMC/MT-2 was obtained from Brendan Larder through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases.
This work was supported in part by the Georgia Veterans Affairs Research Center for AIDS and HIV infections (to R.F.S. and D.R.) and by NIH grant 1-R01-AI41980 (to R.F.S.). Work at ISCIII was supported in part by grants from CAM, ISCII, and AIS (to V.S.).
REFERENCES
- 1.Arts E J, Wainberg M A. Mechanisms of nucleoside analog antiviral activity and resistance during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1996;40:527–540. doi: 10.1128/aac.40.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arts E J, Marois J, Gu Z, Le Grice S F J, Wainberg M A. Effects of 3′-deoxynucleoside 5′-triphosphate concentrations on chain termination by nucleoside analogs during human immunodeficiency virus type 1 reverse transcription of minus-strand strong-stop DNA. J Virol. 1996;70:712–720. doi: 10.1128/jvi.70.2.712-720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher C A B, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caroline M P, Faulds D. Lamivudine: a review of its antiviral activity, pharmacokinetic properties and therapeutic efficacy in the management of HIV infection. Drugs. 1997;53:657–680. doi: 10.2165/00003495-199753040-00008. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter C C, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiviral therapy for HIV infection in 1997: updated recommendations of the International AIDS Society—USA panel. JAMA. 1997;277:1962–1969. [PubMed] [Google Scholar]
- 6.Chou T C, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 7.Conlon C P, Klenerman P, Edwards A, Larder B A, Phillips R E. Heterosexual transmission of human immunodeficiency virus type 1 variants associated with zidovudine resistance. J Infect Dis. 1994;169:411–415. doi: 10.1093/infdis/169.2.411. [DOI] [PubMed] [Google Scholar]
- 8.D’Aquila R T. Nucleosides and foscarnet: clinical aspects. In: Richman D D, editor. Antiviral drug resistance. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 191–224. [Google Scholar]
- 9.Erice A, Mayers D L, Strike D G, Sannerud K J, McCutchan F E, Henry K, Balfour H H. Brief report: primary infection with zidovudine-resistant human immunodeficiency virus type 1. N Engl J Med. 1993;328:1163–1165. doi: 10.1056/NEJM199304223281605. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson B F H, Chu C K, Schinazi R F. Phosphorylation of 3′-azido-2′,3′-dideoxyuridine and preferential inhibition of human and simian immunodeficiency virus reverse transcriptases by its 5′-triphosphate. Antimicrob Agents Chemother. 1989;33:1729–1734. doi: 10.1128/aac.33.10.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgibbon J E, Howell R M, Haberzettl C A, Sperber S J, Gockle D J, Dubin D T. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1992;36:153–157. doi: 10.1128/aac.36.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia Lerma J G, Yamamoto S, Gomez-Cano M, Soriano V, Green T A, Busch M P, Folks T M, Heneine W. Measurement of human immunodeficiency virus type 1 plasma virus load based on reverse transcriptase (RT) activity: evidence of variabilities in levels of virion-associated RT. J Infect Dis. 1998;177:1221–1229. doi: 10.1086/515272. [DOI] [PubMed] [Google Scholar]
- 13.Gulick R M, Mellors J W, Havlir D, Eron J E, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 14.Heneine W, Yamamoto S, Switzer W M, Spira T J, Folks T M. Detection of reverse transcriptase by a highly sensitive assay in sera from persons infected with human immunodeficiency virus type 1. J Infect Dis. 1995;171:1210–1216. doi: 10.1093/infdis/171.5.1210. [DOI] [PubMed] [Google Scholar]
- 15.Heneine W, Garcia Lerma J G, Vazquez-Rosales J G, Qari S, Juodawlkis A, Havlir D, Schinazi R F, Richman D D, Folks T M. Program and abstracts of the 2nd International Workshop on Drug Resistance and Treatment Strategies. 1998. A novel non culture-based approach for the rapid analysis of phenotypic resistance to reverse transcriptase inhibitors of HIV-1 from plasma, abstr. 63; p. 42. [Google Scholar]
- 16.Hertogs K, de Bethune M, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imrie A, Beveridge A, Genn W, Vizzard J, Cooper D A the Sydney Primary HIV Infection Study Group. Transmission of human immunodeficiency virus type 1 resistant to nevirapine and zidovudine. J Infect Dis. 1997;175:1502–1506. doi: 10.1086/516487. [DOI] [PubMed] [Google Scholar]
- 18.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jellinger R M, Shafer R W, Merigan T C. A novel approach to assessing the drug susceptibility and replication of human immunodeficiency virus type 1 isolates. J Infect Dis. 1997;175:561–566. doi: 10.1093/infdis/175.3.561. [DOI] [PubMed] [Google Scholar]
- 20.Kavlik M F, Wyvill K, Yarchoan R, Mitsuya H. Emergence of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 variants, viral sequence variation, and disease progression in patients receiving antiretroviral chemotherapy. J Infect Dis. 1998;177:1506–1513. doi: 10.1086/515324. [DOI] [PubMed] [Google Scholar]
- 21.Kavlick M F, Shirasaka T, Kojima E, Pluda J M, Hui F, Yarchoan R, Mitsuya H. Genotypic and phenotypic characterization of HIV-1 isolated from patients receiving (−)-2′,3′-dideoxy-3′-thiacytidine. Antivir Res. 1995;28:133–146. doi: 10.1016/0166-3542(95)00044-m. [DOI] [PubMed] [Google Scholar]
- 22.Kellam P, Larder B A. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusumi K, Conway B, Cunningham S, Berson A, Evans C, Iversen A K, Colvin D, Gallo M V, Coutre S, Shpaer E G. Human immunodeficiency virus type 1 envelope gene structure and diversity in vivo and after cocultivation in vitro. J Virol. 1992;66:875–885. doi: 10.1128/jvi.66.2.875-885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 25.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 26.Larder B A, Kemp A P, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT+3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′3′-cyclophosphorothioates using 2-chlore-4H-1,2,3-benzodioxaphosphorin-4-one. J Org Chem. 1989;54:631–635. [Google Scholar]
- 29.Sarafinos S G, Pandey V N, Kaushik N, Modak M J. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry. 1995;34:7207–7216. doi: 10.1021/bi00021a036. [DOI] [PubMed] [Google Scholar]
- 30.Schinazi R F, McMillan A, Cannon D, Mathis R, Lloyd R M, Peck A, Sommadossi J, St. Clair M, Wilson J, Furman P A, Painter G, Choi W, Liotta D C. Selective inhibition of human immunodeficiency viruses by racemates and enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]cytosine. Antimicrob Agents Chemother. 1992;36:2423–2431. doi: 10.1128/aac.36.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schinazi R F, Lloyd R M, Nguyen M H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5:129–142. [Google Scholar]
- 33.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C A. Rapid changes in HIV-1 RNA load and appearance of drug-resistant virus population in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 34.Shafer R W, Iversen A K N, Winters M A, Aguiniga E, Katzenstein D A, Merigan T C the AIDS Clinical Trials Group 143 Virology Team. Drug resistance and heterogeneous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. J Infect Dis. 1995;172:70–78. doi: 10.1093/infdis/172.1.70. [DOI] [PubMed] [Google Scholar]
- 35.Shirasaka T, Kavlick M F, Ueno T, Gao W, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 37.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for the rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tisdale N, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veenstra J, Schuurman R, Cornelissen M, van’t Wout A B, Boucher C A B, Schuitemaker H, Goudsmit J, Coutinho R A. Transmission of zidovudine-resistant human immunodeficiency virus type 1 variants following deliberate injection of blood from a patient with AIDS: characteristics and natural history of the virus. J Infect Dis. 1995;21:556–560. doi: 10.1093/clinids/21.3.556. [DOI] [PubMed] [Google Scholar]
- 40.Wainberg M A, Salomon H, Gu Z, Montaner J S G, Cooley T P, MaCaffrey R, Ruedy J, Hirst H M, Cammack N, Cameron J, Nicholson W. Development of HIV-1 resistance to (−) 2′-deoxy-3′-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS. 1995;9:351–357. [PubMed] [Google Scholar]
- 41.Wainberg M A, Hsu M, Gu Z, Borkow G, Parniak M A. Effectiveness of 3TC in HIV clinical trials may be due in part to the M184V substitution in 3TC-resistant HIV-1 reverse transcriptase. AIDS. 1996;10:S3–S10. doi: 10.1097/00002030-199612005-00002. [DOI] [PubMed] [Google Scholar]
- 42.Wainberg M A, Miller M D, Quan Y, Salomon H, Cherrington J. Program and abstracts of the 5th Conference on Retroviruses and Opportunistic Infections. 1998. The M184V substitution in reverse transcriptase moderately increases sensitivity of HIV-1 to PMPA, abstr. 680; p. 207. [Google Scholar]
- 43.Yamamoto S, Folks T M, Heneine W. Highly sensitive qualitative and quantitative detection of reverse transcriptase activity: optimization, validation, and comparative analysis with other detection systems. J Virol Methods. 1996;61:135–143. doi: 10.1016/0166-0934(96)02078-2. [DOI] [PubMed] [Google Scholar]