Abstract
Despite advances in high-throughput sequencing and bioinformatics, molecular investigations of snail intermediate hosts that transmit parasitic trematodes are scant. Here, we report the first transcriptome for Bulinus truncatus – a key intermediate host of Schistosoma haematobium – a blood fluke that causes urogenital schistosomiasis in humans. We assembled this transcriptome from short- and long-read RNA-sequence data. From this transcriptome, we predicted 12,998 proteins, 58% of which had orthologs in Biomphalaria glabrata – an intermediate host of Schistosoma mansoni – a blood fluke that causes hepato-intestinal schistosomiasis. We predicted that select protein groups are involved in signal transduction, cell growth and death, the immune system, environmental adaptation and/or the excretory/secretory system, suggesting roles in immune responses, pathogen defence and/or parasite-host interactions. The transcriptome of Bu. truncatus provides a useful resource to underpin future molecular investigations of this and related snail species, and its interactions with pathogens including S. haematobium. The present resource should enable comparative investigations of other molluscan hosts of socioeconomically important parasites in the future.
Keywords: Transcriptome, Bulinus truncatus, Intermediate host, Schistosoma haematobium, Pathogen-host interactions
Graphical abstract
Highlights
-
•
First transcriptome to represent Bulinus truncatus – a snail intermediate host of Schistosoma haematobium.
-
•
Select protein groups of Bu. truncatus are inferred to associate with innate immune responses against pathogens.
-
•
Transcriptome provides a resource for future studies of parasite-host interactions and snail-host resistance to pathogens.
1. Introduction
Substantial advances in high-throughput nucleic acid sequencing technologies and bioinformatics have enabled transcriptomic and genomic studies to elucidate molecular aspects of parasites and parasitism (Young et al., 2010; Schwarz et al., 2013; Anstead et al., 2015; Korhonen et al., 2016; Howe et al., 2017; International Helminth Genomes Consortium, 2019; Stroehlein et al., 2019). However, there has been relatively limited investigation of parasite systems which involve snail intermediate hosts, such as digenean trematodes and their amphibious or aquatic gastropod hosts (Adema et al., 2012). Understanding the molecular basis of interactions between a snail intermediate host and invading, asexually replicating stages of a trematode species is not only fundamentally interesting but could assist in finding ways of breaking or interfering with the parasite’s life-cycle (Adema et al., 2017; Castillo et al., 2020).
The foundation for such work is the development of essential molecular ‘omics’ resources (e.g. genome, transcriptome and proteome) for intermediate hosts. Most effort, to date, has focused on Biomphalaria glabrata – the key intermediate host of the socioeconomically-important human blood fluke (schistosome) Schistosoma mansoni. This Neotropical species has acclimatised well to long-term culture and continues to maintain excellent compatibility with natural or laboratory-adapted isolates of S. mansoni. Recent work (Kenny et al., 2016; Adema et al., 2017) has created a useful molecular toolbox (genome and transcriptomes) for Bi. glabrata, enabling investigations of the regulation of micro-RNA (miRNA) and piwi-interacting RNA (piRNA) processing pathway genes (Queiroz et al., 2017) and providing select insights into the snail neurophysiology, metabolism, immunity and key aspects of asexual reproduction of the digenean within the snail host (Adema et al., 2017). Surprisingly, such ‘omics’ resources for other snail species that transmit blood flukes causing a similar, or possibly greater, disease burden in humans (van der Werf et al., 2003; Rollinson, 2009), are scarce.
By contrast, there has been limited scrutiny, at the molecular level, of the planorbid snail Bulinus truncatus, which is a major intermediate host of Schistosoma haematobium – the agent of urogenital schistosomiasis. Bulinus truncatus commonly occurs in North-West Africa, lower Egypt and Sudan, extending as far South as Malawi. The northern limit includes Portugal, Sardinia and Corsica. Urogenital schistosomiasis affects more than 200 million people worldwide (Murray et al., 2012; Colley et al., 2014), and S. haematobium is a bio-carcinogen of malignant bladder cancer (Palumbo, 2007). In consideration of the 36 snail species within the genus Bulinus, Bu. truncatus is remarkable in being a tetraploid species and thought to have arisen from ancestral hybridisation by alloploidy of more common diploid species stocks (Brown, 1994). Although a hermaphrodite, the species is unusual in having environmentally determined phally, with the balance between aphallic and euphallic snails being temperature-dependent. Given the major socioeconomic importance of S. haematobium, and its recently acknowledged epidemiology links with HIV/AIDS (Kjetland et al., 2014), here we focused our attention on developing a novel molecular resource (a representative transcriptome) for Bu. truncatus to underpin detailed future investigations of the biology of this snail and its interplay with asexual larval stages of S. haematobium.
2. Materials and methods
2.1. Procurement of parasite material, RNA isolation, library preparation and sequencing
Two adult specimens of Bu. truncatus (Egyptian strain) maintained in artificial pond water at 26 °C at the NIH-NIAID Schistosomiasis Resource Center, Biomedical Resource Institute (Rockville, MD 20850, USA) following standard operating procedures (Tucker et al., 2013) were obtained: one specimen had been exposed to S. haematobium and frozen at patency (54 days), and the other (‘naïve’) specimen had not been exposed. RNA was isolated from each specimen using the TriPure Isolation Reagent (Sigma Aldrich, St. Louis, Missouri, USA) and DNase-treated using a TURBO DNA-freeTM kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Messenger RNA (mRNA) was purified from total RNA using the Dynabeads® mRNA Purification Kit (Thermo Fisher Scientific). The size, integrity (expressed using an ‘RNA integrity number’ [RIN] of > 7.5 – consistent with a cleaved 28S band characteristic for lophotrochozoan organisms – cf. Natsidis et al., 2019) and concentration of RNA were determined using a 4200 TapeStation System RNA ScreenTape Assay (Agilent Technologies, Waldbronn, Germany) and a Qubit® 3.0 flourometer RNA High Sensitivity Assay (Life Technologies, Carlsbad, California, USA).
A TruSeq Stranded mRNA (Illumina, San Diego, California, USA) short-read library (150 bp, paired-end) was prepared from the mRNA sample derived from the snail infected with S. haematobium, according to the manufacturer’s instructions and sequenced on an Illumina NextSeq 500 sequencer. For the mRNA sample isolated from the naïve snail, a long-read library was prepared using the Oxford Nanopore direct RNA-sequencing kit (SQK-RNA002; Oxford Nanopore Technologies, Oxford, UK), according to the manufacturer’s instructions. The prepared library was sequenced on a MinION sequencer (Oxford Nanopore Technologies) for 48 h until no more active pores were available, using an EXP-FLP002 flow cell priming kit and a R9.4.1 flow cell (FLO-MIN106).
2.2. Processing of sequence data and transcriptome assembly
Short-read data were quality-filtered, and adapters removed using the program fastp v.0.20.1 (Chen et al., 2018) using the -m option to collapse read pairs that overlapped by ≥ 30 nucleotides (nt) into a single read. Then, reads were mapped to publicly available S. haematobium genome scaffolds (Stroehlein et al., 2019) using HISAT2 v.2.1.0 (Kim et al., 2019) employing the –fr and –rna-strandness RF options. All unmapped reads were retained by filtering the alignment file using the -f4 flag employing the program samtools v.1.9 (Li et al., 2009). The program centrifuge v.1.0.4 (Kim et al., 2016) was used to remove potential contamination; only unclassified reads or those assigned to the phylum Mollusca were retained. Using the khmer software package v.3.0.0a3 (Crusoe et al., 2015), all remaining reads were digitally normalised to a coverage of 20 (-C option), and reads containing low-abundance k-mers (k = 26) were trimmed or removed. Long-reads produced from the SQK-RNA002 library were base-called from raw fast5 files using a GPU-enabled version of the program Guppy v.3.2.4 and providing the configuration file rna_r9.4.1_70bps_hac.cfg. Reads that did not pass the quality filter within Guppy (i.e. Q < 7) were removed.
Decontaminated and trimmed, high-quality short-reads and quality-filtered long-reads (–nanopore option) were then assembled using the program Spades v.3.14.1 (Bushmanova et al., 2019) in RNA mode using the –ss rf option (strand-specific assembly). The redundancy of the resultant ‘hard-filtered’ (within Spades) transcript dataset was reduced employing the program cd-hit-est v.4.6 (Fu et al., 2012), collapsing transcripts with ≥ 99% nucleotide identity into a single representative transcript. Subsequently, candidate coding regions were identified within transcript sequences using the TransDecoder software v.5.5.0 (Haas et al., 2013).
2.3. Functional annotation of inferred protein sequences
Protein sequences conceptually translated from the Bu. truncatus transcriptome were compared with those of Bi. glabrata and orthologs were inferred using the program orthoFinder v.2.2.6 (Emms & Kelly, 2019). The completeness of the inferred proteome was assessed employing the program BUSCO v.4.1.2 (Waterhouse et al., 2017) using the -l metazoa_odb10 (accessed 5 August 2020) and –update-data options. We then employed complementary homology-based and sequence domain-based approaches for functional annotation; first, functional protein domains, families and superfamilies were inferred using the Pfam (Finn et al., 2010), PANTHER (Mi et al., 2013) and SUPERFAMILY (Wilson et al., 2009) databases within the program InterProScan v.5.44-79.0 (Jones et al., 2014). Next, protein groups and pathways were inferred based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) terms (Kanehisa et al., 2016) using a hierarchical representation of protein function (i.e. KO terms, KEGG BRITE hierarchy and KEGG pathway hierarchy). Protein descriptions were assigned using the program eggNOG-mapper (Huerta-Cepas et al., 2017) using the -m diamond option and the eggNOG database v.5.0 (eukaryotes) (Huerta-Cepas et al., 2019). The sub-cellular localisation of protein sequences was predicted computationally using the program MultiLoc2 v.2.2.25 (Blum et al., 2009) employing a stringent cut-off confidence score of ≥ 0.8. The structures of select unannotated (‘orphan’) protein sequences were inferred using the program I-TASSER v.5.1 (Yang et al., 2015).
3. Results
3.1. Sequence data sets
A total of 76,372,022 paired-end sequences were obtained; 72,879,800 of these sequences were retained after trimming and quality-filtering, of which 68,689,594 (89.9%) overlapped with their mate pair and were collapsed into single, representative reads. Of the remaining 4,190,206 read pairs and 35,699,314 single-end reads, 166,296 and 2,228,343 (2,394,639 reads in total; 6.0%) mapped to the S. haematobium genome, respectively, and were removed. Of the resultant 37,494,881 reads, 32,374,724 (86.3%) represented the phylum Mollusca or were not assigned to a taxon (“unclassified”) and were retained. The final short-read data set represented 1,314,020 read pairs and 4,640,861 single-end reads after digital normalisation and removal of low-abundance k-mers. Complementing this data set, 2,442,455 long-reads were sequenced from the SQK-RNA002 library. Of those, 2,109,546 reads (N50 length of 983 nt; median quality of 10.2) were retained after quality-filtering.
3.2. Characteristics of the Bu. truncatus transcriptome
The initial transcriptome (Table 1) assembled from short- and long-read data, consisted of 28,743 high-quality (i.e. ‘hard-filtered’, cf. Bushmanova et al., 2019) sequences, 13,615 of which had an open reading frame (ORF) and were inferred to be protein-coding. After removing redundancy, 12,792 transcripts containing 12,998 ORFs represented the final transcriptome (Table 2, Supplementary Table S1), 4,188 of which represented complete, 2,178 internal, 5,693 5′-partial and 939 3′-partial ORFs, respectively. Totals of 4,817 5′- and 9,499 3′-untranslated regions (UTRs; respective mean lengths: 152 nt and 310 nt) were identified for this set of transcripts (Table 2; Supplementary Table S1).
Table 1.
Quality metrics for sequence data and the assembled transcriptome for Bulinus truncatus
| Count | |
|---|---|
| Raw paired Illumina reads | 76,372,022 |
| Quality-filtered Illumina reads (paired/single) | 8,380,412/35,699,314 |
| Decontaminated and normalised Illumina reads (paired/single) | 2,628,040/4,640,861 |
| Raw Nanopore long-reads | 2,442,455 |
| Quality-filtered long-reads | 2,109,546 |
| Long-read N50 | 983 |
| Mean/median length of long-reads | 821/699 |
| Number of assembled transcript sequences | 28,743 |
| Protein-coding transcripts | 13,615 |
| Non-redundant protein-coding transcripts (transcriptome) | 12,792 |
| Transcriptome N50 | 1,136 |
| Mean/median length of transcripts | 1,014/833 |
| Shortest/longest transcript | 404/13,929 |
Table 2.
Key sequence features and completeness metrics of coding sequences and inferred protein sequences in the transcriptome of Bulinus truncatus
| Count | Percentage of transcriptome | |
|---|---|---|
| Non-redundant coding sequences | 12,998 | 100 |
| Complete ORFs | 4,188 | 32 |
| Transcripts with inferred 5′ UTR | 4,817 | 37 |
| Transcripts with inferred 3′ UTR | 9,499 | 73 |
| ≥ 1 ortholog in Bi. glabrata | 7,500 | 58 |
| Single-copy orthologs in Bi. glabrata | 4,460 | 34 |
| Annotated via eggNOG database | 9,275 | 71 |
| Annotated via Pfam database | 7,809 | 60 |
| Annotated via PANTHER database | 8,689 | 67 |
| Annotated via SUPERFAMILY database | 6,761 | 52 |
| Gene Ontology (GO) term annotation (eggNog and InterPro) | 7,767 | 60 |
| KEGG Orthology (KO) annotation | 8,460 | 65 |
| Pathway annotation via KEGG database | 4,865 | 37 |
| Annotated by ≥ 1 method/database | 10,349 | 80 |
| Unannotated ‛orphansʼ | 2,649 | 20 |
| Unannotated ‛orphansʼ with complete ORF | 959 | 7 |
| Unannotated ‛orphansʼ with complete ORF and ≥ 1 Bi. glabrata ortholog | 312 | 2 |
3.3. Comparison of inferred proteomes
A comparison of protein sequences (n = 12,998) inferred from the transcriptome of Bu. truncatus with those conceptionally translated from predicted genes in the Bi. glabrata genome identified 7,500 (57.7%) orthologous sequences (Table 2), 4,460 of which represented one-to-one (‘single-copy’) orthologs (Supplementary Tables S2 and S3). In total, 5,498 of all sequences (42.3%) had no ortholog in Bi. glabrata. Conversely, 25,937 of a total of 36,943 (70.2%) sequences of Bi. glabrata had no ortholog in Bu. truncatus.
3.4. Functional annotation
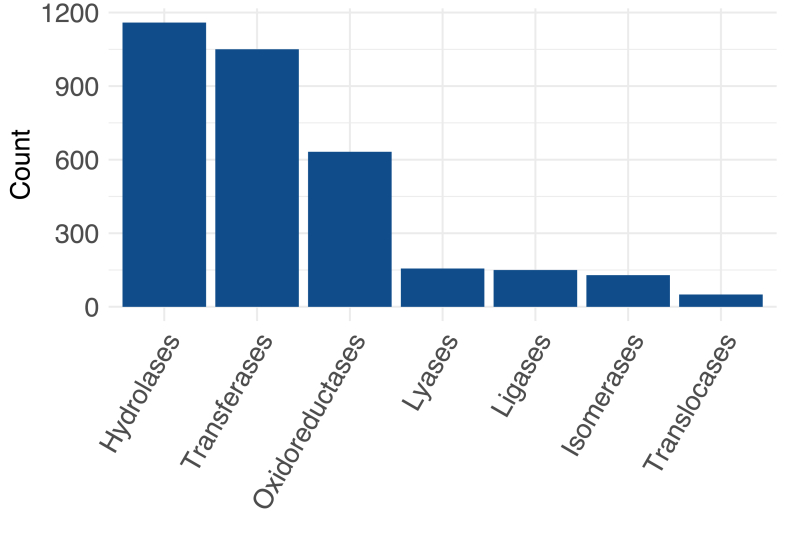
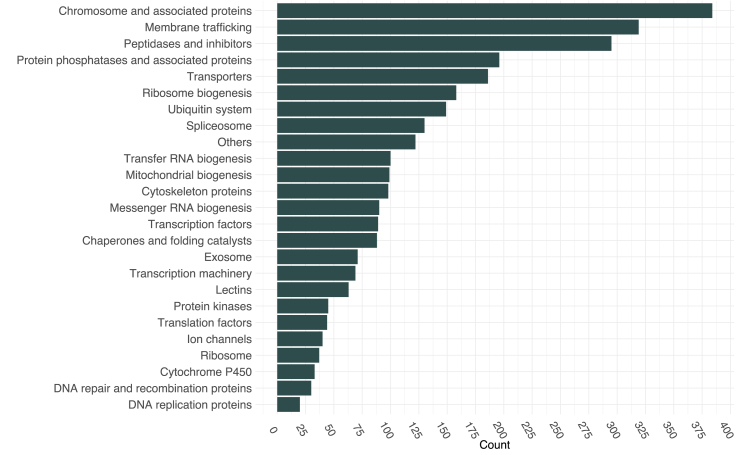
In total, 653 of 954 (68.4%) metazoan Benchmarking Universal Single Copy Orthologs (BUSCOs) were identified (Table 3), 445 of which (46.6%; comprising 399 single-copy and 46 duplicated BUSCOs) were complete and 208 (21.8%) fragmented (Supplementary Table S4). We inferred annotations for 9,542 amino acid sequences based on information from one or more of the three databases Pfam (n = 7,809), PANTHER (n = 8,689) and SUPERFAMILY (n = 6,761) (Supplementary Table S5), and 9,275 (71.4%) sequences were annotated using the eggNOG database (Supplementary Table S6). Based on these analyses, Gene Ontology (GO) terms were assigned to 7,767 of all 12,998 sequences (59.8%). Most sequences (8,460 of all sequences; 81.7%) had KEGG orthology terms (Supplementary Table S7), and of those 3,317 (39.2%) were assigned to seven enzyme sub-classes (KEGG Enzymes, Fig. 1); 2,902 (34.3%) sequences were linked to 25 protein groups/functions (KEGG BRITE; Fig. 2), with most of them assigned to the categories “chromosome and associated proteins” (n = 384), “membrane trafficking” (n = 319), “peptidases and inhibitors” (n = 295), “protein phosphatases and associated proteins” (n = 196) and “transporters” (n = 186). More than half (57.7%) of the 8,460 sequences with KO terms were linked to 32 distinct KEGG pathway modules (Fig. 3), including “signal transduction” (n = 1,041), “cell growth and death” (n = 478), “immune system” (n = 458), “carbohydrate metabolism” (n = 455), “environmental adaptation” (n = 260) and “excretory system” (n = 86), and a total of 310 distinct pathways (Supplementary Table S7). Sub-cellular localisations were inferred for 5,870 sequences, with “cytoplasmic” (n = 4,189; 71.4%), extracellular (n = 598; 10.2%) and nuclear (n = 570; 9.7%) being predominant (Supplementary Table S8). Overall, 10,349 (79.6%) proteins were annotated, and 2,649 (20.4%) were not and were thus called ‘orphan’ (unknown) proteins (Supplementary Table S9). Of these orphans, 959 had complete ORFs.
Table 3.
Benchmarking Universal Single Copy Orthologs (BUSCOs) detected in the transcriptome of Bulinus truncatus
| Count | Percentage of BUSCOs | |
|---|---|---|
| Total number of metazoan BUSCOs | 954 | 100 |
| BUSCOs detected in Bu. truncatus transcriptome | 653 | 68 |
| Complete, single-copy BUSCOs | 399 | 42 |
| Complete, duplicated BUSCOs | 46 | 5 |
| Fragmented BUSCOs | 208 | 22 |
| Missing BUSCOs | 301 | 32 |
Fig. 1.
Protein sequences (n = 3,317; 39.2%) encoded by the Bulinus truncatus transcriptome that represent one of seven enzyme sub-classes (KEGG Ontology, KO)
Fig. 2.
KEGG BRITE annotation for 2,902 (34.3%) protein sequences encoded in the Bulinus truncatus transcriptome
Fig. 3.
Protein sequences of Bulinus truncatus (n = 4,865; 37.4%) that were linked to one or more of 32 distinct KEGG pathway modules
3.5. Protein groups inferred to be involved in parasite-host interplay
Subsequently, we inferred key protein groups in Bu. truncatus with likely or suggested roles in the immune/defence system of the snail, interactions with pathogens and/or susceptibility/resistance to schistosome infection(s), and associated signalling (Supplementary Tables S7, S10 and S11), supported by published evidence or information (Table 4).
Table 4.
Key protein groups in Bulinus truncatus with likely or suggested roles in the immune/defence system of the snail, interactions with pathogens and/or susceptibility/resistance to schistosome infection(s), and associated signalling, supported by published evidence or information
| Protein group | No. of sequences | Known or suggested role(s) in | Key module/pathway association(s) (KEGG pathway terms) | Reference |
|---|---|---|---|---|
| Methyltransferases | 137 | Parasite-host interactions, environmental signalling | Mitogen-activated protein kinase (MAPK) signalling pathway, thermogenesis | Geyer et al. (2017) |
| Lectins | 123 | Immune response | C-type lectin receptor signalling pathway, extracellular matrix (ECM)-receptor interaction, focal adhesion, lysosome, phagosome | Dheilly et al. (2015); Buddenborg et al. (2017) |
| Kinases | 113 | Signalling in response to environmental and pathogenic stressors | Apoptosis, autophagy, calcium signalling pathway, cyclic adenosine monophosphate (cAMP) signalling pathway, C-type lectin receptor signalling pathway, ECM-receptor interaction, focal adhesion, MAPK signalling pathway, necroptosis, NF-kappa B signalling pathway, olfactory transduction, peroxisome, pathogen interaction, thermogenesis, Toll-like receptor signalling pathway | Adema et al. (2017) |
| FREPs/FReDs | 69 | Susceptibility/resistance to schistosome infection, immune response | ECM-receptor interaction, focal adhesion | Lockyer et al. (2012); Dheilly et al. (2015); Gordy et al. (2015); Lu et al. (2020) |
| Phosphatases | 59 | Signalling in response to environmental and pathogenic stressors | cAMP signalling pathway, C-type lectin receptor signalling pathway, focal adhesion, MAPK signalling pathway, NF-kappa B signalling pathway, Toll-like receptor signalling pathway | Adema et al. (2017) |
| BIRs/IAPs | 58 | Drug response, apoptosis, innate immune responses | Toll and Imd signalling pathway, NF-kappa B signalling pathway | Adema et al. (2017); Buddenborg et al. (2019) |
| Chitinases | 47 | Susceptibility/resistance to schistosome infection, excretory/secretory product | Amino sugar and nucleotide sugar metabolism | Tennessen et al. (2015); Fogarty et al. (2019) |
| Calmodulins | 31 | Stress response, drug susceptibility | C-type lectin receptor signalling pathway, environmental adaptation | Buddenborg et al. (2019) |
| Cathepsins | 23 | Susceptibility/resistance to schistosome infection, excretory/secretory product | Antigen processing and presentation, apoptosis, autophagy, lysosome, phagosome | Fogarty et al. (2019) |
| Toll-/IL-1-related proteins | 14 | Susceptibility/resistance to schistosome infection, immune response | Cytokine–cytokine receptor interaction, MAPK signalling pathway, necroptosis, NF-kappa B signalling pathway, phagosome, Toll and Imd signalling pathway, Toll-like receptor signalling pathway | Kenny et al. (2016); Pila et al. (2016) |
| Guadeloupe Resistance Complex (GRC) | 9 (15 in Bi. glabrata) | Susceptibility/resistance to schistosome infection | Lysosome, endocytosis | Tennessen et al. (2015) |
| Polymorphic Transmembrane Cluster 2 (PTC2) | 3 (11 in Bi. glabrata) | Susceptibility/resistance to schistosome infection | na | Tennessen et al. (2020) |
Abbreviation: na, not available.
Sets of kinases (n = 113), phosphatases (n = 59) and Toll-/IL-1-related proteins (n = 14) showed the highest degree of connectivity to key pathways (6–15), whereas the remaining protein groups were less connected (1–5; Fig. 4). Of 137 methyltransferases inferred, most (n = 96; 70.0%) were S-adenosyl-l-methionine-dependent methyltransferases (IPR029063); 73 of them had single-copy orthologs, and 31 had multiple (2–9) orthologs in Bi. glabrata. Of the 38 proteins predicted to be involved in olfactory transduction, 31 were calcium-modulated proteins (calmodulins) and the remaining seven were kinases, receptors, channels or calcium-binding proteins. Calmodulins and kinases (STE group; BTRUNC_01512.1 and BTRUNC_06865.1) were also linked to roles in interactions with pathogens (Supplementary Table S7; Fig. 4).
Fig. 4.
Pathway network for key protein groups encoded in the Bulinus truncatus transcriptome that have suggested roles in parasite-host interactions, environmental adaptation and/or immune defence. Associations between protein groups (orange), pathways (light blue) and functional categories (dark blue) are indicated by grey edges. Node size and edge thicknesses represent the numbers of associated sequences and connections, respectively
We identified 58 transcripts encoding baculovirus inhibitor of apoptosis (IAP) repeat proteins (BIRs) consistent with earlier reports of an expansion of this protein family in molluscs (Adema et al., 2017). All BIRs were linked to roles in apoptosis and 33 of them were related to necroptosis and Toll-, Imd-, and NF-kappa B-signalling pathways (Fig. 4). Among other proteins related to Toll-signalling were 13 sequences with a Toll/interleukin-1 receptor homology domain (IPR000157) of which four were annotated as Toll-like receptors (TLRs). We did not observe an expansion of TLRs as has been reported for Bi. glabrata (see Adema et al., 2017) which has 56 TLR genes encoding 27 complete TLRs (compared with ∼10 TLR genes in insects and mammals).
We detected transcripts encoding 121 lectins, including 42 and 27 annotated as ficolins and tenascins, respectively, and 26 representing one of nine distinct C-type lectin family members. In addition, we identified 69 other, lectin-like molecules that were annotated as fibrinogen-related proteins (FREPs/FReDs; InterPro identifier IPR036056) but lacked the IgSF domain(s) identified in FREPs of Bi. glabrata (cf. Lu et al., 2020). These lectins and lectin-like molecules were mainly assigned to the C-type lectin receptor signalling pathway and associated with roles in focal adhesion, interaction with the extracellular matrix (ECM) as well as the phagosome and lysosome (Fig. 4). The latter two pathways were also assigned to cathepsins (n = 23). FREPs/FReDs and cathepsins have been linked to resistance/susceptibility to schistosome infection of Bi. glabrata, among other families such as chitinases (Table 4). We identified 47 chitinases of which 24 formed a cluster of paralogs/isoforms and 23 represented single copies. Of all 47 sequences, 14 did not have an ortholog in Bi. glabrata and 33 were orthologous to up to four sequences (total number of orthologs in Bi. glabrata: 17), forming ten orthologous clusters.
We investigated orthologs of members of two resistance gene clusters in Bi. glabrata, the Guadeloupe Resistance Complex (GRC; cf. Tennessen et al., 2015) and the Polymorphic Transmembrane Cluster 2 (PTC2; cf. Tennessen et al., 2020) (Supplementary Table S11). Of the 15 proteins encoded by the GRC cluster in Bi. glabrata, nine had orthologs in Bu. truncatus. For the PTC2 cluster, we identified one ortholog via OrthoFinder (BTRUNC_12767.1, BGLB016855) and two additional sequence matches via blastp (BTRUNC_01870.1, BGLB029318 and BTRUNC_08140.1, BGLB027019, BGLB030379). Six out of those 12 orthologous sequences could not be annotated based on sequence features. These six sequences were thus considered orphan sequences of which three had a complete ORF and three had a 5′-partial ORF (Supplementary Table S9). Two sequence products were predicted to be extracellular, whereas two others were inferred to be nuclear and cytoplasmic, respectively.
3.6. Functional predictions for select, unannotated proteins
Of all 959 orphans inferred, 312 (32.5%) had an ortholog in Bi. glabrata (lengths: 95–897 amino acids (aa); mean: 216.2 aa, median: 192.5 aa) and were translated from high-confidence, full-length transcripts with complete ORFs, thus representing bona fide transcripts (Supplementary Table S9). We investigated the likely subcellular localisation of the protein products encoded by these transcripts and predicted that 58 are localised either to the plasma membrane (n = 2) or are extracellular (n = 56). Of the orthologs for these 58 sequences in Bi. glabrata (n = 69), none were annotated in VectorBase (release 49 beta; BglaB1.6.; https://vectorbase.org/vectorbase/app/record/organism/TMPTX_bglaBB02; Accessed 13 August 2020), but 45 were associated with one or more GO terms. The most-assigned GO terms for these sequences were “integral component of membrane” (GO:0016021; n = 32), “protein binding” (GO:0005515; n = 6), “membrane” (GO:0016020; n = 5) and “transmembrane signalling receptor activity” (GO:0004888; n = 5), as well as four GO terms related to extracellular ligand-gated ion channel activity (GO:0034220, GO:0006811, GO:0005230, GO:0005216; each n = 4). One of the 56 orphan proteins predicted to be localised extracellularly (BTRUNC_07650.1) represented an ortholog of three sequences belonging to the resistance gene cluster GRC in Bi. glabrata (BGLB003681, BGLB017395 and BGLB033905) (Tennessen et al., 2015).
To further investigate the potential function of all 58 sequences, we predicted their three-dimensional structure (Supplementary Table S12). For five sequences we predicted a high confidence structural alignment (normalised Z-score > 1) to entries in the Protein Data Bank (PDB), with sequence identity values ranging from 8% to 29%, alignment coverage between 64% and 99% and topology modelling (TM) scores between 0.21 and 0.63. One of the five structural matches was discarded due to structural similarity to a synthetic construct. The other four sequences were annotated as “tumor necrosis factor receptor” (BTRUNC_14698.1), “Dickkopf-related protein 2” (BTRUNC_16137.1), “angiopoietin-1 receptor” (BTRUNC_27016.1) and “acyl-CoA dehydrogenase” (BTRUNC_03469.1), based on their closest structural match.
4. Discussion
The characterisation of the first transcriptome for Bu. truncatus and the functional annotation of inferred proteins provide a foundation for future molecular investigations of Bu. truncatus and comparative investigations of snail intermediate hosts of other schistosomes, such as Bi. glabrata. This effort helps to set a new line of investigation of snail hosts that transmit human and animal schistosomiasis in Africa, with a bearing on recently described hybrid schistosomes within the S. haematobium group and zoonotic transmission (Leger & Webster, 2017).
The present findings show that most conserved metazoan proteins (BUSCOs) are encoded within the Bu. truncatus transcriptome, and that most sequences have at least one ortholog in Bi. glabrata. The analyses provided evidence for the presence of expansion events in some protein groups, consistent with findings for Bi. glabrata (see Simpson et al., 2005; Adema et al., 2017; Geyer et al., 2017). In this context, the large number of S-adenosyl-l-methionine-dependent methyltransferases and their link to roles in thermogenesis pathways may indicate a role for methylation in the snail's interactions with its environment and/or pathogens (Liebsch & Becker, 1990; Nelson et al., 2016), as has been suggested previously (Geyer et al., 2017). Similarly, we indicate an expansion of BIRs, as reported earlier for Bi. glabrata (see Adema et al., 2017). While the exact role of these molecules is not yet known, a larger number of copies present in snails compared with other invertebrates might relate to regulatory roles in apoptosis and innate immune responses in molluscs (Adema et al., 2017).
Calcium-modulated proteins (calmodulins) also appear to be expanded. These proteins transduce signals in response to increases in intracellular Ca2+ and represent a major component of calcium-dependent signalling pathways (Racioppi & Means, 2008). The expansion seen in Bu. truncatus is in accord with findings in other snail species (Simpson et al., 2005): in Bi. glabrata, 28 genes encoding 38 calmodulin isoforms have been identified and linked to roles in shell formation and defence to bacteria, yeast and S. mansoni (see Buddenborg et al., 2019), and in Lymnaea stagnalis, a Ca2+/calmodulin-dependent nitric oxide synthase is reported to associate with nitric oxide production in haemocytes – as a defence mechanism of this snail against pathogens (Wright et al., 2006). Given these findings for other snail species, it is possible that the apparent expansion of calmodulins encoded in the Bu. truncatus transcriptome represents a functional and subcellular diversification, and that these molecules play a role in biotic defence responses (McCormack & Braam, 2003). This proposal is supported by the finding that most calmodulins associate with the KEGG pathway “pathogen-interaction” (KEGG pathway identifier: ko04626).
However, a number of kinase groups and families linked to this pathway in the KEGG database, including Ca2+/calmodulin-dependent protein kinases (CAMKs), myosin light-chain kinases (MLCKs), phosphorylase kinases (PhKs), S locus receptor kinase (SRKs) and creatine phosphokinase (CPKs), were not detected in the present Bu. truncatus transcriptome. Comprehensive transcriptomic and proteomic studies of all developmental stages of Bu. truncatus should establish whether these molecules are absent from this snail species, and should facilitate studies of snail-schistosome interactions.
Other proteins encoded in the present Bu. truncatus transcriptome could be linked to roles in signal transduction, cell growth and death, the immune system, environmental adaptation and/or the excretory/secretory system. For example, kinases and phosphatases linked to NF-kappa B-, Toll-, IL-1- and C-type lectin receptor-signalling pathways could be elements of the innate immune response in the snail (cf. Schultz & Adema, 2017; Castillo et al., 2020; Li et al., 2020). Select lectins and FREPs/FReDs as well as cathepsins might also be linked to snail defence(s) against pathogens including schistosomes (cf. Lockyer et al., 2012; Gordy et al., 2015; Lu et al., 2020).
A substantial number of transcripts in Bu. truncatus encoded orphan molecules which could not be functionally annotated. Given that these transcripts contained complete ORFs and were assembled based on sequenced RNA molecules (rather than predicted de novo, without evidence of being transcribed), they are unlikely to represent technical artifacts. The lack of homology to non-molluscan sequences may be explained by rapid evolution, leading to taxon- or lineage-specific neo-functionalisation of these proteins (Tautz & Domazet-Loso, 2011). Its alloploidy and potential for other chromosomal-level events may also facilitate accelerated change of gene content over time. The predicted two- and three-dimensional structures of some of these molecules indicate that they are located to the plasma membrane or are extracellular, which may relate to interaction of the snail with the environment and/or pathogens. Other orphans had orthologs in Bi. glabrata that were encoded by members of resistance gene clusters, suggesting functions related to resistance to infection by schistosomes. Such orphans deserve detailed investigation, as they might represent snail-specific molecules involved in pathogen recognition and defence.
In conclusion, the transcriptome and inferred proteome characterised in the present study provide novel and important resources that will assist future efforts to decode the entire genome of Bu. truncatus, underpin proteomic and metabolomic studies, and aid in elucidating the molecular biology of this snail and how it interacts with larval stages of schistosome parasites. The present work also creates opportunities for similar investigations of invertebrate intermediate hosts of socioeconomically important parasitic trematodes.
Funding
This project was supported through grants LP180101334 (N.D.Y. and P.K.K.) and LP180101085 (R.B.G.) from the Australian Research Council (ARC). P.K.K. held an Early Career Research Fellowship from the National Health and Medical Research Council (NHMRC) of Australia.
CRediT author statement
Andreas J. Stroehlein: Conceptualisation, Methodology, Formal analysis, Writing - Original Draft, Writing - Review & Editing. Pasi K. Korhonen: Methodology, Data Curation, Writing - Review & Editing, Funding acquisition. David Rollinson: Writing - Review & Editing, Funding acquisition. J. Russell Stothard: Writing - Review & Editing, Funding acquisition. Ross S. Hall: Methodology, Data Curation, Writing - Review & Editing. Robin B. Gasser: Conceptualisation, Writing - Review & Editing, Supervision, Project administration, Funding acquisition. Neil D. Young: Conceptualisation, Methodology, Writing - Review & Editing, Supervision, Project administration, Funding acquisition. All authors read and approved the final manuscript.
Data availability
Sequence data has been submitted to the Sequence Read Archive (SRA) with the accession numbers SRR13147979 (Illumina short-reads) and SRR13164753 (Oxford Nanopore long-reads). The transcriptome assembly has been submitted to the Transcriptome Shotgun Assembly (TSA) Sequence Database with the NCBI BioProject accession number PRJNA680620. All other data generated or analysed in this study are included in this article and its supplementary files.
Declaration of competing interests
The authors declare that they have no competing interests.
Acknowledgements
Bulinus truncatus snails were kindly provided by Dr Margaret Mentink-Kane of the NIH-NIAID Schistosomiasis Resource Center, Biomedical Resource Institute, Rockville, MD 20850, USA.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crpvbd.2021.100015.
Contributor Information
Andreas J. Stroehlein, Email: astroehlein@unimelb.edu.au.
Neil D. Young, Email: nyoung@unimelb.edu.au.
Abbreviations
- aa
amino acid
- BIR
baculovirus IAP repeat
- bp
base pair
- BUSCO
Benchmarking Universal Single Copy Orthologs
- CAMK
Ca2+/calmodulin-dependent protein kinase
- cAMP
cyclic adenosine monophosphate
- CPKs
creatine phosphokinase
- ECM
extracellular matrix
- FREP/FReD
fibrinogen-related protein
- GO
Gene Ontology
- GRC
Guadeloupe Resistance Complex
- IAP
Inhibitor of Apoptosis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG orthology
- MAPK
mitogen-activated protein kinase
- miRNA
micro-RNA
- MLCKs
myosin light-chain kinase
- mRNA
messenger RNA
- nt
nucelotides
- ORF
open reading frame
- PDB
Protein Data Bank
- PhKs
phosphorylase kinase
- piRNA
piwi-interacting RNA
- PTC2
Polymorphic Transmembrane Cluster 2
- RNA-Seq
RNA-sequencing
- SRKs
S locus receptor kinase
- TLRs
Toll-like receptor
- TM
topology modelling
- UTR
untranslated region
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adema C.M., Bayne C.J., Bridger J.M., Knight M., Loker E.S., Yoshino T.P., et al. Will all scientists working on snails and the diseases they transmit please stand up? PLoS Negl. Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema C.M., Hillier L.W., Jones C.S., Loker E.S., Knight M., Minx P., et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 2017;8:15451. doi: 10.1038/ncomms15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstead C.A., Korhonen P.K., Young N.D., Hall R.S., Jex A.R., Murali S.C., et al. Lucilia cuprina genome unlocks parasitic fly biology to underpin future interventions. Nat. Commun. 2015;6:7344. doi: 10.1038/ncomms8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum T., Briesemeister S., Kohlbacher O. MultiLoc2: Integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. CRC Press; London: 1994. Freshwater snails of Africa and their medical importance. [Google Scholar]
- Buddenborg S.K., Bu L., Zhang S.M., Schilkey F.D., Mkoji G.M., Loker E.S. Transcriptomic responses of Biomphalaria pfeifferi to Schistosoma mansoni: Investigation of a neglected African snail that supports more S. mansoni transmission than any other snail species. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddenborg S.K., Kamel B., Bu L., Zhang S.M., Mkoji G.M., Loker E.S. Transcriptional responses of Biomphalaria pfeifferi and Schistosoma mansoni following exposure to niclosamide, with evidence for a synergistic effect on snails following exposure to both stressors. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0006927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmanova E., Antipov D., Lapidus A., Prjibelski A.D. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience. 2019;8 doi: 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M.G., Humphries J.E., Mourao M.M., Marquez J., Gonzalez A., Montelongo C.E. Biomphalaria glabrata immunity: Post-genome advances. Dev. Comp. Immunol. 2020;104:103557. doi: 10.1016/j.dci.2019.103557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley D.G., Bustinduy A.L., Secor W.E., King C.H. Human schistosomiasis. Lancet. 2014;383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusoe M.R., Alameldin H.F., Awad S., Boucher E., Caldwell A., Cartwright R., et al. The Khmer software package: Enabling efficient nucleotide sequence analysis. F1000Research. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly N.M., Duval D., Mouahid G., Emans R., Allienne J.F., Galinier R., et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev. Comp. Immunol. 2015;48:234–243. doi: 10.1016/j.dci.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty C.E., Zhao M., McManus D.P., Duke M.G., Cummins S.F., Wang T. Comparative study of excretory-secretory proteins released by Schistosoma mansoni-resistant, susceptible and naive Biomphalaria glabrata. Parasit. Vectors. 2019;12:452. doi: 10.1186/s13071-019-3708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer K.K., Niazi U.H., Duval D., Cosseau C., Tomlinson C., Chalmers I.W., et al. The Biomphalaria glabrata DNA methylation machinery displays spatial tissue expression, is differentially active in distinct snail populations and is modulated by interactions with Schistosoma mansoni. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordy M.A., Pila E.A., Hanington P.C. The role of fibrinogen-related proteins in the gastropod immune response. Fish Shellfish Immunol. 2015;46:39–49. doi: 10.1016/j.fsi.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.L., Bolt B.J., Shafie M., Kersey P., Berriman M. WormBase ParaSite - a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017;215:2–10. doi: 10.1016/j.molbiopara.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., von Mering C., et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernandez-Plaza A., Forslund S.K., Cook H., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Helminth Genomes Consortium Comparative genomics of the major parasitic worms. Nat. Genet. 2019;51:163–174. doi: 10.1038/s41588-018-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny N.J., Truchado-Garcia M., Grande C. Deep, multi-stage transcriptome of the schistosomiasis vector Biomphalaria glabrata provides platform for understanding molluscan disease-related pathways. BMC Infect. Dis. 2016;16:618. doi: 10.1186/s12879-016-1944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Song L., Breitwieser F.P., Salzberg S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjetland E.F., Hegertun I.E., Baay M.F., Onsrud M., Ndhlovu P.D., Taylor M. Genital schistosomiasis and its unacknowledged role on HIV transmission in the STD intervention studies. Int. J. STD AIDS. 2014;25:705–715. doi: 10.1177/0956462414523743. [DOI] [PubMed] [Google Scholar]
- Korhonen P.K., Young N.D., Gasser R.B. Making sense of genomes of parasitic worms: Tackling bioinformatic challenges. Biotechnol. Adv. 2016;34:663–686. doi: 10.1016/j.biotechadv.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Leger E., Webster J.P. Hybridizations within the genus Schistosoma: Implications for evolution, epidemiology and control. Parasitology. 2017;144:65–80. doi: 10.1017/S0031182016001190. [DOI] [PubMed] [Google Scholar]
- Liebsch M., Becker W. Comparative glucose tolerance studies in the freshwater snail Biomphalaria glabrata: Influence of starvation and infection with the trematode Schistosoma mansoni. J. Comp. Physiol. B. 1990;160:41–50. doi: 10.1007/BF00258761. [DOI] [PubMed] [Google Scholar]
- Li H., Hambrook J.R., Pila E.A., Gharamah A.A., Fang J., Wu X., et al. Coordination of humoral immune factors dictates compatibility between Schistosoma mansoni and Biomphalaria glabrata. eLife. 2020;9 doi: 10.7554/eLife.51708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer A.E., Emery A.M., Kane R.A., Walker A.J., Mayer C.D., Mitta G., et al. Early differential gene expression in haemocytes from resistant and susceptible Biomphalaria glabrata strains in response to Schistosoma mansoni. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Loker E.S., Adema C.M., Zhang S.M., Bu L. Genomic and transcriptional analysis of genes containing fibrinogen and IgSF domains in the schistosome vector Biomphalaria glabrata, with emphasis on the differential responses of snails susceptible or resistant to Schistosoma mansoni. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E., Braam J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003;159:585–598. doi: 10.1046/j.1469-8137.2003.00845.x. [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C.J., Vos T., Lozano R., Naghavi M., Flaxman A.D., Michaud C., et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- Natsidis P., Schiffer P.H., Salvador-Martinez I., Telford M.J. Computational discovery of hidden breaks in 28S ribosomal RNAs across eukaryotes and consequences for RNA Integrity Numbers. Sci. Rep. 2019;9:19477. doi: 10.1038/s41598-019-55573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.K., Cruz B.C., Buena K.L., Nguyen H., Sullivan J.T. Effects of abnormal temperature and starvation on the internal defense system of the schistosome-transmitting snail Biomphalaria glabrata. J. Invertebr. Pathol. 2016;138:18–23. doi: 10.1016/j.jip.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo E. Association between schistosomiasis and cancer: A review. Infect. Dis. Clin. Pract. 2007;15:145–148. [Google Scholar]
- Pila E.A., Tarrabain M., Kabore A.L., Hanington P.C. A novel Toll-like receptor (TLR) Influences compatibility between the gastropod Biomphalaria glabrata, and the digenean trematode Schistosoma mansoni. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz F.R., Silva L.M., Jeremias W.J., Baba E.H., Caldeira R.L., Coelho P.M.Z., et al. Differential expression of small RNA pathway genes associated with the Biomphalaria glabrata/Schistosoma mansoni interaction. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racioppi L., Means A.R. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: Novel routes for an ancient traveller. Trends Immunol. 2008;29:600–607. doi: 10.1016/j.it.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Rollinson D. A wake up call for urinary schistosomiasis: Reconciling research effort with public health importance. Parasitology. 2009;136:1593–1610. doi: 10.1017/S0031182009990552. [DOI] [PubMed] [Google Scholar]
- Schultz J.H., Adema C.M. Comparative immunogenomics of molluscs. Dev. Comp. Immunol. 2017;75:3–15. doi: 10.1016/j.dci.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E.M., Korhonen P.K., Campbell B.E., Young N.D., Jex A.R., Jabbar A., et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.J., Wilding C.S., Grahame J. Intron analyses reveal multiple calmodulin copies in Littorina. J. Mol. Evol. 2005;60:505–512. doi: 10.1007/s00239-004-0232-3. [DOI] [PubMed] [Google Scholar]
- Stroehlein A.J., Korhonen P.K., Chong T.M., Lim Y.L., Chan K.G., Webster B., et al. High-quality Schistosoma haematobium genome achieved by single-molecule and long-range sequencing. GigaScience. 2019 doi: 10.1093/gigascience/giz108. 8, giz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Domazet-Loso T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Tennessen J.A., Bollmann S.R., Peremyslova E., Kronmiller B.A., Sergi C., Hamali B., et al. Clusters of polymorphic transmembrane genes control resistance to schistosomes in snail vectors. eLife. 2020;9 doi: 10.7554/eLife.59395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J.A., Theron A., Marine M., Yeh J.Y., Rognon A., Blouin M.S. Hyperdiverse gene cluster in snail host conveys resistance to human schistosome parasites. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.S., Karunaratne L.B., Lewis F.A., Freitas T.C., Liang Y.S. Schistosomiasis. Curr. Protoc. Immunol. 2013;103 doi: 10.1002/0471142735.im1901s103. 19 11 11-19 11 58. [DOI] [PubMed] [Google Scholar]
- VectorBase release 49 beta; BglaB1.6. https://vectorbase.org/vectorbase/app/record/organism/TMPTX_bglaBB02
- Waterhouse R.M., Seppey M., Simao F.A., Manni M., Ioannidis P., Klioutchnikov G., et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf M.J., de Vlas S.J., Brooker S., Looman C.W., Nagelkerke N.J., Habbema J.D., et al. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86:125–139. doi: 10.1016/s0001-706x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- Wilson D., Pethica R., Zhou Y., Talbot C., Vogel C., Madera M., et al. SUPERFAMILY - sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright B., Lacchini A.H., Davies A.J., Walker A.J. Regulation of nitric oxide production in snail (Lymnaea stagnalis) defence cells: A role for PKC and ERK signalling pathways. Biol. Cell. 2006;98:265–278. doi: 10.1042/BC20050066. [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: Protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.D., Jex A.R., Cantacessi C., Campbell B.E., Laha T., Sohn W.M., et al. Progress on the transcriptomics of carcinogenic liver flukes of humans - unique biological and biotechnological prospects. Biotechnol. Adv. 2010;28:859–870. doi: 10.1016/j.biotechadv.2010.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data has been submitted to the Sequence Read Archive (SRA) with the accession numbers SRR13147979 (Illumina short-reads) and SRR13164753 (Oxford Nanopore long-reads). The transcriptome assembly has been submitted to the Transcriptome Shotgun Assembly (TSA) Sequence Database with the NCBI BioProject accession number PRJNA680620. All other data generated or analysed in this study are included in this article and its supplementary files.