Abstract
Babesia gibsoni is considered as an emerging protozoan parasite of dogs in North America and Europe. However, no data have been published on its prevalence, molecular-phylogenetic characteristics and associated co-infections in dogs used for illegal fighting (i.e. predisposed to acquiring this piroplasm via biting) in Europe. In this study, blood samples from 79 American Staffordshire Terrier dogs, confiscated for illegal dog fights, were molecularly analyzed for tick-borne pathogens. Babesiagibsoni was detected in 32 dogs, i.e. with a prevalence of 40.5%. In addition, Babesia vulpes was found in 8 samples (prevalence of 10.1%), for the first time in dogs in Hungary. Canine hemoplasmas were also identified in 49 samples (62%): only Mycoplasma haemocanis in 32 (40.5%) dogs, only “Candidatus Mycoplasma haematoparvum” in 9 (11.4%) dogs, and both hemoplasmas in 8 (10.1%) dogs. Thus, hemoplasma infections also showed a particularly high prevalence in this dog population. Based on a partial fragment of the 18S rRNA gene, B. gibsoni from Hungary exhibited complete sequence identity with conspecific strains reported from Europe and Asia. The cytochrome c oxidase subunit 1 (cox1) gene sequence of this isolate showed the closest identity with B. gibsoni reported from Japan but had a nonsynonymous mutation (M33I). Furthermore, the 11 B. gibsoni-positive samples analyzed for sequence variants of the cytochrome b (cytb) gene showed the presence of a common mutation (P310S). Most importantly, B. gibsoni had two further nonsynonymous mutations, M121I and F258L, in a dog with severe and relapsing anemia following atovaquone treatment. Phylogenetically, both cytb sequence variants clustered together, with a clear geographical pattern showing the closest relationship of both haplotypes identified in Hungary with those from China and Japan. To the best of our knowledge, this is the first cox1 and cytb characterization of B. gibsoni in Europe, as well as the first report on the emergence of this piroplasm and hemoplasmas with high prevalence among “fighting dogs” north of the Mediterranean Basin.
Keywords: Piroplasms, cox1, cytb, Babesia vulpes, Canine hemoplasmas
Graphical abstract
Highlights
-
•
High prevalence of Babesia gibsoni and canine hemoplasmas in “fighting dogs”.
-
•
Molecular evidence for infection with Babesia vulpes in dogs in Hungary.
-
•
Closest identity with sequences from Asia in two of the three genetic markers tested.
-
•
Two unique nonsynonymous mutations of B. gibsoni in the case of a dog.
-
•
First report of atovaquone resistance for B. gibsoni in Europe.
1. Introduction
Piroplasms (Apicomplexa: Piroplasmida), i.e. species of Babesia, Theileria and Cytauxzoon, are obligate intracellular protozoan parasites, transmitted cyclically to their vertebrate hosts by hard ticks (Schreeg et al., 2016). Among them, members of the group Babesia (sensu stricto) infect erythrocytes, thus affecting humans, wildlife, livestock and companion animals (Homer et al., 2000; Schreeg et al., 2016). Canine babesiosis is caused by several Babesia spp., which are classified based on their size. Thus, large Babesia species of the dog include Babesia canis, Babesia vogeli and Babesia rossi, all from the group Babesia (s.s.) (Schreeg et al., 2016). On the other hand, B. gibsoni (Babesia (s.s.)), Babesia conradae (Western Babesia group) and B. vulpes (Babesia microti group) belong to the category of small Babesia species (Solano-Gallego et al., 2016; Schreeg et al., 2016). Among small Babesia spp., B. gibsoni is regarded as an emerging pathogen, with increasing significance in dogs both in North America and Europe (Boozer and Macintire, 2005; Solano-Gallego et al., 2016). This species may induce severe clinical signs such as lethargy, fever, hemolytic anemia, splenomegaly, although it may also cause subclinical infection depending on such factors, as the pathogenicity of its strain, the immune status or age of the host (Solano-Gallego and Baneth, 2011; Kirk et al., 2017).
Considering the epidemiology of B. gibsoni infection, this piroplasm can also be acquired transplacentally (Fukumoto et al., 2005). However, the predominant mode of its transmission appears to be through bite wounds (Kirk et al., 2017), in which cases usually illegal dog “fighting” and relevant breeds (American Staffordshire Terrier and American Pit Bull Terrier) are involved (Kirk et al., 2017; Birkenheuer et al., 2018). These dogs frequently have co-infections with other hemotropic pathogens which are also able to spread via dog bites, as exemplified by B. vulpes and canine hemotropic mycoplasmas (Mycoplasma haemocanis and “Candidatus Mycoplasma haematoparvum”) (Cannon et al., 2016; Barash et al., 2019). In addition, similar to other Babesia spp., B. gibsoni can be transmitted by hard ticks. Babesia gibsoni has a worldwide distribution, and is endemic to North America, Asia, Africa, Australia and Europe. Hard tick vectors of this piroplasm include Haemaphysalis longicornis and H. bispinosa in Asia, probably H. leachi and/or Rhipicephalus sanguineus (s.s.) in Africa (Matjila et al., 2007), and the latter tick species in North America and Europe (Boozer and Macintire, 2005; Solano-Gallego and Baneth, 2011; Prakash et al., 2018; Jain Jose et al., 2018).
In Europe, the prevalence of B. gibsoni tends to be higher in countries of the Mediterranean Basin and the Balkan Peninsula, where its biological vector, R. sanguineus occurs, ranging between 0.7–5.7% in random sample groups and 2.0–28.6% among symptomatic and “fighting dogs” (Solano-Gallego et al., 2016). However, owing to the possibility of its spread by other means, the sporadic, autochthonous occurrence of B. gibsoni has also been documented north of the Mediterranean Basin (Solano-Gallego et al., 2016). In particular, clinical cases have been reported from Mediterranean countries (Croatia, Italy and Spain), Serbia, Romania and, in addition, from Germany, Poland, Slovakia, and the UK (Smith and Wall, 2013; Solano-Gallego et al., 2016; Víchová et al., 2016; Adaszek et al., 2018). However, in Europe, data are incomplete in terms of geographical distribution and prevalence. In addition, the mutation (M121I) which is thought to be responsible for atovaquone (ATV) resistance (Iguchi et al., 2012) has not hitherto been reported on this continent. In Hungary, the first reports on the occurrence of small Babesia spp. in dogs were based on studying piroplasms in blood smears and splenic impression smears (Farkas et al., 2004; Dékay, 2013). However, their species status remains uncertain due to the lack of molecular identification, especially since B. canis can also exhibit small Babesia spp.-like morphology depending on the conditions of sampling (Demeter et al., 2011) and B. vulpes is known to occur in the country (Farkas et al., 2015). Additionally, B. gibsoni was molecularly identified in a stray dog imported from Hungary to Germany (Hamel et al., 2012).
In summary, all above (suspected or proved) past findings of B. gibsoni infections in Hungary represented single and isolated cases, partly due to the absence of an indigenous status of the biological vector, R. sanguineus (s.s.). However, recently the emergence and establishment of this tick species has been reported in the country (Hornok et al., 2020). In light of these circumstances, when B. gibsoni has recently been discovered in two “fighting dogs”, confiscated together with 84 others during a nationwide police operation against illegal dog fighting, the present study was initiated to investigate: (i) the molecular characteristics of B. gibsoni focusing on the cytochrome c oxidase subunit 1 (cox1) and cytochrome b (cytb) genes; (ii) hematological parameters and co-infections in the dogs; and (iii) the occurrence of any cytb mutations that were reported to be relevant in the context of ATV resistance.
2. Materials and methods
2.1. Origin of samples and hematological examination
In this study, samples of 86 American Staffordshire Terrier dogs were used. These dogs were confiscated by the police because they had been involved in illegal dog fights. The dogs came from the North Central region of Hungary. The mean age of dogs was 16.57 ± 16.73 months according to estimated data, and they included 30 puppies (mean age: 5.52 ± 2.87 months). The sexes were nearly evenly distributed, and no ticks were found on the dogs.
Although circumstances did not allow a detailed physical examination to be carried out, at the time of confiscation most dogs were clinically normal, but in a poor condition and with fresh or healing wounds on their bodies. However, two dogs showed lethargy and signs of anemia: in these B. gibsoni was detected via PCR during preliminary testing in a commercial laboratory. A third dog had pica and gastrointestinal symptoms. Four dogs were found dead during the police operation, at their original keeping place. From the rest of the confiscated dogs, EDTA blood samples were collected, drawn from the cephalic vein in December 2020 (prior to any treatment). Subsequently, blood smears were prepared, fixed with ethanol, stained with Giemsa and examined by a Leica light microscope (Leica Microsystems, Wetzlar, Germany).
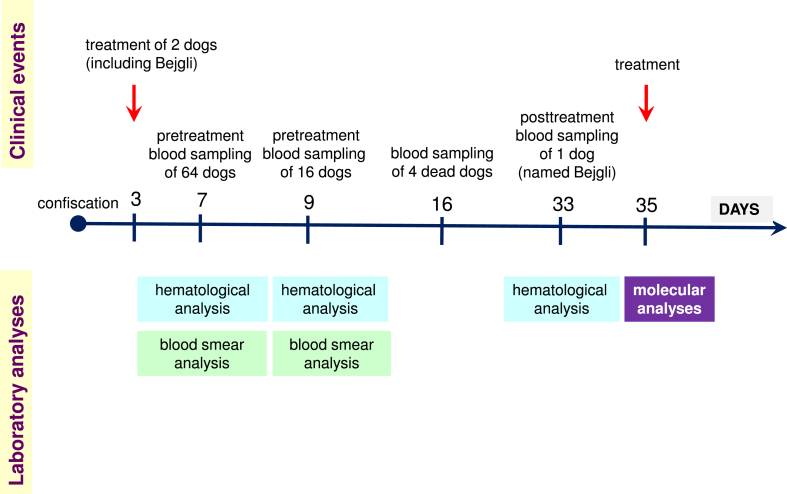
Overall, blood samples were available from 79 dogs for molecular biological analyses, and from 70 of them for hematological examination, as some samples could not be evaluated due to in vitro clotting caused by sampling error or other reasons. Hematological parameters of one dog (named Bejgli) were re-evaluated after treatment due to its relapsing state. These events are summarized in Fig. 1.
Fig. 1.
Timeline illustrating the sequence of events relevant to the study, including clinical observations/interventions, as well as laboratory analyses.
Hematological parameters were evaluated by ADVIA 2120 Hematology System (Siemens Healthineers, Erlangen, Germany). The follow-up of hematological parameters was conducted with URIT-3000 Vet Plus Hematology Analyzer (Medex Worldwide, Bucheon-si, Province, Gyeonggi-do, South Korea). Pre-treatment blood analyses were performed on the day of sampling, thereafter the remainder EDTA-anticoagulated blood samples were stored at −20 °C until further study.
2.2. Treatment
All infected dogs which had anemia (hematocrit (HCT) < 30%) were treated with ATV at a dose of 17 mg/kg BID (bis in die) and azithromycin at a dose of 13 mg/kg SID (semel in die) for 10 days. Moreover, one dog (named Bejgli) remained PCR-positive for B. gibsoni following ATV treatment and developed secondary immune-mediated hemolytic anemia. Therefore, this dog received prednisolone and pantoprazole at a dose of 2 mg/kg SID and 1 mg/kg SID, respectively. It responded well to the latter treatment.
2.3. DNA extraction, PCR and sequencing
DNA was extracted from 200 μl blood samples of 79 dogs using the blood protocol of NucleoSpin® Tissue Kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) following the manufacturer's instructions. All DNA extracts were screened for piroplasms and canine hemoplasmas, but for molecular characterization of B. gibsoni, 10–11 samples were selected to represent dogs with mono- and co-infections, including all anemic dogs for which blood sample was available.
For each PCR method, 5 μl of extracted DNA was added to 20 μl of reaction mixture containing 1.0 U HotStar Taq Plus DNA Polymerase (5 U/μl) (Qiagen, Hilden, Germany), 0.5 μl dNTP Mix (10 mM), 0.5 μl of each primer (50 μM), 2.5 μl of 10 × Coral Load PCR buffer (15 mM MgCl2 included) and distilled water (DW). The volume of DW was 15.8 μl and 14.8 μl in the 18S rDNA PCR to detect piroplasms, and in the PCRs specific for B. gibsoni, respectively. In the latter tests 1.0 μl extra MgCl2 (25 mM) was used. Further details of the PCRs are summarized in Table 1. In all PCRs sequence-verified positive controls were included (in the 18S rDNA PCR: B. vogeli DNA; in the cox1 and cytb PCRs: B. gibsoni DNA). Purification and sequencing of the PCR products were performed by Biomi Ltd. (Gödöllő, Hungary). The newly generated sequences were submitted to GenBank under the accession numbers: MW805762 and MW805763 for the 18S rRNA gene of B. gibsoni and B. vulpes, respectively; MW816918 for the cox1 gene and MW816919-MW816920 for the cytb gene of B. gibsoni.
Table 1.
Primers and details for conventional PCR methods used in this study.
| Target group | Target gene | Primer name | Primer sequence (5′-3′) | Amplicon length (bp) | Thermocycling profile | Reference |
|---|---|---|---|---|---|---|
| Piroplasms | 18S rRNA | BJ1 | GTCTTGTAATTGGAATGATGG | 500 | 95 °C for 10 min; 40 × (95 °C for 30 s; 54 °C for 30 s; 72 °C for 40 s); 72 °C for 5 min | Casati et al. (2006) |
| BN2 | TAGTTTATGGTTAGGACTACG | |||||
| Babesia gibsoni | cox1 | BgCOX1F | ATGCTTCAGAGTTATAATTCAG | 700 | 95 °C for 5 min; 35 × (95 °C for 40 s; 49 °C for 40 s; 72 °C for 90 s); 72 °C for 5 min | Guo et al. (2019) |
| MHR1 | GCTGATACAATATAGGATCTCC | Wickramasekara Rajapakshage et al. (2012) | ||||
| Babesia gibsoni | cytb | 427F | GCATTCTTAGGTTATGTTTTACCAA | 800 | 95 °C for 5 min; 35 × (95 °C for 40 s; 53 °C for 40 s; 72 °C for 90 s); 72 °C for 7 min | Birkenheuer et al. (2018) |
| CYTbR1 | ATATGCAAACTTCCCGGCTA | Wickramasekara Rajapakshage et al. (2012) |
2.4. Phylogenetic analyses
Sequences were aligned and compared to reference GenBank sequences by nucleotide BLASTn program (https://blast.ncbi.nlm.nih.gov). All sequences retrieved from GenBank and included in the phylogenetic analysis had 97–100% coverage (i.e. aligned with a near-identical length and starting position) as sequences from this study. This dataset was resampled 1000 times to generate bootstrap values. Phylogenetic analysis was conducted by using the Maximum-Likelihood method, and Jukes-Cantor model according to the best-fit selection with the program MEGA 7.0 (Kumar et al., 2016).
2.5. Real-time PCR to detect canine hemoplasmas
DNA samples were screened for canine hemoplasmas via two species-specific TaqMan real-time qPCRs, which detect part of the 16S rRNA gene of either M. haemocanis or “Ca. Mycoplasma haematoparvum” (Wengi et al., 2008). In these assays, plasmid DNA containing the cloned 16S rRNA gene of M. haemocanis or “Ca. M. haematoparvum” were used as positive controls.
2.6. Statistical analyses
Prevalence data were compared using Fisherʼs exact test. Exact 95% confidence intervals (CI) were calculated. Normality was evaluated using Kolmogorov-Smirnov test, and mean values were compared with Studentʼs t-test. Differences were regarded significant at P < 0.05.
3. Results
3.1. Phylogenetic analyses
In the PCR for detecting piroplasms, 40 of the 79 dogs were positive. DNA of B. gibsoni was identified by sequencing in 32 samples (prevalence of 40.5%; CI: 29.6–52.2%), whereas B. vulpes was present in 8 samples (prevalence of 10.1%; CI: 4.5–19.0%).
However, single or double co-infections with canine hemoplasmas were detected in 49 of the 79 dogs (62.03%, CI: 50.41–72.72%) (Table 2). Hemotropic mycoplasmas occurred with a prevalence of 62.45% (25 of 40 dogs, CI: 45.80–77.27%) in Babesia-infected dogs, and in 65.63% (21 of 32 dogs, CI: 46.81–81.43%) of B. gibsoni-infected dogs. Interestingly, out of the 30 puppies tested, 13 were PCR-negative, whereas 2 had B. gibsoni, 2 had B. vulpes and 10 had hemoplasma mono-infection. In addition, 3 of them had co-infections.
Table 2.
Occurrence of vector-borne pathogens and pretreatment hematocrit (HCT) values in fighting dogs.
| Pathogen | No. of dogs positive by PCR |
Mean HCT ± SDa in dogs with: |
||||||
|---|---|---|---|---|---|---|---|---|
| Mhc co-infection | CMhp co-infection | Mhc and CMhp co-infection | No co-infection | Mhc co-infection | CMhp co-infection | Mhc and CMhp co-infection | No co-infection | |
| Babesia gibsoni (n = 32) | 9 | 8 | 4 | 11 | 38.13 ± 9.77 | 40.86 ± 3.53 | 33.00 ± 7.26 | 39.44 ± 6.75 |
| Babesia vulpes (n = 8) | 1 | 1 | 2 | 4 | 44b | 48b | 42.50 ± 3.53 | 42b |
Abbreviations: HCT, hematocrit; SD, standard deviation; Mhc, Mycoplasma haemocanis; CMhp, “Candidatus Mycoplasma haematoparvum”.
To assess this parameter, blood samples were available from 28 B. gibsoni-infected and from 5 B. vulpes-infected dogs.
Blood sample from one dog was available for hematological evaluation.
Babesiagibsoni from Hungary showed sequence identity of 100% (413/413 bp) with conspecific strains reported from various geographical regions, including Europe (Romania: KY433318), as well as eastern and southern Asia, i.e. genotype Asia-1 from Japan (AF175300) and genotype Asia-2 from Malaysia and Sri-Lanka (AF175301). Similarly, B. vulpes from dogs in Hungary showed identity of 100% (436/436 bp) with sequences previously reported from foxes in Hungary (KM232513) or other parts of Europe (e.g. Croatia: HM212628).
Two additional genetic markers were analyzed to reveal genetic characteristics and geographical relationships of B. gibsoni that emerged in Hungary. The cox1 gene sequence of this isolate (which was identical to all the 10 analyzed Hungarian samples) showed the closest, but only 99.5–99.7% (647/650 to 648/650 bp) identity with B. gibsoni reported on GenBank from Japan (AB685182-AB685185 and AB499087, respectively). The sequence homology was lower, 99.4% (646/650 bp) in comparison with further isolates from China (KP666169) and Japan (AB685188). Based on the closest match (AB685184), one synonymous and a nonsynonymous mutation were identified, the latter representing an exchange of methionine to isoleucine at position 33 in the amino-acid chain (M33I).
Regarding the cytb gene, B. gibsoni from Hungary had two sequence variants. The most prevalent haplotype (occurring in 10 out of 11 tested dogs) was 99.9% (732/733 bp) identical to the most closely related sequence available on GenBank, reported from Japan (AB685184). This single nucleotide polymorphism, characteristic of all haplotypes of B. gibsoni sequenced here (including the most divergent one outlined below) is a nonsynonymous mutation, i.e. exchange of proline to serine at position 310 in the amino-acid sequence (P310S). Despite ATV treatment, the most divergent B. gibsoni cytb haplotype was found in the dog Bejgli that had persistent anemia. This variant differed from the same reference sequence (AB685184) at three positions, amounting to only 99.6% (730/733) sequence identity. All differences represented nonsynonymous mutations, i.e. in addition to the above (P310S) there were two further changes: methionine to isoleucine at position 121 (M121I) and phenylalanine to leucine at position 258 (F258L) (Fig. 2).
Fig. 2.
Protein BLAST comparison of Babesia gibsoni cytb amino-acid sequences for the GenBank reference sequence AB685184 and the sequence from the dog named Bejgli. Yellow color indicates mutations.
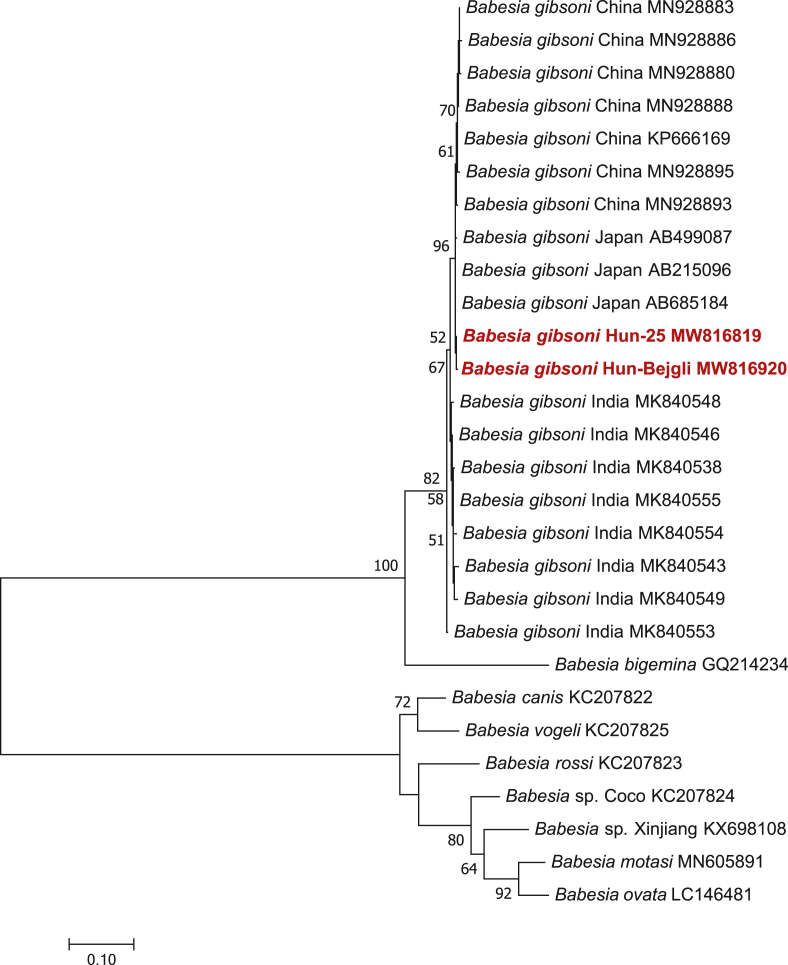
Phylogenetically, the two B. gibsoni cytb haplotypes from Hungary were associated and most closely related to haplotypes from Japan (Fig. 3). Regarding the overall relationships of Eurasian B. gibsoni cytb sequences, the topology of the phylogenetic tree reflected that their clustering is coherent according to geographical region. However, although B. gibsoni cytb sequences reported from eastern Asia (Xiʼan city and Wuhan in eastern China, as well as Japan) or southern Asia (India) formed two sister groups, their separation received low (52%) bootstrap support (Fig. 3).
Fig. 3.
Phylogenetic tree for haplotypes of Babesia gibsoni and closely related piroplasms based on the cytb gene. The tree was generated with the Maximum Likelihood method and Jukes-Cantor model in MEGA 7.0. Nucleotide sequences obtained in this study are indicated in red. There were 648 positions in the final dataset. Branch lengths represent the number of substitutions per site inferred according to the scale shown.
3.2. Hematology
Intraerythrocytic merozoites of small Babesia spp. were microscopically detected in the blood smears of all dogs which were PCR-positive for piroplasms. Hemoplasmas were not seen in the blood smears. Prior to treatment of B. gibsoni-infected dogs, their mean hematocrit (HCT) was 38.50 ± 7.29%, the red blood cell count (RBC) was (5.34 ± 1.20) × 1012/l, and the hemoglobin concentration (HGB) was 12.55 ± 2.68 g/dl. On the other hand, in B. vulpes-infected dogs the mean HCT was 43.80 ± 3.03%, the RBC was (6.10 ± 0.59) × 1012/l and the HGB was 14.50 ± 1.38 g/dl. Comparisons with dogs PCR-negative for piroplasms (HCT: 44.43 ± 5.01%; RBC: (6.38 ± 0.72) × 1012/l; HGB: 14.78 ± 1.75 g/dl), revealed that all these hematological parameters were significantly lower in B. gibsoni-infected dogs (Studentʼs t-test, HCT: t(63) = 3.89, P = 0.0002; RBC: t(63) = 4.39, P < 0.0001; HGB; t(63) = 4.06, P = 0.0001) unlike in the case of B. vulpes-infected dogs (Studentʼs t-test, HCT: t(40) = 0.27, P = 0.79; RBC: t(40) = 0.32, P = 0.75; HGB; t(40) = 0.67, P = 0.51).
In Table 2, pretreatment HCT values are summarized according to co-infections of dogs. Considering B. gibsoni-infected dogs, co-infection with both “Ca. Mycoplasma haematoparvum” (CMhp) and M. haemocanis (Mhc) resulted in significantly lower HCT (33.00 ± 7.26%) than CMhp co-infection alone (40.86 ± 3.53%; Studentʼs t-test, HCT: t(9) = 2.46, P = 0.036). Among B. gibsoni-infected dogs, 8 had mild anemia (30% < HCT< 38%), 3 had moderate anemia (HCT < 30%), and in one severe anemia developed (HCT < 20%). In the latter two groups, two dogs had co-infection with both hemoplasmas, and two had co-infection only with Mhc. The most severely affected dog (Bejgli) was concurrently infected with B. gibsoni and M. haemocanis: its HCT level was 12% and, based on follow-up, the hematological values of this dog declined despite ATV treatment. At this point, hemoplasma positivity was not yet known (Fig. 1), therefore specific treatment against M. haemocanis was not given to this dog.
4. Discussion
To the best of our knowledge, this is the first report of B. gibsoni as an emerging pathogen with remarkably high prevalence among “fighting dogs” in Hungary. The results are also relevant in a broader geographical context, because in countries north of the Mediterranean Basin hitherto only sporadic occurrence of this piroplasm has been reported. In addition, to the best of our knowledge, no data on ATV resistance of B. gibsoni have been reported in Europe, i.e. the most relevant mutation has only been reported from North America and Asia (Sakuma et al., 2009; Iguchi et al., 2012; Birkenheuer et al., 2018).
It is a major advantage of this study that, apart from the availability of blood smears and hematological data, molecular identification of piroplasms was also possible, unlike in previous reports on small Babesia spp. in dogs in Hungary (Farkas et al., 2004; Dékay, 2013). Considering the results of hematological analyses, the mean levels of the most important parameters (HCT, RBC and HGB) were significantly lower in B. gibsoni-infected dogs than in dogs PCR-negative for piroplasms. On the other hand, in the case of B. vulpes these were non-significant associations, implying that B. gibsoni caused anemia, whereas B. vulpes did not cause anemia in dogs of this study. Therefore, although the number of B. vulpes-infected dogs was too low to draw any general conclusions here, the clinical importance of this piroplasm appeared to be lower in Hungary compared to what was reported from Spain, where the majority of B. vulpes-infected dogs (79.6%) showed anemia (Miró et al., 2015). Regarding dogs harboring multiple pathogens that infected their red blood cells concurrently, pretreatment HCT values revealed that B. gibsoni co-infection with both CMhp and Mhc induced more severe anemia than CMhp co-infection alone. However, the lowest HCT value was measured in a dog infected simultaneously with B. gibsoni and Mhc. While these results indicate that co-infections may exacerbate the manifestation of babesiosis-associated clinical signs, especially if three pathogens affect red blood cells simultaneously, anemia was still less frequently observed than in a similar study in North America (49.07%; Cannon et al., 2016).
It is worth noting that hemotropic mycoplasmas may be transmitted via dog bites, similarly to B. gibsoni and probably also to B. vulpes (Cannon et al., 2016; Barash et al., 2019), thereby making the chance of co-infection more likely. It can be assumed that in the dogs studied here, the predominant mode of infection was through dog bites, since most of them took part in dog fights, presumably except for the puppies. This is confirmed by the scars found on their bodies as also described in the North American study (Cannon et al., 2016). Vertical transmission could also occur (Fukumoto et al., 2005), because several puppies were found infected among the seized dogs, without a history of dog fight. Furthermore, tick-borne transmission cannot be completely ruled out here, since R. sanguineus (s.s.), the vector of B. gibsoni (Boozer and Macintire, 2005) and canine hemoplasmas (Seneviratna et al., 1973) has been recently reported to emerge in Hungary (Hornok et al., 2020). However, the lack of any ticks on dogs of this study argues against a significant role of the latter.
Regarding molecular analyses, B. gibsoni was identified in the present study with a prevalence of 40.5%. This is considered remarkable for Europe, because similarly high prevalence (39%) has only been reported in the USA, where 269 confiscated “fighting dogs” were examined (Cannon et al., 2016). In Europe, a relatively high prevalence (28.6%) was reported in western Romania among “fighting dogs” (Imre et al., 2013). However, prevalence rates of B. gibsoni infections are usually much lower among dogs kept as pet animals. In particular, B. gibsoni occurred in 0.7% of asymptomatic dogs in Croatia (Beck et al., 2009), in 2% of dogs in Spain (Tabar et al., 2009) and in 3.3% of dogs in Serbia (Davitkov et al., 2015). In case of dogs imported from Romania and Hungary to Germany, B. gibsoni was identified with a prevalence of only 1.9% (Hamel et al., 2012). The lowest prevalence (0.2%) of B. gibsoni-infected dogs was documented in the Czech Republic (Mitkova et al., 2017).
The present survey also confirmed B. vulpes infection with a prevalence of 10.1%. To the best of our knowledge, this is the first report on the occurrence of B. vulpes in dogs in Hungary, where previously this piroplasm has only been reported in red foxes (Vulpes vulpes) (Farkas et al., 2015). Babesia vulpes has been reported in three dogs in Portugal (Simões et al., 2011), as well as in France (prevalence of 0.7%; René-Martellet et al., 2015), Croatia (prevalence of 3%; Beck et al., 2009) and Serbia (prevalence of 10.1%; Gabrielli et al., 2015). Among European countries, B. vulpes was identified with the highest prevalence (62.5%) in dogs in north-western Spain (Miró et al., 2015). Regarding North American studies, B. vulpes was either not detected in “fighting dogs” infected with B. gibsoni (Cannon et al., 2016), or these two species occurred simultaneously in some of the dogs (Barash et al., 2019). However, in the present study no co-infection was found with both piroplasms, which is most likely a consequence of the low prevalence of B. vulpes.
Interestingly, hemoplasmas were detected with a high prevalence among Babesia spp.-infected (62.45%) and B. gibsoni-infected dogs (65.63%). Both CMhp and Mhc were already reported in non-pet dogs in the southern part of Hungary (Hornok et al., 2013). It is important to note that hemoplasma co-infection compared to B. gibsoni mono-infection occurred at a higher rate in this study (Table 2) than in a previous study from North America which also focused on confiscated “fighting dogs” (28%; Cannon et al., 2016). Moreover, in the latter study strong association was observed between B. gibsoni and CMhp infections. Therefore, based on the present study, it can be assumed that dogs involved in dog fights are at an increased risk of infection with B. gibsoni, B. vulpes and hemoplasmas in Europe, similarly to what was reported in North America (Cannon et al., 2016; Barash et al., 2019).
Three genetic markers were targeted for sequence analyses. Based on the 18S rRNA gene of piroplasms, all sequences of B. gibsoni (as well as those of B. vulpes) were identical to each other and to several conspecific sequences available on GenBank from Europe and Asia. However, the cox1 sequences of B. gibsoni obtained from 10 dogs had a single nucleotide polymorphism in comparison with Asian genotypes (from Japan and China), i.e. possessed a nonsynonymous mutation (methionine to isoleucine) at position 33 in the amino-acid chain (M33I). The significance of this finding remains unknown, although mutation in cox1 gene at position 310 has been examined in connection with diminazene aceturate resistance, with lack of evidence (Wickramasekara Rajapakshage et al., 2012).
Cytochrome b gene and related mutations have been recently studied in Asia and in the USA. However, to the best of our knowledge, in Europe this is the first reported molecular study of the B. gibsoni cytb gene, in which three different mutations were detected. The mutation P310S was reported in a Japanese survey, in which it was assumed that this mutation may not contribute to the development of ATV resistance (Sakuma et al., 2012). However, in our study two other nonsynonymous mutations were also found in the cytb gene in a dog which showed relapsing anemia despite treatment with ATV. The mutation M121I was found for the first time in Europe being reported so far only in Japan (Sakuma et al., 2009; Iguchi et al., 2012) and in the USA (Birkenheuer et al., 2018). This mutation is thought to be responsible for ATV resistance (Iguchi et al., 2012). On the other hand, in this study a F258L mutation was also found, presumably for the first time in a worldwide context. Since ATV is a widely used drug in combination with azithromycin against B. gibsoni (Birkenheuer et al., 2004), the above results suggest that the possibility of therapeutic resistance should also be expected in Europe.
Considering the suitability of various genetic markers to analyze the geographical origin of Babesia isolates, the 18S rRNA gene did not reveal a clear pattern when sequences were analyzed in a geographical context (Guo et al., 2019). Similarly, according to differences in the cox1 gene among Babesia isolates, there was no phylogenetic grouping by geographical origin (Garrett et al., 2019). However, the cytb gene proved to be successful in showing geographically consistent clustering of B. gibsoni isolates from China and Japan (Guo et al., 2019). Therefore, the latter genetic marker was chosen here in an attempt to trace back the origin (or at least the geographical relationships) of the two “Hungarian” cytb strains of B. gibsoni. The phylogenetic analysis including several B. gibsoni sequences from Japan, China and India available on GenBank showed that B. gibsoni from Hungary is most closely related to conspecific haplotypes from Japan (Fig. 3). This is in line with previous, repeated findings of the Asian genotype of B. gibsoni in Europe (in Germany: Hartelt et al., 2007; in the UK: Smith and Wall, 2013) and argues for a single or a limited number of introduction event(s) from that direction. The similar phylogenetic affiliation of the Hungarian and other European B. gibsoni variants (with Chinese and Japanese, and other Asian isolates, respectively) also suggests that the typical mode of transmission in Europe is via dog bites, implying predominantly asexual reproduction of Babesia spp. in dogs (i.e. in the absence of sexual recombination in biological vector ticks, these isolates may show long-term genetic consistency in most loci).
5. Conclusions
Babesiagibsoni is an emerging pathogen in the southern part of central Europe, mainly in “fighting dogs” among which transmission probably takes place via dog bites. Co-infections with B. vulpes and hemotropic mycoplasmas may have exacerbated the symptoms. The cytb mutations found in the present study revealed the most likely geographical origin of these two B. gibsoni mitochondrial lineages in China and Japan. One of these mutations was reported to be relevant in the context of ATV resistance in North America and Asia.
Funding
This study was funded by Project no. TKP2020-NKA-01 implemented with the support provided from the National Research, Development and Innovation Fund of Hungary, financed under the “Tématerületi Kiválósági Program 2020” (2020–4.1.1-TKP2020) funding scheme.
Ethical approval
Not applicable. Sampling was performed as part of the regular veterinary care, therefore no separate ethical permission was required.
CRediT author statement
Barbara Tuska-Szalay: Conceptualization, Methodology, Investigation, Writing - Original Draft. Zsuzsanna Vizi: Methodology, Formal analysis. Regina Hofmann-Lehmann: Methodology, Investigation, Resources. Péter Vajdovich: Methodology, Validation. Nóra Takács: Methodology. Marina L. Meli: Methodology, Validation. Róbert Farkas: Formal analysis, Writing - Review & Editing. Viktória Stummer-Knyihár: Methodology. Ákos Jerzsele: Conceptualization, Funding acquisition. Jenő Kontschán: Methodology. Sándor Szekeres: Formal analysis. Sándor Hornok: Conceptualization, Methodology, Investigation, Writing - Original Draft, Supervision. All authors read and approved the final manuscript.
Data availability
The sequences generated in the study have been deposited in the GenBank database under the accession numbers MW805762 and MW805763 for the 18S rRNA gene of B. gibsoni and B. vulpes, respectively; MW816918 for the cox1 gene and MW816919-MW816920 for the cytb gene of B. gibsoni.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful for the help by the staff at the Clinical Laboratory, Department of Clinical Diagnostics and Services, Vetsuisse Faculty, University of Zurich, Switzerland. Part of the laboratory work was performed using the logistics of the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich.
References
- Adaszek L., Lyp P., Poblocki P., Skrzypczak M., Mazurek L., Winiarczyk S. The first case of Babesia gibsoni infection in a dog in Poland. Vet. Med. 2018;63:225–228. doi: 10.17221/155/2017-VETMED. [DOI] [Google Scholar]
- Barash N.R., Thomas B., Birkenheuer A.J., Breitschwerdt E.B., Lemler E., Qurollo B.A. Prevalence of Babesia spp. and clinical characteristics of Babesia vulpes infections in North American dogs. J. Vet. Intern. Med. 2019;33:2075–2081. doi: 10.1111/jvim.15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R., Vojta L., Mrljak V., Marinculić A., Beck A., Zivicnjak T., Cacciò S.M. Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int. J. Parasitol. 2009;39:843–848. doi: 10.1016/j.ijpara.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Birkenheuer A.J., Levy M.G., Breitschwerdt E.B. Efficacy of combined atovaquone and azithromycin for therapy of chronic Babesia gibsoni (Asian genotype) infections in dogs. J. Vet. Intern. Med. 2004;18:494–498. doi: 10.1892/0891-6640(2004)18%3C494:eocaaa%3E2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Birkenheuer A.J., Marr H.S., Wilson J.M., Breitschwerdt E.B., Qurollo B.A. Babesia gibsoni cytochrome b mutations in canine blood samples submitted to a US veterinary diagnostic laboratory. J. Vet. Intern. Med. 2018;32:1965–1969. doi: 10.1111/jvim.15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boozer L., Macintire D. Babesia gibsoni: an emerging pathogen in dogs. Compendium. 2005;2:33–42. [Google Scholar]
- Cannon S.H., Levy J.K., Kirk S.K., Crawford P.C., Leutenegger C.M., Shuster J.J., et al. Infectious diseases in dogs rescued during dogfighting investigations. Vet. J. 2016;211:64–69. doi: 10.1016/j.tvjl.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati S., Sager H., Gern L., Piffaretti J.C. Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Ann. Agric. Environ. Med. 2006;13:65–70. [PubMed] [Google Scholar]
- Davitkov D., Vucicevic M., Stevanovic J., Krstic V., Tomanovic S., Glavinic U., Stanimirovic Z. Clinical babesiosis and molecular identification of Babesia canis and Babesia gibsoni infections in dogs from Serbia. Acta Vet. Hung. 2015;63:199–208. doi: 10.1556/avet.2015.017. [DOI] [PubMed] [Google Scholar]
- Dékay V. A Babesia gibsoni infection in a pit bull terrier. (In Hungarian) Kisállatpraxis. 2013;14:214–218. [Google Scholar]
- Demeter Z., Palade E.A., Balogh E., Jakab C., Farkas R., Tánczos B., Hornok S. Postmortem small Babesia-like morphology of Babesia canis - short communication. Acta Vet. Hung. 2011;59:427–432. doi: 10.1556/avet.2011.029. [DOI] [PubMed] [Google Scholar]
- Farkas R., Földvári G., Fenyves B., Kótai I., Szilágyi A., Hegedüs G.T. First detection of small babesiae in two dogs in Hungary. Vet. Rec. 2004;154:176–178. doi: 10.1136/vr.154.6.176. [DOI] [PubMed] [Google Scholar]
- Farkas R., Takács N., Hornyák Á., Nachum-Biala Y., Hornok S., Baneth G. First report on Babesia cf. microti infection of red foxes (Vulpes vulpes) from Hungary. Parasit. Vectors. 2015;8:55. doi: 10.1186/s13071-015-0660-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S., Suzuki H., Igarashi I., Xuan X. Fatal experimental transplacental Babesia gibsoni infections in dogs. Int. J. Parasitol. 2005;35:1031–1035. doi: 10.1016/j.ijpara.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Gabrielli S., Otašević S., Ignjatović A., Savić S., Fraulo M., Arsić-Arsenijević V., et al. Canine babesioses in noninvestigated areas of Serbia. Vector Borne Zoonotic Dis. 2015;15:535–538. doi: 10.1089/vbz.2015.1797. [DOI] [PubMed] [Google Scholar]
- Garrett K.B., Hernandez S.M., Balsamo G., Barron H., Beasley J.C., Brown J.D., et al. Prevalence, distribution, and diversity of cryptic piroplasm infections in raccoons from selected areas of the United States and Canada. Int. J. Parasitol. Parasites Wildl. 2019;9:224–233. doi: 10.1016/j.ijppaw.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Miao X., He P., Li M., Wang S., Cui J., et al. Babesia gibsoni endemic to Wuhan, China: mitochondrial genome sequencing, annotation, and comparison with apicomplexan parasites. Parasitol. Res. 2019;118:235–243. doi: 10.1007/s00436-018-6158-2. [DOI] [PubMed] [Google Scholar]
- Hamel D., Silaghi C., Lescai D., Pfister K. Epidemiological aspects on vector-borne infections in stray and pet dogs from Romania and Hungary with focus on Babesia spp. Parasitol. Res. 2012;110:1537–1545. doi: 10.1007/s00436-011-2659-y. [DOI] [PubMed] [Google Scholar]
- Hartelt K., Rieker T., Oehme R.M., Brockmann S.O., Müller W., Dorn N. First evidence of Babesia gibsoni (Asian genotype) in dogs in western Europe. Vector Borne Zoonotic Dis. 2007;7:163–166. doi: 10.1089/vbz.2006.0580. [DOI] [PubMed] [Google Scholar]
- Homer M.J., Aguilar-Delfin I., Telford S.R., 3rd, Krause P.J., Persing D.H. Babesiosis. Clin. Microbiol. Rev. 2000;13:451–469. doi: 10.1128/cmr.13.3.451-469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornok S., Dénes B., Meli M.L., Tánczos B., Fekete L., Gyuranecz M., et al. Non-pet dogs as sentinels and potential synanthropic reservoirs of tick-borne and zoonotic bacteria. Vet. Microbiol. 2013;167:700–703. doi: 10.1016/j.vetmic.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Hornok S., Kováts D., Horváth G., Kontschán J., Farkas R. Checklist of the hard tick (Acari: Ixodidae) fauna of Hungary with emphasis on host associations and the emergence of Rhipicephalus sanguineus. Exp. Appl. Acarol. 2020;80:311–328. doi: 10.1007/s10493-019-00461-6. [DOI] [PubMed] [Google Scholar]
- Iguchi A., Matsuu A., Ikadai H., Talukder M.H., Hikasa Y. Development of in vitro atovaquone-resistant Babesia gibsoni with a single-nucleotide polymorphism in cytb. Vet. Parasitol. 2012;185:145–150. doi: 10.1016/j.vetpar.2011.09.031. [DOI] [PubMed] [Google Scholar]
- Imre M., Farkas R., Ilie M.S., Imre K., Dărăbuş G. Survey of babesiosis in symptomatic dogs from Romania: occurrence of Babesia gibsoni associated with breed. Ticks Tick Borne Dis. 2013;4:500–502. doi: 10.1016/j.ttbdis.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Jain Jose K., Lakshmanan B., Wahlang L., Syamala K., Aravindakshan T.V. Molecular evidence of haemoparasites in ixodid ticks of dogs - first report in India. Vet. Parasitol. Reg. Stud. Reports. 2018;13:177–179. doi: 10.1016/j.vprsr.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Kirk S.K., Levy J.K., Crawford P.C. Efficacy of azithromycin and compounded atovaquone for treatment of Babesia gibsoni in dogs. J. Vet. Intern. Med. 2017;31:1108–1112. doi: 10.1111/jvim.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matjila P.T., Penzhorn B.L., Leisewitz A.L., Bhoora R., Barker R. Molecular characterisation of Babesia gibsoni infection from a pit-bull terrier pup recently imported into South Africa. J. S. Afr. Vet. Assoc. 2007;78:2–5. doi: 10.4102/jsava.v78i1.277. [DOI] [PubMed] [Google Scholar]
- Miró G., Checa R., Paparini A., Ortega N., González-Fraga J.L., Gofton A., et al. Theileria annae (syn. Babesia microti-like) infection in dogs in NW Spain detected using direct and indirect diagnostic techniques: clinical report of 75 cases. Parasit. Vectors. 2015;8:217. doi: 10.1186/s13071-015-0825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkova B., Hrazdilova K., Novotna M., Jurankova J., Hofmannova L., Forejtek P., Modry D. Autochthonous Babesia canis, Hepatozoon canis and imported Babesia gibsoni infection in dogs in the Czech Republic. Vet. Med. 2017;62:138–146. doi: 10.17221/152/2016-VETMED. [DOI] [Google Scholar]
- Prakash B.K., Low V.L., Vinnie-Siow W.Y., Tan T.K., Lim Y.A., Morvarid A.R., et al. Detection of Babesia spp. in dogs and their ticks from Peninsular Malaysia: emphasis on Babesia gibsoni and Babesia vogeli infections in Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) J. Med. Entomol. 2018;55:1337–1340. doi: 10.1093/jme/tjy072. [DOI] [PubMed] [Google Scholar]
- René-Martellet M., Moro C.V., Chêne J., Bourdoiseau G., Chabanne L., Mavingui P. Update on epidemiology of canine babesiosis in Southern France. BMC Vet. Res. 2015;11:223. doi: 10.1186/s12917-015-0525-3. https://doi.org/10.1186/s12917-015-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma M., Setoguchi A., Endo Y. Possible emergence of drug-resistant variants of Babesia gibsoni in clinical cases treated with atovaquone and azithromycin. J. Vet. Intern. Med. 2009;23:493–498. doi: 10.1111/j.1939-1676.2009.0300.x. [DOI] [PubMed] [Google Scholar]
- Sakuma M., Fukuda K., Takayama K., Kobayashi Y., Shimokawa Miyama T., Setoguchi A., Endo Y. Molecular epidemiological survey of the Babesia gibsoni cytochrome b gene in western Japan. J. Vet. Med. Sci. 2012;74:1341–1344. doi: 10.1292/jvms.12-0140. [DOI] [PubMed] [Google Scholar]
- Schreeg M.E., Marr H.S., Tarigo J.L., Cohn L.A., Bird D.M., Scholl E.H., et al. Mitochondrial genome sequences and structures aid in the resolution of Piroplasmida phylogeny. PloS One. 2016;11 doi: 10.1371/journal.pone.0165702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratna P., Weerasinghe N., Ariyadasa S. Transmission of Haemobartonella canis by the dog tick, Rhipicephalus sanguineus. Res. Vet. Sci. 1973;14:112–114. [PubMed] [Google Scholar]
- Simões P.B., Cardoso L., Araújo M., Yisaschar-Mekuzas Y., Baneth G. Babesiosis due to the canine Babesia microti-like small piroplasm in dogs-first report from Portugal and possible vertical transmission. Parasit. Vectors. 2011;4:50. doi: 10.1186/1756-3305-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F.D., Wall L.E. Prevalence of Babesia and anaplasma in ticks infesting dogs in great britain. Vet. Parasitol. 2013;198:18–23. doi: 10.1016/j.vetpar.2013.08.026. [DOI] [PubMed] [Google Scholar]
- Solano-Gallego L., Baneth G. Babesiosis in dogs and cats - expanding parasitological and clinical spectra. Vet. Parasitol. 2011;181:48–60. doi: 10.1016/j.vetpar.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Solano-Gallego L., Sainz Á., Roura X., Estrada-Peña A., Miró G. A review of canine babesiosis: the European perspective. Parasit. Vectors. 2016;9:336. doi: 10.1186/s13071-016-1596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabar M.D., Francino O., Altet L., Sánchez A., Ferrer L., Roura X. PCR survey of vector-borne pathogens in dogs living in and around Barcelona, an area endemic for leishmaniasis. Vet. Rec. 2009;164:112–116. doi: 10.1136/vr.164.4.112. [DOI] [PubMed] [Google Scholar]
- Víchová B., Horská M., Blaňarová L., Švihran M., Andersson M., Petʼko B. First molecular identification of Babesia gibsoni in dogs from Slovakia, central Europe. Ticks Tick Borne Dis. 2016;7:54–59. doi: 10.1016/j.ttbdis.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Wengi N., Willi B., Boretti F.S., Cattori V., Riond B., Meli M.L., et al. Real-time PCR-based prevalence study, infection follow-up and molecular characterization of canine hemotropic mycoplasmas. Vet. Microbiol. 2008;126:132–141. doi: 10.1016/j.vetmic.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Wickramasekara Rajapakshage B.K., Yamasaki M., Hwang S.J., Sasaki N., Murakami M., Tamura Y., et al. Involvement of mitochondrial genes of Babesia gibsoni in resistance to diminazene aceturate. J. Vet. Med. Sci. 2012;74:1139–1148. doi: 10.1292/jvms.12-0056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences generated in the study have been deposited in the GenBank database under the accession numbers MW805762 and MW805763 for the 18S rRNA gene of B. gibsoni and B. vulpes, respectively; MW816918 for the cox1 gene and MW816919-MW816920 for the cytb gene of B. gibsoni.