ABSTRACT
Coronavirus infections induce the expression of multiple proinflammatory cytokines and chemokines. We have previously shown that in cells infected with gammacoronavirus infectious bronchitis virus (IBV), interleukin 6 (IL-6), and IL-8 were drastically upregulated, and the MAP kinase p38 and the integrated stress response pathways were implicated in this process. In this study, we report that coronavirus infection activates a negative regulatory loop that restricts the upregulation of a number of proinflammatory genes. As revealed by the initial transcriptomic and subsequent validation analyses, the anti-inflammatory adenine-uridine (AU)-rich element (ARE)-binding protein, zinc finger protein 36 (ZFP36), and its related family members were upregulated in cells infected with IBV and three other coronaviruses, alphacoronaviruses porcine epidemic diarrhea virus (PEDV), human coronavirus 229E (HCoV-229E), and betacoronavirus HCoV-OC43, respectively. Characterization of the functional roles of ZFP36 during IBV infection demonstrated that ZFP36 promoted the degradation of transcripts coding for IL-6, IL-8, dual-specificity phosphatase 1 (DUSP1), prostaglandin-endoperoxide synthase 2 (PTGS2) and TNF-α-induced protein 3 (TNFAIP3), through binding to AREs in these transcripts. Consistently, knockdown and inhibition of JNK and p38 kinase activities reduced the expression of ZFP36, as well as the expression of IL-6 and IL-8. On the contrary, overexpression of mitogen-activated protein kinase kinase 3 (MKK3) and MAPKAP kinase-2 (MK2), the upstream and downstream kinases of p38, respectively, increased the expression of ZFP36 and decreased the expression of IL-8. Taken together, this study reveals an important regulatory role of the MKK3-p38-MK2-ZFP36 axis in coronavirus infection-induced proinflammatory response.
IMPORTANCE Excessive and uncontrolled induction and release of proinflammatory cytokines and chemokines, the so-called cytokine release syndrome (CRS), would cause life-threatening complications and multiple organ failure in severe coronavirus infections, including severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) and COVID-19. This study reveals that coronavirus infection also induces the expression of ZFP36, an anti-inflammatory ARE-binding protein, promoting the degradation of ARE-containing transcripts coding for IL-6 and IL-8 as well as a number of other proteins related to inflammatory response. Furthermore, the p38 MAP kinase, its upstream kinase MKK3 and downstream kinase MK2 were shown to play a regulatory role in upregulation of ZFP36 during coronavirus infection cycles. This MKK3-p38-MK2-ZFP36 axis would constitute a potential therapeutic target for severe coronavirus infections.
KEYWORDS: coronavirus, p38 MAPK, MK2 kinase, MKK3 kinase, ZFP36, ARE-binding proteins, proinflammatory cytokines and chemokines
INTRODUCTION
Coronaviruses (CoVs) are a group of enveloped viruses with single-stranded, positive-sense RNA genomes, infecting many animal and human species (1). Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory coronavirus (MERS-CoV), coronavirus disease-19 (COVID-19), and coronavirus (SARS-CoV-2) have caused epidemics or pandemics in humans (2–4). Some animal coronaviruses, such as porcine transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), and avian infectious bronchitis viruses (IBV), are important veterinary pathogens (5–7).
In severe cases of SARS, MERS, and COVID-19, the massive and uncontrolled release of cytokines causes life-threatening complications and multiple organ failure, a phenomenon known as cytokine release syndrome (CRS) (8, 9). Elevated levels of some proinflammatory cytokines and chemokines, especially interleukin-6 (IL-6) and IL-8 were detected in SARS patients (10), and severe pneumonia associated with increasing IL-6 and IL-8 plasma levels was reported in COVID-19 patients (11). In PEDV-infected newborn and suckling piglets, both IL-6 and IL-8 were upregulated and contributed to an intense inflammatory response (12). IL-6 and IL-8 were also significantly induced in cells infected with IBV (13). Elucidating the regulatory mechanisms of these cytokines is, therefore, essential for fully understanding the immunopathogenesis of SARS-CoV-2 and other CoVs.
Cytokines and chemokines are small peptides or glycoproteins, synthesized and secreted by a variety of cells, primarily immune cells (14). They mediate the interactions between cells and have a variety of biological functions (14, 15). Among them, IL-8 and IL-6 are two major mediators of the inflammatory response (16). IL-8 is primarily secreted by neutrophils and acts as a chemotactic factor to attract neutrophils, basophils, T cells, but not monocytes to the site of infection (17). IL-6 is involved in the coordination of the innate and acquired immune systems. In addition, IL-6 also plays an important role in metabolic regulation, neurodevelopment, and the occurrence and maintenance of various cancers (18, 19). The core promoter of IL-6 and IL-8 is regulated by the binding of key transcription factors, including cyclic AMP response-element binding protein (CREB), C/EBP homologous protein (CHOP), activating protein-1 (AP-1) and nuclear factor-kappa B (NF-κB). Expression of IL-6 and IL-8 was also regulated post-transcriptionally by altering the mRNA stability.
Zinc finger protein 36 (ZFP36, also known as tristetraprolin, or TTP) is an RNA-binding protein, belonging to the ZFP36 family that also includes two other members, ZFP36 ring finger protein like 1 (ZFP36L1) and ZFP36 ring finger protein like 2 (ZFP36L2) (20, 21). ZFP36 is a well-characterized cytoplasmic mRNA regulator with two tandem CCCH zinc finger domains (22). By binding to adenine-uridine (AU) rich elements (AREs) in the 3′-UTR of mRNA, ZFP36 reduces the stability of the target mRNA (23, 24). The transcripts of numerous genes related to the inflammatory response have been shown to contain AREs and can be bound by ZFP36. These include IL-6, IL-8, IL-17A, IL-33, TNF-α (tumor necrosis factor-alpha) (25), CXCL2 (CXC motif chemokine ligand 2) (26), PTGS2 (prostaglandin-endoperoxide synthase 2) (26), CSF2 (colony-stimulating factor 2) (27), NLRP3 (NOD-, LRR- and pyrin domain-containing 3) (28), PFKFB3 (6-phosphofructose-2-kinase/fructose-2,6-biphosphatase 3) (29), LIF (Leukemia inhibitory factor) (26), VEGF (vascular endothelial growth factor) (30), and so on.
ZFP36 is implicated in the regulation of cytokine and chemokine expression during infection with various viruses. For example, when ZFP36 was ectopically expressed, the stability of IL-10 mRNA was significantly reduced in bone-marrow-derived macrophages (BMDMs) infected with murine cytomegalovirus (MCMV) (31); and the expression of ZFP36 was markedly reduced in the lymphocytes of chronic hepatitis B patients, presumably contributing to the inflammation associated with chronic HBV infection (32). Additionally, in cells infected with influenza A virus, the secretion of IL-6, TNF-α and other cytokines were elevated when ZFP36 expression was suppressed (33). Interestingly, a CCCH zinc-binding motif similar to those in ZFP36 was found in the M2-1 protein of respiratory syncytial virus (RSV) and was essential for M2-1 to enhance readthrough of intergenic junctions during RSV transcription (34). However, little is known about the induction and the role of ZFP36 and its related proteins in coronavirus replication and pathogenesis.
Our previous studies have shown that IBV infection drastically upregulated the expression of IL-6 and IL-8 at the mRNA level, and the MAP kinase p38 and the integrated stress response pathways play a regulatory role (13). In this study, we investigate the induction and function of ZFP36 during coronavirus infection. Expression of ZFP36 family proteins, but not other ARE-binding proteins, was significantly induced in cells infected with IBV, PEDV and two low-pathogenic human coronaviruses. More importantly, we found that ZFP36 served a vital role in regulating the mRNA levels of IL-8 and several other genes related to inflammatory response in IBV-infected cells. The induction of ZFP36 by IBV infection was shown to be dependent on the activation of the p38 mitogen activated protein kinase (MAPK) signaling pathway. Taken together, this study reveals a negative regulatory loop that may limit the excess induction and expression of genes related to the inflammatory response during coronavirus infection.
RESULTS
ZFP36 and related ARE-binding proteins are induced by coronavirus infection.
A general mRNA expression profile of host genes differentially regulated by IBV infection was obtained by transcriptomic analysis (35). As summarized in Table 1, in H1299 cells infected with IBV at MOI∼2 or mock-treated for 16 h, the transcription levels of most ARE-binding proteins were not significantly affected by IBV infection. However, the mRNA level of ZFP36 was induced by ∼23.4-fold in IBV-infected cells compared with the mock control, whereas mRNA levels of ZFP36L1 and ZFP36L2 were also induced by ∼4.2- and ∼2.3-fold, respectively (Table 1).
TABLE 1.
Transcriptomic analysis of ARE-binding proteins in H1299 cells infected with IBV
| Mock (FPKM) |
IBV infection (FPKM) |
Fold change | FDR | |||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |||
| AUF1 | 222.62 | 234.20 | 236.57 | 184.81 | 180.92 | 172.45 | 0.74 | < 0.001 |
| BRF1 | 19.61 | 18.49 | 18.64 | 20.91 | 21.00 | 22.42 | 1.07 | 0.134 |
| TIA-1 | 17.37 | 16.75 | 16.20 | 16.41 | 16.76 | 18.62 | 0.72 | < 0.001 |
| TIAR | 45.69 | 43.93 | 40.01 | 45.30 | 46.44 | 48.04 | 1.06 | 0.227 |
| ZFP36 | 24.19 | 23.81 | 24.59 | 551.08 | 585.54 | 576.21 | 23.43 | < 0.001 |
| ZFP36L1 | 86. 64 | 85.74 | 88.96 | 353.09 | 378.17 | 373.91 | 4.18 | < 0.001 |
| ZFP36L2 | 36.03 | 30.23 | 33.90 | 71.86 | 78.71 | 75.12 | 2.27 | < 0.001 |
| ELAVL1 | 80.65 | 81.22 | 83.97 | 64.78 | 64.37 | 63.27 | 0.80 | < 0.001 |
| ELAVL2 | 0.42 | 0.42 | 0.42 | 0.60 | 0.57 | 1.12 | 1.02 | 0.880 |
Note: FPKM, fragments per kilobase of transcript per million mapped reads; FDR, false discovery rate; AUF1, AU-rich binding factor 1; BRF1, TF IIB-related factor 1; TIA-1, T-cell restricted intracellular antigen-1; TIAR, T-cell intracellular antigen 1 related protein; ZFP36, zinc finger protein 36; ZFP36L1, ZFP36 ring finger protein like 1; ZFP36L2, ZFP36 ring finger protein like 2; ELAVL1, embryonic lethal, abnormal vision, Drosophila-like 1; ELAVL2, embryonic lethal, abnormal vision, Drosophila-like 2.
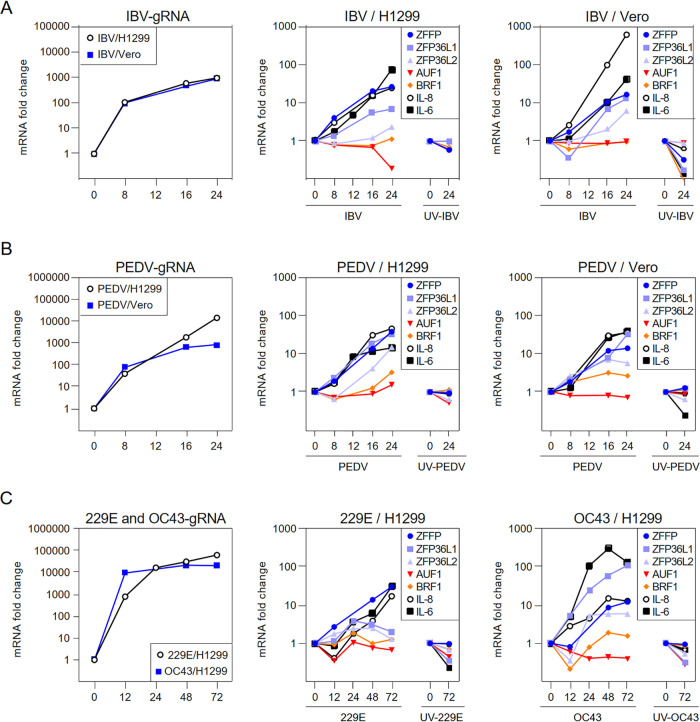
The temporal mRNA expression profiles of selected ARE-binding proteins were then analyzed by time course infection experiments in cells infected with gammacoronavirus IBV, alphacoronavirus PEDV and HCoV-229E, and betacoronavirus HCoV-OC43, respectively. As shown in Fig. 1, the expression of ZFP36 at the mRNA level was induced by 10–40-folds at late stages of coronavirus infection. The induction of ZFP36L1 and ZFP36L2 was more recognizable in cells infected with PEDV, compared with cells infected with other coronaviruses. The mRNA levels of AUF1 (AU-rich binding factor 1) and BRF1 (TF IIB-related factor 1) were not significantly affected by all coronaviruses tested. Notably, both IL-6 and IL-8 were drastically induced at the mRNA level in cells infected with all four coronaviruses (Fig. 1). This and all following experiments were repeated three times, and the result of one representative experiment is shown. Taken together, these data suggest that coronavirus infection induced the expression of ZFP36 and related ARE-binding proteins at late stages of the infection cycle.
FIG 1.
ZFP36 and related ARE-binding proteins are induced by coronavirus infection. H1299 and Vero cells were infected with IBV, PEDV, HCoV-OC43, and HCoV-229E at MOI∼2 or mock-treated with UV-inactivated viruses. Cells were harvested at the indicated time points and total RNAs were extracted for RT-qPCR. The levels of IBV, PEDV, HCoV-OC43, and HCoV-229E genomic RNA (CoV gRNA), and the mRNA levels of ZFP36 family (ZFP36, ZFP36L1, and ZFP36L2), other ARE-binding proteins (AUF1/BRF1), IL-6 and IL-8 were determined by the ΔΔCt method using the GAPDH mRNA from virus-infected cells harvested at 0 hpi for normalization.
ZFP36 regulates the expression of IL-6, IL-8, and other genes related to proinflammatory response during coronavirus infection.
ZFP36 is a well-characterized ARE-binding protein that regulates the stability of transcripts harboring AREs, including mRNAs of IL-8, IL-6, dual-specificity phosphatase 1 (DUSP1), prostaglandin-endoperoxide synthase 2 (PTGS2), and tumor necrosis factor-α-induced protein 3 (TNFAIP3). To test the effect of ZFP36-knockdown on the mRNA levels of these genes, Vero and H1299 cells were transfected with siRNA duplexes targeting EGFP (negative control) or siZFP36, before being infected with respective coronaviruses at MOI∼2. The replication of IBV, PEDV, HCoV-229E and HCoV-OC43 was not significantly affected by the knockdown of ZFP36, as similar levels of IBV titers and gRNA levels of PEDV, HCoV-229E and HCoV-OC43 were determined in the ZFP36-knockdown cells compared with the siEGFP control (Fig. 2A and 3A). RT-qPCR analysis revealed that the endogenous ZFP36 mRNA level was effectively reduced in cells transfected with siZFP36 (Fig. 2A and 3A). Notably, mRNA levels of IL-8, IL-6 and DUSP1 were significantly higher in ZFP36-knockdown cells infected with IBV, compared with the siEGFP control (Fig. 2A). Transcription of PTGS2 and TNFAIP3 was also moderately upregulated by the knockdown of ZFP36 (Fig. 2A). Consistently, upregulation of IL-8, IL-6, DUSP1, PTGS2 and TNFAIP3 at the mRNA levels was also observed in ZFP36-knockdown cells infected with PEDV, HCoV-229E and HCoV-OC43, respectively, comparied with the siEGFP control (Fig. 3A).
FIG 2.
ZFP36 regulates the expression of IL-6, IL-8 and other genes related to proinflammatory response during IBV infection. A. H1299 and Vero cells were transfected with siEGFP and siZFP36 before being infected with IBV at an MOI of ∼2. Cells were harvested at the indicated time points and lysates were prepared for RNA extraction or by three freeze/thaw cycles. Equal amounts of total RNA were reverse-transcribed, and the mRNA levels of ZFP36, IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from siEGFP-transfected and mock-infected cells harvested at 16 hpi for normalization. Virus titers were expressed in units of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. B. H1299 and Vero cells were transfected with pXJ40 and pXJ40-ZFP36 before being infected with IBV at an MOI of ∼2 or mock-infected (M). Cells were harvested at the indicated time points and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left. C. Total RNA samples were extracted from cells in (B) and subjected to RT-qPCR. The levels of IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40-transfected and mock-infected cells at 16 hpi for normalization. Total lysates were prepared from cells harvested in (B) at the indicated time points by three freeze/thaw cycles. Virus titers were determined and expressed in units of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. D. Vero and H1299 cells were either untransfected or transfected with siEGFP, siZFP36, pXJ40, or pXJ40-ZFP36, respectively. Cells were then infected with IBV at an MOI of 2, and treated with actinomycin D (Act D) (10 μM/ml) and DMSO at 16 h postinfection. Total RNA samples were extracted at the indicated time points and subjected to RT-qPCR analysis. The levels of IBV gRNA and ZFP36 were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40 or siEGFP-transfected and IBV-infected cells at 0 h before treatment with actinomycin D for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. E. Total RNA samples were extracted from cells in (D) and subjected to RT-qPCR. The levels of IL-8 were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40 or siEGFP-transfected and IBV-infected cells at 0 h before treatment with actinomycin D for normalization.
FIG 3.
ZFP36 regulates the expression of IL-6, IL-8 and other genes related to proinflammatory response during PEDV, HCoV-229E and HCoV-OC43 infection. A. H1299 and Vero cells were transfected with siEGFP and siZFP36 before being infected with PEDV, HCoV-229E and HCoV-OC43 at MOI∼2. Total RNA samples were extracted at the indicated time points. Equal amounts of total RNA were reverse-transcribed, and the mRNA levels of gRNA, ZFP36, IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from siEGFP-transfected and mock-infected cells harvested at 36 hpi for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. B. H1299 and Vero cells were transfected with pXJ40 and pXJ40-ZFP36 before being infected with PEDV, HCoV-229E, and HCoV-OC43 at MOI∼2 or mock-infected (M). Total RNA samples were extracted at the indicated time points. Equal amounts of total RNA were reverse-transcribed, and the mRNA levels of gRNA, ZFP36, IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from siEGFP-transfected and mock-infected cells harvested at 36 hpi for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined.
To complement the loss-of-function experiment, ZFP36 was cloned to a eukaryotic expression vector pXJ40 and transiently expressed in H1299 and Vero cells, respectively, before being infected with IBV, PEDV, HCoV-229E and HCoV-OC43, respectively. Expression of the ectopic ZFP36, but not the endogenous protein, was detected by Western blotting (Fig. 2B). Overexpression of ZFP36 did not significantly affect IBV, PEDV, HCoV-229E, and HCoV-OC43 replication, as suggested by the comparable levels of IBV N protein and supernatant IBV titer as well as the gRNA levels of PEDV, HCoV-229E, and HCoV-OC43 relative to the vector control (Fig. 2B, 2C, and 3B). Importantly, overexpression of ZFP36 markedly reduced the mRNA levels of IL-8, DUSP1, PTGS2, and TNFAIP3 in Vero and H1299 cells infected with IBV or PEDV-infected, and in H1299 cells infected with HCoV-229E or HCoV-OC43 (Fig. 2C and 3B). Taken together, these results suggest that ZFP36 downregulates the mRNA levels of IL-8 and other genes related to proinflammatory response, presumably by binding to AREs in the transcripts to reduce the mRNA stability.
The mRNA stability of ZFP36 target genes in Vero and H1299 cells infected with IBV was further studied by treatment with actinomycin D or DMSO (negative control). As shown in Fig. 2D and E, the mRNA level of IL-8 was reduced by 65% after treatment with actinomycin D within 2 h, while much less reduction was detected in DMSO-treated cells. In ZFP36-knockdown cells, treatment with actinomycin D showed a slightly higher level of IL-8 mRNA, while a significantly lower level of IL-8 was detected in cells overexpressing ZFP36 in the presence of actinomycin D (Fig. 2D and E). Overall, these results further confirm that ZFP36 affects the mRNA stability of ARE-containing IL-8 genes during IBV infection.
The RNA-binding activity of ZFP36 is required for its regulatory function in gene expression.
ZFP36 contains two C3H1-type zinc finger RNA-binding domains. To confirm that the RNA-binding activity is essential for its function during coronavirus infection, two point mutations, C124R and C147R, were introduced to each of the two zinc finger domains. Both C124R and C147R have been previously shown to abolish the ARE-binding activity of ZFP36 (27). Vero or H1299 cells were transfected with the two mutants, along with the pXJ40 vector and wild type pXJ40-ZFP36. As shown in Fig. 4A, compared with wild type ZFP36, the protein level of ZFP36-C124R was moderately reduced in Vero cells, whereas protein levels of both ZFP36-C124R and ZFP36-C147R were increased in H1299 cells. Overexpression of wild type and mutant ZFP36 did not affect IBV replication, as indicated by the comparable levels of IBV N protein and supernatant IBV titers (Fig. 4A and B). Notably, compared with the vector control, the IBV-induced upregulation of IL-8, DUSP1, PTGS2 and TNFAIP3 mRNA was markedly reduced in cells transfected with wild type ZFP36 but not the two mutants (Fig. 4B). Taken together, these results suggest that the RNA-binding activity of both zinc fingers of ZFP36 was required for its regulatory function in the expression of ARE-containing genes during IBV infection.
FIG 4.
The RNA-binding activity of ZFP36 is required for its regulatory function on gene expression. A. H1299 and Vero cells were transfected with pXJ40, pXJ40-ZFP36, pXJ40-ZFP36-C124R, and pXJ40-ZFP36-C174R, respectively, before being infected with IBV at MOI∼2 or mock-infected (M). Cells were harvested at the indicated time points and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. B. Total RNA samples were extracted from cells in (A) and subjected to RT-qPCR. The levels of IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40-transfected and mock-infected cells at 16 hpi for normalization. The lysates were prepared from cells harvested in (A) at the indicated time points by three freeze/thaw cycles. Virus titers were determined and expressed in the unit of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined.
Induction of ZFP36, IL-6 and IL-8 by IBV infection was dependent on the activation of p38 and JNK.
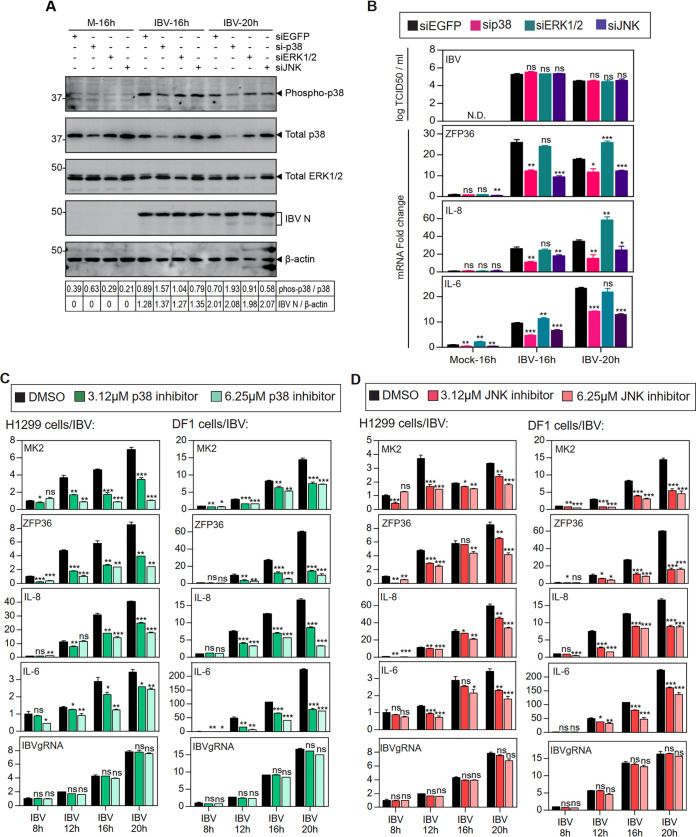
In our previous studies, the induction of IL-6 and IL-8 expression in IBV-infected cells was mediated by the p38 MAPK (36); whereas IBV-induced apoptosis was differentially regulated by JNK and ERK1/2 (37, 38). To see whether MAPKs are also implicated in the induction of ZFP36, H1299 cells were transfected with siRNA targeting EGFP (negative control), p38, ERK1/2 and JNK, before being infected with IBV. As shown in Fig. 5A, the protein levels of phosphorylated p38, total p38 and ERK1/2 were moderately reduced in cells transfected with the respective siRNA. Consistent with our previous observation, knockdown of p38, ERK1/2 and JNK did not affect IBV replication, as shown by the similar levels of IBV N protein and supernatant IBV titers compared with the siEGFP control (Fig. 5A and B). It was noted that induction of ZFP36, IL-6 and IL-8 mRNA was significantly reduced in p38-knockdown and JNK-knockdown cells, but was unaffected or slightly increased in ERK1/2-knockdown cells (Fig. 5A and B).
FIG 5.
Induction of ZFP36 and IL-8 by IBV infection was dependent on the activation of p38 and JNK. A. H1299 cells were transfected with siEGFP, sip38, siJNK and siERK before being infected with IBV at MOI∼2, cells were harvested at the indicated time points and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. B. Total RNA samples were extracted from cells in (A) and subjected to RT-qPCR. The levels of ZFP36, IL-6 and IL-8 were determined by the ΔΔCt method using the GAPDH mRNA extracted from siEGFP-transfected and mock-infected cells at 16 hpi for normalization. The cells lysates were harvested from cells in (A) at the indicated time points by three freeze/thaw cycles, and virus titers were determined and expressed in the unit of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. C. H1299 and DF1 cells were infected with IBV at MOI∼2 and treated with indicated concentrations of SB-203580 or the same volume of DMSO at 2 hpi. Total RNAs were extracted and subjected to RT-qPCR. The levels of MK2, ZFP36, IL-6, IL-8, and IBV genomic RNA were determined by the ΔΔCt method using the GAPDH mRNA extracted from infected and DMSO-treated cells at 0 hpi for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. D. H1299 and DF1 cells were treated with SP600125 as in (C). Total RNAs were extracted and subjected to RT-qPCR. The levels of MK2, ZFP36, IL-6, IL-8, and IBV genomic RNA were determined by the ΔΔCt method using the GAPDH mRNA extracted from infected and DMSO-treated cells at 0 hpi for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined.
To complement the RNAi experiments, we adopted two commonly used p38 and JNK inhibitors, SB203580 and SP600125, respectively. H1299 cells or chicken fibroblast DF1 cells were first inoculated with IBV. At 2 h postinfection, unabsorbed viruses were removed, and cells were treated with medium containing 3.12 μM or 6.25 μM inhibitors or the same volume of DMSO (solvent control). As shown in Fig. 5C, IBV replication was marginally reduced in DF1 cells treated with 6.25 μM p38 inhibitor, but not significantly affected in other groups. However, ZFP36, IL-6 and IL-8 were significantly reduced at the mRNA level by the treatment with the p38 inhibitor in a dosage-dependent manner (Fig. 5C). The induction of MK2, a protein substrate of p38 known to upregulate ZFP36 expression, was also reduced by p38 inhibition. Similarly, the expression levels of MK2, ZFP36, IL-8, and IL-6 mRNA in IBV-infected cells were all suppressed by the treatment of the JNK inhibitor (Fig. 5D). Taken together, these results suggest that among the three MAPKs studied, both p38 and JNK contributed to the induction of ZFP36, IL-6, and IL-8 during IBV infection, and transcriptional control mechanisms would play a major regulatory role in these proinflammatory cytokine expression during coronavirus infection.
ZFP36 acts as a negative regulator controlling the expression of IL-6, IL-8, and other genes related to proinflammatory response during coronavirus infection.
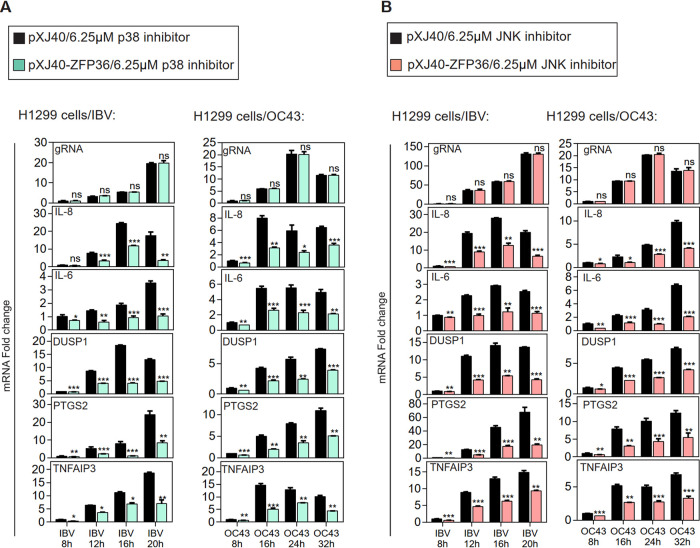
The function of ZFP36 in regulating proinflammatory cytokine expression during coronavirus infection was further studied in ZFP36-overexpressing H1299 cells infected with IBV and HCoV-OC43, respectively, and treated with 6.25 μM p38 or JNK inhibitors. As shown in Fig. 6A, IBV and HCoV-OC43 replication were not affected by ZFP36 overexpression and p38 inhibitors treating, as similar levels of IBV and HCoV-OC43 gRNA were determined in the ZFP36-overexpressed cells compared with the vector control; the mRNA levels of IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 were significantly reduced in the ZFP36-overexpressed cells, compared with the vector control. Similarly, the expression levels of IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 mRNA in IBV- or HCoV-OC43-infected cells were all suppressed by the treatment with the JNK inhibitor (Fig. 6B). Taken together, these results confirm that ZFP36 negatively regulates the expression of IL-6, IL-8 and other genes related to proinflammatory response during coronavirus infection.
FIG 6.
ZFP36 acts as a negative regulator controlling the expression of IL-6, IL-8 and other genes related to proinflammatory response during IBV and HCoV-43 infections. A. H1299 cells were transfected with pXJ40 and pXJ40-ZFP36 before being infected with IBV or HCoV-OC43 at MOI∼2 and treated with indicated concentrations of SB-203580 at 2 hpi. Total RNA samples were extracted from cells at the indicated time points and subjected to RT-qPCR. The levels of IL-6, IL-8, DUSP1, PTGS2, TNFAIP3, and IBV or HCoV-OC43 gRNA were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40-transfected and infected as well as p38 inhibitor-treated cells at 8 hpi for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. B. H1299 cells were treated with SP600125 as in (A). Total RNA samples were extracted from cells at the indicated time points and subjected to RT-qPCR. The levels of IL-6, IL-8, DUSP1, PTGS2, TNFAIP3, and IBV or HCoV-OC43 genomic RNA were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40-transfected and infected as well as JNK inhibitor-treated cells at 8 hpi for normalization. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined.
MK2 (MAPKAP kinase-2) activates the expression of ZFP36, IL-6, IL-8, and other genes related to proinflammatory response during IBV infection.
We next looked at the p38 substrate protein MK2 and how it regulates the expression of these inflammation-related genes. Vero and H1299 cells were transfected with siEGFP or siMK2, before being infected with IBV. As shown in Fig. 7A, the mRNA expression of MK2 was significantly reduced in cells transfected with siMK2. IBV replication was not affected by MK2 knockdown, as indicated by the comparable supernatant IBV titers compared with the siEGFP control. Importantly, mRNA levels of ZFP36, IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 were markedly reduced in the MK2-knockdown Vero cells and moderately reduced in the MK2-knockdown H1299 cells, compared with their respective siEGFP controls (Fig. 7A).
FIG 7.
MK2 activates the expression of ZFP36, IL-8 and other genes related to proinflammatory response during IBV infection. A. H1299 and Vero cells were transfected with siEGFP and siMK2 before being infected with IBV at MOI∼2. Cells were harvested at the indicated time points for RNA extraction or by three freeze/thaw cycles. Equal amounts of total RNA were reverse-transcribed, and the mRNA expression levels of MK2, ZFP36, IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from siEGFP-transfected and mock-infected cells at 16 hpi for normalization. Virus titers were determined and expressed in the unit of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined. B. H1299 and Vero cells were transfected with pXJ40-FLAG and pXJ40-FLAG-MK2 before being infected with IBV at MOI∼2 or mock infected (M). Cells were harvested at the indicated time points and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. C. Total RNA samples were extracted from cells in (B) and subjected to RT-qPCR. The levels of ZFP36, IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40-transfected and mock-infected cells at 16 hpi for normalization. Virus titers were determined from cells lysates harvested in (B) at the indicated time points by three freeze/thaw cycles and expressed in the unit of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined.
MK2 was also cloned to the pXJ40-FLAG expression plasmid for gain-of-function experiments. As shown in Fig. 7B, Vero and H1299 cells were first transfected with pXJ40-FLAG and pXJ40-FLAG-MK2, respectively, before being infected with IBV. Overexpression of MK2 did not affect IBV replication, as indicated by the comparable levels of IBV N protein and supernatant IBV titers but resulted in moderate increase in the IBV-induced mRNA expression of ZFP36, IL-8, DUSP1, PTGS2, and TNFAIP3 in Vero cells, and a significant increased at the mRNA level of these genes in H1299 cells (Fig. 7B and C). Taken together, these results suggest that MK2 activates the expression of ZFP36, IL-8 and several other genes related to proinflammatory response during IBV infection.
MKK3 (Mitogen-activated protein kinase kinase 3) activates the expression of ZFP36, IL-8, and other genes related to proinflammatory response.
Finally, we looked at MKK3, an upstream kinase of p38 MAPK, and how it regulates the expression of ZFP36 and these inflammation-related genes. MKK3 was cloned into the expression vector pXJ40 with an N-terminal FLAG tag. In addition, both S218 and T222 phosphorylation sites were mutated to alanine, generating the inactive mutant MKK3-AA. Mutations of the same two residues to glutamate were also made to generate the phosphomimetic constitutively active mutant MKK3-EE. Vero and H1299 cells were transfected with the vector plasmid, wild type MKK, MKK3-AA and MKK3-EE, respectively, before being infected with IBV. As shown in Fig. 8A, the ectopic wild type and mutant MKK3 proteins were expressed at comparable levels. Overexpression of either wild type or the two mutant forms of MKK3 did not affect IBV replication, as indicated by the similar levels of IBV N protein and supernatant IBV titers (Fig. 8A and B). Compared with the respective vector controls, the mRNA levels of ZFP36, IL-8, DUSP1, PTGS2 and TNFAIP3 were significantly increased in cells transfected with wild type MKK3 or MKK3-EE but were unaffected in cells transfected with MKK3-AA (Fig. 8A and B). Taken together, these results suggest that MKK3 activates the expression of ZFP36 and other genes during IBV infection, presumably by activating the p38-MK2 signaling pathway.
FIG 8.
MKK3 activates the expression of ZFP36, IL-8 and other genes related to proinflammatory. A. H1299 and Vero cells were transfected with pXJ40-FLAG, pXJ40-FLAG-MKK3, pXJ40-FLAG-MKK3-AA, and pXJ40-FLAG-MKK3-EE, before being infected with IBV at MOI∼2 or mock-infected (M). Cells were harvested at the indicated time points and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left. B. Total RNAs were extracted from cells in (A) and subjected to RT-qPCR. The levels of ZFP36, IL-8, DUSP1, PTGS2, and TNFAIP3 were determined by the ΔΔCt method using the GAPDH mRNA extracted from pXJ40-FLAG-transfected and mock-infected cells at 16 hpi for normalization. Virus titers were determined from cell lysates harvested in (A) at the indicated time points by three freeze/thaw cycles and expressed in the unit of log TCID50 per ml. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). N.D., non-determined.
DISCUSSION
Severe coronavirus infections are often associated with life-threatening cytokine release syndromes (2, 4, 6). Extensive research has been focused on the host innate and adaptive immune systems, but the critical roles of some highly conserved cellular stress responses have been overlooked. In this study, we characterized the regulatory functions of the ARE-binding protein ZFP36 in proinflammatory response during coronavirus infection. Our results have shown that in coronavirus-infected cells, ZFP36 is induced by the p38/JNK-MK2 pathway and regulates the mRNA levels of IL-6, IL-8 and several genes related to the inflammatory response (Fig. 9).
FIG 9.
Diagram illustrating the current working model. The working model showing the induction of proinflammatory cytokines and chemokines by MKK3-p38-MK2-ZFP36 axis during coronavirus infection. Pointed and blunt arrows denote activation and suppression, respectively. Dotted lines denote processes that are not fully characterized.
The dysregulated and excessive release of proinflammatory cytokines is one of the clinical hallmarks underlying the immunopathogenesis of severe and critical COVID-19, as well as SARS, MERS and other lethal coronavirus diseases (4, 6, 39). Proinflammatory cytokines, especially IL-6, can activate the acute-phase response in the hepatocytes of severe COVID-19 patients (40). Lung CT scan of COVID-19 patients revealed multiple bilateral lobular pneumonia that was associated with initial increased plasma concentrations of IL-1β, IL-7, IL-8, and IL-9 (11). In this study, we showed that IL-6 and IL-8 were drastically increased in H1299 cells infected with IBV, HCoV-229E, and HCoV-OC43, and in Vero cells infected with PEDV and IBV, suggesting that induction of proinflammatory cytokines/chemokines is likely a mechanistic generality during coronavirus infection. However, this induction was limited by the simultaneous induction of ZFP36 in the infected cells.
ZFP36 is an anti-inflammatory ARE-binding protein and a member of the zinc-finger family that can either stabilize or destabilize mRNA transcripts (20). Rapid induction of ZFP36 has been reported in cells infected with several viruses. In porcine kidney PK-15 cells infected with foot-and-mouth disease virus (FMDV), ZFP36 mRNA level increased by ∼50-folds within 1 h postinfection (41). Similarly, ZFP36 mRNA was also significantly induced in caprine endometrial epithelial cells (EECs) infected with peste des petits ruminant’s virus (PPRV) (42), and in human foreskin fibroblasts (HFFs) and HeLa cells infected with Herpes Simplex Virus 1 (HSV-1) (43). However, ZFP36 mRNA level was reduced by 70% in CD8+ T lymphocytes from chronic hepatitis B virus (CHB) patients, while no significant change was detected in the mRNA levels of ZFP36L1 and ZFP36L2 (32). In this study, significant induction of ZFP36 mRNA expression was observed in H1299 and Vero cells infected with IBV, PEDV, HCoV-229E, and HCoV-OC43, respectively, at late stages of infection. Upregulation of the homologous ZFP36L1 and ZFP36L2 was also detected in some coronavirus-infected cells, but no significant induction of other ARE-binding proteins (such as AUF1 and BRF1) was detected. Thus, among the well characterized ARE-binding proteins, only ZFP36 family proteins are markedly induced by coronavirus infection.
Previous studies have shown that ZFP36 regulates the mRNA stability of various cytokines/chemokines (such as TNF-α, IL-6 and IL-8) and other genes related to the inflammatory response (such as PTGS2 and TNFAIP3) (26, 44, 45). The binding of the two C3H1-type zinc fingers ZFP36 to the ARE in the 3′-UTR of these mRNAs delivers these transcripts to processing bodies for mRNA degradation (46). In fact, ZFP36-deficient mice developed inflammatory phenotypes that resemble transgenic mice overexpressing TNF-α (47). Consistently, we have observed that knockdown of ZFP36 significantly increased the mRNA levels of IL-6, IL-8, DUSP1, PTGS2 and TNFAIP3 induced by IBV infection in H1299 and Vero cells, whereas overexpression of ZFP36 markedly reduced the mRNA levels of these genes. Furthermore, the RNA-binding activity of ZFP36 was required for this regulatory function, as mutation of the essential cysteine residues in the zinc fingers abolished its effect on the above ARE-containing mRNAs. Some recent studies have shown that the regulation of cytokine transcripts by ZFP36 also involves microRNA (miRNA) and its processing machinery. For example, ARE-mediated instability of the TNF-α mRNA is also dependent on miR16 (a human miRNA containing a UAAAUAUU sequence complementary to ARE) and components involved in miRNA processing (48). Further experiments are required to explore these mechanisms in the context of coronavirus infection.
It has been reported that MK2, the prime target of p38, serves as the master regulator of RNA-binding proteins that modulates the transcript stability (49). When MK2 is phosphorylated and activated by p38, its nuclear localization signal is masked and MK2 is retained in the cytoplasm (49). Mechanistically, MK2 not only stimulates the transcription of ZFP36, but also increases its cellular protein abundance by enhancing its cytoplasmic retention and protecting it from proteasomal degradation (50). However, it has also been shown that MK2 can directly phosphorylates ZFP36 in activated macrophages and promote the assembly of ZFP36 with the protein chaperone 14-3-3, thereby excluding ZFP36 from stress granules and inhibiting ZFP36-dependent degradation of ARE-containing transcripts (51). Phosphorylation of ZFP36 by MK2 also prevents the recruitment of mRNA deadenylase, but it does not impair the RNA binding ability of ZFP36 (52). In this study, chemical inhibition and knockdown of p38 and JNK significantly reduced the mRNA levels of MK2 and ZFP36 in IBV-infected cells, whereas overexpression of p38 upstream kinase MKK3 or MK2 increased the induction of ZFP36. These results confirm the essential role of the MKK-p38/JNK-MK2 signaling axis in activating ZFP36 expression during coronavirus infection. Meanwhile, the induction of IL-6, IL-8, DUSP1, PTGS2, and TNFAIP3 was highly dependent on MKK-p38/JNK-MK2 signaling but was also subjected to the negative feedback regulation of ZFP36.
Several viral proteins have been shown to modulate ZFP36 expression or function. For example, during the lytic reactivation of Kaposi's sarcoma associated herpesvirus (KSHV), the viral RNA-binding protein ORF57 binds to and stabilizes specific host pre-mRNAs, including that of ZFP36 (53). In another study, the Tax oncoprotein encoded by delta retroviruses was found to colocalize and physically interact with ZFP36, thereby inhibiting ZFP36-mediated destabilization/degradation of TNF-α (54). Whether coronavirus-encoded proteins engage in similar interactions with ZFP36 remains an open question.
Our previous studies have shown that DUSP1, induced by p38 in IBV-infected cells, acts as a negative regulator to dephosphorylate p38, thereby modulating the induction of IL-6 and IL-8 (13, 55). In terms of TNFAIP3, it is induced in cells infected with HCoV-229E and suppresses the NF-κB-dependent cytokine production by inhibiting the activation of IKKβ (56). TNFAIP3 was also identified as a signature gene that was upregulated in various immune cells from severe COVID-19 patients compared to moderate cases or healthy controls (57). PTGS2, also known as COX2, catalyzes the rate-limiting step of prostaglandin production in response to injury and inflammation, thereby mediating the fever response (58). PTGS2 is upregulated by SARS-CoV-2 in human cells and in a mouse model (59). Although treatment with PTGS2 inhibitor had no effect on viral replication, it reduced the production of proinflammatory cytokines and impaired the humoral immune response by reducing neutralizing antibody titers in a mouse model (59). Our results suggest that ZFP36 is activated by p38 MAPK and downregulates the mRNA expression levels of DUSP1, TNFAIP3, and PTGS2. Therefore, ZFP36 can be considered a negative feedback regulator that modulates proinflammatory response during coronavirus infection.
In conclusion, we have demonstrated the regulatory role of ZFP36 in the proinflammatory response in coronavirus-infected cells. By binding to these ARE-containing transcripts, ZFP36 modulates the mRNA levels of critical cytokines and other important genes implicated in the proinflammatory and host innate immune response. It is of hope that further studies on ZFP36 and other ARE-binding proteins will reveal their potential as therapeutic targets in severe diseases caused by coronaviruses.
MATERIALS AND METHODS
Cell culture and virus.
Vero cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, Carlsbad, CA, USA) supplemented with 6% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. H1299 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 8% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (Gibco). All cells were grown in a 37°C incubator supplied with 5% CO2.
The egg-adapted Beaudette strain of IBV (ATCC VR-22) was obtained from the American Type Culture Collection (ATCC) and adapted to Vero cells as previously described (60, 61). This Vero-adapted strain was named IBV-p65, and the complete genome sequence was uploaded (accession No. DQ001339) (62). The HCoV-229E (accession No. KU291448.1) (63) and HCoV-OC43 (accession No. KU131570.1) (64) were also obtained from ATCC, and the two complete genome sequences were uploaded. PEDV virulent strain DR13 (PEDV-vDR13) was isolated in Korea in 1999 (accession No. JQ023162) as previously reported (65).
To prepare the virus stocks, monolayers of Vero cells were infected with IBV-p65 or PEDV-vDR13 and monolayers of MRC-5 cells were infected with HCoV-229E and HCoV-OC43 at a multiplicity of infection (MOI) of approximately 0.1 and cultured in plain DMEM at 37°C, until almost the entire monolayers exhibited cytopathic effects (CPE) in the form of multinucleated syncytia. After three rounds of freeze-thaw, the cell lysates were clarified by centrifugation at 3000×rpm at 4°C for 10 min, aliquoted in 1.5 ml screw-cap vials and stored at −80°C.
The titers of the virus stocks were determined by plaque assays and 50% tissue culture infective dose (TCID50) assay. IBV, PEDV, HCoV-229E, and HCoV-OC43 stocks were used for infection of cells at an MOI of approximately 2 in all the experiments, whereas the mock control was incubated with same amounts of uninfected cell lysates.
Transcriptomic analysis.
Transcriptomic analysis was carried out by the Biomarker Technologies Co, LTD, Beijing, China. Briefly, RNA was independently prepared from IBV-infected H1299 cells harvested at 20 h postinfection and sequenced using Illumina HiSeq sequencing technology platform to construct transcriptome libraries and obtain sequencing data.
Plasmid constructions and transfection.
The complementary DNA (cDNA) of human ZFP36 (NM_003407.5) was amplified from total RNA of H1299 cells by reverse transcription-PCR (RT-PCR), using the forward primers CTATAGGGCGAATTCGGATCCACCATGGATCTGACTGCCATCTAC and reverse primers AAGATCTGGTACCGAGCTCCTCACTCAGAAACAGAGATGCGATTG. The cDNA of human MKK3 (NM_145109) was amplified in the same way using the forward primer CGATGATAAGTCCGGATCTGAGTCGCCCGCCTCGAGCCA and reverse primers AAGATCTGGTACCGAGCTCCTGCAGCTATGAGTCTTCTCCCAGGATCTCC. The cDNA of human MK2 (NM_032960.4) was also amplified similarly using the forward primer CGATGATAAGTCCGGATCCATGCTGTCCAACTCC CAG and the reverse primers AAGATCTGGTACCGAGCTCCTCAGTGGGCCAG AGCCGCA. The PCR product of ZFP36 was inserted to the vector pXJ40 linearized with BamHI and pstI by homologous recombination, whereas PCR products of MKK3 and MK2 were inserted to pXJ40-FLAG linearized with BamHI and PstI by homologous recombination. These constructs were named pXJ40-ZFP36, pXJ40-FLAG-MKK3 and pXJ40-FLAG-MK2, respectively.
The ZFP36-C124R mutant was generated by site-directed mutagenesis to replace cysteine 124 with arginine in pXJ40-ZFP36 using the forward primer CAAGAGACAGTTTGCCCATGGCCTGG and reverse primer TGGGCAAACTGTCTCTTGGCCCCGTAG. The ZFP36-C174R mutant was generated by replacing cysteine 174 with arginine in pXJ40-ZFP36 using the forward primer CGGAACTCAGACACAAGTTCTACCTCCAGGGC and the reverse primer ACTTGTGTCTGAGTTCCGTCTTGTATTTGGGGT. Both C124R and C174R have been shown to abolish the binding of ZFP36 to AU-rich elements (66).
The MKK3-AA mutant was generated by replacing serine 218 and threonine 222 with alanine in pXJ40-FLAG-MKK3 using the forward primer GATGCAGTGGCAAAGGCTATGGATGCCGGCTGC and the reverse primer ATAGCCTTTGCCACTGCATCCACCAAGTAGCCACTGATGCCAA. The MKK3-EE mutant was generated by replacing serine 218 and threonine 222 with glutamate in pXJ40-FLAG-MKK3 using the forward primer GAAGTGGCCAAGGAAATGGATGCCGGCTG and reverse primer CATTTCCTTGGCCACTTCGTCCACCAAGTAGCCAC. The MKK3-AA mutant cannot be phosphorylated by upstream kinases, whereas the MKK3-EE serve as a phosphomimetic mutant (67).
Plasmids DNA were transfected into H1299 cells using TransIntro EL transfection regent (Transgen biotech), while transfection of Vero cell was done using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction. Briefly, cells were seeded on the 12-well plate the day before transfection. Transfection was performed when cells approached about 60–70% confluence. 2 μg of plasmid DNA and 4 μl of TransIntro EL/Lipofectamine 3000 was each diluted with 100 μl of Opti-MEM (Gibco) and incubated for 5 min. The diluted plasmid and transfection regent were mixed by brief vortex and further incubated for 15 min. Cells were cultured with medium containing 5% FBS and the transfection mixture was added dropwise to each well. The medium was replaced with normal complete medium after 6–8 h. At 24h posttransfection, cells were infected with IBV at an MOI of 2 or mock infected. RNA and/or protein samples were then harvested at the different time points.
RNA interference.
ZFP36 siRNA (+): GGGAUCCGACCCUGAUGAAUA, p38 siRNA (+): GAACUGCGGUUACUUAAAC, JNK siRNA (+): AAAGAAUGUCCUACCUUCU, ERK1/2 siRNA (+): GACCGGAUGUUAACCUUUA, MK2 siRNA (+): GGAGAACUCUUUAGCCGAA were purchased from Sangon Biotech. Transfection of siRNA was performed using TransIntro EL (Transgen biotech) according to the manufacturer’s instructions. Briefly, monolayers of the H1299 or Vero cells were seeded on 12 wells plate and cultured to 40–50% confluence. 5 μl siRNA (20 μM) were mixed with 2.5 μl TransIntro EL for each well. At 36–48 h posttransfection, cells were infected with IBV at an MOI of 2 or mock infected. RNA and/or protein samples were then harvested at the different time points.
Three sets of siRNA duplexes were used in all knockdown experiments, and the representative data from the siRNA duplex with the best knockdown efficiency are presented.
RNA extraction and RT-qPCR analysis.
Total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were lysed with 1 ml TRIzol per 10 cm2 effective growth area, and the lysates were vigorously mixed with one-fifth volume of chloroform. The mixture was then centrifuged at 12,000 × g at 4°C for 15 min, and the aqueous phase was mixed with an equal volume of isopropanol. The RNA was precipitated by centrifugation at 12,000 × g at 4°C for 15 min, washed twice with 70% ethanol, and dissolved in 30–50 μl RNase-free water.
The total RNA was reverse transcribed using the FastKing gDNA Dispelling RT SuperMix kit (Tiangen) according to the manufacturer’s instructions. Briefly, 2 μg total RNA was mixed with 4 μl 5× FastKing-RT SuperMix (containing RT enzyme, RNase inhibitor, random primers, oligo dT primer, dNTP and reaction buffer) in a 20 μl reaction mixture. Using a thermo cycler, reverse transcription was performed at 42°C for 15 min and the RT enzyme was then inactivated at 95°C for 3 min. The cDNA was then diluted 20-fold with RNase-free water for quantitative PCR (qPCR) analysis, using the Talent qPCR PreMix SYBR green kit (Tiangen) according to the manufacturer’s instructions. Briefly, 8.4 μl diluted cDNA was mixed with 10 μl 2× qPCR PreMix, 0.4 μl 50× ROX, 0.6 μl 10 μM forward primer, and 0.6 μl 10 μM reverse primer for a 20 μl reaction mixture. The qPCR analysis was performed using a QuantStudio 3 real-time PCR system (Applied Biosystems). The standard protocol included enzyme activation at 50°C for 3 min, initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturing (95°C, 5 sec) and annealing/extension (60°C, 30 sec) with fluorescent acquisition at the end of each cycle. The results were obtained in the form of cycle threshold (CT) values. Using the ΔΔCT method, the relative abundance of a transcript was calculated using GAPDH as an internal control and normalized to the respective reference sample in each experiment.
The following qPCR primers (forward and reverse) were used: GAPDH, CTGGGCTACACTGAGCACC and AAGTGGTCGTTGAGGGCAATG; IBVgRNA, GTTCTCGCATAAGGTCGGCTA and GCTCACTAAACACCACCAGAAC; IBVsgRNA2, GCCTTGCGCTAGATTTTTAACTG and AGTGCACACAAAAGAGTCACTA; PEDV gRNA, CTGAGCAAATTCGCTGGCG and AACACCCTCAGTACGAGTCC; 229E gRNA, TAGGTTTTGACAAGCCTCAGGAAAAAGA and ACGAGCAAGACTCTTGGCAG; OC43 gRNA, CTATCTGGGAACAGGACCGC and TTGGGTCCCGATCGACAATG; ZFP36, GACTGAGCTATGTCGGACCTT and GAGTTCCGTCTTGTATTTGGGG; ZFP36L1, TCCAGCATAGCTTTAGCTTTGC and GGTCATCGGCGCTCAGAATAG; ZFP36L2, GAGAACAAATTCCGGGACCG and GCGTGGAGTTGATCTGGGAG; AUF1, GCGTGGGTTCTGCTTTATTACC and TTGCTGATATTGTTCCTTCGACA; BRF1, CTGAGGCTCCTACAGAGGATG and CACACACTTTGACCACACTGA; MK2, CCAGGAGAAATTCGCCCTCAA and TCGTACACATCCACGATCCGT; TNFAIP3, TCCTCAGGCTTTGTATTTGAGC and TGTGTATCGGTGCATGGTTTTA; DUSP1, GCCTTGCTTACCTTATGAGGAC and GGGAGAGATGATGCTTCGCC; PTGS2, TAAGTGCGATTGTACCCGGAC and TTTGTAGCCATAGTCAGCATTGT; ELAV1, TTGGGCGGATCATCAACTCG and TCAAACCGGATAAACGCAACC; ELAV2, GTCCAACCACCATAAACAACAAC and GCTCTGCCCTGTTATTTTGTCT; IL-6, GTGCAAATGAGTACAAAAGTCCTGA and GTTCTGCGCCTGCAGCTTC; IL-8, AAGACGTACTCCAAACCTATCCAC and TCTGTATTGACGCAGTGTGGTC.
SDS-PAGE and Western blot analysis.
To obtain whole-cell lysates for protein analysis, cells were harvested at the indicated time points using cell scrapers (Corning) and collected by centrifugation at 16,000g for 1 min. The supernatant was discarded, and the cell pellet was lysed in RIPA buffer (10 mM Tris-HCl pH 8.0, 140 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA, and 0.5 mM EGTA). After clarified by centrifugation, the protein concentration of the cell lysate was determined. The cell lysate was then mixed with 5× Laemmli sample buffer (0.3125 M Tris-HCl pH 6.8, 10% SDS, 50% glycerol, 25% β-mercaptoethanol, and 0.025% bromophenol blue) (68). The culture supernatant was clarified by brief centrifugation and mixed with 5× Laemmli sample buffer.
Protein samples were boiled at 90°C for 5 min and centrifuged at 16,000g for 5 min. Equal amounts of protein samples were loaded to each well and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using the Bio-Rad Mini-PROTEAN Tetra cell system. The resolved proteins were then transferred to a 0.2 μm nitrocellulose membrane using the Bio-Rad Trans-Blot protein transfer system. To block off nonspecific binding sites, the membrane was incubated with 5% skim milk in TBST buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween 20) at room temperature for 1 h. The membrane was then incubated with 1 μg/ml specific primary antibody dissolved in TBST with 3% BSA (wt/vol) at 4°C overnight. The membrane was washed three times with TBST and incubated with 1:10000 diluted goat anti-rabbit or goat anti-mouse IgG secondary antibodies (Licor) at room temperature for 2 h. The membrane was washed three times with TBST, and fluorescence imaging was performed using the Azure c600 Imager according to the manufacturer’s instruction. Densitometric measurement was performed using the AzureSpot software. All experiments were repeated at least three times with similar results, and one of the representative results was shown.
Virus titration by tissue culture infective dose 50 method.
Supernatant samples were harvested from IBV-infected cells and clarified by centrifugation at 16,000 × g at 4°C for 5 min. Cell lysates were prepared by subjecting IBV-infected cells to three freeze-thaw cycles and clarified by centrifugation at 16,000 × g at 4°C for 5 min. Virus samples were kept at –80°C for less than 2 weeks before the titration experiment. Virus titer was determined by the TCID50 assay. Briefly, were. Confluent monolayers of Vero cells seeded on 96-well plates were washed once with plain DMEM, and 100 μl of 10-fold serially diluted sample was added to each well, with 8 wells used for each dilution. Cells were incubated at 37°C for 3–5 days and examined with a phase-contrast microscope. Wells were determined as either positive (with CPE) or negative (without CPE), and TCID50 was calculated using the Reed and Muench method (69). The virus titer was expressed in the unit of the logarithm of TCID50 per ml. Each sample was titrated in duplicate or triplicate in each experiment.
Statistical analysis.
The two-way ANOVA method was used to analyze the significant difference between the indicated sample and the respective control sample. Significance levels were presented by the P value (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
This work was partially supported by National Natural Science Foundation of China grants 31972660, 31900135, and 32170152 and by the Zhaoqing Xijiang Innovative Team Foundation of China.
We have no conflicts of interest to declare.
D.X.L., T.S.F., R.A.C., and S.M.L. designed and organized the study. S.M.L., S.Y.L., and M.H. did all of the experimental work. S.M.L., T.S.F., and D.X.L. wrote the manuscript.
Contributor Information
To Sing Fung, Email: tosingfung@qq.com.
Ding Xiang Liu, Email: dxliu0001@scau.edu.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Fung TS, Liu DX. 2019. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol 73:529–557. 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 2.Fung TS, Liu DX. 2021. Similarities and dissimilarities of COVID-19 and other coronavirus diseases. Annu Rev Microbiol 75:19–47. 10.1146/annurev-micro-110520-023212. [DOI] [PubMed] [Google Scholar]
- 3.Mackay IM, Arden KE. 2015. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J 12:222. 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atzrodt CL, Maknojia I, McCarthy RDP, Oldfield TM, Po J, Ta KTL, Stepp HE, Clements TP. 2020. A guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J 287:3633–3650. 10.1111/febs.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia L, Yang Y, Wang J, Jing Y, Yang Q. 2018. Impact of TGEV infection on the pig small intestine. Virol J 15:102. 10.1186/s12985-018-1012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung K, Saif LJ, Wang Q. 2020. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res 286:198045. 10.1016/j.virusres.2020.198045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu DX, Ng YL, Fung TS. 2019. Infectious bronchitis virus. In Avian virology: current research and future trends. (ed S. K. Samal). 133–178. Caister Academic Press. 10.21775/9781912530106.05. [DOI] [Google Scholar]
- 8.Hu B, Huang S, Yin L. 2021. The cytokine storm and COVID-19. J Med Virol 93:250–256. 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Channappanavar R, Perlman S. 2017. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 39:529–539. 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang K-J, Su I-J, Theron M, Wu Y-C, Lai S-K, Liu C-C, Lei H-Y. 2005. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol 75:185–194. 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.wang s, wu j, wang f, wang h, wu z, wu s, bao w. 2020. expression pattern analysis of antiviral genes and inflammatory cytokines in PEDV-infected porcine intestinal epithelial cells. Front Vet Sci 7:75. 10.3389/fvets.2020.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Y, Wang X, Huang M, Tam JP, Liu DX. 2011. Regulation of the p38 mitogen-activated protein kinase and dual-specificity phosphatase 1 feedback loop modulates the induction of interleukin 6 and 8 in cells infected with coronavirus infectious bronchitis virus. Virology 420:106–116. 10.1016/j.virol.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplansky G, Bongrand P. 2001. Cytokines and chemokines. Cell Mol Biol (Noisy-le-Grand ) 47:569–574. [PubMed] [Google Scholar]
- 15.Opal SM, DePalo VA. 2000. Anti-inflammatory cytokines. Chest 117:1162–1172. 10.1378/chest.117.4.1162. [DOI] [PubMed] [Google Scholar]
- 16.Kishimoto T. 2005. Interleukin-6: from basic science to medicine–40 years in immunology. Annu Rev Immunol 23:1–21. 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 17.Nordsieck K, Baumann L, Hintze V, Pisabarro MT, Schnabelrauch M, Beck-Sickinger AG, Samsonov SA. 2018. The effect of interleukin-8 truncations on its interactions with glycosaminoglycans. Biopolymers 109:e23103. 10.1002/bip.23103. [DOI] [PubMed] [Google Scholar]
- 18.Wallenius V, Wallenius K, Ahrén B, Rudling M, Carlsten H, Dickson SL, Ohlsson C, Jansson J-O. 2002. Interleukin-6-deficient mice develop mature-onset obesity. Nat Med 8:75–79. 10.1038/nm0102-75. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt S, Schumacher N, Schwarz J, Tangermann S, Kenner L, Schlederer M, Sibilia M, Linder M, Altendorf-Hofmann A, Knösel T, Gruber ES, Oberhuber G, Bolik J, Rehman A, Sinha A, Lokau J, Arnold P, Cabron A-S, Zunke F, Becker-Pauly C, Preaudet A, Nguyen P, Huynh J, Afshar-Sterle S, Chand AL, Westermann J, Dempsey PJ, Garbers C, Schmidt-Arras D, Rosenstiel P, Putoczki T, Ernst M, Rose-John S. 2018. ADAM17 is required for EGF-R-induced intestinal tumors via IL-6 trans-signaling. J Exp Med 215:1205–1225. 10.1084/jem.20171696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackshear PJ. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans 30:945–952. 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 21.Sanduja S, Blanco FF, Dixon DA. 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA 2:42–57. 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiedje C, Diaz-Muñoz MD, Trulley P, Ahlfors H, Laaß K, Blackshear PJ, Turner M, Gaestel M. 2016. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res 44:7418–7440. 10.1093/nar/gkw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciais D, Cherradi N, Feige J-J. 2013. Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cell Mol Life Sci 70:2031–2044. 10.1007/s00018-012-1150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275:17827–17837. 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 25.Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005. 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 26.Qiu L-Q, Lai WS, Bradbury A, Zeldin DC, Blackshear PJ. 2015. Tristetraprolin (TTP) coordinately regulates primary and secondary cellular responses to proinflammatory stimuli. J Leukoc Biol 97:723–736. 10.1189/jlb.3A0214-106R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891–1899. 10.1182/blood.V95.6.1891. [DOI] [PubMed] [Google Scholar]
- 28.Bertesi M, Fantini S, Alecci C, Lotti R, Martello A, Parenti S, Carretta C, Marconi A, Grande A, Pincelli C, Zanocco-Marani T. 2020. Promoter methylation leads to decreased ZFP36 expression and deregulated NLRP3 inflammasome activation in psoriatic fibroblasts. Front Med (Lausanne) 7:579383. 10.3389/fmed.2020.579383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang JH, Kim DJ, Ham S-Y, Vo M-T, Jeong SY, Choi SH, Park SS, Jeon DY, Lee BJ, Ko BK, Cho WJ, Park JW. 2020. Tristetraprolin posttranscriptionally downregulates PFKFB3 in cancer cells. Biochem Biophys Res Commun 521:389–394. 10.1016/j.bbrc.2019.10.128. [DOI] [PubMed] [Google Scholar]
- 30.Xiao J, Gao H, Jin Y, Zhao Z, Guo J, Liu Z, Zhao Z. 2014. The abnormal expressions of tristetraprolin and the VEGF family in uraemic rats with peritoneal dialysis. Mol Cell Biochem 392:229–238. 10.1007/s11010-014-2033-3. [DOI] [PubMed] [Google Scholar]
- 31.Ehlting C, Trilling M, Tiedje C, Le-Trilling VTK, Albrecht U, Kluge S, Zimmermann A, Graf D, Gaestel M, Hengel H, Häussinger D, Bode JG. 2016. MAPKAP kinase 2 regulates IL-10 expression and prevents formation of intrahepatic myeloid cell aggregates during cytomegalovirus infections. J Hepatol 64:380–389. 10.1016/j.jhep.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Jin W-J, Chen C-F, Liao H-Y, Gong L-L, Yuan X-H, Zhao B-B, Zhang D, Feng X, Liu J-J, Wang Y, Chen G-F, Yan H-P, He Y-W. 2012. Downregulation of the AU-rich RNA-binding protein ZFP36 in chronic HBV patients: implications for anti-inflammatory therapy. PLoS One 7:e33356. 10.1371/journal.pone.0033356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberger CM, Podyminogin RL, Navarro G, Zhao G-W, Askovich PS, Weiss MJ, Aderem A. 2012. miR-451 regulates dendritic cell cytokine responses to influenza infection. J Immunol 189:5965–5975. 10.4049/jimmunol.1201437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S-R, Jin H, Kim W-T, Kim W-J, Kim SZ, Leem S-H, Kim SM. 2018. Tristetraprolin activation by resveratrol inhibits the proliferation and metastasis of colorectal cancer cells. Int J Oncol 53:1269–1278. 10.3892/ijo.2018.4453. [DOI] [PubMed] [Google Scholar]
- 35.Yuan LX, Liang JQ, Zhu QC, Dai G, Li S, Fung TS, Liu DX. 2020. A gammacoronavirus, avian infectious bronchitis virus, and an alphacoronavirus, porcine epidemic diarrhea virus, exploit a cell-survival strategy via upregulation of cFOS to promote viral replication. J Virol 95:e02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammaker D, Boyle DL, Topolewski K, Firestein GS. 2014. Differential regulation of anti-inflammatory genes by p38 MAP kinase and MAP kinase kinase 6. J Inflamm 11:14. 10.1186/1476-9255-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fung TS, Liu DX. 2017. Activation of the c-Jun NH2-terminal kinase pathway by coronavirus infectious bronchitis virus promotes apoptosis independently of c-Jun. Cell Death Dis 8:3215. 10.1038/s41419-017-0053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Fung TS, Huang M, Fang SG, Zhong Y, Liu DX. 2013. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J Virol 87:8124–8134. 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libby P, Lüscher T. 2020. COVID-19 is, in the end, an endothelial disease. Eur Heart J 41:3038–3044. 10.1093/eurheartj/ehaa623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang T, Chen H, Qi L, Zhang J, Wu R, Zhang Y, Sun Y. 2018. Transcript profiling identifies early response genes against FMDV infection in PK-15 Ccells. Viruses 10:364. 10.3390/v10070364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang B, Qi X, Chen Z, Chen S, Xue Q, Jia P, Wang T, Wang J. 2018. Binding and entry of peste des petits ruminants virus into caprine endometrial epithelial cells profoundly affect early cellular gene expression. Vet Res 49:8. 10.1186/s13567-018-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esclatine A, Taddeo B, Roizman B. 2004. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-Rich RNAs. J Virol 78:8582–8592. 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suswam E, Li Y, Zhang X, Gillespie GY, Li X, Shacka JJ, Lu L, Zheng L, King PH. 2008. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res 68:674–682. 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- 45.Brahma PK, Zhang H, Murray BS, Shu F, Sidell N, Seli E, Kallen CB. 2012. The mRNA-binding protein Zfp36 is upregulated by β-adrenergic stimulation and represses IL-6 production in 3T3-L1 adipocytes. Obesity (Silver Spring) 20:40–47. 10.1038/oby.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franks TM, Lykke-Andersen J. 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21:719–735. 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu L-Q, Stumpo DJ, Blackshear PJ. 2012. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. Ji 188:5150–5159. 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin S-C, Gram H, Han J. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120:623–634. 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 49.Soni S, Anand P, Padwad YS. 2019. MAPKAPK2: the master regulator of RNA-binding proteins modulates transcript stability and tumor progression. J Exp Clin Cancer Res 38:121. 10.1186/s13046-019-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deleault KM, Skinner SJ, Brooks SA. 2008. Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol 45:13–24. 10.1016/j.molimm.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 51.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WFC, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin:14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23:1313–1324. 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. 2011. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol 31:256–266. 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sei E, Wang T, Hunter OV, Xie Y, Conrad NK. 2015. HITS-CLIP analysis uncovers a link between the Kaposi’s sarcoma-associated herpesvirus ORF57 protein and host pre-mRNA metabolism. PLoS Pathog 11:e1004652. 10.1371/journal.ppat.1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Twizere J-C, Kruys V, Lefèbvre L, Vanderplasschen A, Collete D, Debacq C, Lai WS, Jauniaux J-C, Bernstein LR, Semmes OJ, Burny A, Blackshear PJ, Kettmann R, Willems L. 2003. Interaction of retroviral tax oncoproteins with tristetraprolin and regulation of tumor necrosis factor-alpha expression. J Natl Cancer Inst 95:1846–1859. 10.1093/jnci/djg118. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, Liu DX, Sun Y, Meng C, Tan L, Song C, Qiu X, Liu W, Ding C, Liao Y. 2021. Upregulation of DUSP6 impairs infectious bronchitis virus replication by negatively regulating ERK pathway and promoting apoptosis. Vet Res 52:7. 10.1186/s13567-020-00866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poppe M, Wittig S, Jurida L, Bartkuhn M, Wilhelm J, Müller H, Beuerlein K, Karl N, Bhuju S, Ziebuhr J, Schmitz ML, Kracht M. 2017. The NF-κB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog 13:e1006286. 10.1371/journal.ppat.1006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Duche A, Sayer MR, Roosan D, Khalafalla FG, Ostrom RS, Totonchy J, Roosan MR. 2021. SARS-CoV-2 early infection signature identified potential key infection mechanisms and drug targets. BMC Genomics 22. 10.1186/s12864-021-07433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morham SG, Langenbach R, Loftin CD, Tiano HF, Vouloumanos N, Jennette JC, Mahler JF, Kluckman KD, Ledford A, Lee CA, Smithies O. 1995. Prostaglandin synthase 2 gene disruption causes severe renal pathology in the mouse. Cell 83:473–482. 10.1016/0092-8674(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 59.Chen JS, Alfajaro MM, Chow RD, Wei J, Filler RB, Eisenbarth SC, Wilen CB. 2021. Non-steroidal anti-inflammatory drugs dampen the cytokine and antibody response to SARS-CoV-2 infection. J Virol 95. 10.1128/JVI.00014-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim KP, Liu DX. 1998. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology 245:303–312. 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen S, Wen ZL, Liu DX. 2003. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology 311:16–27. 10.1016/s0042-6822(03)00117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang SG, Shen S, Tay FPL, Liu DX. 2005. Selection of and recombination between minor variants lead to the adaptation of an avian coronavirus to primate cells. Biochem Biophys Res Commun 336:417–423. 10.1016/j.bbrc.2005.08.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corman VM, Eckerle I, Memish ZA, Liljander AM, Dijkman R, Jonsdottir H, Juma Ngeiywa KJZ, Kamau E, Younan M, Al Masri M, Assiri A, Gluecks I, Musa BE, Meyer B, Müller MA, Hilali M, Bornstein S, Wernery U, Thiel V, Jores J, Drexler JF, Drosten C. 2016. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci USA 113:9864–9869. 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morfopoulou S, Brown JR, Davies EG, Anderson G, Virasami A, Qasim W, Chong WK, Hubank M, Plagnol V, Desforges M, Jacques TS, Talbot PJ, Breuer J. 2016. Human Coronavirus OC43 Associated with Fatal Encephalitis. N Engl J Med 375:497–498. 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 65.Park S-J, Kim H-K, Song D-S, An D-J, Park B-K. 2012. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J Virol 86:5964. 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 19:4311–4323. 10.1128/MCB.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raingeaud J, Whitmarsh AJ, Barrett T, Dérijard B, Davis RJ. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol 16:1247–1255. 10.1128/MCB.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 69.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]