ABSTRACT
There is a continuously expanding gap between predicted phage gene sequences and their corresponding functions, which has largely hampered the development of phage therapy. Previous studies reported several phage proteins that could interfere with the intracellular processes of the host to obtain efficient infection. But few phage proteins that protect host against phage infection have been identified and characterized in detail. Here, we isolate a phage, vB_Pae_QDWS, capable of infecting Pseudomonas aeruginosa PAO1 and report that its encoded Gp21 protein protects PAO1 against phage infection. Expression of Gp21 regulates bacterial quorum sensing with an inhibitory effect in low cell density and an activation effect in high cell density. By testing the type IV pilus (TFP)-mediated twitching motility and transmission electron microscopy analysis, Gp21 was found to decrease the pilus synthesis. Further, by constructing the TFP synthesis gene pilB mutant and performing adsorption and phage resistance assay, we demonstrated that the Gp21 protein could block phage infection via decreasing the TFP-mediated phage adsorption. Gp21 is a novel protein that inhibits phage efficacy against bacteria. The study deepens our understanding of phage-host interactions.
IMPORTANCE The majority of the annotated phage genes are currently deposited as “hypothetical protein” with unknown function. Research has revealed that some phage proteins serve to inhibit or redirect the host intracellular processes for phage infection. Conversely, we report a phage encoded protein Gp21 that protects the host against phage infection. The pathways that Gp21 involved in antiphage defense in Pseudomonas aeruginosa PAO1 interfere with quorum sensing and decrease type IV pilus-mediated phage adsorption. Gp21 is a novel protein with a low sequence homology with other reported twitching inhibitory proteins. As a lytic phage-derived protein, Gp21 expression protects P. aeruginosa PAO1 from reinfection by phage vB_Pae_QDWS, which may explain the well-known pseudolysogeny caused by virulent phages. Our discoveries provide valuable new insight into phage-host evolutionary dynamics.
KEYWORDS: Pseudomonas aeruginosa PAO1, adsorption, phage, quorum sensing, type IV pilus
INTRODUCTION
Phages are known to be highly abundant in environments, affecting the bacterial communities through killing of bacteria and horizontal gene transfer (1, 2). Phage therapy is being reevaluated as a means to treat or prevent bacterial infections due to the phenomenon of antibiotic resistance that has grown into a global public health concern. However, bacteria have evolved numerous mechanisms that actively stop phage attacks, including the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas system (3), abortive infection (ABI) system (4), restriction-modification (R-M) system (5), and bacteriophage exclusion (BREX) system (6). Besides these, the quorum sensing (QS) system was also reported to play key roles in antiphage defenses via reducing the amount of phage receptors on cell surface (7), increasing the expression of the CRISPR-Cas immune system, (8) or changing the cell physiological state (9). Hence, understanding the complex dynamics interactions between phages and bacteria is an important prerequisite for ensuring the efficacy of phage therapy (10, 11).
Our knowledge on phage gene functions is still limited, considering the enormous genetic diversity of both bacteria and phages. There are about 70% of phage genes whose functions have not been annotated (12, 13). More works on the functional characterization of “hypothetical protein” in phage genome are urgently needed. It has been reported that some phage proteins serve to redirect intracellular processes of the host, including blocking the action of the CRISPR-Cas complexes (14) and silencing the QS system (15), to obtain efficient production of phage progeny (16). In addition, some proteins also serve to inhibit growth of the host and act as potential antimicrobial agents (12). Evidence demonstrates that the phage hypothetical protein plays a role in adapting the phage to the host. However, bacteria are always changing their strategies to prevent phage infection in the coevolutionary arms race (10, 17). It is not very clear whether these hypothetical proteins are involved in bacterial counterattack on phages.
Pseudomonas aeruginosa is an important opportunistic pathogen and a potential target for phage therapy (18–20). P. aeruginosa performs several types of motilities, such as swimming (flagellar mediated), swarming (multiple mechanisms mediated), and twitching (type IV pilus [TFP] mediated), in order to properly attach and colonize a desired surface (21–23). In addition, P. aeruginosa possesses an arsenal of virulence determinants, such as the production of pyocyanin and biofilm, which are regulated by QS. LasIR, RhlIR, and the Pseudomonas quinolone signal (PQS) system are the typical QS systems in P. aeruginosa and are organized in a hierarchical manner with LasIR at the top of the hierarchy, controlling several biological processes (24, 25).
Here, we provide the functional analysis of a P. aeruginosa phage-encoded protein Gp21. Results revealed that the Gp21 protein involved in QS gene expression changed with growth period. Since bacterial QS modulates defense against phage attack, Gp21 dynamically affected phage infection. In addition, we found that Gp21 expression blocked phage infection via decreasing the TFP-mediated phage adsorption. Our study investigates the roles of the Gp21 protein in bacterial defense from the aspects of quorum sensing and the receptor TFP synthesis, which may help in filling several gaps in the field of complex phage-host evolutionary dynamics.
RESULTS
Characteristics of phage vB_Pae_QDWS.
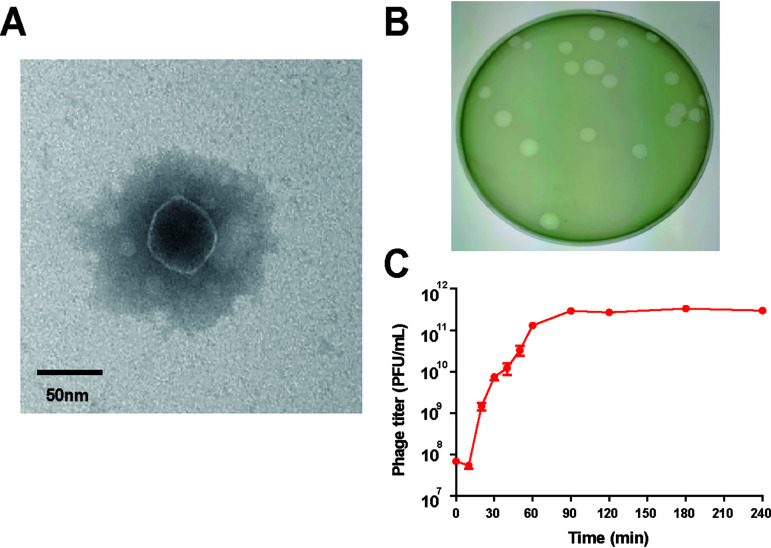
BLASTn analysis of the whole genome sequence showed Pseudomonas phage vB_Pae_QDWS shared high similarity with vB_PaeP_PAO1_1-15pyo (query coverage, 97%; identity, 93.94%) (26) and MPK7 (query coverage, 92%; identity, 98.05%) (27), which are members of the subfamily of Autographivirinae in the Podoviridae family. Transmission electron micrograph (TEM) imaging showed that the phage had an approximately 50-nm-diameter head and a nearly invisible short tail (Fig. 1A). Hence, the phage vB_Pae_QDWS was classified as a member of the Podoviridae family. It was separated from sewage, formed clear, bright plaques on soft agar plates (Fig. 1B), and was named vB_Pae_QDWS. Results of a one-step growth curve showed that the latent period of phage vB_Pae_QDWS was about 10 min, followed by a burst period of 50 min, and then the PFU per milliliter stabilized after 70 min. In the rise period, the phage titer increased from 5.4 × 107 to 1.3 × 1011 PFU/mL in 50 min and then remained at approximately 2.92 × 1011 PFU/mL (Fig. 1C). Since no lysogeny module or lysogens were found in the phage genome, phage vB_Pae_QDWS is lytic.
FIG 1.
Characteristics of phage vB_Pae_QDWS. (A) TEM image of phage vB_Pae_QDWS. (B) Plaque morphology of phage vB_Pae_QDWS. (C) One-step growth curve of phage vB_Pae_QDWS on P. aeruginosa strain PAO1. All data are averages of six samples with standard deviations (error bars).
The effects of Gp21 on bacteria quorum sensing.
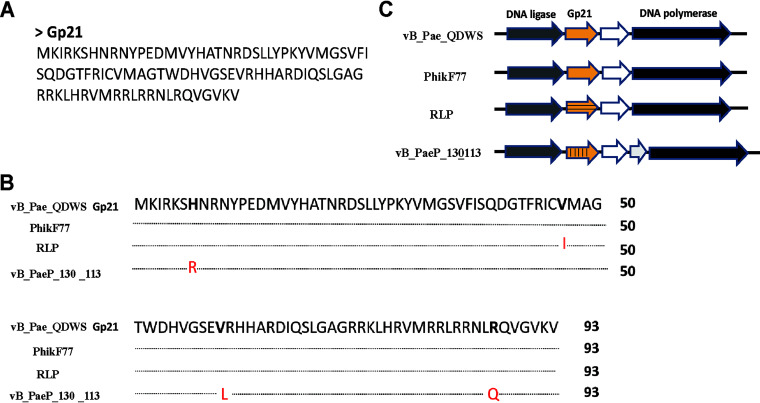
The gene product of open reading frame (ORF) 21 (gp21) of phage vB_Pae_QDWS was predicted to consist of 94 amino acids (Fig. 2A). A similarity search was run in BLASTP against nonredundant protein sequences, and no putative conserved domains were detected. In the BLASTP analysis, 3 sequences (hits) with high similarity (>98%) (Fig. 2B) were found in phages PhikF77, RLP, and vB_PaeP_130_113 infecting P. aeruginosa, and they shared a common genomic position between the phage DNA ligase and DNA polymerase (Fig. 2C).
FIG 2.
Sequence analysis of Gp21. (A) Amino acid sequence of Gp21. (B) Amino acid sequence alignment of Gp21 and its homologs from phages PhikF77, RLP, and vB_PaeP_130_113. (C) The genomic context of gp21 from phages vB_Pae_QDWS, PhikF77, RLP, and vB_PaeP_130_113. The white arrow designates a hypothetical gene.
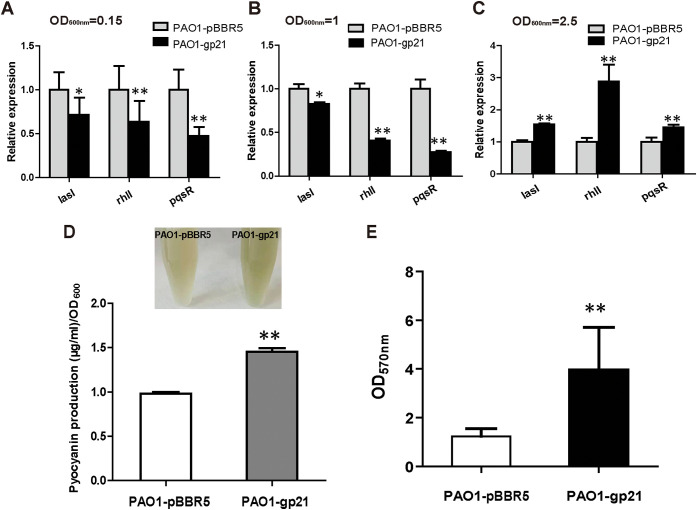
We investigated three QS key genes, lasI, rhlI, and pqsR, and found that Gp21 decreased their expression when cells grow to an optical density at 600 nm (OD600) of 0.15 and 1, especially for rhlI and pqsR genes (Fig. 3A and B). However, when cells grow to an OD600 of 2.5, the expression of the three genes increased in PAO1-gp21, especially for rhlI (Fig. 3B). We also detected the QS-mediated phenotypic changes and found the production of pyocyanin and biofilm increased significantly in PAO1-gp21 strains compared to that in PAO1-pBBR5 (Fig. 3C and D). Overexpression of Gp21 formed different effects on P. aeruginosa QS at the different growth phases.
FIG 3.
Effects of Gp21 on quorum sensing. (A to C) The transcripts of QS-related genes in PAO1-pBBR5 and PAO1-gp21 strains were analyzed by RT-qPCR. Strains were grown in LB medium until the OD600 reached 0.15, 1, and 2.5 for RNA extraction. The rplS transcript was used as the internal standard. (D) Pyocyanin production by PAO1-pBBR5 and PAO1-gp21 strains cultured at 37°C in LB medium for 24 h. (E) Biofilm formation by PAO1-pBBR5 and PAO1-gp21 strains grown at 37°C for 24 h in the 96-well microtiter plate. Data are averages and standard deviations of three experiments. t tests were performed to calculate the P values. *, The sample is different (P < 0.1); **, the sample is significantly different (P < 0.01) from the control PAO1-pBBR5.
Gp21 inhibits TFP-mediated twitching motility.
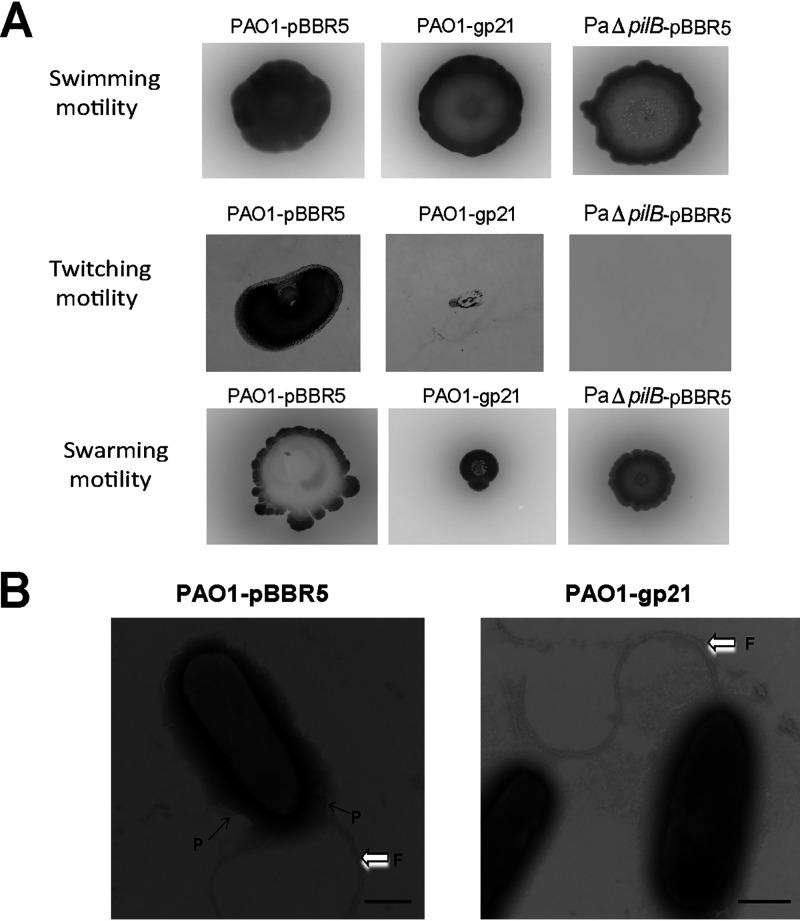
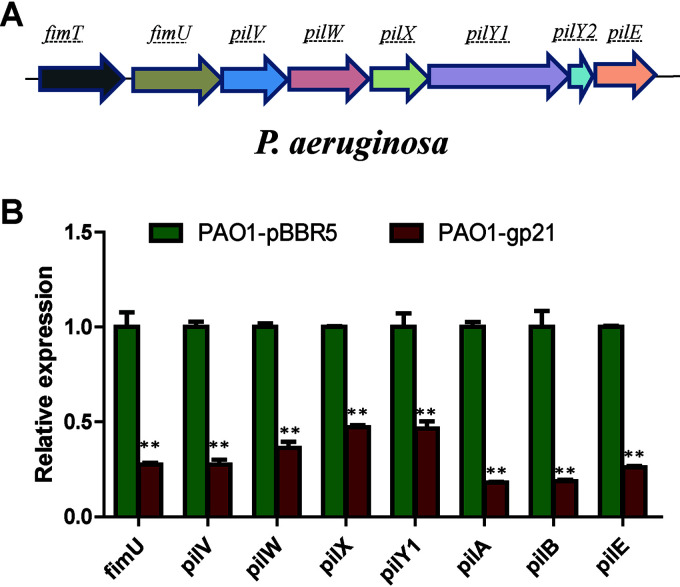
Swimming, twitching, and swarming are three modes of motility found in P. aeruginosa (28). PilB is necessary for the assembly of TFP, which has been known to be responsible for twitching and swarming motility (28, 29). When tested for swimming motility, no significant difference was observed among PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5. For twitching and swarming tests, expression of Gp21 resulted in a significant inhibition as PaΔpilB-pBBR5 (Fig. 4A). We then employed TEM to visualize pilus structure. PAO1-gp21 expressed less functional pilus than PAO1-pBBR5, while no difference was found in the assembly of flagella (Fig. 4B). A series of genes have been verified to be required for TFP assembly, such as pilA (the major structural element of TFP), pilB (TFP extension), fimU, and pilVWXY1E (minor pilins prime pilus assembly) (30, 31). Real-time quantitative reverse transcription (RT-qPCR) was used to quantify the expression of TFP-related genes in PAO1. Results showed that the expression level of TFP-related genes in PAO1-gp21 was significantly lower than in PAO1-pBBR5, especially for pilA and pilB (Fig. 5). These data suggest that Gp21 affects TFP formation.
FIG 4.
Gp21 affects TFP-mediated twitching and swarming phenotypes. (A) The motility assays of PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5 were performed for 60 h at 37°C. (B) TEM of PAO1-pBBR5 and PAO1-gp21. Strains were grown on LB-containing gentamicin (30 mg/L) at 37°C for 6 h and then were harvested for TEM analysis. Pilus and flagella were indicated by the small and large arrows, respectively. P, pilus; F, flagella. Scale bar, 500 nm.
FIG 5.
TFP-related genes were downregulated in P. aeruginosa-expressed Gp21. (A) Gene organization of TFP minor components in P. aeruginosa PAO1. (B) RT-qPCR analysis of PAO1-pBBR5 and PAO1-gp21. The RNA sampling point was OD600 = 1. The rplS transcript was used as the internal standard. The values were the averages of three measures with standard deviation. t tests were performed to calculate the P values. **, The sample is significantly different (P < 0.01) from the control PAO1-pBBR5.
Gp21 increases the resistance of P. aeruginosa to phage attack.
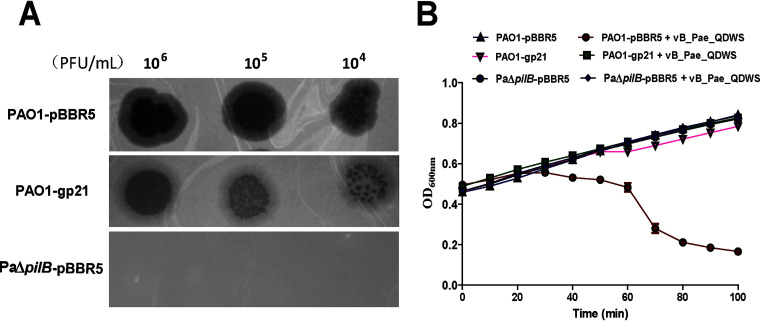
Expression of Gp21 caused a marked increase in phage resistance, as the transparency of plaque decreased significantly compared with that of PAO1-pBBR5 and PaΔpilB-pBBR5 (Fig. 6A). Upon further inspection of growth curves of PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5 infected with phage vB_Pae_QDWS, we found PAO1 exhibited a much steeper drop in optical density, but PAO1-gp21 and PaΔpilB-pBBR5 did not. Additionally, there was no difference in growth between PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5 (Fig. 6B).
FIG 6.
The P. aeruginosa expressing Gp21 gains the ability to defend against phage attacks. (A) Spot assay was used to evaluate phage resistance. Three microliters each of serial 10-fold dilution (106 to 104) of phage vB_Pae_QDWS was spotted onto PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5 for assays. (B) Antiphage activity assays of PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5. Growth curves of P. aeruginosa strains infected with phage vB_Pae_QDWS at an MOI of 0.1 in a 96-well microtiter plate containing 200-μL cultures were detected by a multimode reader. The values were the averages of three measures with standard deviation.
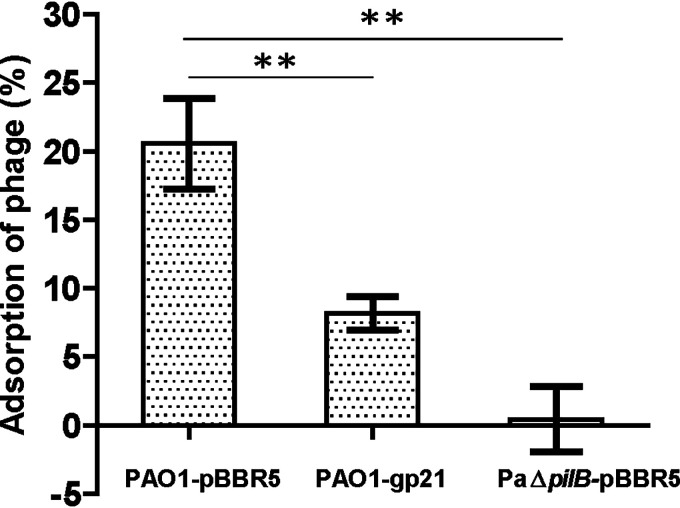
The pilB gene was reported to be necessary for TFP assembly (32). Through adsorption rate assay, we found almost no phage adsorption happened on PaΔpilB-pBBR5, which indicated phage vB_Pae_QDWS could use TFP as its receptor. In addition, there is an obviously reduction in phage adsorption on PAO1-gp21 compared with that of the control PAO1-pBBR5 (Fig. 7). Combined with the results of Fig. 4, we propose that Gp21 could block phage infection by decreasing the TFP-mediated phage adsorption.
FIG 7.
Gp21 decreases the TFP-mediated phage adsorption. The rates of adsorption of phage vB_Pae_QDWS to PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5 were determined by mixing phage and host cultures at an API of 0.001 at 37°C for 5 min without shaking. The values were the averages of three measures with standard deviation. t tests were performed to calculate the P values. **, The sample is significantly different (P < 0.01) from the control PAO1-pBBR5.
A quite low sequence homology was found in Gp21 (Fig. 8) compared to that of other twitching inhibitory proteins, Aqs1 (15), JBD24-4 (33), and Tip (32), that were previously reported. However, the strategies of these small phage proteins are similar to that which allowed the host's survival by blocking TFP-mediated phage adsorption.
FIG 8.
Multiple sequence alignment of Gp21 and other known phage twitching inhibitory proteins. Protein sequences shown include P. aeruginosa phage DMS3 Aqs1, P. aeruginosa phage JBD24 JBD24-4, P. aeruginosa phage D3112 Tip, and phage vB_Pae_QDWS Gp21 (in this study).
DISCUSSION
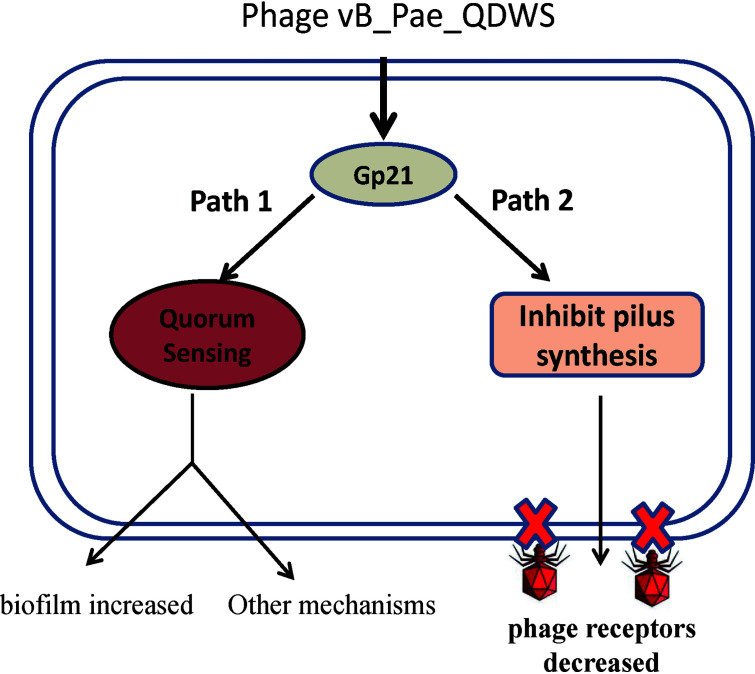
A model that Gp21 is involved in P. aeruginosa antiphage defenses is proposed (Fig. 9). Expression of vB_Pae_QDWS Gp21 has a dual effect on P. aeruginosa QS. Gp21 inhibited QS gene transcription in the early stage of growth and activated QS at high population densities (Fig. 3). Since QS is closely related to phage infection (7, 8, 34, 35), Gp21 could influence bacterial antiphage defenses by dynamically regulating QS. In addition, expression of Gp21 is sufficient for twitching inhibition due to blocking the synthesis of pilus (Fig. 4), which is the important receptor of phage vB_Pae_QDWS (Fig. 7). Preventing TFP synthesis is another way that Gp21 affects phage infection.
FIG 9.
Schematic representation of phage vB_Pae_QDWS Gp21 participate in antiphage defenses in P. aeruginosa PAO1.
Preventing phage adsorption is probably the main mechanism by which Gp21 dampens phage attacks. On the one hand, QS was reported to regulate phage susceptibility by interfering with phage adsorption or limiting CRISPR-based immunity (36). However, PAO1 does not contain the CRISPR-Cas system (37). Gp21 may regulate phage vB_Pae_QDWS infection efficiency mainly by preventing phage adsorption. On the other hand, we found TFP is the cell surface receptor for phage vB_Pae_QDWS. Gp21 expression reduced the number of pili (Fig. 4), which are required for phage infection. TFP are also critical for biofilm formation (38). The formation of biofilm was significantly enhanced in PAO1-gp21 (Fig. 3E), which further increases the barrier to prevent phage adsorption by masking phage receptors (39, 40).
The Gp21 protein is from the lytic phage vB_Pae_QDWS. Comparisons based on amino acid sequences confirmed that Gp21 is also distributed in P. aeruginosa phages PhikF77, RLP, and vB_PaeP_130_113 (Fig. 2). It is unclear whether the Gp21 protein with similar functions is also encoded by other phages due to diversity of phage genomes. However, Gp21 expression may help to resist several phages other than phage vB_Pae_QDWS, as many Pseudomonas phages could use TFP as their receptor (15, 27). Genomic analysis indicated that gp21 is likely an early or middle gene, since it is between DNA ligase and DNA polymerase, which may function to take over the host metabolism after infection (41). The lyase gene, which is required for breaking down the cell wall in order to release progeny phages, is known to be expressed late in the infection cycle (42, 43). Before bacterial lysis, Gp21 protein presumably shuts down pilus assembly quite rapidly and limits the number of phages that infect a single cell. Gp21 also likely serves to affect cell-cell communication, informing the surrounding cells in time to respond to phage invasion. More importantly, pseudolysogeny is a frequent outcome of infection by virulent phages, and bacteria with a pseudolysogeny state can further resist the infection of the same phage (44, 45). The effects of Gp21 on host physiology may explain why Gp21 expression blocked phage vB_Pae_QDWS infection. More detailed mechanisms by which Gp21 functions will be elucidated by means of omics.
The phage-mediated QS interference and loss of twitching act as the strategy of the host bacterium to prevent efficient phage infection, which also represents a trade off in terms of potential benefits for the host bacteria. Though Gp21 interfered with P. aeruginosa QS, Gp21 expression promoted the production of pyocyanin in the late growth stage (Fig. 3D). Pyocyanin is closely associated with pathogenicity (46). TFP and TFP-mediated twitching are essential for attachment and virulence of P. aeruginosa (47). Although Gp21 inhibited TFP and twitching, the loss of motility could enhance the fitness of the bacterium in chronic infection (48). We also observed the significant enhancement in biofilm formation (Fig. 3E), which could increase the resistance of microorganisms in biofilms toward biocides and various stresses (49).
In summary, numerous phages are sequenced so far, and most ORFs coded by phages are functionally unknown. Our understanding of phage gene products is only the tip of the iceberg. Gp21, we proposed, is a novel phage-derived protein that protects P. aeruginosa PAO1 from phage infection. Our study is particularly remarkable for uncovering a phage-encoded protein and evaluating its roles in host physiology. Further identification of the Gp21 receptor and screening competitive inhibitors of Gp21 are needed and will help to confer phage resistance or promote P. aeruginosa phage therapy in the future.
MATERIALS AND METHODS
Bacterial strains and phages.
Phage vB_Pae_QDWS was isolated from Qingdao sewage and stored in Food Safety Laboratory, Ocean University of China. P. aeruginosa and Escherichia coli were cultured in LB medium at 37°C. PAO1 was also grown in minimal salt medium (MM) supplemented with 1% sodium gluconate during the process of genes inactivation. The detailed information of strains and plasmids used in this work are given in Table S1 in the supplemental material. All primers are given in Table S2 in the supplemental material. The following antibiotics (concentrations) were used when required: gentamicin (30 μg/mL), kanamycin (50 μg/mL), and tetracycline (30 μg/mL).
Isolation and purification of phages.
Phages specific to P. aeruginosa PAO1 were isolated from sewage samples in Qingdao, China, and were centrifuged at 2,348 × g for 10 min. Then, the samples were filtered through a 0.22-μm pore size filter (Millipore, Burlington, MA, USA) and mixed with 50 mL of log-phase P. aeruginosa PAO1 cells, incubating at 37°C and 200 rpm rotary agitation for 12 h. The resulting culture suspension was filtered as above, and the phages were cultured using the double-layer agar plate method. Single plaques were separated by stinging with a pipette tip into the plaque followed by resuspending the phages in SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl, pH 7.5). After multiple rounds of purification, the phage was verified by electron microscopy.
One-step growth curve.
The one-step growth curve was determined according to a reported method (50). P. aeruginosa PAO1 was grown in LB medium until the log-phase (OD600 = 0.4∼0.6). Then, the phage vB_Pae_QDWS was added to the PAO1 culture at a multiplicity of infection (MOI) of 0.01 and incubated at 37°C. Using the double-layer agar plate method, we determined the free bacteriophage count at each time point.
Transmission electron microscopy.
Transmission electron microscopy was performed as previously described (51). The bacteria particles or phage pellets were deposited on carbon-coated copper grids and negatively stained with 2% phosphotungstic acid (pH 6.8). Electron microscopy pictures were taken with a transmission electron microscope (TEM) (JEM-1200EX; JEOL, Japan) operating at an accelerating voltage of 100 kV.
Gene cloning and expression.
PCR-amplified gp21 genes were cloned into the broad-host-range plasmid pBBR1MCS5 (linearized via PCR) by using a modified In-Fusion method (52). The verified plasmid was then transformed into the PAO1 cells via electroporation. Transformants were selected using 30 μg/mL gentamicin. PAO1 with the empty plasmid pBBR1MCS5 was used as the control.
Physiological and biochemical tests of bacteria.
Motility assays were performed as previously described, with modifications (28). For swimming assay, bacterial culture was grown to an OD600 of 2. Three microliters of the culture was spotted onto the semisolid medium consisting of 0.8% nutrient broth, 0.5% glucose, and 0.3% agarose. After incubation at 37°C for 60 h, diameter of the swimming zones was measured. For twitching motility, fresh colonies were stab-inoculated through the 1% LB agar layer. After incubation at 37°C for 60 h, the agar was carefully removed, and the motility zone was measured by staining the petri dish with 2% crystal violet for 1 h. For swarming assay, 3 μL of the bacterial culture (OD600 = 2) was spotted onto the semisolid medium consisting of 0.8% nutrient broth, 0.5% glucose, and 0.5% agarose. After incubation at 37°C for 60 h, the motility zone observed was measured.
Biofilm assays were performed in the wells of a 96-well microassay plate (53). Briefly, 10 μL of overnight culture was transferred into a well containing 200 μL LB fresh medium in a flat-bottom 96-well microplate. After incubation at 37°C for 24 h without shaking, cultures were gently removed, washed, and then stained with 0.1% crystal violet solution for 30 min. Bound crystal violet was extracted from the stained biofilm with 200 μL 30% glacial acetic acid and measured the absorbance at the wavelength of 570 nm using a multimode reader (Synergy LX; BioTek Instruments, Inc., Winooski, VT).
For the pyocyanin production assay, P. aeruginosa strains were incubated at 37°C with shaking at 200 rpm rotary agitation for 24 h. The supernatants (6 mL) were collected by centrifugation and extracted with 6 mL of chloroform. Then, 4 mL of the pyocyanin-chloroform solution was extracted again with 1 mL 0.2 M HCl, leading to the development of a pink color, which was measured at 520 nm by a multimode reader (54). The concentration was calculated and normalized to the cell density (OD600).
Estimation of phage resistance.
Phage resistance was detected by spot assay. Briefly, the overnight P. aeruginosa PAO1-pBBR5 and PAO1-gp21 strains were inoculated in LB medium to an OD600 of 2. Then, 100 μL of the cultures were mixed with 5 mL of the melted 1% agar LB medium, forming the double-layer agar. Three microliters of diluted phages were spotted to the plate, and the plates were incubated at 37°C for 12 h before examination. Bacterial growth assay was also used for estimating phage resistance. At an OD600 of 0.5, PAO1-pBBR5 and PAO1-gp21 cultures were mixed with the appropriate phage (MOI = 0.1) in 96-well plates. Infection dynamics were monitored in three replicates by a multimode reader (Synergy LX; BioTek Instruments, Inc., Winooski, VT).
Adsorption rate assay.
The phage adsorption rate was assayed as described previously with modifications (50). In brief, overnight cultures (OD600 of 0.05) of the PAO1-pBBR5, PAO1-gp21, and PaΔpilB-pBBR5 strains were inoculated in fresh LB medium. The cells were cultured until the OD600 reached 2.5, followed by 10-fold dilution in LB medium. The cells were then mixed with the phage vB_Pae_QDWS solution at an MOI of 0.001 and then allowed to adsorb for 5 min at 37°C. The cells were centrifuged at 7,378 × g for 2 min at 4°C to obtain the free phages, which were detected using a soft-agar overlay assay. The adsorption rate was calculated as follows: adsorption rate (%) = [(initial phage titer − phage titer in the supernatant)/(initial phage titer)] × 100.
Construction of the pilB deletion strain of Pseudomonas aeruginosa.
To construct a pilB deletion mutant, upstream (1,016 bp) and downstream (1,021 bp) regions of the pilB ORF were amplified from the P. aeruginosa PAO1 genomic DNA and then subcloned into the pK18mobsacBtet vector at the EcoRI site. The deletion plasmid pK18mobsacBtet-ΔpilB was transformed into E. coli S17-1 and then transferred to P. aeruginosa PAO1 by conjugation. After two rounds of screening by using colony PCR, the correct deletion strain was obtained.
Real-time quantitative reverse transcription-PCR.
For RT-qPCR, the cells were collected at a defined incubation time, and RNA was extracted. RNA was purified using the TRIzol RNA purification kit (12183555; Invitrogen). Total cDNA was synthesized by the HiScript II reverse transcriptase (Vazyme). RT-qPCR was performed by using the SYBR green realtime PCR master mix. The StepOnePlus real-time PCR system (ABI) was used to determine the melting curve and specificity of the PCR products. rplS was selected as the reference gene for normalization.
Data availability.
All data within the paper and its supplementary information file are available from the authors upon request. The complete genome sequence of phage vB_Pae_QDWS has been deposited in GenBank under accession number MZ687409.1.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program (2017YFC1600703 and 2016YFD0400105) and China Agriculture Research System (CARS-47).
G.X. acquired and analyzed the data; H.L. supervised research; J.W. designed the study and wrote the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Jingxue Wang, Email: snow@ouc.edu.cn.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Rohwer F. 2003. Global phage diversity. Cell 113:141. 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 2.Brussow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16. 10.1016/S0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 3.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194:5728–5738. 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blower TR, Chai R, Przybilski R, Chindhy S, Fang X, Kidman SE, Tan H, Luisi BF, Fineran PC, Salmond GPC. 2017. Evolution of Pectobacterium bacteriophage ΦM1 to escape two bifunctional type III toxin-antitoxin and abortive infection systems through mutations in a single viral gene. Appl Environ Microbiol 83:e03229-16. 10.1128/AEM.03229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tock MR, Dryden DT. 2005. The biology of restriction and anti-restriction. Curr Opin Microbiol 8:466–472. 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan D, Svenningsen SL, Middelboe M. 2015. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio 6:e00627. 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, Bassler BL. 2017. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci USA 114:131–135. 10.1073/pnas.1617415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin X, Sun Q, Yang B, Pan X, He Y, Yang H. 2017. Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J Basic Microbiol 57:162–170. 10.1002/jobm.201600510. [DOI] [PubMed] [Google Scholar]
- 10.Safari F, Sharifi M, Farajnia S, Akbari B, Karimi Baba Ahmadi M, Negahdaripour M, Ghasemi Y. 2020. The interaction of phages and bacteria: the co-evolutionary arms race. Crit Rev Biotechnol 40:119–137. 10.1080/07388551.2019.1674774. [DOI] [PubMed] [Google Scholar]
- 11.Torres-Barcelo C. 2018. Phage therapy faces evolutionary challenges. Viruses 10:323. 10.3390/v10060323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van den Bossche A, Ceyssens PJ, De Smet J, Hendrix H, Bellon H, Leimer N, Wagemans J, Delattre AS, Cenens W, Aertsen A, Landuyt B, Minakhin L, Severinov K, Noben JP, Lavigne R. 2014. Systematic identification of hypothetical bacteriophage proteins targeting key protein complexes of Pseudomonas aeruginosa. J Proteome Res 13:4446–4456. 10.1021/pr500796n. [DOI] [PubMed] [Google Scholar]
- 13.Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) Program, KwaZulu-Natal Research Institute for Tuberculosis and HIV (K-RITH) Mycobacterial Genetics Course, University of California—Los Angeles Research Immersion Laboratory in Virology, Phage Hunters Integrating Research and Education (PHIRE) Program. 2013. Complete genome sequences of 63 mycobacteriophages. Genome Announc 1:e00847-13. 10.1128/genomeA.00847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhary K, Chattopadhyay A, Pratap D. 2018. Anti-CRISPR proteins: counterattack of phages on bacterial defense (CRISPR/Cas) system. J Cell Physiol 233:57–59. 10.1002/jcp.25877. [DOI] [PubMed] [Google Scholar]
- 15.Shah MG, Taylor VL, Bona D, Tsao Y, Stanley SY, Pimentel-Elardo SM, McCallum M, Bondy-Denomy J, Howell PL, Nodwell JR, Davidson AR, Moraes TF, Maxwell KL. 2021. A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa. Mol Cell 81:571–583. 10.1016/j.molcel.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Roucourt B, Lavigne R. 2009. The role of interactions between phage and bacterial proteins within the infected cell: a diverse and puzzling interactome. Environ Microbiol 11:2789–2805. 10.1111/j.1462-2920.2009.02029.x. [DOI] [PubMed] [Google Scholar]
- 17.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 18.Xuan G, Lu C, Xu H, Chen Z, Li K, Liu H, Liu H, Xia Y, Xun L. 2020. Sulfane sulfur is an intrinsic signal activating MexR-regulated antibiotic resistance in Pseudomonas aeruginosa. Mol Microbiol 114:1038–1048. 10.1111/mmi.14593. [DOI] [PubMed] [Google Scholar]
- 19.Al-Wrafy F, Brzozowska E, Gorska S, Gamian A. 2017. Pathogenic factors of Pseudomonas aeruginosa - the role of biofilm in pathogenicity and as a target for phage therapy. Postepy Hig Med Dosw (Online) 71:78–91. 10.5604/01.3001.0010.3792. [DOI] [PubMed] [Google Scholar]
- 20.Yadav J, Kumari RM, Verma V, Nimesh S. 2021. Recent development in therapeutic strategies targeting Pseudomonas aeruginosa biofilms—a review. MaterialsToday Proc 46:2359–2373. 10.1016/j.matpr.2021.05.245. [DOI] [Google Scholar]
- 21.Patrick JE, Kearns DB. 2012. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol Microbiol 83:14–23. 10.1111/j.1365-2958.2011.07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You J, Sun L, Yang X, Pan X, Huang Z, Zhang X, Gong M, Fan Z, Li L, Cui X, Jing Z, Jin S, Rao Z, Wu W, Yang H. 2018. Regulatory protein SrpA controls phage infection and core cellular processes in Pseudomonas aeruginosa. Nat Commun 9:1846. 10.1038/s41467-018-04232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Zhang L. 2015. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6:26–41. 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siriken B, Oz V, Erol I. 2021. Quorum sensing systems, related virulence factors, and biofilm formation in Pseudomonas aeruginosa isolated from fish. Arch Microbiol 203:1519–1528. 10.1007/s00203-020-02159-5. [DOI] [PubMed] [Google Scholar]
- 26.Essoh C, Latino L, Midoux C, Blouin Y, Loukou G, Nguetta SP, Lathro S, Cablanmian A, Kouassi AK, Vergnaud G, Pourcel C. 2015. Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Cote d'Ivoire. PLoS One 10:e0130548. 10.1371/journal.pone.0130548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae HW, Cho YH. 2013. Complete genome sequence of Pseudomonas aeruginosa podophage MPK7, which requires type IV pili for infection. Genome Announc 1:e00744-13. 10.1128/genomeA.00744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G, Liu H, Ma LZ. 2014. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl Environ Microbiol 80:6724–6732. 10.1128/AEM.01237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain R, Sliusarenko O, Kazmierczak BI. 2017. Interaction of the cyclic-di-GMP binding protein FimX and the type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog 13:e1006594. 10.1371/journal.ppat.1006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 112:7563–7568. 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen Y, Harvey H, Sugiman-Marangos S, Bell SD, Buensuceso RN, Junop MS, Burrows LL. 2015. Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE. J Biol Chem 290:26856–26865. 10.1074/jbc.M115.683334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung IY, Jang HJ, Bae HW, Cho YH. 2014. A phage protein that inhibits the bacterial ATPase required for type IV pilus assembly. Proc Natl Acad Sci USA 111:11503–11508. 10.1073/pnas.1403537111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsao YF, Taylor VL, Kala S, Bondy-Denomy J, Khan AN, Bona D, Cattoir V, Lory S, Davidson AR, Maxwell KL. 2018. Phage morons play an important role in Pseudomonas aeruginosa phenotypes. J Bacteriol 200:e00189-18. 10.1128/JB.00189-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4:e00362-12. 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silpe JE, Bassler BL. 2019. A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 176:268–280. 10.1016/j.cell.2018.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broniewski JM, Chisnall MAW, Hoyland-Kroghsbo NM, Buckling A, Westra ER. 2021. The effect of quorum sensing inhibitors on the evolution of CRISPR-based phage immunity in Pseudomonas aeruginosa. ISME J 15:2465–2473. 10.1038/s41396-021-00946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez D, Gomila M, Bennasar A, Lalucat J, Garcia-Valdes E. 2014. Genome analysis of environmental and clinical P aeruginosa isolates from sequence type-1146. PLoS One 9:e107754. 10.1371/journal.pone.0107754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Parsek MR, Wozniak DJ, Ma LZ. 2013. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ Microbiol 15:2238–2253. 10.1111/1462-2920.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. 10.1016/S0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- 40.Samson JE, Magadan AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 41.Pavlova O, Lavysh D, Klimuk E, Djordjevic M, Ravcheev DA, Gelfand MS, Severinov K, Akulenko N. 2012. Temporal regulation of gene expression of the Escherichia coli bacteriophage phiEco32. J Mol Biol 416:389–399. 10.1016/j.jmb.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong M, Ryu S. 2015. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl Environ Microbiol 81:2274–2283. 10.1128/AEM.03485-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berdygulova Z, Westblade LF, Florens L, Koonin EV, Chait BT, Ramanculov E, Washburn MP, Darst SA, Severinov K, Minakhin L. 2011. Temporal regulation of gene expression of the Thermus thermophilus bacteriophage P23-45. J Mol Biol 405:125–142. 10.1016/j.jmb.2010.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Los M, Wegrzyn G, Neubauer P. 2003. A role for bacteriophage T4 rI gene function in the control of phage development during pseudolysogeny and in slowly growing host cells. Res Microbiol 154:547–552. 10.1016/S0923-2508(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 45.Latino L, Midoux C, Hauck Y, Vergnaud G, Pourcel C. 2016. Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa. Microbiology (Reading) 162:748–763. 10.1099/mic.0.000263. [DOI] [PubMed] [Google Scholar]
- 46.Manago A, Becker KA, Carpinteiro A, Wilker B, Soddemann M, Seitz AP, Edwards MJ, Grassme H, Szabo I, Gulbins E. 2015. Pseudomonas aeruginosa pyocyanin induces neutrophil death via mitochondrial reactive oxygen species and mitochondrial acid sphingomyelinase. Antioxid Redox Signal 22:1097–1110. 10.1089/ars.2014.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilmury SLN, Burrows LL. 2018. The Pseudomonas aeruginosa PilSR two-component system regulates both twitching and swimming motilities. mBio 9:e01310-18. 10.1128/mBio.01310-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahenthiralingam E, Campbell ME, Speert DP. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic-fibrosis. Infect Immun 62:596–605. 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masák J, Čejková A, Schreiberová O, Rezanka T. 2014. Pseudomonas biofilms: possibilities of their control. FEMS Microbiol Ecol 89:1–14. 10.1111/1574-6941.12344. [DOI] [PubMed] [Google Scholar]
- 50.Li P, Lin H, Mi ZQ, Xing SZ, Tong YG, Wang JX. 2019. Screening of polyvalent phage-resistant Escherichia coli strains based on phage receptor analysis. Front Microbiol 10:850. 10.3389/fmicb.2019.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan XW, Cui XL, Zhang FJ, He Y, Li LY, Yang HJ. 2016. Genetic evidence for O-specific antigen as receptor of Pseudomonas aeruginosa phage K8 and its genomic analysis. Front Microbiol 7:252. 10.3389/fmicb.2016.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okegawa Y, Motohashi K. 2015. A simple and ultra-low cost homemade seamless ligation cloning extract (SLiCE) as an alternative to a commercially available seamless DNA cloning kit. Biochem Biophys Rep 4:148–151. 10.1016/j.bbrep.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Smet J, Wagemans J, Hendrix H, Staes I, Visnapuu A, Horemans B, Aertsen A, Lavigne R. 2021. Bacteriophage-mediated interference of the c-di-GMP signalling pathway in Pseudomonas aeruginosa. Microb Biotechnol 14:967–978. 10.1111/1751-7915.13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeBritto S, Gajbar TD, Satapute P, Sundaram L, Lakshmikantha RY, Jogaiah S, Ito SI. 2020. Isolation and characterization of nutrient dependent pyocyanin from Pseudomonas aeruginosa and its dye and agrochemical properties. Sci Rep 10:1542. 10.1038/s41598-020-58335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2<br>. Download jvi.01769-21-s0001.pdf, PDF file, 0.2 MB (158.9KB, pdf)
Data Availability Statement
All data within the paper and its supplementary information file are available from the authors upon request. The complete genome sequence of phage vB_Pae_QDWS has been deposited in GenBank under accession number MZ687409.1.