ABSTRACT
PA-X is a nonstructural protein of influenza A virus (IAV), which is encoded by the polymerase acidic (PA) N-terminal region that contains a C-terminal +1 frameshifted sequence. IAV PA-X protein modulates virus-induced host innate immune responses and viral pathogenicity via suppression of host gene expression or cellular shutoff, through cellular mRNA cleavage. Highly pathogenic avian influenza viruses (HPAIV) of the H5N1 subtype naturally infect different avian species, they have an enormous economic impact in the poultry farming, and they also have zoonotic and pandemic potential, representing a risk to human public health. In the present study, we describe a novel bacterium-based approach to identify amino acid residues in the PA-X protein of the HPAIV A/Viet Nam/1203/2004 H5N1 that are important for its ability to inhibit host protein expression or cellular shutoff activity. Identified PA-X mutants displayed a reduced shutoff activity compared to that of the wild-type A/Viet Nam/1203/2004 H5N1 PA-X protein. Notably, this new bacterium-based screening allowed us to identify amino acid residues widely distributed over the entire N-terminal region of PA-X. Furthermore, we found that some of the residues affecting A/Viet Nam/1203/2004 H5N1 PA-X host shutoff activity also affect PA polymerase activity in a minigenome assay. This information could be used for the rational design of new and more effective compounds with antiviral activity against IAV. Moreover, our results demonstrate the feasibility of using this bacterium-based approach to identify amino acid residues important for the activity of viral proteins to inhibit host gene expression.
IMPORTANCE Highly pathogenic avian influenza viruses continue to pose a huge threat to global animal and human health. Despite of the limited genome size of Influenza A virus (IAV), the virus encodes eight main viral structural proteins and multiple accessory nonstructural proteins, depending on the IAV type, subtype, or strain. One of the IAV accessory proteins, PA-X, is encoded by the polymerase acidic (PA) protein and is involved in pathogenicity through the modulation of IAV-induced host inflammatory and innate immune responses. However, the molecular mechanism(s) of IAV PA-X regulation of the host immune response is not well understood. Here, we used, for the first time, a bacterium-based approach for the identification of amino acids important for the ability of IAV PA-X to induce host shutoff activity and describe novel residues relevant for its ability to inhibit host gene expression, and their contribution in PA polymerase activity.
KEYWORDS: baloxavir, H5N1, influenza A virus, minigenome, PA-X protein, shutoff, avian viruses
INTRODUCTION
Influenza are enveloped viruses belonging to the Orthomyxoviridae family and are the causative agents of flu (1–3). There are four recognized influenza types: A, B, C, and D (1–6). Influenza A viruses (IAV) are divided into subtypes based on the antigenic properties of the two glycoproteins on the virus envelope, hemagglutinin (HA) and neuraminidase (NA). To date, 18 different HA subtypes (H1 through H18) and 11 different NA subtypes (N1 through N11) have been described (1–3, 7). All IAV subtypes (with the exception of H17N10 and H18N11 identified in fruit bats) have been isolated from wild aquatic birds, which are considered their natural reservoirs (1–3, 7). IAVs are able to infect a wide range of avian and mammalian species, including humans, pigs, dogs, cats, and horses (1–3, 7–9).
Based on the World Organization for Animal Health (OIE), avian influenza viruses (AIVs) are classified as low pathogenic and highly pathogenic (LPAIV and HPAIV, respectively), depending on the severity of the disease that they induce in poultry. H5N1 HPAIVs infect different avian species, are highly contagious among individuals, and particularly deadly for poultry, causing 90 to 100% deaths in domestic flocks (10). H5N1 HPAIVs were detected for the first time in 1996 in geese in China (11) and continue to infect avian, causing important problems to animal and human health. Indeed, since January 2003 until now, the cumulative number of laboratory‐confirmed human cases for H5N1 viral infections is 861, causing death in 53% of them, in 17 different countries (https://www.who.int/influenza/human_animal_interface/2020_01_20_tableH5N1.pdf?ua=1). Although H5N1 viruses do not effectively transmit among humans, a pandemic caused by H5N1 infections in humans remains a possibility. Patients infected with H5N1 HPAIV developed severe respiratory disease that frequently progressed to acute respiratory distress syndrome (ARDS) and multiorgan failure (12). Systemic spread (13) and cytokine storm (14) have been described as possible disease-aggravating factors of H5N1 HPAIV, but the reasons for their high virulence in humans or poultry need further investigation.
IAV segment 3 encodes both the polymerase acidic (PA) and the PA-X proteins (15). PA-X is translated as a +1 frameshifting from the viral PA mRNA (15). During translation, the ribosome shifts at a specific sequence in the PA mRNA, a U-rich region, followed by a rare codon, which usually promotes ribosomal shifting because they are typically decoded more slowly (16). PA-X protein shares the same first N-terminal 191 amino acids with PA protein, including the endonuclease domain, and a unique short C-terminal sequence (15, 17, 18). Most of the human IAVs and the avian H5N1 strains encode a 252-amino-acid PA-X, with 61 C-terminal amino acids that result from the frameshift (19). However, some IAVs, including the 2009 human pandemic H1N1 (pH1N1), canine, and certain swine influenza viruses, encode a stop codon at position 42 of the X open reading frame (ORF), leading to a C-terminal domain of 41 amino acids (20). Influenza PA-X selectively degrades RNAs transcribed by host RNA polymerase II but not other polymerases (21). Complete degradation of host mRNAs by influenza PA-X is dependent on the host 5′→3′-exonuclease Xrn1 (21). This process leads to cellular shut down and inhibition of host antiviral responses (15, 22, 23). Although PA and PA-X share the N-terminal endonuclease domain, PA shows lower shutoff activity than PA-X, suggesting that the C-terminal PA-X-specific region is also important for its ability to inhibit host gene expression (15, 17, 18, 24–29).
Interestingly, influenza PA-X modulates host inflammation, antiviral responses, cell death, cell differentiation, and tissue remodeling (15). However, the role of PA-X in viral pathogenesis is likely strain specific (30). Loss of PA-X expression increased viral replication, host inflammatory response, and virulence in both pH1N1 and H5N1-infected mice (31, 32), as well as in H5N1-infected ducks and chickens (33). Furthermore, PA mRNA and protein synthesis was upregulated in PA-X-deficient pH1N1 and H5N1 virus-infected cells (31). Using strains of pH1N1 (A/Beijing/16/2009), a HPAIV H5N1 (A/tree sparrow/Jiangsu/1/2008), and a LPAIV H9N2 (A/chicken/Hebei/LC/2008) virus, it was shown that viruses encoding the 252-amino-acid PA-X protein replicated more efficiently and were more pathogenic in mice than viruses encoding a truncated 232 amino acid PA-X protein (20, 23).
Different residues have been shown to be important for PA-X’s endonuclease activity, including the bivalent cation-binding residue D108 and the catalytic residue K134 (15, 26). In addition, at the C-terminal PA-X-specific region, six basic amino acid residues (R195, K198, R199, K202, K203, and K206) play a key role in PA-X’s ability to inhibit host gene expression (34–36). Using a yeast screening, it has been shown that amino acid substitutions F4S, F9L, Y24S, D27G, C39Y, C45W, A87V, I94N, L106P/S184I, P107S, D108E, D108N, E119N, I120F, T123I, R124S, R125K, H146Y, E154K, E154A, D160Y, L163R, R168M, and I171M decrease influenza A/WSN/33 H1N1 PA-X’s inhibition of host gene expression (37), highlighting that there are many amino acid changes which can affect PA-X’s activity. Although residues important for PA-X’s shutoff activity have been identified in A/WSN/33 H1N1, amino acid residues involved in the ability of HPAIV H5N1 PA-X protein to inhibit host gene expression have not been yet determined. Moreover, differences among IAV subtypes (e.g., H1N1 and H5N1) will help us to understand the mechanism of PA-X’s inhibition of host gene expression. Here, we developed a new approach based on the ability of PA-X protein to inhibit bacterial host gene expression—and therefore bacterial growth—to identify amino acid residues important for H5N1 PA-X protein host shutoff activity. Importantly, we have been able to identify several novel amino acid residues involved in the shutoff activity of H5N1 PA-X. These results also demonstrate the feasibility of using the same bacterium-based approach to identify amino acid residues involved in the ability of other IAV PA-X proteins or in viral proteins with similar inhibition of host gene expression than that of H5N1 PA-X to induce host cellular shutoff.
RESULTS
Inhibition of host gene expression by A/Viet Nam/1203 H5N1 PA-X.
To determine whether the PA-X protein from HPAIV A/Viet Nam/1203/2004 H5N1 is able to inhibit host gene expression, as described for other strains (19, 30, 33), human HEK293T or avian DF-1 cells were cotransfected with pCAGGS plasmids expressing green fluorescent protein (GFP) and Gaussia luciferase (Gluc), together with increasing concentrations of pCAGGS plasmids expressing A/Viet Nam/1203/2004 H5N1 PA-X wild-type (WT) protein with an HA epitope tag (Fig. 1). Empty pCAGGS plasmid was included as a negative control. The amount of GFP and Gluc expression was determined at 24 h posttransfection (hpt). Similar to the PA-X protein from other viral strains (21, 30, 34, 35, 38–40), A/Viet Nam/1203/2004 H5N1 PA-X protein inhibited host gene expression in both human HEK293T and avian DF-1 cells as observed by decreased levels of GFP (Fig. 1A) or Gluc (Fig. 1B) expression. Notably, inhibition of host gene expression mediated by A/Viet Nam/1203/2004 H5N1 PA-X was dose dependent (Fig. 1A and B). Upon protein expression analysis by Western blotting (Fig. 1C), A/Viet Nam/1203/2004 H5N1 PA-X expression was only detected at the largest amount of transfected plasmid, whereas GFP expression decreased in a dose-dependent manner with increasing concentrations of A/Viet Nam/1203/2004 H5N1 PA-X expression plasmids. These results demonstrate that A/Viet Nam/1203/2004 H5N1 PA-X is able to inhibit host gene expression in human and avian cells, similar to previous studies with other PA-X proteins (21, 30, 34, 35, 38–40).
FIG 1.
Inhibition of host gene expression by A/Viet Nam/1203/2004 PA-X. Human HEK293T or avian DF-1 cells (96-well plate format, 104 cells/well, triplicates) were cotransfected with 0, 1, 10, or 100 ng of pCAGGS expression plasmids encoding A/Viet Nam/1203/2004 H5N1 PA-X fused to an HA epitope tag, together with 250 ng of pCAGGS plasmids encoding Gluc or GFP under the control of the human polymerase II promoter. Empty plasmid was included as a control. At 24 hpt, GFP was observed under a fluorescence microscope (A), and Gluc activity was quantified using a luciferase plate reader (B). Scale bars, 300 μm. The activity of Gluc was normalized to cells transfected with the empty plasmid control (100%). Results represent means and SD of triplicates. The data are representative of three independent experiments. ****, P < 0.0001 (0 ng of PA-X WT plasmid versus other concentrations), determined using one-way ANOVA. (C) PA-X and GFP expression levels by Western blotting from total cell lysates were detected using specific PAbs against the HA epitope tag (PA-X) or GFP, respectively. A MAb against β-actin was included as a loading control. Sizes of molecular markers (kDa) are noted on the left.
Identification of PA-X mutants affecting its ability to inhibit host gene expression in bacteria.
Next, we sought to identify A/Viet Nam/1203/2004 H5N1 PA-X mutants affected in their ability to inhibit host gene expression in yeast. To that end, we attempted to clone A/Viet Nam/1203/2004 H5N1 PA-X into the pGBKT7 yeast expression plasmid using DH5α bacterium-competent cells. Surprisingly, only a few bacterial colonies were found compared to same ligation and transformation experimental approach using influenza A/Puerto Rico/8/34 H1N1 NS1 (data not shown) (Fig. 2A). We and others have previously found that proteins cloned under a T7 promoter in plasmids were expressed in bacteria (data not shown) (41, 42). Therefore, we hypothesize that we were not able to clone a WT form of A/Viet Nam/1203/2004 H5N1 PA-X in the pGKT7 plasmid because the low levels of expression in bacteria from the T7 promoter were deleterious or toxic to bacteria (41, 42), and only those containing mutations affecting the ability of A/Viet Nam/1203/2004 H5N1 PA-X to induce cellular shutoff resulted in the recovery of bacterial colonies.
FIG 2.
Schematic representation of the bacterium-based assay to identify PA-X mutants affected in inhibition of host gene expression and identified mutants. (A) A bacterium-based assay for the identification of amino acid residues of A/Viet Nam/1203/2004 H5N1 PA-X involved in inhibition of host gene expression: steps involved in the molecular cloning of A/Viet Nam/1203/2004 H5N1 PA-X into pGBKT7 plasmid, under the control of T7 polymerase promoter (blue arrow). After ligation of PA-X into pGBT7, the mixture is transformed in bacteria, and individual colonies are isolated and sequenced. Leaked PA-X expression from the pGBKT7 promoter results in inhibition of host gene expression in bacteria and therefore no bacterial growth. PA-X mutants affected in inhibition of host gene expression allow bacteria to grow. Sequencing bacterial clones identify A/Viet Nam/1203/2004 H5N1 PA-X mutants affected in host shutoff. (B) A/Viet Nam/1203/2004 H5N1 PA-X and PA ORFs and amino acid mutations in PA-X identified in the bacterial-based assay: the +1 frameshift motif (UCC UUU CGU C) is indicated by a striped bar. The amino acid substitutions identified are indicated with black (affecting both PA-X and PA proteins) or gray (affecting only PA-X protein) lines. The superscript 1 indicates that mutations E101G and R125K were found in the same clone; the superscript 2 indicates that mutations D111N and E196G were found in the same clone. “##” symbols indicate that the clones were found twice. (C) Deletions and/or insertions of nucleotides (NT) leading to no protein expression (i), translation of short peptides (ii to v), and translation of a protein encoding the first 250 residues of WT PA-X and a short tail (vi). The #, ##, and ### symbols indicate that the clones were found one, two, and three times, respectively. Other mutations were found in one clone. *, Stop codons. Amino acids in boldface indicate WT PA-X sequence.
Initial sequencing analysis of pGBKT7 plasmids containing A/Viet Nam/1203/2004 H5N1 PA-X extracted from these few bacterial colonies revealed single or double mutations leading to amino acid changes in A/Viet Nam/1203/2004 H5N1 PA-X protein (Fig. 2B), together with early or late stop codons as a consequence of deletions and/or insertions within the PA-X gene (Fig. 2C). A total of 30 bacterial colonies, all containing mutations in A/Viet Nam/1203/2004 H5N1 PA-X were isolated, 15 of them containing unique single (L16P, N33S, K34S, E43K, F76L, P107S, L132P, H146Y, I147T, E153V, R170G, L187P, and F191L) or double (E101G/R125K and D111N/E196G) amino acid mutations (Fig. 2B). We noted that amino acid substitution E196G does not change A/Viet Nam/1203/2004 H5N1 PA sequence, only PA-X, whereas the other amino acid changes affected both A/Viet Nam/1203/2004 PA-X and PA protein sequences. Taking into account that there were sequence changes in 30 different isolated bacterial colonies and that the changes affected the protein sequence either by generating truncated/elongated PA-X proteins or by introducing amino acid mutations, we hypothesized that the identified amino acid changes were most probably not random and likely to be important for A/Viet Nam/1203/2004 PA-X host shutoff activity.
To evaluate whether these amino acid changes in the PA and PA-X proteins were also found in H5N1 viruses circulating globally, sequences available in the Influenza Research Database (https://www.fludb.org/) were examined. Notably, the residues found in the PA-X WT proteins from H5N1 viruses are highly conserved, and almost 100% of the H5N1 viruses isolated from avian and human hosts encode the WT sequence (Table 1), with the only exception of residue E101 that in our assay was found together with the R125K mutation (Fig. 2B), although amino acid R125 was also highly conserved in H5N1 viruses. In addition, we also observed small variability at residue L132 (Table 1).
TABLE 1.
Amino acid conservation in PA-X and PA proteins
| Amino acid change | Amino acid(s) (%)a |
|
|---|---|---|
| Human sequences (n = 279) | Avian sequences (n = 2,264) | |
| L16P | L (99.63), P (0.36) | L (100) |
| N33S | N (100) | N (99.92), D (0.04), S (0.04) |
| K34E | K (100) | K (99.54), R (0.4), T (0.04) |
| E43K | E (100) | E (99.92), G (0.04), D (0.04) |
| F76L | F (100) | F (99.92), C (0.04), Y (0.04) |
| E101G | E (66.18), D (31.29), N (1.8), G (0.36), V (0.36) | E (66.11), D (36.42), N (13.73), G (0.83), S (0.22), V (0.08) |
| P107S | P (100) | P (100) |
| D111N | D (100) | D (99.92), G (0.04), Y (0.04) |
| R125K | R (100) | R (99.96), K (0.04) |
| L132P | L (94. 6), M (5.4) | L (99.5), M (0.45), I (0.04) |
| H146Y | H (100) | H (100) |
| I147T | I (100) | I (99.88), M (0.08), T (0.04) |
| E153V | E (100) | E (99.96), G (0.04) |
| R170G | R (100) | R (99.96), G (0.04) |
| L187P | L (100) | L (99.76), I (0.24) |
| F191L | F (100) | F (100) |
| E196Gb | E (100) | E (99.88), K (0.04), V (0.04), D (0.04) |
Amino acid residues in A/Viet Nam/1203/2004 H5N1 PA-X WT are underlined. The frequencies of IAV H5N1 PA-X proteins containing the indicated residue are shown.
The E196G amino acid substitution does not change the A/Viet Nam/1203/2004 H5N1 PA amino acid sequence.
PA-X mutants displayed reduced shutoff activity.
Based on our previous results, we hypothesized that A/Viet Nam/1203/2004 PA-X mutations identified in bacteria were responsible for a decrease in host shutoff ability. To demonstrate this hypothesis, we evaluated the ability of the identified A/Viet Nam/1203/2004 PA-X mutants to inhibit host gene expression as previously described with WT PA-X (Fig. 3). Inhibition of GFP (Fig. 3A) and Gluc (Fig. 3B) expression by PA-X mutants was significantly reduced compared to that of A/Viet Nam/1203/2004 PA-X WT. The reduced PA-X’s ability to inhibit host gene expression was more evident for P107S, H146Y, L187P, and D111N/E196G A/Viet Nam/1203/2004 PA-X mutants (Fig. 3A and B). To further validate these findings and since PA-X inhibit its own expression when expressed from plasmids under the control of a polymerase II promoter (30, 37, 38), we evaluated PA-X protein expression levels by Western blotting (Fig. 3C). Notably, all A/Viet Nam/1203/2004 PA-X mutants were expressed to higher levels than those of PA-X WT (Fig. 3C). Moreover, the levels of GFP expression, measured by Western blotting, were higher in cells transfected with the A/Viet Nam/1203/2004 PA-X mutants (Fig. 3C), specifically those more affected in the inhibition of host gene expression (P107S, H146Y, L187P, and D111N/E196G).
FIG 3.
Mutations in A/Viet Nam/1203/2004 H5N1 PA-X identified in the bacterium-based assay are affected in inhibition of host gene expression. Human HEK293T cells (24-well plate format, 2.5 × 105 cells/well, triplicates) were cotransfected with 0, 1, 10, or 100 ng of pCAGGS expression plasmids encoding A/Viet Nam/1203/2004 H5N1 PA-X WT and mutants fused to an HA epitope tag, together with 250 ng of pCAGGS plasmids encoding Gluc or GFP under the control of the human polymerase II promoter. Empty plasmid was included as a control. After 24 hpt, GFP was observed under a fluorescence microscope (A), and Gluc activity in tissue culture supernatants was determined using a luciferase reader (B). Scale bars, 300 μm. The activity of Gluc was normalized to the empty plasmid control transfected cells (100%). The results are means and SD of triplicates. The data are representative of three independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001 (PA-X WT versus PA-X mutants for each amount of plasmid), determined using one-way ANOVA. (C) PA-X and GFP expression levels were detected by Western blotting from total cell lysates using specific PAbs against the HA epitope tag (PA-X) or GFP, respectively. A MAb against β-actin was included as a loading control. Western blots were quantified by densitometry using the Fiji software and the amounts of PA-X and GFP proteins were normalized to the amounts of β-actin (numbers below PA-X and GFP blots, respectively). PA-X and GFP expression levels in cells transfected with the pCAGGS plasmid expressing the PA-X WT protein were considered as 1. Sizes of molecular markers (kDa) are noted on the left.
We further evaluated levels of A/Viet Nam/1203/2004 PA-X mutant expression in HEK293T cells transfected with pCAGGS PA-X expression plasmids by immunofluorescence by using an antibody against the HA epitope tag (Fig. 4). As expected, A/Viet Nam/1203/2004 PA-X WT was not detected in transfected cells because it suppresses its own expression (Fig. 4) (21, 30, 34, 35, 38–40). However, we were able to detect the expression of L16P, E43K, F76L, P107S, L132P, H146Y, L187P, and D111N/E196G PA-X mutants previously identified to be most affected in shutoff activity (Fig. 3). Notably, all of the PA-X mutants that were positive by immunofluorescence were detected in a limited number of cells. This could be due to PA-X’s shutoff activity, protein conformation changes that result in hiding the HA epitope tag, or different posttranslational modifications that could also hide the HA epitope tag. Although these hypotheses were not directly addressed, this could explain why PA-X mutants K34E, I147T and E101G/R125K were detected by Western blotting (Fig. 3C) but not in the immunofluorescence assay (Fig. 4). Overall, these results demonstrate that A/Viet Nam/1203/2004 PA-X mutants identified in our bacterium-based assay are all affected in their ability to inhibit host gene expression in HEK29T cells.
FIG 4.
Expression of A/Viet Nam/1203/2004 H5N1 PA-X WT and mutants by immunofluorescence. Human HEK293T cells (24-well plate format, 2.5 × 105 cells/well, triplicates) were transiently transfected with 1 μg of the pCAGGS plasmids encoding PA-X WT or the indicated mutations. At 24 hpt, cells were fixed, permeabilized, and stained with an Ab against the HA epitope tag (green). Cell nuclei were stained with DAPI (blue). Representative images are shown. Scale bar, 10 μm.
Effect of PA-X amino acid changes on inhibition of IFN responses.
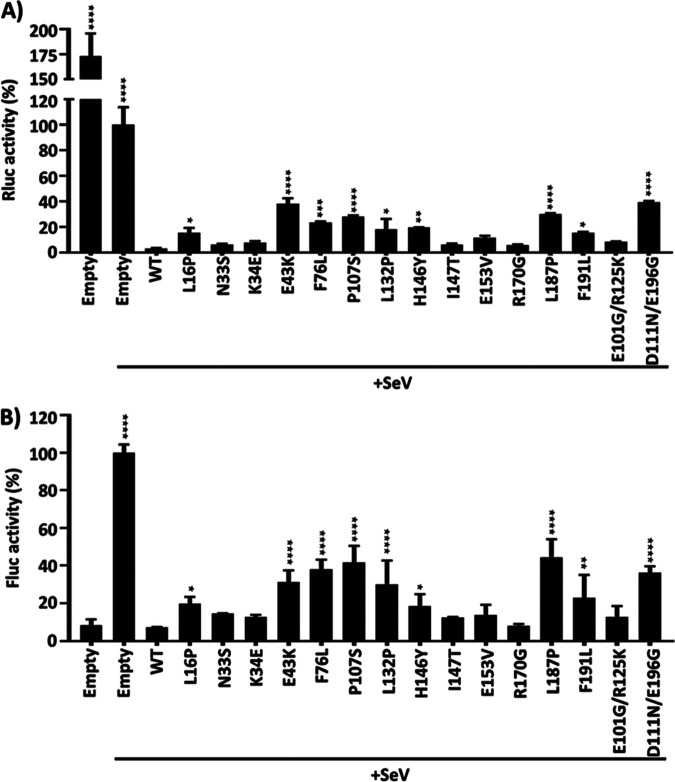
IAV PA-X protein has been shown to thwart the innate immune system (19, 30, 38). To investigate the ability of the identified amino acid changes in PA-X to modulate innate immune responses, we used a well-established IFN assay (6, 8, 43). For that, HEK293T cells were cotransfected with pCAGGS plasmids expressing A/Viet Nam/1203/2004 PA-X WT or mutants, together with a plasmid constitutively expressing Renilla luciferase under a polymerase II-driven simian virus 40 (SV40) promoter (SV40-Rluc) to evaluate shutoff activity (Fig. 5A), and a plasmid expressing Firefly luciferase (Fluc) under the control of an interferon (IFN)-stimulated response element (ISRE) promoter to evaluate inhibition of IFN responses (Fig. 5B). At 24 hpt, cells were mock infected or infected (multiplicity of infection [MOI] of 3) with Sendai virus (SeV; Cantell strain), and reporter expression was evaluated at 21 h postinfection (hpi) by assessing Renilla luciferase (Rluc) (Fig. 5A) and Fluc (Fig. 5B) expression levels. Notably, the shutoff activity for PA-X WT or the identified mutants, measured by Rluc levels, showed similar results than those previously in Fig. 3 and 5A. As expected, SeV infection induced high levels of Fluc expression driven by the ISRE promoter in cells transfected with an empty plasmid (Fig. 5B). Importantly, ISRE promoter activation in cells transfected with PA-X proteins was significantly reduced (Fig. 5B), consistent with previous data showing that PA-X efficiently counteracts IFN responses (17, 21, 23, 30, 38). Importantly, PA-X proteins showing higher shutoff activity were also able to inhibit ISRE promoter activation to higher extents. These data indicate that there is a functional correlation between PA-X’s ability to inhibit host gene expression and PA-X’s ability to block IFN responses.
FIG 5.
Effect of A/Viet Nam/1203/2004 H5N1 PA-X mutants on IFN responses induced by SeV infection. (A and B) HEK293T cells were transiently cotransfected, using calcium phosphate, with the indicated HA-tagged PA-X pCAGGS expression plasmids, together with plasmids expressing Rluc under an SV40 promoter (A), or Fluc under the control of an ISRE promoter (B). An empty pCAGGS plasmid was included as internal control. At 24 hpt, cells were mock infected or infected (MOI of 3) with the SeV, Cantell strain (+SeV) to induce ISRE promoter activation (B). At 21 h postinfection, cell lysates were prepared for reporter gene expression. Rluc (A) and Fluc (B) expression levels were measured by luminescence. Activity of Rluc and Fluc were normalized to the empty plasmid control (+SeV) transfected cells (100%). The results are the means and SDs of triplicates. The data are representative of three independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001 (PA-X WT versus PA-X mutants for each amount of plasmid), determined using one-way ANOVA.
Effect of identified PA-X amino acid changes on A/Viet Nam/1203/2004 PA polymerase activity.
Given that IAV PA and PA-X proteins share the same N-terminal amino acid region, implying that with the exception of amino acid change E196G all the other amino acid changes also affect PA amino acid sequence and that PA-X residue changes affecting also PA protein are very conserved in H5N1 viruses circulating in nature, with the exception of E101G (Table 1), it is possible to speculate that amino acid conservation in the identified residues is due to their potential negative impact on PA’s polymerase activity. To analyze whether these amino acid changes decrease PA polymerase activity, we performed a minigenome assay to assess whether the identified amino acid substitutions could influence A/Viet Nam/1203/2004 polymerase activity (Fig. 6). To that end, pCAGGS plasmids encoding A/Viet Nam/1203/2004 PB2, PB1, PA (WT or mutants), and NP fused to a HA epitope tag were transiently cotransfected in human HEK293T cells, together with two viral (v)RNA-like expression plasmids encoding GFP and Gluc driven by a human RNA polymerase I promoter, and a reporter plasmid constitutively expressing Cypridina luciferase (Cluc) under an SV40 polymerase II-driven promoter, to normalize transfection efficiencies. GFP or Gluc expression levels were determined at 24 hpt. For most of the examined A/Viet Nam/1203/2004 PA mutants (L16P, N33S, K34E, E43K, F76L, P107S, L132P, I147T, E153V, R170G, F191L, and E101G/R125K), we did not observe significant differences in GFP (Fig. 6A) or Gluc (Fig. 6B) expression, when normalized to Cluc expression levels. Conversely, A/Viet Nam/1203/2004 polymerase activity was significantly reduced, compared to that of WT PA, with PA mutants H146Y, L187P, and D111N/E196G (Fig. 6A and B).
FIG 6.
Effect of A/Viet Nam/1203/2004 H5N1 PA-X mutations in PA polymerase activity. Human HEK293T cells (96-well plate format, 1 × 104 cells/well, triplicates) were transiently cotransfected with 31.25 ng of A/Viet Nam/1203/2004 H5N1 pCAGGS expression plasmids encoding the minimal components for viral genome replication and gene transcription (PB2, PB1, and PA) and NP containing a HA epitope tag, together with 62.5 ng of pPOL-I vRNA-like expression plasmids encoding Gluc or GFP under the control of the human polymerase I promoter, and 12.5 ng of the SV40-Cluc plasmid to normalize transfection efficiencies. At 24 hpt, viral replication and transcription were analyzed by GFP expression under a fluorescence microscope (A) and quantified by luminescence (B). Gluc activity was normalized to that of Cluc, and the data are represented as relative activity to that of PA WT (100%). Data represent the means and SDs of the results determined from triplicate wells. ****, P < 0.0001 (PA WT versus PA mutants), determined using one-way ANOVA. Experiment was performed three times with similar results. (C) PB2, PB1, PA, and NP protein expression levels from cell lysates were evaluated by Western blotting with a specific PAb against the HA epitope tag. A MAb against β-actin was included as a loading control. The sizes of molecular markers (kDa) are noted on the left.
Moreover, the protein expression levels for A/Viet Nam/1203/2004 viral polymerase complex (PB2, PB1, and PA) and NP were evaluated by Western blotting with an antibody against the HA epitope tag (Fig. 6C). We did not observe significant differences in PB2, PB1, PA, and NP expression levels when the minigenome assay was performed with the different PA constructs (Fig. 6C). These data indicated that most of the amino acid changes identified in A/Viet Nam/1203/2004 PA-X did not affect viral polymerase activity, with the exception of H146Y, L187P, and D111N/E196G. Although the mechanism involving the drastic reduction in the activity of the viral polymerase of A/Viet Nam/1203/2004 observed with H146Y, L187P, and D111N/E196G PA mutants is unknown, it could involve its endonuclease activity and/or the interaction with other viral proteins or cellular host factors (21, 44, 45).
Baloxavir marboxil (BXM) is a potent inhibitor of influenza virus replication, with antiviral effects specific for the four types of influenza viruses (A, B, C, and D) (46), that acts over the cap-dependent endonuclease within the polymerase PA subunit (47, 48). To assess whether the activity of IAV polymerase in the presence of BXM could be evaluated using our minigenome assay, human HEK293T cells were transiently cotransfected with the polymerase complex (PB2, PB1, and PA) and the NP of A/Viet Nam/1203/2004, as indicated above, in the presence of different concentrations of BXM (Fig. 7A to C). BXM was able to inhibit A/Viet Nam/1203/2004 polymerase activity in the minigenome assay, as determined by GFP (Fig. 7A) and Gluc (Fig. 7B) expression in a concentration-dependent manner. The expression of A/Viet Nam/1203/2004 polymerase complex components and NP was confirmed by Western blotting with an antibody against the HA epitope tag (Fig. 7C). These results indicated that BXM inhibits the activity of A/Viet Nam/1203/2004 polymerase complex in our minigenome assay.
FIG 7.
Effect of BXM in A/Viet Nam/1203/2004 H5N1 PA polymerase activity. (A to C) Effect of BXM in polymerase activity of replication complexes containing the WT PA. Human HEK293T cells were transiently cotransfected (96-well plate format, 1 × 104 cells/well, triplicates) with 31.25 ng of A/Viet Nam/1203/2004 H5N1 pCAGGS expression plasmids encoding HA-tagged PB2, PB1, PA, and NP, together with 62.5 ng of vRNA-like Gluc or GFP expression plasmids (pPOL-I) under the control of the human polymerase I promoter; and 12.5 ng of the SV40-Cluc plasmid to normalize transfection efficiencies. Viral replication and transcription were analyzed at 24 hpt by GFP expression (A) and quantified by luminescence (B). Gluc activity was normalized to that of Cluc, and the data are represented as the relative activity of PA without (w/o) BXM and with (w) DMSO (100%). Data represent the means and SD of the results determined from triplicate wells. ****, P < 0.0001, determined using one-way ANOVA. The experiment was performed twice with similar results. (C) PB2, PB1, PA, and NP protein expression levels from cell lysates were evaluated by Western blotting with a specific PAb against the HA epitope tag. A MAb against β-actin was included as a loading control. The sizes of molecular markers (kDa) are noted on the left. The higher DMSO concentration in this assay was 0.1%. (D to F) Effect of the identified PA mutations in PA polymerase activity in the presence of BXM. Human HEK293T cells were transiently cotransfected as described in Fig. 6. Cells were incubated with 0.01 μM BXM (based on results from Fig. 6) and, at 24 hpt, viral replication and transcription were analyzed by GFP (D) and Gluc (E) expression. Gluc activity was normalized to that of Cluc (B). Data are represented as the relative activity to PA WT (100%). Data represent the means and SDs of the results determined from triplicate wells. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001; (PA WT versus PA mutants), determined using one-way ANOVA. (E) PB2, PB1, PA, and NP protein expression levels from cell lysates were evaluated by Western blotting with a specific PAb against the HA epitope tag. A MAb against β-actin was included as a loading control. Sizes of molecular markers (kDa) are noted on the left.
Based on these results, we next evaluated the ability of the identified A/Viet Nam/1203/2004 PA-X mutations to affect the polymerase activity in the presence of BXM to determine the impact of these amino acid substitutions in BXM susceptibility (Fig. 7D to F). To that end, we conducted a similar minigenome assay in the presence of 0.01 μM BXM and evaluated GFP (Fig. 7D) and Gluc (Fig. 8E) expression at 24 hpt. When A/Viet Nam/1203/2004 PA WT protein was used to reconstitute the polymerase complex, a significant reduction in the polymerase activity was detected in the presence of BXM, as was previously observed (Fig. 7). Interestingly, we found that all the PA substitutions negatively affected A/Viet Nam/1203/2004 polymerase activity in the presence of BXM (Fig. 7A and B). However, A/Viet Nam/1203/2004 polymerase activity with H146Y, L187P, and D111N/E196G mutations was reduced to a higher extent compared to the other A/Viet Nam/1203/2004 mutants, as previously observed in the minigenome assay in the absence of BXM (Fig. 6).
FIG 8.
Location of identified amino acid mutations in A/Vietnam/1203/2004 H5N1 PA. The N-terminal region of A/Vietnam/1203/2004 H5N1 PA protein (accession no. AY818132) is shown in two different views using the same coloring code. PA is indicated in gray. Locations of amino acids L16, N33, K34, E43, F76, P107, L132, H146, I147, E153, R170, L187, and F191 (red), E101/R125 (blue), and or D111/E196 (green) are shown on the background of the PA. The crystal structure figure was adapted from PDB 4E5E using PyMOL.
DISCUSSION
Influenza PA-X protein plays a major role in the suppression of host protein synthesis during viral infection (19–21, 23, 30, 31, 33–35, 37, 38, 49, 50). In the present study, we demonstrate that A/Viet Nam/1203/2004 H5N1 PA-X protein, similar to the PA-X of other IAV strains (18–21, 30, 31, 33–35, 37, 38, 49, 50), is able to inhibit host protein expression in human HEK293T and avian DF-1 cells (Fig. 1). Then, we used a novel approach based in the use of the T7 promoter in bacteria to identify residues important for A/Viet Nam/1203/2004 H5N1 PA-X shutoff activity (Fig. 2). Since genes under the control of a T7 promoter are expressed in bacterial cells due to T7 promoter leakage (41, 42) and low levels of A/Viet Nam/1203/2004 H5N1 PA-X expression were likely deleterious or toxic for growth, bacteria were forced to introduce mutations affecting PA-X inhibition of host gene expression to grow. Sequencing results from the pGBKT7 plasmids isolated from 30 bacterial colonies identified 13 A/Viet Nam/1203/2004 H5N1 PA-X mutants containing single amino acid changes and 2 mutants containing two amino acid changes, all affecting the ability of A/Viet Nam/1203/2004 H5N1 PA-X to inhibit host gene expression (Fig. 2B). In addition, we also identified insertion and/or deletions of nucleotides leading to early or late stop codons within the A/Viet Nam/1203/2004 H5N1 PA-X gene, all affecting PA-X’s host shutoff activity (Fig. 2C). Although it has been described that amino acid residues located at the C-terminal unique region of influenza PA-X are highly relevant for its shutoff activity (34), amino acid substitutions in the C-terminal region of A/Viet Nam/1203/2004 H5N1 PA-X, with the exception of E196G, were not identified (Fig. 2B). Notably, with the only exception of mutant E101G (which was found together with R125K mutation, being R125 a highly conserved residue in the PA protein from H5N1 viruses), amino acid residues present in A/Viet Nam/1203/2004 H5N1 PA-X WT are highly conserved (>94%) in avian and human H5N1 viruses (Table 1). These data suggest that H5N1 viruses encoding PA-X proteins affected in inhibition of host gene expression are not fit in replication in avian or mammalian hosts and therefore not present in nature. Given that IAV PA-X shares its N-terminal 191 amino acids with PA (15), we mapped the identified amino acids at positions onto the N-terminal structure of PA. Interestingly, amino acid mutations identified in our screening were widely distributed in the N-terminal region of A/Vietnam/1203/2004 H5N1 PA protein (Fig. 8).
The relevance for PA-X shutoff activity of three of the amino acid substitutions identified in our screening (P107S, H146Y, and R125K) was recently reported in another study in which the authors used a yeast-based assay to identify amino acid changes involved in A/WSN/33 H1N1 PA-X inhibition of host gene expression (37), further validating our results (Fig. 3). Moreover, these results suggest that the contribution of these amino acid residues to inhibit host gene expression are not subtype specific. In the case of the PA-X double mutants E101G/R125K and D111N/E196G, we have not determined whether one or both amino acid residues are required for A/Viet Nam/1203/2004 H5N1 PA-X shutoff activity. However, among the basic amino acids in A/Viet Nam/1203/2004 H5N1 PA-X C-terminal region, three residues (K195, K198, and R199) have been shown to be important for its host cellular shutoff activity (35, 36). Therefore, we could speculate that amino acid E196G, which is in between those residues, could affect A/Viet Nam/1203/2004 H5N1 PA-X ability to inhibit host gene expression. In the case of the other double mutant (E101G/R125K), it has been shown that amino acid change R125K reduces PA-X-mediated inhibition of host gene expression (37).
In the immunofluorescence studies, we could not detect expression of A/Viet Nam/1203/2004 H5N1 PA-X WT and mutants containing amino acid changes N33S, K34E, I147T, E153V, R170G, F191L, and E101G/R125K (Fig. 4). This is most likely due to the fact that in this experiment, in which genes are transcribed by the RNA polymerase II, PA-X is inhibiting its own expression, as previously shown (21, 34). Notably, amino acid mutants L16P, E43K, F76L, P107S, L132P, H146Y, L187P, and D111N/E196G were detected by immunofluorescence and correspond to those more severely affected in inhibition of host gene expression (Fig. 3).
Notably, PA-X mutants with reduced ability to inhibit host gene expression blocked ISRE promoter activation less efficiently (Fig. 5). This result indicates that PA-X shutoff activity is linked to its ability to counteract the innate immune responses.
In addition to the effect of these amino acid mutations on PA-X shutoff activity or blocking IFN responses, we also evaluated their effect on A/Vietnam/1203/2004 H5N1 polymerase activity using a minigenome assay (Fig. 6). This information could be relevant to understand the reason because the PA-X WT sequence is highly conserved in proteins from H5N1 viruses (Table 1). Moreover, this study could help us to predict the possibility that viruses containing the identified amino acid changes could occur in nature and indirectly infer the importance of those residues in viral fitness.
A/Vietnam/1203/2004 H5N1 PA mutants H146Y, L187P, and D111N/E196G showed the lower levels of polymerase activity, while other mutations were not significantly affected in viral genome replication and gene transcription (Fig. 6). Regan et al. determined that A/WSN/33 H1N1 PA double mutant D111A/Y112A yielded nonfunctional polymerases (51). Thus, PA D111N mutation could be responsible, at least in part, of the lack of polymerase activity in our minigenome assay. BXM showed broad-spectrum inhibition of influenza A, B, C, and D viruses. In addition, BXM also is able to inhibit IAV of avian and swine origin, which are considered to have pandemic potential (46). Notably, when minigenome assays were conducted in the presence of 0.01 μM BXM, all A/Vietnam/1203/2004 H5N1 PA mutants displayed reduced polymerase activity compared to WT PA. Moreover, H146Y, L187P, and D111N/E196G PA mutants showed the lower levels of activity in viral replication and transcription (Fig. 7). This information could be important to develop novel BXM-like compounds or to predict potential changes involving BXM resistance, since it has been reported IAV BXM resistance due to changes at amino acid position I38 of the viral PA protein (47, 48, 52, 53).
In summary, we describe here a novel strategy, based on the use of bacteria, for identifying residues relevant for PA-X inhibition of host gene expression. Using this system, we identified A/Vietnam/1203/2004 H5N1 PA-X amino acid residues important for its shutoff activity. Some of the identified A/Vietnam/1203/2004 H5N1 PA-X residues have been described with other influenza subtypes, validating the use of this bacterium-based approach to identify mutations affecting PA-X inhibition of host gene expression. In addition, we also demonstrate that some of the identified residues were important for A/Vietnam/1203/2004 H5N1 polymerase activity.
MATERIALS AND METHODS
Cells.
Human embryonic kidney 293T, HEK293T (American Type Culture Collection [ATCC], CRL-11268) and DF-1 chicken embryo fibroblast (ATCC CRL-12203) cells were grown and maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% PSG (penicillin, 100 U/ml; streptomycin 100 μg/ml; l-glutamine, 2 mM) at 37°C in air enriched with 5% CO2.
Bacterial screening assay.
The PA-X gene of A/Viet Nam/1203/2004 H5N1 was cloned into pGBKT7 plasmid (TaKaRa, catalog no. 630443) using standard molecular biology techniques and specific primers (VT/NdeI/F, 5′-aattcatatggaagactttgtgcgacaa-3′; VT/SalI/R, 5′-aattgtcgactcacttcttttgacattt-3′). DH5α competent E. coli bacteria (Thermo Fisher Scientific) were then transformed with the ligation reactions and plated on Luria broth (LB) agar plates supplemented with 100 μg/ml kanamycin before incubation at 30°C for 24 h. A separate transformation of DH5α competent Escherichia coli cells involving pGBKT7 plasmid with no insert was included as a control. Likewise, the PA-X gene of A/Viet Nam/1203/2004 H5N1 was cloned into pCAGGS (8, 54) as an internal control (VT/SmaI/F, 5′-aattcccgggatggaagactttgtgcgacaa-3′; VT/XhoI/R, 5′-aattctcgagtcacttcttttgacattt-3′). Single bacterial colonies were grown in LB medium at 30°C for 24 h before plasmid extraction. Plasmid DNA preparations were carried out using the E.Z.N.A plasmid mini extraction kit (Omega Bio-Tek) according to the manufacturer’s recommendations. PA-X mutants were confirmed by sequencing (ACGT, Inc.).
Plasmids.
The nucleotide sequences of PA-X mutants were cloned into a mammalian expression plasmid pCAGGS containing an HA epitope tag (8, 54, 55). To generate pCAGGS plasmids encoding the full-length PA WT protein, a pCAGGS-PA plasmid containing an HA epitope tag and a unique MluI restriction site in the PA ORF at position 517, before the frameshift, was generated by introducing silent mutations by performing two PCRs with the primers ClaI-PA-VS (5′-catgcaatcgataccatgggaagactttgtgcgacaatg-3′, including a ClaI restriction site) and MluI-PA-RS (5′-ctgccttatggtgaacagAcGCgttttaattcttgccctgc-3′; capital letters indicate the silent mutations to introduce a MluI restriction site) for PCR1 and the primers MluI-PA-VS (5′-gcagggcaagaattaaaacGCgTctgttcaccataaggcag-3′; capital letters indicate the silent mutations to introduce a MluI restriction site) and PA-1140-RS (5′-gtctactttctctggtgccatgttc-3′) for PCR2. Then, an overlapping PCR using as the templates PCR1 and PCR2 products and primers ClaI-PA-VS and PA-1140-RS was performed. The final PCR product was digested with ClaI and DraIII restriction enzymes and cloned in the pCAGGS PA plasmid. The pCAGGS PA plasmids encoding mutations L16P, N33S, K34E, F76L, P107S, L132P, H146Y, I147T, and E153V and R170G were obtained by subcloning the fragment from the pCAGGS PA-X plasmids using ClaI and MluI restriction sites. pCAGGS PA plasmids encoding mutations L187P, F191L and E196G were obtained by overlapping PCR with the primer ClaI-PA-VS (indicated above) and the reverse primers PA-L187P-RS (5′-gacgaaaggaatcccatGgacccctactggcc-3′), PA F191L-RS (5′-cctctctcggattgacgaaGggaatcccatagacc-3′), and PA-E196G-RS (5′-caattgtctcttcgccCctctcggattgacgaaagg-3′), showing in capital letters the mutations introduced to generate the amino acid changes L187P, F191L, and E196G, for PCR1 and the forward primers PA-L187P-VS (5′-ggccagtaggggtcCatgggattccttcgtc-3′), PA F191L-VS (5′-ggtctatgggattccCttcgtcaatccgagagagg-3′), and PA-E196G-VS (5′-cctttcgtcaatccgagagGggcgaagagacaattg-3′), showing in capital letters the mutations introduced to generate the amino acid changes L187P, F191L, and E196G, and the reverse primer PA-1140-RS (indicated above) for PCR2. Then, overlapping PCRs using as the templates the respective PCR1 and PCR2 products and the primers ClaI-PA-VS and PA-1140-RS were performed. The final PCR products were digested with ClaI and DraIII restriction sites and cloned in the pCAGGS PA plasmid digested with the same restriction sites. All constructs were confirmed by sequencing (ACGT, Inc.). All primers used for the generation of plasmids constructs are available upon request.
Host shutoff assays.
To determine the shutoff activity of A/Viet Nam/1203/2004 (H5N1) PA-X protein on host protein synthesis (8, 43, 56), HEK293T and DF-1 cells (24-well plate format, 2.5 × 105 cells/well, triplicates) were cotransfected with 0, 1, 10, or 100 ng of pCAGGS expression plasmids encoding WT H5N1 PA-X fused to an HA epitope tag, together with 250 ng of pCAGGS plasmids expressing GFP or Gaussia luciferase (Gluc) under the control of the human polymerase II promoter, using Lipofectamine 2000 (LPF2000; Invitrogen). An empty pCAGGS plasmid was used as a control and to normalize the amount of total plasmid DNA used in each transfection. At 24 hpi, GFP expression was evaluated under a fluorescence microscope and Gluc activity was determined from tissue culture supernatants using a Biolux Gaussia luciferase assay kit (New England BioLabs) and a Synergy LX multimode microreader (BioTek). Experiments to evaluate the ability of H5N1 PA-X mutants to inhibit host gene expression were carried out similarly to those described with H5N1 PA-X WT protein.
Inhibition of ISRE promoter activation.
To evaluate inhibition of ISRE promoter activation, HEK293T cells (5 × 104 cells/well, 96-well plate format, triplicates) were transiently cotransfected, using a calcium phosphate mammalian transfection kit (Agilent Technologies), with 10 ng/well of pCAGGS plasmids encoding the indicated PA-X proteins fused to an HA epitope tag, or an empty plasmid as a control, together with 20 ng/well of a SV40 Renilla luciferase (Rluc) expression plasmid and 50 ng/well of a plasmid expressing Fluc under the control of the ISRE promoter (pISRE-Fluc) (57). At 24 hpt, cells were washed and infected (MOI of 3) with the Sendai virus (SeV), Cantell strain, for ISRE promoter activation (57). At 21 h postinfection, cells were harvested and lysed using passive lysis buffer (Promega). Luciferase expression in the cell lysates was determined using a dual-luciferase kit (Promega) according to the manufacturer’s instructions. Measurements were recorded with a microplate reader (Apliskan; Thermo Scientific), and the mean values and standard deviations (SD) were calculated using Microsoft Excel software.
Minigenome assays.
To evaluate the effect of WT and mutant PA proteins on viral polymerase activity, a minigenome assay was performed as previously described (38, 58). Briefly, HEK293T cells (1 × 104 cells/well, 96-well plate format, triplicates) were transiently cotransfected in suspension, using Lipofectamine 3000 (LPF3000; Invitrogen), with 31.25 ng of each pCAGGS plasmids encoding the PB2, PB1, PA (WT or mutants), and NP containing an HA epitope tag, together with 62.5 ng of two reporter viral (v)RNA-like expression pPOL-I plasmids encoding GFP and Gluc driven by a human RNA polymerase I promoter. A Cypridina luciferase (Cluc)-expressing plasmid (12.5 ng) under the control of the SV40 promoter was included to normalize transfection efficiencies (38, 58). HEK293T transfected cells in the absence of pCAGGS PA plasmid were used as a negative control. At 6 hpt, medium was substituted by fresh medium. Barloxavir marboxil (MCE; HY-109025) was prepared as a 10 mM stock solution in dimethyl sulfoxide (DMSO) and kept at −80°C until experimental use. MCE was added to the medium at the indicated concentrations at 6 hpt. At 24 hpt, the Gluc and Cluc expression levels were determined using a Gaussia or a Cypridina luciferase glow assay kit (Thermo Scientific) and a plate reader. Polymerase activity was calculated by standardization of the Gluc activity to the Cluc activity.
Protein gel electrophoresis and Western blot analysis.
Transfected HEK293T and DF-1 cells were lysed in passive lysis buffer (Promega) for 20 min. Proteins from transfected cell lysates were separated by denaturing electrophoresis using 8 to 16% (nUView; minigenome assays) or 12% (shutoff assays) SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (Bio-Rad) with a Bio-Rad Mini Protean II electroblotting apparatus at 100 V for 2 h. Membranes were blocked for 1 h with 5% dried skim milk in 1× phosphate-buffered saline (PBS) containing 0.1% Tween 20, followed by incubation overnight at 4°C with a primary rabbit polyclonal antibody (P)Ab against the HA epitope tag (Sigma, H6908) or GFP (Sigma, GSN149). Mouse monoclonal antibody (MAb) against β-actin (Sigma, A1978) was used as an internal loading control. Bound primary antibodies (Abs) were detected with secondary horseradish peroxidase-conjugated Abs (Sigma) against the different primary (mouse or rabbit) Abs. Proteins were detected by chemiluminescence (Thermo Fisher Scientific) according to the manufacturer’s recommendations and photographed using a ChemiDoc Imaging System (Bio-Rad).
Immunofluorescence assay.
To evaluate PA-X protein expression by indirect immunofluorescence, HEK293T cells (24-well plate format, 2.5 × 105 cells/well) were transfected with 1 μg of the indicated pCAGGS expression plasmids containing HA-tagged PA-X construct, using LPF3000 (Invitrogen). At 24 hpt, the cells were fixed with 4% (vol/vol) formaldehyde and permeabilized with 0.5% (vol/vol) Triton X-100 (Sigma). The cells were then blocked with 10% FBS diluted in 1× PBS. After blocking, cells were incubated with a polyclonal antibody (PAb) against the HA epitope tag (Sigma, H6908) and probed with a fluorescein isothiocyanate-conjugated donkey anti-rabbit secondary antibody (Invitrogen). DAPI (4′,6′-diamidino-2-phenylindole) staining was used to visualize cell nuclei. Images were obtained by using a Zeiss AX10 (Carl Zeiss).
Structure analysis.
Amino acid positions were plotted on the crystal structure of the N-terminal region of PA protein (PDB accession: 4E5E) from A/Vietnam/1203/2004 H5N1 (accession no. AY818132) (59) using the PyMOL molecular graphics system.
Statistical analysis.
Microsoft Excel (Microsoft Corporation) and GraphPad Prism software were used to analyze the data. Microsoft Excel was necessary to perform some of the calculations, and to visualize the raw data. One-way analysis of variance (ANOVA) was performed using GraphPad Prism software.
ACKNOWLEDGMENTS
This study was supported with funds from Comunidad de Madrid (Spain), reference 2017-T1/BMD-5155, and the Spanish Ministry of Science, Innovation, and Universities (RTI-2018-094213-A-I00) to M.L.D. and from a Ramon y Cajal Incorporation grant (RYC-2017) from the Spanish Ministry of Science, Innovation, and Universities to A.N. D.L.-G. received a JAE-INTRO 2020 fellowship from the Spanish National Research Council (CSIC; JAEINT-20-01805). This research was partially funded by the New York Influenza Center of Excellence (NYICE), a member of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400005C (NYICE).
We thank members at our institutes for their efforts in keeping them fully operational during the COVID-19 pandemic.
Contributor Information
Marta L. DeDiego, Email: marta.lopez@cnb.csic.es.
Aitor Nogales, Email: nogales.aitor@inia.es.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Shaw MP. 2007. Orthomyxoviridae: the viruses and their replication. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA (ed), Fields virology, 5th ed. Lippincott/Williams & Wilkins, Co, Philadelphia, PA. [Google Scholar]
- 2.Nogales A, DeDiego ML. 2020. Influenza virus and vaccination. Pathogens 9:220. 10.3390/pathogens9030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wille M, Holmes EC. 2020. The ecology and evolution of influenza viruses. Cold Spring Harb Perspect Med 10:a038489. 10.1101/cshperspect.a038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu R, Sheng Z, Huang C, Wang D, Li F. 2020. Influenza D virus. Curr Opin Virol 44:154–161. 10.1016/j.coviro.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu R, Sreenivasan C, Yu H, Sheng Z, Newkirk SJ, An W, Smith DF, Chen X, Wang D, Li F. 2020. Influenza D virus diverges from its related influenza C virus in the recognition of 9-O-acetylated N-acetyl- or N-glycolyl-neuraminic acid-containing glycan receptors. Virology 545:16–23. 10.1016/j.virol.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nogales A, Aydillo T, Avila-Perez G, Escalera A, Chiem K, Cadagan R, DeDiego ML, Li F, Garcia-Sastre A, Martinez-Sobrido L. 2019. Functional characterization and direct comparison of influenza A, B, C, and D NS1 proteins in vitro and in vivo. Front Microbiol 10:2862. 10.3389/fmicb.2019.02862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nogales A, Chauche C, DeDiego ML, Topham DJ, Parrish CR, Murcia PR, Martinez-Sobrido L. 2017. The K186E amino acid substitution in the canine influenza virus H3N8 NS1 protein restores its ability to inhibit host gene expression. J Virol 91:e00877-17. 10.1128/JVI.00877-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrish CR, Murcia PR, Holmes EC. 2015. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J Virol 89:2990–2994. 10.1128/JVI.03146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander DJ. 2000. A review of avian influenza in different bird species. Vet Microbiol 74:3–13. 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Subbarao Cox NJ, Guo Y. 1999. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19. 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 12.Hui DS. 2008. Review of clinical symptoms and spectrum in humans with influenza A/H5N1 infection. Respirology 13(Suppl 1):S10–S13. 10.1111/j.1440-1843.2008.01247.x. [DOI] [PubMed] [Google Scholar]
- 13.Gu J, Xie Z, Gao Z, Liu J, Korteweg C, Ye J, Lau LT, Lu J, Gao Z, Zhang B, McNutt MA, Lu M, Anderson VM, Gong E, Yu ACH, Lipkin WI. 2007. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet 370:1137–1145. 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth AE, Jagger BW, Wise HM, Nelson CC, Parsawar K, Wills NM, Napthine S, Taubenberger JK, Digard P, Atkins JF. 2012. Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol 2:120109. 10.1098/rsob.120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XH, Gong XQ, Wen F, Ruan BY, Yu LX, Liu XM, Wang Q, Wang SY, Wang J, Zhang YF, Zhou YJ, Shan TL, Tong W, Zheng H, Kong N, Yu H, Tong GZ. 2020. The role of PA-X C-terminal 20 residues of classical swine influenza virus in its replication and pathogenicity. Vet Microbiol 251:108916. 10.1016/j.vetmic.2020.108916. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Song S, Shen Y, Ma C, Wang T, Tong Q, Sun H, Pu J, Iqbal M, Liu J, Sun Y. 2020. Truncation of PA-X contributes to virulence and transmission of H3N8 and H3N2 canine influenza viruses in dogs. J Virol 94:e00949-20. 10.1128/JVI.00949-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol 86:12411–12413. 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao H, Sun H, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Pu J, Chang KC, Liu X, Liu J, Sun Y. 2015. Twenty amino acids at the C terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol 96:2036–2049. 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khaperskyy DA, Schmaling S, Larkins-Ford J, McCormick C, Gaglia MM. 2016. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog 12:e1005427. 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reavill C, Stolerman IP. 1990. Locomotor activity in rats after administration of nicotinic agonists intracerebrally. Br J Pharmacol 99:273–278. 10.1111/j.1476-5381.1990.tb14693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clements AL, Peacock TP, Sealy JE, Lee HM, Hussain S, Sadeyen JR, Shelton H, Digard P, Iqbal M. 2021. PA-X is an avian virulence factor in H9N2 avian influenza virus. J Gen Virol 10.1099/jgv.0.001531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913. 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 27.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 28.Sanz-Ezquerro JJ, Zurcher T, de la Luna S, Ortin J, Nieto A. 1996. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J Virol 70:1905–1911. 10.1128/JVI.70.3.1905-1911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz-Ezquerro JJ, de la Luna S, Ortin J, Nieto A. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol 69:2420–2426. 10.1128/JVI.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. 2018. Modulation of innate immune responses by the influenza A NS1 and PA-X proteins. Viruses 10:708. 10.3390/v10120708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J, Mo Y, Gao Z, Wang X, Gu M, Liang Y, Cheng X, Hu S, Liu W, Liu H, Chen S, Liu X, Peng D, Liu X. 2016. PA-X-associated early alleviation of the acute lung injury contributes to the attenuation of a highly pathogenic H5N1 avian influenza virus in mice. Med Microbiol Immunol 205:381–395. 10.1007/s00430-016-0461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu J, Mo Y, Wang X, Gu M, Hu Z, Zhong L, Wu Q, Hao X, Hu S, Liu W, Liu H, Liu X, Liu X. 2015. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol 89:4126–4142. 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oishi K, Yamayoshi S, Kawaoka Y. 2015. Mapping of a region of the PA-X protein of influenza A virus that is important for its shutoff activity. J Virol 89:8661–8665. 10.1128/JVI.01132-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi T, Chaimayo C, McGuinness J, Takimoto T. 2016. Critical role of the PA-X C-terminal domain of influenza A virus in its subcellular localization and shutoff activity. J Virol 90:7131–7141. 10.1128/JVI.00954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y, Hu Z, Zhang X, Chen M, Wang Z, Xu G, Bi Y, Tong Q, Wang M, Sun H, Pu J, Iqbal M, Liu J. 2020. An R195K mutation in the PA-X protein increases the virulence and transmission of influenza A virus in mammalian hosts. J Virol 94:e01817-19. 10.1128/JVI.01817-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oishi K, Yamayoshi S, Kawaoka Y. 2018. Identification of novel amino acid residues of influenza virus PA-X that are important for PA-X shutoff activity by using yeast. Virology 516:71–75. 10.1016/j.virol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogales A, Rodriguez L, DeDiego ML, Topham DJ, Martinez-Sobrido L. 2017. Interplay of PA-X and NS1 proteins in replication and pathogenesis of a temperature-sensitive 2009 pandemic H1N1 influenza A virus. J Virol 91:e00720-17. 10.1128/JVI.00720-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilimire TA, Nogales A, Chiem K, Ortego J, Martinez-Sobrido L. 2020. Increasing the safety profile of the master donor live attenuated influenza vaccine. Pathogens 9:86. 10.3390/pathogens9020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nogales A, Martinez-Sobrido L, Chiem K, Topham DJ, DeDiego ML. 2018. Functional evolution of the 2009 pandemic H1N1 influenza virus NS1 and PA in humans. J Virol 92:e01206-18. 10.1128/JVI.01206-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosano GL, Ceccarelli EA. 2014. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rybkin I, Gorin D, Sukhorukov G, Lapanje A. 2019. Thickness of polyelectrolyte layers of separately confined bacteria alters key physiological parameters on a single cell level. Front Bioeng Biotechnol 7:378. 10.3389/fbioe.2019.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez L, Nogales A, Iqbal M, Perez DR, Martinez-Sobrido L. 2018. Identification of amino acid residues responsible for inhibition of host gene expression by influenza A H9N2 NS1 targeting of CPSF30. Front Microbiol 9:2546. 10.3389/fmicb.2018.02546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci U S A 99:10736–10741. 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishin VP, Patel MC, Chesnokov A, De La Cruz J, Nguyen HT, Lollis L, Hodges E, Jang Y, Barnes J, Uyeki T, Davis CT, Wentworth DE, Gubareva LV. 2019. Susceptibility of influenza A, B, C, and D viruses to baloxavir. Emerg Infect Dis 25:1969–1972. 10.3201/eid2510.190607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Hanlon R, Shaw ML. 2019. Baloxavir marboxil: the new influenza drug on the market. Curr Opin Virol 35:14–18. 10.1016/j.coviro.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, Kawaguchi K, Uehara T, Shishido T, Naito A, Cusack S. 2018. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep 8:9633. 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Yu H, Li Y, Ma J, Lang Y, Duff M, Henningson J, Liu Q, Li Y, Nagy A, Bawa B, Li Z, Tong G, Richt JA, Ma W. 2017. Impacts of different expressions of PA-X protein on 2009 pandemic H1N1 virus replication, pathogenicity and host immune responses. Virology 504:25–35. 10.1016/j.virol.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao H, Xu G, Sun Y, Qi L, Wang J, Kong W, Sun H, Pu J, Chang KC, Liu J. 2015. PA-X is a virulence factor in avian H9N2 influenza virus. J Gen Virol 96:2587–2594. 10.1099/jgv.0.000232. [DOI] [PubMed] [Google Scholar]
- 51.Regan JF, Liang Y, Parslow TG. 2006. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J Virol 80:252–261. 10.1128/JVI.80.1.252-261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abed Y, Saim-Mamoun A, Boivin G. 2020. Fitness of influenza A and B viruses with reduced susceptibility to baloxavir: a mini-review. Rev Med Virol :e2175. [DOI] [PubMed] [Google Scholar]
- 53.Takashita E. 2020. Influenza polymerase inhibitors: mechanisms of action and resistance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038687.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 55.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. 2017. NS1 protein amino acid changes D189N and V194I affect interferon responses, thermosensitivity, and virulence of circulating H3N2 human influenza A viruses. J Virol 91:e01939-16. 10.1128/JVI.01930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nogales A, Rodriguez L, Chauche C, Huang K, Reilly EC, Topham DJ, Murcia PR, Parrish CR, Martinez-Sobrido L. 2017. Temperature-sensitive live-attenuated canine influenza virus H3N8accine. J Virol 91:e02211-16. 10.1128/JVI.02211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DuBois RM, Slavish PJ, Baughman BM, Yun MK, Bao J, Webby RJ, Webb TR, White SW. 2012. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog 8:e1002830. 10.1371/journal.ppat.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]