Abstract
During a 6-month period, 21 pairs of Pseudomonas aeruginosa isolates susceptible (pretherapy) and resistant (posttherapy) to antipseudomonal β-lactam antibiotics were isolated from hospitalized patients. In vivo emergence of β-lactam resistance was associated with the overexpression of AmpC β-lactamase in 10 patients. In the other 11 patients, the posttherapy isolates produced only low, basal levels of β-lactamase and had increased levels of resistance to a variety of non-β-lactam antibiotics (e.g., quinolones, tetracyclines, and trimethoprim) compared with the levels of β-lactamase production and resistance of their pretherapy counterparts. These data suggested the involvement of the MexA-MexB-OprM active efflux system in the multidrug resistance phenotype of the posttherapy strains. Immunoblotting of the outer membrane proteins of these 11 bacterial pairs with a specific polyclonal antibody raised against OprM demonstrated the overexpression of OprM in all the posttherapy isolates. To determine whether mutations in mexR, the regulator gene of the mexA-mexB-oprM efflux operon, could account for the overproduction of the efflux system, sequencing experiments were carried out with the 11 bacterial pairs. Eight posttherapy isolates were found to contain insertions or deletions that led to frameshifts in the coding sequences of mexR. Two resistant strains had point mutations in mexR that yielded single amino acid changes in the protein MexR, while another strain did not show any mutation in mexR or in the promoter region upstream of mexR. Introduction of a plasmid-encoded wild-type mexR gene into five posttherapy isolates partially restored the susceptibility of the bacteria to selected antibiotics. These results indicate that in the course of antimicrobial therapy multidrug-resistant active efflux mutants overexpressing the MexA-MexB-OprM system may emerge as a result of mutations in the mexR gene.
Active efflux pump systems have recently been recognized to play a major role in the intrinsic and acquired resistances of Pseudomonas aeruginosa to antimicrobial drugs. Because of its wide substrate specificity and its constitutive expression in wild-type P. aeruginosa cells, the efflux pump system encoded by the mexA-mexB-oprM operon contributes significantly to the elevated resistance that this opportunistic pathogen naturally displays to a variety of antibiotics (24). Substrates for MexA-MexB-OprM include compounds as structurally diverse as β-lactams, β-lactamase inhibitors, quinolones, tetracyclines, trimethoprim, chloramphenicol, macrolides, and novobiocin (11, 16, 17, 19, 25, 28). When overproduced, the efflux system allows bacterial cells to reach higher levels of resistance to the substrate antibiotics listed above. In the nalB multidrug-resistant mutant OCR1, overexpression of mexA-mexB-oprM has been found to be associated with a point mutation in mexR, the regulator gene of the efflux operon, leading to a predicted substitution of Trp for Arg at position 69 in the encoded peptide, MexR (26). Such an amino acid change has been proposed to alter the function of MexR in vivo. In addition, inactivation of mexR in several knockout mutants was found to result in overexpression of mexA-mexB-oprM and elevated multidrug resistance, although to a lower extent than in the nalB strain OCR1 (26). Altogether these results highlighted the key role of MexR in controlling the expression of the efflux system but also raised questions about the presence of a second mutation in the nalB mutant OCR1.
Evidence for the involvement of the MexA-MexB-OprM system in the multidrug resistance of P. aeruginosa has essentially been obtained with laboratory mutants selected by quinolones, β-lactams, or tetracyclines (13, 14, 19, 22, 35). The relevance of the efflux mechanism in clinical strains showing a multidrug resistance phenotype, however, has not been clearly established, mainly because of the lack of an accurate method for measuring the efflux rates of antibiotics in intact bacteria. Some data strongly suggest that at least a part of the strains commonly designated “intrinsically resistant” to carbenicillin (because they produce only low, basal amounts of β-lactamase) may consist of MexA-MexB-OprM efflux mutants (15, 19). These strains, which represent up to 69 to 74% of the β-lactam-resistant isolates found in British hospitals (3, 38), show typical cross-resistance to a variety of structurally unrelated antimicrobial agents (18).
By comparing posttherapy isolates resistant to β-lactams to their pretherapy susceptible counterparts, we establish here the role of the MexA-MexB-OprM pump system as a multiple-drug resistance mechanism in clinical strains. Furthermore, we demonstrate that the emergence of resistance associated with MexA-MexB-OprM in vivo is mostly due to mutations affecting the regulator gene mexR.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Clinical strains of P. aeruginosa were recovered from urine (n = 3), transtracheal aspirate (n = 4), or stool (n = 1) specimens or from surgical wounds (n = 3) from patients hospitalized at the University Hospital of Besançon, Besançon, France, during a 6-month survey in 1996. The isolates were identified by conventional methods (2). The other P. aeruginosa strains used in the study were PAO1 (a wild-type strain obtained from B. W. Holloway), 4098 (a mutant of PAO1 producing basal, noninducible levels of AmpC β-lactamase) (15), 4098E (a single step-mutant of 4098 overexpressing the MexA-MexB-OprM efflux system) (16), 4098ET (an oprM::Ω-Hg transductant of 4098E) (22), PT75 (a mutant of 4098ET overproducing the MexE-MexF-OprN efflux system) (12), and ERYRT (a mutant of oprM::Ω-Hg PAO1 overexpressing the MexC-MexD-OprJ pump system) (22). Escherichia coli INFα was used as a host for DNA cloning and plasmid preparation. All bacterial strains were routinely cultured at 37°C on Mueller-Hinton agar medium (Becton Dickinson) or grown in brain heart infusion broth (Difco Laboratories), unless otherwise stated.
Identification of susceptible-resistant bacterial pairs.
Repetitive isolates of P. aeruginosa showing different levels of susceptibility to β-lactams (designated pre- and posttherapy strains) were first typed by O agglutination, according to the Habs classification scheme, with specific antisera provided by Sanofi Diagnostics Pasteur, Marnes La Coquette, France. Bacterial pairs were further characterized by analyzing the restriction banding patterns obtained by pulsed-field gel electrophoresis (PFGE) (CHEF apparatus; Bio-Rad Laboratories) of chromosomal DNA cleaved with the endonuclease DraI (Boehringer) (7). According to commonly accepted guidelines, two isolates are considered clonally related if their DNA fingerprints differ by less than three bands (36). Banding patterns were found to be strictly identical between the pre- and posttherapy isolates except for isolates 12 and 14, which differed by two bands.
DNA methods and PCR.
The mexR-coding regions of the P. aeruginosa isolates were amplified with primers M1 (5′-GGTTTACTCGGCCAAACC-3′) and M2 (5′-CTTCGAAAAGAATGTTC-3′), which anneal upstream of and downstream of mexR, respectively. In some strains, an additional set of primers (M3, 5′-CATAGCGTTGTCCTCATG-3′; M4, 5′-GTTCGTCGATAAGCTTCA-3′) was used to amplify the intergenic region between mexA and mexR that contains the putative −10 and −35 promoter sequences (26). Approximately 100 ng of chromosomal DNA served as a template for PCRs. The PCR mixtures contained 1 μM each primer, each deoxynucleoside triphosphate at a concentration of 250 μM in 1× PCR buffer, and 1.25 U of Taq polymerase (Perkin-Elmer) in a final volume of 150 μl. The reaction mixtures were subjected to a 6 min of initial denaturation at 92°C, followed by 35 cycles of 1 min at 92°C, 1 min at 50°C, and 1 min at 72°C and a 10-min final extension at 72°C. PCR products were run on 0.7% agarose gels, purified with the Wizard PCR Preps kit (Promega), and sequenced with an ABI 373A automatic sequencer (Perkin-Elmer Division, Applied Biosystems) at the Institut d’Etude et de Transfert de Gènes of Besançon. Both strands were sequenced, and the reactions were repeated at least once to confirm the sequencing data. Nucleotide and deduced amino acid sequences were analyzed with the BLAST software package (1).
Complementation with the mexR gene.
The mexR gene of PAO1 was cloned on a 4.2-kbp KpnI-BamHI fragment derived from cosmid pOM1 (11) in vector pUC19, yielding plasmid pMXA13 (10a). A 2.1-kbp SalI DNA fragment containing mexR was subsequently recloned into the broad-host-range vector pAK1900 which codes for amoxicillin-carbenicillin resistance (26). One recombinant clone in which the mexR gene was transcribed in the same orientation as the lac promoter of the plasmid was named pAKR4. Strains of P. aeruginosa were transformed by electroporation (34) with pAKR4 or pAK1900 (purified with the Qiagen plasmid kit), and the transformants were selected on Mueller-Hinton agar containing ticarcillin at concentrations equal to four times the MIC for the resistant isolates.
Analysis of outer membrane components.
The outer membrane proteins of the P. aeruginosa strains were extracted and were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (22). Protein bands were stained with 0.1% Coomassie blue solution or were transferred as such onto nitrocellulose membranes prior to immunoblotting (see below). Isolation of lipopolysaccharides (LPSs) from bacterial envelopes was carried out by following the method of Hitchcock and Brown (9). The LPS preparations were separated on standard SDS–15% (wt/vol) polyacrylamide gels and were visualized by silver staining as reported by Tsai and Frasch (37).
Production of an anti-OprM polyclonal antiserum.
To facilitate the purification of OprM, the oprM gene previously cloned on the recombinant plasmid pOMC1 (11) was recloned on a 1.5-kbp SphI-NruI DNA fragment into the expression vector pMMB207, which codes for chloramphenicol resistance (23). The resultant construct, named pOMI5, was then transferred from E. coli INFα to P. aeruginosa 4098ET by triparental mating by using E. coli HB101 (pRK2013) for plasmid mobilization (4). Overnight cultures of 4098ET(pOMI5) diluted 1:100 in Luria-Bertani broth containing 100 μg of chloramphenicol ml−1 and 1 mM (final concentration) isopropyl-β-d-galactopyranoside were incubated at 37°C up to an A650 of 0.6 to 0.7. The cell envelopes were then extracted and subjected to SDS-PAGE (22). The electrophoretic bands corresponding to OprM (ca. 50 μg of protein per gel) were excised thoroughly and were used to raise specific polyclonal antibodies in a rabbit (Eurogentec).
Western blotting (immunoblotting).
Ten-microgram fractions of outer membrane proteins were separated by SDS-PAGE and transferred electrophoretically to nitrocellulose filters. These filters were subsequently blocked with 3% (wt/vol) gelatin and hybridized for 1 h with the whole anti-OprM serum diluted 1:5,000 in phosphate-buffered saline. Development of membranes was carried out with alkaline phosphatase conjugated to an anti-rabbit secondary antibody by using the AP color reagent kit from Bio-Rad. The specificity of the anti-OprM antibody was established as a result of the following immunoblotting results: detection of a protein band of ca. 50 kDa in the outer membrane extracts of strains PAO1, 4098, and 4098E (the band was overexpressed in the last strain) and loss of the band in strains 4098ET, ERYRT, and PT75 (data not shown). These data demonstrated the absence of cross-reactivity of the anti-OprM antibody with OprJ and OprN, the outer membrane proteins of the multidrug efflux systems MexC-MexD-OprJ and MexE-MexF-OprN, respectively.
Susceptibility tests.
The MICs were determined by the standard serial twofold dilution technique in agar (2) with a Steers replicator that inoculated 104 CFU per spot. The following antibiotic powders were kindly provided by the indicated companies: ceftazidime and trimethoprim, Glaxo Wellcome; aztreonam and cefepime, Bristol-Myers Squibb; ticarcillin and cloxacillin, SmithKline Beecham; piperacillin, Wyeth Lederle; imipenem, Merck Sharp & Dohme; meropenem, Zeneca Pharma; ciprofloxacin, Bayer Pharma; tetracycline, Roussel Diamant; and sulfadiazine, Doms Chibret. Nalidixic acid and chloramphenicol were obtained from Sigma Chemical Co. Routine determination of bacterial susceptibilities to β-lactam antibiotics was performed by the agar diffusion method (33) with disks from Sanofi Diagnostics Pasteur. Posttherapy isolates selected for further investigation were resistant to one or more of the following drugs at the indicated concentrations: ticarcillin, 16 μg ml−1; ticarcillin with clavulanic acid, 16 and 2.13 μg ml−1, respectively; piperacillin, 16 μg ml−1; piperacillin with tazobactam, 16 and 2.13 μg ml−1, respectively; cefsulodin, 8 μg ml−1; ceftazidime, 4 μg ml−1; cefepime, 4 μg ml−1; and aztreonam, 4 μg ml−1.
Detection of β-lactamases in the clinical strains.
Pre- and posttherapy isolates of P. aeruginosa were first screened for β-lactamase production by the nitrocefin (BBL, Cockeysville, Md.) method performed with EDTA-treated cells reported by Williams et al. (38). The β-lactamase contents of strains yielding a negative or a very weak positive reaction by this method were subsequently reexamined by isoelectric focusing on pH 3.5 to 9.5 Ampholine PAGplate gels (Pharmacia Biotech) with crude cell lysates (21). Bands of β-lactamases were detected by overlaying the gels with a 0.05% (wt/vol) solution of nitrocefin in 0.1 M phosphate buffer (pH 7.0).
RESULTS
Emergence of multidrug-resistant mutants in vivo.
During a period of 6 months in 1996, 835 consecutive isolates of P. aeruginosa were collected from patients hospitalized at the University Hospital of Besançon. A survey of the susceptibility patterns of the repetitive isolates allowed us to identify 21 pairs of strains susceptible and then resistant to at least one of the eight antipseudomonal β-lactams to which susceptibility is routinely tested by the disk diffusion method (listed in Materials and Methods). The identity of the pre- and posttherapy isolates of each pair was first established by O serotyping and was then confirmed by comparing the restriction banding patterns obtained by PFGE after cleavage of total DNA with endonuclease DraI (data not shown). Analysis of the β-lactamase contents of the clonally related bacteria (i) by isoelectric focusing with cell lysates and (ii) by a nitrocefin test performed with EDTA-treated cells (38) demonstrated that 10 of the 21 β-lactam-resistant strains overproduced the chromosomally encoded AmpC β-lactamase. The other 11 resistant strains, like their susceptible counterparts, produced only barely detectable amounts of β-lactamase. Table 1 shows that these minimal β-lactamase producers not only were more resistant to β-lactams than the respective pretherapy isolates (two- to eightfold increased MICs) but were also more resistant to a variety of structurally unrelated antibiotics. Interestingly, the mutants displayed higher levels of resistance to meropenem but not to imipenem compared with the susceptible pretherapy strains (except isolate 283, which was more resistant to imipenem than strain 284). Altogether, these data suggested that the posttherapy mutants might overexpress the MexA-MexB-OprM active efflux pump system.
TABLE 1.
Relative antibiotic susceptibilities of pre- and posttherapy strains of P. aeruginosa
| Pretherapy/post-therapy strains | MIC (μg/ml) for pretherapy/posttherapy strainsa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tic | Fep | Caz | Atz | Ipm | Mer | Nal | Chl | Tmp | |
| 287/273 | 16/128 | 4/8 | 2/4 | 4/32 | 4/4 | 4/32 | >1,024/>1,024 | 64/256 | 128/1,024 |
| 96/109 | 16/128 | 8/8 | 2/4 | 4/32 | 1/0.5 | 0.5/4 | 128/1,024 | ≤32/256 | 64/512 |
| 70/92 | 32/128 | 8/16 | 2/4 | 4/32 | 16/8 | 4/32 | 128/1,024 | 64/256 | ≤32/256 |
| 284/283 | 32/128 | 8/32 | 4/16 | 8/64 | 1/4 | 0.5/4 | >1,024/>1,024 | ≤32/256 | 128/>1,024 |
| 141/128 | 16/128 | 8/8 | 2/8 | 4/32 | 1/0.5 | 0.5/1 | 128/>1,024 | 64/256 | ≤32/512 |
| 14/12 | 16/128 | 4/8 | 2/8 | 4/32 | 1/0.5 | 0.5/4 | 128/1,014 | 64/256 | ≤32/512 |
| 18/17 | 16/64 | 1/2 | 1/4 | 4/32 | 4/4 | 0.5/4 | 256/>1,024 | 32/256 | 64/512 |
| A1/A1 | 16/64 | 4/16 | 2/4 | 2/16 | 2/1 | 2/1 | 128/512 | 128/256 | 256/512 |
| M1/M2 | 32/256 | 2/8 | 4/16 | 8/64 | 1/0.5 | 0.25/1 | 128/512 | 128/512 | 128/256 |
| C1/C2 | 16/128 | 4/16 | 2/8 | 2/64 | 4/4 | 1/8 | ≤64/512 | ≤32/512 | 128/>1,024 |
| R1/R2 | 16/64 | 4/8 | 2/4 | 4/32 | 16/16 | 4/16 | >1,024/512 | 64/256 | 128/1,024 |
Tic, ticarcillin; Fep, cefepime; Caz, ceftazidime; Atz, aztreonam; Ipm, imipenem; Mer, meropenem; Nal, nalidixic acid; Chl, chloramphenicol; Tmp, trimethoprim.
Overexpression of OprM.
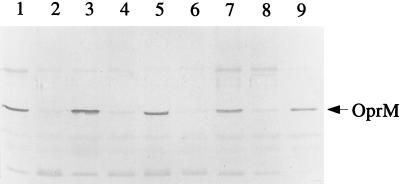
The involvement of the MexA-MexB-OprM system in the resistance phenotype of the minimal β-lactamase-producing strains was studied by comparing the amounts of OprM, the outer membrane component of the system, in the susceptible-resistant clonally related bacteria. Outer membrane proteins were separated by SDS-PAGE, transferred to nitrocellulose films, and hybridized with a polyclonal rabbit antibody raised against OprM. The Western blots unambiguously showed that all the posttherapy isolates contained greater amounts of OprM in their outer membranes than their respective pretherapy counterparts (Fig. 1). In an attempt to see if some other alterations were present in the resistant mutants, we also examined the electrophoretic profiles of outer membrane proteins stained with Coomassie blue and the banding patterns of the LPSs for every bacterial pair. However, no marked difference was noticed between the pre- and posttherapy strains except for the expression of the ca. 50-kDa protein band corresponding to OprM (as identified by Western blotting).
FIG. 1.
Western immunoblots of outer membrane proteins (10 μg per lane) developed with a rabbit polyclonal antibody raised against OprM. Posttherapy resistant isolates 273, 92, 128, and A2 (lanes 1, 3, 5, and 7, respectively) and their respective pretherapy susceptible counterparts (isolates 287, 70, 141, and A1; lanes 2, 4, 6, and 8, respectively) were tested. Lane 9, strain 4098E overexpressing the active efflux system MexA-MexB-OprM. The arrow indicates the position of OprM (ca. 50 MDa).
Mutations in mexR.
MexR, the product of the regulator gene mexR, has been reported to negatively control the transcription of the mexA-mexB-oprM operon in reference strain PAO1 (26). We carried out sequencing experiments to determine whether overexpression of the MexA-MexB-OprM pump system in the posttherapy resistant strains was associated with mutations in mexR. A 480-bp DNA region encompassing the mexR gene was amplified by PCR as described in Materials and Methods. Direct sequencing of the PCR products from the 11 susceptible strains showed that 6 of them had a mexR sequence strictly identical to that published by Poole et al. (26), while a Val→Glu substitution at codon 126 was present in the remaining five strains due to a T→A transversion at position 377 (starting with position 1 at the A of the start codon of mexR). Comparison of the sequences of post- and pretherapy-derived amplicons revealed the presence of deletions (6 of 11 strains) or insertions (2 of 11 strains) of one or more nucleotides in the mexR gene of the resistant isolates, resulting in the production of altered peptides (Table 2). While the deletions were confined to two distinct regions of mexR (around nucleotide 47 and between positions 366 to 388), the two insertions occurred in a small region between nucleotides 65 to 70. Two posttherapy strains were found to possess base pair substitutions in mexR leading to single amino acid changes in the encoded protein. These mutations (Asp8→Glu and Ala66→Val) were different from the Arg69→Trp substitution reported for nalB strain OCR1 (26). In contrast, no mutation was detected for strain 92 either in mexR or in the 273-bp intergenic promoter region extending between mexR and mexA (data not shown).
TABLE 2.
mexR mutations in the multidrug-resistant isolates of P. aeruginosa
| Resistant/ susceptible strains | mexR mutation in the posttherapy resistant strainsa |
|---|---|
| 273/287 | 11-bp deletion from nucleotides 47 to 57 |
| 109/96b | 11-bp deletion from nucleotides 367 to 377 |
| 92/70b | None |
| 283/284 | T deletion at position 47 |
| 128/141b | Ala 66→Val |
| 12/14b | Asp 8→Glu |
| 17/18 | A deletion at position 388 |
| A2/A1b | C insertion between nucleotides 69 and 70 |
| M2/M1 | G deletion at position 366 |
| C2/C1 | 6-bp deletion from nucleotide 382 to 387 |
| R2/R1 | CG insertion between nucleotides 65 and 66 |
Nucleotides were numbered from the start codon of the mexR gene.
Both susceptible and resistant isolates differed from PAO1 by substitution of T to A at position 377 of mexR (see reference 26).
Complementation of mutants.
To further investigate whether the mutations in mexR were solely responsible for the observed multidrug resistance phenotype, plasmid pAKR4 carrying the mexR gene on the broad-host-range vector pAK1900 was constructed. Attempts at introducing these two plasmids into the resistant strains by electroporation were successful for only 5 of the 11 posttherapy strains. The effect of the mexR gene in the posttherapy strains was determined by measuring the MICs of aztreonam, meropenem, nalidixic acid, chloramphenicol, and trimethoprim and was expressed as ratios of the MICs for bacterial cells harboring pAK1900 and the MICs for those transformed with the mexR carrying plasmid pAKR4 (Table 3). The presence of plasmid pAKR4 decreased the MICs (ratios of 4 to 8) of all the antibiotics listed above for isolate 128, a result that indicates that the intact mexR gene successfully complemented the defect of that strain. The introduction of the cloned gene into the other posttherapy mutants was followed by a substantial decrease in the levels of resistance to only some of the tested drugs (see data for strains 109, 283, and 17 in Table 3). As expected, isolate 92 (for which no mutation was found in mexR), when transformed with pAKR4, showed unchanged susceptibilities to aztreonam and meropenem, two substrates of the MexA-MexB-OprM efflux system (11, 15, 19), but became more susceptible to nalidixic acid (fourfold). The levels of resistance of the reference strain PAO1 were not notably affected by the presence of pAKR4.
TABLE 3.
Complementation of posttherapy efflux mutants with mexR
| Strain | MIC ratioa
|
||||
|---|---|---|---|---|---|
| Atz | Mer | Nal | Chl | Tmp | |
| PAO1 | 0.5 | 0.5 | 1 | 0.5 | 0.5 |
| 109 | 8 | 4 | 8 | 4 | 2 |
| 92 | 1 | 1 | 4 | 1 | 1 |
| 283 | 2 | 4 | NDb | 2 | 2 |
| 128 | 4 | 4 | 8 | 8 | 4 |
| 17 | 4 | 4 | 8 | 1 | 2 |
The MIC ratio is the MIC for pAK1900/MIC for pAKR4. Resistant strains were each transformed with the mexR gene cloned on plasmid pAK1900 (construct named pAKR4) and with the vector alone. MIC ratios of ≥4 are in boldface. For definitions of abbreviations, see footnote a of Table 1.
ND, not determined because the pretherapy strain was highly resistant to nalidixic acid (MIC, ≥1,024 μg per ml), suggesting the presence of a gyrA-type mutation.
Antibiotic treatments received by the patients.
The emergence of bacterial resistance in vivo is known to depend on multiple and rather complex factors (5). In an attempt to better understand the therapeutic circumstances that may lead to the emergence of MexA-MexB-OprM efflux mutants, we retrospectively examined the antibiotic treatments given to the patients from whom pre- and posttherapy bacterial pairs had been isolated. It was found that most patients (9 of 11) had received combination therapies involving either β-lactams and fluoroquinolones (5 patients), β-lactams and aminoglycosides (3 patients), or aminoglycosides and fluoroquinolones (1 patient), while 2 patients had been treated by monotherapy with ticarcillin and clavulanic acid, respectively. Interestingly, in four of four patients receiving aminoglycosides, the resistant mutants were isolated once the aminoglycosides were no longer administered (to avoid nephrotoxicity) during a period of single-drug therapy with a β-lactam or a fluoroquinolone. With the notable exception of three antibiotic regimens containing amoxicillin (in combination with a fluoroquinolone) given empirically, all other treatments included drugs known for their antipseudomonal activities (data not shown).
DISCUSSION
The importance of antibiotic efflux in multidrug-resistant strains of P. aeruginosa is becoming more and more recognized in the hospital setting. Previous reports have shown that, in particular, nfxB- and nfxC-type mutants, which overexpress the MexC-MexD-OprJ and MexE-MexF-OprN efflux systems, respectively, occur in clinical isolates of various origins (6, 10, 39).
In the present report, we document for the first time that nalB-type mutants, which overexpress the mexA-mexB-oprM efflux operon, have been selected in vivo during the course of antibiotic therapy. Several studies have pointed out the fact that many clinical isolates of P. aeruginosa resistant to carbenicillin actually produce only low, basal amounts of β-lactamase (3, 38). Furthermore, the development of such resistant strains in patients receiving β-lactam antibiotics has repeatedly been reported since the early days of antipseudomonal chemotherapy (27, 31, 32). These so-called intrinsically resistant or β-lactamase-negative strains often show cross-resistance to a variety of unrelated drugs such as tetracyclines, chloramphenicol, and fluoroquinolones (18), a multidrug resistance phenotype similar or close to that of MexA-MexB-OprM-overproducing mutants selected in vitro. Although this similarity tends to suggest that most clinical isolates intrinsically resistant to carbenicillin overproduce the MexA-MexB-OprM pump system, the relevance of the efflux mechanism in the resistance of clinical strains has rarely been demonstrated (15), mainly because of the lack of an experimental approach for accurate measurement of the rates of export of antibiotics in bacterial cells. In this study, all 11 resistant isolates that emerged under therapy turned out to be MexA-MexB-OprM efflux mutants, on the basis of differences in OprM expression compared with that of their susceptible pretherapy counterparts. This shows that such efflux mutants have a clinical significance and that their possible emergence under antimicrobial therapy must be taken into account in the management of patients infected with P. aeruginosa.
Most of MexA-MexB-OprM mutants obtained in vitro have been selected by fluoroquinolones, which have been used alone (28, 30) or in combination with β-lactams (14, 19, 20). Clearly, the small number of patients in this study does not allow us to draw general conclusions about therapeutic factors that may affect the emergence of such efflux mutants in vivo. However, it is interesting that nearly all mutants developed under combination therapy. The role of aminoglycosides (which are not substrates for the MexA-MexB-OprM pump system) (8, 20, 22) in preventing the emergence of MexA-MexB-OprM mutants is unclear. Four of our patients did receive an aminoglycoside combined with a β-lactam or a fluoroquinolone, but the emergence of MexA-MexB-OprM-associated resistance occurred after the completion of aminoglycoside therapy. Because of its important clinical implications, the latter point warrants further assessment.
The MexR protein is the transcriptional repressor of the mexA-mexB-oprM operon (26). Knockout mutants of mexR show a modest twofold increase in mexA transcription with a concomitant two- to fourfold decrease in antibiotic susceptibility. The only mutation so far reported to lead to overexpression of the efflux system is a single amino acid change at position 70 of MexR (position 69 in reference 26) in the nalB-type mutant strain OCR1. In the present study, alterations of the mexR gene were found in 10 of the 11 posttherapy isolates. Mutants with frameshift mutations in the mexR gene were predominant, while only two strains (strains 128 and 12) presented amino acid substitutions. Comparison of the amounts of OprM on Western blots did not show any significant difference in amounts between mutants with frameshift mutations and the two mutants with amino acid substitutions. Similarly, MIC ratios (Table 1), which were rather homogeneous for β-lactams but which varied quite strongly for chloramphenicol and trimethoprim (2- to ≥16-fold), did not enable us to establish a correlation between the type of mexR mutation and the levels of mexA-mexB-oprM expression. This suggests that the point mutations seen in isolates 128 and 12 do not result in mutants with a gain of function, as in strain OCR1.
Complementation with the cloned wild-type mexR gene significantly decreased the MICs of all antibiotics tested for strain 128 but reduced the MICs of only some of them for the other mexR mutants, mutants 17, 109, and 283. It was noted that for all the other posttherapy isolates, resistance levels decreased consistently only for nalidixic acid, even for strain 92, which did not show any mutation in mexR. One explanation for this puzzling observation might be that the presence of an additional mutation(s) outside of the mexR gene could interfere with the regulation of the mexA-mexB-oprM operon or affect other efflux pumps or antibiotic resistance mechanisms. In support of this, it has been reported that clinical strains of P. aeruginosa resistant to quinolones may exhibit alterations in DNA gyrase along with LPS defects (29) or decreased levels of drug accumulation (39). Alternatively, it is possible that some of the resistant mutants might require adjusted expression of the intact mexR gene to recover a wild-type phenotype. Further investigation is needed to better understand the regulation of the mexA-mexB-oprM operon and the emergence of multidrug resistance in P. aeruginosa.
ACKNOWLEDGMENTS
We thank C. Godard and C. Bailly for technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balows A, Hausler W J, Jr, Herrmann K L, Isenberg H D, Shadomy H J. Manual of clinical microbiology. 5th ed. Washington, D.C: ASM Press; 1991. [Google Scholar]
- 3.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 4.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fish D N, Piscitelli S C, Danziger L H. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy. 1995;15:279–291. [PubMed] [Google Scholar]
- 6.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godard C, Plésiat P, Michel-Briand Y. Persistence of Pseudomonas aeruginosa strains in seven cystic fibrosis patients followed over 20 months. Eur J Med. 1993;2:117–120. [PubMed] [Google Scholar]
- 8.Gotoh N, Itoh N, Tsujimoto H, Yamagishi J, Oyamada Y, Nishino T. Isolation of OprM-deficient mutants of Pseudomonas aeruginosa by transposon insertion mutagenesis: evidence of involvement in multiple antibiotic resistance. FEMS Microbiol Lett. 1994;122:267–273. doi: 10.1111/j.1574-6968.1994.tb07179.x. [DOI] [PubMed] [Google Scholar]
- 9.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakics E B, Iyobe S, Hirai K, Fukuda H, Hashimoto H. Occurrence of the nfxB type mutations in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2562–2565. doi: 10.1128/aac.36.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Köhler, T. Unpublished data.
- 11.Köhler T, Kok M, Michéa Hamzehpour M, Plésiat P, Gotoh N, Nishino T, Curty L K, Pechère J C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Köhler T, Michéa Hamzehpour M, Henze U, Gotoh N, Kocjancic Curty L, Pechère J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 13.Köhler T, Michéa Hamzehpour M, Plésiat P, Kahr A L, Pechère J C. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X Z, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X Z, Srikumar R, Poole K. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore D M. Penicillin-binding proteins, porins and outer-membrane permeability of carbenicillin-resistant and -susceptible strains of Pseudomonas aeruginosa. J Med Microbiol. 1984;18:261–270. doi: 10.1099/00222615-18-2-261. [DOI] [PubMed] [Google Scholar]
- 19.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–175. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 22.Michéa Hamzehpour M, Pechère J C, Plésiat P, Köhler T. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:2392–2396. doi: 10.1128/aac.39.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales V M, Bäckman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole K, Krebes K, Mcnally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poole K, Tetro K, Zhao Q X, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preheim L C, Penn R G, Sanders C C, Goering R V, Giger D K. Emergence of resistance to β-lactam and aminoglycoside antibiotics during moxalactam therapy of Pseudomonas aeruginosa infection. Antimicrob Agents Chemother. 1982;22:1037–1041. doi: 10.1128/aac.22.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rella M, Haas D. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of β-lactam antibiotics: mapping of chromosomal genes. Antimicrob Agents Chemother. 1982;22:242–249. doi: 10.1128/aac.22.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robillard N J. Broad-host-range gyrase A gene probe. Antimicrob Agents Chemother. 1990;34:1889–1894. doi: 10.1128/aac.34.10.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robillard N J, Scarpa A L. Genetic and physiological characterization of ciprofloxacin resistance in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1988;32:535–539. doi: 10.1128/aac.32.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders C C, Sanders W E., Jr Emergence of resistance during therapy with the newer β-lactam antibiotics: role of the inducible β-lactamases and implications for the future. Rev Infect Dis. 1983;5:639–648. doi: 10.1093/clinids/5.4.639. [DOI] [PubMed] [Google Scholar]
- 32.Shannon K, King A, Phillips I. Development of resistance to beta-lactam antibiotics during therapy of Pseudomonas aeruginosa infections. Lancet. 1982;i:1466. doi: 10.1016/s0140-6736(82)92473-4. [DOI] [PubMed] [Google Scholar]
- 33.Sirot J, Courvalin P, Soussy C J. Definition and determination of in vitro antibiotic susceptibility breakpoints for bacteria. Technical recommendations for in vitro susceptibility testing. Clin Microbiol Infect. 1996;2:S5–S25. doi: 10.1111/j.1469-0691.1996.tb00870.x. [DOI] [PubMed] [Google Scholar]
- 34.Smith A W, Iglewski B H. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 1989;17:10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumita Y, Fukasawa M. Meropenem resistance in Pseudomonas aeruginosa. Chemotherapy (Basel) 1996;42:47–56. doi: 10.1159/000239421. [DOI] [PubMed] [Google Scholar]
- 36.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai C M, Frash C E. A sensitive silver staining method for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 38.Williams R J, Livermore D M, Lindridge M A, Said A A, Williams J D. Mechanisms of beta-lactam resistance in British isolates of Pseudomonas aeruginosa. J Med Microbiol. 1984;17:283–293. doi: 10.1099/00222615-17-3-283. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida T, Muratani T, Iyobe S, Mitsuhashi S. Mechanisms of high-level resistance to quinolones in urinary tract isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1994;38:1466–1469. doi: 10.1128/aac.38.7.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]