Abstract
RNA molecules can fold into complex and stable 3D structures, allowing them to carry out important genetic, structural, and regulatory roles inside the cell. These complex structures often contain 3D pockets made up of secondary structural motifs that can be potentially targeted by small molecule ligands. Indeed, many RNA structures in PDB contain bound small molecules, and high-throughput experimental studies have generated a large number of interacting RNA and ligand pairs. There is considerable interest in developing small molecule lead compounds targeting viral RNAs or those RNAs implicated in neurological diseases or cancer. We hypothesize that RNAs that have similar secondary structural motifs may bind to similar small molecule ligands. Toward this goal, we established a database collecting RNA secondary structural motifs and bound small molecule ligands. We further developed a computational pipeline, which takes as input an RNA sequence, predicts its secondary structure, extracts structural motifs, and searches the database for similar secondary structure motifs and interacting small molecule. We demonstrated the utility of the server by querying α-synuclein mRNA 5′ UTR sequence and finding potential matches which were validated as correct. The server is publicly available at http://RNALigands.ccbr.utoronto.ca. The source code can also be downloaded at https://github.com/SaisaiSun/RNALigands.
Keywords: RNA structure, database, bioinformatics, ligands
INTRODUCTION
RNA molecules not only carry genetic information; they also have other important structural, regulatory, and catalytic roles. From a structural aspect, RNAs such as ribosomal RNA (rRNA) or small nuclear RNAs (snRNA) can form large and complex ribonuclear particles (RNP), for example, ribosome and spliceosome. From a regulatory aspect, riboswitches, microRNAs, piRNAs can regulate gene expression post-transcriptionally. From a catalytic aspect, ribosomal RNAs, ribozymes, self-splicing introns can carry out complex enzymatic functions. To carry out these functions, RNA molecules are often required to fold into specific secondary and tertiary structures and interact with other nucleic acids, proteins, or small molecules inside cells. It has been increasingly appreciated in the research and pharmaceutical community that, similar to proteins, RNA molecules can also serve as important therapeutic targets, targeted by small RNAs or small molecule ligands. It has been proposed that RNAs behave similarly to proteins as drug targets and contain many potential binding pockets that can be targeted by small molecules (Warner et al. 2018; Hewitt et al. 2019). Indeed, RNAs such as riboswitches bind to a wide variety of metabolites (Mironov et al. 2002; Mandal et al. 2003; Winkler et al. 2003; Warner et al. 2007; Garst et al. 2011); other examples of interactions between RNA and small molecules have also been well documented (Li and Disney 2018; Velagapudi et al. 2018; Costales et al. 2019; Matarlo et al. 2019). In addition, a number of studies have focused on finding small molecules that can target viruses and oncogenes in tumors (Lozano et al. 2016; Velagapudi et al. 2016; Costales et al. 2017). All of these point to exciting and timely research opportunities in cataloging and studying RNA and small molecular interactions. Toward this goal, we have built a database and web server, RNALigands, which can facilitate these research endeavors.
There have been previous efforts in cataloging and searching for RNA sequence and structural motifs, notably RNAMotif (Macke et al. 2001), RNAProfile (Pavesi et al. 2004), CMfinder (Yao et al. 2006), RNAMotifScan (Zhong et al. 2010), and BEAR (Mattei et al. 2014; Pietrosanto et al. 2018; Adinolfi et al. 2019), among others. These tools are useful in finding enriched secondary or tertiary RNA motifs, for example, unique types of bulges or loops. There is another class of databases or algorithms which were designed to study the structural properties of RNA sequences bound by trans-regulators such as RNA binding proteins (RBPs). These tools include MEMERIS (Hiller et al. 2006), RNAContext (Kazan et al. 2010), GraphProt (Maticzka et al. 2014), and ssHMM (Heller et al. 2017). Another class of software attempts to directly conduct docking experiments between RNA 3D structures and small molecule ligands; notable programs include rDOCK (RiboDock) (Morley and Afshar 2004; Ruiz-Carmona et al. 2014), AutoDock (Moitessier et al. 2006; Morris et al. 2009), DrugScoreRNA (Pfeffer and Gohlke 2007), MORDOR (Guilbert and James 2008), Dock6 (Lang et al. 2009), DARS-RNP and QUASI-RNP (Tuszynska and Bujnicki 2011), LigandRNA (Philips et al. 2013), SPA-LN (Yan and Wang 2017), and RNAPosers (Chhabra et al. 2020). Despite the usefulness of these databases and servers, to the best of our knowledge there has not been very active research in cataloging and predicting RNA and small molecule ligand interactions. A database, LigandRNA, was previously published in 2013, but has not been updated and is currently not accessible as of early 2021 (Philips et al. 2013). At the time when we were submitting this paper, another computational method, RNAmigos, was published (Oliver et al. 2020). RNAmigos represents RNA 3D pockets as networks and uses deep learning to classify and predict bound ligands. There are major differences between RNAmigos and RNALigands as RNALigands do not require 3D structure as direct input. In addition to the computational approaches, there have been experimental high-throughput approaches investigating RNA and small molecule ligand interactions. Perhaps the most relevant resource is the Inforna database developed by Disney et al. (2016), but it primarily focuses on microRNA and ligand interactions.
We have taken a novel approach in tackling this problem from the perspective of RNA secondary structures. We reasoned that, similar to ligand binding sites in proteins, the recurring RNA secondary or tertiary structure motifs form the basis of interactions between RNA and small molecule ligands. We first searched and collected instances of RNA secondary structure motif–small molecule ligand interactions through three databases, PDB, R-BIND, and Inforna (Berman et al. 2000; Disney et al. 2016; Morgan et al. 2019). We further developed a computational pipeline, which takes an RNA sequence as input, predicts its secondary structure, extracts secondary structural motifs, and searches our RNALigands database for similar secondary structural motifs. Our rationale is that RNA sequences that have similar secondary structural motifs are also likely to bind to similar small molecular ligands; such information is very useful in screening for a small molecule compound that potentially targets these RNAs (Disney et al. 2016). We demonstrated the utility of the server by querying α-synuclein mRNA 5′ UTR sequence and showing potential matches. The server is publicly available at http://rnaligands.ccbr.utoronto.ca/.
RESULTS AND DISCUSSION
Collection of RNA–small molecular ligand interactions
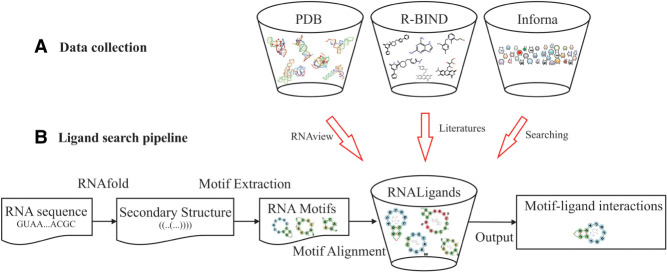
Figure 1A shows a flowchart of how we collected the RNAs and the bound small molecules from three sources: PDB (386 pairs), R-BIND (67 pairs), and Inforna (388 pairs) (Berman et al. 2000; Disney et al. 2016; Morgan et al. 2019). The R-Bind database manually curates RNA–small molecule interacting pairs from the literature, while the Inforna database contains microRNA and small molecule interactions determined from two-dimensional combinatorial screening (2DCS). Although R-Bind and Inforna do not contain 3D structure information, they are still useful for our purpose. The details of data collection are described in Materials and Methods.
FIGURE 1.
Flowchart of the data collection and query pipelines implemented in RNALigands. (A) Data collection procedures, (B) ligand query pipeline.
RNAmigos is a recently published method that models a ligand binding pocket as a graph and uses deep learning to predict RNA and small molecule ligand binding preferences (http://rnamigos.cs.mcgill.ca) (Oliver et al. 2020).There are several differences between RNAmigos and RNALigands in how the respective training set was collected: (i) RNALigands excludes PDB entries that contain peptides, for example, ribosomes and spliceosomes; (ii) RNALigands only keeps nucleotides from a single RNA chain while RNAmigos allows 3D ligand binding pockets consisting of nucleotides from multiple RNA chains; and (iii) RNAmigos only curates 3D structures from PDB while RNALigands curates secondary structures. There are also subtle differences in how the ligand-interacting nucleotides are identified as RNAmigos and RNALigands, as these two softwares use different distance thresholds. In the end, the training set of RNAmigos contains 773 ligand–RNA pairs while RNALigands has 841 pairs.
Classification of RNA secondary structural motifs
Figure 2 shows an example of RNA secondary structure, consisting of four different types of secondary structure motifs: hairpin, internal loop, bulge, and multibranch loop. A hairpin is a single-stranded loop closed by a base pair. An internal loop or a bulge is a single-stranded loop surrounded by stems: An internal loop contains unpaired nucleotides on both sides of the stem, while a bulge only has unpaired nucleotides on one side of the stem. A multibranch loop is the junction region of multiple stems, which is composed of multiple closing base pairs and unpaired nucleotides. The current version of RNALigands database (January 2021) contains 156 hairpins, 176 bulges, 313 internal loops, and 80 multibranch loops. We further classified these loops according to criteria such as the length of the unpaired nucleotides in the loop region, types of unpaired nucleotides, and types of base pairs. Details are listed in Supplemental Material and on the server website.
FIGURE 2.
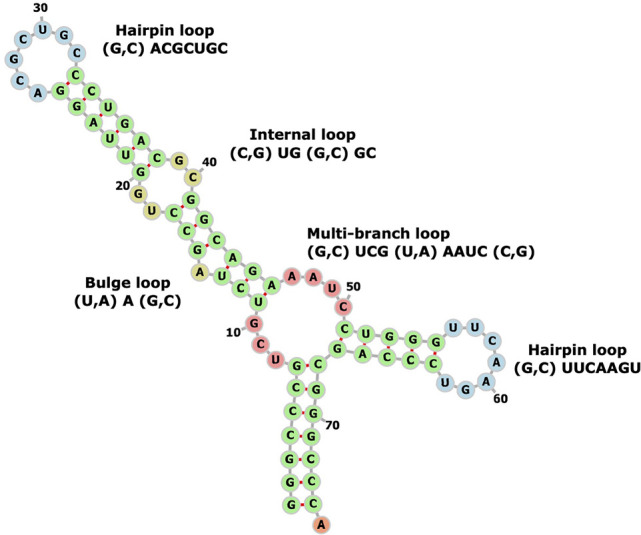
Example of RNA secondary structural motifs (GenBank: X06054.1:c711-638 Sulfolbus solfataricus genes for tRNA-Val).
Query the database for potential RNA motif–ligand interactions
Figure 1B shows the query pipeline of RNALigands, which takes as input an RNA sequence and predicts its secondary structure by using RNAfold (Gruber et al. 2008). The default parameters are used and G:U base pairing is allowed. Users also have the option of providing their own secondary structure predictions as input. At this moment, only a single structure with minimum energy is considered from the input sequence, but structure ensembles will be considered in the future. The predicted secondary structure is then examined by an in-house developed algorithm to identify secondary structure motifs in an approach similar to what was described in a previous study (Liu et al. 2016). These secondary structural motifs are then compared to the motifs stored in the database and similarity scores are calculated, which requires the motifs to have identical topology. A detailed definition of motif alignment scores between RNA motifs can be found in Materials and Methods. The matched RNA motifs and curated small molecule ligands stored in the database are provided as output.
Testing on a benchmark data set
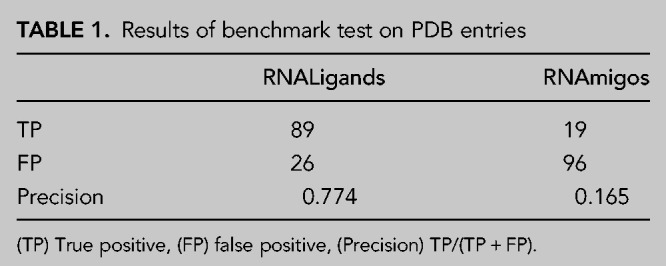
We reexamined the RNA–ligand structure entries in the PDB and grouped these entries into groups that have the same ligand. Next, in each group, we removed the RNA–ligand pairs that were present in our database. Note that in our curation step we removed those RNAs that have >95% sequence identity, therefore these previously removed RNA–ligand pairs could serve as a good benchmark test set to compare RNALigands with other methods. This benchmark test set contained 115 entries and is listed in Supplemental Table S3. We were able to correctly predict the bound ligands for 89 of these 115 entries (77%) by using RNALigands software and the curated RNA ligand database, which compares favorably with RNAmigos (Table 1).
TABLE 1.
Results of benchmark test on PDB entries
Case studies
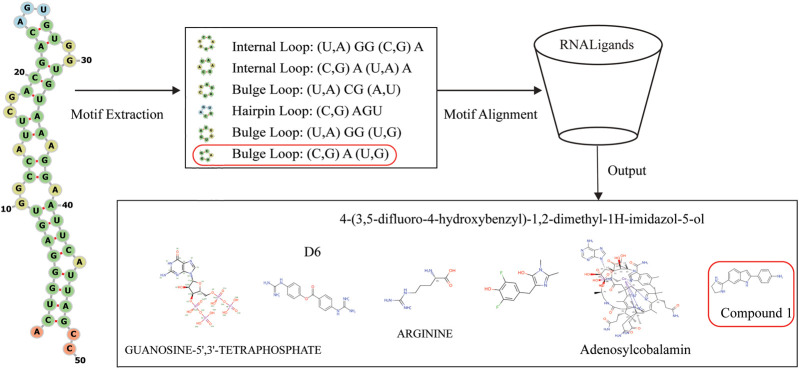
Having demonstrated the effectiveness of RNALigands on the benchmark data set, we next tested the RNALigands server by choosing α-synuclein mRNA 5′ UTR as a worked example. It was reported in early 2020 that the iron-responsive element (IRE) in the 5′ UTR of this mRNA transcript is targeted by a small molecule (Zhang et al. 2020). However, due to the recency of this publication, as of December 2020 this interaction had not been curated in any of the RNA–ligand databases, which made it an ideal test case. We used RNALigands and identified six secondary structural motifs from this RNA sequence, including one hairpin, two internal loops, and three bulge loops (Fig. 3). Querying RNALigands with these motifs returned a potential ligand for each of these secondary structure motifs, including a small molecule “Compound 1” for the bulge motif. This candidate RNA–small molecule interaction was based on an entry in our database curated from an earlier publication in which “Compound 1” was found to bind to a bulge loop on the mRNA of gene MAPT (mutated microtubule-associated protein tau) (Luo and Disney 2014). It is worth noting that this “Compound 1” was validated binding to IRE of the α-synuclein mRNA 5′ UTR, as we described above. We subsequently verified that these two RNA sequences, α-synuclein and MAPT, indeed share a similar RNA bulge structure, which are potentially targeted by the same small molecule ligand.
FIGURE 3.
Test case on α-synuclein mRNA.
We also tested the 5′UTR of the SARS-CoV-2 genome sequence on the RNALigands server. The SARS-CoV-2 virus genome sequence and annotations were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/sars-cov-2/). A total of 30 RNA secondary structure motifs were identified from the 10 stem–loop regions (SL) in the viral genome and were queried against the database. As shown in Supplemental Figure S3, the server returns matched secondary structure motifs and ligand pairs for 19 of the 30 input motifs. Interestingly, ligand SAM (S-adenosylmethionine) is involved in viral mRNA 5′ capping and binds to two non-structure proteins, nsp10 and nsp16 (Lin et al. 2020). Further investigation and molecular modeling on these potential ligands are warranted.
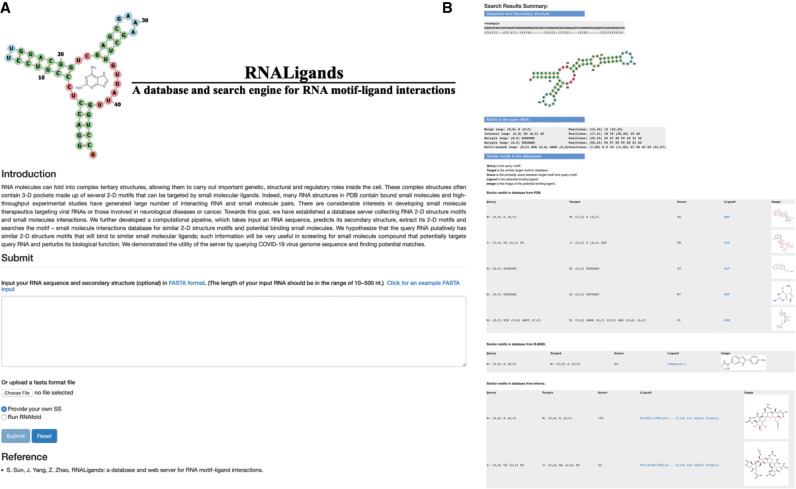
Interface of RNALigands server
Figure 4 shows the web interface and results page of the RNALigands server. Users have the option of providing RNA sequence alone or RNA sequence together with secondary structural information. If the secondary structure is not provided, the RNALigands server provides RNAfold as an optional prediction method (Gruber et al. 2008). The input sequence is limited to 10–500 nt long. The matched RNA loops are presented in both graphic and dot-bracket format. The entire database and a standalone package of the ligand query pipeline can be downloaded in bulk from the website.
FIGURE 4.
The interface page (A) and results page (B) of RNALigands server.
MATERIALS AND METHODS
Curation of RNA–small molecule interactions
Figure 1 shows a flowchart of how we collected the RNAs and the bound small molecules from three sources: PDB (386 pairs), R-BIND (67 pairs), and Inforna (388 pairs). We downloaded the entire PDB database in August 2020 and identified 544 entries (949 RNA chains) that contain an RNA molecule (between 10 and 500 nt in length) and small molecule ligands. In the current version, we excluded those PDB entries that contained both proteins and RNAs since the binding of small molecule ligands may be influenced by the proteins. We next conducted several filtering steps to derive the final RNA secondary structural motif–small molecule set. (i) We first removed the PDB entries that did not contain any biologically relevant small molecules (Yang et al. 2013). (ii) We next removed those entries in which the minimal atomic distance between RNA and small molecule was greater than 4 Å; this reduced the collection to 379 RNA chains. (iii) We used CD-HIT to remove those RNA chains that have sequence identity >95% (Fu et al. 2012); identical small molecules in identical binding modes were also removed. We also removed those RNA fragments that did not have any base pairings. After these filtering steps, we arrived at a final set of 125 unique and representative RNA chains and 149 small molecules from PDB. A summary of these RNA structures and small molecule ligands is provided in Supplemental Tables S1 and S2. We next generated RNA secondary structures from 3D structures by using RNAview (Yang et al. 2003) and used an in-house software to identify RNA secondary structural motifs that have physical interaction with the small molecule ligands, which requires minimal distance between motifs and ligand to be less than 4 Å (Yang et al. 2003). A final set of 386 RNA secondary structural motif and small molecule pairs was obtained from PDB.
R-Bind is a literature-curated database, containing 104 RNA and small molecule pairs that have demonstrated biological activity (Morgan et al. 2019). It provides the name and class of the RNA, the name and molecular properties of the small molecule, and the binding region on the RNA. After removing overlapping entries with PDB and Inforna, we derived 67 RNA secondary structure motif and small molecule interaction pairs from R-Bind.
The Inforna database contains 1936 experimentally determined RNA secondary structural motif–small molecule interactions, including 244 unique small molecules and 1331 RNA secondary structure motifs (Disney et al. 2016). After removing redundant and identical entries, we retained 388 unique RNA secondary structural motif and small molecule pairs.
Classify RNA 3D structures into secondary structural motifs
The key to our approach is to represent the ligand binding pockets as RNA secondary structural motifs so that we can reliably compare an input query sequence with the database and identify potential ligand binding sites. We first used RNAview to generate secondary structures from tertiary structures. A locally developed Perl script was used to examine the dot-bracket file to extract secondary structural motifs. As shown in Figure 2, these motifs were classified into four categories including hairpin loop, internal loop, bulge loop, and multibranch loop, following the convention previously described (Liu et al. 2016). When classifying different secondary structure motifs, we only included the nucleotide pairs immediately adjacent to the opening of the loop. The code of the motif extraction algorithm can be found on the RNALigands website and on GitHub (https://github.com/SaisaiSun/RNALigands).
RNA secondary structure motif alignment algorithm
Our RNA secondary structure motif alignment algorithm is motivated by the Needleman–Wunsch algorithm and considers both RNA sequence identity and secondary structure information (Needleman and Wunsch 1970). The nucleotide substitution matrix, the base pair substitution matrix, and the cost of unpaired loops are derived from FOLDALIGN (Havgaard et al. 2005). Only motifs belonging to the same type are aligned. The motif alignment algorithm consists of three components: alignment of unpaired nucleotides based on dynamic programming, scoring of substitutions of base pairs, and scoring of alignment of loops. In Equation 1, SNW represents the Needleman–Wunsch alignment score, where Ssingle is the nucleotide substitution and GapPenalty is the gap cost in the loop of two aligned motifs. For hairpin loop, internal loop, bulge loop, and multibranch loop, we used Equations 2, 3, 4, and 5 to calculate the similarity score of two motifs, respectively. Sbp is the closing base pair substitution in the two motifs; Δcostlength is the deviation of the costs of two loops.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
Z.Z. acknowledges support from the Nature Science and Engineering Research Council of Canada (NSERC, RGPIN-2017-06743), J.Y. acknowledges support from the National Science Foundation of China (NSFC, 11871290). S.S. is supported by a scholarship from the China Scholarship Council.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078889.121.
MEET THE FIRST AUTHOR
Saisai Sun.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Saisai Sun is the first author of this paper, “RNALigands: a database and web server for RNA–ligand interactions.” Saisai is currently a faculty member in the School of Computer Science and Technology at Xidian University, China. Saisai obtained her PhD in Bioinformatics in 2021 under the supervision of Dr. Jianyi Yang at the School of Mathematical Sciences, Nankai University, China, focusing on the prediction of RNA tertiary structural features. In 2019, Saisai was a visiting student in the laboratory of Dr. Zhaolei Zhang at the Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, working on RNA and small molecule interactions.
What are the major results described in your paper and how do they impact this branch of the field?
We built the first RNA motif and small molecule interaction database and proposed a searching pipeline for RNA ligands based on motif alignment. The database can be a resource for RNA drug discovery research. Furthermore, the web server can provide hit ligands of any query (RNA only) with sequence information, which can provide a reference for RNA drug screening.
What led you to study RNA or this aspect of RNA science?
In the beginning, I was attracted by the mystery of the RNA folding mechanism. Then, after I got to know a little about RNA secondary and tertiary structures, the complexity of the RNA structure kept me curious, despite the fact that it only contains four types of nucleotides.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
Professor Jianyi Yang has influenced me the most. He works hard and has a strict attitude to research. He taught me to be patient and think more when results are not as expected. In addition, Professor Zhaolei Zhang influenced my research perception. He made me realize that doing research should be one thing significant to our lives.
What are your subsequent near- or long-term career plans?
I will keep exploring RNA structures and studying the molecular interactions important for RNA structures.
REFERENCES
- Adinolfi M, Pietrosanto M, Parca L, Ausiello G, Ferre F, Helmer-Citterich M. 2019. Discovering sequence and structure landscapes in RNA interaction motifs. Nucleic Acids Res 47: 4958–4969. 10.1093/nar/gkz250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28: 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra S, Xie J, Frank AT. 2020. RNAPosers: machine learning classifiers for ribonucleic acid-ligand poses. J Phys Chem B 124: 4436–4445. 10.1021/acs.jpcb.0c02322 [DOI] [PubMed] [Google Scholar]
- Costales MG, Haga CL, Velagapudi SP, Childs-Disney JL, Phinney DG, Disney MD. 2017. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J Am Chem Soc 139: 3446–3455. 10.1021/jacs.6b11273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costales MG, Hoch DG, Abegg D, Childs-Disney JL, Velagapudi SP, Adibekian A, Disney MD. 2019. A designed small molecule inhibitor of a non-coding RNA sensitizes HER2 negative cancers to herceptin. J Am Chem Soc 141: 2960–2974. 10.1021/jacs.8b10558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Winkelsas AM, Velagapudi SP, Southern M, Fallahi M, Childs-Disney JL. 2016. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem Biol 11: 1720–1728. 10.1021/acschembio.6b00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. 2011. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol 3: a003533. 10.1101/cshperspect.a003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74. 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbert C, James TL. 2008. Docking to RNA via root-mean-square-deviation-driven energy minimization with flexible ligands and flexible targets. J Chem Inf Model 48: 1257–1268. 10.1021/ci8000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havgaard JH, Lyngso RB, Stormo GD, Gorodkin J. 2005. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%. Bioinformatics 21: 1815–1824. 10.1093/bioinformatics/bti279 [DOI] [PubMed] [Google Scholar]
- Heller D, Krestel R, Ohler U, Vingron M, Marsico A. 2017. ssHMM: extracting intuitive sequence-structure motifs from high-throughput RNA-binding protein data. Nucleic Acids Res 45: 11004–11018. 10.1093/nar/gkx756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt WM, Calabrese DR, Schneekloth JS. 2019. Evidence for ligandable sites in structured RNA throughout the Protein Data Bank. Bioorg Med Chem 27: 2253–2260. 10.1016/j.bmc.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Pudimat R, Busch A, Backofen R. 2006. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res 34: e117. 10.1093/nar/gkl544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. 2010. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comput Biol 6: e1000832. 10.1371/journal.pcbi.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. 2009. DOCK 6: combining techniques to model RNA–small molecule complexes. RNA 15: 1219–1230. 10.1261/rna.1563609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Disney MD. 2018. Precise small molecule degradation of a noncoding RNA identifies cellular binding sites and modulates an oncogenic phenotype. ACS Chem Biol 13: 3065–3071. 10.1021/acschembio.8b00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Chen H, Ye F, Chen Z, Yang F, Zheng Y, Cao Y, Qiao J, Yang S, Lu G. 2020. Crystal structure of SARS-CoV-2 nsp10/nsp16 2′-O-methylase and its implication on antiviral drug design. Signal Transduct Target Ther 5: 131. 10.1038/s41392-020-00241-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Childs-Disney JL, Znosko BM, Wang D, Fallahi M, Gallo SM, Disney MD. 2016. Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinformatics 17: 112. 10.1186/s12859-016-0960-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano G, Fernandez N, Martinez-Salas E. 2016. Modeling three-dimensional structural motifs of viral IRES. J Mol Biol 428: 767–776. 10.1016/j.jmb.2016.01.005 [DOI] [PubMed] [Google Scholar]
- Luo Y, Disney MD. 2014. Bottom-up design of small molecules that stimulate exon 10 skipping in mutant MAPT pre-mRNA. Chembiochem 15: 2041–2044. 10.1002/cbic.201402069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29: 4724–4735. 10.1093/nar/29.22.4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577–586. 10.1016/S0092-8674(03)00391-X [DOI] [PubMed] [Google Scholar]
- Matarlo JS, Krumpe LRH, Heinz WF, Oh D, Shenoy SR, Thomas CL, Goncharova EI, Lockett SJ, O'Keefe BR. 2019. The natural product butylcycloheptyl prodiginine binds pre-miR-21, inhibits dicer-mediated processing of pre-miR-21, and blocks cellular proliferation. Cell Chem Biol 26: 1133–1142.e4. 10.1016/j.chembiol.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maticzka D, Lange SJ, Costa F, Backofen R. 2014. GraphProt: modeling binding preferences of RNA-binding proteins. Genome Biol 15: R17. 10.1186/gb-2014-15-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei E, Ausiello G, Ferre F, Helmer-Citterich M. 2014. A novel approach to represent and compare RNA secondary structures. Nucleic Acids Res 42: 6146–6157. 10.1093/nar/gku283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111: 747–756. 10.1016/S0092-8674(02)01134-0 [DOI] [PubMed] [Google Scholar]
- Moitessier N, Westhof E, Hanessian S. 2006. Docking of aminoglycosides to hydrated and flexible RNA. J Med Chem 49: 1023–1033. 10.1021/jm0508437 [DOI] [PubMed] [Google Scholar]
- Morgan BS, Sanaba BG, Donlic A, Karloff DB, Forte JE, Zhang Y, Hargrove AE. 2019. R-BIND: an interactive database for exploring and developing RNA-targeted chemical probes. ACS Chem Biol 14: 2691–2700. 10.1021/acschembio.9b00631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley SD, Afshar M. 2004. Validation of an empirical RNA–ligand scoring function for fast flexible docking using Ribodock. J Comput Aided Mol Des 18: 189–208. 10.1023/B:JCAM.0000035199.48747.1e [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48: 443–453. 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- Oliver C, Mallet V, Gendron RS, Reinharz V, Hamilton WL, Moitessier N, Waldispühl J. 2020. Augmented base pairing networks encode RNA–small molecule binding preferences. Nucleic Acids Res 48: 7690–7699. 10.1093/nar/gkaa583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Stefani M, Pesole G. 2004. RNAProfile: an algorithm for finding conserved secondary structure motifs in unaligned RNA sequences. Nucleic Acids Res 32: 3258–3269. 10.1093/nar/gkh650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer P, Gohlke H. 2007. DrugScoreRNA–knowledge-based scoring function to predict RNA–ligand interactions. J Chem Inf Model 47: 1868–1876. 10.1021/ci700134p [DOI] [PubMed] [Google Scholar]
- Philips A, Milanowska K, Lach G, Bujnicki JM. 2013. LigandRNA: computational predictor of RNA–ligand interactions. RNA 19: 1605–1616. 10.1261/rna.039834.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrosanto M, Adinolfi M, Casula R, Ausiello G, Ferre F, Helmer-Citterich M. 2018. BEAM web server: a tool for structural RNA motif discovery. Bioinformatics 34: 1058–1060. 10.1093/bioinformatics/btx704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carmona S, Alvarez-Garcia D, Foloppe N, Garmendia-Doval AB, Juhos S, Schmidtke P, Barril X, Hubbard RE, Morley SD. 2014. rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput Biol 10: e1003571. 10.1371/journal.pcbi.1003571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuszynska I, Bujnicki JM. 2011. DARS-RNP and QUASI-RNP: new statistical potentials for protein–RNA docking. BMC Bioinformatics 12: 348. 10.1186/1471-2105-12-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR, Phinney DG, Disney MD. 2016. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci 113: 5898–5903. 10.1073/pnas.1523975113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Costales MG, Vummidi BR, Nakai Y, Angelbello AJ, Tran T, Haniff HS, Matsumoto Y, Wang Z-F, Chatterjee AK, et al. 2018. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem Biol 25: 1086–1094.e1087. 10.1016/j.chembiol.2018.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DF, Savvi S, Mizrahi V, Dawes SS. 2007. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J Bacteriol 189: 3655–3659. 10.1128/JB.00040-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner KD, Hajdin CE, Weeks KM. 2018. Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov 17: 547–558. 10.1038/nrd.2018.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707. 10.1038/nsb967 [DOI] [PubMed] [Google Scholar]
- Yan Z, Wang J. 2017. SPA-LN: a scoring function of ligand–nucleic acid interactions via optimizing both specificity and affinity. Nucleic Acids Res 45: e110. 10.1093/nar/gkx255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. 2003. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res 31: 3450–3460. 10.1093/nar/gkg529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Roy A, Zhang Y. 2013. BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res 41: D1096–D1103. 10.1093/nar/gks966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo WL. 2006. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22: 445–452. 10.1093/bioinformatics/btk008 [DOI] [PubMed] [Google Scholar]
- Zhang P, Park HJ, Zhang J, Junn E, Andrews RJ, Velagapudi SP, Abegg D, Vishnu K, Costales MG, Childs-Disney JL, et al. 2020. Translation of the intrinsically disordered protein α-synuclein is inhibited by a small molecule targeting its structured mRNA. Proc Natl Acad Sci 117: 1457–1467. 10.1073/pnas.1905057117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Tang H, Zhang S. 2010. RNAMotifScan: automatic identification of RNA structural motifs using secondary structural alignment. Nucleic Acids Res 38: e176. 10.1093/nar/gkq672 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.