Abstract
Background and Objectives
CDKL5 deficiency disorder (CDD) is a neurodevelopmental encephalopathy characterized by early-onset epilepsy and impaired psychomotor development. Variations in the X-linked CDKL5 gene coding for a kinase cause CDD. Molecular genetics has proved that almost all pathogenic missense substitutions localize in the N-terminal catalytic domain, therefore underlining the importance for brain development and functioning of the kinase activity. CDKL5 also features a long C-terminal domain that acts as negative regulator of the enzymatic activity and modulates its subcellular distribution. CDD is generally attributed to loss-of-function variations, whereas the clinical consequences of increased CDKL5 activity remain uncertain. We have identified a female patient characterized by mild epilepsy and neurologic symptoms, harboring a novel c.2873C>G nucleotide substitution, leading to the missense variant p.(Thr958Arg). To increase our comprehension of genetic variants in CDKL5-associated neurologic disorders, we have characterized the molecular consequences of the identified substitution.
Methods
MRI and video EEG telemetry were used to describe brain activity and capture seizure. The Bayley III test was used to evaluate the patient development. Reverse transcriptase PCR was used to analyze whether the identified nucleotide variant affects messenger RNA stability and/or splicing. The X chromosome inactivation pattern was analyzed determining the DNA methylation status of the androgen receptor (AR) gene and by sequencing of expressed alleles. Western blotting was used to investigate whether the novel Thr958Arg substitution affects the stability and/or enzymatic activity of CDKL5. Immunofluorescence was used to define whether CDKL5 subcellular distribution is affected by the Thr958Arg substitution.
Results
Our data suggested that the proband tends toward a skewed X chromosome inactivation pattern in favor of the novel variant. The molecular investigation revealed that the p.(Thr958Arg) substitution leads to a significant increase in the autophosphorylation of both the TEY motif and residue Tyr171 of CDKL5, as well as in the phosphorylation of the target protein MAP1S, indicating an hyperactivation of CDKL5. This occurs without evidently affecting the kinase subcellular distribution.
Discussion
Our data provide a strong indication that the c.2873C>G nucleotide substitution represents an hypermorphic pathogenic variation of CDKL5, therefore highlighting the importance of a tight control of CDKL5 activity in the brain.
CDKL5 variations are responsible for a large spectrum of neuropsychiatric disorders among which CDKL5 deficiency disorder (CDD) (OMIM 300203) represents the most common one. CDD is a severe X-linked early infantile epileptic encephalopathy, representing one of the most common genetic causes of epilepsy in early childhood associated with a full host of comorbidities.1 The epidemiology of CDD is not well documented, but the estimated incidence is 1 in 40,000–60,000 live births.2,3 Epilepsy occurs within the first 3 months of life (median age at onset at 6 weeks) in over 90% of the patients, although age at seizure onset may vary from 1 to 78 weeks of life.4 Epilepsy in CDD is characterized by different seizure types, such as absences and tonic and myoclonic seizures, which are invariably refractory to antiepileptic medications.5 With time, global psychomotor delay, profound hypotonia, and stereotypical behaviors ensue, often accompanied with diverse comorbidities,4,6 namely dysregulation in breathing (hyperventilation and breath holding spells), dysautonomia, gastrointestinal problems, and cortical visual impairment.7,8
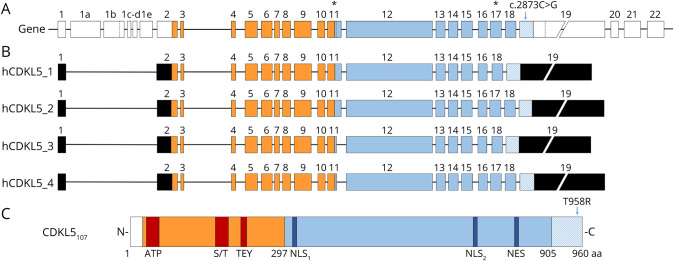
CDD is caused by variations in the X-linked CDKL5 gene, encoding a serine/threonine kinase expressed in multiple tissues, including the brain.9 CDKL5 gene structure and the exon composition of the coding isoforms have been exhaustively characterized in the mouse, rat, and human.10,11 Human CDKL5 (hCDKL5) gene contains 27 distinct exons that through alternative splicing generate 5 different transcriptional isoforms (Figure 1, A and B). The first 4 isoforms are widely expressed in various tissues including the brain, whereas hCDKL5_5 is expressed in the adult testes and fetal brain.10 hCDKL5_1 encodes a protein of 960 amino acids (aa) (CDKL5107) and represents the most abundant isoform in the brain, whereas transcripts hCDKL5_2–4 account for less than 15% of brain CDKL5 messenger RNAs (mRNAs). CDKL5 expression is developmentally regulated with low levels in the human fetal brain and high levels in the adult brain.10 All CDKL5 isoforms have a N-terminal catalytic domain, containing an ATP-binding region, a serine/threonine kinase active site, and a Thr-Xaa-Tyr (TEY) motif, whose phosphorylation plays an active role; a long C-terminal domain plays a crucial role in regulating the catalytic activity, the interaction with interactive partners, and subcellular distribution of CDKL5 protein9,12 (Figure 1C).
Figure 1. hCDKL5 Gene, Transcript Isoforms, and Protein Containing the c.2873C>G Substitution.
(A) Schematic representation of the human CDKL5 gene displaying the position of c.2873C>G substitution. Coding exons are indicated by colored boxes, whereas noncoding exons are in white. The c.2873C>G variant is contained in the coding portion (striped) of exon 19. Dotted lines within exons 1b, 11, and 19 indicate alternative splicing sites. Asterisks above exon number highlight where different mRNA isoforms might diverge. Exons forming the catalytic domain are represented in orange, whereas the C-terminus is depicted in light blue. (B) Diagrams of the 4 transcript isoforms mainly expressed in the human brain10 (hCDKL5_1 to hCDKL5_4). hCDKL5_1 and hCDKL5_3 lack the portion codified by exon 17, which is instead present in hCDKL5_2 and hCDKL5_4. All these isoforms present a long 3′-UTR codified by exon 19. UTR regions are depicted in black. (C) Schematic representation of CDKL5 isoform_1 (107 kDa) displaying the position of the p.(Thr958Arg) variant. The catalytic domain is shown in orange, with its functional domains highlighted in red (ATP-binding site (aa 19–43); S/T: Ser/Thr kinase active site (aa 131–143); TEY motif: Thr/Glu/Tyr motif (aa 169–171). The C-terminus domain is depicted in light blue, with the NLSs (aa 312–315 and aa 784–789) and the NES (aa 836–845) shown in blue. Thr958Arg variant is indicated in the last portion of the C-terminal domain. aa = amino acid; NES = nuclear export signal; NLS = nuclear localization signal.
Numerous pathogenic variants have been reported so far, including missense, nonsense, and frameshift variations,12,13 mainly in females. CDD is caused by CDKL5 haploinsufficiency in females, whereas CDKL5 variants in males are scarcer, often linked with a more severe phenotype unless somatic mosaicism has occurred.14–17 Missense substitutions are mainly distributed in the catalytic domain, highlighting the importance of CDKL5 activity for brain development and function; conversely, nonsense and frameshift variations are scattered across the gene.9 As the quest to correlate genotypes and phenotypes continues, some reports suggest that C-terminal vaiants cause milder phenotypes, whereas others imperceptibly report that no correlation exists between the nature of the variation and the clinical severity. Yet, most of the pathogenic variants are loss-of-function variations, leading to the lack or reduction of functional CDKL5 protein. In contrast, the clinical consequences of increased dosage of CDKL5 remain poorly understood. Few studies reported neurologic features in patients with CDKL5 duplications. However, the involved large-sized duplications made it difficult to understand to what extent CDKL5 contributes to the observed phenotypes.18–21 By describing the phenotypes of 7 patients with a CDKL5 duplication smaller than 1 Mb, it has been established that increased dosage of CDKL5 affects neurodevelopment.17
We report the identification of a novel c.2873C>G nucleotide substitution in a female patient affected by a mild phenotype with regard to both epilepsy and psychomotor development. The alteration leads to the p.(Thr958Arg) missense variant in the very last C-terminus of CDKL5. We demonstrate that this missense variant leads to a hypermorphic kinase, thus providing evidence of an activating CDKL5 missense substitution and reinforcing the importance of a tight control of CDKL5 activity in the brain.
Methods
Antibodies
Used primary antibodies were used: mouse monoclonal anti-GFP (1814460; Roche Diagnostics, Basel, Switzerland); polyclonal serum (rabbit) recognizing the mitogen-activated protein kinase (MAPK) phosphorylated TEY motif (V8031; Promega, Madison, WI)13; sheep polyclonal anti-phosphoTyr171 CDKL522 and sheep polyclonal anti-phosphoSer900 MAP1S22 (both obtained from MRC-PPU Reagents, University of Dundee, Dundee, United Kingdom); monoclonal anti-CDKL5 (mouse; sc-376314; Santa Cruz Biotechnologies, Dallas, TX); and polyclonal anti-RFP (rabbit; PM005; MBL International, Woburn, MA).
Generation of the Mutated CDKL5 Expression Vectors
The pGFP-CDKL5-Thr958Arg vector was generated by inserting the human variation into the pEGFPC1-hCDKL5107 vector, expressing the hCDKL5_1 isoform.10,23 Site-directed mutagenesis was performed using the Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA) using the following primers: forward 5′-TTAAAAGAGAGAGCCTTGTAAATG-3; reverse 5′-ATCATTCAGATTTGGCATTC-3′. To obtain the pCMV-FLAG2B-hCDKL5-Thr958Arg vector, a fragment containing the Thr958Arg variant was excised using EcoRI from pGFP-CDKL5-Thr958Arg and subcloned into an EcoRI digested pCMV-FLAG2B-hCDKL5 vector.23
The kinase dead pCMV-FLAG2B-Lys42ArgCDKL5-Thr958Arg was obtained through site-directed mutagenesis of the pCMV-FLAG2B-hCDKL5-Thr958Arg vector using the following primers: forward 5′-GTGGCGATCAGGAAATTCAAG-3′; reverse 5′-AATTTCATGTGTTTCCTTGTG-3′. All PCR-generated constructs were verified by sequencing.
RNA Isolation and cDNA Synthesis
Total RNA was extracted from human whole blood collected into PAXgene Blood RNA tubes and purified using the PAXgene Blood RNA Kit (Qiagen, Hilden, Germany). RNA concentration was spectroscopically quantified and integrity assessed on agarose gel. Complementary DNA (cDNA) for quantitative reverse transcriptase PCR (RT-PCR) was obtained by reverse transcribing 1 μg of RNA (Thermo Fisher, Waltham, MA).
Analysis of Splicing
The presence of putative cryptic splice sites was evaluated by exploiting the Human Splicing Finder tool (umd.be/HSF/). Splicing was evaluated by standard PCR with the following primers: forward 5′-ATGTGTCCTCTGTGACCAGGAG-3′; reverse 5′-CCACTGGCTTGTCTGTCCAC-3′. PCR conditions were: 94°C for 2 minutes for the initial denaturation, 94°C for 30 seconds, 56°C for 30 seconds, 72°C for 20 seconds for 40 cycles, and 72°C for 2 minutes for the final extension. PCR products were resolved on 3% agarose gel.
XCI Analysis
X chromosome inactivation (XCI) was tested through 2 different approaches that led to comparable results. Canonical XCI was performed analyzing the methylation status of the androgen receptor (AR) locus. Briefly, whole blood was collected into EDTA blood tubes, and the genomic DNA was isolated with the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD). One hundred fifty nanograms of genomic DNA was digested for 16 hours with HpaII, and one-tenth of the resulting digestion, heat inactivated at 80°C for 20 minutes, was used for PCR amplification of the CAG-repeat-containing AR locus. The resulting products were run on an ABI 373A automated sequencer and analyzed by GeneScan software (Applied Biosystems, Waltham, MA). Comparison of the peak areas and analysis of the CAG-repeat length of the 2 AR alleles allowed the determination of the X-inactivation as well as allele origin, respectively.
Furthermore, by RT-PCR, we amplified a CDKL5 fragment of 596 bp spanning the substitution site. The following primers were used: forward 5′-AGCAGCAGACCAAAGGAGTGGC-3′ and reverse 5′-TCAACGGCCCCAACACATGCAA-3′. PCR products were cloned into the pGEM-T Easy Vector (Promega). Insert was PCR amplified and gel extracted from 36 independent colonies using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA) and sequenced with a canonical T7 primer. The ratio of colonies expressing the wild-type (WT) vs variant allele reflected the inactivation status of the CDKL5 locus.
Quantitative RT-PCR
Quantitative RT-PCR was performed using SYBR Selected Master Mix (Applied Biosystem) and the following CDKL5 primers: forward 5ʹ-GGGTTGTAGGTGAAGGAGCC-3ʹ and reverse 5ʹ-CCTCCGACGAAATGCTTCCT-3ʹ.13 Each sample was analyzed in triplicate; GAPDH served as internal standard (forward primer 5′-CCACATCGCTCAGACACCAT-3′; reverse primer 5′-CCAGGCGCCCAATACG-3′) as internal standard. Fold change in gene expression was calculated using the 2−ΔΔCt method.13
Cell Cultures and Transfection
HEK293T and HeLa cells were maintained as described in reference 13. HEK293T were seeded in 12-well plates for Western blotting, and HeLa (50,000 cells/well) were seeded on glass coverslips in 24-well plates for immunofluorescence experiments. Seventy percent to 80% confluent cells were transfected with 600 ng of plasmid DNA for HEK293T and 350 ng for HeLa using Lipofectamine 3000 (Life Technologies, Carlsbad, CA). Transfection efficiency was estimated by cotransfecting a pcDNA3-mCherry-XhoI-XbaI vector kindly donated by Professor Giacomo Consalez (San Raffaele Scientific Institute, Milan, Italy).
Western Blotting
Transfected HEK293T cells were lysed in sample buffer (120 µL) and sonicated at 30% amplitude for 10 seconds. Proteins were resolved on a 10% SDS-PAGE before blotting on nitrocellulose membrane using the TransBlot SD apparatus from Bio-Rad (Hercules, CA). Filters were blocked for 1 hour in 5% nonfat dry milk in TBS-T (Tris-buffered saline, 0.1% Tween-20), followed by an overnight incubation (4°C) with the following antibodies: anti-active MAPK (1:2,000 in 0.1% BSA in TBS-T),13 anti-phosphoTyr171 CDKL5 (1:1,000 in 5% milk in TBS-T),22 anti-CDKL5 (1:1,000 in 5% milk in TBS-T),13 anti-phosphoSer900 MAP1S (1:1,000 in 5% milk in TBS-T),22 and anti-RFP (1:1,000 in 5% milk in TBS-T). Total CDKL5 was detected by stripping the phospho-TEY signal using the StripAblot solution (Euroclone; 15 minutes at room temperature) and reprobing with anti-CDKL5 antibody. Immunocomplexes were visualized as described in reference 13.
The mean value derived from the control group was set at 100% and data presented as percentage of control value. As an index of CDKL5 activity, both the signal intensity of phospho-TEY or -Tyr171 divided by the corresponding signal of CDKL5 and the phosphorylation level of its target MAP1S Ser900 (reference 22) normalized to total protein content (visualized by TGX stain-free technology; Bio-Rad) were measured.
Immunofluorescence
Immunofluorescence on transfected HeLa cells was performed as described in reference 13 with the exception of anti-CDKL5 primary antibody dilution (1:100). Confocal fluorescence microscope (excitation wavelength of 488 nm with a 63× objective; Nikon, Tokyo, Japan) was used to acquire images from at least 3 coverslips for experimental group. Fiji software was used to evaluate CDKL5 subcellular distribution.
Statistical Analysis
GraphPad software 7.0 was used to statistically analyze data that are expressed as mean ± SE. Normal distribution of data was verified by the D'Agostino-Pearson normality test, and the significance of results was assessed by the Student t test or Mann-Whitney test, considering the p value <0.05 as significant.
Patient Consent
Standard protocol approvals, registration and patient consents were obtained from the parents of the patients.
Data Availability
Data will be made available on request to qualified investigators.
Results
One 6-week-old White Caucasian British female patient presented with clusters of focal onset seizures with secondary generalization, each cluster lasting several seconds. Episodes of excessive startle were also noted, and glabellar tap sign was negative. Apart from an intermittent limb hypertonia, neurologic and developmental examinations were normal. Prenatal and perinatal history was uneventful, parents are nonconsanguineous, and there is no family history of epilepsy or intellectual disability. The child responded partially to IV levetiracetam, and subsequently, carbamazepine, biotin, and pyridoxine were added due to ongoing seizures. Brain MRI with dedicated sequences for epilepsy (volume fluid-attenuated inversion recovery and volume T1) was normal at 8 weeks, whereas routine interictal EEG showed normal organization, with faster activity and occasional sharp waves over central regions. Video EEG telemetry study captured 4 episodes of focal-onset seizures. Bifrontal and central discharges were noted during each episode. Abdominal ultrasound and ophthalmologic evaluation were normal. Further blood investigations with copper, ceruloplasmin, serum vitamin B12, folate, and ammonia were all normal. Complete neurometabolic workup in serum and CSF, including amino acids, organic acids, VLCFA, carnitine, acylcarnitine, and neurotransmitters, was also negative. Evaluation of visual evoked potentials and electroretinography was normal.
Exome sequencing was used to investigate 569 genes (EPIDASD569 Panel) associated with epilepsy, intellectual disability, or autism spectrum disorder; rare or low-frequency variants were evaluated according to the ACMG guidelines, also including literature and database searches such as the Human Genome Mutation Database and the Genome Aggregation Database (gnomAD, release 2.0.1). A CDKL5 variant in heterozygous state was identified. The genetic finding was within the alternative CDKL5 transcript NM_001323289.2 described as a c.2873C>G missense variation, leading in the hCDKL5_1 isoform10 to the missense substitution p.(Thr958Arg).
The prediction software MutationTaster and PolyPhen-2 Polymorphism Phenotyping v2 indicated a damaging effect of the variant, whereas Sorting Intolerant From Tolerant (SIFT) was unable to provide a prediction on the current transcript. Based on the bioinformatic evaluation, the variant was classified into class IV: likely pathogenic. Parental analysis (Sanger sequencing) revealed a de novo appearance of the variant.
The child had 6 monthly developmental assessments using the Bayley III test, which revealed cognitive, language, and motor scores close to the normative mean. By age 4 months, she was able to hold her head against gravity when in the prone position, was fixing on and following objects with her eyes, responding to sounds, and even beginning to grab at toys and interact socially. By 5 months, she was sitting unsupported; at 7, she was crawling; and at 9, she was able to pick up toys and transfer them from hand to hand and to pick up very small objects between the thumb and first finger. Babbling consonants and vowels were present. By age 18 months, the child was walking and running and could throw and kick a ball. At 21 months, she had mild delay on fine and gross motor areas and in some behavioral scores (Home Living, Self-care, and Social interaction). Since 24 months, she scored normally in all areas of development, i.e., she could walk up stairs and was able to say 10–20 words and understand more complex phrases. Her latest assessment was at the chronological age of 41 months. At that time, the cognitive scale was at a developmental age of 42 months. The language scale, expressive and receptive, was as expected for her age of 41–42 months. She stammered a little, but this was not consistent. She is being brought up trilingual. Concerning fine motor skills, she was not very keen on activities with hands and was functioning at the level of a 35 months. Gross motor skills, despite her flexibility, hypermobility, and hypotonia, were at 39–40 months. The score for emotional development was well above her own age of 41 months at home. At school, her scores were consistently low, with an age equivalent of 15 months. Some subtle sensory processing issues (visual, tactile, and auditory) were described at school but not at home (Table 1).
Table 1.
Developmental Scores of Our Patient Through Time
Seizures persist, once per week during light sleep with eye opening, versive eye movements to either side with blinking, clenching fist, flexion of the arms, and symmetrical rhythmic jerking movements of the hands lasting up to 45 seconds. She may make a heaving sound or grind her teeth or cycle the limbs. During 10%–25% of seizures, she alerts from sleep and cries briefly in distress. She remains on monotherapy with levetiracetam. She is proportionately growing (weight 78th percentile, height 77th percentile, orbifrontal cortex (OFC) 69th percentile, and body mass index 67th percentile).
Her latest (at age 3 years) interictal video telemetry EEG shows quite frequent interictal epileptiform discharges mainly over the left hemisphere and with a central or fronto-central field (3 years). The previous telemetry, performed at 16 months, also showed moderately frequent interictal epileptiform discharges over both hemispheres, involving the fronto-central or centro-parietal fields. In that earlier study, there were more right-sided or bilateral discharges than in the subsequent telemetry where they were mainly left sided. Indeed, the 2 studies are quite similar in terms of morphology of the spike and wave discharges, but in the most recent study, they predominantly involved the left hemisphere (Table 2).
Table 2.
Features Manifested by the Patient Recorded in Our Case
With respect to the hCDKL5_1 isoform described by reference 10, the identified nucleotide variation, that leads to the de novo p.(Thr958Arg) missense substitution, occurs just 3 residues before the canonical translational termination codon and has never been reported in the gnomAD v3 database nor in almost 140,000 exomes containing CDKL5. This substitution modifies the CDKL5 C-terminus of all brain isoforms (Figure 1B), but not the tail of the testis-specific hCDKL5_5 isoform that diverges before the mutant residue.
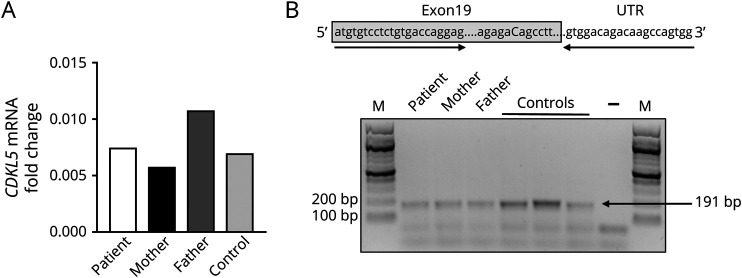
Because missense variations are generally found in the kinase domain of CDKL5,13 we found it relevant to investigate the molecular consequences of this substitution. Initially, we tested whether the nucleotide variant might affect mRNA stability and/or splicing. Quantitative RT-PCR, performed on blood RNA extracted from the proband, her nonconsanguineous parents, and an unrelated control, did not reveal any significant difference in CDKL5 mRNA abundance, suggesting that the identified transversion is not affecting mRNA stability (Figure 2A). Bioinformatic prediction of splice sites revealed that the c.2873C>G nucleotide variant potentially creates a new 3′ acceptor splice site (predicted score 70/100 based on HSF tool at umd.be/HSF/) that, if used, would lead to a transcript lacking most of the CDKL5 exon 19. However, inspection of splicing pattern by RT-PCR did not reveal any effect on exon 19 alternative splicing (Figure 2B).
Figure 2. c.2873C>G Substitution Does Not Affect mRNA Stability and Splicing.
(A) Quantitative RT-PCR reported no alteration in the expression levels of CDKL5 mRNA in the blood of the patient compared with that of her unaffected mother and father and an unrelated control. The graph shows the fold change of CDKL5 mRNA, calculated using the 2−ΔΔCt (B) CDKL5 splicing patterns in leukocytes. Primers (indicated by arrows) amplifying the missense variant (in capital letter) are used to evaluate the splicing pattern including exon 19. Gray box indicates the CDKL5 coding sequence. Amplified products were separated on 3% agarose gel. M = 100-bp molecular-weight marker. mRNA = messenger RNA; RT-PCR = reverse transcriptase PCR.
Because a skewed XCI pattern might contribute to the clinical manifestations of the patient, by determining the DNA methylation status of the AR gene, we deduced a 30:70 ratio of expression of the WT allele vs the variated one. However, it has been proposed that this assay does not always properly reflect XCI24; we thus exploited sequencing of the expressed alleles (see Methods) to confirm the result. In this case, we measured a 25:75 ratio of expression in favor of the novel allele. Because XCI ratios of 50:50 to 79:21 are generally considered indicative of random XCI, our data suggest a trend of the patient's pattern of XCI toward a skewed one. Then, we analyzed whether the C-terminal p.(Thr958Arg) missense variant affects CDKL5 activity and/or subcellular distribution.
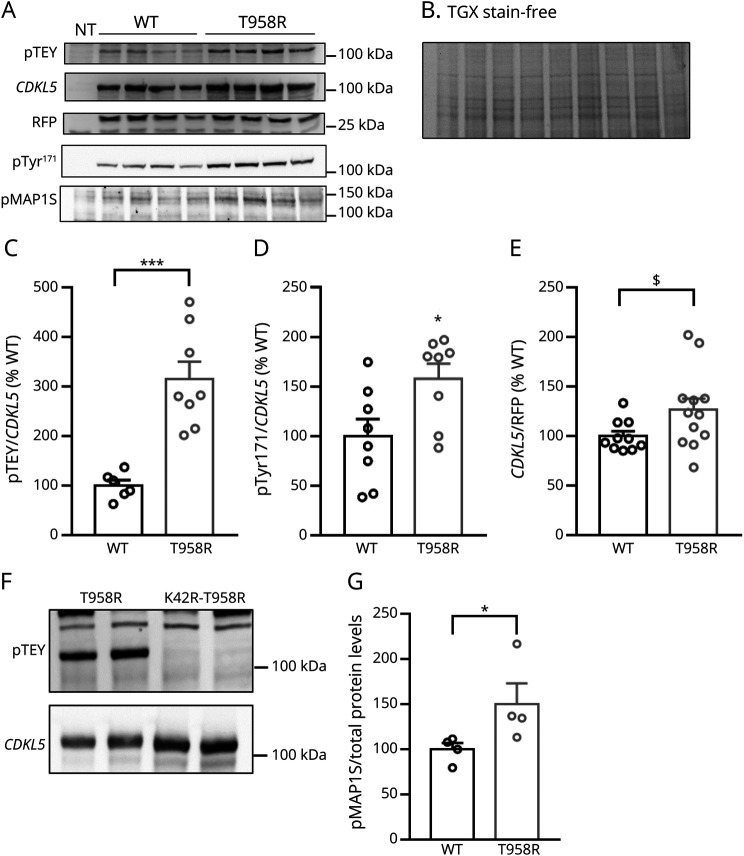
Previously, we demonstrated that CDKL5 activity can be tested measuring its TEY motif phosphorylation.25 Thus, we expressed the CDKL5-Thr958Arg derivative in human cells and evaluated the phosphorylation level of its TEY motif with respect to the WT protein (Figure 3, A and C). Quantitative analyses demonstrated that the variant leads to a significant hyperactivation of CDKL5 kinase activity (Figure 3C). It has been recently proposed that CDKL5 Tyr171 represents a major site of CDKL5 autophosphorylation.22 We confirmed the increased activity of the CDKL5-Thr958Arg variant using the phospho-specific Tyr171 antibody (Figure 3, A and D). Furthermore, normalization of the CDKL5 signal to the cotransfected RFP showed a tendency toward increased protein levels of the mutant CDKL5, although it did not reach statistical significance (Figure 3, A and E). By introducing the missense variant into the kinase dead CDKL5 Lys42Arg mutant, we also proved that the observed hyperactivation was not caused by an aberrant interaction of the mutated kinase with an unknown kinase25 (Figure 3F). Altogether, these data suggested that the CDKL5-Thr958Arg variant might represent a gain-of-function substitution. To confirm this hypothesis, we compared the phosphorylation level of MAP1 Ser900, a recently identified substrate of CDKL5.22 Importantly, Western blotting analyses indicated a significant increase in phosphorylation of the CDKL5 substrate in cells transfected with the mutated construct relative to the WT one (Figure 3, A and G).
Figure 3. Thr958Arg Substitution Leads to a Hyperfunctional CDKL5 Kinase.
(A) Representative Western blotting of phospho-TEY, CDKL5, RFP, phosphoTyr171 CDKL5, and phosphoSer900 MAP1S in HEK293T cells transfected with the WT hCDKL5_1 cDNA or its Thr958Arg variant. NT: untransfected control cells. (B) Total protein content, visualized by a TGX stain-free technology (Bio-Rad). (C–E) Histograms show the mean ± SE of the percentage of TEY phosphorylation of WT and Thr958Arg-mutant CDKL5, calculated as ratio of phospho-TEY or phosphoTyr171 to total CDKL5 normalized to WT CDKL5 (C, D), and CDKL5 expression levels relative to RFP normalized to WT CDKL5 (E). *p < 0.05, ***p < 0.001 by the Mann-Whitney test and $p = 0.06 by the Student t test. (F) No phosphorylation of the TEY motif of Thr958Arg CDKL5 is detected when the missense substitution is inserted into a cDNA coding for a kinase dead CDKL5 (Lys42Arg-Thr958Arg CDKL5). Representative Western blot of phospho-TEY and CDKL5 in HEK293T cells transfected with a plasmid expressing the Thr958Arg or Lys42Arg-Thr958Arg hCDKL5_1 variant. (G) Histogram shows the mean ± SE of the percentage of phosphoSer900 MAP1S expression levels normalized to total protein content, visualized by TGX stain-free technology. *p < 0.05 by the Mann-Whitney test. cDNA = complementary DNA; WT = wild type.
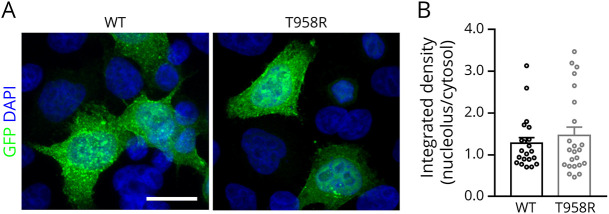
Eventually, we verified whether the Thr958Arg missense variant of CDKL5 affects the kinase subcellular distribution. For this purpose, as previously performed, we transfected WT GFP-CDKL5 and its Thr958Arg derivative into HeLa cells and assessed their relative distribution between nucleus and cytoplasm.13,25 As shown in Figure 4, A and B, the WT and Thr958Arg CDKL5 proteins manifested a coinciding subcellular distribution.
Figure 4. Thr958Arg Substitution Does Not Affect CDKL5 Subcellular Distribution.
(A) Representative immunofluorescence of CDKL5 demonstrating its subcellular distribution in HeLa cells transfected with a plasmid expressing the WT GFP-hCDKL5 or its Thr958Arg variant (green). Nuclei are labeled by DAPI staining (blue). Scale bar 20 µm. (B) Histograms depict the nuclear/cytosol distribution of WT and Thr958Arg-mutant CDKL5, calculated by measuring the integrated density of GFP fluorescence signal in the 2 cellular compartments and presented as mean ± SE. DAPI = 4′,6-diamidino-2-phenylindole; WT = wild type.
Discussion
CDKL5 variations are generally investigated in patients having drug-resistant epileptic seizures and severe psychomotor and intellectual delay. Indeed, the clinical features of CDD include epilepsy that typically develops within the first 3 months of life, hypotonia, hand stereotypes, and severe psychomotor retardation. Gastrointestinal problems, bruxism, breathing, and sleep disorders are less common features that should also be considered in the diagnosis.26,27 However, some clinical variability in the degree of impairment and disability is generally recognized, with females generally less severely affected than males. Furthermore, although rare, the absence of epilepsy has also been reported.4,28,29 The nature of the CDKL5 alteration, its position in the gene, and the pattern of XCI in female patients seem to affect clinical severity of CDD. Epigenetic and environmental factors might also be involved.27 It has been suggested that genetic variations beyond aa 938 in human CDKL5 protein may have minor or no significance.30 A more recent study suggested that compared with patients with no functional CDKL5 protein, those with truncating variants after aa 781 show milder symptoms.4 Analysis of CDKL5 functions and the involved molecular domains might provide an explanation for these observations.
CDKL5 encodes for a serine-threonine kinase widely expressed, but with the highest expression level in the brain, particularly in neurons.12,31 Although the exact roles of the kinase remain to be identified, the protein is certainly involved in neuronal migration, axon outgrowth, dendritic morphogenesis, and synaptogenesis.32 Its functions appear regulated by the kinase subcellular localization, its autophosphorylation, synthesis, and degradation. CDKL5 protein is distributed in both cytoplasmic and nuclear compartments, and its nuclear export and subsequent degradation in neurons are modulated by specific stimuli, such as glutamate treatment.9 Importantly, cytoplasmic CDKL5 affects neuronal arborization, whereas the nuclear protein might affect gene expression including splicing.33 At the molecular level, CDKL5 contains an ATP-binding domain followed by a catalytic portion and a TEY motif, which together constitute the first 297 residues. The autophosphorylation of the TEY motif on the conserved tyrosine 171 appears to be associated with an active conformation of the kinase. The remaining 663 aa, that are expressed by all main brain CDKL5 isoforms, define the C-terminal domain that acts as negative regulator of the kinase activity and modulator of its subcellular localization. The most C-terminal portion of the kinase is required to localize it into the cytoplasm; indeed, the pathogenic Leu879X and Arg781X CDKL5 truncations have been found exclusively localized into the nucleus.12 This result, together with the aforementioned evidence that truncating variations downstream of aa 781 are less severe, highlights the importance of CDKL5 cytoplasmic localization. Similarly, the importance of the kinase activity is underlined by the distribution of pathogenic missense substitutions that almost exclusively localize in the catalytic domain. CDKL5 variants are generally considered loss of function; however, few studies reporting pathogenic duplications of X chromosome regions including CDKL517 have led to debate if deregulation of the levels of CDKL5 activity can lead to neurologic symptoms.
In this work, we have described a new case of female patient harboring a hypermorphic CDKL5 missense variant that resides in a fully conserved domain of 15 residues among mammals, reptiles, and birds, a strong indication for its biological relevance. The patient is characterized by mild neurologic symptoms that do not meet common clinical characteristics of patients with CDD.2 She was referred to us when she was 6 weeks old because of focal seizures. She is now 4 years old. Although seizures persist, she presents normal cognitive and fine motor skills and little alterations in social, emotional, and adaptive behaviors. Milder defects in gross motor skills together with hypotonia and stereotypic hand movements are present. Molecular testing has identified the de novo CDKL5 c.2873C>G nucleotide substitution, which has never been described before and not identified in more than 100,000 exomes containing CDKL5; we thus considered it a potential novel pathogenic mutation. In keeping with the mild clinical symptoms, the identified genetic alteration causes the missense variant p.(Thr958Arg) occurring in the most extreme portion of the C-terminal domain, a region that is generally not taken into account for understanding CDKL5 molecular functions. Our molecular data suggest that the identified variation modifies CDKL5 protein properties, with no effect on splicing and mRNA abundance. In particular, the Thr958Arg CDKL5 variant exhibits an evident increment in the phosphorylation of both the TEY motif and Tyr171, therefore suggesting the acquirement of a more active enzymatic activity.22 Accordingly, we used endogenous MAP1S, a recognized target of CDKL5 activity, as a molecular readout of the hypermorphic functions of the mutated kinase,22 to reveal higher phosphorylation in cells expressing the mutant CDKL5 relative to the WT control. Furthermore, the mutated CDKL5 exogenously expressed in cultured cells exhibited a trend toward an increase of protein levels, which might mirror a slight increase in protein stability. No effect on the subcellular distribution of CDKL5 was observed, possibly suggesting that the most extreme portion of the protein is involved in regulating the catalytic activity.
Collectively, our data provide a strong indication that the Thr958Arg missense variant is causative of the child's symptoms. Although this hypothesis could only be confirmed by identifying additional patients with similar variants and clinical presentations, we believe that our data provide strong indication of a hypermorphic CDKL5 pathogenic variant, therefore corroborating the hypothesis that although loss-of-function variants are expected to be more severe for neurodevelopment, gain-of-function variations might also be detrimental. The demonstration that in the patient the pattern of XCI leads the variant allele to be more frequently expressed than the WT also suggests that gain-of-function variations might be particularly detrimental in the male hemizygous brain, a hypothesis that will have to be supported by additional cases.
Acknowledgment
The authors are grateful to the family of the patient for allowing them to describe the case of their daughter and for their financial support to the molecular studies. They also acknowledge the technical help provided by Eleonora Spiombi and Clarissa Butti. Finally, they thank Dr. Angela Huertas for her detailed neurodevelopmental evaluation of the patient and Dr. Valeria Ruggiero for the study of the evolutionary conservation of Thr958.
Glossary
- aa
amino acid
- AR
androgen receptor
- CDD
CDKL5 deficiency disorder
- cDNA
complementary DNA
- GFP
green fluorescent protein
- gnomAD
Genome Aggregation Database
- hCDKL5
human CDKL5
- MAPK
mitogen-activated protein kinase
- mRNA
messenger RNA
- RFP
red fluorescent protein
- RT-PCR
reverse transcriptase PCR
- SIFT
Sorting Intolerant From Tolerant
- WT
wild type
- XCI
X chromosome inactivation
Appendix. Authors

Contributor Information
Angelisa Frasca, Email: angelisa.frasca@unimi.it.
Efterpi Pavlidou, Email: efterpi.pavlidou@gmail.com.
Matteo Bizzotto, Email: matteo.bizzotto@humanitasresearch.it.
Yunan Gao, Email: y.gao13@imperial.ac.uk.
Dario Balestra, Email: blsdra@unife.it.
Mirko Pinotti, Email: pnm@unife.it.
Hans Atli Dahl, Email: atli@amplexa.com.
Nicholas D. Mazarakis, Email: n.mazarakis@imperial.ac.uk.
Maria Kinali, Email: m.kinali@imperial.ac.uk.
Study Funding
Molecular studies and the salary of M.B. were supported by the family of the patient.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosures.
References
- 1.Lindy AS, Stosser MB, Butler E, et al. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia. 2018;59(5):1062-1071. [DOI] [PubMed] [Google Scholar]
- 2.Olson HE, Demarest ST, Pestana-knight EM, et al. Cyclin-dependent kinase-like 5 deficiency disorder: clinical review. Pediatr Neurol. 2019;97:18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demarest S, Olson H, Moss A, et al. CDKL5 deficiency disorder: relationship between genotype, epilepsy, cortical visual impairment, and development. Epilepsia. 2019;60(8):1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fehr S, Wong K, Chin R, et al. Seizure variables and their relationship to genotype and functional abilities in the CDKL5 disorder. Neurology. 2016;87(21):2206-2213. [DOI] [PubMed] [Google Scholar]
- 5.Arican P, Gencpinar P, Dundar NO. A new cause of developmental and epileptic encephalopathy with continuous spike-and-wave during sleep: CDKL5 disorder. Neurocase. 2019;25(1-2):59-61. [DOI] [PubMed] [Google Scholar]
- 6.Bahi-Buisson N, Bienvenu T. CDKL5-related disorders: from clinical description to molecular genetics. Mol Syndromol. 2012;2(3-5):137-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangatt M, Wong K, Anderson B, et al. Prevalence and onset of comorbidities in the CDKL5 disorder differ from Rett syndrome. Orphanet J Rare Dis. 2016;11:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazziotti R, Lupori L, Sagona G, et al. Searching for biomarkers of CDKL5 disorder: early-visual impairment in CDKL5 mutant mice. Hum Mol Genet. 2017;26(12):2290-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilstrup-Nielsen C, Rusconi L, La Montanara P, et al. What we know and would like to know about CDKL5 and its involvement in epileptic encephalopathy. Neural Plast. 2012;2012:728267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hector RD, Dando O, Landsberger N, et al. Characterisation of CDKL5 transcript isoforms in human and mouse. PLoS One. 2016;11(6):e0157758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hector RD, Dando O, Ritakari TE, Kind PC, Bailey ME, Cobb SR. Characterisation of Cdkl5 transcript isoforms in rat. Gene. 2017;603:21-26. [DOI] [PubMed] [Google Scholar]
- 12.Rusconi L, Salvatoni L, Giudici L, et al.J CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. Biol Chem. 2008;283(44):30101-30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fazzari M, Frasca A, Bifari F, Landsberger N. Aminoglycoside drugs induce efficient read-through of CDKL5 nonsense mutation, slightly restoring its kinase activity. RNA Biol. 2019;16:1414-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mei D, Darra F, Barba C, et al. Optimizing the molecular diagnosis of CDKL5 gene-related epileptic encephalopathy in boys. Epilepsia. 2014;55(11):1748-1753. [DOI] [PubMed] [Google Scholar]
- 15.Mirzaa GM, Paciorkowski AR, Marsh ED, et al. CDKL5 and ARX mutations in males with early-onset epilepsy. Pediatr Neurol. 2013;48(5):367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neupauerová J, Štěrbová K, Vlčková M, et al. Two novel variants affecting CDKL5 transcript associated with epileptic encephalopathy. Genet Test Mol Biomarkers. 2017;21:613-618. [DOI] [PubMed] [Google Scholar]
- 17.Szafranski P, Golla S, Jin W, et al. Neurodevelopmental and neurobehavioral characteristics in males and females with CDKL5 duplications. Eur J Hum Genet. 2015;23(7):915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froyen G, Van Esch H, Bauters M, et al. Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: important role for increased gene dosage of XLMR genes. Hum Mutat. 2007;28(10):1034-1042. [DOI] [PubMed] [Google Scholar]
- 19.Tzschach A, Chen W, Erdogan F, et al. Characterization of interstitial Xp duplications in two families by tiling path array CGH. Am J Med Genet A. 2008;146A(2):197-203. [DOI] [PubMed] [Google Scholar]
- 20.Thorson L, Bryke C, Rice G, et al. Clinical and molecular characterization of overlapping interstitial Xp21-p22 duplications in two unrelated individuals. Am J Med Genet A. 2010;152A(4):904-915. [DOI] [PubMed] [Google Scholar]
- 21.Sismani C, Anastasiadou V, Kousoulidou L, et al. 9 Mb familial duplication in chromosome band Xp22.2-22.13 associated with mental retardation, hypotonia and developmental delay, scoliosis, cardiovascular problems and mild dysmorphic facial features. Eur J Med Genet. 2011;54(5):e510-e515. [DOI] [PubMed] [Google Scholar]
- 22.Muñoz IM, Morgan ME, Peltier J, et al. Phosphoproteomic screening identifies physiological substrates of the CDKL5 kinase. EMBO J. 2018;37(24):e99559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williamson SL, Giudici L, Kilstrup-Nielsen C, et al. A novel transcript of cyclin-dependent kinase-like 5 (CDKL5) has an alternative C-terminus and is the predominant transcript in brain. Hum Genet. 2012;131(2):187-200. [DOI] [PubMed] [Google Scholar]
- 24.Swierczek SI, Piterkova L, Jelinek J, et al. Methylation of AR locus does not always reflect X chromosome inactivation state. Blood. 2012;119(13):e100-e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertani I, Rusconi L, Bolognese F, et al. Functional consequences of mutations in CDKL5, an X-linked gene involved in infantile spasms and mental retardation. J Biol Chem. 2006;281(42):32048-32056. [DOI] [PubMed] [Google Scholar]
- 26.Fehr S, Wilson M, Downs J, et al. The CDKL5 disorder is an independent clinical entity associated with early-onset encephalopathy. Eur J Hum Genet. 2013;21(3):266-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakimiec M, Paprocka J, Śmigiel R. CDKL5 deficiency disorder—a complex epileptic encephalopathy. Brain Sci. 2020;10(2):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacKay CI, Bick D, Prokop JW, et al. Expanding the phenotype of the CDKL5 deficiency disorder: are seizures mandatory? Am J Med Genet. 2020;182(5):1217. [DOI] [PubMed] [Google Scholar]
- 29.Fehr S, Leonard H, Ho G, et al. There is variability in the attainment of developmental milestones in the CDKL5 disorder. J Neurodev Disord. 2015;7(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diebold B, Delépine C, Gataullina S, Delahaye A, Nectoux J, Bienvenu T. Mutations in the C-terminus of CDKL5: proceed with caution. Eur J Hum Genet. 2014;22(2):270-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin C, Franco B, Rosner MR. CDKL5/Stk9 kinase inactivation is associated with neuronal developmental disorders. Hum Mol Genet. 2005;14(24):3775-3786. [DOI] [PubMed] [Google Scholar]
- 32.Zhu YC, Xiong ZQ. Molecular and synaptic bases of CDKL5 disorder. Dev Neurobiol. 2019;79(1):8-19. [DOI] [PubMed] [Google Scholar]
- 33.Chen Q, Zhu YC, Yu J, et al. CDKL5, a protein associated with rett syndrome, regulates neuronal morphogenesis via Rac1 signaling. J Neurosci. 2010;30(38):12777-12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request to qualified investigators.