Abstract
A clinical strain of Vibrio cholerae non-O1 non-O139 isolated in France produced a new β-lactamase with a pI of 5.35. The purified enzyme, with a molecular mass of 33,000 Da, was characterized. Its kinetic constants show it to be a carbenicillin-hydrolyzing enzyme comparable to the five previously reported CARB β-lactamases and to SAR-1, another carbenicillin-hydrolyzing β-lactamase that has a pI of 4.9 and that is produced by a V. cholerae strain from Tanzania. This β-lactamase is designated CARB-6, and the gene for CARB-6 could not be transferred to Escherichia coli K-12 by conjugation. The nucleotide sequence of the structural gene was determined by direct sequencing of PCR-generated fragments from plasmid DNA with four pairs of primers covering the whole sequence of the reference CARB-3 gene. The gene encodes a 288-amino-acid protein that shares 94% homology with the CARB-1, CARB-2, and CARB-3 enzymes, 93% homology with the Proteus mirabilis N29 enzyme, and 86.5% homology with the CARB-4 enzyme. The sequence of CARB-6 differs from those of CARB-3, CARB-2, CARB-1, N29, and CARB-4 at 15, 16, 17, 19, and 37 amino acid positions, respectively. All these mutations are located in the C-terminal region of the sequence and at the surface of the molecule, according to the crystal structure of the Staphylococcus aureus PC-1 β-lactamase.
Clinical strains of Vibrio cholerae are naturally susceptible in vitro to broad-spectrum penicillins and cephalosporins. Some isolates that exhibit plasmid-mediated resistance to β-lactams that are susceptible to the effects of β-lactamase inhibitors produce TEM-1 (9, 11, 35) or SAR-1, a carbenicillin-hydrolyzing β-lactamase with an isoelectric point of 4.9 (29).
Until 1992, V. cholerae of serogroup O1 was the only known causative agent of pandemic cholera. Strains termed non-O1 had never been shown to generate epidemic cholera, although some were responsible for sporadic gastrointestinal or extraintestinal diseases, such as pyogenic infection and septicemia in susceptible hosts (24, 30). However, a non-O1 strain is causing the current cholera pandemic described in Bangladesh. The strain is V. cholerae O139, which was initially isolated in southern Asia (34).
We report here the biochemical and structural properties of a new β-lactamase produced by a clinical strain of V. cholerae non-O1, non-O139 that is resistant to β-lactams and that was responsible for the death of a cirrhotic patient after a lower-limb infection (3). The strain was isolated at Bellevue Hospital (CHU, Saint-Etienne, France).
In preliminary studies, the β-lactamase was shown to have a pI of 5.35 and to be inhibited by an anti-CARB-3 serum (3) that inactivates the five previously described enzymes (27). We report here the physical and biochemical properties of the purified enzyme. The nucleotide sequence of the gene and the deduced amino acid composition of this β-lactamase were determined and compared to those of other β-lactamases.
(This work was presented, in part, at the 16th and 17th Interdisciplinary Meetings of Anti-Infectious Chemotherapy, Paris, France, in 1996 [5] and 1997 [6], respectively.)
MATERIALS AND METHODS
Clinical strain.
The clinical strain of V. cholerae non-O1, non-O139 used in this study was resistant to aminopenicillins and carboxypenicillins and was isolated from a cirrhotic French farmer at the Bellevue Hospital (CHU, Saint-Etienne, France). This strain was responsible for a fatal septic shock as a consequence of a severe infection of the right leg that developed within 2 weeks after a stay on the Mediterranean coast (3).
β-Lactamase purification and analytical IEF.
The β-lactamase extract was prepared from 24 liters (92 g [wet weight]) of an overnight culture grown in nutrient broth (Difco) with amoxicillin (50 μg/ml). The cells were collected by centrifugation and broken by sonication at 4°C (100 W at 20 kHz) in 50 mM Tris HCl buffer (pH 7) containing 2 mM EDTA, 7 mM β-mercaptoethanol, and 10% sucrose. The sample was centrifuged at 20,000 rpm (Sorvall RC2B) for 20 min, and the supernatant containing the β-lactamase was collected. The enzyme was purified by ammonium sulfate precipitation to between 40 and 80% saturation, ion-exchange chromatography (DE 52), and gel filtration on 5% acrylamide and 4% agarose (Ultrogel AcA54; IBF-BIOSEPRA). The purified enzyme was concentrated by ultrafiltration (26). Purity was verified by sodium dodecyl sulfate (SDS)-electrophoresis (19). Analytical isoelectric focusing (IEF) was carried out in a polyacrylamide gel with an Ampholine (Pharmacia) gradient (pH 3.5 to 9.5) (20). The β-lactamase in the gel was revealed by the iodine procedure in the presence of benzylpenicillin (17). The pI of the enzyme was determined by using TEM-1 (R111) and PSE-4 (16, 23) as reference β-lactamases.
Molecular weight.
The molecular weight was determined by SDS-gel electrophoresis (19).
Substrate and inhibitor profiles of the β-lactamase.
The following antibiotics and β-lactamase inhibitors were used to determine kinetic constants: benzylpenicillin (Sarbach); amoxicillin, ticarcillin, clavulanic acid, and cloxacillin (Smith Kline Beecham); piperacillin and tazobactam (Lederlé); methicillin and oxacillin (Bristol-Myers); cephaloridine (Glaxo); sulbactam (Pfizer); cephalothin (Panpharma); and mezlocillin (Bayer).
Kinetic parameters (Km and relative Vmax) were determined and inhibitor studies were performed at pH 7 and 37°C with a pH Stat apparatus by the microacidimetric method (15). One β-lactamase unit is defined as the amount of enzyme that hydrolyzes 1 μmol of benzylpenicillin per min at pH 7 and 37°C.
Preparation of DNA for PCR.
Plasmid DNA was extracted and purified by the X-Trax procedure (Medgene Science).
PCR amplification and nucleotide sequencing.
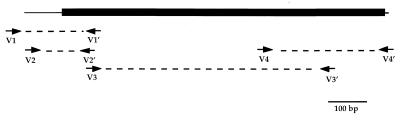
The nucleotide sequence was determined by direct sequencing of PCR-generated DNA fragments with four pairs of synthetic primers (Table 1) covering the whole reference sequence of CARB-3 gene (18). The first fragment (V2-V2′) was obtained by the nested PCR method (Fig. 1).
TABLE 1.
Oligonucleotide primers used for PCR amplification
| Primer pair | Location (5′-3′)a | Sequence (5′-3′) |
|---|---|---|
| V1 | 44–65 | TTGATGTTATGGAGCAGCAACG |
| V1′ | 266–248 | CAATTGCCTTAACGTCTTG |
| V2 | 75–88 | AGCAGGGCAGTCGC |
| V2′ | 254–236 | CGTCTTGTTCAACTTGCTG |
| V3 | 235–254 | TCAGCAAGTTGAACAAGACG |
| V3′ | 899–878 | CAACTGCTGTAATACTCCGAGC |
| V4 | 684–708 | TGAGGGATACGACAACTCCTAAGGC |
| V4′ | 1047–1027 | TGTTAGCCTTATCAGCGCGAC |
Oligonucleotide positions are given according to their location on the CARB-3 gene sequence as described elsewhere (4).
FIG. 1.
Sequencing strategy for CARB-6 β-lactamase gene. The CARB-6 gene is represented by the black box. The arrows indicate the oligonucleotide positions.
Amplification by PCR with specific primers was performed with 10 μl of the plasmid DNA preparation and 2 μl of each primer solution (100 ng/μl) in a reaction mixture with a total volume of 100 μl. The reaction mixture contained 10 μl of PCR buffer (GIBCO BRL), each deoxynucleoside triphosphate at a concentration of 0.2 mM, 4.5 mM MgCl2, and 2.5 U of Taq DNA polymerase (GIBCO BRL). The PCR program used was an initial denaturation step at 96°C for 5 min, followed by 30 cycles (50 s of denaturation at 96°C, 50 s of annealing at 55°C, and 50 s of extension at 72°C) and a final extension at 72°C for 7 min. Nested PCR amplification was performed with V1 and V1′ as the outer primer pair and V2 and V2′ as the inner primer pair (Table 1). One microliter of the PCR product obtained with the outer primer pair was used for the PCR with the inner primer pair under the conditions described above. Amplifications were carried out on a Perkin-Elmer Cetus apparatus, and the products were analyzed on 2% agarose gels.
The PCR products were sequenced with an automatic sequencer (ABI 377) by using a dye terminator cycle sequencing kit (Ready Reaction 402080) with Ampli Taq polymerase (Fluorescent Sequencing).
Sequence analysis.
The BLASTN program for nucleic acid databases (GenBank, EMBL) and the BLASTP program for amino acid database (SwissProt) were used to search for related β-lactamases with sequences homologous to that of the purified β-lactamase through the World Wide Web BLAST server of the National Center for Biotechnology Information (1). Multiple sequence alignments were performed with the CLUSTALW facilities of the BISANCE software package in the INFOBIOGEN server (8, 33).
The InsightII program was used for molecular modeling of the Staphylococcus aureus PC-1 β-lactamase molecule.
Nucleotide sequence accession number.
The CARB-6 β-lactamase sequence has been submitted to GenBank. Its accession no. is AF 030945.
RESULTS
Purification and molecular weight determination.
The enzyme was purified 200-fold, and the yield was 10%. Its molecular weight was estimated to be 33,000 (Table 2).
TABLE 2.
Physicochemical properties of the CARB-6 β-lactamase of V. cholerae and comparison with TEM-1 and other carbenicillin-hydrolyzing enzymes
| β-Lactamase | Mol wt | pIa |
|---|---|---|
| CARB-6 | 33,000b | 5.35 |
| SAR-1 | 33,700b | 4.9 |
| TEM-1 | 28,911c | 5.4 |
| CARB-1 | 31,405c | 5.3 |
| CARB-2 | 31,347c | 5.7 |
| CARB-3 | 31,313c | 5.75 |
pI was determined in a polyacrylamide gel with an Ampholine gradient (pH 3.5 to 9.5; Pharmacia).
Molecular weight determined by electrophoresis in an SDS–12% polyacrylamide gel by the method of Laemmli (19) with low-range prestained SDS-polyacrylamide gel electrophoresis standards.
Molecular weight as estimated according to the amino acid sequence. The molecular mass of CARB-6 calculated according to the amino acid composition is 31,429 Da (Bio-Oriented Web Server Facilities, ABIM-Aix Marseille University).
pI.
The β-lactamase, both as a crude extract and after purification, was subjected to IEF. In both cases a single band was obtained at pI 5.35, which is between those of TEM-1 (pI 5.4) and PSE-4 (CARB-1) (pI 5.3) (Table 2).
Substrate and inhibition profiles.
The substrate and inhibition profiles of the purified enzyme were determined (Tables 3 and 4). They indicate that it is a penicillinase which hydrolyzes ticarcillin and mezlocillin (59 and 100% hydrolysis relative to that for penicillin, respectively). The Km values indicate that the purified enzyme has a higher affinity for carboxypenicillins such as ticarcillin (55 μM) and ureidopenicillins such as mezlocillin (58.5 μM) than for benzylpenicillin (96 μM) and amoxicillin (157 μM) (Table 3).
TABLE 3.
Kinetic parameters for V. cholerae CARB-6 β-lactamase in comparison with the published values for β-lactamases TEM-1, SAR-1, and CARB-1 to CARB-3a
| Antibiotic |
Km (μM)
|
Vm (relative)b
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CARB-6 | SAR-1 | TEM-1 | CARB-1 | CARB-2 | CARB-3 | CARB-6 | SAR-1 | TEM-1 | CARB-1 | CARB-2 | CARB-3 | |
| Penicillin G | 96 | 42 | 24 | 13 | 7.3 | 9.8 | 100 | 100 | 100 | 100 | 100 | 100 |
| Amoxicillin | 157 | —c | 43 | 13.6 | 16.4 | 21.8 | 266 | — | 84 | 63 | 128 | 97 |
| Ampicillin | — | 68 | 35 | 30 | 20.9 | 24 | — | 63 | 111 | 94.5 | 93 | 100 |
| Ticarcillin | 55 | — | 10 | 40 | 18.8 | 21 | 59 | — | 3 | 88 | 126 | 97 |
| Carbenicillin | — | 190 | 14 | 122 | 72 | 95 | — | 122 | 10 | 126 | 343 | 147 |
| Piperacillin | 7 | — | 43 | — | — | — | 15.9 | — | 86 | — | — | — |
| Mezlocillin | 58.5 | — | 246 | 15.5 | 14 | 20 | 100 | — | 100 | 86 | 83 | 111 |
| Oxacillin | 37 | — | 6 | 58 | 31 | 24.5 | 5.25 | — | 5 | 5.4 | 11.4 | 12.5 |
| Cephaloridine | 526 | 93 | 950 | 40.5 | 208 | 418 | 59.5 | 21 | 76 | 14.6 | 16 | 44.2 |
| Cephalothin | — | — | — | — | — | — | <0.5 | — | 20 | 1.25 | 0.8 | 0.5 |
TABLE 4.
Effects of chemical and β-lactam inhibitors on CARB-6, SAR-1, and TEM-1 β-lactamasesa
| Substrates | % Inhibition (concn [μM] of inhibitor)a
|
||
|---|---|---|---|
| CARB-6 | SAR-1 | TEM-1 | |
| NaCl | 3 (5.104) | —b | 0 |
| Cloxacillin | 100 (100) | 50 (7) | 70 (100) |
| Oxacillin | 50 (100) | — | — |
| Methicillin | 100 (100) | — | — |
| Tazobactam | 50 (0.06) | — | 50 (0.74) |
| Sulbactam | 50 (1.4) | — | 50 (19) |
| Clavulanic acid | 50 (0.075) | 50 (0.005) | 50 (0.034) |
| PCMBc | 52 (20) | 50 (>100) | 50 (200) |
Data for SAR-1 (29) and TEM-1 (21) have been published previously. Results are expressed as percentage of inhibited activity at the indicated concentration of inhibitor in the presence of benzylpenicillin. The inhibitory effects of clavulanic acid and sulbactam were determined after preincubation with the enzyme for 10 min at 37°C.
—, not determined.
PCMB, p-hydroxymercuribenzoate.
The inhibition profile is compatible with that of a class A β-lactamase (Table 4).
Sequence analysis.
The nucleotide sequence of the PCR product obtained with consecutive primers (967 nucleotides) contains an open reading frame of 962 nucleotides. The coding region (CDS) of 867 nucleotides (positions 96 to 962) encodes a protein of 288 amino acids (Fig. 2 and 3).
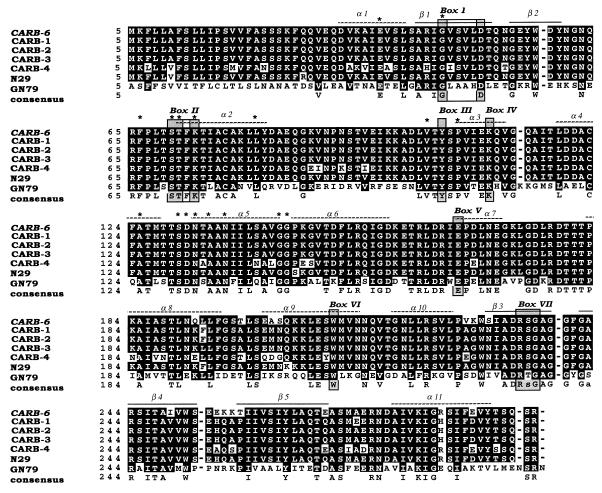
FIG. 2.
Gene sequence and deduced amino acid sequence of the V. cholerae β-lactamase (CARB-6). The deduced amino acid sequence is designated by the one-letter code. The active site, STFK, is boxed, and the differences relative to CARB-1 to CARB-3 are underlined.
FIG. 3.
Multiple sequence alignment of the amino acid sequences of CARB-1, CARB-2, CARB-3, CARB-4, P. mirabilis N29, P. mirabilis GN79, and CARB-6 β-lactamases. The shadowed boxes (I to VII) correspond to amino acid boxes conserved in all penicillin-recognizing enzymes, as identified by Joris et al. (14). Alpha-helix and beta-barrel motifs are indicated from the PC-1 crystal structure (4, 12). Asterisks indicate the conserved residues specific for class A β-lactamases. Amino acid changes are written as black letters in white boxes. Sequences are numbered as described by Ambler (2).
The deduced amino acid sequence is very similar to those of class A β-lactamases: a bla active-site (STFK) tetrad at positions 65 to 68 (positions 70 to 73 according to the standard numbering scheme of Ambler [2]), cysteine residues at positions 72 and 118 (Ambler positions 77 and 123), and all seven conserved amino acid boxes (4, 14) and specific conserved residues (Fig. 3).
Nucleic and amino acid analyses with the BLAST and FASTA programs showed the purified enzyme has substantial homology (94%) with the CARB-1, CARB-2, and CARB-3 β-lactamases of the same group. The starting codon and the length of the coding region (288 amino acids) are strictly conserved (Fig. 2).
Multiple sequence alignment of the CARB-6 amino acid sequence with the sequences of the previously described CARB-1, CARB-2, and CARB-3 (16, 18), CARB-4 (28, 32), and Proteus mirabilis N29 (13) β-lactamases confirms that it is a carbenicillinase. Moreover, CARB-6 possess an RSG box VII that is encountered only in this class of β-lactamases. It appears to be more closely related to CARB-3, CARB-2, CARB-1, and N29 than to CARB-4 (Fig. 3). The nucleotide and amino acid sequences of CARB-6 and CARB-3 are very similar. CARB-6 differs from CARB-3 at 15 amino acid positions (Table 5) located in the downstream third of the sequence (Fig. 2). By molecular modeling with the known crystal coordinates of the S. aureus PC-1 enzyme (12), these mutations were localized on the surface of the molecule.
TABLE 5.
Amino acid point mutations in β-lactamases CARB-1, CARB-2, CARB-3, and CARB-6
| Amino acid positiona | Amino acid changeb
|
|||
|---|---|---|---|---|
| CARB-6 | CARB-1 | CARB-2 | CARB-3 | |
| 192 | Gln | Lys | Lys | Lys |
| 193 | Lys | Phe | Phe | Lys |
| 198 | Thr | Ala | Ala | Ala |
| 202 | Ala | Met | Met | Met |
| 203 | Ser | Asn | Asn | Asn |
| 227 | Val | Ala | Ala | Ala |
| 228 | Lys | Gly | Gly | Gly |
| 230 | Ser | Asn | Asn | Asn |
| 249 | Ile | Val | Val | Val |
| 255 | Glu | His | His | His |
| 256 | Lys | Gln | Gln | Gln |
| 257 | Lys | Ala | Ala | Ala |
| 258 | Thr | Pro | Pro | Pro |
| 269 | Glu | Gln | Gln | Gln |
| 273 | Ala | Glu | Ala | Ala |
| 284 | Arg | His | His | His |
| 288 | Glu | Asp | Asp | Asp |
The numbering of the positions of the mutations is according to Ambler (2).
Boldface italics indicate mutated amino acids relative to CARB-3 β-lactamase.
DISCUSSION
V. cholerae strains belonging to antigenic group O1 were the etiologic agents of the first seven cholera pandemics, and O139 strains are the etiologic agents of the current pandemic. Some non-O1, non-O139 isolates of V. cholerae are responsible for sporadic intestinal or extraintestinal noncholera infections. Generally, wild-type strains of V. cholerae are naturally susceptible to antibiotics that are active against gram-negative bacteria, but biotype El Tor is resistant to polymyxins. However, acquired multiple-drug resistance, presumably plasmid mediated, may occur (9, 11, 25, 29, 35). Moreover, blaCARB and blaPSE genes are known to be plasmid mediated (22) and to be part of transposons and integrons (4).
The phenotypic expression of plasmid-mediated β-lactamase production by strains is resistance to aminopenicillins and carboxypenicillins, but no interference with cephalosporin activities occurs. β-Lactamase inhibitors such as clavulanic acid can restore susceptibility to inactive penicillins. These characteristics are consistent with the presence of TEM-1 or SAR-1, as reported previously for resistant strains. However, a pyogenic strain of V. cholerae non-O1, non-O139 expressed the same resistance phenotype, but its phenotype was associated with the production of a new β-lactamase of pI 5.35. Indeed, the pI was clearly different from that of SAR-1 (pI 4.9) but was similar to that of TEM-1 (pI 5.4) (29). Conversely, its kinetic constants identify this enzyme as a carbenicillin-hydrolyzing enzyme similar to SAR-1 but different from TEM-1 (16, 21). Finally, the same substrates are similarly hydrolyzed by CARB-6, CARB-1 to CARB-3, and SAR-1, but the affinity of CARB-6 for most substrates except for piperacillin (50% inhibition) is slightly lower than those of the CARB β-lactamases. The inhibition profile reveals that clavulanic acid, an inhibitor of class A β-lactamases, inhibits CARB-6, as well as SAR-1 and TEM-1. If CARB-6 is compared to β-lactamases not yet found in V. cholerae strains, the enzyme with the nearest pI (pI 5.3), molecular mass (33,000 Da) (Table 2), and substrate profile is CARB-1 (PSE-4) (16, 21).
The resistance gene could not be transferred by conjugation to the recipient Escherichia coli K-12 strain, despite several attempted mating experiments. This inability to be transferred by conjugation is a character often reported for the V. cholerae and other CARB or CARB-like genes (35).
Initially, only the TEM-1 enzyme was found in ampicillin-resistant V. cholerae (9, 10). Later, a novel β-lactamase designated SAR-1 was reported in a strain of V. cholerae (29). Twenty-nine strains isolated in Africa have been studied in order to assess the distribution of resistance genes in V. cholerae (25). None of the β-lactam-resistant isolates studied cross-hybridized with oligonucleotide probes specific for TEM-1 or OXA-1 β-lactamases. This confirms the presence of β-lactamases other than TEM-1 in V. cholerae.
Most CARB β-lactamases were described as being Pseudomonas-specific enzymes, hence the designation PSE. This indicates that they were originally identified in Pseudomonas aeruginosa strains (28). Indeed, these enzymes have rarely been reported in members of the family Enterobacteriaceae (22) and have been reported only once in Achromobacter xylosoxidans (7). No structural information is available for SAR-1, which is found in V. cholerae, or for CARB-5, which is found in members of the Acinetobacter genus (27). The two carbenicillin-hydrolyzing enzymes described in Japan from P. mirabilis are antigenically and structurally unrelated (Fig. 3): that from strain N29 is a variant of the CARB group of enzymes (13), while that from strain GN79 has a distant relationship with these enzymes but contains the important Arg-234 CARB signature (31). The AER-1 β-lactamase produced by a strain of Aeromonas hydrophila is also a structurally unique carbenicillin-hydrolyzing enzyme (32). A feature common to all bacterial species that produce these β-lactamases is their distribution in an aquatic environment.
Nucleotide sequencing, analysis, and comparison of the amino acid sequence of CARB-6 with those of other members of the β-lactamase family showed the presence of the seven boxes described by Joris et al. (14) and the active-site tetrad characteristic of class A β-lactamases, according to the classification scheme of Ambler (2). Multiple sequence alignment with class A β-lactamase amino acid sequences confirmed the relatedness of CARB-6 to carbenicillinases. The amino acid substitutions that characterize the CARB-6 sequence make this enzyme more similar to P. mirabilis N29, CARB-1, CARB-2, and CARB-3 β-lactamases (19, 17, 16, and 15 amino acid differences, respectively) than to the CARB-4 β-lactamase (38 amino acid differences) (Fig. 3 and Table 5). Identification of these mutated positions on the 2-Å crystal structure of the S. aureus penicillinase showed that all of them are located on the surface of the protein. These changes would therefore be expected to modify the function of the enzyme rather than its structure and are consistent with faster amoxicillin hydrolysis (higher Vmax) and a lower affinity for all β-lactam substrates (higher Km) (Table 3). Further analysis may elucidate the mechanisms by which these mutations affect enzyme function.
In conclusion, CARB-6, described for the first time in V. cholerae, appears to be a new carbenicillin-hydrolyzing enzyme with a unique combination of enzymatic and physicochemical properties.
ACKNOWLEDGMENTS
We thank I. Siebert for efficient technical assistance, F. Letourneur (ICGM) for the nucleotide sequencing, and L. Camoin (UPR 415, ICGM) for fruitful help with and discussions about the modeling experiments.
REFERENCES
- 1.Altschul S F, Thomas L M, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc London Biol. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Aubert G, Zambardi G, Zeni F, Paul G, Fournier J M, Oulahal N, Vermesch R. Abstract presented at the 7th International Congress for Infectious Diseases. 1996. [Google Scholar]
- 4.Boissinot M, Levesque R C. Nucleotide sequence of the PSE-4 carbenicillinase gene and correlations with the Staphylococcus aureus PC-1 β-lactamase crystal structure. J Biol Chem. 1990;265:1225–1230. [PubMed] [Google Scholar]
- 5.Choury D, Aubert G, Siebert I, Assous M, Névot P, Paul G. Abstract presented at the 16th Interdisciplinary Meeting of Anti-Infectious Chemotherapy. 1996. [Google Scholar]
- 6.Choury D, Aubert G, Szajnert M F, Assous M, Blanchard H, Delpech M, Paul G. Abstract presented at the 17th Interdisciplinary Meeting of Anti-Infectious Chemotherapy. 1997. [Google Scholar]
- 7.Decré D, Arlet G, Bergogue-Bérézin E, Philippon A. Identification of a carbenicillin-hydrolyzing β-lactamase in Alcaligenes denitrificans subsp. xylosoxidans. Antimicrob Agents Chemother. 1995;39:771–774. doi: 10.1128/AAC.39.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dessen P, Fondrat C, Valencien C, Mugnier C. BISANCE: a French service for access to biomolecular sequence databases. Comput Appl Biosci. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- 9.Dupont M J, Jouvenot M, Couetdic G, Michel-Briand Y. Development of plasmid-mediated resistance in Vibrio cholerae during treatment with trimethoprim-sulfamethoxazole. Antimicrob Agents Chemoter. 1985;27:280–281. doi: 10.1128/aac.27.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedges R W, Jacobs A E. A 98-megadalton R factor of compatibility group C in a Vibrio cholerae El Tor isolated from southern USSR. J Gen Microbiol. 1975;89:383–386. doi: 10.1099/00221287-89-2-383. [DOI] [PubMed] [Google Scholar]
- 11.Hedges R W, Vialard J L, Pearson N J, O’Grady F. R plasmids from asian strains of Vibrio cholerae. Antimicrob Agents Chemother. 1977;11:585–588. doi: 10.1128/aac.11.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzberg O. Refined crystal structure of β-lactamase from Staphylococcus aureus PC-1 at 2Å resolution. J Mol Biol. 1991;217:701–719. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- 13.Ito Y, Hirano T. Carbenicillin-hydrolyzing penicillinase mediated by a plasmid of Proteus mirabilis and its relationship to the PSE-type enzymes of Pseudomonas aeruginosa. J Appl Microbiol. 1997;83:175–180. doi: 10.1046/j.1365-2672.1997.00203.x. [DOI] [PubMed] [Google Scholar]
- 14.Joris B, Ghuysen J M, Dive G, Renard A, Dideberg O, Charlier P, Frère J M, Kelly J A, Boyington J C, Moews P C, Knox J R. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 dd-peptidase family. Biochem J. 1988;250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazmierczak A, Philippon A, Chardon H, Labia R, Le Goffic F. Constantes cinétiques (Km et Vmax) des β-lactamases mesurées par une méthode micro-acidimétrique couplée à l’ordinateur. Ann Microbiol (Inst Pasteur) 1973;124B:259–268. [PubMed] [Google Scholar]
- 16.Labia R, Guionie M, Barthélémy M, Philippon A. Properties of three carbenicillin-hydrolyzing β-lactamases (CARB) from Pseudomonas aeruginosa: identification of a new enzyme. J Antimicrob Chemother. 1981;7:49–56. doi: 10.1093/jac/7.1.49. [DOI] [PubMed] [Google Scholar]
- 17.Labia R, Barthélémy M. L’enzymogramme des β-lactamases: adaptation en gel de la méthode iodométrique. Ann Microbiol (Inst Pasteur) 1979;130B:295–304. [PubMed] [Google Scholar]
- 18.Lachapelle J, Dufresnes J, Levesque R C. Characterization of the blaCARB-3 gene encoding the carbenicillinase-3 β-lactamase of Pseudomonas aeruginosa. Gene. 1991;102:7–12. doi: 10.1016/0378-1119(91)90530-o. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 21.Medeiros A A. β-Lactamases. Br Med Bull. 1984;40:18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- 22.Medeiros A A, Hedges R W, Jacoby G A. Spread of “Pseudomonas-specific” β-lactamase to plasmids of enterobacteria. J Bacteriol. 1982;149:700–707. doi: 10.1128/jb.149.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel-Briand Y, Bruant L. Etude de la production de β-lactamase pour la carbénicilline chez trente souches de Pseudomonas aeruginosa (mise en évidence d’une souche hautement résistante) C R Acad Sci Ser D. 1972;275:503–506. [PubMed] [Google Scholar]
- 24.Morris J G J. Non O group 1 Vibrio cholerae strains not associated with epidemic didease. In: Wachsmuth I K, Blake P A, Olsvik R, editors. Vibrio cholerae and cholera—molecular to global perspectives. Washington D.C: American Society for Microbiology; 1994. pp. 103–115. [Google Scholar]
- 25.Ouellette M, Gerbaud G, Courvalin P. Genetic, biochemical and molecular characterization of strains of Vibrio cholerae multiresistant to antibiotics. Ann Microbiol (Inst Pasteur) 1988;139:105–113. [PubMed] [Google Scholar]
- 26.Paul G, Gerbaud G, Buré A, Philippon A M, Paugon B, Courvalin P. TEM-4, a new plasmid-mediated β-lactamase that hydrolyzes broad-spectrum cephalosporins in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1989;33:1958–1963. doi: 10.1128/aac.33.11.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul G, Joly-Guillou M L, Bergogue-Bérézin E, Nevot P, Philippon A. Novel carbenicillin-hydrolyzing β-lactamase (CARB-5) from Acinetobacter calcoaceticus var. anitratus. FEMS Microbiol Lett. 1989;59:45–50. doi: 10.1111/j.1574-6968.1989.tb03080.x. [DOI] [PubMed] [Google Scholar]
- 28.Philippon A, Paul G, Thibaut A, Jacoby G. Properties of a novel carbenicillin-hydrolyzing β-lactamase (CARB-4) specified by an IncP-2 plasmid form. Antimicrob Agents Chemother. 1986;24:362–369. doi: 10.1128/aac.29.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid A J, Amyes S G B. Plasmid penicillin resistance in Vibrio cholerae: identification of new β-lactamase SAR-1. Antimicrob Agents Chemother. 1986;30:245–247. doi: 10.1128/aac.30.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safrin S, Morris J G, Adams M, Pons V, Jacobs R, Conte J E. Non O1 Vibrio cholerae bacteremia: a case report and review. Rev Infect Dis. 1987;10:1012–1017. doi: 10.1093/clinids/10.5.1012. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai Y, Tsukamoto K, Sawai T. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J Bacteriol. 1991;173:7038–7041. doi: 10.1128/jb.173.21.7038-7041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanschagrin F, Bejaoui N, Levesque R C. Structure of CARB-4 and AER-1 carbenicillin-hydrolyzing beta-lactamases. Antimicrob Agents Chemother. 1998;42:1966–1972. doi: 10.1128/aac.42.8.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. Epidemic diarrhea due to Vibrio cholerae non-O1. Weekly Epidemiol Rec. 1993;68:141–142. [PubMed] [Google Scholar]
- 35.Young H K, Nandivada L S, Amyes G B. Antibiotic resistance in the tropics. 1. The genetics of bacterial ampicillin resistance in tropical areas. Trans R Soc Trop Med Hyg. 1989;83:38–41. doi: 10.1016/0035-9203(89)90699-8. [DOI] [PubMed] [Google Scholar]