Abstract
Bacterial colonization has been already demonstrated in heart valve tissues of patients without cardiovascular infections. However, the evidence of a valvular microbiome is still scarce. The next-generation sequencing method was carried out on 34 specimens of aortic (n = 20) and mitral valves (n = 14) explanted from 34 patients having neither evidence nor history of infectious diseases, particularly infective endocarditis. While no bacteria were demonstrated using standard culture methods, bacterial deoxyribonucleic acid (DNA) sequences were found using next-generation sequencing in 15/34 (44%) cases. Escherichia coli was present in 6 specimens and was the most frequently identified bacterium. There was a trend towards a higher rate of bacterial DNA positivity in specimens of calcific valves than in those of non-calcific valves (10/17 vs 5/17, P = 0.17). Based on a quantitative test, E. coli accounted for 0.7% ± 1% in calcific valvular tissue and 0.3% ± 0.3% in non-calcific valvular tissue (P = 0.2), and for 11% ± 27% in the valvular tissue of diabetic patients and 0.3% ± 1% in the valvular tissue of non-diabetic patients (P = 0.08). Detection of bacterial DNA in non-endocarditis valvular tissues could be a relatively common finding. There could be an association between the valvular microbiome and certain models of valve degeneration and common metabolic disorders.
Keywords: Bacterial colonization, Calcific degeneration, Diabetes mellitus, Heart valve diseases, Microbiome
The link between the microbiome and the endothelial injury underlying cardiovascular diseases has been investigated with increasing interest throughout the last 2 decades.
INTRODUCTION
The link between the microbiome and the endothelial injury underlying cardiovascular diseases has been investigated with increasing interest throughout the last 2 decades. Recently, using the next-generation sequencing (NGS) method, bacterial sequences have been found inside the atheromatous plaques and heart valve tissues of patients without cardiovascular infections [1, 2]. Although the role of several microorganisms as causative pathogens of cardiovascular diseases has long been speculated, the current evidence remains scarce.
Two years ago, for clinical purposes, the present authors used 16S ribosomal ribonucleic acid gene sequencing to evaluate specimens of tissue derived from explanted heart valves of patients without infective endocarditis. Given that multiple and various bacterial deoxyribonucleic acid (DNA) sequences were identified, further patients undergoing heart valve surgery were evaluated for valvular microbiome. The results were retrospectively reviewed in this brief communication.
METHODS
Between November 2017 and September 2019, 34 tissue specimens were collected from 34 native cardiac valves of subjects (mean age, 69 ± 12.8 years; 13 females) undergoing valve repair or replacement at the Cardio-Thoracic and Vascular Department of the University Hospital of Trieste, Italy. Among the cardiovascular risk factors, hypertension, dyslipidaemia and diabetes mellitus were present in 24, 21 and 5 cases, respectively. The patients had no history of bacteraemia and were not suffering from any infectious disease, nor received antibiotics during the last 3 months prior to surgery. The tissue specimens derived from 20 aortic (stenotic = 17, regurgitant = 3) and 14 mitral (stenotic = 2, regurgitant myxomatous = 7 and regurgitant non-myxomatous = 5) valves were explanted.
Heart valves leaflets were removed, stored in a sterile container with saline solution, and sent for examination to microbiology laboratory. While part of each sample was homogenized, inoculated on Columbia agar, Schaedler agar and into liquid Wilkins-Chalgren broth and incubated at 37°C both in aerobic and anaerobic conditions, the remaining part of valvular tissue was used for NGS analysis. The V3 region of the 16S ribosomal ribonucleic acid gene was sequenced using the Ion Personal Genome Machine™ System technology (Life Technologies, New York, NY). Negative controls, including no template control, were processed with the clinical specimens. NGS data were processed by Quantitative Insights Into Microbial Ecology 2 software [3], retaining reads with Q ≥ 20 and read length of 180 bp after the Divisive Amplicon Denoising Algorithm [4], and were aligned to the SILVA database (release 132) [5] with a Basic Local Alignment Search Tool plus consensus method and with a similarity threshold of 99%. Linear discriminating analysis effect size was used to test differentially abundant taxa between groups, using a rarefaction depth of 2400 reads/sample. The Fisher's exact test was used for associations with clinical metadata.
At the presenting authors’ institution (Trieste University Hospital), microbiome analysis has been implemented in clinical practice for selected patients. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
RESULTS
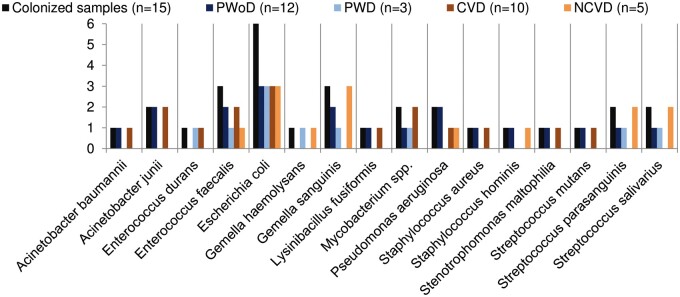
No bacteria have been isolated by standard cultures of the valvular tissue. With regard to bacterial DNA, the negative controls produced no sequencing output after the quality filtering. In the remaining specimens, NGS produced a total of 1 188 820 reads (Q score >20, range 2438–394 689) and 3089 features. There was a microbial colonization of 15/34 (44%) explanted valves. Acinetobacter spp., Enterococcus spp., Escherichia coli, Gemella spp., Lysinibacillus fusiformis, Mycobacterium spp., Pseudomonas aeruginosa, Staphylococcus spp., Stenotrophomonas maltophilia and Streptococcus spp. were identified. Escherichia coli was present in 6 specimens and was the most frequently identified bacterium. Enterococcus faecalis and Gemella sanguinis were present in 3 specimens. Other bacteria such as Acinetobacter baumannii, Enterococcus durans, Gemella haemolysans, L. fusiformis, Staphylococcus aureus, Staphylococcus hominis, S. maltophilia and Streptococcus mutans were found less frequently (Fig. 1).
Figure 1:
Characterization of the heart valve microbiome. The bacteria identified in the explanted cardiac valves are reported here according to the number of positive specimens for a given bacterium out of all positive specimens (black bars, n = 15), according to the presence of calcific degeneration of the valvular tissue (blue and light blue bars) and according to the presence of diabetes mellitus (orange and light orange bars). CVD: calcific valve degeneration; NCVD: non-calcific valve degeneration; PWD: patients with diabetes; PWoD: patients without diabetes.
There was a trend towards a higher rate of bacterial DNA positivity in specimens of tissue from aortic than mitral valves (11/20 vs 4/14, P = 0.17), and in specimens of tissue from calcific valves than in those of tissue from non-calcific valves (10/17 vs 5/17, P = 0.17). Based on the linear discriminating analysis effect size test, E. coli accounted for 0.7% ± 1% in calcific valvular tissue and for 0.3% ± 0.3% in non-calcific valvular tissue (P = 0.2); for 11% ± 27% in valvular tissue of diabetic patients and for 0.3% ± 1% in valvular tissue of non-diabetic subjects (P = 0.08) (Fig. 1 and Table 1).
Table 1:
Bacteria identified as biomarkers
| Species | NCVD versus CVD | LDA score | P-value | |
|---|---|---|---|---|
| Escherichia coli | 0.3% ± 0.3% | 0.7% ± 1% | −5.43 | 0.2 |
| PWoD versus PWD | LDA score | P-value | ||
|---|---|---|---|---|
| Escherichia coli | 0.3% ± 1% | 11% ± 27% | −5.86 | 0.08 |
LEfSe test showing the bacterial species with a trend of significance. Negative values of LDA scores (log 10) are indicative for enriched taxa in the calcific valve degeneration group and in the diabetic patient group.
CVD: calcific valve degeneration; LDA: linear discriminative analysis; LEfSe: linear discriminative analysis effect size; NCVD: non-calcific valve degeneration; PWD: patients with diabetes; PWoD: patients without diabetes.
DISCUSSION
The relationship between infectious diseases and cardiovascular disorders has long been theorized. While recent studies emphasized the potential role of periodontal microorganisms in the inflammatory process leading to atheromatous plaque, other investigations showed bacterial DNA sequences in tissue specimens derived from diseased cardiac valves, with similar rates to those of saprophytes of the human skin and oral cavity [1].
In the present study, the detection of bacterial DNA using NGS in the valvular tissue of patients with non-infective endocarditis heart valve diseases occurred in 44% of the cases. To the authors’ knowledge, no study patient suffered from any, acute or subacute, infection. Escherichia coli and Enterococcus, both common gut inhabitants, were the most frequently detected bacteria in the valvular tissue of this series.
Two other important findings of this communication are the followings: (i) bacterial sequences of DNA were found more frequently in aortic than in mitral valves. As calcific degeneration is more common in fragments of tissue from aortic than from mitral valves of patients undergoing heart valve surgery, it could be hypothesized that some relationship prevails between the bacterial colonization (when present) and the model of valvular lesion, with reference to calcific degeneration; (ii) E. coli was found more commonly inside valve tissues of diabetic subjects. This finding is in agreement with the literature, where a microbiome shift towards Proteobacteria, namely the phylum that includes E. coli, is significantly more frequent among hyperglycaemic patients [6]. This fact could suggest a relationship between the intestinal barrier impairment related to intestinal dysbiosis and to a leaky gut, which are present by definition in diabetic patients, and the increased rates of degenerative (and infective) valvular diseases of diabetic subjects [7, 8]. Consequently, every study highlighting the role of gut dysbiosis as risk factor for bacterial colonization of the heart valve tissues would increase comprehension of the aetiology and complex mechanisms that underlie heart valve diseases.
In conclusion, the present preliminary report would prove that the detection of bacterial DNA sequences in non-infective endocarditis valvular tissues could be a relatively common finding. There could be a possible role of the valvular microbiome in the date of cardiac valves, through modulation of chronic inflammatory processes.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Gianfranco Sinagra (Cardio-Thoracic and Vascular Department, Trieste University Hospital) for intellectual support and funding acquisition and the NGS Facility at the Institute for Maternal and Child Health-IRCCS Burlo-Garofolo, Trieste, Italy.
Funding
This study was funded by the University of Trieste, the University of Udine and the International School for Advanced Studies (SISSA)—Accordo Quadro project (U11SPECCARDIO).
Conflict of interest: none declared.
Reviewer information
Interactive CardioVascular and Thoracic Surgery thanks Dinakaran Vasudevan and the other, anonymous reviewer(s) for their contribution to the peer review process of this article.
REFERENCES
- 1. Chalupova M, Skalova A, Hajek T, Geigerova L, Kralova D, Liska P. et al. Bacterial DNA detected on pathologically changed heart valves using 16S rRNA gene amplification. Folia Microbiol 2018;63:707–11. [DOI] [PubMed] [Google Scholar]
- 2. Szulc M, Kustrzycki W, Janczak D, Michalowska D, Baczynska D, Radwan-Oczko M.. Presence of periodontopathic bacteria DNA in atheromatous plaques from coronary and carotid arteries. Biomed Res Int 2015;2015:1– 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP.. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2012;41:D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Demmer RT, Trinh P, Rosenbaum M, Li G, LeDuc C, Leibel R. et al. Subgingival microbiota and longitudinal glucose change: the Oral Infections, Glucose Intolerance and Insulin Resistance Study (ORIGINS). J Dent Res 2019;98:1488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Kort S, Keszthelyi D, Masclee AAM.. Leaky gut and diabetes mellitus: what is the link? Obes Rev 2011;12:449–58. [DOI] [PubMed] [Google Scholar]
- 8. Slyepchenko A, Maes M, Machado-Vieira R, Anderson G, Solmi M, Sanz Y. et al. Intestinal dysbiosis, gut hyperpermeability and bacterial translocation: missing links between depression, obesity and type 2 diabetes. CPD 2016;22:6087–106. [DOI] [PubMed] [Google Scholar]