Abstract
During normal vision, our eyes provide the brain with a continuous stream of useful information about the world. How visually specialized areas of the cortex, such as face-selective patches, operate under natural modes of behavior is poorly understood. Here we report that, during the free viewing of movies, cohorts of face-selective neurons in the macaque cortex fractionate into distributed and parallel subnetworks that carry distinct information. We classified neurons into functional groups on the basis of their movie-driven coupling with functional magnetic resonance imaging time courses across the brain. Neurons from each group were distributed across multiple face patches but intermixed locally with other groups at each recording site. These findings challenge prevailing views about functional segregation in the cortex and underscore the importance of naturalistic paradigms for cognitive neuroscience.
Natural visual experience reveals parallel functional subnetworks of neurons embedded within the macaque face patch system.
INTRODUCTION
Face patches in the temporal cortex of macaques are spatially separate, densely interconnected (1, 2) regions, defined by their shared preference for faces (3, 4). Similar regions exist in the cortex of humans (5) and marmosets (6), suggesting that this functional network is a conserved feature in primates (7–9). Individual patches differ in their neural response selectivity to facial attributes, such as viewpoint, identity, movement, and expression (10–12). These differences have prompted speculation that face patches are arranged in one or more functional hierarchies (13–15).
Knowledge about the organization of the macaque face network derives principally from neural responses to briefly presented images. However, a few studies have begun to characterize the brain’s responses to freely viewed natural movies (16–20), with some evidence suggesting that the basic layout of the face patch network is preserved under these conditions (17). While naturalistic paradigms sacrifice precise control over visual stimulation, they are able to more closely approximate the conditions under which the visual specialization evolved in the primate brain (21). An important question is whether the principles derived from image responses will extend to more natural modes of vision (22).
Here, we approach this question using a combination of methods that offers a fresh perspective on the visual responses of face-selective neurons. Briefly, we describe the activity of isolated single neurons in multiple nodes of the macaque face patch network during the free and repeated viewing of movies depicting natural social behaviors. Rather than taking the conventional approach of analyzing a neuron’s representation of visual features, we instead compare each neuron’s activity to corresponding functional magnetic resonance imaging (fMRI) responses across the brain obtained to the same movie (Fig. 1A). Thus, each neuron is characterized through its cortical network profile, summarized as a map of positive and negative covariations, rather than through its explicit stimulus selectivity. This approach provides a robust, data-driven method to assess the functional organization of the face patch system amid the complexity of naturalistic stimuli, in the context of the whole-brain network.
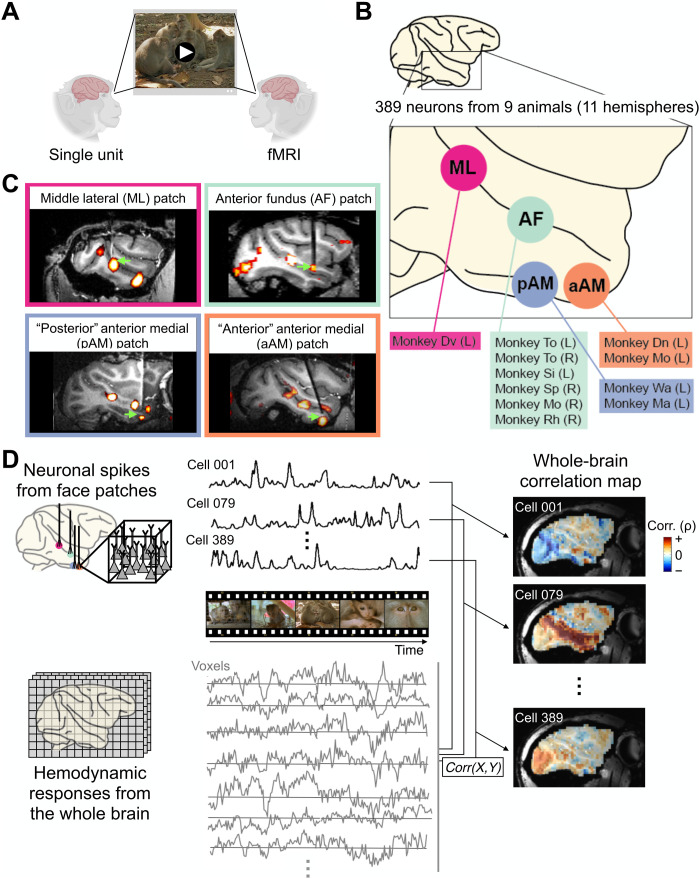
Fig. 1. Computing whole-brain correlation maps for individual neurons from four face patches.
(A) Schematic diagram of experimental paradigm. Two cohorts of animals freely and repeatedly viewed the same set of naturalistic movies (shown here is an example scene from the actual movie stimuli, available at https://doi.org/10.5281/zenodo.4044578), during single-unit recording and fMRI acquisition, respectively. The two measures of neural activity could be directly correlated, as the responses of both were synchronized to the same movie content. (B) Schematic diagram of microwire bundle recording locations in this study, in all cases introduced into fMRI-identified face patches. Animal identifications for each face patch recording are listed with hemispheric information in parenthesis. Abbreviation for face patches: pAM, posterior part of AM; aAM, anterior part of AM. (C) Representative MR images showing recording electrode in each face patch. (D) Basic multimodal analyses underlying single-unit fMRI mapping. The time series corresponding to each neuron’s spiking fluctuations was correlated with that of fMRI voxels throughout the brain, after adjusting for the hemodynamic response profile (see Materials and Methods). Representative examples of single-unit correlation maps from three different neurons are shown on a sagittal section on the right.
RESULTS
To begin, we identified face patches in 11 behaving macaques using standard fMRI methods (4, 17). In nine animals, chronic microwire bundles were inserted into one or two of the four patches featured in the present study (see Materials and Methods). Each bundle consisted of flexible microwires that covered a small volume of tissue (<1 mm3) and allowed us to record the activity of isolated neurons across multiple daily sessions (16, 23). We then recorded responses from 389 single neurons across four fMRI-defined face patches (Fig. 1, B and C; n = 33 from middle lateral (ML), n = 224 from anterior fundus (AF), n = 77 from posterior portion of anterior medial (pAM), n = 55 from anterior portion of AM (aAM) face patches; see Materials and Methods for distinction between pAM and aAM). For each neuron, we recorded the spiking time courses elicited during the repeated free viewing of naturalistic movies. For 254 of the recorded neurons, we independently measured basic category selectivity to briefly presented images of faces, objects, and other categories. Of these neurons, 166 (65%) were determined to be face selective (see Materials and Methods).
Throughout the population, neighboring neurons in each face patch exhibited diverse response time courses to the same movie content (see fig. S1A for the time courses of all neurons in the study). In agreement with previous recordings from face patch AF (16), the time courses of neighboring neurons in all patches often diverged markedly, despite sharing some temporal features, even among the 166 neurons demonstrated to be face selective in separate testing (fig. S1, A and B, and fig. S2). Neurons sampled from different face patches, typically several millimeters apart, often responded with very similar time courses (fig. S1, A and C). Together, these initial observations suggested that face-selective neurons exhibit a diversity of responses during natural modes of vision that are not segregated into distinct patches.
Cortical fMRI readout of single neurons
To explore this finding further, we applied the intermodal technique of single-unit fMRI mapping developed previously (24), which summarizes complex movie-driven single-unit activity as a functional map of activity correlations across the brain (Fig. 1D and fig. S3; see Materials and Methods). Each neuron’s unique pattern of positive and negative activity correlations sheds light on its participation in different functional brain networks under natural viewing conditions. Methodologically, this approach overlaps with paradigms mapping spontaneous neural activity using intermodal methods (25–27). However, in this case, correlations are driven by an external movie stimulus, followed by extensive averaging across presentations, and experiments were carried out across the brains of different individuals. Thus, our method is designed to eliminate components of spontaneous or otherwise trial-unique activity and is distinctly different from conventional functional connectivity.
The temporal structure of the 15-min natural movie served as a shared timeline for electrophysiological and fMRI data, which were collected in two separate sets of animals (see Materials and Methods). Response time courses were averaged across multiple movie presentations, both for individual neurons (8 to 15 presentations) and for the fMRI responses (28 to 40 presentations). This averaging, combined with the inherent reproducibility observed in both modalities (16, 17), provided excellent estimations of the mean response time course for both neurons and voxels (also see fig. S1C for examples). We focus here on the correlation of average spiking time courses with cortical voxels (figs. S3 and S4), only noting that structures outside the cerebral cortex, including the superior colliculus, pulvinar, striatum, amygdala, and cerebellum, were also engaged [see (24)].
Mixed neural populations, shared broadly
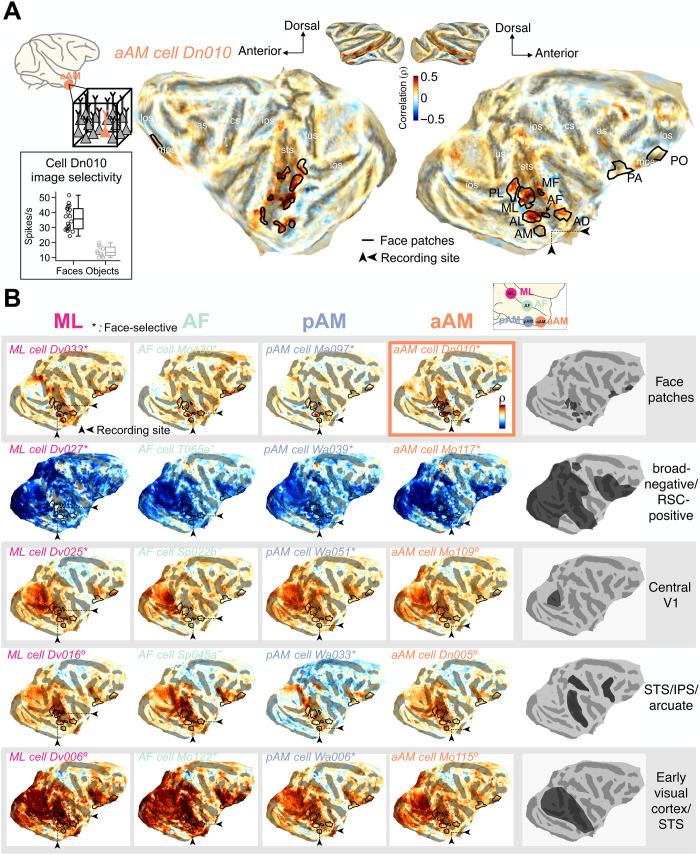
The correlation maps derived from each face patch were notably diverse, consistent with a previous application of this method to face patch AF (24). An example of one neuron’s map is shown in Fig. 2A, where the fMRI coupling is depicted on a flattened cortical hemisphere. This neuron was recorded from face patch aAM and determined to be face selective in separate testing (Fig. 2A, inset). This cortical correlation profile exhibited peaks that roughly matched the location of face patches, mapped in separate experiments (Fig. 2A, black lines; see Materials and Methods), suggesting that the visual operation performed by this neuron can be related to modes of visual analysis that are largely restricted to other face patches.
Fig. 2. Examples of single-unit fMRI maps from neurons in four face patches.
Cortical fMRI correlation pattern of example neurons recorded from four face patches projected onto the flattened cortical surface of one animal’s brain. Small black triangles on each map point to the location of recording site of the featured neuron. Location of face patches (black lines) are based on separate fMRI block design experiments. Abbreviation for face patches: PL, posterior lateral; MF, middle fundus; AL, anterior lateral; AD, anterior dorsal; PA, prefrontal arcuate; PO, prefrontal orbital face patch. (A) Correlation map of an example neuron from face patch aAM. Inset: Spiking responses to brief presentations of faces (n = 20) and nonface objects (n = 10) demonstrate that this neuron is face selective. Center line, median; box limits, upper and lower quartiles; whiskers, range. lus, lunate sulcus; ios, inferior occipital sulcus; sts, superior temporal sulcus; ips, intraparietal sulcus; cs, central sulcus; as, arcuate sulcus; los, lateral orbital sulcus; mos, medial orbital sulcus. (B) Maps of 20 example neurons (cell identity is written in italics) from four face patch recording sites, presented in a matrix format. Each column of the matrix contains five neurons from each local recording site, face patches ML, AF, pAM, and aAM (from left to right), whose fMRI maps differ substantially. Each row shows fMRI maps with similar features that were common to neurons across all four recording sites, with the specific features summarized on the right. On the basis of separate testing, asterisks indicate neurons that were face selective, circles those that were not face selective, and dashes those that were not tested (see Materials and Methods). Map of the example neuron in (A) is marked with an outline. RSC, retrosplenial cortex.
The cortical correlation pattern shown in the example in Fig. 2A was relatively common among face patch neurons, with similar examples shown from each of the four face patches in the top row of Fig. 2B. At the same time, the columns in Fig. 2B show that neurons with very different cortical correlation profiles were found within each of the four face patches. Dissimilar functional profiles were, for example, commonly observed among pairs of neurons recorded simultaneously from the same patch and even from the same electrode (see fig. S5 for examples). Together, the rows and columns in Fig. 2B illustrate that distinct neural subtypes were intermixed locally but shared across different face patches. For instance, each face patch contained neurons whose visual responses were broadly anticorrelated with visual areas but positively correlated with the retrosplenial cortex (Fig. 2B, second row), potentially suggesting that these neurons are influenced by the default mode network (28, 29). Each face patch also contained neurons whose activity was correlated with the foveal retinotopic cortex (Fig. 2B, third row) and others whose activity was correlated with dorsal stream visual areas in caudal superior temporal sulcus (STS), intraparietal sulcus (IPS), and arcuate sulcus (Fig. 2B, fourth row), possibly linking to attentional network (30, 31). Notably absent in our analysis was any indication of elevated local fMRI correlation in the vicinity of the neural recording sites. Thus, multiple, highly distinct functional profiles were shared across each of the four face patches.
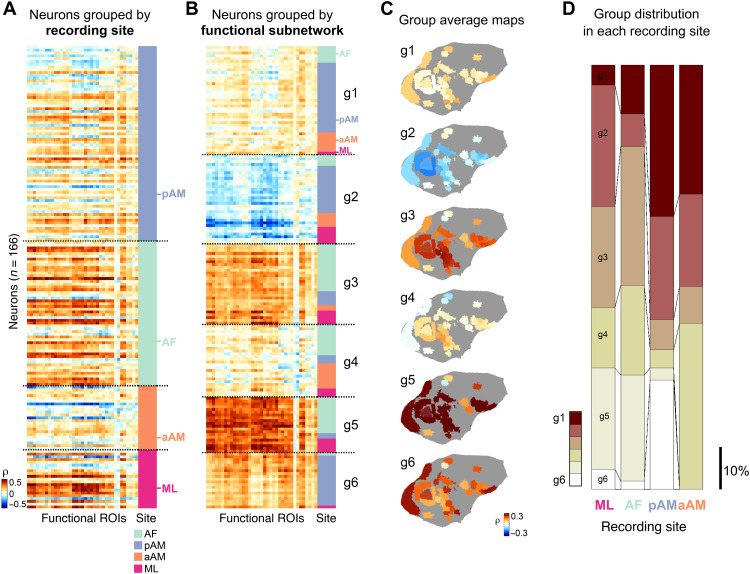
We next performed a population analysis of functional profiles obtained across the four face patches. We focused on neurons determined to be face selective in separate testing (Fig. 3; see fig. S8 for the results of similar analysis applied to the entire population). To succinctly summarize the important information derived from each neuron’s correlation map, we delineated a set of 37 functional regions of interest (fROIs). For this, we identified continuous clusters of voxels across the cortical sheet that exhibited substantial correlations across all recorded neurons (see Materials and Methods; see fig. S6 for correspondence with atlas). This step allowed the correlation profile of each neuron to be expressed as a 37-element vector of value, with each element pertaining to the average correlation value within one fROI.
Fig. 3. Distinct cortical correlation patterns distributed across spatially remote face patches.
(A) fMRI correlation profiles of spiking activity from verified face-selective neurons (row; n = 166) across 37 fROIs in the cerebral cortex (see Materials and Methods and fig. S3) (column; n = 37 from fROI 1 to fROI 37, from left to right). Each row is a 37-element vector of fROI correlation coefficients stemming from one neuron. Neurons are grouped by face patch recording location, indicated on the right in different colors. (B) Same format as (A), but with neurons sorted into groups g1 to g6 according to their functional clustering (see Materials and Methods). (C) Neural fMRI correlation profiles from (B) averaged across neurons in each group and mapped spatially onto the original fROIs on the cortical surface. (D) Proportion of neurons belonging to each cell group found at each of the recorded face patches. Color indicates identity of cell groups from (B) and (C). Scale bar indicates 10% of proportion.
Correlation profiles for all face-selective neurons are shown in Fig. 3A, sorted by their recording locations. Consistent with Fig. 2B, this figure highlights the heterogeneous composition of face cells within each patch, with marked local intermixing of neural responses to the movie stimulus. To better highlight the functional groups within this matrix, we applied unsupervised (K-means) clustering to the fMRI correlation maps of the population of face-selective cells (see Materials and Methods and fig. S7). The optimization of the clustering suggested six functional subpopulations, which we termed cell groups g1 to g6. When the neural profiles were reordered on the basis of this clustering (Fig. 3B), it was plain that each functional subpopulation was contributed by neurons located in multiple face patches.
To better aid visualization of the functional profiles of the different cell groups, we remapped the average fROI correlation vectors from each cell group back onto the flattened brain (Fig. 3C). The resulting maps revealed each cell group’s specific regional coupling with the fMRI signal during naturalistic viewing. For example, one group of neurons showed relatively selective coupling to a subset of face patches, face patch anterior lateral, anterior dorsal, and orbitofrontal areas (group g1), whereas neurons in another group (group g4) showed networks that appear to be complementary to the ones of group g1, featuring selective correlation to areas in the lower bank of STS. Another group was correlated strongly with areas in the STS and primary visual areas (group g3), while another (group g6) correlated strongly with eccentric portions of primary visual cortex in addition to the areas coupled with the group g1. While individual neurons composing each group differed somewhat in their specific profiles, this grouping provided an effective summary of the fMRI correlation patterns across the face-selective neural population.
Across the four patches, most functional cell groups were represented at each location, although they were not evenly distributed (Fig. 3D), likely reflecting a degree of specialization of individual patches. For example, functional group g5 with a broad, strong positive correlation profile was not observed among face-selective neurons recorded in patch aAM, nor was group g6. Groups g3 and g5, two groups coupled strongly with STS regions, were more prominent among face-selective neurons recorded from STS face patches (ML and AF), whereas group g1 with selective face patch correlation was observed more in face neurons from the more ventral patches (pAM and aAM).
DISCUSSION
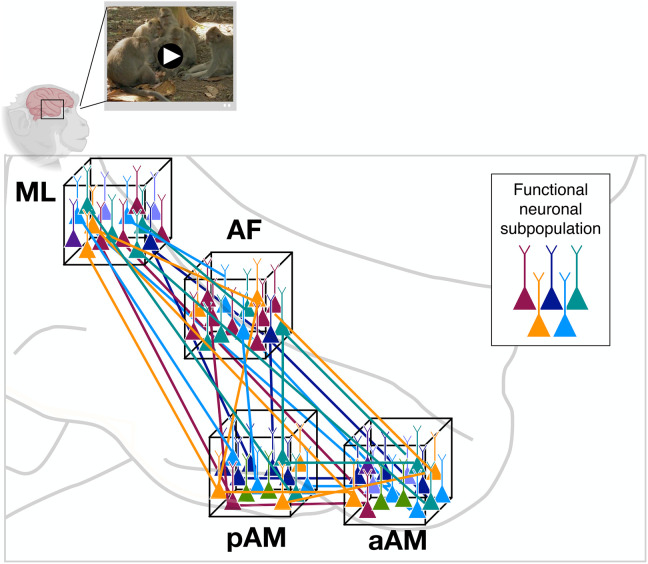
These findings shed new light on the division of labor among single neurons within a functionally defined cortical network. They show that, under stimulation conditions that approximate natural vision, face patches overlap markedly in their functional composition. The data suggest the face patch system is composed of parallel threads of neural subnetworks, intermixed at a local level and carrying out distinct types of visual operations across multiple nodes of the network (Fig. 4).
Fig. 4. Parallel functional subnetworks pervading the face patch system.
Schematic diagram depicting functionally distinct subnetworks operating in parallel within the face processing network. Local cell populations are indicated by neurons within each of the cubes, corresponding to the four face patches recorded in the present study. Coloration indicates functionally distinct classes of neurons. The sharing of distinct functional operations across face patches, and the mixing of these signals locally, were central observations in the present study. The selective long-range connections among functionally similar classes of neurons (colored lines) are hypothesized. An example scene from the movie stimuli is shown at the top.
The extreme mixing of signals in each face patch is unexpected in light of the domain specificity normally associated with these regions. While this traditional view is supported by many experiments, it does not make strong predictions for how neurons in this area respond during natural modes of visual behavior. The functional diversity observed here might be attributed to the richness of the movie stimuli, which include dynamic events, spatial layouts, color contrasts, and social interactions. This natural visual combination of features has seldom been incorporated into the study of face patch responses. It is also possible that active elements of vision that accompany free viewing, including exploration and attention, may unmask the different contribution of neural subpopulations (32). Regardless, the pronounced mixed selectivity observed during movie viewing bears some resemblance to that measured in other regions across the brain (33, 34). It also presents practical challenges for systems and cognitive neuroscience, which often makes the tacit assumption of functional segregation in the brain.
At a more mechanistic level, it is interesting to speculate that the contrast between conventional and naturalistic testing derives from the engagement of different synaptic subpopulations providing input to individual cortical neurons. For example, during brief image presentation, a given neuron’s selectivity to faces might be conferred by the simultaneous activation of a locally shared set of inputs concerned with the analysis of facial structure. These inputs are likely concentrated within the face patches and serve as the basis for their functional definition. However, under more naturalistic conditions, the same neuron’s activity may be dominated by other, long-range connections with a sparser pattern of input among face patch neurons [see (35) for similar ideas in a different context], as well as the effects of neuromodulation during active behaviors (36).
The analysis of neural coupling in the present study is shaped by three important factors. First, the averaging of responses across many trials isolates the stimulus-entrained components of the neural activity. Trial-unique patterns of eye movements or attentional shifts cannot account for the observed response diversity, which was expressed among neurons recorded simultaneously during the same trials. Future studies using simultaneous electrophysiological and fMRI measurement may separately assess the contribution of these factors, along with forms of intrinsic activity, to single-unit fMRI coupling across the brain. Second, the fMRI mapping method is based on comparison to hemodynamic responses, which restricts analysis to time scales of seconds. We found that the basic diversity of neural responses during natural stimulation was evident at both short and long time scales (fig. S2). Nonetheless, to assess the nature of millisecond-scale interaction of face patch neurons, for example, among neurons occupying different patches but belonging to the same subnetwork, simultaneous electrophysiological recordings are required. Third, all the neurons analyzed in this study were recorded from fMRI-defined face patches. Future studies using this method to examine neurons from a broader set of areas, such as cortical object- and body-selective patches, can shed further light on the degree of functional overlap among high-level visual areas.
Last, combining natural conditions with novel readouts pushes researchers to explore previously untapped conceptual spaces for studying brain function. In departing from the usual practice of linking neural responses to specific visual features, less emphasis is placed on local representation and encoding and more on the physiological coordination of brain regions involved in a wide range of sensory and motor functions. The present findings underscore the value of intermodal mapping as a robust tool for characterizing neural activity during complex natural behavior, as well as the inherent value of combining controlled and naturalistic paradigms for the development of hypotheses and conceptual frameworks in systems neuroscience.
MATERIALS AND METHODS
The electrophysiological and the fMRI responses analyzed in the present study were recorded separately using two separate cohorts of animals. A full description of each set of methods, including surgical procedures, tasks, and behaviors, is given in previous publications that detail electrophysiology (16) and fMRI investigations (17) that generated part of the movie-driven datasets studied here. Technical details of the chronic microwire bundles and their targeted implantation into face patches are described in previous work (37). Last, the method of single-unit fMRI mapping, comparing movie-driven single-unit activity and fMRI responses across the brain, is described in a previous publication (24).
Subjects
Eleven rhesus monkeys (Macaca mulatta, six females and five males, 5.0 to 11.3 kg, 6 to 14 years old) participated in this study. All monkeys were surgically implanted with a fiberglass headpost for head immobilization during experiments. All monkeys participated in a separate sessions of face patch localizer fMRI scans using on one or more of the following methods: a standard block design with images (16, 17, 38), a block design with clips of different categories (39), and a movie watching paradigm (17, 38, 40). The locations of functionally defined face patches had some variation across animals as often observed in previous literature (2), especially for AM patch. In two of the animals (monkeys Dn and Mo), we found that their AM patches are located approximately 2 mm anterior and lateral to the AM patches in the other animals. To differentiate these two areas, we use the term “aAM” (anterior portion of AM) to describe the anterior AM patch locations and “pAM” (posterior portion of AM) to describe the posterior AM patch locations in the other two animals (Fig. 1, B and C). Two monkeys participated in multiple fMRI scans of free viewing of movies, whereas nine monkeys received uni- or bilateral implants of chronic microwire electrode bundles in one or two of the face patches functionally localized from the fMRI localizer scans: ML, AF, pAM, and aAM face patches (Fig. 1, B and C). All procedures were approved by the Animal Care and Use Committee of the U.S. National Institutes of Health [National Institute of Mental Health (NIMH)] and followed U.S. National Institutes of Health guidelines.
Experimental design
Movie stimulus
For visual experiments involved free viewing of naturalistic movies, we used three 5-min movies depicting nonhuman primates (primarily macaque monkeys) interacting with conspecifics and humans, which are the set of movies used in previous studies (16, 17, 41). Each movie was edited from a set of nature documentaries. Individual scenes are approximately 40 s in length with cinematic cuts throughout the scenes. Across three movies, 78 to 88% of the movie contained faces. All movies were encoded at 30 frames per second and a frame resolution of 640 × 480 pixels. Various visual features of the movie contents are described and quantified previously (17) and are available along with the movies (https://doi.org/10.5281/zenodo.4044578).
fMRI: Free viewing of movies
During the functional echo planar imaging (EPI) scans, subjects viewed the visual stimuli projected onto a screen above their head through a mirror inside of the MRI scanner. Three 5-min movies depicting macaque monkeys interacting with conspecifics and humans were presented in 640 × 480 pixel resolution with a frame rate of 30 frames per second. Each session started with an eye calibration, and the calibration was repeated every two 5-min trials. Each movie viewing began with a 500-ms presentation of a white central point surrounded by a 10° diameter annulus indicating the gaze-position tolerance window, which was approximately the size of the movie stimulus. Subjects received a drop of juice reward every 2 s as long as their eyes were within the movie frame.
Electrophysiology: Free viewing of movies
Nine electrophysiology subjects viewed the same three 5-min movies used in fMRI movie watching experiments during the electrophysiological recordings. The movies were presented on a liquid-crystal display or an organic light-emitting diode TV within a rectangular frame of 10.4° wide and 7.6° high. Eye position was calibrated at the beginning of each session and as needed during subsequent intertrial intervals. One monkey viewed the movie without liquid reward, whereas the other eight received a water or juice reward for directing gaze to the movie, defined as a large “fixation window” of 10.5°.
Electrophysiology: Fixation on flashing images
Six of nine electrophysiology subjects underwent another set of passive fixation experiment with images of different categories on each day before their movie watching experiments. Subjects viewed a stimulus set consisting of 60 images containing sets of face categories (monkey faces and human faces) and nonface categories (objects and scenes). Images were 12° in width and randomly presented for 100 ms with 200-ms blanks. Subjects were rewarded every six presentations of images for maintaining their gaze within 1.2° to 2° in radius of the fixation point (size, 0.2° radius).
Data acquisition
Whole-brain fMRI
Structural and functional images were acquired in a 4.7-T, 60-cm vertical scanner (Bruker Biospec, Ettlingen, Germany) equipped with a Bruker S380 gradient coil in the Neurophysiology Imaging Facility Core [NIMH, National Institute of Neurological Disorders and Stroke (NINDS), National Eye Institute (NEI)]. From two female monkeys, we collected whole-brain images with an eight-channel transmit and receive radiofrequency coil system (Rapid MR International, Columbus, OH). Functional EPI scans were collected as 40 sagittal slices covering the whole brain with an in-plane resolution of 1.5 mm by 1.5 mm and a slice thickness of 1.5 mm. Repetition time and echo time were 2.4 s and 12 ms, respectively. Monocrystalline iron oxide nanoparticles, a T2* contrast agent, was administered before the start of EPI data acquisition. Subjects participated in multiple fMRI scans (28–40) of free viewing of three 5-min movies across multiple days. All aspects of the task related to timing of stimulus presentation, eye position monitoring, and reward delivery were controlled by custom software courtesy of David Sheinberg (Brown University, Providence, RI) running on a QNX computer. Horizontal and vertical eye positions were recorded using an MR-compatible infrared camera (MRC Systems, Heidelberg, Germany) fed into an eye-tracking system (SensoMotoric Instruments GmbH, Teltow, Germany) at the sampling rate of 200 Hz.
Electrophysiological recordings from cortical face patches
From nine monkeys, electrophysiological recordings were obtained from bundles of 32, 64, or 128 NiCr microwires chronically implanted in one or more face patches. The microwire electrodes were designed and initially constructed by I. Bondar (Institute of Higher Nervous Activity and Neurophysiology, Moscow, Russia) and subsequently manufactured commercially (Microprobes). Data were collected using either a Multichannel Acquisition Processor with 32-channel capacity (Plexon) or a neural recording system with 128-channel capacity (Tucker-Davis Technologies). All aspects of the task related to timing of stimulus presentation, eye position monitoring, and reward delivery were controlled either by a custom software running on a QNX computer [courtesy of D. Sheinberg (Brown University, Providence, RI)] or by NIMH MonkeyLogic (42). Horizontal and vertical eye positions were recorded using an infrared camera-tracking system (EyeLink; SR Research). Event codes, eye positions, and a photodiode signal were stored to a hard disk using OpenEX software (Tucker Davis Technologies).
Data analysis
Preprocessing of fMRI data
All fMRI data were analyzed using the AFNI/SUMA software package (43) (https://afni.nimh.nih.gov/afni) and custom-written MATLAB code (MathWorks, Natick, MA). Raw images were first converted into AFNI data file format. Each scan underwent slice-timing correction, correction for static magnetic field inhomogeneities using the PLACE algorithm (44), motion correction (AFNI function 3dvolreg), and between-session spatial registration to a single EPI reference scan. The first seven time frames of each scan were discarded for T1 stabilization, followed by removal of low-order temporal trends (polynomials up to the fifth order). The time series of each voxel was converted to units of percent signal change by subtracting, and then dividing by, the mean intensity at each voxel over the course of each 5-min scan. The high-resolution anatomy volume was skull-stripped and normalized using AFNI and then imported into CARET (45). Within CARET, surface and flattened maps were generated from a white matter mask and exported to SUMA for viewing.
Preprocessing of electrophysiology data
Individual spikes were extracted off-line from the broadband signal (2.5 to 8 kHz) after band-pass filtering between 300 and 5000 Hz for spikes, using the OfflineSorter software package (Plexon) or WaveClus spike-sorting package (46). We identified single units longitudinally across 2 to 5 days based on multiple criteria, including the similarity in waveform shapes and interspike interval histograms across days, and the distinctive visual response pattern generated by isolated spikes as a neural “fingerprint,” which we previously found in this area of cortex. Further details on spike sorting and longitudinal identification of neurons across days can be found in (23) and (37). In total, we isolated 389 single units from four face patches: n = 33 from the patch ML (monkey Dv), n = 224 from AF (48 from monkey To, 5 from Ro, 16 from Si, 106 from Sp, and 49 from Mo), n = 77 from pAM (27 from monkey Ma, 50 from Wa), and n = 55 from aAM (13 from monkey Dn, 42 from Mo).
Computation of single-unit functional maps of individual neurons
For each of the 389 neurons recorded from four face patches, we computed whole-brain functional maps, where the value of each voxel was the correlation coefficient between its fMRI time course and the single neuron’s time course (Fig. 1D). The detailed methods are described in a previous publication (24). Briefly, for single-unit responses, we down-sampled each single-unit response to match the fMRI temporal resolution (2.4 s), averaged time courses across trials for a given 5-min movie, convolved a generic hemodynamic impulse response function to the averaged and normalized (i.e., centered to the mean) time course, and then concatenated the convolved neuronal time courses from the three different movies, resulting in a single 15-min time series for each neuron. A separate analysis of interneuronal correlation confirmed that this fMRI temporal resolution did not deteriorate the nascent differences across neurons existing at a finer neuronal time scale (fig. S2). For fMRI responses, we averaged the time series across all trials for each 5-min movie and then concatenated across three movies, resulting in 15-min (i.e., 375 volumes) time series of each voxel in the whole brain. After this preparation of the time courses, for each single unit, we computed the Spearman’s rank correlation coefficients between the neuronal time course and fMRI time courses of all the voxels (n = 27,113) in the whole brain (fig. S3; see also fig. S4 for comparison of the maps using each 5-min movie and using concatenated movies).
Principal components analysis on neuronal time series
To assess the diverse nature of movie-driven responses of the population of neurons, we applied the principal components analysis on neuronal time series of 15 min. Specifically, for each neuron, we computed the number of spikes in a nonoverlapping time bin of 2.4 s from a single trial for a 5-min movie (i.e., 125 data points) and then averaged the time series across trials. Then, we concatenated the averaged time course across three movies, generating a single 15-min time series for each neuron (i.e., 375 data points). Each neuron’s time series was then converted into z score by subtracting its mean and dividing by SD. The matrix of the normalized time series from the whole population (neuron × time) was then fed into the pca function of MATLAB (pca.m), where singular value decomposition was applied. The projection value on the first principal component axis was used to sort the neurons along the first principal component as shown in fig. S1A.
Computation of face selectivity
For 254 neurons (n = 33 from ML, n = 89 from AF, n = 77 from pAM, and n = 55 from aAM) recorded from six of the nine electrophysiology subjects that underwent passive fixation experiments on different categories of images, we evaluated the categorical face selectivity (3, 47–49). First, responses to each image exemplar were calculated as the average spike rate in a fixed window of 0 to 200 ms after stimulus onset and baseline-subtracted using the average firing rate in a window of −50 to 0 ms from stimulus onset across all image exemplars. Then, we computed a face selectivity index (FSI) from the mean responses to faces and nonface objects following the conventions of previous studies (48, 49) as below
where FSI varies between −1 and 1, with additional condition of FSI = −FSI when and to incorporate inhibitory face-selective response. When the FSI of a neuron is larger than 0.33 (i.e., twice larger excitatory or inhibitory responses to the images of faces than the images of nonface objects), the neuron is defined as “face selective” (e.g., neurons marked with black in figs. S1A and S8D, example neurons marked with asterisks in Fig. 2B and fig. S5, and neurons included for Fig. 3). In total, 166 of 254 neurons (65%) were determined face selective (n = 21 from ML, n = 52 from AF, n = 70 from pAM, and n = 23 from aAM).
Clustering of single units based on the whole-brain correlation maps
To group the neurons on the basis of their whole-brain movie-driven correlation pattern irrespective of their recording location, we applied a K-means clustering algorithm on the matrix of correlation values between the face-selective neurons (n = 166) and within-brain voxels (n = 27,113). We used the kmeans.m function in MATLAB’s Statistics Toolbox with the squared Euclidean distance metric. We repeated the clustering procedure 100 times each for K values ranging from 2 to 40, and for each repetition for each K, we computed the percentage of variance explained by the clustering (i.e., the between-clusters sum of squares relative to the total sum of squares) (fig. S7A). As the K value increased, there was an asymptotic increase in explained variance as expected, but no clear point of concavity. By computing the amount of increase in explained variance that we gain as we increase K (i.e., difference in explained variance between K − 1 and K; fig. S7B), we found that there was much less gain after increasing K from 5 to 6 (arrow in fig. S7B). Because K-means clustering is not a deterministic algorithm but a stochastic one, we evaluated the stability of the clustering by computing the SD of explained variance across the 100 repetitions (fig. S7C). The variability of the explained variance showed an asymptotic increase as K increases as expected. However, for the case of K = 6, it started to become more stable (i.e., less variable) and showed the most stability comparing to the cases of K’s around it. On the basis of these two additional factors (fig. S7), we determined that six clusters, or groups of neurons, were appropriate to summarize this population of neurons. For the entire population of neurons (n = 389), we performed identical procedure on clustering and selection of the number of clusters (fig. S8, A to C), resulted in determining 10 clusters were appropriate to summarize the entire population of all the neurons irrespective of their selectivity for faces (fig. S8, D and E).
Delineation of fROIs on cortex
To summarize the correlation profile of each neuron with cortical regions, we delineated a set of 37 fROIs on the cortex of one fMRI animal (fig. S6). For a broad and fair inclusion of cortical regions that are correlated with face patch neurons regardless of their recording sites, we first computed which cortical regions are correlated with face patch neurons higher than the chance level (24). More specifically, for each voxel, the fraction of neurons that exhibit an absolute correlation value higher than 0.3 is computed and mapped as shown in the fig. S6A. The population map of fraction of highly correlated neurons, where the fraction is computed using the total number of neurons (n = 389), provided a useful and complementary guidance on which cortical regions are correlated with the cells that we recorded. Because the fraction of neuron map has no bearing on which voxels are correlated together (i.e., specific patterns of correlation or which voxels can be grouped together as one fROI), we additionally considered the repeated patterns of correlation observed across individual neurons. We first used SUMA to draw the ROIs on the cortical surface (fig. S6B) and then used AFNI for a necessary correction of the voxels for each ROI in three-dimensional slices (fig. S6C). The resulting 37 fROIs covered the occipital, temporal, parietal, and frontal lobes. Most of the fROIs overlap with multiple cytoarchitectonic areas, the correspondence of which has been established by a separate registration of this fMRI animal’s brain to D99 atlas using AFNI (see fig. S6C for correspondence with atlas), as our main purpose of the ROI delineation was an effective summary or description of each neuron’s cortical correlation profiles.
Acknowledgments
We thank K. Smith and J. Hong for assistance with data collection and members of the Leopold laboratory for helpful comments. Functional and anatomical MRI scanning was carried out in the Neurophysiology Imaging Facility Core (NIMH, NINDS, NEI). This work used the computational resources of the NIH High Performance Computing Core (https://hpc.nih.gov/). Parts of figures are created with BioRender.com.
Funding: This study was supported by the Intramural Research Program of the NIMH (ZIAMH002838 and ZIAMH002898) and Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare grant HI14C1220 (S.H.P.).
Author contributions: Conceptualization: S.H.P. and D.A.L. Data curation: S.H.P. Formal analysis: S.H.P., K.W.K., B.E.R., E.N.W., and D.B.T.M. Funding acquisition: D.A.L. Investigation: K.W.K., B.E.R., E.N.W., and D.B.T.M. Methodology: S.H.P., K.W.K., B.E.R., and D.B.T.M. Supervision: D.A.L. Visualization: S.H.P. Writing—original draft: S.H.P. and D.A.L. Writing—review and editing: S.H.P., K.W.K., B.E.R., E.N.W., D.B.T.M., and D.A.L.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The fMRI data used in this study are part of the shared NIMH dataset (D.A.L. and B.E.R.) through the Primate Data Exchange (PRIME-DE: http://fcon_1000.projects.nitrc.org/indi/PRIME/nimh1.html). Movies used in the study along with hand-coded visual features are available for download at https://doi.org/10.5281/zenodo.4044578. Other data used to draw the conclusions in the paper are available for download at https://doi.org/10.6084/m9.figshare.17131649.v2.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
REFERENCES AND NOTES
- 1.Grimaldi P., Saleem K. S., Tsao D., Anatomical connections of the functionally defined "face patches" in the macaque monkey. Neuron 90, 1325–1342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moeller S., Freiwald W. A., Tsao D. Y., Patches with links: A unified system for processing faces in the macaque temporal lobe. Science 320, 1355–1359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao D. Y., Freiwald W. A., Tootell R. B. H., Livingstone M. S., A cortical region consisting entirely of face-selective cells. Science 311, 670–674 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsao D. Y., Freiwald W. A., Knutsen T. A., Mandeville J. B., Tootell R. B. H., Faces and objects in macaque cerebral cortex. Nat. Neurosci. 6, 989–995 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanwisher N., McDermott J., Chun M. M., The fusiform face area: A module in human extrastriate cortex specialized for face perception. J. Neurosci. 17, 4302–4311 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung C. C., Yen C. C., Ciuchta J. L., Papoti D., Bock N. A., Leopold D. A., Silva A. C., Functional mapping of face-selective regions in the extrastriate visual cortex of the marmoset. J. Neurosci. 35, 1160–1172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner K. S., Grill-Spector K., The evolution of face processing networks. Trends Cogn. Sci. 19, 240–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanwisher N., Yovel G., The fusiform face area: A cortical region specialized for the perception of faces. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 2109–2128 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao D. Y., Moeller S., Freiwald W. A., Comparing face patch systems in macaques and humans. Proc. Natl. Acad. Sci. U.S.A. 105, 19514–19519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freiwald W. A., Tsao D. Y., Functional compartmentalization and viewpoint generalization within the macaque face-processing system. Science 330, 845–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher C., Freiwald W. A., Contrasting specializations for facial motion within the macaque face-processing system. Curr. Biol. 25, 261–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taubert J., Japee S., Murphy A. P., Tardiff C. T., Koele E. A., Kumar S., Leopold D. A., Ungerleider L. G., Parallel processing of facial expression and head orientation in the macaque brain. J. Neurosci. 40, 8119–8131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hesse J. K., Tsao D. Y., The macaque face patch system: A turtle’s underbelly for the brain. Nat. Rev. Neurosci. 21, 695–716 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Freiwald W. A., The neural mechanisms of face processing: Cells, areas, networks, and models. Curr. Opin. Neurobiol. 60, 184–191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitcher D., Ungerleider L. G., Evidence for a third visual pathway specialized for social perception. Trends Cogn. Sci. 25, 100–110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon D. B. T., Russ B. E., Elnaiem H. D., Kurnikova A. I., Leopold D. A., Single-unit activity during natural vision: Diversity, consistency, and spatial sensitivity among AF face patch neurons. J. Neurosci. 35, 5537–5548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russ B. E., Leopold D. A., Functional MRI mapping of dynamic visual features during natural viewing in the macaque. Neuroimage 109, 84–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasson U., Nir Y., Levy I., Fuhrmann G., Malach R., Intersubject synchronization of cortical activity during natural vision. Science 303, 1634–1640 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Mantini D., Hasson U., Betti V., Perrucci M. G., Romani G. L., Corbetta M., Orban G. A., Vanduffel W., Interspecies activity correlations reveal functional correspondence between monkey and human brain areas. Nat. Methods 9, 277–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huth A. G., Lee T., Nishimoto S., Bilenko N. Y., Vu A. T., Gallant J. L., Decoding the semantic content of natural movies from human brain activity. Front. Syst. Neurosci. 10, 81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. A. Leopold, J. F. Mitchell, W. A. Freiwald, in Evolution of Nervous Systems, J. H. Kaas, Ed. (Academic Press, ed. 2, 2017), pp. 203–235. [Google Scholar]
- 22.Leopold D. A., Park S. H., Studying the visual brain in its natural rhythm. Neuroimage 216, 116790 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bondar I. V., Leopold D. A., Richmond B. J., Victor J. D., Logothetis N. K., Long-term stability of visual pattern selective responses of monkey temporal lobe neurons. PLOS ONE 4, e8222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park S. H., Russ B. E., McMahon D. B. T., Koyano K. W., Berman R. A., Leopold D. A., Functional subpopulations of neurons in a macaque face patch revealed by single-unit fMRI mapping. Neuron 95, 971–981.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logothetis N. K., Eschenko O., Murayama Y., Augath M., Steudel T., Evrard H. C., Besserve M., Oeltermann A., Hippocampal-cortical interaction during periods of subcortical silence. Nature 491, 547–553 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Barson D., Hamodi A. S., Shen X., Lur G., Constable R. T., Cardin J. A., Crair M. C., Higley M. J., Simultaneous mesoscopic and two-photon imaging of neuronal activity in cortical circuits. Nat. Methods 17, 107–113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholvinck M. L., Maier A., Ye F. Q., Duyn J. H., Leopold D. A., Neural basis of global resting-state fMRI activity. Proc. Natl. Acad. Sci. U.S.A. 107, 10238–10243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra A., Snyder A. Z., Blazey T., Raichle M. E., Lag threads organize the brain’s intrinsic activity. Proc. Natl. Acad. Sci. U.S.A. 112, E2235–E2244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent J. L., Patel G. H., Fox M. D., Snyder A. Z., Baker J. T., van Essen D. C., Zempel J. M., Snyder L. H., Corbetta M., Raichle M. E., Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83–86 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Caspari N., Janssens T., Mantini D., Vandenberghe R., Vanduffel W., Covert shifts of spatial attention in the macaque monkey. J. Neurosci. 35, 7695–7714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sani I., McPherson B. C., Stemmann H., Pestilli F., Freiwald W. A., Functionally defined white matter of the macaque monkey brain reveals a dorso-ventral attention network. eLife 8, e40520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder C. E., Wilson D. A., Radman T., Scharfman H., Lakatos P., Dynamics of active sensing and perceptual selection. Curr. Opin. Neurobiol. 20, 172–176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fusi S., Miller E. K., Rigotti M., Why neurons mix: High dimensionality for higher cognition. Curr. Opin. Neurobiol. 37, 66–74 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Huk A. C., Katz L. N., Yates J. L., The role of the lateral intraparietal area in (the study of) decision making. Annu. Rev. Neurosci. 40, 349–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato T., Uchida G., Lescroart M. D., Kitazono J., Okada M., Tanifuji M., Object representation in inferior temporal cortex is organized hierarchically in a mosaic-like structure. J. Neurosci. 33, 16642–16656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peck C. J., Salzman C. D., The amygdala and basal forebrain as a pathway for motivationally guided attention. J. Neurosci. 34, 13757–13767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon D. B. T., Bondar I. V., Afuwape O. A. T., Ide D. C., Leopold D. A., One month in the life of a neuron: Longitudinal single-unit electrophysiology in the monkey visual system. J. Neurophysiol. 112, 1748–1762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koyano K. W., Jones A. P., McMahon D. B. T., Waidmann E. N., Russ B. E., Leopold D. A., Dynamic suppression of average facial structure shapes neural tuning in three macaque face patches. Curr. Biol. 31, 1–12.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khandhadia A. P., Murphy A. P., Romanski L. M., Bizley J. K., Leopold D. A., Audiovisual integration in macaque face patch neurons. Curr. Biol. 31, 1826–1835.e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russ B. E., Petkov C. I., Kwok S. C., Zhu Q., Belin P., Vanduffel W., Hamed S. B., Common functional localizers to enhance NHP & cross-species neuroscience imaging research. Neuroimage 237, 118203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russ B. E., Kaneko T., Saleem K. S., Berman R. A., Leopold D. A., Distinct fMRI responses to self-induced versus stimulus motion during free viewing in the macaque. J. Neurosci. 36, 9580–9589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang J., Mitz A. R., Murray E. A., NIMH MonkeyLogic: Behavioral control and data acquisition in MATLAB. J. Neurosci. Methods 323, 13–21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox R. W., AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Xiang Q.-S., Ye F. Q., Correction for geometric distortion and N/2 ghosting in EPI by phase labeling for additional coordinate encoding (PLACE). Magn. Reson. Med. 57, 731–741 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Van Essen D. C., Drury H. A., Dickson J., Harwell J., Hanlon D., Anderson C. H., An integrated software suite for surface-based analyses of cerebral cortex. J. Am. Med. Inform. Assoc. 8, 443–459 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quiroga R. Q., Nadasdy Z., Ben-Shaul Y., Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural Comput. 16, 1661–1687 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Bell A. H., Malecek N. J., Morin E. L., Hadj-Bouziane F., Tootell R. B. H., Ungerleider L. G., Relationship between functional magnetic resonance imaging-identified regions and neuronal category selectivity. J. Neurosci. 31, 12229–12240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aparicio P. L., Issa E. B., DiCarlo J. J., Neurophysiological organization of the middle face patch in macaque inferior temporal cortex. J. Neurosci. 36, 12729–12745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Issa E. B., DiCarlo J. J., Precedence of the eye region in neural processing of faces. J. Neurosci. 32, 16666–16682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8