Abstract
Global crop production is greatly reduced by vascular diseases. These diseases include bacterial blight of rice and crucifer black rot caused by Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas campestris pv. campestris (Xcc). The molecular mechanisms that activate vascular defense against such pathogens remains underexplored. Here, we show that an Arabidopsis MAPK phosphatase 1 (MKP1) mutant has increased host susceptibility to the adapted pathogen Xcc and is compromised in nonhost resistance to the rice pathogen Xoo. MKP1 regulates MAPK-mediated phosphorylation of the transcription factor MYB4 that negatively regulates vascular lignification through inhibiting lignin biosynthesis. Induction of lignin biosynthesis is, therefore, an important part of vascular-specific immunity. The role of MKP-MAPK-MYB signaling in lignin biosynthesis and vascular resistance to Xoo is conserved in rice, indicating that these factors form a tissue-specific defense regulatory network. Our study likely reveals a major vascular immune mechanism that underlies tissue-specific disease resistance against bacterial pathogens in plants.
Protein phosphorylation signaling mediates vascular-specific defense in plants.
INTRODUCTION
Plant vascular system that is crucial for plant growth and development (1). A number of pathogens invade the plant vascular system, causing severe disease symptoms and substantial reductions in crop yield. Vascular pathogens include Xanthomonas oryzae pv. oryzae (Xoo), which causes rice bacterial blight (2), Xanthomonas campestris pv. campestris (Xcc) that underlies crucifer black rot, Ralstonia solanacearum, which results in bacterial wilt, and Verticillium dahliae responsible for cotton wilt. These pathogens can infect plants through wounds, leaf hydathode water pores, or roots (3), as observed for Xoo (2), Xcc (4), R. solanacearum (5, 6), and V. dahlia (7, 8). They then multiply in and block xylem vessels, resulting in systemic spread, tissue damage, and plant death (2, 9). However, current understanding of plant-pathogen interactions relies mainly on pathogens that infect leaf mesophyll tissues or broad-spectrum disease resistance to multiple pathogens with different lifestyles [as exampled in (10–12)]. Despite the agricultural impact of vascular pathogens, little is understood about the biochemical and molecular mechanisms underlying plant vascular immunity. Compared to other immune responses (13–15), our knowledge of plant defenses that target pathogens in the vasculature remains limited (16).
Coevolution of plants and vascular pathogens has likely shaped plant vascular immunity so that this defense system has both common and unique features compared to mesophyll pathogen defense (16). Tissue-specific infection has been reported for Xanthomonadaceae (17). For example, vascular xanthomonads colonize water-transporting xylem and extend along with the vascular system, while nonvascular Xanthomonas invade the mesophyll and cause localized symptoms (18, 19). The genomes of Xoo and Xoc (X. oryzae pv. oryzicola), two representative xylem and mesophyll xanthomonads, encode secreted transcription activator–like effectors (20, 21). These effectors can activate either host susceptibility (S) genes, leading to virulence, or resistance (R) genes resulting in tissue-specific resistance. It has been reported that vascular Xanthomonas pathogens secrete cell wall–degrading enzymes during infection, such as cellulases (clsA and cbsA), xylanase (xyn), pectinase (pglA), and esterase (lipA) (22–24). These enzymes also induce host immune responses. cbsA, a cellobiohydrolase that degrades cell walls, was found to be enriched in vascular Xanthomonas subgroups and lost in nonvascular Xanthomonas species (25). The evolution of this enzyme may have promoted a switch from nonvascular to vascular colonization, suggesting that a single locus can shift tissue-specific infection in closely related pathogens (25, 26). This divergence of pathogen lifestyles may have shaped tissue-specific plant immune responses. This raises a fundamental question: How do different resistance genes activate tissue-specific immune responses upon detecting pathogens with different lifestyles?
As one of the most abundant cell wall components and an important secondary metabolite in plant growth and development, lignin has been implicated in effective defense responses in plants. Lignin builds a physical barrier that blocks vessels or fills extracellular spaces, thus impeding bacterial multiplication and movement and acting as a key factor in vascular defense (27–29). Arabidopsis, soybean, and rice genes linked to lignin biosynthesis were up-regulated during pathogen infection, resulting in increased lignin levels and reinforcing the cell wall–based defense (27–29). Consistently, many lignin biosynthesis genes are highly expressed in plant vascular bundles (30). Moreover, previous studies have shown that several MYB family transcription factors regulate expression of lignin biosynthesis genes (30–32). Notably, MYB4 represses lignin biosynthesis, as seen in studies examining the plant response to ultraviolet light (33–35). However, the molecular links between the plant immune machinery and lignin-based vascular defense remain largely unexplored.
The vascular pathogens Xoo and Xcc and the leaf mesophyll cell pathogen Xoc serve as comparative models for investigating vascular-specific resistance. Here, we report that mitogen-activated protein kinase (MAPK) phosphatase 1 (MKP1) and its target kinases MPK3 and MPK6 form a signaling cascade that promotes vascular defense in both Arabidopsis and rice. We found that nonhost resistance (NHR) to Xoo is lost in Arabidopsis mkp1 mutant, leading to Xoo growth in the leaf veins. The corresponding rice Osmkp1 mutant exhibited enhanced host susceptibility to Xoo. Pathogen infection attenuates the MAPK phosphorylation pathway by inducing the MPK1 gene, leading to inactivation of the MYB transcription factors that negatively regulate lignin biosynthesis genes. This regulatory cascade enhances lignin production and, thus, disease resistance against vascular pathogens. Our study uncovers a previously unappreciated molecular mechanism that contributes to plant vascular defense that is conserved in different plant species.
RESULTS
MKP1 controls vascular defense in Arabidopsis
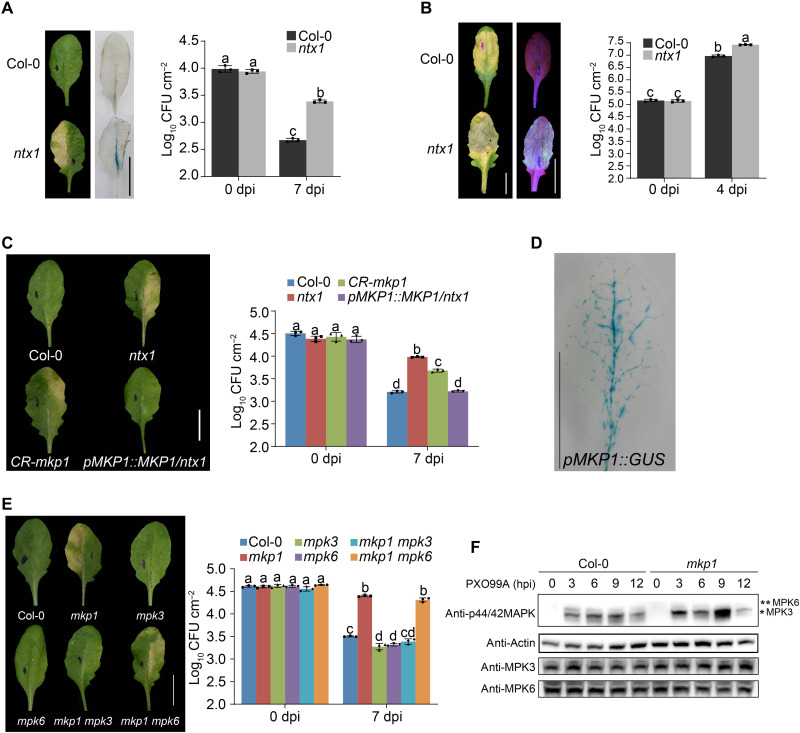
To uncover the mechanism(s) that regulate plant vascular defense, we screened an ethylmethane sulfonate–mutagenized Arabidopsis Col-0 population challenged with Xoo, which does not usually infect Arabidopsis. The ntx1 (nonhost disease resistance to Xoo 1) mutant developed disease symptoms indicating loss of NHR to the Xoo strain PXO99A (Fig. 1A). To examine Xoo growth in ntx1 leaves, we created a beta-glucuronidase (GUS)-expressing Xoo strain. Notably, PXO99A-GUS colonized and proliferated in ntx1 leaf veins (Fig. 1A), suggesting that vascular defense against Xoo is defective in ntx1. In addition to compromised NHR to the nonadapted pathogen Xoo, the ntx1 mutant was more susceptible to the adapted vascular pathogen Xcc (Fig. 1B).
Fig. 1. MKP1 mutants have reduced vascular resistance to Xoo and Xcc.
(A) PXO99A-GUS populates the leaf veins of ntx1 plants. Disease symptoms, GUS staining, and Xoo levels were measured in wild-type Col-0 and ntx1/mkp1 mutant plants 7 days after infiltration with Xoo strain PXO99A and GUS-labeled PXO99A-GUS [2 × 107 colony-forming unit (CFU) ml−1]. (B) Disease symptoms, green fluorescent protein (GFP) fluorescence, and growth of Xcc in Col-0 and ntx1 at 4 dpi with GFP-labeled Xcc8004-GFP (1 × 107 CFU ml−1). (C) Genetic complementation of MKP1 in ntx1 plants. Disease symptoms and bacterial populations were recorded at 7 days post inoculation (dpi) with PXO99A in Col-0, ntx1, CR-mkp1, and pMKP1::MKP1/ntx1. (D) GUS staining of pMKP1::GUS transgenic plants demonstrated that MKP1 was mainly expressed in the vascular bundles. Scale bar, 1 cm. (E) The nonhost Xoo susceptibility phenotype of mkp1 was restored by a mutation in MPK3 but not MPK6, showing that the double-mutant mkp1 mpk3 restored the wild-type NHR to Xoo, 7 dpi with PXO99A. Data were shown as means ± SD (n = 3). Letters indicate significant differences (P < 0.05) determined by two-way analysis of variance (ANOVA) with Tukey’s test. Scale bar, 1 cm. Experiments were repeated three times with similar results. (F) Kinase activation of MPK3 but not MPK6 was induced by Xoo inoculation. Proteins were prepared from leaf samples for in-gel kinase assay at the indicated times. hpi, hours post inoculation.
We next determined whether the loss of vascular resistance in the ntx1 mutant is specific to vascular pathogens rather than loss of common NHR or basal defense. Xanthomonas citri subsp. citri (Xac) and Xanthomonas campestris pv. vesicatoria (Xcv) caused citrus bacterial canker and pepper bacterial spot disease, respectively (36–40), the destructive diseases of citrus and pepper. We found that ntx1 was not more susceptible than wild-type Col-0 to nonhost and nonvascular Xoc, Xac, and Xcv (fig. S1, A to C). Last, we found that ntx1 plants showed enhanced basal resistance to a leaf mesophyll–specific bacterial pathogen Pseudomonas syringae pv. tomato (Pst DC3000) (fig. S2A), with marked increases in transcript abundance of the salicylic acid (SA) pathway genes ICS1 and PR1 (fig. S2, B and C). These results strongly suggest that NTX1 is specifically involved in vascular resistance rather than a more general process.
We cloned the NTX1 gene using polymerase chain reaction (PCR)–based genetic mapping coupled with genome sequencing (fig. S3A). We found that the ntx1 allele carries a G-to-A transition at nucleotide position 1416 in the MKP1 (At3G55270) gene, which encodes MKP1. This G-A transition causes an Arg241-Gln (MKP1R241Q) missense mutation in the dual-specificity phosphatase catalytic site domain (fig. S3B). Consistent with the known functions of MKP1 in plant development (41), the ntx1 plants displayed developmental defects, including decreased biomass, reduced seed setting due to abortive stamens, and larger seeds compared to the wild-type plants (fig. S3C).
To confirm the molecular identity of NTX1, we generated a pMKP1::MKP1 complementation transgene and transformed it into the ntx1 mutant. The pMKP1::MKP1 transgene fully restored vascular resistance to Xoo. We also used CRISPR-Cas9 to generate a MKP1 knockout line (CR-mkp1) in the Col-0 background and found that CR-mkp1 plants phenocopied the ntx1 mutant, displaying a loss of resistance to Xoo (Fig. 1C and fig. S3D). Together, these results link MKP1 function to vascular resistance against Xoo in Arabidopsis.
MKP1 regulates vascular resistance to Xoo by inactivating MPK3
We examined the expression of MKP1 during Xoo infection and found that MKP1 expression was induced by Xoo (fig. S4A). We also generated pMKP1::GUS fusion reporter plants and performed GUS histological analysis. Strong GUS activity was observed in vascular bundles (Fig. 1D), consistent with the idea that MKP1 functions in the vascular defense. We also carefully examined the potential cellular changes in mkp1 using transmission electron microscopy (TEM) and found that vascular bundles were smaller in leaf sections of mkp1 compared to wild-type plants, with no obvious change in cell wall thickness (fig. S4, B and C). These changes may explain the weak growth phenotype of mkp1.
On the basis of its predicted biochemical function, we hypothesized that MKP1 might regulate vascular defense by controlling MAPK activity, which plays a known role in plant immunity (42). Because MKP1 was previously shown to specially deactivate MPK3 and MPK6 via dephosphorylation (43), we analyzed the interaction of MKP1 with MPK3 and MPK6 using both the yeast two-hybrid (Y2H) and split luciferase complementation (SLC) assays. MKP1 interacted with MPK3 and MPK6 in both assays (fig. S5, A and B). Furthermore, we confirmed that MPK3 and MPK6 were dephosphorylated in vitro by MKP1 but not the catalytically inactive mutant variant MKP1R241Q [R241 is the conserved residue of the signature catalytic motif C(X)5R of the tyrosine/dual-specificity protein phosphatases] (fig. S5, C and D).
We generated mkp1 mpk3 and mkp1 mpk6 double mutants by crossing mkp1 with the single mutants mpk3 and mpk6. When inoculated with Xoo, the mkp1 mpk6 double mutant displayed mkp1-like susceptibility to Xoo, whereas the mkp1 mpk3 double mutant exhibited Col-0–like resistance to Xoo (Fig. 1E). These results suggest that MKP1 regulates vascular resistance to Xoo through MPK3, rather than MPK6 inactivation, to regulate the vascular resistance against Xoo. To test this hypothesis, we analyzed phosphorylation of MPK3 and MPK6 during Xoo infection and found that Xoo inoculation strongly induced MPK3 phosphorylation but only slightly induced MPK6 phosphorylation (Fig. 1F). Furthermore, MPK3 phosphorylation was increased in the mkp1 mutant, in contrast to MPK6 (Fig. 1G). These results suggest that MKP1-mediated dephosphorylation and inactivation of MPK3 underlies vascular resistance against Xoo.
Lignin plays a critical role in vascular resistance to Xoo
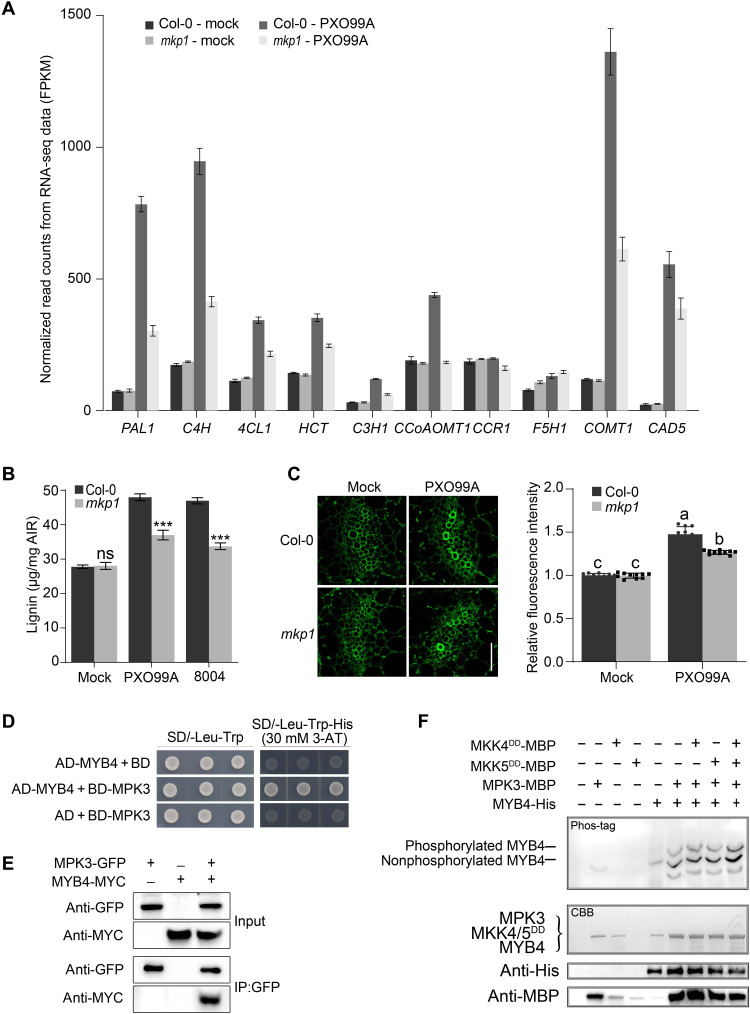
Given that mkp1 plants showed enhanced resistance to Pst DC3000 (fig. S2A), with marked increases in the transcript abundance of SA pathway genes ICS1 and PR1 (fig. S2, B and C), and that SA and ROS (reactive oxygen species) play important roles in disease resistance (44, 45), we measured the SA and ROS levels during Xoo infection. We found that SA and ROS accumulation were not significantly induced by Xoo in mkp1 plants compared to wild-type Col-0 plants (fig. S6, A to C), suggesting that the vascular defense mediated by MKP1 does not involve SA and ROS. Next, we performed an RNA transcriptome analysis of wild-type Col-0 and mkp1 mutant plants infected with PXO99A. We found that genes linked to a range of biological processes showed differential expression in mkp1 compared to the wild-type plants (fig. S7A). Almost all genes of the lignin biosynthesis pathway, including PAL1, C4H, 4CL1, and COMT1, were down-regulated in mkp1 during Xoo infection (Fig. 2A and fig. S7B). Thus, we hypothesized that reduced lignin biosynthesis might contribute to the compromised vascular defense in the mkp1 mutant. We found that, although lignin was induced by PXO99A in both the wild type and mkp1, the degree of lignin accumulation was markedly reduced in mkp1 plants (Fig. 2B). Cell wall autofluorescence and phloroglucinol staining confirmed decreased lignin accumulation in the xylems of mkp1 leaf veins (Fig. 2C and fig. S7C).
Fig. 2. MPK3 phosphorylates MYB4 to regulate lignin biosynthesis.
(A) Differential expression of lignin biosynthetic genes induced by Xoo in wild-type Col-0 and mkp1 as revealed by RNA sequencing (RNA-seq). Many lignin biosynthesis genes were less induced in mkp1 as compared with Col-0. FPKM, Fragments per kilobase of exon per million reads mapped. (B) Lignin quantification in leaves at 12 hours infiltrated with Xoo and Xcc. Data were shown as means ± SD (n = 3). Asterisks represented statistical significance (***P < 0.001, Student’s t test). ns, not significant. Experiments were repeated three times. (C) Cell wall autofluorescence and relative fluorescence intensity of Col-0 and mkp1 infiltrated with Xoo. Note that less lignification of the leaf vessel was induced by Xoo in mkp1 as compared with Col-0. Scale bar, 50 μm. Data were shown as means ± SD (n = 10). Letters indicated significant differences (P < 0.05) determined by two-way ANOVA with Tukey’s test. Experiments were repeated three times. (D and E) MPK3 interacts with MYB4, as revealed by Y2H screen (D) and coimmunoprecipitation (coIP) assay (E). SD/-Trp-Leu-His, synthetic dropout (SD) media lacking leucine, tryptophan, and histidine; SD/-Leu -Trp, SD media lacking leucine and tryptophan; 3AT, 3-aminotriazole. MPK3-GFP and MYB4-MYC in transgenic Arabidopsis plants were purified and immunodetected using anti-GFP or anti-MYC antibody. (F) MPK3 phosphorylates MYB4 in vitro. MPK3-MBP (myelin basic protein), MYB4-His, MKK4DD-MBP (T224D/S230D), and MKK5DD-MBP (T215D/S221D) were expressed in E. coli and purified for in vitro phosphorylation assays. MYB4-His was incubated at 30°C for 1 hour then separated on a Phos-tag gel. CBB, Coomassie brilliant blue staining for loading control.
MPK3-mediated phosphorylation of MYB4 negatively regulates lignin biosynthesis
The transcriptional repressor MYB4 has previously been shown to negatively regulate expression of lignin biosynthesis genes and is specifically expressed in leaf veins (fig. S7D) (30). We therefore hypothesized that MPK3 phosphorylates and activates MYB4 and that the levels of active MPK3 and MYB4 might therefore be higher in mkp1 plants than in wild-type plants. To test this hypothesis, we performed several protein-protein interaction assays, including Y2H, SLC, and in vitro pull-down, and found that MPK3-MYB4 interact (Fig. 2D and fig. S7, E and F). Coimmunoprecipitation (coIP) confirmed that MPK3 and MYB4 interact in planta (Fig. 2E).
Next, we tested whether MPK3 can phosphorylate MYB4 in vitro. The constitutively active MKK4DD (T224D/S230D)/MKK5DD (T215D/S221D) mutant strongly activates endogenous MPK3 and MPK6 (46). Coincubation of recombinant MYB4-His with MPK3–myelin basic protein (MBP) in the presence or absence of MKK4DD/MKK5DD resulted in a slow migrating MYB4-His band on Phos-tag SDS–polyacrylamide gel electrophoresis, indicating that MPK3 can phosphorylate MYB4-His (Fig. 2F).
Phosphorylation of MYB4 suppresses vascular defense by inhibiting lignin biosynthesis
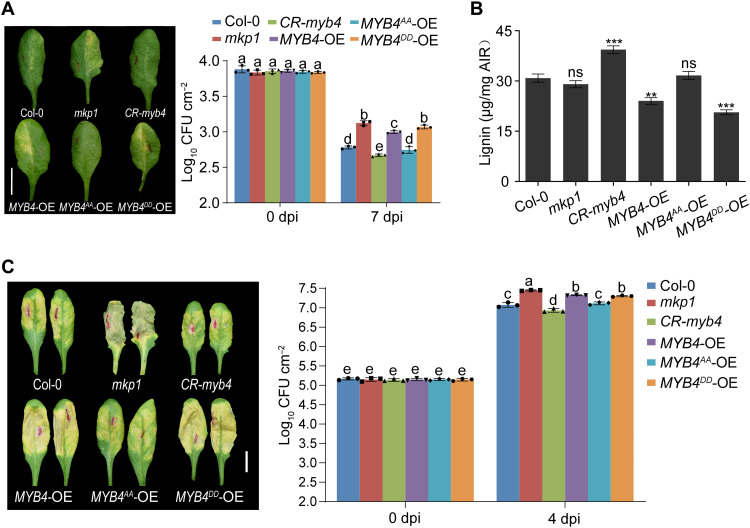
We previously predicted that MPK3 might phosphorylate MYB4 at two sites, Ser4 and Thr221 (47). Thus, we generated loss-of-phosphorylation (MYB4AA, Ser4/Thr221 mutated to Ala) and phosphomimetic (MYB4DD, Ser4/Thr221 mutated to Asp) mutant variants of MYB4. Both MYB4AA and MYB4DD could still interact with MPK3 (fig. S7G). We used CRISPR-Cas9 editing to generate an MYB4 knockout line, CR-myb4, and created transgenic lines overexpressing MYB4-Myc (MYB4-OE), MYB4AA-Myc (MYB4AA-OE), or MYB4DD-Myc (MYB4DD-OE) (fig. S8, A and B). Similar to the mkp1 mutant, NHR was compromised in both MYB4-OE and MYB4DD-OE transgenic plants in response to Xoo inoculation (Fig. 3A). We observed that the phosphorylated/active form MYB4DD was more stable than the nonphosphorylated/inactive form MYB4AA (fig. S8C). This was confirmed by following protein degradation (fig. S8D), suggesting that MYB4 might be subjected to dynamic control by protein phosphorylation and degradation.
Fig. 3. Phosphorylation of MYB4 suppresses vascular defense by inhibiting lignin biosynthesis.
(A) Disease symptoms (left) and bacterial growth (right) in Col-0, mkp1, CR-myb4, MYB4-OE, MYB4AA-OE, and MYB4DD-OE at 7 dpi with PXO99A. The wild-type MYB4 and the site mutation variants (MYB4DD and MYB4AA) were ectopically expressed (OE) as a MYC fusion protein. Scale bar, 1 cm. (B) Leaf lignin quantification indicated that lignin biosynthesis was inhibited by overexpression of MYB4 and MYB4DD but not MYB4AA. (C) MYB4 also negatively regulates resistance to Xcc. Disease symptoms (left) and bacterial growth (right) of Xcc in Col-0, mkp1, CR-myb4, MYB4-OE, MYB4AA-OE, and MYB4DD-OE at 4 dpi with Xcc8004. Scale bar, 1 cm. Data were shown as means ± SD (n = 3). Asterisks represented statistical significance (**P < 0.01 and ***P < 0.001, Student’s t test) (B). Letters indicated significant differences (P < 0.05) determined by two-way ANOVA with Tukey’s test (A to C). Experiments were repeated three times.
MYB4 is known to bind the promoter of C4H, thus repressing expression of critical rate-limiting regulator of lignin biosynthesis (33, 34). We found that C4H was up-regulated in CR-myb4 and down-regulated in MYB4-OE and MYB4DD-OE transgenic lines as compared with wild-type plants (fig. S8E). CR-myb4 lines consistently accumulated more lignin, whereas MYB4-OE and MYB4DD-OE transgenic lines accumulated less lignin than wild type (Fig. 3B). Collectively, these data indicate that the phosphorylation-mediated activation of MYB4 by MPK3 suppress lignin biosynthesis, thus compromising vascular resistance to Xoo in mkp1 plants. Similar results were also observed with Xcc inoculation (Fig. 3C). These data together strongly suggest that MYB4 negatively regulates resistance to both adapted and nonadapted vascular pathogens through inhibiting lignin biosynthesis.
OsMKP1 regulates vascular resistance in rice
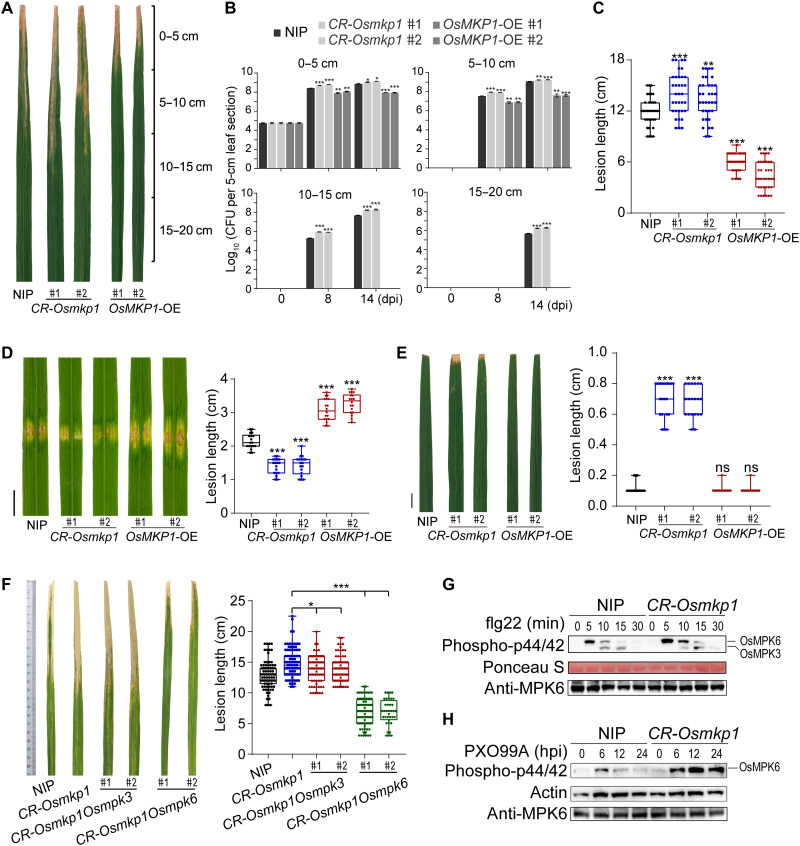
The rice genome encodes an MKP1 homolog, OsMKP1 (fig. S9A) (48). To investigate whether OsMKP1 is also involved in rice vascular defense against Xoo, we generated CRISPR-Cas9 knockout mutant (CR-Osmkp1) and OsMKP1 overexpression (OsMKP1-OE) lines (fig. S9B). Xoo proliferates and spreads within the xylem (fig. S9C). As in Arabidopsis, OsMKP1 expression was induced by Xoo (fig. S9D). Furthermore, CR-Osmkp1 plants exhibited enhanced susceptibility to Xoo in comparison with wild-type Nipponbare (NIP) plants. CR-Osmkp1 plants had also decreased lignin accumulation and decreased PR gene expression during Xoo infection (Fig. 4, A to C, and fig. S9, E and F). In contrast, OsMKP1-OE exhibited enhanced resistance to Xoo (Fig. 4, A to C), although lignin accumulation was only slightly increased (fig. S9E).
Fig. 4. OsMKP1 mediates Xoo resistance through OsMPK6 dephosphorylation.
(A and B) Population size and distribution of Xoo in leaves of 2-month-old wild-type NIP, CR-Osmkp1, and OsMKP1-OE following clip inoculation. Bacterial populations in 5-cm leaf segments were measured at 0, 8, and 14 dpi. (C) Lesion lengths of NIP, CR-Osmkp1, and OsMKP1-OE inoculated with PXO99A [optical density at 600 nm (OD600), 1.0] at 14 dpi. (D) Lesions (left) and statistical analysis (right) of NIP, CR-Osmkp1, and OsMKP1-OE inoculated with Xoc strains RS85. OsMKP1 negatively regulated resistance to Xoc. Scale bar, 1 cm. (E) Lesions and statistical analysis of NIP, CR-Osmkp1, and OsMKP1-OE inoculated with Xcc8004. (F) Disease symptoms of representative leaves (left) and lesion lengths (right) of NIP, CR-Osmkp1, CR-Osmkp1Osmpk3, and CR-Osmkp1Osmpk6 inoculated with PXO99A at 14 dpi. Data were shown as means ± SD; n > 20 (C to F) and n = 3 (B). Asterisks represented statistical significance (*P < 0.05, **P < 0.01, and ***P < 0.001, Student’s t test) (B to F). Experiments were repeated three times with similar results. (G) OsMPK3 and OsMPK6 activation by flg22 treatment. Two-week-old seedlings of NIP and CR-Osmkp1 were treated with flg22 (1 μM) for the indicated times for protein preparation. The phosphorylated OsMPK3 and OsMPK6 were detected using the anti-p44/42 MPK antibody, with Ponceau S staining as loading control. (H) MPK6 was stronger activated in CR-Osmkp1 infected by PXO99A as compared with NIP.
Xoo invades systemically through the xylem tissue, while Xoc is a nonvascular pathogen and colonizes the intercellular spaces of mesophyll parenchyma. To determine whether MKP1-mediated resistance is also specific to vascular pathogens in rice, we inoculated the rice plants with Xoc. CR-Osmkp1 plants showed enhanced resistance, whereas OsMKP1-OE plants displayed decreased resistance to Xoc, compared with wild-type NIP plants (Fig. 4D). In addition, similar to Arabidopsis, CR-Osmkp1 was more susceptible to the nonadapted vascular pathogen Xcc8004 (Fig. 4E). Therefore, OsMKP1 plays a conserved role in the vascular defense in the dicot Arabidopsis and the monocot rice.
Next, we investigated the role of OsMKP3 and OsMPK6 in rice vascular defense. Both factors interact with OsMKP1 (fig. S10, A and B). We used CRISPR-Cas9–based editing to create CR-Osmkp1Osmpk3 and CR-Osmkp1Osmpk6 double knockout mutants (fig. S10C). CR-Osmkp1Osmpk6 plants showed significantly enhanced resistance to Xoo, whereas CR-Osmkp1Osmpk3 had only slightly increased disease resistance in comparison with the CR-Osmkp1 single mutant (Fig. 4F). These data suggest that rice OsMPK3 and OsMPK6 both negatively regulate disease resistance to Xoo, with OsMPK6 playing a more prominent role in regulation. Consistently, an immunoblot analysis of flg22/PXO99A-induced phosphorylation of OsMPK3 and OsMPK6 revealed that OsMPK6 exhibited a greater increase in phosphorylation than OsMPK3 in CR-Osmkp1 plants (Fig. 4, G and H) (49). These results suggest that OsMKP1 regulates resistance to Xoo in rice, mainly through dephosphorylating/inactivating OsMPK6.
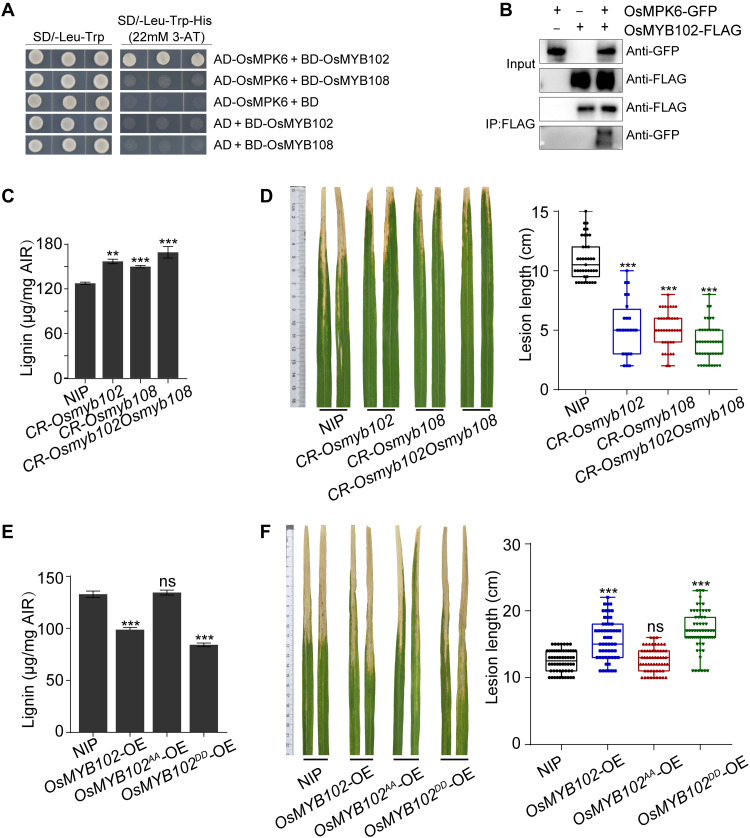
OsMYB102 and OsMYB108 negatively regulate bacterial blight resistance
The rice genome encodes two MYB4 homologs, OsMYB102 and OsMYB108 (fig. S11, A and B), which were reported to negatively regulate lignin biosynthesis (50). We found that the transcript levels of OsMYB102 and OsMYB108 were unchanged in CR-Osmkp1 or during Xoo infection (fig. S11C). Y2H and coIP assays indicated that OsMYB102 interacted with OsMPK6 (Fig. 5, A and B), suggesting that OsMYB102 might be an OsMPK6 substrate. To evaluate the function of OsMYB102 and OsMYB108 in Xoo resistance, we generated single knockout mutants CR-Osmyb102 and CR-Osmyb108 and CR-Osmyb102Osmyb108 double mutant (fig. S11D). These mutants accumulated higher levels of lignin than wild-type NIP plants (Fig. 5C) and exhibited significantly increased resistance to Xoo, with CR-Osmyb102 Osmyb108 showing stronger resistance (Fig. 5D). Thus, OsMYB102 and OsMYB108 function redundantly in the negative regulation of lignin biosynthesis and Xoo resistance in rice.
Fig. 5. OsMYB102 and OsMYB108 negatively regulate lignin biosynthesis and resistance to Xoo.
(A) OsMPK6 interacts with OsMYB102 in a Y2H assay. (B) coIP assay of the OsMPK6-OsMYB102 interaction in N. benthamiana. Fusion proteins were purified and immunodetected using anti-GFP or anti-FLAG antibody. (C) Lignin quantification in leaves of NIP, CR-Osmyb102, CR-Osmyb108, and CR-Osmyb102Osmyb108, indicating that OsMYB102 and OsMYB108 negatively regulate lignin production. (D) Disease resistance of representative lines (left) and lesion lengths (right) of 2-month-old NIP, CR-Osmyb102, CR-Osmyb108, and CR-Osmyb102Osmyb108 at 14 dpi with PXO99A. (E and F) Phosphorylation of OsMYB102 negatively regulates lignin biosynthesis and Xoo resistance. Lesions and bacterial growth of wild-type NIP, OsMYB102-OE, OsMYB102AA-OE, and OsMYB102DD-OE inoculated with PXO99A at 14 dpi. Data were shown as means ± SD; n = 3 (C and E) and n > 20 (D and F). Asterisks represented statistical significance (**P < 0.01 and ***P < 0.001, Student’s t test). Experiments were repeated three times with similar results.
We further generated OsMYB102-OE, OsMYB102AA-OE (Ser4/Ser169 mutated to Ala), and OsMYB102DD-OE (Ser4/Ser169 mutated to Asp) transgenic plants (fig. S11E). Similar to our data in Arabidopsis, OsMYB102-OE and OsMYB102DD-OE accumulated significantly less lignin (Fig. 5E) and were more susceptible to Xoo compared to the wild-type NIP control (Fig. 5F). Thus, we conclude that the MKP1-MPK3/6-MYB cascade plays a conserved role in lignin-based vascular defense in Arabidopsis and rice.
DISCUSSION
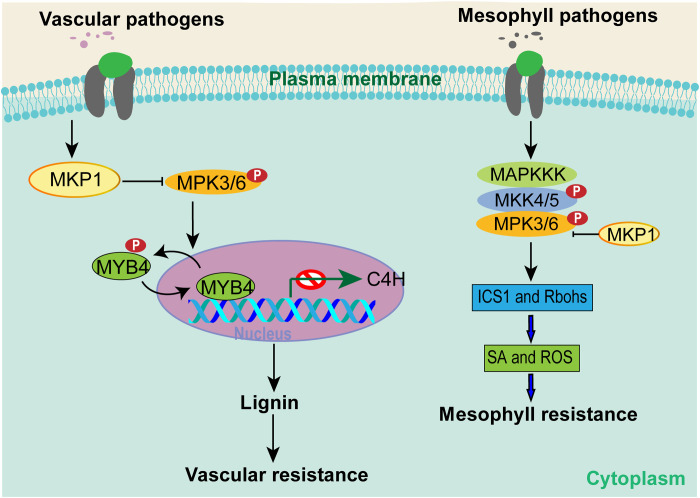
How plants perceive diverse pathogen signals and activate tissue-specific immune responses is an open question. One mechanism could be tissue-specific regulation of defense gene expression and metabolic pathways tailored to specific pathogen lifestyles (e.g., leaf mesophyll versus vascular infections). In this study, we uncover a protein phosphorylation-mediated mechanism that promotes vascular-specific immune: promotion of lignin biosynthesis in vascular tissues by the MKP1-MPK3/6-MYB signaling cascade (Fig. 6). This immune mechanism underlies both NHR against nonadapted vascular pathogens and host resistance to adapted vascular pathogens and is conserved in monocot rice and the dicot Arabidopsis.
Fig. 6. A proposed model for the function of the MKP-MPK protein phosphorylation cascade in vascular defense in plants.
MKP1 is induced by vascular bacterial pathogens, such as Xoo and Xcc, which negatively regulate MPK3/6 through dephosphorylation. MPK3/6, in turn, activates the MYB4 transcription factor that negatively regulates lignin biosynthesis. This cascade orchestrates vascular-specific resistance against vascular pathogens and negatively regulates defense against mesophyll cell pathogens through inhibiting SA and ROS biosynthesis.
The MAPK pathways have been to function in many signaling processes regulating plant development, abiotic stress responses, and immunity (51–53). The MKP1-MAPK cascade has been previously shown to negatively regulate plant resistance against mesophyll pathogens, likely through inhibiting the SA and ROS signaling pathways. In contrast, we find that the MKP1-MAPK cascade positively regulates vascular defense through activating lignin biosynthesis via direct targeting of MYB4 transcription factors. Therefore, our study has provided an example in which the same signaling cascade leads to divergent immune outcomes against pathogens with different lifestyles, shedding light on tissue-specific defense responses in monocot and dicot plants. MPK3 and OsMPK6 are activated by Xoo in mkp1 and Osmkp1 mutant plants, respectively. Therefore, monocot and dicot plants likely adopt different MAPK targets to fine-tune a conserved defense against vascular pathogens.
The xylem, which is developed from procambium and cambium and is composed of four types of cells—tracheids, vessels, xylem fibers, and xylem parenchyma—forms specialized vascular bundles together with the phloem and the cambium, providing structural integrity to plants and transporting water and nutrients from the soil to leaves and stems (27, 54). In particular, lignin is the second most abundant polymer next to cellulose in the cell walls of vascular tissues (55) and is often rapidly deposited through induction of lignin biosynthesis genes in responses to pathogen infection in plants (28–30). We discover that genes regulating lignin synthesis are less induced in the mkp1 mutants when inoculated with Xoo and Xoc, resulting in reduced lignin accumulation in the developing xylem. The lignin-mediated vascular defense is also likely used by race-specific R genes against vascular pathogens in the rice-Xoo pathosystem. For example, the Xa21-mediated Xoo resistance is related to lignin accumulation (56, 57). Similarly, Xa10 stimulates the accumulation of lignin-like phenolic compounds (58). This raises a fundamental question: How do these different resistance genes encoding diverse upstream immune regulators activate lignin-based vascular-specific immune responses upon perceiving Xoo bacteria in rice?
Our discovery of a conserved vascular-specific defense in monocot rice and dicot Arabidopsis will facilitate the identification of other key regulators of vascular defense through future suppressor screening. It would be worthy of further investigating how the plant immune machinery perceives diverse vascular pathogens and activates the MKP1-MAPK cascade to trigger lignin-based vascular defense. With this scenario, whether and how the rice Xa genes confer Xoo resistance are also open questions. Moreover, the discovery of the MKP1-MPK3/6-MYB cascade in vascular defense also opens the door to broadly investigating how diverse plants interact with vascular pathogens, including major crop pathogens, such as Candidatus Liberibacter asiaticus (CLas), Xylella fastidiosa, phytoplasmas, and vasculature-feeding insects (16). Hence, the basic findings from this study have broad implications for the future development of a new generation of crop resistance strategies against many devastating vascular pathogens and insects.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis and rice were cultivated in a growth room and at an experiment station, respectively, as previously described (59, 60). All Arabidopsis thaliana experiments were in Col-0 background, including the mutants mpk3 and mpk6 (61). The transgenic lines CR-mkp1, CR-myb4, pMKP1::MKP1/ntx1, pMYB4::GUS, MYB4-OE, MYB4AA-OE, and MYB4DD-OE were generated in this study. The double mutants mkp1 mpk3 and mkp1 mpk6 were generated by crossing the mkp1 with the mpk3 and mpk6 line, respectively. Rice transgenic lines were generated in a NIP background. Rice mutants and overexpression lines including CR-Osmkp1, CR-Osmkp1Osmpk3, CR-Osmkp1Osmpk6, CR-Osmyb102, CR-Osmyb108, CR-Osmyb102Osmyb108, OsMKP1-OE, OsMYB102-OE, OsMYB102AA-OE, and OsMYB102DD-OE were generated in this study. Four-week-old Arabidopsis and 2-month-old rice plants were used for bacterial inoculation and disease assays.
Plasmid constructs and plant transformation
To make CRISPR-Cas9 knockout constructs, the target sequences of MKP1, MYB4, OsMKP1, OsMPK3, OsMPK6, OsMYB102, and OsMYB108 were generated according to previously reported protocols (62, 63). For overexpression constructs, the coding sequence (CDS) sequences were inserted into pCambia1300-MYC or pCambia1300-GFP (green fluorescent protein) for Arabidopsis transformation or inserted into PUN1301-FLAG for rice transformation with targets. The constructs were introduced into Agrobacterium strain GV3101 (for Arabidopsis) and EHA105 (for rice) and then transformed into different genetic backgrounds to produce more than 15 independent transgenic lines for each construct. Further selection and validation were based on PCR-based sequencing or Western blotting. All primers used for cloning are listed in table S1.
PXO99A-GUS–labeled strain
Pprt+gus cut from the vector pLAFR6 (64) was connected to binary vector pUFR034 (65). The recombination vector pUFR034-Pprt-gus was constructed and transformed to prepared PXO99A competent cells. The GUS that expressed transformants were screened and verified for pathogenicity by plant inoculation.
Pathogen inoculation and disease assay
For the Arabidopsis inoculation assay, Pst DC3000 [105 colony-forming units (CFU) ml−1] was grown on LB medium [tryptone (10 g/liter), yeast extract (6 g/liter), KH2PO4 (1.5 g/liter), NaCl (0.6 g/liter), MgSO4·7H2O (0.4 g/liter), and rifampicin (100 mg/liter); 28°C], PXO99A (2 × 107 CFU ml−1) was grown on PSA medium [tryptone (10 g/liter), sucrose (10 g/liter), glutamate (1 g/liter), and cephalexin (15 mg/liter); pH 7.0; 28°C], and 8004-GFP [107 CFU ml−1] was grown on NYG medium [tryptone (5 g/liter), yeast extract (3 g/liter), and glycerol (20 g/liter); 28°C]. The inoculates were used for syringe infiltration. To quantify bacterial infection, Arabidopsis leaves were surface-sterilized using 75% ethanol, and CFUs were counted in leaf disks by serial dilutions. One technical replicate consists of four-leaf disks, and three technical replicates were included in each biological experiment. Experiments were repeated three to five times with biologically independent samples.
For rice inoculation assay, Xoo strain PXO99A [optical density at 600 nm (OD600), 1.0] and Xoc strain RS85 (OD600, 0.5) grown on NB medium [tryptone (10 g/liter), yeast extract (5 g/liter), and sucrose (10 g/liter); 28°C] were prepared for inoculation. Two-month-old plants were inoculated by leaf-clipping method and syringe infiltration method. Lesion length was recorded at day 14 post inoculation (dpi). To generate growth curves, 10 ml of sterile water resuspended 5 cm of leaf tissues to count CFUs (66, 67). All rice inoculation experiments were repeated for three biological replicates.
RNA analysis and RNA sequencing
Total RNA was extracted using TRIzol reagent and reverse-transcribed into cDNA using ReverTra Ace qPCR RT Master Mix with genomic DNA remover. For quantitative real-time PCR analysis, TB Green Premix Ex Taq and gene-specific primers were used (table S1). Experiments were repeated at least three times with biologically independent samples. For sample preparation, leaves of 4-week-old seedlings (Col-0 and mkp1) after PXO99A incubation for 12 hours was used for RNA sequencing (RNA-seq), with three biological replicates for each treatment. RNA-seq analysis was performed in Shanghai Biotechnology Corporation. The entire RNA-seq dataset was deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE161152.
Y2H assay
The CDSs of the target genes were cloned into pDEST22/32 (Invitrogen) or PGADT7/PGBKT7 (Clontech). Clones were cotransformed into yeast strain AH109, grown on dropout medium (without either Trp and Leu or Trp, Leu, and His) containing 3-aminotriazole (Sigma-Aldrich, A8056). The experiments were repeated three times independently to confirm the interactions.
SLC assay
The CDSs of the genes were cloned into pCAMBIA-35S-nLuc and pCAMBIA-35S-cLuc. The clones were transformed into Agrobacterium strain GV3101, and bacteria solution was collected and resuspended in infiltration buffer [10 mM MgCl2, 10 mM MES, and 150 mM acetosyringone (pH 5.6)] mixed with p19, incubated for 2 to 3 hours at 30°C before being infiltrated into Nicotiana benthamiana as previously described (59, 60). Two days later, LUC signals were measured using the Luciferase Assay System (Promega) and imaged using the Tanon-5200 Chemiluminescent Imaging System. The experiments were repeated three times independently to confirm the in planta interactions.
Protein pull-down assay
Fusion proteins were expressed in Escherichia coli, and 30 mg of protein was bound to beads in buffer [20 mM tris-HCl (pH 7.4), 1 mM EDTA, 200 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM dithiothreitol (DTT)] at 4°C for 2 hours and then washed five times with the washing buffer [50 mM tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, and 0.1% Triton X-100] to remove nonspecifically bound proteins. The binding protein was released by heating at 95°C for 5 min in 100 μl of SDS loading buffer for immunoblot.
Coimmunoprecipitation
Samples were ground in liquid nitrogen; protein was extracted using IP buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM PMSF, and 1× protease inhibitor cocktail (11836153001, Sigma-Aldrich)] and then vortexed and centrifuged at 12,000g for 10 min at 4°C. Supernatant was incubated with beads for 2 hours at 4°C, and the beads were washed four times with washing buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF, and 1× protease inhibitor cocktail]. The binding protein was released by heating at 95°C for 5 min in 100 μl of SDS loading buffer for Western blot. Proteins were resolved on a 4 to 20% Biofuraw Precast Gel (Tanon) by electrophoresis and transferred to polyvinylidene difluoride membranes using Trans-Blot Turbo blotting system (Bio-Rad). The following antibodies were used: anti-MYC (Merck Millipore, 05-724), anti-FLAG (Sigma-Aldrich, F1804), anti-GFP (Abcam, ab290), and anti-ACTIN (CMCTAG, AT0004). Immunodetection was imaged using Tanon-5200 Chemiluminescent Imaging System (Tanon), and ACTIN was used as a loading control.
MAPK activity assay
Proteins were extracted in MAPK extraction buffer [50 mM HEPES (pH 7.5), 75 mM NaCl, 5 mM EDTA, 5 mM EGTA, 50 mM β-glycerophosphate, 2 mM DTT, 0.5% Triton X-100, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail (4906845001, Sigma-Aldrich)]. MAPK activities were detected using Phospho-p44/42 MAPK antibody (1:2000; 9101, Cell Signaling Technology), anti-MPK3 (1:2000; M8318, Sigma-Aldrich), and anti-MPK6 (1:2000; A7104, Sigma-Aldrich).
In vitro protein phosphorylation assay
The purified fusion proteins MKK4DD-MBP (50 ng), MKK5DD-MBP (50 ng), MPK3-MBP (200 ng), and MYB4-His (200 ng) produced in E. coli were used for in vitro protein phosphorylation assay. The proteins were incubated in reaction buffer [25 mM tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 10 mM adenosine triphosphate] at 30°C for 30 min, and the reaction was stopped by heating at 95°C for 5 min with SDS loading buffer. MYB4-His phosphorylation was visualized by immunoblotting with 10% Phos-tag Gel (Wako) as described (68).
GUS reporter assay
The ~2-kb promoter regions of MKP1 and MYB4 were inserted into vector 1300-GUS-Nos and then introduced into Agrobacterium strain GV3101 to develop GUS reporter transgenic plants. For GUS staining, plant tissues were stained in buffer [50 mM NaPO4 (pH 7.0), 5 mM K3Fe (CN)6, 5 mM K4Fe (CN)6, 0.1% Triton X-100, and 1 mM X-Gluc] overnight at 37°C and then dehydrated through a graded ethanol series.
Lignin staining
For lignin staining, plant materials were soaked in FAA (50% ethanol, 5% glacial acetic acid, and 10% formaldehyde) at 4°C and then dehydrated with a graded ethanol series (100, 75, 50, and 25%) for 30 min each. Samples were fixed in resin and cut into slices (6 to 7 μm), which were then stained with 1% phloroglucinol dissolved in 12% HCl for 5 min at room temperature and observed under an optical microscope (Leica EZ4 E) as previously described (69).
Microscopic analysis of lignin
For histochemical observation of the cell wall autofluorescence, leaf tissues were fixed in FAA for 24 hours and then dehydrated in a graded series of ethanol from 50 to 100%. Samples were then cleared with xylene and embedded in paraffin to prepare sections (5 to 10 μm). The sections were observed under a fluorescence microscope (Zeiss LSM 880) at 488/530 nm excitation and emission (70).
Lignin measurement
Lignin was quantified as previously described (71, 72). Briefly, leaf samples (~100 mg) were frozen and ground to powder in liquid nitrogen and washed with 70% ethanol, chloroform/methanol (1:1 v/v), and acetone. Powder samples were then evaporated until dry at 35°C, suspended in 0.1 M sodium acetate buffer (pH 5.0), and heated for 20 min at 80°C. After adding 35 μl of amylase [H2O (50 μg/ml); from Bacillus species; Sigma-Aldrich] and 17 μl of pullulanase (17.8 U from Bacillus acidopullulyticus; Sigma-Aldrich), samples were incubated overnight at 37°C and digested with 100 μl of 25% acetyl bromide in acetic acid at 50°C for 2 hours. The samples were mixed with 2 M NaOH (400 μl), 0.5 M hydroxylamine hydrochloride (70 μl), and acetic acid to 2 ml. Absorbance was measured at 280 nm. The content of acetyl bromide soluble lignin (%ABSL) was calculated using Beer’s law with extinction coefficient of 15.69 mg/cm per liter for Arabidopsis and 17.2 mg/cm per liter for rice.
SA analysis
For total and free SA measurement (12), leaf tissues (100 mg) were ground in liquid nitrogen and extracted with 90% methanol, and 250 ng of o-anisic acid was used as an internal standard. The samples were subjected to phase separation in ethyl acetate/cyclopentane (1:1 v/v) and then evaporated and solubilized in 20% methanol with 5% trichloroacetic acid. Filtered extracts were quantified using high-performance liquid chromatography, and fluorescence was recorded with excitation/emission wavelengths of 305/407 nm for o-anisic acid and SA.
Microscopic observation of bacteria in planta
For scanning electron micrograph (SEM) of bacteria, plant materials inoculated with pathogens were soaked in FAA at 4°C; dehydrated with 50, 70, 85, and 100% ethanol for 5 min each; dried with CO2 critical point drier; and observed under Zeiss Merlin Compact SEM. For TEM of bacteria, plant materials inoculated with pathogens were soaked in 2.5% glutaraldehyde solution at 4°C and embedded with conventional methods, and slices were observed with Hitachi H-7650 TEM.
Statistical analysis
In this study, all values are presented as means ± SD, and the number (n) of samples or replicates are indicated in the figure legends. Significant differences were analyzed using Student’s t test for pairwise comparisons and one-way or two-way analysis of variance (ANOVA) with Tukey’s test between multiple groups comparison indicated with P values or different letters.
Acknowledgments
We thank J. Li for critical reading; S. Zhang for providing the mpk3 and mpk6 mutants; W. Qian for Xcc strain 8004-GFP; F. Dai for help in Arabidopsis mutant screening; X. Wang for rice transformation; X. Zhong, K. Cui, B. Ma, and X. Gong for help in field test; and J. Li and Z. Zhang for help in tissue sections and electron microscope observation.
Funding: This study was supported by grants from the Chinese Academy of Sciences (XDB27040201), National Key Research and Development Program of China (2016YFD0100600), the National Natural Science Foundation of China (32088102), and the National Key Laboratory of Plant Molecular Genetics.
Author contributions: H.L. and Z.H. designed the experiments. H.L., M.W., Y.C., K.N., S.H., J.G., X.Z., and Y.W. performed the experiments. H.L., M.W., Y.C., J.L., Q.L., and Y.D. developed the materials. Z.H., S.W., S.Y.H., L.L., and M.Y. supervised the project. H.L., S.Y.H., and Z.H. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
REFERENCES AND NOTES
- 1.Lucas W. J., Groover A., Lichtenberger R., Furuta K., Yadav S. R., Helariutta Y., He X. Q., Fukuda H., Kang J., Brady S. M., Patrick J. W., Sperry J., Yoshida A., López-Millán A. F., Grusak M. A., Kachroo P., The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 55, 294–388 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Niño-Liu D. O., Ronald P. C., Bogdanove A. J., Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Guo A., Leach J. E., Examination of rice hydathode water pores exposed to Xanthomonas campestris pv. oryzae. Phytopathology 79, 433–436 (1989). [Google Scholar]
- 4.Vicente J. G., Holub E. B., Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 14, 2–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe T. M., Ailloud F., Allen C., Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Mol. Plant Microbe Interact. 28, 286–297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denancé N., Ranocha P., Oria N., Barlet X., Rivière M.-P., Yadeta K. A., Hoffmann L., Perreau F., Clément G., Maia-Grondard A., van den Berg G. C. M., Savelli B., Fournier S., Aubert Y., Pelletier S., Thomma B. P. H. J., Molina A., Jouanin L., Marco Y., Goffner D., Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73, 225–239 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Fradin E. F., Thomma B. P. H. J., Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71, –86 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Montes-Osuna N., Mercado-Blanco J., Verticillium wilt of olive and its control: What did we learn during the last decade? Plants 9, 753 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genin S., Denny T. P., Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Gao M., He Y., Yin X., Zhong X., Yan B., Wu Y., Chen J., Li X., Zhai K., Huang Y., Gong X., Chang H., Xie S., Liu J., Yue J., Xu J., Zhang G., Deng Y., Wang E., Tharreau D., Wang G., Yang W., He Z., Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 184, 5391–5404.e17 (2021). [DOI] [PubMed] [Google Scholar]
- 11.You Q., Zhai K., Yang D., Yang W., Wu J., Liu J., Pan W., Wang J., Zhu X., Jian Y., Liu J., Zhang Y., Deng Y., Li Q., Lou Y., Xie Q., He Z., An E3 ubiquitin ligase-bag protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Yin X., Zou B., Hong X., Gao M., Yang W., Zhong X., He Y., Kuai P., Lou Y., Huang J., Hua J., He Z., Rice copine genes OsBON1 and OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol. J. 16, 1476–1487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J. M., Zhang Y., Plant immunity: Danger perception and signaling. Cell 181, 978–989 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Jones J. D., Vance R. E., Dangl J. L., Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Xin X. F., Kvitko B., He S. Y., Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y., Zhang C. X., Chen R., He S. Y., Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. U.S.A. 116, 23390–23397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee S., Almeida R. P., Lindow S., Living in two worlds: The plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 46, 243–271 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Jacques M. A., Arlat M., Boulanger A., Boureau T., Carrère S., Cesbron S., Chen N. W., Cociancich S., Darrasse A., Denancé N., Fischer-Le Saux M., Gagnevin L., Koebnik R., Lauber E., Noël L. D., Pieretti I., Portier P., Pruvost O., Rieux A., Robène I., Royer M., Szurek B., Verdier V., Vernière C., Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu. Rev. Phytopathol. 54, 163–187 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Cerutti A., Jauneau A., Auriac Lauber E., Martinez Y., Chiarenza S., Leonhardt N., Berthomé R., Noël L. D., Immunity at cauliflower hydathodes controls systemic infection by Xanthomonas campestris pv campestris. Plant Physiol. 174, 700–716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdier V., Triplett L. R., Hummel A. W., Corral R., Cernadas R. A., Schmidt C. L., Bogdanove A. J., Leach J. E., Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 196, 1197–1207 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Zhou J. H., Peng Z., Long J. Y., Sosso D., Liu B., Eom J. S., Huang S., Liu S. Z., Vera Cruz C., Frommer W. B., White F. F., Yang B., Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Jha G., Rajeshwari R., Sonti R. V., Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant Microbe Interact. 20, 31–40 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Tayi L., Maku R., Patel H. K., Sonti R. V., Action of multiple cell wall-degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Mol. Plant Microbe Interact. 29, 599–608 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Tayi L., Kumar S., Nathawat R., Haque A. S., Maku R. V., Patel H. K., Sankaranarayanan R., Sonti R. V., A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice. Mol. Plant Pathol. 19, 1364–1376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gluck-Thaler E., Cerutti A., Perez-Quintero A. L., Butchacas J., Roman-Reyna V., Madhavan V. N., Shantharaj D., Merfa M. V., Pesce C., Jauneau A., Vancheva T., Lang J. M., Allen C., Verdier V., Gagnevin L., Szurek B., Beckham G. T., De La Fuente L., Patel H. K., Sonti R. V., Bragard C., Leach J. E., Noël L. D., Slot J. C., Koebnik R., Jacobs J. M., Repeated gain and loss of a single gene modulates the evolution of vascular plant pathogen lifestyles. Sci. Adv. 6, eabc4516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samal B., Chatterjee S., New insight into bacterial social communication in natural host: Evidence for interplay of heterogeneous and unison quorum response. PLOS Genet. 15, e1008395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Růžička K., Ursache R., Hejátko J., Helariutta Y., Xylem development—From the cradle to the grave. New Phytol. 207, 519–535 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Lee S., Sharm Y., Lee T. K., Chang M., Davis K. R., Lignification induced by Pseudomonads harboring avirulent genes on Arabidopsis. Mol. Cells 12, 25–31 (2001). [PubMed] [Google Scholar]
- 29.Malinovsky F. G., Fangel J. U., Willats W. G. T., The role of the cell wall in plant immunity. Front. Plant Sci. 5, 178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon J., Choi H., An G., Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 57, 902–912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J., Lee C., Zhong R., Ye Z. H., MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li E., Bhargava A., Qiang W., Friedmann M. C., Forneris N., Savidge R. A., Johnson L. A., Mansfield S. D., Ellis B. E., Douglas C. J., The class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 194, 102–115 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C., Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemm M. R., Herrmann K. M., Chapple C., AtMYB4: A transcription factor general in the battle against UV. Trend. Plant Sci. 6, 135–136 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Mitra M., Agarwal P., Kundu A., Banerjee V., Roy S., Investigation of the effect of UV-B light on Arabidopsis MYB4 (AtMYB4) transcription factor stability and detection of a putative MYB4-binding motif in the promoter proximal region of AtMYB4. PLOS ONE 14, e0220123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye G., Hong N., Zou L. F., Zou H. S., Zakria M., Wang G. P., Chen G. Y., Tale-based genetic diversity of chinese isolates of the citrus canker pathogen Xanthomonas citri subsp citri. Plant Dis. 97, 1187–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Zou L. F., Ye G., Xiong L., Ji Z. Y., Zakria M., Hong N., Wang G. P., Chen G. Y., A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp citri. Mol. Plant 7, 912–915 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Bartetzko V., Sonnewald S., Vogel F., Hartner K., Stadler R., Hammes U. Z., Börnke F., The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: Evidence for interference with cell wall-associated defense responses. Mol. Plant Microbe Interact. 22, 655–664 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Priller J. P. R., Reid S., Konein P., Dietrich P., Sonnewald S., The Xanthomonas campestris pv. vesicatoria type-3 effector XopB inhibits plant defence responses by interfering with ROS production. PLOS ONE 11, e0159107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An C. F., Mou Z. L., Non-host defense response in a novel Arabidopsis-Xanthomonas citri subsp citri pathosystem. PLOS ONE 7, e31130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartels S., Anderson J. C., González Besteiro M. A., Carreri A., Hirt H., Buchala A., Métraux J. P., Peck S. C., Ulm R., MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21, 2884–2897 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J., MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Ulm R., Ichimura K., Mizoguchi T., Peck S. C., Zhu T., Wang X., Shinozaki K., Paszkowski J., Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu Z. Q., Dong X., Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Tsuda K., Sato M., Glazebrook J., Cohen J. D., Katagiri F., Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53, 763–775 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Ren D., Yang H., Zhang S., Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol.Chem. 277, 559–565 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Pitzschke A., Modes of MAPK substrate recognition and control. Trends Plant Sci. 20, 49–55 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Katou S., Kuroda K., Seo S., Yanagawa Y., Tsuge T., Yamazaki M., Miyao A., Hirochika H., Ohashi Y., A calmodulin-binding mitogen-activated protein kinase phosphatase is induced by wounding and regulates the activities of stress-related mitogen-activated protein kinases in rice. Plant Cell Physiol. 48, 332–344 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Helft L., Thompson M., Bent A. F., Directed evolution of FLS2 towards novel flagellin peptide recognition. PLOS ONE 11, e0157155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto T., Takada R., Tobimatsu Y., Takeda Y., Suzuki S., Yamamura M., Osakabe K., Osakabe Y., Sakamoto M., Umezawa T., OsMYB108 loss-of-function enriches p-coumaroylated and tricin lignin units in rice cell walls. Plant J. 98, 975–987 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Bartels S., González Besteiro M. A., Lang D., Ulm R., Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci. 15, 322–329 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Shankar A., Agrawal N., Sharma M., Pandey A., Girdhar K. P. M., Role of protein tyrosine phosphatases in plants. Curr. Genomics 16, 224–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng X., Zhang S., MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Yadeta K. A., Thomma B. P. H. J., The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4, 97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neutelings G., Lignin variability in plant cell walls: Contribution of new models. Plant Sci. 181, 379–386 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Song W., Wang G. L., Chen L. L., Kim H. S., Pi L. Y., Holsten T., Gardner J., Wang B., Zhai W. X., Zhu L. H., Fauquet C., Ronald P., A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Shamsunnaher, Chen X., Zhang X., Wu X., Huang X., Song W., Rice immune sensor XA21 differentially enhances plant growth and survival under distinct levels of drought. Sci. Rep. 10, 16938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reimers P. J., Leach J. E., Race-specific resistance to Xanthomonas oryzae pv oryzae conferred by bacterial blight resistance gene Xa10 in rice (Oryza Sativa) involves accumulation of a lignin-like substance in host tissues. Physiol. Mol. Plant Pathol. 38, 39–55 (1991). [Google Scholar]
- 59.Liu J., Feng L., Gu X., Deng X., Qiu Q., Li Q., Zhang Y., Wang M., Deng Y., Wang E., He Y., Bäurle I., Li J., Cao X., He Z., An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 29, 379–390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhai K., Deng Y., Liang D., Tang J., Liu J., Yan B., Yin X., Lin H., Chen F., Yang D., Xie Z., Liu J. Y., Li Q., Zhang L., He Z., RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 74, 996–1009.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Xu J., Xie J., Yan C., Zou X., Ren D., Zhang S., A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., Xie Y., Shen R., Chen S., Wang Z., Chen Y., Guo J., Chen L., Zhao X., Dong Z., Liu Y. G., A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q., High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Lu G. T., Ma Z. F., Hu J. R., Tang D. J., He Y. Q., Feng J. X., Tang J. L., A novel locus involved in extracellular polysaccharide production and virulence of Xanthomonas campestris pathovar campestris. Microbiology 153, 737–746 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Defeyter R., Kado C. I., Gabriel D. W., Small, stable shuttle vectors for use in Xanthomonas. Gene 88, 65–72 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Bartonwillis P. A., Roberts P. D., Guo A., Leach J. E., Growth dynamics of Xanthomonas campestris pv oryzae in leaves of rice differential cultivars. Phytopathology 79, 573–578 (1989). [Google Scholar]
- 67.Yang B., Bogdanove A., Inoculation and virulence assay for bacterial blight and bacterial leaf streak of rice. Methods Mol. Biol. 956, 249–255 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Jiang L., Anderson J. C., Gonzalez Besteiro M. A., Peck S. C., Phosphorylation of Arabidopsis MAP kinase phosphatase 1 (MKP1) is required for PAMP responses and resistance against bacteria. Plant Physiol. 175, 1839–1852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gui J., Luo L., Zhong Y., Sun J., Umezawa T., Li L., Phosphorylation of LTF1, an MYB transcription factor in populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol. Plant 12, 1325–1337 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Baldacci-Cresp F., Spriet C., Twyffels L., Blervacq A. S., Neutelings G., Baucher M., Hawkins S., A rapid and quantitative safranin-based fluorescent microscopy method to evaluate cell wall lignification. Plant J. 102, 1074–1089 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Foster C. E., Martin T. M., Pauly M., Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: Lignin. J. Vis. Exp. 1745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Cássia Siqueira-Soares R., Finger-Teixeira A., Matias de Oliveira D., Ferro A. P., da Rocha G. J., Ferrarese M. d. L. L., Dantas dos Santos W., Ferrarese-Filho O., The acetyl bromide method is faster, simpler and presents best recovery of lignin in different herbaceous tissues than Klason and thioglycolic acid methods. PLOS ONE 9, e110000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1