Abstract
The factors controlling lignin composition remain unclear. Catechyl (C)–lignin is a homopolymer of caffeyl alcohol with unique properties as a biomaterial and precursor of industrial chemicals. The lignin synthesized in the seed coat of Cleome hassleriana switches from guaiacyl (G)– to C-lignin at around 12 to 14 days after pollination (DAP), associated with a rerouting of the monolignol pathway. Lack of synthesis of caffeyl alcohol limits C-lignin formation before around 12 DAP, but coniferyl alcohol is still synthesized and highly accumulated after 14 DAP. We propose a model in which, during C-lignin biosynthesis, caffeyl alcohol noncompetitively inhibits oxidation of coniferyl alcohol by cell wall laccases, a process that might limit movement of coniferyl alcohol to the apoplast. Developmental changes in both substrate availability and laccase specificity together account for the metabolic fates of G- and C-monolignols in the Cleome seed coat.
Laccase specificity can determine lignin composition irrespective of the pool size of the available monolignol precursors.
INTRODUCTION
Lignin is the second most abundant biopolymer on Earth. Its monomeric composition determines not only its properties as a biomaterial but also its impact on biomass processing and forage digestibility (1–6). Lignin composition is more flexible than previously assumed, with a number of noncanonical “monolignol” building blocks found in recent years (7). Monolignols are polymerized via free radical reactions initiated/catalyzed by laccases and peroxidases (8), both of which are encoded by large gene families in plants (9, 10). Recent studies have suggested that some laccases may show preference for specific monolignols (11–13), although it is unclear whether the final composition of lignin is determined by monomer availability, laccase/peroxidase specificity, or both.
Catechyl (C)–lignin is a recently found homopolymer of caffeyl alcohol that occurs naturally in the seed coats of phylogenetically diverse plant species including orchids, cacti, several oil seed species including Jatropha and castor bean, and the ornamental plant Cleome hassleriana (14–16). In contrast, classical lignin is a complex heteropolymer of two to three different monolignol units found in the cell walls of secondarily thickened plant tissues such as xylem and interfascicular fibers in angiosperms and a guaiacyl (G)–rich polymer in secondary cell walls of gymnosperms (17). Heterogeneity and cross-linking of classical lignin limit its utility as a biomaterial (2). In contrast, C-lignin has been identified as an ideal subject for lignin valorization in the biorefinery. This is because its homogeneity (caffeyl alcohol units linked exclusively through benzodioxane structures) favors its selective reductive catalytic fractionation to simple products for subsequent biological funneling (18–20), and its linearity, with lack of cross-linking either internally or to other cell wall polymers, makes it a good material for production of carbon fibers (21). Furthermore, C-lignin is stable to pretreatment conditions used during processing of lignocellulosic biomass for fermentation to liquid biofuels (18), in contrast to classical lignin that can dissolve and reprecipitate during pretreatment. However, other than limited evidence for the presence of C-lignin in root cultures of genetically modified pine (22), the polymer currently appears to be limited to seed coats.
Lignin biosynthesis in the C. hassleriana seed coat switches markedly from production of G-lignin to production of C-lignin at around 12 to 14 days after pollination (DAP) (16). This makes C. hassleriana an excellent model system to interrogate the molecular mechanisms underlying C-lignin biosynthesis. Synthesis of caffeyl alcohol requires suppression of the O-methylation reactions involved in the biosynthesis of the major monolignol coniferyl alcohol (the G unit of lignin) (16), and C-lignin accumulation in the Cleome seed coat is associated with a rapid decline in the gene expression and enzymatic activities of caffeoyl coenzyme A (CoA) 3-O-methyltransferase (CCoAOMT) and caffeic acid/5-hydroxyconiferaldehyde 3/5-O-methyltransferase (COMT) (23). Although COMT has been ascribed a role primarily in syringyl (S)–lignin biosynthesis (24), it is likely associated with the biosynthesis of G-lignin in the C. hassleriana seed coat, as S-lignin is absent (14).
Although a dehydrogenation polymer with identical composition and linkage type to natural C-lignin can be generated in vitro by peroxidase-mediated coupling of caffeyl alcohol (14), the biosynthesis of caffeyl alcohol and its precursor caffealdehyde has yet to be demonstrated in plant tissues producing C-lignin. Here, we use a combination of metabolite profiling, isotopic labeling analyses, metabolic flux analysis (MFA), and enzyme specificity studies to determine the origin, levels, and fates of the precursor pools for lignin biosynthesis during the development of the C. hassleriana seed coat. Unexpectedly, coniferyl alcohol is still made during the period of C-lignin biosynthesis but not incorporated into lignin. We present a model in which caffeyl alcohol production directed to C-lignin and C-lignans accompanies the maintenance of a pool of coniferyl alcohol, presumably for other cellular functions. This is achieved, in part, through caffeyl alcohol inhibiting the oxidation of coniferyl alcohol by cell wall laccases and peroxidases, potentially preventing its movement to the apoplast.
RESULTS
Targeted profiling of monolignol pathway intermediates during Cleome seed coat development
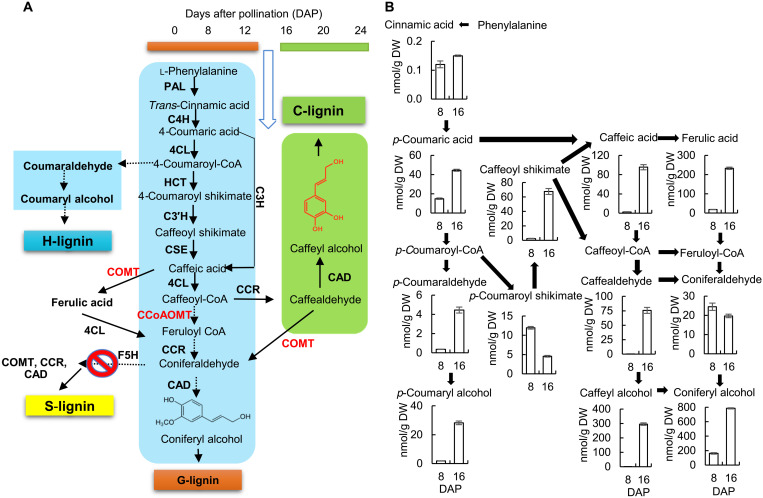
Using liquid chromatography–mass spectrometry (LC-MS) analysis, we profiled the known precursors of G-lignin and predicted precursors of C-lignin (simplified schematic pathway in Fig. 1A), except for the CoA esters of caffeate and ferulate, in extracts from C. hassleriana whole seeds and isolated seed coats during developmental stages (8 to 24 DAP) that spanned the switch from G- to C-lignin deposition. The patterns of metabolite accumulation in whole seeds (fig. S1) followed three main trends: increasing in level up to 16 to 20 DAP and then decreasing (trans-cinnamic acid, p-coumaric acid, ferulic acid, p-coumaraldehyde, p-coumaryl alcohol, coniferaldehyde, and coniferyl alcohol; highlighted in blue); decreasing between 8 and 12 DAP and then increasing again, with a second decrease after 12 or 16 DAP (coumaroyl shikimate, caffeoyl shikimate, and caffeic acid; highlighted in purple); or close to detection limit until 12 DAP, followed by a sharp increase and then a decrease (caffealdehyde and caffeyl alcohol; highlighted in green). Caffealdehyde and caffeyl alcohol were not detected at times before 16 DAP. Unexpectedly, however, levels of coniferyl alcohol, the monomer of G-lignin, were higher during the period of C-lignin biosynthesis (after 12 DAP) than during the period of G-lignin biosynthesis. Levels of coumaroyl shikimate and caffeoyl shikimate, respectively the substrate and product of coumaroyl shikimate 3′-hydroxylase (C3′H), the key enzyme of the shikimate shunt for generation of the caffeoyl moiety, declined steeply during the latter part of the period of G-lignin biosynthesis (between 8 and 12 DAP), only to increase again between 12 and 16 DAP, suggesting the operation of a metabolic switch.
Fig. 1. Monolignol pathway metabolite levels in the C. hassleriana seed coat.
(A) Simplified scheme of pathways to G-, H-, S-, and C-lignins. Lignin synthesis in the Cleome seed coat switches from G-lignin to C-lignin between 12 and 14 DAP. Enzymes are as follows: PAL, l-phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; C3H, coumarate 3-hydroxylase; 4CL, 4-coumarate: CoA ligase; HCT, hydroxycinnamoyl CoA: shikimate/quinate hydroxycinnamoyl transferase; C3′H, 4-coumaroyl shikimate 3′-hydroxylase; CSE, caffeoyl shikimate esterase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; COMT, caffeic acid/5-hydroxyconiferaldehyde 3-O-methyltransferase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; F5H, ferulate/coniferaldehyde 5-hydroxylase. Conversion of phenyalanine to caffeoyl CoA is common to the biosynthesis of both G- and C-lignin. S-lignin is not made as a result of lack of expression of F5H in the seed coat. H-lignin synthesis stops after 12 DAP, resulting in less than 1 μmol/g dry weight (DW) (~0.25% of total lignin). O-methyltransferases that are down-regulated at the onset of C-lignin biosynthesis are shown in red. For a scheme showing all potential routes to G- and C-lignin, see (32). (B) Levels of monolignol pathway intermediates, as determined by targeted LC-MS analysis, at 8 (G-lignin produced) and 16 (C-lignin produced) DAP. Approximately 100 mg of seed coats isolated from seeds harvested from one individual Cleome plant was counted as one biological replicate. Data are means ± SE derived from three biological replicates.
To determine changes in metabolites specific to the seed coat, we repeated the above experiment concentrating on 8 and 16 DAP. The seed coats were removed, and metabolite levels determined separately in the seed coats and the remainder of the seeds. Caffeic acid levels were very low, and caffealdehyde and caffeyl alcohol not present, in the seed coat at 8 DAP (Fig. 1B). However, a substantial proportion of the coniferyl alcohol observed in whole seeds at 16 DAP was present in the seed coat. The level of coniferyl alcohol in the seed coat at 16 DAP was approximately twice that of caffeyl alcohol. A comparison of the metabolite data for both the seed coat and the remainder of the seed is given in fig. S2. There were large amounts of p-coumaric acid, p-coumaroyl shikimate, ferulic acid, coniferaldehyde, and coniferyl alcohol in the remainder of the seed at 16 DAP, suggesting that these compounds may be involved in other biochemical pathways or transported to the seed coat for lignin biosynthesis.
Lignans and proanthocyanidin precursors in the Cleome seed coat
Lignans are dimers of monolignols that play roles in both plant defense and development (25–28). The existence of lignans derived from caffeyl alcohol was recently reported in the seed coat of tung trees (29). To determine whether lignans are present in the Cleome seed coat, we used nontargeted gas chromatography–MS (GC-MS) analysis. The full data, including statistical analysis, are available in dataset S1. The only G-lignan derivative found in seed coats at above trace levels was dehydrodiconiferyl alcohol, at low levels that did not differ substantially during development (Table 1). Pinoresinol, another G-lignan, was also detected at 8 to 20 DAP, but only at trace levels. There was, however, a large accumulation of lignans that contained at least one caffeyl alcohol unit at 16 and 20 DAP (Table 1). The caffeyl lignans were mainly homodimers of caffeyl alcohol or heterodimers of caffeyl alcohol with caffealdehyde, although heterodimers of caffeyl alcohol with 3,4-dihydroxybenzoic acid or caffeic acid were also detected. Whereas the linkages in C-lignin are exclusively through benzodioxane units (14), the units in the caffeyl lignans were joined through multiple linkage types: primarily cis- and trans-benzodioxanes, which constituted 73% of the caffeyl alcohol lignan dimers at 20 DAP, although resinols and lactones were also present. The most abundant C-lignans were trans-isoamericanol A (a benzodioxane homodimer), 4′,4″-dihydroxyenterolactone (a homodimer), and trans-americanin A (a benzodioxane heterodimer) that constituted 59, 20, and 5% of the total lignans present at 20 DAP, respectively (Table 1). The next three most abundant lignans were present at 3 to 4% of the total and identified as iso-caffeyl alcohol-3,4-dihydroxybenzoic acid benzodioxane, (±)-3,3′-bisdemethylpinoresinol, and trans-americanol A. The structures of the major seed coat lignans (fig. S3) were elucidated by in-house condensation synthesis reactions, and further details of the structural confirmation of these and the minor lignan components will be presented elsewhere. Isoamericanol A, isoamericanin A, americanol A, and americanin A, which were all detected in the 16 to 20 DAP samples, have been reported to be the main oxidative coupling products of horseradish peroxidase–catalyzed reactions of caffeic acid via reduction to caffealdehyde and caffeyl alcohol (30, 31). No C-lignan glycosides, homodimers of p-coumaryl alcohol, or heterodimers of caffeyl alcohol and p-coumaryl alcohol were observed.
Table 1. Levels of lignans, flavonoids, and selected primary metabolites in Cleome seed coats at different stages of development (DAP).
Metabolites were measured by nontargeted GC-MS analysis, and data show means and SDs for four biological replicates. See dataset S1 for further details and statistical analysis and fig. S3 for the structures of the six most abundant lignans.
| Metabolite | Metabolite concentration (μg/g fresh weight sorbitol equivalents) | |||
| 8 DAP | 12DAP | 16DAP | 20 DAP | |
| Lignans: | ||||
| Trans-isoamericanol A | 0.0 | 0.0 | 848 ± 116 | 1350 ± 108 |
| 4′,4″-Dihydroxyenterolactone | 0.0 | 0.0 | 290 ± 45 | 468 ± 36 |
| Trans-americanin A | 0.0 | 0.0 | 74.9 ± 9.7 | 114.3 ± 11.1 |
| Iso-caffeyl alcohol-3,4-dihydroxybenzoic acid benzodioxane |
1.8 ± 0.4 | 0.5 ± 0.2 | 33.6 ± 2.9 | 98.7 ± 2.2 |
| (±)-3,3′-Bisdemethylpinoresinol | 0.0 | 0.0 | 37.6 ± 4.4 | 86.3 ± 2.5 |
| Trans-americanol A | 0.0 | 0.0 | 43.9 ± 6.9 | 72.8 ± 4.6 |
| Caffeyl alcohol 2-O-3/6-O-3 lignan | 0.0 | 0.0 | 16.0 ± 3.7 | 29.0 ± 4.2 |
| (3R,4R)-Rel-3,4-bis[(3,4-dihydroxyphenyl) methyl]dihydro-2(3H)-furanone |
0.0 | 0.0 | 15.3 ± 2.6 | 23.9 ± 1.8 |
| Cis-isoamericanol A | 0.0 | 0.0 | 13.7 ± 1.1 | 23.7 ± 1.0 |
| Trans-isoamericanin A | 0.0 | 0.0 | 10.6 ± 1.3 | 15.4 ± 1.9 |
| Trans-isoamericanoic acid | 0.0 | 0.0 | 0.8 ± 0.2 | 4.5 ± 0.4 |
| Trans-caffeyl alcohol-caffealdehyde resinol | 0.0 | 0.0 | 1.2 ± 0.3 | 3.9 ± 0.6 |
| Cis-americanin A | 0.0 | 0.0 | 2.2 ± 0.3 | 3.5 ± 0.5 |
| Trans-iso-caffeyl alcohol-caffeic acid resinol | 0.0 | 0.0 | 0.5 ± 0.1 | 2.4 ± 0.1 |
| Dehydrodiconiferyl alcohol | 2.7 ± 0.4 | 12.8 ± 4.4 | 6.2 ± 0.8 | 4.5 ± 0.8 |
| Pinoresinol | 0.0 | 1.2 ± 0.6 | 1.0 ± 0.2 | 0.4 ± 0.2 |
| Trans-caffeyl alcohol 4-O-glucoside | 0.0 | 0.0 | 7.1 ± 0.4 | 16.8 ± 0.6 |
| Flavonoids: | ||||
| Kaempferol | 40.4 ± 5.5 | 46.3 ± 15.4 | 17.9 ± 3.9 | 9.8 ± 2.1 |
| Aromadendrin (dihydrokaempferol) | 22.61 ± 1.37 | 8.44 ± 2.07 | 2.04 ± 0.27 | 0.98 ± 0.13 |
| Trans-3,4-leucopelargonidin | 57.01 ± 3.55 | 6.24 ± 0.91 | 0.82 ± 0.19 | 0.15 ± 0.06 |
| Afzelechin | 26.47 ± 2.10 | 19.94 ± 6.56 | 2.43 ± 0.41 | 0.53 ± 0.15 |
| Selected primary metabolites: | ||||
| Citric acid | 788 ± 39 | 949 ± 45 | 836 ± 45 | 126 ± 6 |
| Malic acid | 780.7 ± 56.0 | 262.6 ± 16.3 | 45.3 ± 1.0 | 40.0 ± 2.8 |
| Sucrose | 21849 ± 1030 | 11110 ± 1234 | 2245 ± 280 | 1119 ± 204 |
| Glucose | 12008 ± 512 | 4542 ± 511 | 1343 ± 106 | 460 ± 32 |
| Fructose | 2322 ± 204 | 600 ± 35 | 231 ± 7 | 51 ± 4 |
| Oleic acid | 94 ± 11 | 156 ± 72 | 435 ± 92 | 525 ± 124 |
| Linoleic acid | 264 ± 27 | 600 ± 284 | 1053 ± 246 | 985 ± 217 |
| Glyceric acid | 236.7 ± 36.1 | 17.9 ± 1.3 | 6.4 ± 0.4 | 5.5 ± 0.9 |
Flavonoids were also observed in the seed coats by GC-MS analysis (Table 1). These were limited to the flavan-3-ol afzelechin, its precursors trans-3,4-leucopelargonidin and aromadendrin (dihydrokaempferol), and the related flavonol kaempferol (fig. S4A). Levels of these compounds were highest at 8 DAP and generally very low by 16 DAP. Afzelechin is a building block for condensed tannins (CTs), and these flavonoid polymers have been shown to accumulate early during seed development in C. hassleriana (12). The CTs were identified as propelargonidins on the basis of the formation of afzelechin-phloroglucinol conjugate from CT extension units after phloroglucinolysis (fig. S4B).
The other major changes observed from GC-MS analysis of the seed coats during development were strong decreases in 5-oxo-proline, malic acid, sucrose, glucose, galactose, and fructose, consistent with a major decrease in primary metabolism as the seed coats mature (Table 1). Levels of certain fatty acids did, however, increase during seed coat development, concomitant with a decline in glyceric acid over time (Table 1 and dataset S1).
Incorporation of 13C-Phe and 13C–trans-cinnamic acid into lignin and monolignol precursors during seed coat development
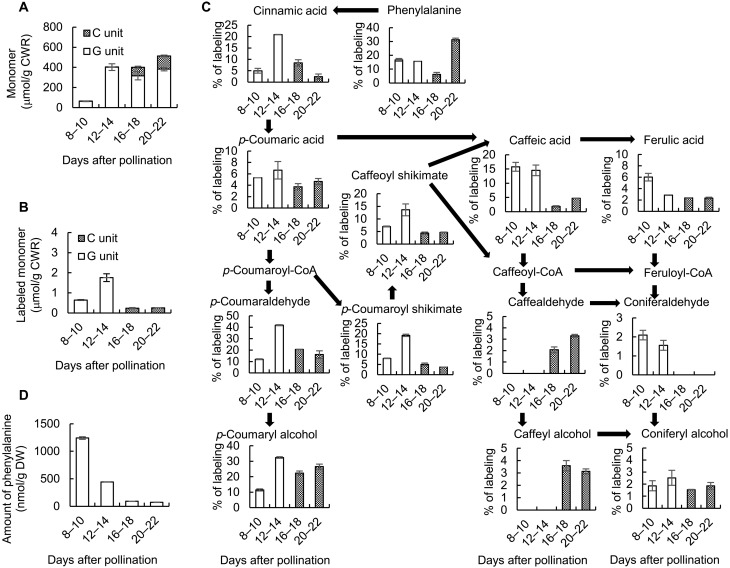
To determine whether the high level of coniferyl alcohol present in the seed coat at later stages of development was a result of continued synthesis or back-up of preexisting compound due to reduced polymerization, we performed a series of labeling experiments. After first testing different labeling periods and treatment regimens (fig. S5), we fed cut half seeds with 13C9–l-phenylalanine (Phe) at 8, 12, 16, or 20 DAP and then harvested the seed coats 2 days later (Fig. 2). Lignin was extracted from cell wall preparations and label-incorporated into thioacidolysis products derived from the constituent monolignols determined as described previously (12). As expected, no C-lignin was synthesized at the two earliest labeling periods (Fig. 2, A and B), and 13C-Phe was only incorporated into G-lignin units (Fig. 2B). Conversely, 13C-Phe was incorporated into C-lignin units at 16 and 20 DAP but not incorporated into G units (Fig. 2B).
Fig. 2. Labeling of lignin and monolignol pathway intermediates with 13C–l-phenylalanine during C. hassleriana seed coat development.
(A) Total lignin monomer thioacidolysis yields from isolated cell walls after feeding seed coats with 13C-Phe for the times shown (DAP). (B) Total thioacidolysis yields of labeled lignin monomers in the preparations in (A). Yields were calculated for the M + 7 ions by the method described previously (12). (C) Percentage labeling of individual pathway intermediates as determined by LC-MS analysis. (D) Total phenylalanine amount as determined by LC-MS analysis. Approximately 100 mg of seed coats isolated from seeds harvested from each individual Cleome plant were counted as one biological replicate. Data are means ± SE derived from three biological replicates. CWR, cell wall residue.
We then determined the incorporation of label from 13C-Phe into monolignol pathway intermediates in the seed coats. There was no measurable incorporation of label into the immediate precursors of C-lignin, caffealdehyde, and caffeyl alcohol, with labeling at 8 to 10 or 12 to 14 DAP (Fig. 2C), consistent with the results in Fig. 2B, indicating no labeling of C-lignin at these stages. Furthermore, although less label appeared in intermediates potentially common to both G- and C-lignin biosynthesis (coumaroyl shikimate, caffeoyl shikimate, and caffeic acid) from label applied at 16 and 20 DAP, there was now a large increase in label in caffealdehyde and caffeyl alcohol. On the basis of the lack of incorporation of label from 13C-Phe into G-lignin at the later labeling periods (Fig. 2B), it was expected that the pool of coniferyl alcohol would remain unlabeled when 13C-Phe was applied at the later times. However, this was not the case, and the incorporation of label into coniferyl alcohol was very similar when 13C-Phe was applied at 8, 12, 16, or 20 DAP. In sharp contrast, there was no incorporation of label into coniferaldehyde during the two later labeling periods (Fig. 2C). This suggests that, at later time points, labeled caffeyl alcohol may be converted to coniferyl alcohol, although this coniferyl alcohol is not converted to G-lignin.
The pool size of L-Phe may vary during seed development in view of its being a substrate for protein synthesis and a product of protein degradation in addition to its involvement as a precursor of lignin. This was confirmed by determining the size of the L-Phe pool, and its percentage of labeling after feeding 13C-Phe, at the four different periods of seed coat development (Fig. 2, C and D). To bypass the Phe pool, we repeated the labeling experiments with 13C–trans-cinnamic acid as primary substrate. The results (fig. S6) indicated that trans-cinnamic acid is incorporated into G- and C-lignin similarly to L-Phe and confirmed that coniferyl alcohol is still being synthesized from upstream precursors at later stages of development.
The extracts from this experiment were also analyzed for label incorporation into C-lignan (benzodioxane dimers). With labeling from 16 to 18 DAP, 46 nmol/g dry weight (DW) of C-lignan benzodioxane dimers were labeled as compared with 370 nmol/g DW of labeled C-lignin. A not insignificant proportion of the caffeyl alcohol synthesized during the later stages of seed coat development is therefore destined for lignan biosynthesis.
Biosynthesis of lignin from 13C-labeled coniferyl and caffeyl alcohols during seed coat development
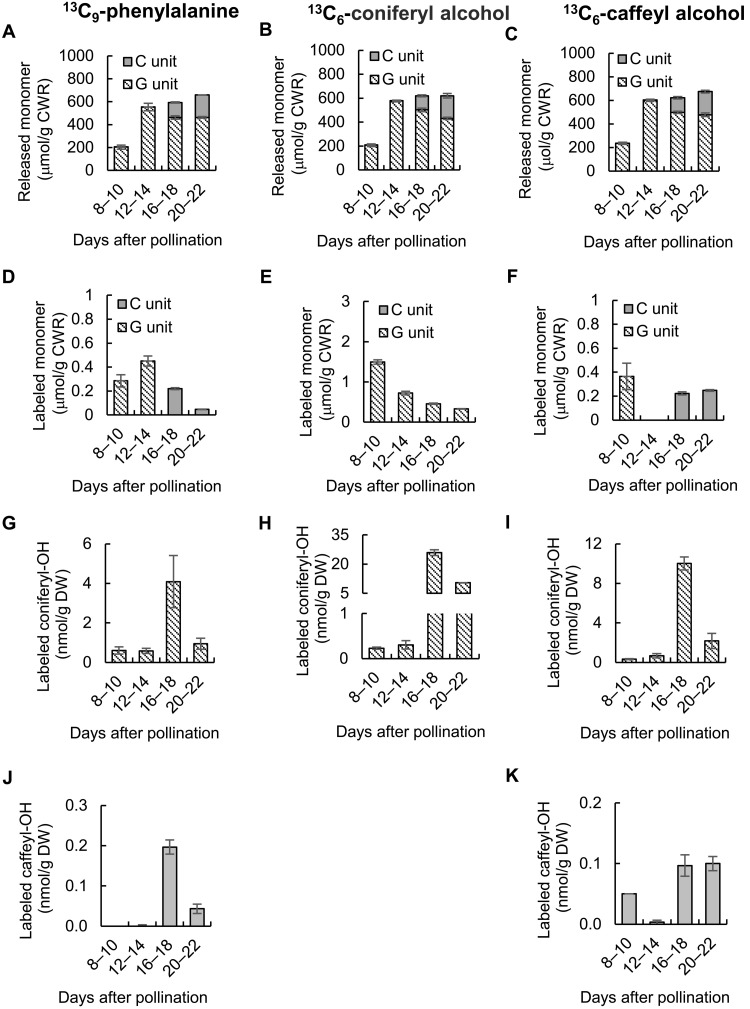
The above results indicate that C-lignin does not accumulate in the Cleome seed coat before around 12 to 14 DAP because its precursor, caffeyl alcohol, is not available. However, the reason for the lack of G-lignin biosynthesis after 12 to 14 DAP is not clear as a relatively large pool of newly synthesized coniferyl alcohol is present at this time. To determine whether factors beyond substrate availability allow seed coats to synthesize G- and C-lignins at different times, we first fed cut half-seeds with 13C6-coniferyl alcohol or 13C6-caffeyl alcohol, chemically synthesized as described previously (12), at 8, 12, 16, and 20 DAP, harvesting the seed coats 2 days later (Fig. 3). 13C-Phe labeling was also repeated with the new samples. Overall lignin contents and compositions were not affected by precursor feeding (Fig. 3, A to C). Label from 13C-coniferyl alcohol was incorporated into G units of lignin; however, although most incorporation was seen when feeding at 8 to 10 DAP, there was still incorporation at 16 to 18 or 22 to 24 DAP, comparable to the incorporation of 13C-Phe at 12 to 14 DAP (Fig. 3, D and E). As expected, 13C-caffeyl alcohol was incorporated into C-lignin at 16 to 18 and 20 to 22 DAP (Fig. 3F) but was not incorporated into G-lignin at these times. However, labeled G-lignin was observed in seed coats fed with 13C-caffeyl alcohol at 8 to 10 DAP (Fig. 3F). This suggests that, during the earlier labeling period, exogenously supplied caffeyl alcohol is converted to coniferyl alcohol by the O-methyltransferases that are not down-regulated until after 12 DAP (23), and this coniferyl alcohol is then incorporated into G-lignin.
Fig. 3. Incorporation of 13C–l-phenylalanine, 13C-coniferyl alcohol, and 13C-caffeyl alcohol into lignin and monolignol pools in developing seed coats of C. hassleriana.
(A to C) Thioacidolysis yields of total G- and C-monolignols from lignin isolated from Cleome seed coats fed 13C-Phe (A), 13C-coniferyl alcohol (B), or 13C-caffeyl alcohol (C) at the times shown. (D to F) Thioacidolysis yields of 13C-labeled G- and C-monolignols from lignin isolated from Cleome seed coats fed 13C-Phe (D), 13C-coniferyl alcohol (E), or 13C-caffeyl alcohol (F) at the times shown. (G to I) 13C-labeled coniferyl alcohol extracted from Cleome seed coats fed 13C-Phe (G), 13C-coniferyl alcohol (H), or 13C-caffeyl alcohol (I) at the times shown. (J and K) 13C-labeled caffeyl alcohol extracted from Cleome seed coats fed 13C-Phe (J) or 13C-caffeyl alcohol (K) at the times shown. Amounts of labeled monomers in cellular pools, or as components of lignin, were calculated for the M + 7 ions (13C-Phe) or M + 6 ions (13C-coniferyl alcohol and 13C-caffeyl alcohol) by the method described previously (12). Approximately 100 mg of seed coats isolated from seeds harvested from each Cleome plant were counted as one biological replicate. Data are means ± SE derived from three biological replicates.
Last, we measured the amounts of labeled caffeyl and coniferyl alcohols in the seed coats after feeding 13C-Phe, 13C-caffeyl alcohol, and 13C-coniferyl alcohol during the four time periods (Fig. 3, G to K). As previously observed, 13C-Phe only labeled caffeyl alcohol during the two later periods (Fig. 3J), whereas coniferyl alcohol was labeled at all the times, with the highest labeling at 16 to 18 DAP (Fig. 3G). When 13C-coniferyl alcohol was fed, it accumulated to higher levels at the later times (Fig. 3H), consistent with the fact that it is not being converted to lignin during the period of C-lignin biosynthesis. In contrast, when 13C-caffeyl alcohol was fed, its levels in the seed coat were higher at the times when C-lignin was being made than at the earlier times when only G-lignin was made, and very little was detected when fed at 12 to 14 DAP (Fig. 3K). At the same time, much larger amounts of labeled coniferyl alcohol appeared, especially at 16 to 18 DAP (Fig. 3I). These results directly confirm that caffeyl alcohol can be converted to coniferyl alcohol; paradoxically, this conversion is greatest at the time of C-lignin biosynthesis when the O-methyltransferases classically involved in the biosynthesis of coniferyl alcohol have been down-regulated (16, 23). This coniferyl alcohol is not, however, converted to G-lignin. Because the initial feeding was to cut half-seeds, with the seed coat being dissected at the end of the labeling period, some of the conversion might occur in the embryo, and ectopic formation of G-lignin at the wounded surface cannot be ruled out.
MFA suggests that different routes through the monolignol pathway operate during the phases of G- and C-lignin synthesis
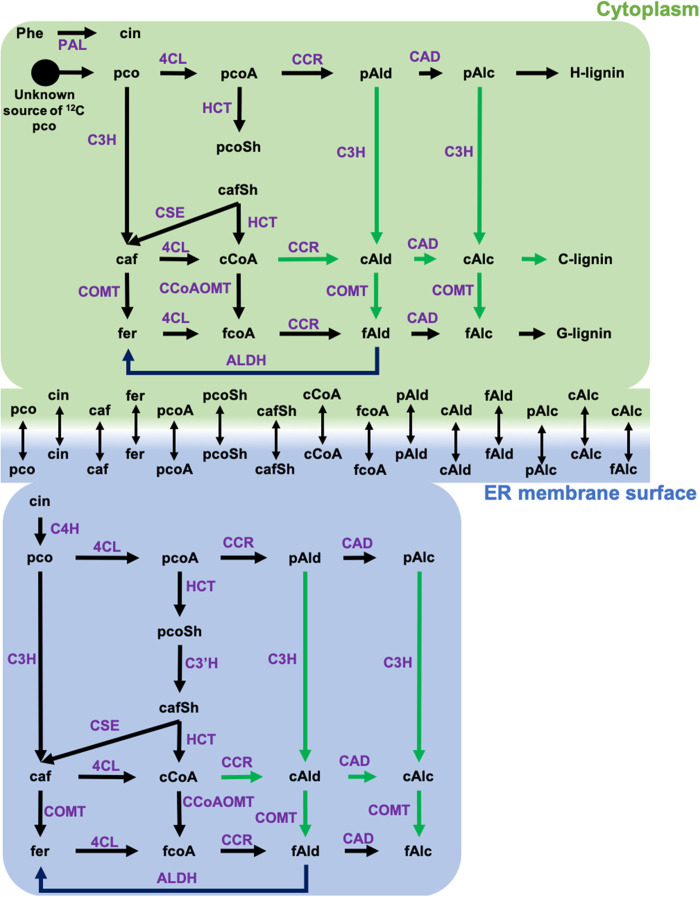
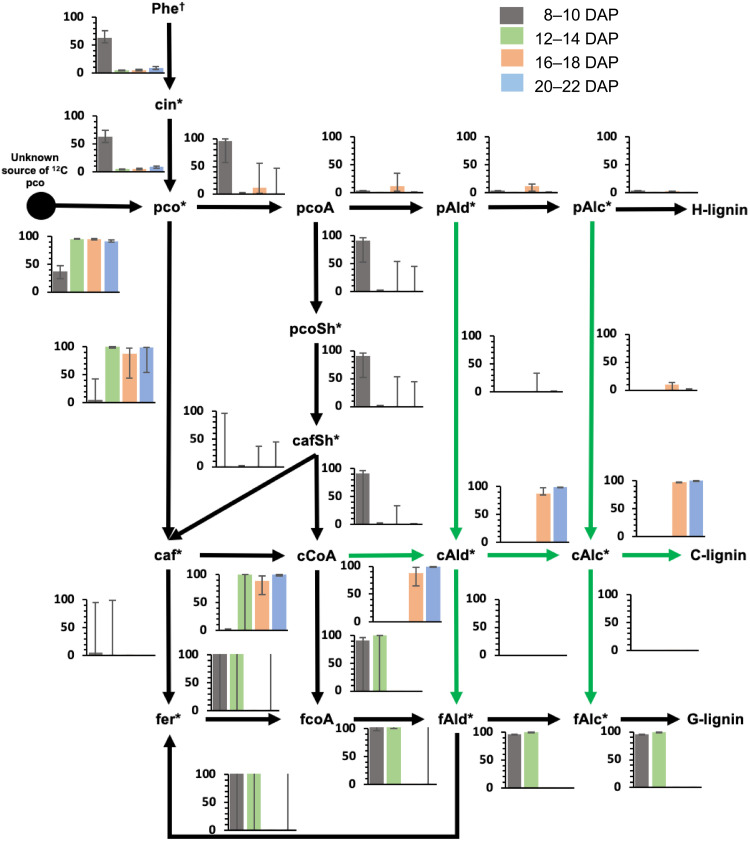
Figure 1A presented a simplified model for the monolignol pathway. In reality, multiple metabolic routes for C- and G-lignin synthesis are theoretically possible, based on the existence of parallel pathways for ring hydroxylation and O-methylation, as well as side chain reduction (23, 32). However, the quantitative extent of involvement of alternative routes is poorly understood for lignin biosynthesis in any species. We used 13C-MFA with labeling data from the 13C-Phe labeling experiments (dataset S2) to derive feasible ranges for the fluxes through the interconnected pathways at different times of labeling during seed coat development, using the soluble and membrane-associated intermediate pool model for the monolignol pathway derived by Faraji et al. (33, 34).
The metabolic network used for the MFA is shown in Fig. 4 and dataset S3. Caffealdehyde and caffeyl alcohol were omitted from the model at the two earlier labeling periods as they are not detected at these times. Obtaining a good fit between measured and calculated labeling data required introducing an additional reaction that introduces unlabeled coumarate. Removing this ad hoc coumarate source from the model increased the sum of squared residuals (SSR) between measured and calculated labeling data by at least 12-fold and resulted in poor resolution of fluxes through the network (fig. S7A). The coumarate source reaction carried a nonzero flux, varying from 0.3 to 19 times the flux carried by l-phenylalanine ammonia-lyase across the different time points. The necessity for this reaction and its higher flux can be explained partially by the low amount of label (4.5%) in the overall output [hydroxyphenyl (H)-lignin + G-lignin or H-lignin + C-lignin] of the pathway at any time. During the 8 to 10 and 12 to 14 DAP labeling periods, the ratio of G- to H-lignin demand was 24:1 and 480:1, respectively. During the 16 to 18 and 20 to 22 DAP labeling periods, the ratio of C- to H-lignin demand was 38:1 and 190:1, respectively. However, the labeling data clearly indicate that the precursor of both C-lignin (<3.6% labeled caffeyl alcohol) and G-lignin (<2.5% labeled coniferyl alcohol) are less enriched in 13C label when compared to H-lignin (11.4 to 32.4% labeled p-coumaryl alcohol) at all time points. Thus, the flux through the pathway is sustained more by an unlabeled precursor than the labeled Phe or cinnamate supplied through feeding. Our assumption in the present case is that unlabeled coumarate in the half-seed, levels of which greatly exceed those of cinnamate (fig. S1,S2), is contributing to this input.
Fig. 4. Schematic showing the metabolic network used for elucidating the fluxes from 13C labeling data.
On the basis of earlier modeling work carried out in B. distachyon (33, 34), the biosynthesis of lignin was assumed to occur at two different locations: cytosol (c) and surface of endoplasmic reticulum (er). Reactions catalyzed by membrane-bound enzymes such as C3′H and C4H were confined to the ER compartment of the model. The remainder of the reactions was included in both the compartments. Exchange reactions were added for compounds present in both compartments to allow for their free exchange. Reactions highlighted with green arrows were included only in the 16 to 18 and 20 to 22 DAP models. caf, caffeic acid; cafSh, caffeoyl shikimate; cAlc, caffeyl alcohol; cAld, caffealdehyde; cCoA, caffeoyl CoA; cin, cinnamic acid; fAlc, coniferyl alcohol; fAld, coniferaldehyde; fCoA, feruloyl CoA; fer, ferulic acid; pco, p-coumaric acid; pcoA, p-coumaroyl CoA; pcoSh, p-coumaroyl shikimate; pAld, p-coumaraldehyde; pAlc, p-coumaryl alcohol.
The flux distribution in seed coats at 8 to 10 DAP (Fig. 5) indicates that the shikimate shunt with the two-step involvement of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) (Fig. 4) acts as the major route to G-lignin. The caffeoyl shikimate node carried at least 54% of the flux directed toward G-lignin biosynthesis. From caffeoyl shikimate, the exact route taken to produce feruloyl-CoA could not be traced due to poor resolution. The activity of the reaction catalyzed by caffeic acid–forming coumarate 3-hydroxylase (C3H) (Fig. 4) (35) was unresolved, and its contribution to G-lignin biosynthesis could not be precisely estimated.
Fig. 5. 13C-MFA reveals different routes to monolignols during the phases of G- and C-lignin biosynthesis.
The bars in the graphs indicate the flux values (in micromoles per gram of DW per day) scaled to total p-coumaric acid synthesis flux of 100 μmol/g DW per day. Confidence intervals (95%) estimated for the fluxes are represented by the error bars. Reactions highlighted by green arrows were included only in the 16 to 18 and 20 to 22 DAP models. Dagger (†) indicates phenylalanine labeling data that were used only for 8 to 10 and 20 to 22 DAP. Asterisks (*) indicate metabolites whose labeling data were used in the flux estimation.
The flux distribution from feeding at 12 to 14 DAP (Fig. 5) was different from that obtained for 8 to 10 DAP in that the shikimate shunt was found to carry a negligible flux and at least 97% of the p-coumaric acid flux toward G-lignin biosynthesis was carried through C3H. However, similar to what was observed for labeling at 8 to 10 DAP, the exact route taken to produce feruloyl-CoA could not be determined because of poor resolution.
The flux distributions obtained for 16 to 18 and 20 to 22 DAP (Fig. 5) indicated that the direct conversion of p-coumaric acid to caffeic acid was active with a nonzero lower bound on its flux interval. However, the activity of the shikimate route was unresolved with zero lower bound and a nonzero upper bound equal to ~50% of flux toward C-lignin formation. Thus, it is likely that both the acids pathway and the shikimate shunt are active during the C-lignin biosynthesis phase. Caffeoyl-CoA acts as the major source of caffealdehyde, which is a major precursor of C-lignin. Theoretical reactions catalyzing ring 3-hydroxylation at the aldehyde and alcohol levels were predicted to be active only in the 20 to 22 DAP labeling period (fig. S7B). They carried negligible fluxes in the 16 to 18 DAP labeling periods, and therefore, it is likely that they are inactive before 20 DAP (fig. S7B). These reactions are currently unsupported by direct enzymatic evidence (35).
Removal of the aldehyde dehydrogenase (ALDH) that can generate ferulic acid from coniferaldehyde (Fig. 4) (36) caused a 2.4- and 2.5-fold increase in SSR in seed coats labeled at 8 to 10 and 20 to 22 DAP (fig. S7C). This suggests that the ALDH is active in the early G-lignin synthesis phase and in the late C-lignin synthesis phase. There was only a marginal increase (1.2-fold) in SSR for the 12 to 14 DAP dataset, making it difficult to conclude on the activity of ALDH during this period. However, in the 12 to 14 DAP labeling period, the percentage labeling of ferulic acid (2.85%) was more similar to that of coniferaldehyde (1.56%) than caffeic acid (14.52%), favoring the hypothesis that ALDH is active during this period. There was no change in SSR on removal of ALDH for the 16 to 18 DAP (early C-lignin synthesis phase) (fig. S7C).
Caffeyl alcohol inhibits the oxidation of coniferyl alcohol by Cleome seed coat cell wall laccases
The oxidation of monolignols to form lignin polymers takes place in the apoplast and involves the concerted activities of laccases (EC 1.10.3.2) and class III peroxidases (EC 1.11.17) (8, 37–39). It has been suggested that laccases are responsible for the initiation of lignin polymerization in planta, because a laccase triple mutant of Arabidopsis essentially produces no lignin in vascular tissues or fibers despite expressing a complete suite of peroxidases (40). Laccases are glycoproteins that are associated with the cell wall, but they can be released from isolated walls by extraction in 1 M calcium chloride (38). We therefore examined the ability of such cell wall extracts to catalyze the oxidation of caffeyl and coniferyl alcohols.
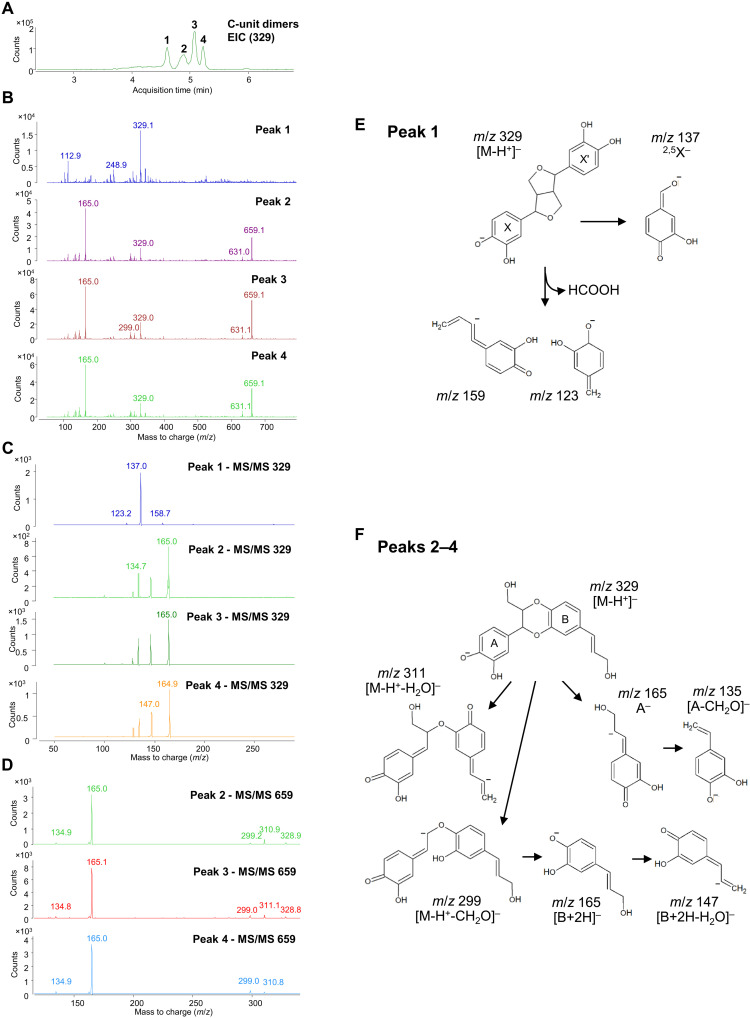
First, we validated the assay for Cleome seed coat laccase activity. The disappearance of the monolignol substrate can be monitored by the reduction in its ultraviolet absorption after separation by high-performance LC (HPLC) (11, 12). However, it is important to confirm the appearance of products in the form of dimers and larger oligomers in these assays (12). We therefore added caffeyl alcohol to protein extracts from Cleome seed coat cell walls in the presence of catalase to inhibit peroxidases, incubated for 4 hours, and analyzed the products by HPLC-MS. Chromatograms from reactions with caffeyl alcohol were scanned and extracted with mass/charge ratio (m/z) = 329 and 493 to detect the corresponding dimer (Fig. 6) and trimer (fig. S8), respectively. Chromatograms revealed the accumulation of the expected products in the extracts, but there was only background noise in the corresponding blanks. As there were multiple peaks for the products, we ran subsequent tandem MS (MS/MS) analyses to confirm their molecular compositions. Among the four peaks of potential dimer (Fig. 6A), the fragment m/z = 329 of peak 1 (Fig. 6B) was broken into ions of m/z = 159, 137, and 123 (Fig. 6C) via the fragmentation pattern shown in Fig. 6E, consistent with this dimer representing the resinol structure. Subsequent MS/MS analysis of peaks 2 to 4 (Fig. 6, C and D) revealed the presence of ions of m/z = 329 and 659, the latter suggesting coupling of two dimers. The m/z = 659 fragment was broken into ions of m/z = 329, 311, 299, 165, and 135 (Fig. 6D) via the fragmentation pathway shown in Fig. 6F, consistent with peaks 2 to 4 being benzodioxane linked dimers. All the fragments of m/z = 329 were broken into ions of m/z = 165,147, and 135 (Fig. 6C), indicating that peaks 2 to 4 were stereoisomers of isoamericanol A and americanol A (fig. S3). In the case of the five peaks of potential trimers, a similar approach was used for identification (fig. S8), revealing the presence of ions of m/z = 165 from the m/z = 493 fragment by MS/MS analysis of all five peaks and the presence of ions of m/z = 137 and 159 from peak 2 (fig. S8C). These results suggest that peak 2 was a combination of resinol- and benzodioxane-linked structures and the other trimers contain benzodioxane structures exclusively. Similar results were obtained monitoring the dimers formed from coniferyl alcohol (m/z = 375 and 357 for β-β– and β-5–, and β-O-4–linked structures, respectively) (fig. S9) according to Morreel et al. (41).
Fig. 6. Identification of caffeyl alcohol–derived dimers generated following incubation of Cleome seed coat cell wall protein extracts with caffeyl alcohol.
(A) Extracted ion chromatogram (EIC) at m/z = 329 showing four peaks of caffeyl alcohol dimers. (B) Mass spectra of peaks 1 to 4 in (A). (C) MS/MS spectra of the m/z = 329 peaks in (B). (D) MS/MS spectra of the m/z = 659 peaks from peaks 2 to 4 in (B). (E) Fragmentation pattern of the compound in peak 1 in (A). (F) Fragmentation patterns of the compounds in peaks 2 to 4 in (A). Potential structures of caffeyl alcohol benzodioxane dimers are shown in fig. S3.
To determine the efficiency of the cell wall protein extraction method, cell wall residues were extracted sequentially with CaCl2, dithiothreitol (DTT), NaCl, and borate, according to the procedure of Robertson et al. (42). No laccase activity was detected in DTT extracts, whereas low activity against both caffeyl and coniferyl alcohols was seen in NaCl and borate extracts (fig. S10, A and B). However, because the initial CaCl2 extraction removed 85 to 91% of cell wall laccase activity, this extract was used for subsequent kinetic experiments.
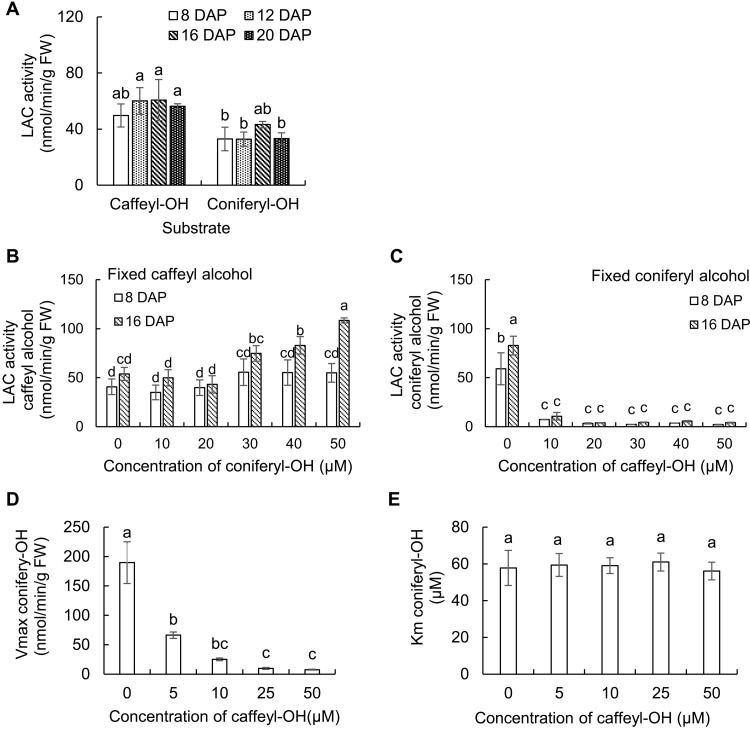
Having confirmed that the cell wall protein extracts catalyzed the oligomerization of monolignols, we next measured the rates of oxidation of caffeyl and coniferyl alcohols, as measured by substrate disappearance, in protein extracts from Cleome seed coat cell walls isolated at 8, 12, 16, and 20 DAP. The relative activities with the different substrates were not significantly different at the different time points. Caffeyl alcohol was slightly preferred with extracts from all time points, and coniferyl alcohol was oxidized at around 63% the rate of caffeyl alcohol overall (Fig. 7A). We then determined the effects of coniferyl alcohol on the oxidation of caffeyl alcohol, and vice versa, in the cell wall protein extracts. At a fixed concentration of 50 μM caffeyl alcohol, addition of increasing concentrations of coniferyl alcohol up to 50 μM unexpectedly resulted in increased oxidation of caffeyl alcohol, and this effect was greater with extracts from seed coats at 16 DAP than at 8 DAP (Fig. 7B). In contrast, inclusion of only 10 μM caffeyl alcohol inhibited the oxidation of 50 μM coniferyl alcohol by over 80% and by over 90% at concentrations of 20 μM and above (Fig. 7C). Using cell wall protein extracts from seed coats at 16 DAP, we determined the kinetics for the inhibition of laccase-mediated coniferyl alcohol oxidation by caffeyl alcohol. The Vmax for coniferyl alcohol was reduced by increasing concentrations of caffeyl alcohol, whereas the Km was unchanged, indicating noncompetitive inhibition (Fig. 7, D and E). The Ki for caffeyl alcohol was 1.9 ± 0.3 μM.
Fig. 7. Laccase activities in the C. hassleriana seed coat.
(A) Laccase activity against caffeyl and coniferyl alcohols in protein extracts from Cleome seed coat cell walls during different developmental stages. Activity was calculated by substrate disappearance monitored by HPLC. Identification of reaction products is provided in Fig. 6 and figs. S8 and S9. (B and C) Effects of coniferyl alcohol on the oxidation of caffeyl alcohol, and vice versa, in cell wall protein extracts from Cleome seed coats at 8 and 16 DAP. Increasing concentrations of one monolignol were added, while the concentration of the other was maintained at 50 μM. (D and E) Kinetic properties of laccases in cell wall protein extracts from Cleome seed coats at 16 DAP against coniferyl alcohol in the presence of caffeyl alcohol. Protein extracts were incubated with 25 to 200 μM coniferyl alcohol and 0 to 50 μM caffeyl alcohol, and kinetic constants were calculated and plotted. Laccase activities were measured in the presence of catalase to inhibit peroxidase as described in Materials and Methods. Approximately 300 mg of seed coats isolated from seeds harvested from each individual Cleome plant were counted as one biological replicate. Data are means ± SE derived from three biological replicates. The different letters above the bars represent statistically significant differences determined by analysis of variance (ANOVA) (Duncan, P ≤ 0.05) with SPSS Statistics (version 27; IBM) FW, fresh weight.
Peroxidase activities in the cell wall extracts were then determined by incubation with monolignol in the absence of catalase and presence of hydrogen peroxide; owing to the short incubation time, laccase activity would be negligible. The peroxidase activities were three orders of magnitude greater than the laccase activity (fig. S10, A to C). Inhibition of peroxidase-mediated coniferyl alcohol oxidation by caffeyl alcohol was observed in CaCl2 extracts of cell wall proteins from Cleome seed coats at 8 and 16 DAP (fig. S10, D and E).
Roles of individual laccases in monolignol oxidation in the Cleome seed coat
Transcriptome analysis indicates that Cleome hassleriana LACCASE 5 (ChLAC5), ChLAC8, and ChLAC15 are the three laccase genes expressed in the Cleome seed coat at the time of C-lignin biosynthesis, and ChLAC8 and ChLAC15 are seed specific (table S1) (12). We have recently shown that ChLAC8 has a substrate preference for caffeyl alcohol and does not accept coniferyl alcohol when expressed in Escherichia coli (12). Genetic analysis indicates that ChLAC8 is both necessary and sufficient for C-lignin polymerization in vivo when ectopically expressed in Arabidopsis thaliana (12). LAC15 (also known as TT10) has not only been implicated in the oxidation of PAs in the Arabidopsis seed coat (43) but also been ascribed a potential role in lignin polymerization (44). ChLAC15, as well as ChLAC4 and ChLAC17 (homologs of Arabidopsis laccases involved in G- and S-lignin biosynthesis and also expressed in stems), is expressed in the seed coat during the period of G-lignin biosynthesis (12). To determine how the different laccases contribute to the oxidation of caffeyl alcohol and coniferyl alcohol, we cloned the ChLAC4, ChLAC5, ChLAC8, and ChLAC15 open reading frames and expressed them transiently in leaves of Nicotiana benthamiana. The 6x histidine (6xHis)–tagged enzymes were purified from crude protein extracts by Ni-affinity chromatography, and analysis by SDS–polyacrylamide gel electrophoresis before and after deglycosylation with peptide N-glycosidase F indicated that the enzymes had retained glycosyl substituents (fig. S11).
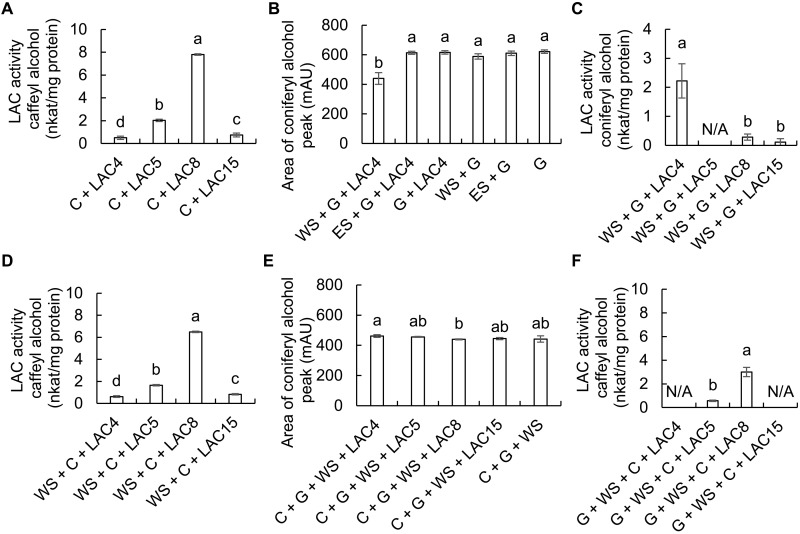
Incubation of the recombinant, plant-expressed Cleome laccases with caffeyl alcohol confirmed that ChLAC8 had the highest activity with this substrate, approximately four times higher than ChLAC5 (Fig. 8A). Lack of any activity of ChLAC8 with coniferyl alcohol (12) was confirmed, consistent with a specific function of ChLAC8 in C-lignin oligomerization. However, initial studies unexpectedly also failed to show any ability of recombinant ChLAC4, ChLAC5, or ChLAC15 to oxidize coniferyl alcohol. This is in contradiction to the oxidation of this monolignol by crude Cleome cell wall preparations from times when these enzymes are being expressed and under conditions in which peroxidase is inhibited (Fig. 7A). We reasoned that there may be a missing factor essential for initiating the oxidation of coniferyl alcohol and therefore generated water- and ethyl acetate–soluble fractions from available lignifying poplar fibers. These fractions were incubated with coniferyl alcohol (G) in the presence and absence of ChLAC4 (Fig. 8B); LAC4 has been shown genetically to be involved in classical lignin polymerization in Arabidopsis (40, 45). Addition of small amounts of the water-soluble fraction (WS) facilitated LAC4-catalyzed oxidation of coniferyl alcohol. We next compared the abilities of plant-expressed LAC4, LAC5, LAC8, and LAC15 to oxidize coniferyl alcohol in the presence of WS. LAC4 had by far the highest activity (Fig. 8C). Addition of WS to reactions with caffeyl alcohol had no effect (Fig. 8, A and D). Together, these results suggest that LAC4 is the major laccase in the Cleome seed coat involved in oxidation/polymerization of coniferyl alcohol (the appearance of products in the form of dimers was confirmed by LC-MS; fig. S12), whereas LAC8 is the laccase largely responsible for polymerization of caffeyl alcohol.
Fig. 8. Activity and competition assays with recombinant seed coat ChLACs expressed in N. benthamiana.
(A) Activity of recombinant Cleome LAC4, LAC5, LAC8, and LAC15 against caffeyl alcohol. (B) Effects of water- and ethyl acetate–soluble extracts from poplar fibers on the oxidation of coniferyl alcohol by ChLAC4. Data are the areas of the coniferyl alcohol peak determined by HPLC. mAU, milli–absorbance units. (C) Activity of recombinant Cleome LAC4, LAC5, LAC8, and LAC15 against coniferyl alcohol in the presence of water-soluble extract (WS) from poplar fibers. (D) Activity of recombinant Cleome LAC4, LAC5, LAC8, and LAC15 against caffeyl alcohol in the presence of WS. (E) Effects of caffeyl alcohol on the oxidation of coniferyl alcohol by recombinant Cleome LAC4, LAC5, LAC8, and LAC15 in the presence of WS. Data are the areas of the coniferyl alcohol peak determined by HPLC. (F) Oxidation of caffeyl alcohol by ChLAC4, ChLAC5, ChLAC8, and ChLAC15 in the presence of coniferyl alcohol and WS. C, caffeyl alcohol; G, coniferyl alcohol; ES, ethyl acetate–soluble fraction from poplar fibers; N/A, no activity. Reactions (100 μl) contained 50 μM of each monolignol substrate and 300 ng of purified recombinant protein. Activity was calculated by substrate disappearance as monitored by HPLC. Water- and ethyl acetate–soluble fractions from poplar fibers were prepared as described in Materials and Methods and added at a level of 1 μl per 100 μl of reaction. Recombinant LAC protein purified from infiltrated tobacco (N. benthamiana) leaves harvested from three plants was counted as one biological replicate. Data are means ± SE derived from three biological replicates. The different letters above the bars represent statistically significant differences determined by ANOVA (Duncan, P ≤ 0.05) with SPSS Statistics (version 27; IBM). nkat, nano katals.
Next, we reexamined the ability of caffeyl alcohol to inhibit the oxidation of coniferyl alcohol, this time using the purified recombinant laccases with addition of WS to facilitate coniferyl alcohol oxidation. The oxidation of coniferyl alcohol observed on addition of LAC4 and WS (Fig. 8B) was not observed in the presence of 50 μM caffeyl alcohol, although coniferyl alcohol oxidation was stimulated by the presence of WS (Fig. 8, B and E). However, the stimulation of caffeyl alcohol oxidation by coniferyl alcohol observed with crude cell wall laccases (Fig. 7C) did not occur with recombinant laccases in the presence of WS; rather, the presence of excess coniferyl alcohol and WS inhibited, but did not block, caffeyl alcohol oxidation by LAC8 (Fig. 8, A and F).
Next, we asked whether afzelechin, a precursor of the CTs made primarily during the first 14 days of seed coat development, might influence the oxidation of caffeyl or coniferyl alcohols. Equimolar concentrations of afzelechin neither inhibited nor stimulated the oxidation of caffeyl alcohol by ChLAC8 nor facilitated the oxidation of coniferyl alcohol by ChLAC4 in the absence of WS (fig. S13, A and B). Last, we examined the ability of ChLAC15, the ortholog of Arabidopsis TT10 that functions in the oxidation of CTs (43) to oxidize caffeyl or coniferyl alcohols in the presence of afzelechin. ChLAC15 neither oxidized afzelechin nor oxidized caffeyl or coniferyl alcohols in the presence of afzelechin (fig. S13C). Among the recombinant laccases, only ChLAC8 could oxidize afzelechin (fig. S13D).
DISCUSSION
Monolignol precursor pools do not predict lignin composition in the Cleome seed coat
LC-MS–based targeted metabolite profiling of both seed coats and whole seeds revealed that the level of the G-lignin precursor coniferyl alcohol was, paradoxically, approximately fourfold higher at 16 DAP, during the period of C-lignin biosynthesis, than at 8 DAP when G-lignin is being made. The C-lignin precursor caffeyl alcohol was not detected at 8 DAP and was present at approximately 300 nmol/g DW (less than half the level of coniferyl alcohol) at 16 DAP. Levels of the H-lignin precursors coumaraldehyde and p-coumaryl alcohol were very low at 8 DAP but substantially increased at 16 DAP, although no H-lignin is produced from phenylalanine at this time. This suggests the possibility of a pathway from p-coumaryl alcohol to caffeyl alcohol in the seed coats. However, such a conversion is not catalyzed by the bifunctional ascorbate peroxidase/C3H (35), and, furthermore, MFA did not support its operation. The metabolic role/fate of p-coumaryl alcohol at the later times of seed coat development therefore remains unclear.
MFA reveals nonclassical routes to monolignol pathway intermediates during Cleome seed coat development
The MFA analysis was based on a model of the monolignol pathway in Brachypodium distachyon that postulates equilibration of pools of metabolites between soluble and endoplasmic reticulum (ER)–associated compartments (33, 34). To obtain minimum SSR values, it was necessary to include an additional flux into unlabeled coumarate. Studies in B. distachyon have shown the presence of more than one pool of coumarate with different metabolic fates (46), associated with the presence of both phenylalanine and tyrosine ammonia lyases in this species (46). To the best of our knowledge, cinnamate, tyrosine, and various coumarate esters are the only direct precursors of coumarate. Our labeling strategy using half seeds was optimal for label incorporation, but it is possible that the large unlabeled pool of coumarate external to the seed coat could provide substantial flux into monolignol biosynthesis.
The 3-hydroxylation of the monolignol aromatic ring can occur either at the level of the shikimate ester through the cytochrome P450 C3′H (47) or at the level of the free acid (coumarate) through C3H (35). The genes encoding the two key enzymes of the classical shikimate esters pathway, HCT and C3′H, are expressed at similar levels throughout seed coat development (23), and, after initially dropping, levels of both caffeoyl shikimate and caffeic acid increase between 12 and 16 DAP. MFA suggested that the esters pathway is the major route for G-lignin biosynthesis at 8 to 10 DAP. However, the acids route via C3H then becomes dominant at the time of the switch to C-lignin formation, with both pathways likely operating at later times.
Accumulation of lignans and CTs is temporally separated in the Cleome seed coat
The Cleome seed coat contains a wide range of lignans derived from caffeyl alcohol. Some of these accumulate to very high levels, but only during the phase of C-lignin biosynthesis. One specific benzodioxane-linked C-lignan, trans-isoamericanol, was the most abundant metabolite detected in the seed coats after sucrose and glucose. Two aspects of these results are unexpected. The first is that no lignans derived from coniferyl alcohol, other than dehydrodiconiferyl alcohol and traces of pinoresinol, or from p-coumaryl alcohol, accumulated to substantial levels in the seed coats at any stage of development. These molecules are commonly found in lignifying tissues (48, 49). It is also unexpected that no mixed G-C lignans were observed, as down-regulation of CCoAOMT results in formation of isomers of benzodioxane-linked coniferyl alcohol-8-O-4-caffeic acid dimer in Arabidopsis vacuoles (48). Second, the diversity of C-lignans, with multiple linkage types, is in notable contrast to the structure of C-lignin, in which all the units are joined through β-O-4 linkages, with subsequent internal cyclization to create benzodioxane units (14). Most commonly, lignans are linked via so-called β-β′ bonds between the central atoms of the respective side chains (position 8); 3-3′, 8-O-4′, or 8-3′ bonds are observed less frequently, and, in these cases, the dimers are called neolignans.
The exact localization of the lignans in the Cleome seed coat remains to be determined. Lignans are thought to be synthesized in the apoplast by the combined activities of a laccase and a dirigent protein (50). However, the presence of at least three different linkage types in the Cleome seed coat, along with different stereochemistries, suggests that C-lignan formation may only be under chemical control and not involve dirigent proteins to control the stereospecificity of the coupling (50). The presence of a wide range of glycosylated monolignol dimers and higher oligomers in the vacuoles of Arabidopsis has led to the hypothesis that lignans may also be made in the cytosol (48). The G-lignan dehydrodiconiferyl alcohol has been shown to exist as its glucoside in Arabidopsis vacuoles (48). It is notable that no C-lignan glycosides were detected in the present work, suggesting that the C-lignans may be present in the Cleome seed coat cell wall, where they might act as defensive compounds. The fact that ChLAC8, with an extracellular transit peptide, can form in vitro the multiple C-monolignol dimer types (resinol and benzodioxane stereoisomers) extracted from seed coat tissues is consistent with these lignans being made and accumulated in the apoplast.
CTs accumulate early during Cleome seed coat development (12) and are shown here to be of the less common propelargonidin type derived from the flavan 3-ol monomer afzelechin. Recent studies indicate that CT polymerization is nonenzymatic, initiated by attack of a carbocationic extension unit at the nucleophilic C8 position of a flavan-3-ol starter unit (51, 52). The laccase TT10 has been ascribed roles in the oxidation of CTs and also in lignin polymerization in the Arabidopsis seed coat, but a role in tannin oligomerization seems unlikely (44, 53). ChLAC15, an ortholog of Arabidopsis TT10, although expressed most highly at 10 DAP, is still strongly expressed during the period of C-lignin biosynthesis. ChLAC15 could oxidize caffeyl alcohol but did not appear to oxidize monomeric afzelechin, and the flavan 3-ol did not appear to affect laccase-mediated monolignol polymerization in vitro. These results suggest that lignin/lignan and CT oligomerization are nonoverlapping processes in the Cleome seed coat.
Different metabolic fates of coniferyl and caffeyl alcohols during the period of C-lignin biosynthesis
The fact that monolignols are made in the cytosol but polymerized in the apoplast is important for interpreting the present labeling experiments. Monolignol synthesized from an upstream precursor such as L-Phe or trans-cinnamic acid will have to transfer across the plasma membrane, whereas monolignols applied to a cut surface with vacuum infiltration could also find their way directly to the apoplast or be involved in ectopic lignification at the cut (wounded) surface. This distinction might explain why there was incorporation of applied coniferyl alcohol into G-lignin at 16 to 18 or 22 to 24 DAP, whereas 13C-Phe did not label G-lignin at these times. The incorporation of 13C-caffeyl alcohol into G-lignin at 8 to 10 DAP, but not at 12 to 14 DAP, is explained by the presence of COMT activity during the earlier labeling period; caffeyl alcohol is an excellent substrate for COMT (54). Bearing these points in mind, the labeling data show that the substrate pool available for polymerization in the apoplast is not simply a reflection of the overall cellular plus extracellular monolignol pool sizes.
The higher level of coniferyl alcohol during the period of C-lignin biosynthesis than during G-lignin biosynthesis might simply reflect the cessation of polymerization of coniferyl alcohol after around 12 DAP, consistent with the inhibition of coniferyl alcohol oxidation by caffeyl alcohol, discussed further below. If this were the only explanation, then the coniferyl alcohol accumulating at late time points would have been synthesized during the phase of G-lignin biosynthesis. However, this model is inconsistent with the continued de novo synthesis of coniferyl alcohol from L-Phe during the phase of C-lignin biosynthesis. The retained commitment for coniferyl alcohol biosynthesis raises two major questions: Why is coniferyl alcohol made when it is not being incorporated into lignin, and how is it made when the methylation machinery for its biosynthesis has been turned off to enable C-lignin biosynthesis (23)?
In addition to lignin, coniferyl alcohol is also a precursor of lignans such as the dehydrodiconiferyl alcohol glucosides that have been proposed to function as factors controlling cell division (55, 56). Coniferyl alcohol can also act as a signal molecule in plant-microbe interactions (57). Thus, it may be critical for more than one reason for plants to maintain a pool of coniferyl alcohol, even under conditions in which the nonmethylated caffeyl alcohol must be made in large amounts to support C-lignin biosynthesis.
The most extreme negative growth phenotypes in monolignol pathway mutants in Arabidopsis and Medicago truncatula result from simultaneous loss of function of COMT and CCoAOMT; the resulting dwarf seedlings die well before reaching maturity (49, 58). Although both classical COMT and CCoAOMT genes are strongly down-regulated at the onset of C-lignin biosynthesis in Cleome, seed coat development continues and coniferyl alcohol is still made. This suggests that the Cleome seed coat has a pathway to coniferyl alcohol that is independent of the activities of classical COMT and CCoAOMT, and this is supported by the relatively high incorporation of caffeyl alcohol into coniferyl alcohol between 16 and 22 DAP. Studies are underway to identify the O-methyltransferase(s) involved.
Caffeyl alcohol is an inhibitor of coniferyl alcohol oxidation by cleome seed coat laccases
In A. thaliana, loss of function of AtLAC4, AtLAC11, and AtLAC17 results in plants in which lignin can only be detected in the Casparian strip in the root (40), a tissue in which laccases are not necessary for lignin polymerization (40, 59). The fact that plants of the lac4, lac11, and lac17 triple mutant express a normal complement of peroxidase genes suggests that laccase is essential for initiating lignification in tissues other than the Casparian strip (40). Several laccases are expressed in the seed coat of Cleome; on the basis of transcript profiles, ChLAC4 and ChLAC17 are expressed mainly during the phase of G-lignin biosynthesis, as is ChLAC15, the ortholog of the TT10 gene of Arabidopsis (43). ChLAC8 expression is more closely correlated with C-lignin accumulation (12), and this enzyme, when expressed in a prokaryotic system that does not allow for glycan substitution of the laccase protein, demonstrates a strong preference for oxidation of caffeyl alcohol as compared to coniferyl alcohol (12). This enzyme is shown here to maintain this substrate preference when expressed transiently as a glycosylated protein in N. benthamiana.
Although the complement of laccases in the Cleome seed coat changes significantly during the switch from G- to C-lignin accumulation, the inhibitory effect of caffeyl alcohol on oxidation of coniferyl alcohol appears similar at all developmental stages. Of the recombinant seed coat-expressed Cleome laccases, only ChLAC4 could effectively oxidize coniferyl alcohol and then only in the presence of a yet-to-be identified water-soluble factor that is unlikely to be a protein/enzyme based on its extraction in methanol, followed by air drying and water/ethyl acetate partitioning. This reaction was totally inhibited by an equimolar concentration of caffeyl alcohol and by much lower concentrations when experiments were conducted with crude cell wall laccase preparations. The demonstration of noncompetitive inhibition of cell wall laccases by caffeyl alcohol with an apparent Ki of only 1.9 μM suggests that levels of caffeyl alcohol are sufficient to block oxidation of coniferyl alcohol in the apoplast and that increasing concentrations of coniferyl alcohol will not easily overcome this. Furthermore, the inhibition of peroxidase-mediated oxidation of coniferyl alcohol by caffeyl alcohol provides a further mechanism to ensure C-homopolymer biosynthesis at late stages of seed coat development.
A model for multilevel regulation of C-lignin biosynthesis
Phylogenetic studies suggest that C-lignin has evolved recently and often in certain plant lineages (15). The underlying mechanism of C-lignin biosynthesis should therefore not require a number of independently controlled events but rather should feature a simple switch that turns on a cascade of downstream events. The defining event is likely the transcriptional switch that turns on the down-regulation of COMT and CCoAOMT (23), resulting in the formation of caffeyl alcohol, a powerful noncompetitive inhibitor of coniferyl alcohol oxidation.
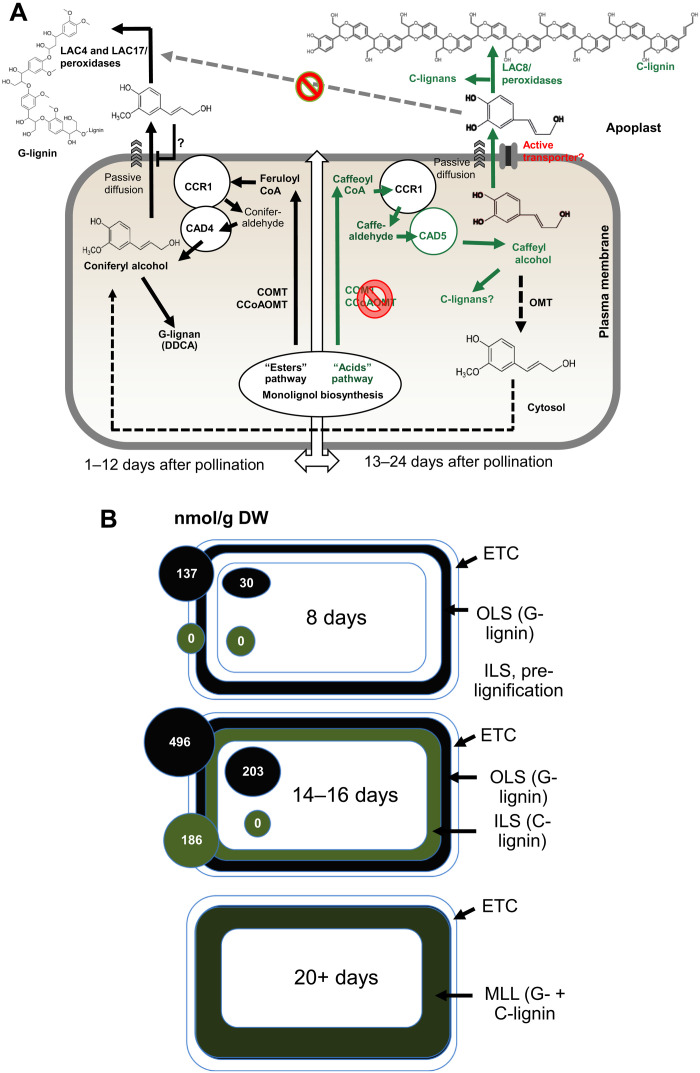
A comparison of the labeling of lignin from coniferyl alcohol generated in vivo or supplied exogenously appears to support control of the pathway at the level of monolignol transport across the apoplast. There is no incorporation of coniferyl alcohol formed in vivo from 13C–l-Phe between 16 and 22 DAP, whereas 13C-coniferyl alcohol supplied exogenously is incorporated at a level similar to that observed when it is fed at 8 to 14 DAP, possibly as a result of ectopic formation of G-lignin at cut surfaces and/or following vacuum infiltration. Figure 9A presents a model to explain the changes of lignin composition during development of the Cleome seed coat based on the present data. Before around 12 to 13 DAP, lignin biosynthesis proceeds by the classical pathway involving the shikimate shunt. No S-lignin accumulates because F5H is not expressed (23). At around 12 DAP, a switch (likely involving transcription factor expression) leads to suppression of the two monolignol O-methyltransferases, COMT and CCoAOMT, resulting in the biosynthesis of caffeyl alcohol, assisted by the expression of CAD5, a form of CAD with preference for caffealdehyde (23). The mechanism behind the shift in the monolignol pathway from the shikimate shunt to the “acids” pathway during the onset of C-lignin biosynthesis requires further study.
Fig. 9. A model for the control of lignin composition in the Cleome seed coat based on substrate availability, transport, and polymerization.
(A) Between 8 and 10 DAP, coniferyl alcohol is synthesized by the classical shikimate esters pathway via caffeoyl shikimate. Feruloyl CoA is converted to coniferaldehyde and thence to coniferyl alcohol by a proposed complex of ChCCR1 and ChCAD4. Formation of coniferyl alcohol close to the plasma membrane may facilitate its passive diffusion to the apoplast, where it is oxidized by ChLAC4 and ChLAC17 to oligomers; further polymerization may involve cell wall peroxidases. To date, there is no evidence for a specific coniferyl alcohol transporter and if the diffusion is passive, it may be facilitated by the “pull” from the reduced apoplastic concentration. After approximately 13 DAP, down-regulation of COMT and CCoAOMT (23) results in formation of caffeoyl CoA but now with a major contribution from the acids pathway involving C3H (35), which accounts for essentially all the flux to caffeyl alcohol at 12 to 14 DAP. Caffeoyl CoA is reduced via ChCCR1 and ChCAD5 (23) to caffeyl alcohol, which may again reach the apoplast by passive diffusion, possibly assisted by active transport. Caffeyl alcohol inhibits the polymerization of coniferyl alcohol, thereby also reducing its potential diffusion to the apoplast, and is itself polymerized to C-lignin via ChLAC8 (12). DDCA, dihydrodiconiferyl alcohol. (B) Spatiotemporal deposition of lignin in the Cleome seed coat. At 8 DAP, G-lignin is being deposited in an outer lignified sublayer (OLS) of cells interior to the extratestal sublayer (ETC). A distinct layer of cells interior to the OLS is visible microscopically but does not start to accumulate lignin (C-lignin) to become the interior lignified sublayer (ILS) until around 14 DAP. By 20 to 24 DAP, the two cell layers have merged (merged lignified layer, MLL) and contain distinct C- and G-lignin homopolymers (16). The concentrations of coniferyl alcohol (black circles) and caffeyl alcohol (green circles) in the seed coat and rest of the seed (internal to the seed coat) are indicated (from fig. S2).
Despite a previous demonstration of adenosine 5′-triphosphate–binding cassette transporters active with monolignol aglycones in plasma membrane vesicles (60), no G-monolignol (or S-monolignol) transporter has yet been identified at the molecular level (61). We therefore suggest that coniferyl alcohol likely arrives in the apoplast via passive diffusion (62). Work is in progress to determine whether there are specific transporters for caffeyl alcohol, although it is predicted to diffuse across a model plasma membrane more readily than coniferyl alcohol (62). Because of the low Ki value for caffeyl alcohol as an inhibitor of coniferyl alcohol oxidation, transporters may not be necessary to generate a sufficiently high concentration of free caffeyl alcohol to maintain inhibition of coniferyl alcohol oxidation, despite the higher concentrations of coniferyl alcohol than caffeyl alcohol in the seed coat (Fig. 9B). Furthermore, the passive diffusion model is suitably parsimonious in removing the need for additional factors for C-lignin accumulation, consistent with evolutionary considerations (15).
Although not critical for the model, it is possible that both cinnamyl alcohol dehydrogenase 4 (CAD4) and CAD5 are located near the plasma membrane through association with cinnamoyl-CoA reductase (CCR), an enzyme that has been shown to interact with a RAC–guanosine triphosphatase that is part of a complex spanning the plasma membrane and orchestrating the localization of an enzyme complex for generation of apoplastic oxidizing equivalents that can support monolignol polymerization [reviewed in (32)] (Fig. 9). Such a localization could help facilitate diffusion of monolignols across the plasma membrane, driven by a negative concentration gradient as the monolignol is polymerized within the apoplast. Within the apoplast, caffeyl alcohol is preferentially oxidized by LAC8 (12). Inhibition of coniferyl alcohol oxidation by caffeyl alcohol might disrupt the concentration gradient and impede the passage of coniferyl alcohol to the apoplast. This feature of the model remains speculative absent a reliable technique to measure the relative concentrations of monolignols in the cytosol and apoplast but is not essential to explain the present data. Furthermore, the model supports either laccase- or peroxidase-mediated lignin polymerization.
Compartmentation of G- and C-lignin biosynthesis to different cell types might obviate the need for controls of the type outlined above. Figure 9B summarizes our knowledge of the sites of lignin biosynthesis in the seed coat. Although C-lignin is initiated in a separate cell layer from G-lignin, this layer is interior to the G-lignin layer and therefore likely to have high levels of coniferyl alcohol represented by that in the “rest of the seed.” Ultimately, the G- and C-lignin layers fuse but retain individual G- and C-homopolymers.
In summary, our results explain how C-lignin biosynthesis proceeds when coniferyl alcohol is still being formed and show that monolignol supply is not itself sufficient to determine lignin composition. The enzyme(s) that support COMT and CCoAOMT-independent formation of coniferyl alcohol remain to be identified, as does the mechanism that allows polymerization of caffeyl alcohol to C-lignin simultaneously with its oligomerization to a range of lignans with different linkage types.
MATERIALS AND METHODS
Chemicals
Coniferyl alcohol was purchased from MilliporeSigma. 13C9–l-Phe and 13C6–trans-cinnamic acid were purchased from Cambridge Isotope Laboratories. Caffeyl alcohol, 13C6-caffeyl alcohol, and 13C6-coniferyl alcohol were synthesized as described previously (12).
Plant material and growth conditions
C. hassleriana plants were grown in the greenhouse at 24° to 28°C with a 16-hour-per-day light regimen of 150 μmol/m2 per second of light intensity, using high pressure sodium (red spectrum) and metal halide (blue spectrum) lamps as supplemental lighting if needed. Flowers were hand pollinated, and seeds were harvested periodically at 8 to 24 DAP at 4-day intervals. Freshly harvested seeds were treated with chemicals or frozen in liquid nitrogen and stored at −80°C until use.
Labeling of seeds with 13C9-phenylalanine, 13C6–trans-cinnamic acid, 13C6-caffeyl alcohol, or 13C6-coniferyl alcohol
Freshly harvested seeds were cut in half under sterile water, transferred into liquid Murashige and Skoog medium supplemented with 100 μM isotope-labeled compound, and vacuum-infiltrated for 10 min. Samples were washed with water three times and harvested after incubation for 2 days. Seed coats were isolated, frozen in liquid nitrogen, and stored at −80°C until use.
Measurement of lignin composition
Samples were ground into powder and sequentially extracted with methanol, chloroform/methanol (2:1, v/v), methanol, and water and then freeze-dried. Lignin composition was determined by a modified thioacidolysis method (63) with 4,4′-ethylidene bisphenol as internal standard. 13C incorporation into lignin monomer units was determined by measurement of the m/z = +7 (from L-Phe) or m/z = +6 ion peaks from the thioacidolysis products as described previously (12). Three biological replicates were measured.
Soluble metabolite extraction and ultra-HPLC–MS/MS analysis
Whole seed, seed coat, and the reminder of the seed samples were freeze-dried and ground into powder. About 10 mg of ground sample was transferred to a 1.5-ml tube and 1 ml of 60% (v/v) methanol containing 10 nmol of [1-13C1]-benzoic acid as an internal standard was added. The mixture was vortexed for 30 s, sonicated for 1 hour in an ice-water ultrasonic bath, rotated for 1 hour at 4°C, and centrifuged at 12,000g for 15 min at 4°C. The supernatant was recovered and stored at −20°C until use.
Quantification of monolignol intermediates was performed by the Bioanalytical Facility of the BioDiscovery Institute, University of North Texas, using an ultra-HPLC–MS/MS method as described by Cocuron et al. (64). Metabolites were identified and quantified using standards of each metabolite. Incorporation of label into metabolites was determined by measurement of the relative proportions of the native and M + x ions, where x is the number of 13C-labeled carbon atoms in the molecule. Three biological replicates were measured.
Determination of CTs composition
About 100 mg of ground sample was extracted three times with 100% ethanol. To the air-dried pellet after extraction, 0.2 ml of phloroglucinol solution [phloroglucinol (50 mg/ml) and ascorbic acid (10 mg/ml) in 0.1 N HCl in methanol] was added, and the solution vortexed and incubated at 70°C for 20 min. After the reaction, 0.2 ml of 0.2 M sodium acetate was added and the mixture centrifuged at 12,000g for 10 min. The supernatant (150 μl) was transferred into an HPLC vial with insert and analyzed by LC-MS analysis according to the method described by Jun et al. (65).
Assay of laccase and peroxidase activities
Cell wall proteins were extracted as described by Ranocha et al. (66) with minor modifications. About 0.3 g of seed coats was ground into powder and rotated for 3 hours at 4°C in 3 ml of 50 mM sodium acetate buffer (pH 5.0) with 10% (v/v) glycerol. Soluble proteins were discarded after centrifugation and cell wall proteins were obtained by reextracting in 1 ml of the same buffer containing 1 M CaCl2 overnight at 4°C. For further extraction of cell wall proteins, the residue after CaCl2 extraction was sequentially extracted for 30 min each with 1 ml of 2 mM DTT (pH 6.5) (for proteins interacting via cysteine disulfide bonds), followed by 2 M NaCl (pH 6.5) (for strongly ionically bound proteins) and 0.2 M borate (pH 7.5) (for proteins interacting via glycoprotein side chains) according to Robertson et al. (42). The supernatants were collected after centrifugation at 13,000g for 15 min. The extract was desalted on a PD-10 desalting column containing Sephadex G-25 resin (GE Healthcare) according to the manufacturer’s manual. Ten microliters (for laccase activity) or 2 μl (for peroxidase activity) of protein extract and 10 μl of monolignol substrate (556 μM for coniferyl alcohol or 595 μM for caffeyl alcohol) were added to 100 μl of 20 mM bis-tris buffer (pH 5.9). For determination of laccase activity, catalase (130 U/ml) was added to remove endogenous H2O2 and thereby inhibit peroxidase activity. For peroxidase activity, 0.5 mM H2O2 was included without addition of catalase. The reaction was incubated at room temperature by shaking for 4 hours for laccase activity and 10 min for peroxidase activity and was stopped by adding sodium azide solution (0.004%, w/v). Reaction products were injected into the HPLC system for analysis as described below. Three biological replicates were measured.
For assay of substrate competition reactions, 10 μl of cell wall protein extract was incubated with combined substrates (fixed 50 μM caffeyl alcohol with 0 to 50 μM coniferyl alcohol, and vice versa) as described above. Three biological replicates were measured.
For assay of laccase kinetics against coniferyl alcohol in the presence of caffeyl alcohol, 10 μl of cell wall protein extracts from Cleome seed coats at 16 DAP were incubated with 25 to 200 μM coniferyl alcohol and 0 to 50 μM caffeyl alcohol. Vmax and Km were calculated from Lineweaver-Burk plots, resulting in noncompetitive inhibition. Therefore, Ki was calculated using the formula Vmax+I = Vmax−I/(1 + [I]/Ki). Three replicates were determined.
HPLC and LC-MS/MS analysis
High-performance reversed phase LC (HPLC) analysis was carried out on an Agilent 1220 Infinity II system equipped with a 250 mm by 4.6 mm, 5 μm, C18 column (Agilent), using the following gradient as described by Wallace and Fry (67) with minor modifications: solvent A [H2O/butanol/acetic acid (98.3:1.2:0.5, v/v)] and B [acetonitrile/butanol/acetic acid (98.3:1.2:0.5, c/v)] at 1 ml/min of flow rate: 0 to 15 min, 0 to 100% B; 15 to 17 min, 100% B; 17 to 18 min, 100 to 0% B; and 18 to 19 min, 100% A. Data were collected at 280 nm, and identifications were based on chromatographic behavior compared to authentic standards. The laccase activity was calculated as the consumption of monolignol. Three biological replicates were measured.
To determine dimers and trimers, LC-MS/MS analysis was carried out on an Agilent 1290 Infinity II LC system with a 2.1 mm by 250 mm, 5 μm, XTerra MS C18 column (Waters) coupled to an Agilent 6400 Series Triple Quadrupole System with electrospray ionization source switching from negative (3500 V) to positive mode (3500 V). Solvent A [0.1% (v/v) formic acid in water] and solvent B [0.1% (v/v) formic acid in acetonitrile] were used for separation with the following gradient at 0.45 ml/min of flow rate: 0 to 1 min, 5% B; 1 to 12 min, 5 to 95% B; 12 to 14 min, 95% B; 14 to 15 min, 95 to 5% B; and 15 to 20 min, 5% B. MS data were recorded in negative ionization mode using full scan detection survey with the range of m/z = 100 to 1000. An MS/MS product ion scan survey was conducted to identify dimers and trimers formed from caffeyl alcohol. Dimers formed from coniferyl alcohol were identified according to Morreel et al. (41).
Metabolite extraction and GC-MS analysis for untargeted metabolomics
Metabolite profiling of methanolic extracts from seed coats was performed as reported previously (68), with 10 ml of the extracts being dried under nitrogen. Sorbitol (15 μg) was added as internal standard, the extracts were silylated for 2 days as described previously (68), and 0.5 μl of the 1-ml reaction volume was analyzed by GC-MS.
Transient expression of laccases in N. benthamiana
The coding sequences of ChLAC4, ChLAC5, ChLAC8, and ChLAC15 lacking the stop codon were amplified using the primers listed in table S2 and introduced into the pCR8/GW/TOPO gateway vector (Invitrogen), followed by recombination to the pEarleyGate103 (69) binary vector with green fluorescent protein (GFP) and 6xHis C-terminal tags driven by the CaMV 35S promoter. Resulting vectors were transformed into Agrobacterium tumefaciens GV3101. Transient gene expression in tobacco (N. benthamiana) leaves by agroinfiltration was performed following the procedures of Sparkes et al. (70). Infiltrated leaves were harvested 3 days after infiltration, and cell wall proteins were extracted as described above. The supernatant was desalted on a PD-10 Desalting Column (GE Healthcare). The LAC-GFP-6xHis fusion proteins were purified on Ni–nitrilotriacetic acid resin according to the manufacturer’s manual (Thermo Fisher Scientific). Protein concentrations were measured using the Bradford Protein Assay (71). About 250 ng of recombinant protein was added to the reaction for laccase activity assay as described above.
Isolation of low molecular weight fractions from poplar fibers
Five grams of poplar (Populus deltoides) fibers were ground into powder and sequentially extracted with 20, 50, and 80% (v/v) methanol. The extracts were combined, dried, and then suspended with shaking in 3 ml of water/ethyl acetate (1:1, v/v). After separation of the phases, the aqueous fraction was retained, and the ethyl acetate fraction was dried and suspended in 1.5 ml of 50% dimethyl sulfoxide. One microliter of fraction was added per 100 μl of laccase reaction assay.
13C–metabolic flux analysis
The lignin biosynthesis pathway in the seed coats of C. hassleriana was modeled on the basis of earlier studies (23), and the metabolic network accounted for the ER membrane–associated metabolic channels as proposed in earlier work on B. distachyon (33, 34). Reactions catalyzed by membrane-associated enzymes such as C3′H and cinnamate 4-hydroxylase (C4H) were assumed to be active only on the ER surface. Remaining enzymatic steps were modeled to occur in both subcellular locations. Metabolites were allowed to exchange between these two compartments. A reaction that drains unlabeled carbon into the p-coumaric acid node was added to account for the large pool of p-coumaric acid compared to cinnamic acid observed in the C. hassleriana seed coats and seeds. The 8 to 10 and 12 to 14 DAP models included sink reactions only for p-coumaryl alcohol and coniferyl alcohol. Since the concentration of caffeyl alcohol and caffealdehyde was below detection limit during this period, the reactions associated with these intermediates were not included in these models. The 16 to 18 and 20 to 22 DAP models included sink reactions for p-coumaryl alcohol and caffeyl alcohol to account for synthesis of H-lignin and C-lignin. In addition, 16 to 18 and 20 to 22 DAP models included sink reactions for ferulic acid and coniferyl alcohol due to their continued synthesis implied by the labeling data.
Flux distributions were calculated from the labeling datasets using 13C-MFA (72). A mathematical model that simulates metabolite labeling pattern for a given flux distribution was constructed from the metabolic network and associated atom transitions using elementary metabolite unit framework (73). Using least square regression, the model-simulated labeling patterns were fitted against experimental data by iteratively updating the fluxes. This regression was repeated 100 times using random initial flux distribution as starting points. The flux distribution with the lowest sum of squared residuals (SSR) was selected as the final solution. The 95% confidence intervals were derived using the method described by Antoniewicz et al. (74). Briefly, confidence intervals were calculated by assessing the sensitivity of SSR for individual fluxes. Flux estimation was performed for four datasets obtained for labeling at 8 to 10, 12 to 14, 16 to 18, and 20 to 22 DAP (dataset S2).
The involvement of alcohol dehydrogenase that can generate ferulic acid from coniferaldehyde [ALDH (36)] and the theoretical aldehyde and alcohol producing variants of 4-C3H were evaluated by comparing the SSR models with and without the corresponding activity. Reactions were predicted if inclusion of a particular activity lowered the SSR (improvement in fit).
Statistical analysis
Statistical analysis was performed by one-way analysis of variance (ANOVA) with Duncan multiple comparisons at 0.05 significance level using SPSS Statistics (version 27; IBM).
Acknowledgments
We thank the UNT BioDiscovery Institute’s BioAnalytical Facility for LC-MS analysis.
Funding: This work was supported by grants to R.A.D. from the National Science Foundation Integrated Organismal Systems program (award number 1456286) and the Center for Bioenergy Innovation (Oak Ridge National Laboratory), a U.S. Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the DOE under contract DE-AC05-00OR22725.
Author contributions: R.A.D., F.C., and C.Z. conceived and designed the research. C.Z., X.W., M.D.-P., B.C.S., N.L.E., T.J.T., J.I.H., C.D.M., and F.C. carried out the experiments and performed the data analyses. R.A.D. and C.Z. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Sequence data from this article can be found in the GenBank libraries under the following accession numbers: ChLAC4 LOC104823387, ChLAC5 LOC104810126, ChLAC8 LOC104823484, and ChLAC15 LOC104800224. All other data needed to support the conclusions of this paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Datasets S1 to S3
REFERENCES AND NOTES
- 1.Chen F., Dixon R. A., Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas A. J., Beckham G. T., Biddy M. J., Chandra R., Chen F., Davis M. F., Davison B. H., Dixon R. A., Gilna P., Keller M., Langan P., Naskar A. K., Saddler J. N., Tschaplinski T. J., Tuskan G. A., Wyman C. E., Lignin valorization: Improving lignin processing in the biorefinery. Science 344, 1246843 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Li M., Pu Y., Ragauskas A. J., Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 4, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barros-Rios J., Temple S., Dixon R. A., Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotechnol. 56, 48–54 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Halpin C., Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. 56, 223–229 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Mahon E. L., Mansfield S. D., Tailor-made trees: Engineering lignin for ease of processing and tomorrow’s bioeconomy. Curr. Opin. Biotechnol. 56, 147–155 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ralph J., Lapierre C., Boerjan W., Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Tobimatsu Y., Schuetz M., Lignin polymerization: How do plants manage the chemistry so well? Curr. Opin. Biotechnol. 56, 75–81 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Duroux L., Welinder K., The peroxidase gene family in plants: A phylogenetic overview. J. Mol. Evol. 57, 397–407 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Turlapati P. V., Kim K. W., Davin L. B., Lewis N. G., The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 233, 439–470 (2011). [DOI] [PubMed] [Google Scholar]
- 11.He F., Machemer-Noonan K., Golfier P., Unda F., Dechert J., Zhang W., Hoffmann N., Samuels L., Mansfield S. D., Rausch T., Wolf S., The in vivo impact of MsLAC1, a Miscanthus laccase isoform, on lignification and lignin composition contrasts with its in vitro substrate preference. BMC Plant Biol. 19, 552 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Zhuo C., Xiao X., Wang X., Chen F., Dixon R. A., Substrate Specificity of LACCASE8 facilitates polymerization of caffeyl alcohol for C-lignin biosynthesis in the seed coat of Cleome hassleriana. Plant Cell 32, 3825–3845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Cao S., Liu X., Li Y., Wang B., Sun Y., Zhang C., Guo X., Li H., Lu H., PtrLAC16 plays a key role in catalyzing lignin polymerization in the xylem cell wall of Populus. Int. J. Biol. Macromol. 188, 983–992 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Chen F., Tobimatsu Y., Havkin-Frenkel D., Dixon R. A., Ralph J., A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. U.S.A. 109, 1772–1777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F., Tobimatsu Y., Jackson L., Nakashima J., Ralph J., Dixon R. A., Novel seed coat lignins in the Cactaceae: Structure, distribution and implications for the evolution of lignin diversity. Plant J. 73, 201–211 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Tobimatsu Y., Chen F., Nakashima J., Escamilla-Trevino L. L., Jackson L., Dixon R. A., Ralph J., Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25, 2587–2600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis N., Yamamoto E., Lignin: Occurrence, biogenesis and biodegradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 455–496 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Shuai L., Kim H., Motagamwala A. H., Mobley J. K., Yue F., Tobimatsu Y., Havkin-Frenkel D., Chen F., Dixon R. A., Luterbacher J. S., Dumesic J. A., Ralph J., An “ideal lignin” facilitates full biomass utilization. Sci. Adv. 4, eaau2968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone M. L., Anderson E. M., Meek K. M., Reed M., Katahira R., Chen F., Dixon R. A., Beckham G. T., Román-Leshkov Y., Reductive catalytic fractionation of C-lignin. ACS Sustain. Chem. Eng. 6, 11211–11218 (2018). [Google Scholar]
- 20.Wang S., Zhang K., Li H., Xiao L.-P., Song G., Selective hydrogenolysis of catechyl lignin into propenylcatechol over an atomically dispersed ruthenium catalyst. Nat. Commun. 12, 416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nar M., Rizvi H. R., Dixon R. A., Chen F., Kovalcik A., D’Souza N., Superior plant based carbon fibers from electrospun poly-(caffeyl alcohol) lignin. Carbon 103, 372–383 (2016). [Google Scholar]
- 22.Wagner A., Tobimatsu Y., Phillips L., Flint H., Torr K., Donaldson L., Pears L., Ralph J., CCoAOMT suppression modifies lignin composition in Pinus radiata. Plant J. 67, 119–129 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Zhuo C., Rao X., Azad R., Pandey R., Xiao X., Harkelroad A., Wang X., Chen F., Dixon R. A., Enzymatic basis for C-lignin monomer biosynthesis in the seed coat of Cleome hassleriana. Plant J. 99, 506–520 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Humphreys J. M., Chapple C., Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5, 224–229 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Lee T. S., Purse J. G., Pryce R. J., Horgan R., Wareing P. F., Dihydroconiferyl alcohol – A cell division factor from Acer species. Planta 152, 571–577 (1981). [DOI] [PubMed] [Google Scholar]
- 26.Cutillo F., D’Abrosca B., DellaGreca M., Fiorentino A., Zarrelli A., Lignans and neolignans from Brassica fruticulosa: Effects on seed germination and plant growth. J. Agric. Food Chem. 51, 6165–6172 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Schroeder F. C., del Campo M. L., Grant J. B., Weibel D. B., Smedley S. R., Bolton K. L., Meinwald M., Eisner T., Pinoresinol: A lignol of plant origin serving for defense in a caterpillar. Proc. Natl. Acad. Sci. U.S.A. 103, 15497–15501 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N., Zhao M., Liu T., Dong L., Cheng Q., Wu J., Wang L., Chen X., Zhang C., Lu W., Xu P., Zhang S., A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 8, 1185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T., Onishi Y., Katagi A., Kim T.-W., Katayama T., A catechol-type lignan and neolignans are specifically present in the seed coat of tung trees. J. Wood Sci. 66, 57 (2020). [Google Scholar]
- 30.Matsumoto K., Takahashi H., Miyake Y., Fukuyama Y., Convenient syntheses of neurotrophic americanol A and isoamericanol A by HRP catalyzed oxidative coupling of caffeic acid. Tetrahedron Lett. 40, 3185–3186 (1999). [Google Scholar]
- 31.Takahashi H., Matsumoto K., Ueda M., Miyake Y., Fukuyama Y., Biomimetic syntheses of neurotrophic americanol A and isoamericanol A by horseradish peroxidase (HRP) catalyzed oxidative coupling. Heterocycles 56, 245–256 (2002). [Google Scholar]
- 32.Dixon R. A., Barros J., Lignin biosynthesis- old roads revisited and new roads explored. Open Biol. 9, 190215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faraji M., Fonseca L. L., Escamilla-Trevino L., Barros-Rios J., Engle N., Yang Z. K., Tschaplinski T. J., Dixon R. A., Voit E. O., Mathematical models of lignin biosynthesis. Biotechnol. Biofuels 11, 34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faraji M., Fonseca L. L., Escamilla-Treviño L., Barros-Rios J., Engle N., Yang Z. K., Tschaplinski T. J., Dixon R. A., Voit E. O., A dynamic model of lignin biosynthesis in Brachypodium distachyon. Biotechnol. Biofuels 11, 253 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barros J., Escamilla-Trevino L., Song L., Rao X., Serrani-Yarce J. C., Palacios M. D., Engle N., Choudhury F. K., Tschaplinski T. J., Venables B. J., Mittler R., Dixon R. A., 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 10, 1994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair R. B., Bastress K. L., Ruegger M. O., Denault J. W., Chapple C., The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16, 544–554 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterjiades R., Dean J. F., Eriksson K. E., Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 99, 1162–1168 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao W., O’Malley D. M., Whetten R., Sederoff R. R., A laccase associated with lignification in loblolly pine xylem. Science 260, 672–674 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Barros-Rios J., Serk H., Granlund I., Pesquet E., The cell biology of lignification in higher plants. Ann. Bot. 115, 1053–1074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.-Y., Dixon R. A., LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morreel K., Kim H., Lu F., Dima O., Akiyama T., Vanholme R., Niculaes C., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W., Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal. Chem. 82, 8095–8105 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Robertson D., Mitchell G. P., Gilroy J. S., Gerrish C., Bolwell G. P., Slabas A. R., Differential extraction and protein sequencing reveals major differences in patterns of primary cell wall proteins from plants. J. Biol. Chem. 272, 15841–15848 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Pourcel L., Routaboul J. M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I., TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17, 2966–2980 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang M., Davis E., Gardner D., Cai X., Wu Y., Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224, 1185–1196 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., Lapierre C., Jouanin L., Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barros-Rios J., Serrani-Yarce J. C., Chen F., Baxter D., Venables B. J., Dixon R. A., Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2, 16050 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Schoch G., Goepfert S., Morant M., Hehn A., Meyer D., Ullmann P., Werck-Reichhart D., CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276, 36566–36574 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Dima O., Morreel K., Vanholme V., Kim H., Ralph J., Boerjan W., Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27, 695–710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha C. M., Fine D., Bahtia A., Rao X., Martin M. Z., Engle N., Wherritt D. J., Tschaplinski T. J., Sumner L. W., Dixon R. A., Ectopic defense gene expression is associated with growth defects in Medicago truncatula lignin pathway mutants. Plant Physiol. 181, 63–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki S., Umezawa T., Biosynthesis of lignans and norlignans. J. Wood Sci. 53, 273–284 (2007). [Google Scholar]
- 51.Liu C., Wang X., Shulaev V., Dixon R. A., A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat. Plants 2, 16182 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Jun J. H., Lu N., Docampo-Palacios M. L., Wang X., Dixon R. A., Dual activity of anthocyanidin reductase supports the dominant proanthocyanidin extension unit pathway. Sci. Adv. 7, eabg4682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J., Pang Y., Dixon R. A., The mysteries of proanthocyanidin transport and polymerization. Plant Physiol. 153, 437–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parvathi K., Chen F., Guo D., Blount J. W., Dixon R. A., Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25, 93–102 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Teutonico R. A., Dudley M. W., Orr J. D., Lynn D. G., Binns A. N., Activity and accumulation of cell division-promoting phenolics in tobacco tissue cultures. Plant Physiol. 97, 288–297 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orr J. D., Lynn D. G., Biosynthesis of dehydrodiconiferyl alcohol glucosides: Implications for the control of tobacco cell growth. Plant Physiol. 98, 343–352 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gafni Y., Levy Y., Coniferyl alcohol, a lignin precursor, stimulates Rhizobium rhizogenes A4 virulence. Curr. Microbiol. 50, 262–265 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Do C. T., Pollet B., Thevenin J., Sibout R., Denoue D., Barriere Y., Lapierre C., Jouanin L., Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226, 1117–1129 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Lee Y., Rubio M. C., Alassimone J., Geldner N., A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Miao Y. C., Liu C.-J., ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U.S.A. 107, 22728–22733 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perkins M., Smith R. A., Samuels L., The transport of monomers during lignification in plants: Anything goes but how? Curr. Opin. Biotechnol. 56, 69–74 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Vermaas J. V., Dixon R. A., Chen F., Mansfield S. D., Boerjan W., Ralph J., Crowley M. F., Beckham G. T., Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. U.S.A. 116, 23117–23123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen F., Zhuo C., Xiao X., Pendergast T. H., Devos K. M., A rapid thioacidolysis method for biomass lignin composition and tricin analysis. Biotechnol. Biofuels 14, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cocuron J. C., Casas M. I., Yang F., Grotewold E., Alonso A. P., Beyond the wall: High-throughput quantification of plant soluble and cell-wall bound phenolics by liquid chromatography tandem mass spectrometry. J. Chromatogr. A 1589, 93–104 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Jun J. H., Xiao X., Rao X., Dixon R. A., Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat. Plants 4, 1034–1043 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Ranocha P., McDougall G., Hawkins S., Sterjiades R. G., Borderies G., Stewart D., Cabanes M., Macheteau M., Boudet A. M., Goffner D., Biochemical characterization, molecular cloning and expression of laccases- a divergent gene family- in poplar. Eur. J. Biochem. 259, 485–495 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Wallace G., Fry S. C., Action of diverse peroxidases and laccases on six cell wall-related phenolic compounds. Phytochemistry 52, 769–773 (1999). [Google Scholar]
- 68.Tschaplinski T. J., Standaert R. F., Engle N. L., Martin M. Z., Sangha A. K., Parks J. M., Smith J. C., Samuel R., Jiang N., Pu Y., Ragauskas A., Hamilton C. Y., Fu C., Wang Z. Y., Davison B. H., Dixon R. A., Mielenz J. R., Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol. Biofuels 5, 71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S., Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Sparkes I. A., Runions J., Kearns A., Hawes C., Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 72.Kruger N. J., Ratcliffe R. G., Insights into plant metabolic networks from steady-state metabolic flux analysis. Biochimie 91, 697–702 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Antoniewicz M. R., Kelleher J. K., Stephanopoulos G., Elementary metabolite units (EMU): A novel framework for modeling isotopic distributions. Metab. Eng. 9, 68–86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antoniewicz M. R., Kelleher J. K., Stephanopoulos G., Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab. Eng. 8, 324–337 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 and S2
Datasets S1 to S3