Abstract
The precise control of growth and maintenance of the retinal ganglion cell (RGC) dendrite arborization is critical for normal visual functions in mammals. However, the underlying mechanisms remain elusive. Here, we find that the N6-methyladenosine (m6A) reader YTHDF2 is highly expressed in the mouse RGCs. Conditional knockout (cKO) of Ythdf2 in the retina leads to increased RGC dendrite branching, resulting in more synapses in the inner plexiform layer. Interestingly, the Ythdf2 cKO mice show improved visual acuity compared with control mice. We further demonstrate that Ythdf2 cKO in the retina protects RGCs from dendrite degeneration caused by the experimental acute glaucoma model. We identify the m6A-modified YTHDF2 target transcripts which mediate these effects. This study reveals mechanisms by which YTHDF2 restricts RGC dendrite development and maintenance. YTHDF2 and its target mRNAs might be valuable in developing new treatment approaches for glaucomatous eyes.
Research organism: Mouse
Introduction
The mammalian retina is an ideal model system to study neuronal development and neural circuit formation. The retinal ganglion cells (RGCs) are the final and only output neurons in the vertebrate retina and their dendrites collect the electrical information concerning the visual signal from all other cells preceding them. One of the major focuses of research in the retina is to understand how RGC dendrite arborization arises during development (Prigge and Kay, 2018). Existing evidences supported that homotypic repulsion controls retinal dendrite patterning (Lefebvre et al., 2015). However, in mice which had most RGCs genetically eliminated, the dendrite size and shape of remaining RGCs appeared relatively normal (Lin et al., 2004). Thus, the fact that the dendrites of remaining RGCs did not expand to neighboring areas by the remaining RGCs supports the existence of the intrinsic limit for RGC dendrite patterning, which cooperates with the homotypic repulsion to determine the dendrite size of RGCs (Lefebvre et al., 2015). However, such intrinsic limiting mechanisms remain elusive.
Glaucoma is one of the leading causes for blindness. The major risk factors for glaucoma include increased intraocular tension. Studies have shown that glaucoma causes pathological changes in RGC dendrites before axon degeneration and soma loss were detected in different model animals (Weber et al., 1998; Shou et al., 2003; Morgan et al., 2006). Thus, elucidation of mechanisms governing RGC dendrite arbor maintenance bears clinical significance.
N6-methyladenosine (m6A) is the most widely distributed and extensively studied internal modification in mRNA (Dominissini et al., 2012; Meyer et al., 2012; Nachtergaele and He, 2018). m6A modification has been shown to regulate brain development and functions in the nervous system (Livneh et al., 2020; Yu et al., 2021a). By effectors, most of these studies have focused on its demethylases (‘m6A erasers’) and methyltransferases (‘m6A writers’). Since the fate of m6A-modified transcripts is decoded by the m6A-binding proteins (‘m6A readers’), how the readers mediate these functions and what are their neural target mRNAs remain to be elucidated. In addition, more precisely controlled spatial-temporal ablation of the m6A readers instead of null knockout is required to elucidate their functions and mechanisms in nervous system.
In this study, we identified an m6A-dependent intrinsic limiting mechanism for RGC dendrite arborization and maintenance. Conditional knockout (cKO) of the m6A reader YTHDF2 in the developing mouse retina increases RGC dendrite branching and improves visual acuity. YTHDF2 also mediates acute ocular hypertension (AOH)-induced RGC degeneration, the experiment model for glaucoma, and Ythdf2 cKO in the retina alleviates AOH-induced RGC dendrite shrinking and neuronal loss. The regulation of RGC dendrite development and maintenance by YTHDF2 is mediated by two distinct groups of m6A-modified target mRNAs which encode proteins that promote dendrite arborization during development and maintain dendrite tree during injury, respectively. Therefore, our study reveals mechanisms by which YTHDF2 restricts RGC dendrite development and maintenance, which sheds light on developing new treatment approaches for glaucomatous eyes.
Results
Knockdown of YTHDF2 leads to a robust increase of RGC dendrite branching
To examine whether m6A modification and its reader proteins play a role in the dendrite development, we utilized the retina as the model system. We first checked their expression patterns in the developing mouse retina. Immunostaining with a widely used m6A antibody demonstrated that RGCs had high m6A modification levels (Figure 1—figure supplement 1A). Consistent with the m6A distribution, the m6A reader YTHDF2 is highly expressed in RGCs (Figure 1A; Figure 1—figure supplement 1B). Conversely, the expression of YTHDF2 in other layers and cells of the retina is much lower or absent (Figure 1A; Figure 1—figure supplement 1B-D). Another two m6A readers YTHDF1 and YTHDF3 show similar expression patterns (Figure 1—figure supplement 1E, F). The strong expression of YTHDFs and high level of m6A modification in RGCs suggest that the m6A reader YTHDFs might play roles in RGC development. We dissected and dissociated the retinal cells and cultured in vitro. We generated lentiviral shRNAs against YTHDFs, which showed similarly efficient knockdown (KD) of YTHDFs in RGC cultures in vitro (Figure 1B; Figure 1—figure supplement 1G,H). In these YTHDF-deficient RGC cultures, the first and most obvious phenotype that we observed is the robust increase of dendrite branching of cultured RGCs treated by shYthdf2 (Figure 1C and D; Figure 1—figure supplement 1I,J). In contrast, the dendrite branching of RGCs with YTHDF1 KD using shYthdf1 was not significantly different from control shRNA (Figure 1—figure supplement 1K), while YTHDF3 KD using shYthdf3 caused a slight (statistically significant in several Sholl radii) decrease of RGC dendrite branching compared with control shRNA (Figure 1—figure supplement 1L). These results suggest that the m6A reader YTHDF2 might play an important role in controlling dendrite branching of RGCs.
Figure 1. Knockdown (KD) of YTHDF2 leads to a robust increase of retinal ganglion cell (RGC) dendrite branching.
(A) Representative confocal images showing high expression of YTHDF2 in RGCs (marked by RBPMS) in P0 retina. Note that all RGCs marked by the pan-RGC marker RBPMS express YTHDF2 while all YTHDF2-expressing cells are RBPMS+ RGCs. GCL, ganglion cell layer. Scale bars: 20 μm. (B) Western blotting (WB) confirming efficient KD of YTHDF2 in cultured RGCs using shYthdf2. Data of WB quantification are mean ± SEM and are represented as dot plots: ***p = 0.00012 (n = 3 replicates); by unpaired Student’s t test. (C) Examination of RGC dendrite development after YTHDF2 KD. As shown, significantly increased branching of dendrites marked by MAP2 immunofluorescence was observed in cultured RGCs marked by Brn3a. Dendrite traces were drawn for the corresponding RGCs. Scale bar: 10 μm. (D) Quantification of dendrite branching (C) using Sholl analysis. As shown, numbers of interactions are significantly greater in shYthdf2 groups (n = 68 RGCs) than shCtrl groups (n = 72 RGCs) in Sholl radii between 10 and 30 μm. Data are mean ± SEM. ****p = 4.32E-05 (10 μm), ***p = 0.00038 (15 μm), ****p = 2.85E-05 (20 μm), ***p = 0.00084 (25 μm), *p = 0.020 (30 μm), by unpaired Student’s t test.
Figure 1—figure supplement 1. Retinal ganglion cells (RGCs) have high level of N6-methyladenosine (m6A) modification and strong expression of YTHDFs.
cKO of Ythdf2 in the retina increases RGC dendrite branching in vivo without disturbing sublaminar targeting
To further explore whether YTHDF2 physiologically regulates RGC dendrite branching in vivo, we generated Ythdf2 cKO mouse (Figure 2A). We used the Six3-cre mouse line (Furuta et al., 2000), which has been widely used in the field to generate retina-specific knockouts (Lefebvre et al., 2012; Riccomagno et al., 2014; Sapkota et al., 2014; Krishnaswamy et al., 2015). YTHDF2 expression is efficiently eliminated in the Ythdf2 cKO retina compared with their littermate controls at E12.5 (Figure 2—figure supplement 1A) and E15.5 (Figure 2B). Retina progenitors, amacrine cells, bipolar cells, photoreceptors, horizontal cells, Müller glia, or astrocytes were not affected in Ythdf2 cKO retina (Figure 2—figure supplement 1B-K; Figure 2—figure supplement 2A-D), suggesting that YTHDF2 is not involved in the generation or development of these cells. This is in line with the low or no YTHDF2 expression in these cells. The RGC number or density was not affected in the Ythdf2 cKO retina (Figure 2C and D), demonstrating that Ythdf2 knockout does not disturb RGC neurogenesis. We then cultured RGCs from the Ythdf2 cKO retina. The dendrite branching of Ythdf2 cKO RGCs was significantly increased compared with littermate controls (Figure 2E and F). RGCs include over 40 subtypes (Sanes and Masland, 2015; Baden et al., 2016). We thus examined the RGC dendrite branching within different subtypes. One of the RGC subgroups responds preferentially to movement in particular directions and is named the ON-OFF directionally selective RGCs (ooDSGCs). Expression of CART (cocaine- and amphetamine-regulated transcript), a neuropeptide, distinguishes ooDSGCs from other RGCs (Kay et al., 2011a). The dendrite branching of ooDSGCs marked by CART/Brn3a co-staining in Ythdf2 cKO retinal cultures also increased compared with control (Figure 2G and H). These data further confirm that the m6A reader YTHDF2 regulates dendrite branching of RGCs.
Figure 2. Dendrite branching is dramatically increased in cultured retinal ganglion cells (RGCs) from Ythdf2 conditional knockout (cKO).
(A) Schematic drawings of the genetic deletion strategy for Ythdf2. Exon 4 which contains YTH domain-coding sequence is deleted after Cre-mediated recombination. (B) Depletion of YTHDF2 protein in retina of Six3-cre+/-;Ythdf2fl/fl cKO mice. Anti-YTHDF2 immunostaining of E15.5 retina vertical sections confirmed cKO of YTHDF2 protein, compared with Ythdf2fl/fl littermate controls. Scale bar: 20 μm. (C, D) RGC neurogenesis not affected in the Ythdf2 cKO retina. Wholemount immunostaining using a Brn3a antibody was carried out in P20 retina (C). Numbers of Brn3a+ RGC per 10,000 μm2 of retina were quantified and showed no difference between the Ythdf2 cKO and their littermate controls (D). n = 12 confocal fields for each genotype. Data are represented as box and whisker plots: ns, not significant (p = 0.79); by unpaired Student’s t test. Scale bar: 25 μm. (E) Examination of RGC dendrite development in Ythdf2 cKO RGCs. As shown, knockout of YTHDF2 was confirmed by YTHDF2 IF (green). Significantly increased branching of dendrites marked by MAP2 IF (red) was observed in cultured RGCs from the Ythdf2 cKO retina compared with their littermate controls. Dendrite traces were drawn for the corresponding RGCs. Scale bar: 10 μm. (F) Quantification of RGC dendrite branching (E) using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in Six3-cre+/-,Ythdf2fl/fl groups (n = 68 RGCs) than Ythdf2fl/fl groups (n = 42 RGCs) in Sholl radii between 10 and 30 μm: ***p = 0.00030 (10 μm), ****p = 1.19E-05 (15 μm), ***p = 0.00018 (20 μm), *p = 0.021 (25 μm), **p = 0.0022 (30 μm), by unpaired Student’s t test. (G) Examination of CART+ (cocaine- and amphetamine-regulated transcript) RGC dendrite development in Ythdf2 cKO RGCs. Cultured CART+ RGCs from the Ythdf2 cKO retina have significantly increased branching of dendrites marked by MAP2 IF (red) compared with their littermate controls. Dendrite traces were drawn for the corresponding RGCs. Scale bar: 10 μm. (H) Quantification of CART+ RGC dendrite branching (G) using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in Six3-cre+/-,Ythdf2fl/fl groups (n = 77 RGCs) than Ythdf2fl/fl groups (n = 90 RGCs) in Sholl radii between 10 and 30 μm: ****p = 3.17E-05 (10 μm), ****p = 6.50E-11 (15 μm), ****p = 5.14E-12 (20 μm), ****p = 5.00E-07 (25 μm), ***p = 0.00020 (30 μm), by unpaired Student’s t test.
Figure 2—figure supplement 1. Ythdf2 conditional knockout (cKO) does not change numbers of retinal progenitors, amacrine cells, bipolar cells, photoreceptors, or horizontal cells.
Figure 2—figure supplement 2. Ythdf2 conditional knockout (cKO) does not change numbers of Müller glia or astrocytes.
Next, we wanted to confirm this phenotype in vivo by checking specific RGC subtypes. Intravitreal injection of an AAV reporter expressing ZsGreen visualized the dendrite morphology of ooDSGCs marked by CART immunostaining (Figure 3A). ooDSGCs showed dramatically increased dendrite branching in Ythdf2 cKO retina compared with control retina by Sholl analysis (Figure 3A and B). The intrinsically photosensitive RGCs (ipRGCs) are unique and melanopsin-expressing cells, which exhibit an intrinsic sensitivity to light (Hattar et al., 2002). We analyzed the morphology of ipRGCs visualized by wholemount immunostaining of melanopsin and found that the dendrite branching of ipRGCs was significantly increased in the Ythdf2 cKO retina (Figure 3C and D; Figure 3—figure supplement 1A-E). A similar trend was observed in the SMI-32+αRGCs (Figure 3E and F). These results strongly indicate that the m6A reader YTHDF2 negatively regulates RGC dendrite branching in vivo and Ythdf2 cKO promotes RGC dendrite arborization.
Figure 3. Dendrite branching of specific retinal ganglion cell (RGC) subtypes increases in Ythdf2 conditional knockout (cKO) in vivo.
(A) Co-labeling of ON-OFF directionally selective RGCs (ooDSGCs) by AAV-ZsGreen and CART (cocaine- and amphetamine-regulated transcript) IF in vivo. Intravitreal injection of AAV-expressing ZsGreen reporter was performed at P17 and retinas were collected at P27. The white arrowheads indicate ooDSGCs co-labeled by ZsGreen and CART IF, which show dramatically increased dendrite branching in Ythdf2 cKO compared with control. Dendrite traces were drawn for the corresponding RGCs shown. Scale bar: 20 μm. (B) Quantification of dendrite branching of ZsGreen+/CART+ ooDSGCs (A) using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in Six3-cre+/-,Ythdf2fl/fl groups (n = 15 RGCs) than Ythdf2fl/fl groups (n = 18 RGCs) in Sholl radii between 50 and 120 μm: **p = 0.0041 (50 μm), **p = 0.0059 (60 μm), ***p = 0.00036 (70 μm), **p = 0.0058 (80 μm), **p = 0.0018 (90 μm), **p = 0.0064 (100 μm), **p = 0.0045 (110 μm), *p = 0.040 (120 μm), by unpaired Student’s t test. (C) Dendrites of intrinsically photosensitive RGCs (ipRGCs) visualized by wholemount immunostaining of P20 retina using a melanopsin antibody in vivo. Dendrite traces were drawn for the corresponding RGCs shown. Scale bar: 50 μm. (D) Quantification of dendrite branching of melanopsin+ ipRGCs (C) using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in Six3-cre+/-,Ythdf2fl/fl groups (n = 18 RGCs) than Ythdf2fl/fl groups (n = 21 RGCs) in Sholl radii between 20 and 100 μm: **p = 0.0083 (20 μm), *p = 0.018 (40 μm), ***p = 0.00068 (50 μm), ***p = 0.00027 (60 μm), *p = 0.048 (70 μm), **p = 0.0048 (80 μm), **p = 0.0023 (100 μm), by unpaired Student’s t test. (E) Dendrites of αRGCs visualized by wholemount immunostaining of P20 retina using an SMI-32 antibody in vivo. Dendrite traces were drawn for the corresponding RGCs shown. Scale bar: 20 μm. (F) Quantification of dendrite branching of SMI-32+αRGCs (E) using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in Six3-cre+/-,Ythdf2fl/fl groups (n = 14 RGCs) than Ythdf2fl/fl groups (n = 22 RGCs) in Sholl radii between 40 and 140 μm: **p = 0.0044 (40 μm), **p = 0.0035 (50 μm), ***p = 0.00021 (60 μm), ****p = 2.63E-05 (70 μm), ****p = 2.38E-06 (80 μm), ****p = 1.68E-06 (90 μm), ****p = 6.76E-06 (100 μm), ****p = 5.72E-05 (110 μm), **p = 0.0011 (120 μm), **p = 0.0032 (130 μm), *p = 0.047 (140 μm), by unpaired Student’s t test.
Figure 3—figure supplement 1. General dendrite density in inner plexiform layer (IPL) is increased without affecting sublaminar targeting.
In the retina, RGCs target their dendrites in different sublaminae of the inner plexiform layer (IPL). Since the IPL sublaminar targeting of RGC dendrites is critical for normal visual functions, we wondered whether the increased dendrite branching caused by Ythdf2 cKO was also accompanied by altered sublaminar patterning of RGC dendrites. We used a Thy1-GFP reporter (line O) which labels a few RGCs (Feng et al., 2000). As shown in Figure 3—figure supplement 1F,G, GFP intensity is generally higher in IPL of the Ythdf2 cKO retina compared with their littermate controls, which further proves the increased RGC dendrite branching and density. However, the sublaminar pattern of GFP signals looks similar between cKO and littermate control (Figure 3—figure supplement 1F,G). Sublaminar dendrite patterning of the ipRGC subtype visualized by immunostaining of melanopsin also demonstrated the similar phenotype (Figure 3—figure supplement 1H,I). These data suggest that YTHDF2 has a general control of RGC dendrite branching but has no striking effect on the sublaminar targeting of RGC dendrite. These results are consistent with the previous findings that the RGC dendrite targeting is determined genetically and several transcription factors controlling laminar choice have been identified in RGCs and amacrine cells (Cherry et al., 2011; Kay et al., 2011b; Lefebvre et al., 2015; Liu et al., 2018).
IPL of Ythdf2 cKO retina is thicker and has more synapses
The increased dendrite branching of RGCs further prompted us to check whether Ythdf2 cKO changes IPL development. Immunostaining of P6 retina vertical sections using a MAP2 antibody demonstrated that IPL thickness significantly increased in Ythdf2 cKO retina (Figure 4A and B). As a control, the thicknesses of other retinal layers showed no difference between the Ythdf2 cKO and control mice (Figure 4—figure supplement 1A-D). Quantification of MAP2 IF intensity in IPL suggested that the IPL of Ythdf2 cKO retina became denser with dendrites (Figure 4A and C). These results suggest that the increased dendrite branching results in a thicker and denser IPL in the Ythdf2 cKO retina.
Figure 4. Inner plexiform layer (IPL) of the Ythdf2 conditional knockout (cKO) retina is thicker and has more synapses.
(A) Cross-sections of P6 Six3-cre+/-,Ythdf2fl/fl retina showing increased IPL thickness by MAP2 staining compared with littermate control. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, granule cell layer. Scale bar: 20 μm. (B, C) Quantification showing increased IPL thickness and MAP2 IF intensity per area in IPL of the Ythdf2 cKO retina (A). Quantification data are represented as box and whisker plots: ****p = 1.28E-07 for B (n = 12 sections for each genotype), by unpaired Student’s t test; **p = 0.0045 for C (n = 12 sections for each genotype), by paired Student’s t test. (D, E) Representative confocal images showing the excitatory synapses labeled by colocalization of Bassoon (presynaptic) and PSD-95 (postsynaptic) in the IPL of P30 retina (D). There are significantly more synapses in the Ythdf2 cKO IPL compared with control. Quantification data are represented as box and whisker plots (E): n = 47 confocal fields for Ythdf2fl/fl, n = 23 confocal fields for Six3-cre+/-,Ythdf2fl/fl; ****p = 1.63E-05; by unpaired Student’s t test. Scale bars: 10 μm (D) and 5 μm (inset in D).
Figure 4—figure supplement 1. Thickness or synapse numbers in outer plexiform layer (OPL) shows no difference between the Ythdf2 conditional knockout (cKO) and control retinas.
The IPL of retina is concentrated with synaptic connections, which contain synapses among and between bipolar-amacrine-ganglion cells. The increased RGC dendrite branching and denser IPL in the Ythdf2 cKO retina prompted us to wonder whether there are changes in synaptic connections in IPL. We used co-staining of the presynaptic marker Bassoon and the postsynaptic marker PSD-95 to count the colocalization puncta of Bassoon+/PSD-95+. We found that the numbers of Bassoon+/PSD-95+ excitatory synapses in IPL of Ythdf2 cKO retina are significantly larger than that of control retina (Figure 4D and E). As a control, the numbers of the excitatory ribbon synapses marked by the colocalization of Bassoon+/PSD-95+ in OPL (outer plexiform layer) show no difference between Ythdf2 cKO and control retinas (Figure 4—figure supplement 1E,F).
All these data verify that the IPL of Ythdf2 cKO retina is thicker and has more synapses.
Visual acuity is improved for the Ythdf2 cKO mice
The features of RGC dendrites, including their size, shape, arborization pattern, and localization, influence the amount and type of synaptic inputs that RGCs receive, which in turn determine how RGCs respond to specific visual stimuli such as the direction of motion (Liu and Sanes, 2017). The increased dendrite branching, the thicker and denser IPL, and the more synapses in the IPL inspired us to further explore whether the visual responses of the Ythdf2 cKO mice were changed or not. Ythdf2 cKO mice looked normal and had similar body weight and size compared with control mice for either sex (male in Figure 5A and B; female in Figure 5C and D). The generally normal development of Ythdf2 cKO mice is consistent with the specific and limited expression of Six3-cre in retina (Figure 5—figure supplement 1A), and only sparse spots in ventral forebrain (Figure 5—figure supplement 1B; Furuta et al., 2000). We used an optomotor response (OMR)-based assay (Prusky et al., 2004; Umino et al., 2008; Shi et al., 2018) to monitor visual functions of Ythdf2 cKO mice (Figure 5E). Surprisingly, the Ythdf2 cKO mice showed modestly improved visual acuity compared with the control mice, measuring spatial frequency threshold as 0.45 ± 0.0043 c/deg (cycle per degree) and 0.43 ± 0.0085 c/deg, respectively (Figure 5F, male mice). Similar phenotype was observed in female mice (Figure 5G). These results suggest that the visual acuity is modestly improved in the Ythdf2 cKO mice.
Figure 5. Visual acuity is improved for the Ythdf2 conditional knockout (cKO) mice.
(A–D) Six3-Cre-mediated Ythdf2 cKO showing normal animal development and body weight (male in A, female in C). Quantification data of body weight (B, D) are represented as box and whisker plots: p = 0.41 in B (male, n = 24 for control, n = 18 for cKO); P = 0.08 in D (female, n = 23 for control, n = 25 for cKO); ns, not significant; by unpaired Student’s t test. (E) The setup of optomotor response assay is illustrated by schematic drawing. (F, G) Optomotor response assay demonstrating improved visual acuity in the Ythdf2 cKO mice. Quantification data are mean ± SEM: *p = 0.048 in F (male, n = 24 control, n = 16 cKO); *p = 0.015 in G (female, n = 21 control, n = 25 cKO); by unpaired Student’s t test.
Figure 5—figure supplement 1. Guidance or central targeting of optic nerves is not affected in Six-Cre-mediated Ythdf2 conditional knockout (cKO).
This phenotype is most likely attributed to the increased RGC dendrite branching and thicker and denser IPL with more synapses because all other parts and processes of retina are not affected except RGC dendrite in the Ythdf2 cKO mediated by Six3-cre (Figure 2—figure supplement 1 and Figure 4—figure supplement 1). The eyes and optic fibers also showed no difference between Ythdf2 cKO and control mice (Figure 5—figure supplement 1C-E). We further checked the targeting of optic nerves to the brain by anterograde labeling with cholera toxin subunit B (CTB) and found no difference of retinogeniculate or retinocollicular projections between Ythdf2 cKO and control mice (Figure 5—figure supplement 1F, G), suggesting the guidance and central targeting of RGC axons are not affected in the Ythdf2 cKO.
YTHDF2 target mRNA were identified with transcriptomic and proteomic analysis
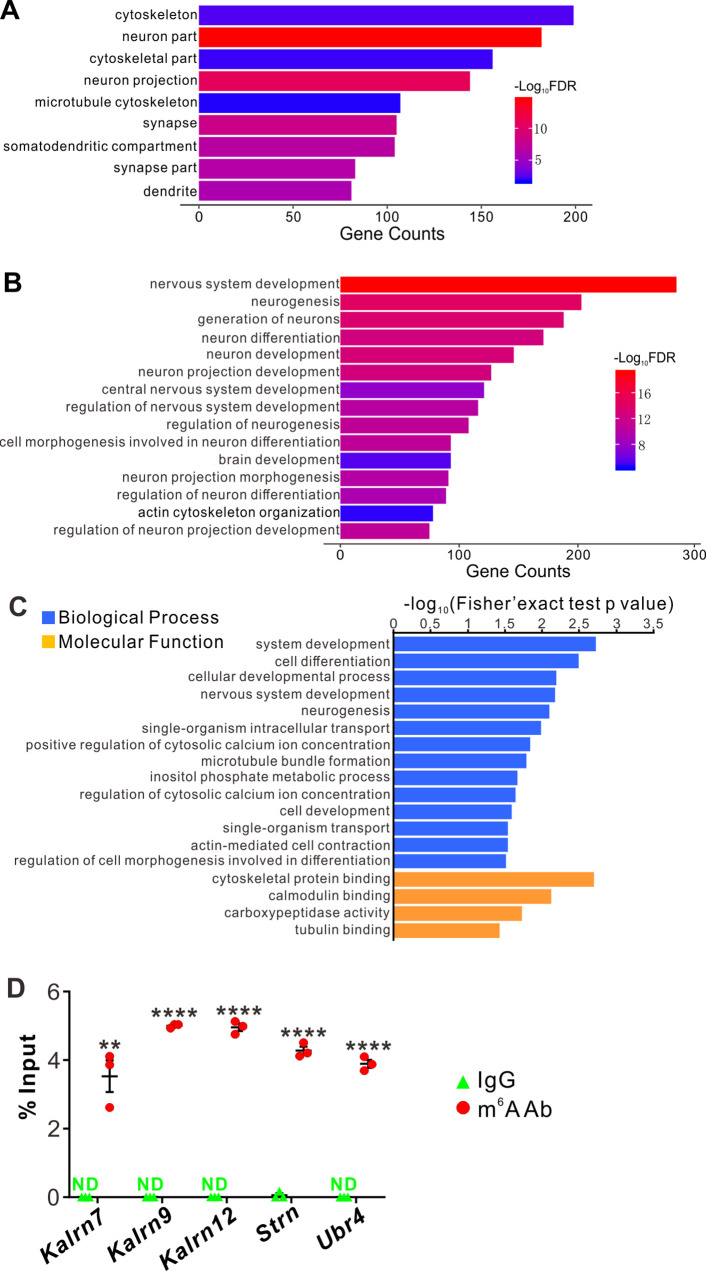
Next, we continued to explore the underlying molecular mechanisms of the effects on dendrite branching caused by Ythdf2 cKO in the retina. First, we wanted to know what transcripts YTHDF2 recognizes and binds. We carried out anti-YTHDF2 RNA immunoprecipitation (RIP) in the retina followed by RNA sequencing of the elute (RIP-Seq). Two biological replicates of anti-YTHDF2 RIP-Seq identified 1638 transcripts (Supplementary file 1). Functional annotation of YTHDF2 RIP targets revealed significant enrichment in cellular component terms such as neuron part and neuron projection, and biological process terms such as cellular component organization and neuron projection development. We further zoomed in to check neural terms in cellular component (Figure 6A) and biological process (Figure 6B). We found that substantial numbers of YTHDF2 target transcripts are involved in cytoskeleton, dendrite, and their organization and development (Figure 6A and B), which is consistent with the dendrite branching phenotype observed in the Ythdf2 cKO retina.
Figure 6. YTHDF2 target mRNAs were identified with transcriptomic and proteomic analysis.
(A, B) Gene Ontology (GO) analysis of YTHDF2 target transcripts identified by anti-YTHDF2 RNA immunoprecipitation (RIP) in the retina followed by RNA sequencing (RIP-Seq). Neural terms were picked out in cellular component (A) and biological process (B). (C) GO analysis of proteins which are upregulated after YTHDF2 knockodwn (KD) by mass spectrometry (MS). (D) Verification of N6-methyladenosine (m6A) modification of YTHDF2 target mRNAs by anti-m6A pulldown followed by RT-qPCR. ND, not detected. Data are mean ± SEM and are represented as dot plots (n = 3 replicates): **p = 0.0016 for Kalrn7; ****p = 1.40E-08 for Kalrn9; ****p = 1.46E-06 for Kalrn12; ****p = 5.46E-06 for Strn; ****p = 4.90E-06 for Ubr4; by unpaired Student’s t test.
The working model for YTHDF2 is that it binds and destabilizes its m6A-modified target transcripts (Wang et al., 2014). Since the destabilization of mRNAs will eventually decrease their protein levels, we carried out proteome analysis using mass spectrometry (MS) in acute shYthdf2-mediated KD of cultured RGCs, in order to identify directly affected targets. Three biological replicates of YTHDF2 KD followed by MS (YTHDF2 KD/MS) identified 114 proteins which were upregulated by YTHDF2 KD (Supplementary file 2). Functional annotation of these proteins revealed significant enrichment in neuron development- and cytoskeleton-related terms (Figure 6C), which is similar to anti-YTHDF2 RIP-Seq results.
By overlapping the two gene lists screened from anti-YTHDF2 RIP-Seq (Supplementary file 1) and YTHDF2 KD/MS_upregulation (Supplementary file 2), we identified a group of potential YTHDF2 target mRNAs in RGCs (Supplementary file 3), including Kalrn, Strn, and Ubr4. m6A modification of these mRNAs was verified by anti-m6A pulldown (Figure 6D). Kalrn (Kalirin) gene generates three alternative splicing isoforms Kalrn7, Kalrn9, and Kalrn12 encoding guanine-nucleotide exchange factors for Rho GTPases, which have been shown to regulate hippocampal and cortical dendritic branching (Xie et al., 2010; Yan et al., 2015), and are required for normal brain functions (Penzes et al., 2001; Xie et al., 2007; Cahill et al., 2009; Russell et al., 2014; Lu et al., 2015; Herring and Nicoll, 2016). Strn (Striatin) was first identified in striatum, and functions as a B subunit of the serine/threonine phosphatase PP2A and is also a core component of a multiprotein complex called STRIPAK (striatin-interacting phosphatase and kinase complex) (Benoist et al., 2006; Li et al., 2018). Strn was reported to regulate dendritic arborization only in striatal neurons but not in cortical neurons (Li et al., 2018). However, whether and how Kalrn and Strn work in the retina was still unknown. Ubr4 (ubiquitin protein ligase E3 component N-recognin 4) is also known as p600 and has been shown to play roles in neurogenesis, neuronal migration, neuronal signaling, and survival (Parsons et al., 2015). However, whether Ubr4 regulates dendrite development remains elusive.
YTHDF2 controls the stability of its target mRNAs which encode proteins regulating RGC dendrite branching
MS analysis after YTHDF2 KD has shown that the protein levels of these target mRNAs were upregulated (Supplementary file 2). IF using antibodies against Strn and Ubr4 detected specific signals in the IPL which were increased in Ythdf2 cKO retina compared with control retina (Figure 7—figure supplement 1A). Enrichment of these proteins in IPL implies that these proteins might function locally in RGC dendrites to regulate dendrite development.
We next wanted to know whether YTHDF2 controlled the protein levels of these m6A-modified target mRNAs through regulation of translation or transcript stability. As shown in Figure 7—figure supplement 1B-E, the mRNA levels of Kalrn7, Kalrn9, Kalrn12, Strn, and Ubr4 were dramatically increased after KD of YTHDF2, KO of Ythdf2, or KD of METTL14, supporting that YTHDF2 might regulate stability of these target mRNAs. We further evaluated potential changes in the stability of these target mRNAs in an m6A-dependent manner. We further verified this by directly measuring the stability of these target mRNAs. As shown in Figure 7A, all the target mRNAs showed significantly increased stability in the Ythdf2 cKO retina compared with controls. These results suggest that YTHDF2 controlled the protein levels of its m6A-modifed target mRNAs by decreasing their stability.
Figure 7. YTHDF2 target mRNAs mediate YTHDF2-controlled retinal ganglion cell (RGC) dendrite branching.
(A) YTHDF2 target mRNAs showing increased stability in the Ythdf2 conditional knockout (cKO) retina. RGCs dissected from E14.5 Ythdf2 cKO and control embryos were cultured, treated with actinomycin D (ActD), and collected at different timepoints. Data are mean ± SEM (n = 3 replicates). For Kalrn7, **p = 0.0057 (3 hr), *p = 0.014 (6 hr), ***p = 0.00039 (12 hr); for Kalrn9, **p = 0.0036 (3 hr), ***p = 0.00090 (6 hr), *p = 0.032 (12 hr); for Kalrn12, **p = 0.0012 (3 hr), *p = 0.010 (6 hr), **p = 0.0069 (12 hr); for Strn, *p = 0.014 (3 hr), *p = 0.012 (6 hr), *p = 0.016 (12 hr); for Ubr4, **p = 0.0077 (3 hr), **p = 0.0059 (6 hr), *p = 0.041 (12 hr); all by unpaired Student’s t test. (B) Knockdown (KD) of the target mRNAs causing decreased dendrite branching of cultured RGCs prepared from wild type (WT) E14.5 retina by Sholl analysis. Brn3a and MAP2 IF were used to mark RGCs and visualize dendrites. Data are mean ± SEM. For Kalrn7 (n = 59 for siCtrl, n = 56 for siKalrn7), ****p = 2.33E-06 (10 μm), ****p = 5.85E-06 (15 μm), ****p = 8.67E-05 (20 μm), **p = 0.0045 (25 μm), **p = 0.0058 (30 μm), *p = 0.010 (35 μm); for Kalrn9 (n = 59 for siCtrl, n = 46 for siKalrn9), ****p = 3.69E-05 (10 μm), ****p = 5.53E-05 (15 μm), ***p = 0.00020 (20 μm), ****p = 3.09E-06 (25 μm), ****p = 4.63E-06 (30 μm), ***p = 0.00059 (35 μm), **p = 0.0010 (40 μm), *p = 0.042 (45 μm); for Kalrn12 (n = 59 for siCtrl, n = 39 for siKalrn12), **p = 0.0031 (10 μm), ***p = 0.00017 (15 μm), ****p = 6.56E-05 (20 μm), **p = 0.0017 (25 μm), *p = 0.017 (30 μm); for Strn (n = 51 for siCtrl, n = 57 for siStrn), ****p = 4.19E-05 (10 μm), ***p = 0.00067 (15 μm), **p = 0.0079 (20 μm), *p = 0.015 (30 μm); for Ubr4 (n = 81 for siCtrl, n = 81 for siUbr4), ****p = 1.26E-08 (10 μm), ****p = 7.61E-10 (15 μm), ****p = 2.35E-08 (20 μm), ****p = 1.39E-05 (25 μm), **p = 0.0061 (30 μm); all by unpaired Student’s t test. (C) Increased dendrite branching of cultured RGCs prepared from E14.5 Ythdf2 cKO (Y2 cKO) retina was rescued by KD of target mRNAs using siRNAs. Data are mean ± SEM. Ctrl, Ythdf2fl/fl; Y2 cKO, Six3-cre+/-,Ythdf2fl/fl. In C1,'Ctrl, siCtrl’ (n = 35 neurons) vs. ‘Y2 cKO, siCtrl’ (n = 52 neurons), *p = 0.038 (15 μm), *p = 0.045 (20 μm), **p = 0.0036 (30 μm); ‘Y2 cKO, siKalrn7’ (n = 55 neurons) vs. ‘Y2 cKO, siCtrl’, *p = 0.020 (15 μm), *p = 0.025 (20 μm), *p = 0.031 (25 μm); ‘Y2 cKO, siKalrn9’ (n = 66 neurons) vs. ‘Y2 cKO, siCtrl’, *p = 0.020 (10 μm), *p = 0.013 (15 μm), *p = 0.031 (20 μm), *p = 0.017 (25 μm), *p = 0.031 (30 μm), *p = 0.031 (45 μm); ‘Y2 cKO, siKalrn12’ (n = 80 neurons) vs. ‘Y2 cKO, siCtrl’, *p = 0.015 (10 μm), **p = 0.0018 (15 μm), *p = 0.015 (20 μm), *p = 0.027 (40 μm). In C2, 'Ctrl, siCtrl’ (n = 50 neurons) vs. ‘Y2 cKO, siCtrl’ (n = 47 neurons), **p = 0.0031 (10 μm), **p = 0.0013 (15 μm), *p = 0.029 (20 μm), **p = 0.0015 (30 μm), *p = 0.014 (35 μm); ‘Y2 cKO, siStrn’ (n = 45 neurons) vs. ‘Y2 cKO, siCtrl’, ***p = 0.00016 (10 μm), **p = 0.0043 (15 μm), *p = 0.010 (20 μm), *p = 0.018 (30 μm); ‘Y2 cKO, siUbr4’ (n = 57 neurons) vs. ‘Y2 cKO, siCtrl’, ***p = 0.00084 (10 μm), ****p = 4.89E-05 (15 μm), **p = 0.0058 (20 μm), **p = 0.0045 (30 μm). All by unpaired Student’s t test. (D) Increased dendrite branching of RGC subtypes in Ythdf2 cKO (Y2 cKO) retina was rescued by KD of target mRNAs through intravitreal injection of AAV shRNAs in vivo. Data are mean ± SEM. Ctrl, Ythdf2fl/fl; Y2 cKO, Six3-cre+/-,Ythdf2fl/fl. In D1 (CART+/eGFP+ ooDSGCs), ‘Ctrl, shCtrl’ (n = 10 neurons) vs. ‘Y2 cKO, shCtrl’ (n = 6 neurons), *p = 0.010 (10 μm), ***p = 0.00049 (20 μm), **p = 0.0021 (30 μm), **p = 0.0047 (40 μm), *p = 0.028 (50 μm), *p = 0.011 (60 μm), *p = 0.030 (90 μm), *p = 0.042 (110 μm); ‘Y2 cKO, shKalrn12’ (n = 8 neurons) vs. ‘Y2 cKO, shCtrl’, *p = 0.012 (20 μm), *p = 0.014 (30 μm); ‘Y2 cKO, shUbr4’ (n = 6 neurons) vs. ‘Y2 cKO, shCtrl’, *p = 0.011 (10 μm), **p = 0.0084 (20 μm), *p = 0.029 (30 μm). In D2 (SMI-32+αRGCs), ‘Ctrl, shCtrl’ (n = 14 neurons) vs. ‘Y2 cKO, shCtrl’ (n = 14 neurons), *p = 0.032 (40 μm), **p = 0.0019 (50 μm), **p = 0.0014 (60 μm), *p = 0.015 (70 μm), *p = 0.044 (90 μm); ‘Y2 cKO, shKalrn12’ (n = 26 neurons) vs. ‘Y2 cKO, shCtrl’, **p = 0.0023 (20 μm), ***p = 0.00076 (30 μm), ***p = 0.00030 (40 μm), ***p = 0.00020 (50 μm), *p = 0.015 (60 μm); ‘Y2 cKO, shUbr4’ (n = 15 neurons) vs. ‘Y2 cKO, shCtrl’, *p = 0.042 (30 μm), *p = 0.024 (40 μm), *p = 0.018 (50 μm). All by unpaired Student’s t test.
Figure 7—figure supplement 1. YTHDF2 target mRNAs were characterized and validated.
Next we explored the functions of these YTHDF2 target mRNAs in RGC dendrite development. We first generated siRNAs against these transcripts (Figure 7—figure supplement 1F). We then checked the effects on RGC dendrite branching after KD of these target mRNAs by siRNAs in cultured RGCs. As shown in Figure 7B, KD of Kalrn7, Kalrn9, Kalrn12, Strn, or Ubr4 led to significant decreases of RGC dendrite branching. Interestingly, the siCocktail against all these target mRNAs further significantly reduced the RGC dendrite branching compared with each individual siRNA (Figure 7—figure supplement 1G), suggesting that these targets may work in different pathways to regulate the RGC dendrite morphology. We further examined whether these target mRNAs mediate YTHDF2-regulated RGC dendrite branching. As shown in Figures 2E–H ,–3, cKO of Ythdf2 led to increased dendrite branching of RGCs both in vitro and in vivo. Transfection of siRNAs against these target mRNAs rescued dendrite branching increases in cultured Ythdf2 cKO RGCs (Figure 7C). We continued to generate and performed intravitreal injection of AAV viral shKalrn12 and shUbr4, which significantly rescued dendrite branching increases of CART+ ooDSGCs and SMI-32+αRGCs in Ythdf2 cKO retina in vivo (Figure 7D).
Taken together, we identified a group of YTHDF2 target mRNAs that encode proteins regulating RGC dendrite branching, which mediate YTHDF2-controlled RGC dendrite branching.
Ythdf2 cKO retina is more resistant to AOH
The glaucomatous eyes are symptomatized with progressive neurodegeneration and vision loss (Agostinone and Di Polo, 2015). High intraocular pressure is a major risk factor in glaucoma and has been shown to cause pathological changes in RGC dendrites before axon degeneration or soma loss is detected in different model animals (Weber et al., 1998; Shou et al., 2003; Morgan et al., 2006). Our findings that Ythdf2 cKO in retina promotes RGC dendrite branching during development inspired us to wonder whether YTHDF2 also regulates RGC dendrite maintenance in the acute glaucoma model caused by AOH. We utilized the AOH model made with control and Ythdf2 cKO mice to check whether Ythdf2 cKO in the retina could alter the pathology in the glaucomatous eyes. RGC dendrite branching is significantly decreased after AOH operation compared with non-AOH in either genotype (Figure 8—figure supplement 1A,B). Interestingly, the Ythdf2 cKO retina with AOH operation maintains significantly higher dendrite complexity compared with the glaucomatous eyes of Ythdf2fl/fl control mice (Figure 8A and B). In addition, there are significant RGC neuron losses in both genotypes after AOH (Figure 8C and D). However, the reduction of RGC number in the Ythdf2 cKO retina is less than control retina (Figure 8C and D). These results support that Ythdf2 cKO protects retina from RGC dendrite degeneration and soma loss caused by AOH.
Figure 8. Ythdf2 conditional knockout (cKO) retina is more resistant to acute ocular hypertension (AOH).
(A, B) Better maintenance of retinal ganglion cell (RGC) dendrite arborization in Ythdf2 cKO retina after AOH operation. AOH was performed using adult mice, and retinas were collected after AOH for wholemount immunostaining of melanopsin and SMI-32 to visualize the dendrite arbors of corresponding RGC subtype, respectively. Dendrite traces were drawn as previously shown and quantification of dendrite branching was done using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in Six3-cre+/-,Ythdf2fl/fl retina than Ythdf2fl/fl control retina in both RGC subtypes after AOH: for melanopsin+ intrinsically photosensitive RGCs (ipRGCs) in A, Ythdf2fl/fl/AOH (n = 51 RGCs) vs. cKO/AOH (n = 64 RGCs), ***p = 0.00015 (10 μm), **p = 0.0017 (20 μm), *p = 0.034 (30 μm), ***p = 0.00035 (40 μm), ****p = 3.02E-05 (50 μm), ****p = 2.63E-05 (60 μm), **p = 0.0029 (70 μm), **p = 0.0028 (80 μm), ***p = 0.00035 (90 μm), **p = 0.0032 (100 μm), **p = 0.0014 (110 μm), **p = 0.0043 (120 μm), **p = 0.0014 (130 μm), *p = 0.023 (140 μm), *p = 0.013 (150 μm); for SMI-32+αRGCs in B, Ythdf2fl/fl/AOH (n = 21 neurons) vs. cKO/AOH (n = 15 neurons), **p = 0.0052 (40 μm), **p = 0.0057 (50 μm); all by unpaired Student’s t test. (C, D) Ythdf2 cKO retina showing less severe RGC loss after AOH. AOH was performed using adult mice and retinas were collected after AOH for wholemount immunostaining using a Brn3a antibody (C). Numbers of Brn3a+ RGCs per 10,000 μm2 of retina were quantified for different genotypes and conditions (confocal fields for analysis: n = 117 for Ythdf2fl/fl/Ctrl; n = 98 for Ythdf2fl/fl/AOH; n = 110 for cKO/Ctrl; n = 104 for cKO/AOH). Data are represented as box and whisker plots (D): ns, not significant (p = 0.16; Ythdf2fl/fl/Ctrl vs. cKO/Ctrl); **p = 0.0077 (Ythdf2fl/fl/AOH vs. cKO/AOH); by unpaired Student’s t test. Scale bar: 25 μm. (E, F) Overexpression (OE) of YTHDF2 targets Hspa12a and Islr2 protecting retina from RGC dendrite degeneration in the AOH model. Wild type (WT) mice were intravitreally injected with AAV overexpressing Hspa12a or Islr2 and then operated with AOH. Wholemount immunostaining of CART/ZsGreen and SMI-32/ZsGreen was carried out to visualize the dendrite arbors of corresponding RGC subtype, respectively. Dendrite traces were drawn as previously shown and quantification of dendrite branching was done using Sholl analysis. Data are mean ± SEM. Numbers of interactions are significantly greater in retina with OE of Hspa12a or Islr2 than control retina in both RGC subtypes after AOH. For CART+ ON-OFF directionally selective RGCs (ooDSGCs) in E: OE-Ctrl/AOH (n = 11 RGCs) vs. OE-Hspa12a/AOH (n = 11 RGCs), *p = 0.014 (20 μm), **p = 0.0025 (30 μm), *p = 0.018 (40 μm); OE-Ctrl/AOH vs. OE-Islr2/AOH (n = 6 RGCs), *p = 0.024 (10 μm), ***p = 0.00031 (20 μm), **p = 0.0038 (30 μm), *p = 0.013 (40 μm). For SMI-32+αRGCs in F: OE-Ctrl/AOH (n = 49 neurons) vs. OE-Hspa12a/AOH (n = 46 neurons), **p = 0.0023 (30 μm), ***p = 0.00080 (40 μm), **p = 0.0059 (50 μm), **p = 0.0051 (60 μm), **p = 0.0036 (70 μm), ***p = 0.00070 (80 μm), **p = 0.0015 (90 μm), *p = 0.016 (100 μm), *p = 0.011 (110 μm); OE-Ctrl/AOH vs. OE-Islr2/AOH (n = 13 RGCs), *p = 0.010 (30 μm), **p = 0.0093 (40 μm), **p = 0.0019 (50 μm), ***p = 0.00085 (60 μm), ***p = 0.00067 (70 μm), ****p = 4.25E-05 (80 μm), ****p = 2.54E-05 (90 μm), ***p = 0.00020 (100 μm) , **p = 0.0016 (110 μm). All by unpaired Student’s t test. (G) OE of YTHDF2 targets Hspa12a and Islr2 alleviating RGC loss in the AOH model. WT mice were intravitreally injected with AAV overexpressing Hspa12a or Islr2 and then operated with AOH. Wholemount immunostaining of Brn3a was performed to label RGCs. Numbers of Brn3a+ RGCs per 10,000 μm2 of retina were quantified for different conditions (confocal fields for analysis: n = 19 for OE-Ctrl; n = 26 for OE-Hspa12a; n = 24 for OE-Islr2). Data are represented as box and whisker plots: *p = 0.034 (OE-Hspa12a vs. OE-Ctrl; *p = 0.029 (OE-Islr2 vs. OE-Ctrl); by unpaired Student’s t test.
Figure 8—figure supplement 1. Hspa12a and Islr2 are two target mRNAs of YTHDF2 in adult retina.
Next we wanted to know whether and how YTHDF2 target mRNAs mediate these effects in the AOH models. We first checked the expression of YTHDF2 target mRNAs identified in the developing retina (Supplementary file 3) in the adult Ythdf2 cKO and control retina. We found that two target mRNAs Hspa12a and Islr2 show upregulation in the adult Ythdf2 cKO retina compared with control (Figure 8—figure supplement 1C). m6A modification of Hspa12a and Islr2 mRNAs was further verified by anti-m6A pulldown (Figure 8—figure supplement 1D). Hspa12a encodes heat shock protein A12A which is an atypical member of the heat shock protein 70 family and has been shown to be downregulated in diseases such as ischemic stroke, schizophrenia, and renal cell carcinoma (Pongrac et al., 2004; Mao et al., 2018; Min et al., 2020). Islr2 encodes immunoglobulin superfamily containing leucine-rich repeat protein two and is poorly studied. Here, we found that Hspa12a and Islr2 are downregulated in the retina after AOH operation (Figure 8—figure supplement 1E), which is likely caused by upregulation of YTHDF2 in the AOH-treated retina (Figure 8—figure supplement 1F-H). We therefore hypothesized that AOH upregulates YTHDF2 which in turn downregulates its targets Hspa12a and Islr2, thus causing RGC dendrite degeneration and soma loss. If this is the case, overexpression of Hspa12a and Islr2 might protect RGC dendrite from AOH-triggered degeneration. We thus generated AAV harboring overexpression constructs of Hspa12a and Islr2 which were intravitreally injected to wild type retinas. After the AOH induction, the retinas overexpressing Hspa12a and Islr2 maintain significantly more complex RGC dendrite arbor and show better RGC survival compared with control AAV (Figure 8E–G).
These data verify that loss-of-function of YTHDF2 and gain-of-function of its targets Hspa12a and Islr2 have neuroprotective roles in the glaucomatous retina.
Discussion
Functions and mechanisms of mRNA m6A modification in the dendrite development were not known. Here, we revealed a critical role of the m6A reader YTHDF2 in RGC dendrite development and maintenance. YTHDF2 have two phases of function to control RGC dendrite development first and then maintenance through regulating two sets of target mRNAs. In early postnatal stages, the target mRNAs Kalrn7, Kalrn9, Kalrn12, Strn, and Ubr4 mediate YTHDF2 functions to regulate RGC dendrite development. In the adult mice, another set of target mRNAs Hspa12a and Islr2 mediate YTHDF2 function to regulate RGC dendrite maintenance.
Positive and negative regulators for dendrite development
The general principle for dendrite arborization is that the dendrite arbor cannot be either too big or too small in order to precisely sample a presynaptic target area during neural circuit formation (Lefebvre et al., 2015). Numerous extrinsic and intrinsic mechanisms have been found to regulate dendritic arbor patterning, which involves both positive and negative factors to achieve balanced control of dendritic growth (Jan and Jan, 2010; Dong et al., 2015; Ledda and Paratcha, 2017). For the secreted and diffusible cues, BDNF promotes dendrite branching and complexity (Cheung et al., 2007); the non-canonical Wnt7b/PCP pathway is a positive regulator of dendrite growth and branching (Rosso et al., 2005); the non-canonical Wnt receptor Ryk works as a negative regulator by limiting the extent of dendritic branching (Lanoue et al., 2017). For the contact-mediated signals, the cadherins Celsr2 and Celsr3 regulate dendrite growth in an opposite manner in cortical pyramidal and Purkinje neurons, and hippocampal neurons, respectively (Shima et al., 2004; Shima et al., 2007). For the transcription factors, studies have shown that manipulation of Cux1 and Cux2 levels has distinct effects on apical and basal arbors of cortical dendrites (Cubelos et al., 2015); interestingly, the functions of Sp4 in dendrite development are dependent on the cellular context of its expression, for example, Sp4 promotes dendrite growth and branching in hippocampal dentate granule cells but limits dendrite branching in cerebellar granule cells (Ramos et al., 2007; Zhou et al., 2007). Here, we identified another negative regulator YTHDF2 which works posttranscriptionally, and loss-of-function of YTHDF2 increased dendrite complexity during development and protected RGC degeneration from AOH. We have validated this effect on several RGC subtypes. Since there are dozens of RGC subtypes, it is technically challenging but still interesting to test whether this effect is universal to all subtypes or there is specificity for RGC subtypes.
Posttranscriptional regulation of dendrite development
It is well established that mRNAs can be transported and targeted to specific neuronal compartments such as axons and dendrites. Local translation of these mRNAs enables exquisite and rapid control of local proteome in specific subcellular compartments (Ledda and Paratcha, 2017). Local translation is known to play roles in controlling dendrite arborization (Chihara et al., 2007), and is regulated by specific RNA-binding proteins (Jan and Jan, 2010). In Drosophila, the RNA-binding proteins Pumilio (Pum), Nanos (Nos), Glorund (Glo), and Smaug (Smg) regulate morphogenesis and branching of specific classes of dendritic arborization neurons through controlling translation of their target mRNAs including nanos mRNA itself (Ye et al., 2004; Brechbiel and Gavis, 2008). The mouse homologue of another RNA-binding protein Staufen, Stau1, regulates dendritic targeting of ribonucleoprotein particles and dendrite branching (Vessey et al., 2008). Here, we found that the m6A reader and RNA-binding protein YTHDF2 control stability of its target mRNAs and regulate dendrite branching in RGCs. It would be interesting to see whether these target mRNAs are localized into dendrites and whether YTHDF2 works in dendrites to control their stability and translation. Actually, Strn4 mRNA has been shown to be present in dendrites and locally translated (Lin et al., 2017). In addition, how the proteins encoded by these target mRNAs regulate RGC dendrite branching during development and maintenance remains to be explored and will be important future directions.
Neuroprotective genes in retinal injuries and degeneration
Transcriptome analyses have revealed differentially expressed genes after retinal injuries such as AOH-induced glaucoma and optic nerve crush (ONC), and the upregulated genes are of importance for discovering new treatment approaches (Jakobs, 2014; Tran et al., 2019). One of the previous studies has identified Mettl3, encoding the m6A writer, as an upregulated gene after ONC (Agudo et al., 2008). Here, we found Ythdf2, encoding an m6A reader, was also upregulated in the retina after AOH. We further found that Hspa12a and Islr2, two targets of YTHDF2 in adult retina, were downregulated in glaucomatous retinas. Overexpression of Hspa12a and Islr2 protected retina from AOH-caused RGC dendrite degeneration. Our findings in this study suggest that YTHDF2 and its neuroprotective target mRNAs might be valuable in developing novel therapeutic approaches to treat neurodegeneration caused by glaucoma and other retinal injuries.
Materials and methods
Key resources table.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (mouse) | Mouse: Ythdf2fl/fl | Yu et al., 2021b | N/A | |
| Strain, strain background (mouse) | Mouse: Tg(Six3-cre)69Frty/GcoJ | Jackson Laboratory | Cat#: JAX_019755RRID: IMSR_JAX:019755 | |
| Strain, strain background (mouse) | Mouse: B6.Cg-Tg(Thy1-EGFP)OJrs/GfngJ | Jackson Laboratory | Cat#: JAX_007919RRID: IMSR_JAX:007919 | |
| Strain, strain background (mouse) | Mouse: B6.129 × 1-Gt(ROSA)26Sortm1(EYFP)Cos/J | Jackson Laboratory | Cat#: JAX_006148RRID: IMSR_JAX:006148 | |
| Antibody | Anti-GFP (Chicken polyclonal) | Abcam | Cat#: ab13970, RRID: AB_300798 |
IF (1:1000) |
| Antibody | Anti-MAP2 (Chicken polyclonal) | Abcam | Cat#: ab5392, RRID: AB_2138153 |
IF (1:10,000) |
| Antibody | Anti-RBPMS (Guinea pig polyclonal) | PhosphoSolutions | Cat#: 1832-RBPMS, RRID: AB_2492226 |
IF (1:1000) |
| Antibody | Anti-VAChT (Goat polyclonal) | Millipore | Cat#: ABN100, RRID: AB_2630394 | IF (1:1000) |
| Antibody | Anti-β Actin (Mouse monoclonal) | Abcam | Cat#: ab6276, RRID: AB_2223210 |
WB (1:30,000) |
| Antibody | Anti-β Actin (Mouse monoclonal) | ABclonal | Cat#: AC004, RRID: AB_2737399 |
WB (1:30,000) |
| Antibody | Anti-AP2α (Mouse monoclonal) | DSHB | Cat#: 3B5, RRID: AB_2313947 | IF (1:1000) |
| Antibody | Anti-Bassoon (Mouse monoclonal) | Enzo Life Sciences | Cat#: ADI-VAM-PS003, RRID: AB_10618753 | IF (1:2500) |
| Antibody | Anti-Brn3a (Mouse monoclonal) | Millipore | Cat#: MAB1585, RRID: AB_94166 | IF (1:300) |
| Antibody | Anti-Calbindin-D-28K (Mouse monoclonal) | Sigma-Aldrich | Cat#: C9848, RRID: AB_476894 | IF (1:200) |
| Antibody | Anti-PKCα (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc-8393, RRID: AB_628142 | IF (1:500) |
| Antibody | Anti-SMI-32 (Mouse monoclonal) | BioLegend | Cat#: 801701, RRID: AB_2564642 | IF (1:200) |
| Antibody | Anti-Strn (Striatin) (Mouse monoclonal) | BD Biosciences | Cat#: 610838, RRID: AB_398157 | IF (1:500) |
| Antibody | Anti-CART (Rabbit polyclonal) | Phoenix Pharmaceuticals | Cat#: H-003–62, RRID: AB_2313614 | IF (1:2000) |
| Antibody | Anti m6A (Rabbit polyclonal) | Synaptic Systems | Cat# 202003, RRID: AB_2279214 |
IF (1:200) |
| Antibody | Anti-melanopsin (Rabbit polyclonal) | Thermo Fisher Scientific | Cat#: PA1-780, RRID: AB_2267547 | IF (1:1000) |
| Antibody | Anti-PKCα (Rabbit polyclonal) | Cell Signaling | Cat#: CST-2056 | IF (1:1000) |
| Antibody | Anti-PSD-95 (Mouse monoclonal) | Abcam | Cat#: ab2723, RRID: AB_303248 |
IF (1:500) |
| Antibody | Anti-Recoverin (Rabbit polyclonal) | Millipore | Cat#: AB5585, RRID: AB_2253622 |
IF (1:1000) |
| Antibody | Anti-YTHDF2 (Rabbit polyclonal) | Proteintech | Cat#: 24744–1-AP, RRID: AB_2687435 | IF (1:1000) |
| Antibody | Anti-YTHDF1 (Rabbit polyclonal) | Proteintech | Cat#: 17479–1-AP, RRID: AB_2217473 | IF (1:1000) |
| Antibody | Anti-YTHDF3 (Rabbit polyclonal) | Abcam | Cat#: ab103328, RRID: AB_10710895 | IF (1:1000) |
| Antibody | Anti-Ubr4 (Rabbit polyclonal) | Abcam | Cat#: ab86738, RRID: AB_1952666 | IF (1:300) |
| Antibody | Anti-Chx10 (Sheep polyclonal) | Exalpha | Cat#: X1179P | IF (1:1000) |
| Antibody | Anti-GFAP (Chicken polyclonal) | Millipore | Cat#: AB5541, RRID: AB_177521 | IF (1:500) |
| Antibody | Anti-Lhx2 (Goat polyclonal) | Santa Cruz Biotechnology | Cat#: sc-19344, RRID: AB_2135660 | IF (1:200) |
| Antibody | Anti-Lhx2 (Rabbit monoclonal) | Abcam | Cat#: ab184337 | IF (1:500) |
| Antibody | Anti-chicken IgY (Alexa 488 donkey) | Jackson Immunoresearch | Cat#: 703-545-155, RRID: AB_2340375 | IF (1:500) |
| Antibody | Anti-G. pig IgG (Alexa 488 donkey) | Jackson Immunoresearch | Cat#: 706-545-148, RRID: AB_2340472 | IF (1:500) |
| Antibody | Anti-mouse IgG (Alexa 488 donkey) | Thermo Fisher Scientific | Cat#: A-21202, RRID: AB_141607 | IF (1:500) |
| Antibody | Anti-rabbit IgG (Alexa 488 donkey) | Thermo Fisher Scientific | Cat#: A-21206, RRID: AB_141708 | IF (1:500) |
| Antibody | Anti-goat IgG (Alexa 555 donkey) | Thermo Fisher Scientific | Cat#: A-21432, RRID: AB_2535853 | IF (1:1000) |
| Antibody | Anti-mouse IgG (Alexa 555 donkey) | Thermo Fisher Scientific | Cat#: A-31570, RRID: AB_2536180 | IF (1:1000) |
| Antibody | Anti-rabbit IgG (Alexa 555 donkey) | Thermo Fisher Scientific | Cat#: A-31572, RRID: AB_162543 | IF (1:1000) |
| Antibody | Anti-sheep IgG (Alexa 555 donkey) | Thermo Fisher Scientific | Cat#: A-21436, RRID: AB_2535857 | IF (1:1000) |
| Antibody | Anti-chicken IgY (Alexa 555 goat) | Thermo Fisher Scientific | Cat#: A-21437, RRID: AB_2535858 | IF (1:1000) |
| Antibody | Anti-mouse IgG (Alexa 647 donkey) | Thermo Fisher Scientific | Cat#: A-31571, RRID: AB_162542 | IF (1:200) |
| Antibody | Anti-mouse IgG (HRP donkey) | Abcam | Cat#: ab97030, RRID: AB_10680919 | WB (1:2500) |
| Antibody | Anti-rabbit IgG (HRP donkey) | Abcam | Cat#: ab16284, RRID: AB_955387 | WB (1:2500) |
| Antibody | Anti-mouse IgG (HRP VHH) | AlpaLife | Cat#: KTSM1321 | WB (1:5000) |
| Antibody | Anti-rabbit IgG (HRP VHH) | AlpaLife | Cat#: KTSM1322 | WB (1:5000) |
| Recombinant DNA reagent | Plasmid: pLKO.1-TRC | Addgene | Addgene plasmid #10878, RRID: Addgene_10878 | |
| Sequence-based reagent | shRNA targeting sequence of negative control |
This paper | N/A | GCATCAAGGTG AACTTCAAGA |
| Sequence-based reagent | shRNA targeting sequence of mouse Ythdf2 |
Yu et al., 2018 | N/A | GGACGTTCCC AATAGCCAACT |
| Sequence-based reagent | shRNA targeting sequence of mouse Ythdf1 |
This paper | N/A | GGACATTGGT ACTTGGGATAA |
| Sequence-based reagent | shRNA targeting sequence of mouse Ythdf3 |
This paper | N/A | GGATTTGGCAA TGATACTTTG |
| Sequence-based reagent | shRNA targeting sequence of mouse Mettl14#6 | This paper | N/A | GCTGGACCTGG GATGATATTA |
| Sequence-based reagent | shRNA targeting sequence of mouse Mettl14#7 | This paper | N/A | CCCAGCTTGT ACTTTGCTTTA |
| Sequence-based reagent | shRNA targeting sequence of negative control (AAV) | This paper | N/A | TTCTCCGAAC GTGTCACGTAA |
| Sequence-based reagent | shRNA targeting sequence of mouse Kalrn12 | This paper | N/A | TGATGAGCTGA TGGAAGAA |
| Sequence-based reagent | shRNA targeting sequence of mouse Ubr4 | This paper | N/A | AATGATGAGC AGTCATCTC |
| Sequence-based reagent | siRNA targeting sequence of negative control | Yu et al., 2018 | N/A | UUCUCCGAAC GUGUCACGUTT |
| Sequence-based reagent | siRNA targeting sequence of mouse Kalrn7 | Xie et al., 2007 | N/A | AGUACAAUCCU GGCCAUGUTT |
| Sequence-based reagent | siRNA targeting sequence of mouse Kalrn9 | Yan et al., 2015 | N/A | ACUGGACUGG ACUUCUAUUTT |
| Sequence-based reagent | siRNA targeting sequence of mouse Kalrn12 | Yan et al., 2015 | N/A | CGAUGAGCUG AUGGAAGAATT |
| Sequence-based reagent | siRNA targeting sequence of mouse Strn | Breitman et al., 2008 | N/A | GGUGAAGAUCG AGAUACAATT |
| Sequence-based reagent | siRNA targeting sequence of mouse Ubr4 | Shim et al., 2008 | N/A | AAUGAUGAGC AGUCAUCUATT |
| Sequence-based reagent | qPCR primers of mouse 18s | Wang et al., 2018 | N/A | Fwd: GCTTAATTTGACT CAACACGGGARev: AGCTATCAATCTG TCAATCCTGTC |
| Sequence-based reagent | qPCR primers of mouse Gapdh | Mains et al., 2011 | N/A | Fwd: TTGTCAGCAATG CATCCTGCACCACCRev: CTGAGTGGCAGT GATGGCATGGAC |
| Sequence-based reagent | qPCR primers of mouse Ythdf2 | This paper | N/A | Fwd: GAGCAGAGA CCAAAAGGTCAAGRev: CTGTGGGCTC AAGTAAGGTTC |
| Sequence-based reagent | qPCR primers of mouse Kalrn7 | Mains et al., 2011 | N/A | Fwd: GATACCATATCCAT TGCCTCCAGGACCRev: CCAGGCTGCGC GCTAAACGTAAG |
| Sequence-based reagent | qPCR primers of mouse Kalrn9 | Mains et al., 2011 | N/A | Fwd: GCCCCTCGCC AAAGCCACAGCRev: CCAGTGAGT CCCGTGGTGGGC |
| Sequence-based reagent | qPCR primers of mouse Kalrn12 | Mains et al., 2011 | N/A | Fwd: CAGCAGCCA CGTGCCTGCAGCRev: TCTTGACATTGGG AATGGGCCGCAC |
| Sequence-based reagent | qPCR primers of mouse Strn |
This paper | N/A | Fwd: TGAAGCCTG GAATGTGGACCRev: CTATTGGGC CTCTTCACCCC |
| Sequence-based reagent | qPCR primers of mouse Ubr4 |
This paper | N/A | Fwd: TGAGTGAGG ACAAGGGCAACRev: GGGTTGGAT CGAACGAAGGT |
| Sequence-based reagent | qPCR primer for mouse Hspa12a | This paper | N/A | Fwd: GGGTTTGCACA GGCTAAGGARev: TCTGATGGACG GTCAGGTCT |
| Sequence-based reagent | qPCR primer for mouse Islr2 | This paper | N/A | Fwd: GAAGCTCCCTTA GACTGTCACCRev: CCCCATCGTGA CTCCTGCTG |
| Sequence-based reagent | PCR primer for mouse Hspa12a CDS | This paper | N/A | Fwd: ATGGCGGACAA GGAAGCTGGRev: GTAATTTAAGAA GTCGATCCCC |
| Sequence-based reagent | PCR primer for mouse Islr2 CDS |
This paper | N/A | Fwd: ATGGGGCC CTTTGGAGCRev: GCCCGCTGTC TGCCTGTAG |
| Sequence-based reagent | Mouse genotyping primers for Ythdf2 loxp site 1 | This paper | N/A | GCTTGTAGTTATG TTGTGTACCAC and GCAGCTCTGACT ATTCTAAAACCTCC |
| Sequence-based reagent | Mouse genotyping primers for Ythdf2 loxp site 2 | This paper | N/A | CTCATAACATCC ATAGCCACAGG and CCAAGAGATAG CTTTCCTAATG |
| Sequence-based reagent | Mouse genotyping primers for Six3-cre | Chunqiao Liu’s lab | N/A | CCTTCCTCCCT CTCTATGTG and GAACGAACCT GGTCGAAATC |
| Sequence-based reagent | Mouse genotyping primers for Thy1-GFP | The Jackson Laboratory website | N/A | CGGTGGTGC AGATGAACTT and ACAGACACAC ACCCAGGACA |
| Sequence-based reagent | Mouse genotyping primers for Rosa-YFP mutant site | The Jackson Laboratory website | N/A | AGGGCGAGG AGCTGTTCA and TGAAGTCGAT GCCCTTCAG |
| Sequence-based reagent | Mouse genotyping primers for Rosa-YFP wild type site | The Jackson Laboratory website | N/A | CTGGCTTCT GAGGACCG and CAGGACAAC GCCCACACA |
| Peptide, recombinant protein | Insulin | Sigma | Cat#: I6634 | |
| Peptide, recombinant protein | Recombinant Human/Murine/Rat BDNF |
PeproTech | Cat#: 450–02 | |
| Peptide, recombinant protein | Recombinant Human NT-3 | PeproTech | Cat#: 450–03 | |
| Peptide, recombinant protein | Recombinant Murine EGF | PeproTech | Cat#: 315–09 | |
| Peptide, recombinant protein | Recombinant Human FGF-basic | PeproTech | Cat#: 100-18B | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#: 23227 | |
| Commercial assay or kit | GeneSilencer Transfection Reagent | Genlantis | Cat#: T500750 | |
| Commercial assay or kit | Magna MeRIP m6A Kit | Millipore | Cat#: 17–10499 | |
| Commercial assay or kit | EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit | Millipore | Cat#: 17–701 | |
| Chemical compound, drug | cpt-cAMP, 8-(4-Chlorophenylthio) Adenosine 3':5'-CY |
Sigma | Cat#: C3912 | |
| Chemical compound, drug |
N-acetyl-L-cysteine (NAC) |
Sigma | Cat#: A8199 | |
| Chemical compound, drug | Forskolin | Sigma | Cat#: F6886 | |
| Chemical compound, drug | Puromycin | Thermo Fisher Scientific | Cat#: A11138-03 | |
| Chemical compound, drug | Puromycin | Sigma | Cat#: P8833 | |
| Chemical compound, drug | Paraformaldehyde | Vetec | Cat#: V900894-100G | |
| Chemical compound, drug | Triton X-100 | Sigma | Cat#: V900502 | |
| Software, algorithm | GraphPad Prism 7.0 | GraphPad |
https://www.graphpad.com, RRID: SCR_002798 |
|
| Software, algorithm | STAR v2.5 | Dobin et al., 2013 |
https://github.com/alexdobin/STAR/ RRID:SCR_004463 |
|
| Software, algorithm | HTSeq | Anders et al., 2015 | https://pypi.org/project/HTSeq/ | |
| Software, algorithm | ImageJ (Fiji) | Schindelin et al., 2012 | http://fiji.sc, RRID:SCR_002285 | |
| Software, algorithm | Matlab | Matlab | https://ww2.mathworks.cn | |
| Other | TRIzol Reagent | Life | Cat#: 15596018 | |
| Other | PrimeScript RT Master Mix | Takara | Cat#: RR036B | |
| Other | 2× ChamQ Universal SYBR qPCR Master Mix | Vazyme | Cat#: Q711-02 | |
| Other | DMEM, high glucose | Gibco | Cat#: 11965–092 | |
| Other | Dulbecco’s Modified Eagle’s Medium, 10×, low glucose | Sigma | Cat#: D2429 | |
| Other | DMEM, high glucose | Hyclone | Cat#: SH30022.01 | |
| Other | Fetal Bovine Serum (FBS) | Gibco | Cat#: 10270–106 | |
| Other | Dulbecco’s Phosphate-Buffered Saline, 1× without calcium and magnesium (DPBS) | Corning | Cat#: 21–031-CVR | |
| Other | Poly-D-lysine, Cultrex | Trevigen | Cat#: 3439-100-01 | |
| Other | Laminin (mouse), Culrex | Trevigen | Cat#: 3400-010-01 | |
| Other | DMEM/F-12, GlutaMAX | Gibco | Cat#: 10565–018 | |
| Other | Neurobasal Medium, minus phenol red | Gibco | Cat#: 12348–017 | |
| Other | Penicillin-Streptomycin | Life | Cat#: 15140–122 | |
| Other | B27 serum-free supplement, 50× |
Life | Cat#: 17504044 | |
| Other | N-2 Supplement, 100× | Gibco | Cat#: 17502–048 | |
| Other | OCT Compound and Cryomolds, Tissue-Tek | SAKURA | Cat#: 4583 | |
| Other | ChemiBLOCKER | Millipore | Cat#: 2170 | |
| Other | CTB (Cholera Toxin Subunit B) conjugated by Alexa Fluor 555 |
Invitrogen | Cat#: C34776 | |
| Other | VECTASHIELD Antifade Mounting Medium with DAPI |
Vector Laboratory | Cat#: H-1200 | |
| Other | Mounting Medium, antifading (with DAPI) |
Solarbio | Cat#: S2110 | |
| Other | Normal Goat Serum | Novus | Cat#: NBP2-23475 |
Animals and generation of the Ythdf2 cKO mice
Ythdf2fl/fl mice were reported previously (Yu et al., 2021b). Six3-cre (Furuta et al., 2000), Thy1-GFP (Feng et al., 2000), and Rosa26-eYFP (Srinivas et al., 2001) mice were from Jackson Laboratory. For timed pregnancy, embryos were identified as E0.5 when a copulatory plug was observed. Genotyping primers are as following: the first Ythdf2-loxP site, 5’-GCTTGTAGTTATGTTGTGTACCAC-3’ and 5’-GCAGCTCTGACTATTCTAAAACCTCC-3’; the second Ythdf2-loxP site, 5’-CTCATAACATCCATAGCCACAGG-3’, and 5’-CCAAGAGATAGCTTTCCTAATG-3’.
Six3-cre site, 5’-CCTTCCTCCCTCTCTATGTG-3’ and 5’-GAACGAACCTGGTCGAAATC-3’.
Rosa26-eYFP wild type site, 5’-CTGGCTTCTGAGGACCG-3’ and 5’-CAGGACAACGCCCACACA-3’; the mutant site, 5’-AGGGCGAGGAGCTGTTCA-3’ and 5’-TGAAGTCGATGCCCTTCAG-3’. All experiments using mice were carried out following the animal protocols approved by the Laboratory Animal Welfare and Ethics Committee of Southern University of Science and Technology.
Retinal neuronal culture
Retinal neurons were dissociated from E14.5 to 15.5 mouse embryos by papain in DPBS (1× Dulbecco’s phosphate-buffered saline [PBS], Corning, NY) following the previously described methods (Kechad et al., 2012), and neuronal suspension was plated on acid-washed glass coverslips pre-coated with poly-D-lysine (Trevigen, 100 μg/ml) for 1 hr and laminin (Trevigen, 5 μg/ml) overnight at 37°C. Culture medium was made up of half DMEM/F12 medium (Gibco) and half neurobasal medium (Gibco), supplemented with B27 supplement (Life, 0.5×), penicillin-streptomycin (Life, 1×), N-2 supplement (Gibco, 0.5×), N-acetyl-L-cysteine (Sigma, NAC 0.6 mg/ml), cpt-cAMP (Sigma, 100 μM), forskolin (Sigma, 10 μM), and insulin (Sigma, 25 μg/ml). EGF (PeproTech, 50 ng/ml), BDNF (PeproTech, 50 ng/ml), NT-3 (PeproTech, 25 ng/ml), and FGF-basic (PeproTech, 10 ng/ml) were freshly added before using.
KD using lentiviral shRNA, siRNA or AAV shRNA, and overexpression using AAV system
Lentiviral KD plasmids encoding shRNA (shCtrl: 5’-GCATCAAGGTGAACTTCAAGA-3’; shYthdf2: 5’-GGACGTTCCCAATAGCCAACT-3’; shYthdf1: 5’- GGACATTGGTACTTGGGATAA-3’; shYthdf3: 5’- GGATTTGGCAATGATACTTTG-3’; shMettl14#6: 5’-GCTGGACCTGGGATGATATTA-3’; shMettl14#7: 5’-CCCAGCTTGTACTTTGCTTTA-3’) were generated from pLKO.1-TRC and lentivirus preparation process was described previously (Yu et al., 2018). All siRNAs were chosen from previous studies and the target sequences of siRNA are as following: siCtrl (RNAi negative control): 5’- UUCUCCGAACGUGUCACGUTT-3’ (Yu et al., 2018); siKalrn7: 5’- AGUACAAUCCUGGCCAUGUTT-3’ (Xie et al., 2007); siKalrn9: 5’-ACUGGACUGGACUUCUAUUTT-3’ (Yan et al., 2015); siKalrn12: 5’-CGAUGAGCUGAUGGAAGAATT-3’ (Yan et al., 2015); siStrn: 5’-GGUGAAGAUCGAGAUACAATT-3’ (Breitman et al., 2008); siUbr4: 5’-AAUGAUGAGCAGUCAUCUATT-3’ (Shim et al., 2008). AAV KD plasmids encoding shRNA (shCtrl: 5’-TTCTCCGAACGTGTCACGTAA-3’; shKalrn12: 5’-TGATGAGCTGATGGAAGAA-3’; shUbr4: 5’-AATGATGAGCAGTCATCTC-3’) were generated using pHBAAV-U6-MCS-CMV-EGFP and packaged in serotype-9 by Hanbio (1.5 × 1012 genomic copies per ml). AAV overexpression plasmids of Hspa12a (NM_175199.3; PCR primer for mouse Hspa12a: 5’-ATGGCGGACAAGGAAGCTGG-3’ and 5’-GTAATTTAAGAAGTCGATCCCC-3’) and Islr2 (NM_001161541.1; PCR Primer for mouse Islr2: 5’-ATGGGGCCCTTTGGAGC-3’ and 5’-GCCCGCTGTCTGCCTGTAG-3’) were generated from pHBAAV-CMV-MCS-3flag-T2A-ZsGreen and packaged serotype-9 by Hanbio (1.2 × 1012 genomic copies per ml).
GeneSilencer Transfection Reagent (Genlantis) was used in siRNA transfection following the manufacturer’s protocols. Culture medium was changed after 1 day of lentiviral shRNA infection or siRNA transfection. For lentiviral shRNA assay, puromycin (Thermo or Sigma, 1 μg/ml) was added after 2 days of infection. Immunofluorescence, RNA, or protein preparation was performed after shRNA or siRNA worked for 3 days. For AAV intravitreal injection, P0-P1 mouse pups were anesthetized in ice and then eyes were pierced at the edge of corneal by 30G × 1/2 needle (BD, 305106) under stereomicroscope. Then 1 μl AAV was intravitreally injected with 10 µl Syringe (Hamilton, 80330) following the pinhole. P15 or adult mice were anesthetized with 2.5% Avertin and then eyes were pierced at the side of corneal and the outer segment of sclera by 30G × 1/2 needle successively. Two μl AAV was intravitreally injected with 10 µl Syringe following the pinhole on the sclera. All subsequent experiments such as AOH operation and immunostaining were carried out after at least 3 weeks (10 days for ZsGreen/CART labeling of ooDSGCs in Ythdf2 cKO and control mice in Figure 3A).
RT-qPCR
Total RNA was extracted from cells or tissues with TRIzol Reagent (Life) and then used for reverse transcription by PrimeScript RT Master Mix (TaKaRa). Synthesized cDNA was used for qPCR by 2× ChamQ Universal SYBR qPCR Master Mix (Vazyme) on StepOnePlus Real-Time PCR System (ABI) or BioRad CFX96 Touch Real-Time PCR system. Primers used for qPCR are as following: mouse Gapdh: 5’-TTGTCAGCAATGCATCCTGCACCACC-3’ and 5’-CTGAGTGGCAGTGATGGCATGGAC-3’ (Mains et al., 2011); mouse Kalrn7: 5’- GATACCATATCCATTGCCTCCAGGACC-3’ and 5’-CCAGGCTGCGCGCTAAACGTAAG-3’ (Mains et al., 2011); mouse Kalrn9: 5’- GCCCCTCGCCAAAGCCACAGC-3’ and 5’-CCAGTGAGTCCCGTGGTGGGC-3’ (Mains et al., 2011); mouse Kalrn12: 5’- CAGCAGCCACGTGCCTGCAGC-3’ and 5’-TCTTGACATTGGGAATGGGCCGCAC-3’ (Mains et al., 2011); mouse Strn: 5’-TGAAGCCTGGAATGTGGACC-3’ and 5’-CTATTGGGCCTCTTCACCCC-3’; mouse Ubr4: 5’- TGAGTGAGGACAAGGGCAAC-3’ and 5’-GGGTTGGATCGAACGAAGGT-3’; mouse Ythdf2: 5’-GAGCAGAGACCAAAAGGTCAAG-3’and 5’-CTGTGGGCTCAAGTAAGGTTC-3’; 18 s: 5’-GCTTAATTTGACTCAACACGGGA-3’ and 5’-AGCTATCAATCTGTCAATCCTGTC-3’ (Wang et al., 2018); mouse Hspa12a: 5’-GGGTTTGCACAGGCTAAGGA-3’ and 5’-TCTGATGGACGGTCAGGTCT-3’; mouse Islr2: 5’-GAAGCTCCCTTAGACTGTCACC-3’ and 5’-CCCCATCGTGACTCCTGCTG-3’.
Immunofluorescence and immunostaining
For tissue sections, mouse embryonic eyes were fixed with 4% PFA (Sigma) in 0.1 M phosphate buffer (PB) for 30–45 min at room temperature (RT); eyes of mouse pups (<P10) were pre-fixed briefly and then eyecups were dissected and fixed for 45 min-1 hr at RT; for P20-30 or adult mice, eyecups were dissected after myocardial perfusion with 0.9% NaCl, followed by fixation for 1 hr. After PBS (3 × 5 min) washing, tissues were dehydrated with 30% sucrose in 0.1 M PB overnight at 4°C, then embedded with OCT (SAKURA) and cryosectioned at 12 μm (20 μm for Thy1-GFP section analysis) with Leica CM1950 Cryostat. Tissue sections were permeabilized and blocked with 10% ChemiBLOCKER (Millipore) and 0.5% Triton X-100 (Sigma) in PBS (PBST) for 1 hr at RT and incubated in PBST overnight at 4°C with following primary antibodies: chicken anti-GFP (1:1000, Abcam ab13970), chicken anti-MAP2 (1:10,000, Abcam ab5392), goat anti-VAChT (1:1000, Millipore ABN100), guinea pig anti-RBPMS (1:1000, PhosphoSolutions 1832-RBPMS), mouse anti-AP2α (1:1000, DSHB 3B5), mouse anti-Bassoon (1:2500, Enzo Life Sciences ADI-VAM-PS003), mouse anti-Brn3a (1:300, Millipore MAB1585), mouse anti-Calbindin-D-28K (1:200, Sigma C9848), mouse anti-PKCα (1:500, Santa Cruz sc-8393), rabbit anti-Strn (Striatin) (1:500, BD Biosciences 610838), rabbit anti-CART (1:2000, Phoenix Pharmaceuticals H-003–62), rabbit anti-m6A (1:200, Synaptic Systems 202003), rabbit anti-melanopsin (1:1000, Thermo PA1-780), rabbit anti-PKCα (1:1000, Cell Signaling CST-2056), rabbit anti-PSD95 (1:1000, Abcam ab18258), rabbit anti-Recoverin (1:1000, Millipore AB5585), rabbit anti-YTHDF2 (1:1000, Proteintech 24744–1-AP), rabbit anti-YTHDF1 (1:1000, Proteintech 17479–1-AP), rabbit anti-YTHDF3 (1:1000, Abcam ab103328), rabbit anti-Ubr4 (1:300, Abcam ab86738), sheep anti-Chx10 (1:1000, Exalpha X1179P), chicken anti-GFAP (1:500, Millipore AB5541), goat anti-Lhx2 (1:200, Santa Cruz Biotechnology sc-19344), rabbit anti-Lhx2 (1:500, Abcam ab184337). After three times of PBS washing, sections were incubated in PBST for 1 hr at RT with secondary antibodies: Alexa 488 donkey anti-chicken (1:500, Jackson 703-545-155), Alexa 488 donkey anti-guinea pig (1:500, Jackson 706-545-148), Alexa 488 donkey anti-mouse (1:500, Thermo A21202), Alexa 488 donkey anti-rabbit (1:500, Thermo A21206), Alexa 555 donkey anti-goat (1:1000, Thermo A21432), Alexa 555 donkey anti-mouse (1:1000, Thermo A31570), Alexa 555 donkey anti-rabbit (1:1000, Thermo A31572), Alexa 555 donkey anti-sheep (1:1000, Thermo A21436), Alexa 555 goat anti-chicken (1:1000, Thermo A21437), or Alexa 647 donkey anti-mouse (1:200, Thermo A31571) and then mounted with the VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratory).
For cultured neurons, after twice of PBS washing, cells were fixed for 15 min with 4% PFA in 0.1 M PB at RT, then washed with PBS three times and blocked in PBST for 20 min at RT. Antibody incubation conditions are the same as tissue sections.
For wholemount immunostaining of retina, eyes were dissected after myocardial perfusion with 0.9% NaCl. Then retinas were separated from sclera and fixed with 4% PFA in 0.1 M PB for 1 hr at RT. Then retinas were blocked with 5% normal goat serum (Novus), 0.4% Triton X-100 in PBS overnight at 4°C. Primary antibodies such as chicken anti-GFP (1:1000, Abcam ab13970), mouse anti-Brn3a (1:300, Millipore MAB1585), mouse anti-SMI-32 (1:200, BioLegend 801701), or rabbit anti-Melanopsin (1:1000, Thermo PA1-780), rabbit anti-CART (1:2000, Phoenix Pharmaceuticals H-003–62) were diluted in 5% normal goat serum, 0.4% Triton X-100 in PBS and incubated overnight at 4°C. Then retinas were incubated with Alexa 488 donkey anti-chicken (1:500, Jackson 703-545-155), Alexa 488 donkey anti-mouse (1:500, Thermo A21202), Alexa 555 donkey anti-mouse (1:1000, Thermo A-31570) and Alexa 555 donkey anti-rabbit (1:1000, Thermo A31572) secondary antibodies in 5% normal goat serum (Novus), 0.4% Triton X-100 in PBS and finally mounted with the VECTASHIELD Antifade Mounting Medium with DAPI.
All images were captured on Nikon A1R confocal microscope or Zeiss LSM 800 confocal microscope with identical settings for each group in the same experiment. A region of interest, length or thickness in immunofluorescence experiments were obtained with ImageJ. The number of neurons in specific area was counted blindly and manually. To quantify RGC dendrite lamination in IPL with Thy1-GFP, z-stack and maximum projection were performed during the analysis. GFP intensity values across IPL depth were measured by ImageJ/Analyze/Plot Profile function (Liu et al., 2018). To quantify the numbers of Bassoon+/PSD-95+ excitatory synapses in IPL, the colocalization puncta was measured by ImageJ/Analyze/Puncta Analyzer as described previously (Ippolito and Eroglu, 2010).
Sholl analysis
For confocal images of cultured RGCs, MAP2 signals in original format were analyzed with simple neurite tracer and then quantified with Sholl analysis (5 μm per distance from soma center) which was a widely used method in neurobiology to quantify the complexity of dendritic arbors using ImageJ (Schindelin et al., 2012; Binley et al., 2014). Retina wholemount data were captured in z-stack mode (0.5–1 μm per slide) with confocal microscopes. ZsGreen, eGFP, and SMI-32 signals were directly analyzed with simple neurite tracer and then z projection of all tracers was quantified with Sholl analysis (10 μm per distance from soma center), while melanopsin signals were maximum-projected before tracing.
OMR assay
Ythdf2 cKO and control mice aged about 6 weeks were dark-adapted overnight before experiment and used in the OMR assay following the previously reported protocols (Douglas et al., 2005; Sergeeva et al., 2018). Using the Matlab program, 0.2 c/deg (15 s per direction of rotation) was first used for mice to adapt this experiment, and 0.3, 0.35, 0.4, 0.43, 0.45, 0.47, 0.5 and 0.55 c/deg (30 s per direction of rotation) were used in the following recordings. Mouse behaviors were analyzed in real time during the experiment and re-checked with video recordings. Finally, data for each mouse were determined by the minimal spatial frequency between left and right OMR.
CTB labeling of optic nerve
To label RGC axon terminals in mouse brain, RGC axons were anterogradely labeled by CTB conjugated with Alexa Fluor 555 (Invitrogen, C34776) through intravitreal injection 48 hr before sacrifice. After PFA perfusion, the brains were fixed with 4% PFA in 0.1 M PB overnight, dehydrated with 15% sucrose and 30% sucrose in 0.1 M PB overnight at 4°C sequentially, embedded with OCT for coronal section, and cryosectioned at 12 μm with Leica CM1950 Cryostat. After PBS washing, the sections were mounted with VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratory). The images were captured on Tissue Genostics with identical settings for each group in the same experiment with the TissueFAXS 7.0 software.
RIP and sequencing
For RIP experiment, we used the EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manual with minor modifications. Briefly, 1 × 107 retinal neurons were subjected to each 100 μl lysis buffer. The amount of YTHDF2 antibody (Proteintech, 24744–1-AP) and control IgG used for immunoprecipitation is 5 μg, respectively. RIP experimental steps, RNA sample preparation and sequencing, and sequence data analysis followed the procedures reported previously (Yu et al., 2021a).
MS analysis
E15.5 retinal neurons were cultured and infected with lentiviral shYthdf2 or shCtrl. Sample collection and lysis, protein and peptide preparation were performed following procedures reported previously (Yu et al., 2021b). Proteins with fold changes greater than 1.3 and p values less than 0.05 were considered to be regulated by YTHDF2 KD with statistical significance.
Anti-m6A immunoprecipitation
Total retinal RNA was extracted from P0 WT mouse pups. Immunoprecipitation of m6A-modified transcripts was carried out with Magna MeRIP m6A Kit (Merck-Millipore, 17–10499) following the manual. m6A antibody (Synaptic Systems, 202003) and corresponding control IgG were used in this experiment. The RNA samples pulled down from the experiment were used for RT-qPCR.
AOH model
Mice were anesthetized with 5% chloral hydrate in normal saline (10 μl/g) based on body weight and the Compound Tropicamide Eye Drops were used to scatter pupil. The anterior chamber was penetrated using the 32G × 1/2’’ needles (TSK) and filled with the BBS Sterile Irrigating Solution (Alcon) which was hung at a high position to provide proper pressure. Intraocular pressure was measured with the Tonolab tonometer (icare) for every 10 min and maintained at 85–90 mmHg for 1 hr. Levofloxacin hydrochloride was used after the operation and mice were revived in a 37°C environment. Retinas were analyzed for gene expression of YTHDF2 1 day after AOH, gene expression of Hspa12a and Islr2 3 days after AOH, dendritic complexity and RGC number 3–7 days after AOH.
Statistical analysis
All experiments were conducted at a minimum of three independent biological replicates (two biological replicates for the RIP assay) or three mice/pups for each genotype/condition in the lab. Data are mean ± SEM. Statistical analysis was preformed using GraphPad Prism 7.0. When comparing the means of two groups, an unpaired or paired t test was performed on the basis of experimental design. The settings for all box and whisker plots are: 25th-75th percentiles (boxes), minimum and maximum (whiskers), and medians (horizontal lines). A p value less than 0.05 was considered as statistically significant: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Acknowledgements
We thank Ke Wang and Kwok-Fai So (Jinan University) for help on the OMR assay. We thank Mengqing Xiang and Suo Qiu (Zhongshan Ophthalmic Center, Sun Yat-sen University) for help on AAV experiments. We thank other members of Ji laboratory for technical support, helpful discussions, and comments on the manuscript. This work was supported by National Natural Science Foundation of China (31871038 and 32170955 to S-JJ; 31922027 and 32170958 to BP), Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2021SHIBS0002, 2019SHIBS0002), High-Level University Construction Fund for Department of Biology (internal grant no. G02226301), Science and Technology Innovation Commission of Shenzhen Municipal Government (ZDSYS20200811144002008), Program of Shanghai Subject Chief Scientist (21XD1420400), and the Innovative Research Team of High-Level Local University in Shanghai (BP).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Bo Peng, Email: peng@fudan.edu.cn.
Sheng-Jian Ji, Email: jisj@sustech.edu.cn.
Carol A Mason, Columbia University, United States.
Catherine Dulac, Harvard University, United States.
Funding Information
This paper was supported by the following grants:
National Natural Science Foundation of China 31871038 to Sheng-Jian Ji.
National Natural Science Foundation of China 31922027 to Bo Peng.
Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions 2021SHIBS0002 to Sheng-Jian Ji.
High-Level University Construction Fund for Department of Biology G02226301 to Sheng-Jian Ji.
Science and Technology Innovation Commission of Shenzhen Municipality ZDSYS20200811144002008 to Sheng-Jian Ji.
Program of Shanghai Subject Chief Scientist 21XD1420400 to Bo Peng.
Innovative Research Team of High-Level Local University in Shanghai to Bo Peng.
Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions 2019SHIBS0002 to Sheng-Jian Ji.
National Natural Science Foundation of China 32170955 to Sheng-Jian Ji.
National Natural Science Foundation of China 32170958 to Bo Peng.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing.
Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization.
Formal analysis, Investigation, Methodology.
Formal analysis, Investigation, Methodology.
Formal analysis, Investigation, Methodology.
Formal analysis, Investigation, Methodology.
Investigation, Methodology.
Investigation, Methodology.
Investigation.
Investigation.
Investigation, Resources.
Conceptualization, Methodology, Resources, Software, Supervision, Writing – review and editing.
Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review and editing.
Ethics
All experiments using mice were carried out following the animal protocols approved by the Laboratory Animal Welfare and Ethics Committee of Southern University of Science and Technology (approval numbers: SUSTC-JY2017004, SUSTC-JY2019081).
Additional files
Data availability
The RIP-seq data have been deposited to the Gene Expression Omnibus (GEO) with accession number GSE145390. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017775.
The following datasets were generated:
Niu F, Yang L, Ji S. 2022. Anti YTHDF2 RIP-seq to identify YTHDF2 target mRNAs in P0 mouse retinas. NCBI Gene Expression Omnibus. GSE145390
Niu F, Ji S. 2022. Proteome analysis using mass spectrometry (MS) in acute shYthdf2-mediated knockdown of cultured RGCs. PRIDE. PXD017775
References
- Agostinone J, Di Polo A. Retinal ganglion cell dendrite pathology and synapse loss: Implications for glaucoma. Progress in Brain Research. 2015;220:199–216. doi: 10.1016/bs.pbr.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Agudo M, Pérez-Marín MC, Lönngren U, Sobrado P, Conesa A, Cánovas I, Salinas-Navarro M, Miralles-Imperial J, Hallböök F, Vidal-Sanz M. Time course profiling of the retinal transcriptome after optic nerve transection and optic nerve crush. Molecular Vision. 2008;14:1050–1063. [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, Euler T. The functional diversity of retinal ganglion cells in the mouse. Nature. 2016;529:345–350. doi: 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist M, Gaillard S, Castets F. The striatin family: a new signaling platform in dendritic spines. Journal of Physiology, Paris. 2006;99:146–153. doi: 10.1016/j.jphysparis.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Binley KE, Ng WS, Tribble JR, Song B, Morgan JE. Sholl analysis: a quantitative comparison of semi-automated methods. Journal of Neuroscience Methods. 2014;225:65–70. doi: 10.1016/j.jneumeth.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Brechbiel JL, Gavis ER. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Current Biology. 2008;18:745–750. doi: 10.1016/j.cub.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman M, Zilberberg A, Caspi M, Rosin-Arbesfeld R. The armadillo repeat domain of the APC tumor suppressor protein interacts with Striatin family members. Biochimica et Biophysica Acta. 2008;1783:1792–1802. doi: 10.1016/j.bbamcr.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Cahill ME, Xie Z, Day M, Photowala H, Barbolina MV, Miller CA, Weiss C, Radulovic J, Sweatt JD, Disterhoft JF, Surmeier DJ, Penzes P. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. PNAS. 2009;106:13058–13063. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry TJ, Wang S, Bormuth I, Schwab M, Olson J, Cepko CL. NeuroD factors regulate cell fate and neurite stratification in the developing retina. The Journal of Neuroscience. 2011;31:7365–7379. doi: 10.1523/JNEUROSCI.2555-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ZH, Chin WH, Chen Y, Ng YP, Ip NY. Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLOS Biology. 2007;5:e63. doi: 10.1371/journal.pbio.0050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nature Neuroscience. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Briz CG, Esteban-Ortega GM, Nieto M. Cux1 and Cux2 selectively target basal and apical dendritic compartments of layer II-III cortical neurons. Developmental Neurobiology. 2015;75:163–172. doi: 10.1002/dneu.22215. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dong X, Shen K, Bülow HE. Intrinsic and extrinsic mechanisms of dendritic morphogenesis. Annual Review of Physiology. 2015;77:271–300. doi: 10.1146/annurev-physiol-021014-071746. [DOI] [PubMed] [Google Scholar]
- Douglas RM, Alam NM, Silver BD, McGill TJ, Tschetter WW, Prusky GT. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Visual Neuroscience. 2005;22:677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis (New York, N.Y. 2000;26:130–132. doi: 10.1002/(SICI)1526-968X(200002)26:2<130::AID-GENE9>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science (New York, N.Y.) 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Nicoll RA. Kalirin and Trio proteins serve critical roles in excitatory synaptic transmission and LTP. PNAS. 2016;113:2264–2269. doi: 10.1073/pnas.1600179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito DM, Eroglu C. Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. Journal of Visualized Experiments. 2010:2270. doi: 10.3791/2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs TC. Differential gene expression in glaucoma. Cold Spring Harbor Perspectives in Medicine. 2014;4:a020636. doi: 10.1101/cshperspect.a020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nature Reviews. Neuroscience. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, Sanes JR. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. The Journal of Neuroscience. 2011a;31:7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, Chu MW. Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nature Neuroscience. 2011b;14:965–972. doi: 10.1038/nn.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechad A, Jolicoeur C, Tufford A, Mattar P, Chow RWY, Harris WA, Cayouette M. Numb is required for the production of terminal asymmetric cell divisions in the developing mouse retina. The Journal of Neuroscience. 2012;32:17197–17210. doi: 10.1523/JNEUROSCI.4127-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, Sanes JR. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524:466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoue V, Langford M, White A, Sempert K, Fogg L, Cooper HM. The Wnt receptor Ryk is a negative regulator of mammalian dendrite morphogenesis. Scientific Reports. 2017;7:5965. doi: 10.1038/s41598-017-06140-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda F, Paratcha G. Mechanisms regulating dendritic arbor patterning. Cellular and Molecular Life Sciences. 2017;74:4511–4537. doi: 10.1007/s00018-017-2588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Sanes JR, Kay JN. Development of dendritic form and function. Annual Review of Cell and Developmental Biology. 2015;31:741–777. doi: 10.1146/annurev-cellbio-100913-013020. [DOI] [PubMed] [Google Scholar]
- Li D, Musante V, Zhou W, Picciotto MR, Nairn AC. Striatin-1 is a B subunit of protein phosphatase PP2A that regulates dendritic arborization and spine development in striatal neurons. The Journal of Biological Chemistry. 2018;293:11179–11194. doi: 10.1074/jbc.RA117.001519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–485. doi: 10.1016/j.neuron.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lin L, Lo LH-Y, Lyu Q, Lai K-O. Determination of dendritic spine morphology by the striatin scaffold protein STRN4 through interaction with the phosphatase PP2A. The Journal of Biological Chemistry. 2017;292:9451–9464. doi: 10.1074/jbc.M116.772442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sanes JR. Cellular and Molecular Analysis of Dendritic Morphogenesis in a Retinal Cell Type That Senses Color Contrast and Ventral Motion. The Journal of Neuroscience. 2017;37:12247–12262. doi: 10.1523/JNEUROSCI.2098-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Reggiani JDS, Laboulaye MA, Pandey S, Chen B, Rubenstein JLR, Krishnaswamy A, Sanes JR. Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nature Neuroscience. 2018;21:659–670. doi: 10.1038/s41593-018-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nature Reviews. Neuroscience. 2020;21:36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- Lu J, Luo C, Bali KK, Xie RG, Mains RE, Eipper BA, Kuner R. A role for Kalirin-7 in nociceptive sensitization via activity-dependent modulation of spinal synapses. Nature Communications. 2015;6:6820. doi: 10.1038/ncomms7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains RE, Kiraly DD, Eipper-Mains JE, Ma XM, Eipper BA. Kalrn promoter usage and isoform expression respond to chronic cocaine exposure. BMC Neuroscience. 2011;12:20. doi: 10.1186/1471-2202-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Kong Q, Li R, Zhang X, Gui Y, Li Y, Li C, Zhao Y, Liu L, Ding Z. Heat shock protein A12A encodes a novel prosurvival pathway during ischaemic stroke. Biochimica et Biophysica Acta. Molecular Basis of Disease. 2018;1864:1862–1872. doi: 10.1016/j.bbadis.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min X, Zhang X, Li Y, Cao X, Cheng H, Li Y, Li C, Kong Q, Mao Q, Peng P, Ni Y, Li J, Duan Y, Liu L, Ding Z. HSPA12A unstabilizes CD147 to inhibit lactate export and migration in human renal cell carcinoma. Theranostics. 2020;10:8573–8590. doi: 10.7150/thno.44321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JE, Datta AV, Erichsen JT, Albon J, Boulton ME. Retinal ganglion cell remodelling in experimental glaucoma. Advances in Experimental Medicine and Biology. 2006;572:397–402. doi: 10.1007/0-387-32442-9_56. [DOI] [PubMed] [Google Scholar]
- Nachtergaele S, He C. Chemical Modifications in the Life of an mRNA Transcript. Annual Review of Genetics. 2018;52:349–372. doi: 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons K, Nakatani Y, Nguyen MD. p600/UBR4 in the central nervous system. Cellular and Molecular Life Sciences. 2015;72:1149–1160. doi: 10.1007/s00018-014-1788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29:229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Pongrac JL, Middleton FA, Peng L, Lewis DA, Levitt P, Mirnics K. Heat shock protein 12A shows reduced expression in the prefrontal cortex of subjects with schizophrenia. Biological Psychiatry. 2004;56:943–950. doi: 10.1016/j.biopsych.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Prigge CL, Kay JN. Dendrite morphogenesis from birth to adulthood. Current Opinion in Neurobiology. 2018;53:139–145. doi: 10.1016/j.conb.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Investigative Ophthalmology & Visual Science. 2004;45:4611–4616. doi: 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]
- Ramos B, Gaudillière B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. PNAS. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Sun LO, Brady CM, Alexandropoulos K, Seo S, Kurokawa M, Kolodkin AL. Cas adaptor proteins organize the retinal ganglion cell layer downstream of integrin signaling. Neuron. 2014;81:779–786. doi: 10.1016/j.neuron.2014.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nature Neuroscience. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- Russell TA, Blizinsky KD, Cobia DJ, Cahill ME, Xie Z, Sweet RA, Duan J, Gejman PV, Wang L, Csernansky JG, Penzes P. A sequence variant in human KALRN impairs protein function and coincides with reduced cortical thickness. Nature Communications. 2014;5:4858. doi: 10.1038/ncomms5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annual Review of Neuroscience. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Sapkota D, Chintala H, Wu F, Fliesler SJ, Hu Z, Mu X. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. PNAS. 2014;111:E4086–E4095. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva EG, Espinosa-Garcia C, Atif F, Pardue MT, Stein DG. Neurosteroid allopregnanolone reduces ipsilateral visual cortex potentiation following unilateral optic nerve injury. Experimental Neurology. 2018;306:138–148. doi: 10.1016/j.expneurol.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Yuan X, Chang K, Cho KS, Xie XS, Chen DF, Luo G. Optimization of Optomotor Response-based Visual Function Assessment in Mice. Scientific Reports. 2018;8:9708. doi: 10.1038/s41598-018-27329-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SY, Wang J, Asada N, Neumayer G, Tran HC, Ishiguro K, Sanada K, Nakatani Y, Nguyen MD. Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. The Journal of Neuroscience. 2008;28:3604–3614. doi: 10.1523/JNEUROSCI.5278-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Kengaku M, Hirano T, Takeichi M, Uemura T. Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Developmental Cell. 2004;7:205–216. doi: 10.1016/j.devcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kawaguchi S, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, Hirano T, Uemura T. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nature Neuroscience. 2007;10:963–969. doi: 10.1038/nn1933. [DOI] [PubMed] [Google Scholar]
- Shou T, Liu J, Wang W, Zhou Y, Zhao K. Differential dendritic shrinkage of alpha and beta retinal ganglion cells in cats with chronic glaucoma. Investigative Ophthalmology & Visual Science. 2003;44:3005–3010. doi: 10.1167/iovs.02-0620. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology. 2001;1:4. doi: 10.1186/1471-213x-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, Levin JZ, Lin D, Wang C, Lieber CM, Regev A, He Z, Sanes JR. Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron. 2019;104:1039–1055. doi: 10.1016/j.neuron.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umino Y, Solessio E, Barlow RB. Speed, spatial, and temporal tuning of rod and cone vision in mouse. The Journal of Neuroscience. 2008;28:189–198. doi: 10.1523/JNEUROSCI.3551-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F, Aparicio SAJ, Abel T, Kiebler MA. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. PNAS. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, Zhao JC. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nature Neuroscience. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AJ, Kaufman PL, Hubbard WC. Morphology of single ganglion cells in the glaucomatous primate retina. Investigative Ophthalmology & Visual Science. 1998;39:2304–2320. [PubMed] [Google Scholar]
- Xie Z, Srivastava DP, Photowala H, Kai L, Cahill ME, Woolfrey KM, Shum CY, Surmeier DJ, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Cahill ME, Penzes P. Kalirin loss results in cortical morphological alterations. Molecular and Cellular Neurosciences. 2010;43:81–89. doi: 10.1016/j.mcn.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Eipper BA, Mains RE. Kalirin-9 and Kalirin-12 Play Essential Roles in Dendritic Outgrowth and Branching. Cerebral Cortex (New York, N.Y. 2015;25:3487–3501. doi: 10.1093/cercor/bhu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Current Biology. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, Park J, Ji SJ. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Research. 2018;46:1412–1423. doi: 10.1093/nar/gkx1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, She Y, Ji SJ. m6A Modification in Mammalian Nervous System Development, Functions, Disorders, and Injuries. Frontiers in Cell and Developmental Biology. 2021a;9:679662. doi: 10.3389/fcell.2021.679662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, She Y, Yang L, Zhuang M, Han P, Liu J, Lin X, Wang N, Chen M, Jiang C, Zhang Y, Yuan Y, Ji SJ. The m6 A Readers YTHDF1 and YTHDF2 Synergistically Control Cerebellar Parallel Fiber Growth by Regulating Local Translation of the Key Wnt5a Signaling Components in Axons. Advanced Science (Weinheim, Baden-Wurttemberg, Germany) 2021b;8:e2101329. doi: 10.1002/advs.202101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Qyang Y, Kelsoe JR, Masliah E, Geyer MA. Impaired postnatal development of hippocampal dentate gyrus in Sp4 null mutant mice. Genes, Brain, and Behavior. 2007;6:269–276. doi: 10.1111/j.1601-183X.2006.00256.x. [DOI] [PubMed] [Google Scholar]