Abstract
Purpose
Ferroptosis is an iron-dependent and reactive oxygen species (ROS)-reliant form of cell death, exhibiting cellular, molecular, and gene-level characteristics distinct from those of necrosis, autophagy, apoptosis, and pyroptosis. Solute carrier family 7 member 11 (SLC7A11), which encodes a cystine/glutamate antiporter transmembrane protein, inhibits ferroptosis by importing cystine and promoting glutathione (GSH) biosynthesis and was found to be overexpressed in multiple human cancers. However, the specific role and underlying mechanism of SLC7A11 in cancers remains poorly characterized. This research aimed to identify the relationship between SLC7A11 expression and tumor microenvironment and visualize its prognostic value in pan-cancer.
Patients and Methods
Transcriptomic data for 6313 tumors and normal samples across 20 cancer types were acquired from The Cancer Genome Atlas (TCGA) database. Besides, we presented a novel bioinformatics pipeline that uncovered the impacts of SLC7A11 on cancer prognosis, tumor mutational burden (TMB), immune cell infiltration in tumor microenvironment, and drug responses. The Genotype-Tissue Expression (GTEx), cBioportal, TCGA and Connectivity Map (CMap) databases were used to explore the expression, genetic alterations, immune microenvironment, and drug responses of SLC7A11. A series of deconvolution algorithms, including EPIC, CIBERSORT and GSEA, were utilized for multidimensional analyses of the cancer transcriptomic data.
Results
SLC7A11 was found to be highly expressed in the 20 types of cancer, especially in solid tumors. Survival analysis uncovered that most cancer patients with up-regulated expression of SLC7A11 showed poor prognosis, suggesting that SLC7A11 is a potential oncogene in most cancer types. Furthermore, the expression level of SLC7A11 was confirmed to be associated with immune cell infiltration in tumor microenvironment, TMB, and drug responses. Gene set enrichment analysis (GESA) revealed that dysregulation of SLC7A11 was associated with metabolic and immunity-related signaling pathways in the cancers.
Conclusion
The comprehensive pan-cancer analyses identified SLC7A11 as an attractive biomarker for immune infiltration and poor prognosis in cancers, shedding new light on the therapeutics of cancers.
Keywords: ferroptosis, SLC7A11, pan-cancer, immunotherapy, drug sensitivity
Introduction
The application of immune checkpoint inhibitors (ICIs) has achieved significant success in improving cancer patients’ prognosis and is likely to be a crucial part of treatments for patients with a variety of malignancies.1–4 However, it should be noted that only a subset of cancer patients are responsive to ICIs.5 Several biological indices have been reported as possible biomarkers to identify cancer patients who may benefit from ICIs, such as tumor mutation burden, PD-L1 expression level, and immune cell infiltration in tumor microenvironment.6
Ferroptosis is a special form of programmed cell death that shows dependence on active ferrous ions and the accumulation of phospholipid hydroperoxides on the cytoplasmic membrane.7–9 Emerging evidence suggests that ferroptosis exhibits suppressive effects on the development of diverse types of cancer, indicating its potential as a therapeutic target for cancers.10–12 Similar to apoptosis, increasing evidence suggests that ferroptosis is an essential mechanism for tumor suppression.13,14 However, the clinical application of putative indicator of ferroptosis in cancer therapy is limited, mainly because the specific mechanisms underlying ferroptosis remain unclarified.
SLC7A11 is a cystine/glutamate antiporter transmembrane protein that plays a critical role in mediating cystine uptake in negative modulation of ferroptosis and maintaining cell survival under oxidative stress conditions inhibits and is found to be overexpressed in multiple human cancers.15,16 Consistent with this, it was shown that SLC7A11 inactivation has induced potent ferroptosis in many cancer cells; conversely, overexpression of SLC7A11 in cancer cells contributes to the ferroptosis resistance.13,17,18 Importantly, recent studies have also demonstrated that immunotherapy and radiotherapy can induce cell ferroptosis partly through increased SLC7A11 expression.19–22 Correspondingly, considerable attention has been paid to find out the potential regulatory mechanisms of SLC7A11 in ferroptosis and therapeutically targeting SLC7A11 in cancer therapy. Nevertheless, the role of SLC7A11 in cancers and the potential molecular mechanisms have not been elucidated, and no studies have been carried out to provide comprehensive pan-cancer analyses for SLC7A11.
In the present study, we summarized the possible mechanisms of ferroptosis in cancers and tested the potential of ferroptosis as a putative indicator for cancer immunotherapy via pan-cancer analyses. We further explored the expression patterns and genetic mutations of SLC7A11 to evaluate their influence on the prognosis of patients with various types of cancer. In addition, we sought to investigate the possible correlation between SLC7A11 expression and immune cell infiltration in tumor microenvironment. The comprehensive pan-cancer analyses identified SLC7A11 as an attractive biomarker for tumor microenvironment immune infiltration and poor prognosis in cancers, shedding new light on the therapeutics of cancers.
Materials and Methods
Data Acquisition and Preprocessing
The mRNA expression profiles, genetic data, and related clinical information of pan-cancer cohorts from The Cancer Genome Atlas (TCGA) were downloaded from UCSC xena (http://xena.ucsc.edu/), an online database that incorporates gene expression matrix and clinical information from the TCGA and Genotype-Tissue Expression (GTEx) databases. Of the 20 cancer types involved in these two databases, 10 cancer types, including thyroid carcinoma, rectum adenocarcinoma, diffuse large B-cell lymphoma, pheochromocytoma and paraganglioma, thymoma, testicular germ cell tumors, breast invasive carcinoma, lower-grade glioma, chromophobe kidney cancer, and prostate adenocarcinoma, were excluded because the death events were too little for overall survival (OS) analyses.23 Cases with sarcoma, glioblastoma multiforme, and acute myeloid leukemia were also excluded. Finally, 6313 samples from 20 cancer types, including adrenocortical carcinoma (ACC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), bladder urothelial carcinoma (BLCA), head and neck squamous cell carcinoma (HNSC), colon adenocarcinoma (COAD), kidney renal papillary cell carcinoma (KIRP), esophageal carcinoma (ESCA), kidney renal clear cell carcinoma (KIRC), lung squamous cell carcinoma (LUSC), liver hepatocellular carcinoma (LIHC), ovarian serous cystadenocarcinoma (OV), lung adenocarcinoma (LUAD), mesothelioma (MESO), stomach adenocarcinoma (STAD), pancreatic adenocarcinoma (PAAD), skin cutaneous melanoma (SKCM), uterine carcinosarcoma (UCS), uterine corpus endometrial carcinoma (UCEC), and uveal melanoma (UVM) with detailed clinical information of the patients from the TCGA and GTEx databases were included in the current study.
Oncomine, an online cancer microarray data-mining platform (http://www.oncomine.org/)24 was applied to further compare SLC7A11 expression in tumor tissues with that in corresponding normal tissues.25 A fold change of 1.5 and a P value of 0.05 were applied as the threshold.
Analyses of the Expression and Prognostic Value of SLC7A11
To uncover the correlation between SLC7A11 expression and OS, the patients were divided into a high-expression group and a low-expression group based on the median level of SLC7A11 expression in each type of cancer. OS was defined as the duration (days) from the date of diagnosis of cancer to the date of death or the date of the last follow-up for each of the patients. P values were calculated using the Log rank test. Survival curves were generated by “survivaal” and “survminer” R packages, and a p value <0.05 was considered statistically significant. Immunostaining analysis of SLC7A11 protein level was carried out with a rabbit monoclonal anti-SLC7A11 antibody (1:2000, 26864-1-AP, Proteintech). Two experienced pathologists independently assessed the staining of SLC7A11 in one formalin-fixed paraffin-embedding (FFPE) slide. Staining intensity was analyzed and scored from 0 to 3 as follows: 0 = no staining; 1 = weak staining; 2 = moderate staining; and 3 = strong staining. Staining extent was also graded from 0 to 4, according to the percentage of immunoreactive tumor cells (0 = 0%, 1 = 1–25%, 2 = 26–50%, 3 = 51–75%, 4 = 76–100%). The scores of staining intensity were then multiplied by the staining extent scores to yield a total immunohistochemistry (IHC) score (from 0 to 12). A total IHC score of 0 to 3 signified negative staining and a total IHC score of 4 to 12 signified positive staining.
Analyses of SLC7A11 Expression and Clinical Phenotypes
In this study, tumor stages were defined as clinical phenotypes and their relationship with SLC7A11 expression was explored. Statistical analyses (Kruskal–Wallis) for tumor stages were conducted using R software v4.0.3 and plots were generated by the “ggpubr” package in R. P < 0.05 was considered statistically significant.
Gene Set Expression Analyses
Gene set enrichment analyses (GSEAs) were performed with the Molecular Signature database (MSigDB) “c2.cp.kegg.v7.2.symbols.gmt” gene set26 to assess enrichment of canonical pathways between the SLC7A11 high-expression and low-expression groups, using Gene Set Enrichment Analyses software (Java version 4.0).27 Signaling pathways with a normalized p < 0.05 and a false discovery rate (FDR) q < 0.25 were considered to be significantly enriched pathways.
Analyses of Genetic and Epigenetic Alterations
The open-source cBioPortal database (http://www.cbioportal.org) contains cancer genomics data, including information on somatic mutations and copy-number variations (CNVs) for genes, derived from the TCGA and GEO databases.28 Therefore, we utilized this database to analyze single-nucleotide variations in the SLC7A11 gene in our pan-cancer cohorts. OncoPrints was used to describe the distributions of mutants and copy number alterations (CNAs) in the samples, which were presented in the form of heat map, according to the online instructions at c-BioPortal.
Relationship Between SLC7A11 Expression and Cancer Immunity
Tumor mutational burden (TMB) was defined as the number of somatic, coding, indel mutations and base substitutions per megabases of a genome examined. The TMB in our pan-cancer cohorts was determined based on the TCGA dataset. First, we tested the relationship between TMB and SLC7A11 gene expression by Spearman correlation analysis. Next, we quantified the proportions of immune cells using the CIBERSORTx29 and EPIC30 algorithms, separately. Spearman correlation analysis was conducted for the relationships among TMB, Immune cell infiltration, and SLC7A11 expression in multiple tumor tissues.
Drug Responses
Connectivity Map (CMap)31 is a public database that contains more than 7000 transcriptional expression profiles from cultured human cells treated with 1309 bioactive small molecules. The expression of SLC7A11 gene was used to query CMap to predict potential therapeutic drugs for cancer patients. We ranked differentially expressed genes (DEGs) identified between the SLC7A11 high-expression and low-expression groups according to their p values and the top 500 up-regulated and downregulated genes were deemed eligible for querying against CMap. Drugs with positive connectivity scores are able to induce the input signature in human cell lines. Conversely, drugs with negative connectivity scores are able to induce reverse effects against the input signature in human cell lines.
Results
Expression Patterns of SLC7A11 in Our Pan-Cancer Cohorts
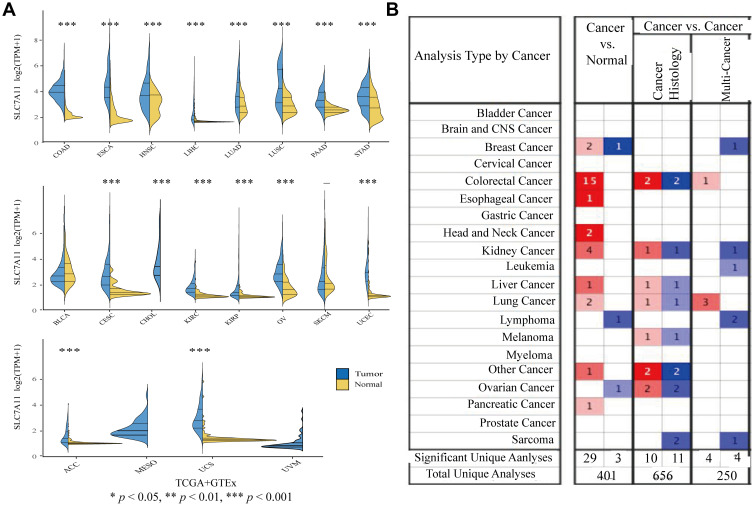
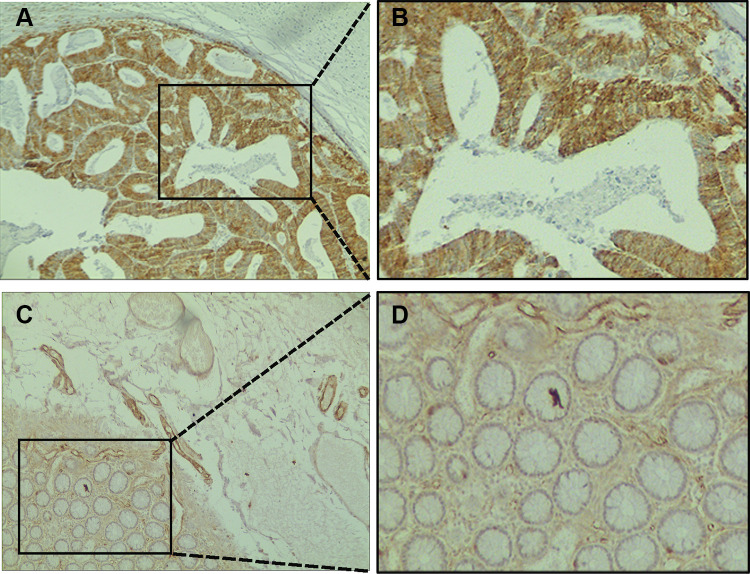
To determine the expression levels of SLC7A11 in the 20 evaluated cancer types, a total of 6313 samples were enrolled in the study. We found significant differences in the expression pattern of SLC7A11 between cancer and para-carcinoma tissues in almost all the solid tumors (except for BLCA, SKCM, MESO, and UVM) (Figure 1A), with the expression level of SLC7A11 being universally higher in almost all the solid tumors (except for BLCA, SKCM, MESO, and UVM) than in the corresponding normal tissues. Moreover, analyses based on the Oncomine database also revealed that, compared with normal tissues, SLC7A11 expression was up-regulated in solid tumors, such as COAD, ESCA, LUSC, PAAD, BRCA, HNSC, and KIRP (Figure 1B). After comprehensively considering the results of the expression pattern analyses based on different databases, we suggested that SLC7A11 might be an oncogene for the 20 evaluated types of cancer. To establish if SLC7A11 is expressed in COAD, we carried out IHC assays on CRC and adjacent normal tissues. As shown in Figure 2, a diffuse pattern of SLC7A11 expression was observed in both the cytoplasm and the nucleus of colon cancer tissues. Compared with normal adjacent tissues, SLC7A11 was statistically significantly overexpressed in cancerous tissues (p<0.001).
Figure 1.
Pan-cancer analysis of SLC7A11 expression. (A) Expression levels of SLC7A11 in tumor and normal tissues were assessed based on the TCGA and GTEx databases (Wilcoxon test was used for statistical analyses, *P < 0.05, **P < 0.01, ***P < 0.001). (B) Expression levels of SLC7A11 in tumor and normal tissues were assessed based on Oncomine. A fold change of 2 and a P value of 0.01 were designed as the cutoff values.
Figure 2.
Immunohistochemical staining of SLC7A11 in colon cancer tissues and normal adjacent tissues. (A) Primary cancer tissues magnification 100×. (B) Magnification 400×. (C) Normal adjacent tissues magnification 100×. (D) Magnification 400×.
Prognostic Value of SLC7A11 in Our Pan-Cancer Cohorts
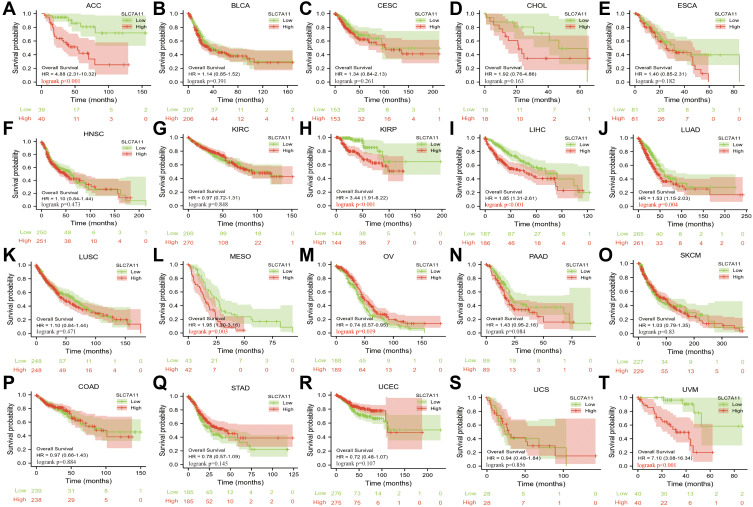
The correlation between SLC7A11 expression and prognosis (OS) in our pan-cancer cohorts was examined by survival analysis of the TCGA datasets. As shown in Figure 3, we found that SLC7A11 expression significantly affected patients’ OS in seven cancer types, including ACC (HR=4.88, P<0.001), KIRP (HR=3.44, P<0.001), LIHC (HR=1.85, P<0.001), LUAD (HR=1.53, P=0.004), MESO (HR=1.95, P=0.003), OV (HR=0.74, P=0.019), UVM (HR=7.10, P<0.001). For most of these cancer types, high SLC7A11 expression predicted a poor prognosis. However, in OV, high SLC7A11 expression was associated with better OS (Figure 3B). Overall, these results indicated that SLC7A11 was an important prognostic factor in various cancers and might be a potential pan-cancer prognostic marker.
Figure 3.
Kaplan-Meier analysis of OS of SLC7A11 high expression versus low expression patients among the 20 cancer types.(A) ACC; (B) BLCA; (C) CESC; (D) CHOL; (E) ESCA; (F) HNSC; (G) KIRC; (H) KIRP; (I) LIHC; (J) LUAD; (K) LUSC; (L) MESO; (M) OV; (N) PAAD; (O) SKCM; (P) COAD; (Q) STAD; (R) UCEC; (S) UCS; (T) UVM.
Analyses of SLC7A11 Expression and Clinical Phenotypes
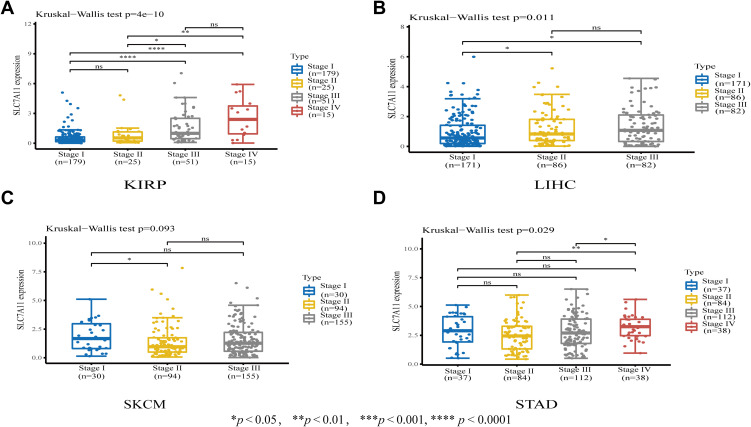
Next, we performed subgroup analyses of the differential expression of SLC7A11 according to tumor stage and found that SLC7A11 expression significantly correlated with tumor stage in KIRP, LIHC, STAD, and SKCM (Figure 4). Notably, the majority of significant differences in SLC7A11 expression were observed between tumor stages II, III and IV. Interestingly, SLC7A11 expression increased significantly in stages III and IV compared with stages I and II in KIRP and LIHC, but not in SKCM and STAD.
Figure 4.
Associations between SLC7A11 expression and tumor stages in (A) KIRP, (B) LIHC, (C) SKCM, and (D) STAD. *P < 0.05, **P < 0.01, **** P < 0.0001.
Mechanisms Underlying the Prognostic Value of SLC7A11 in Our Pan-Cancer Cohorts
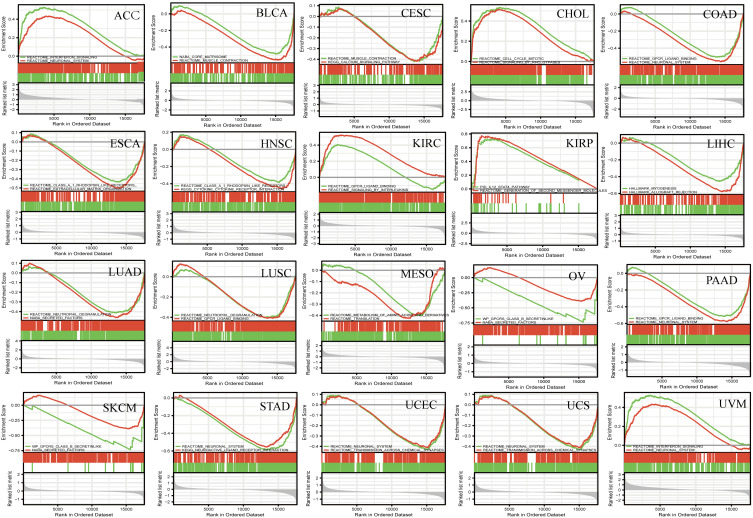
Given the important role of SLC7A11 in ferroptosis and the interesting results we found, we determined to explore the role of SLC7A11 dysregulation in the pathogenesis of the 20 investigated types of cancer. We examined the enriched gene sets in samples with high- and low-SLC7A11 mRNA expression, according to the SLC7A11 median expression level, and the first two most significant pathways are shown in Figure 5. The GSEA analysis demonstrated that several cancer-associated pathways and immune-related pathways were hyperactivated in the high SLC7A11 expression group in most tumors. INTERFERON SIGNALING and IL_12 STAT4 PATHWAY were found in ACC, KIRP, and UVM. GPCR LIGAND BINDING was found in COAD, PAAD, LUSC, and KIRC. CELL CYCLE MITOTIC, and CYTOKINE_CYTOKINE RECEPTOR INTERACTION PATHWAY were found in HNSC, and CHOL. The specific function of SLC7A11 in the pathogenesis of pan-cancer is needed to be experimentally verified.
Figure 5.
GSEA of SLC7A11 RNA-seq data in 20 cancer types. GSEA results and GSEA plots of representative data are shown. Normalized enrichment scores (NESs), nominal P values (NOMs), and false discovery rates (FDRs) are indicated. Peaks on the upward curve indicate positive regulation and peaks on the downward curve indicate negative regulation.
Analyses of Genetic Alterations in SLC7A11 in Our Pan-Cancer Cohorts
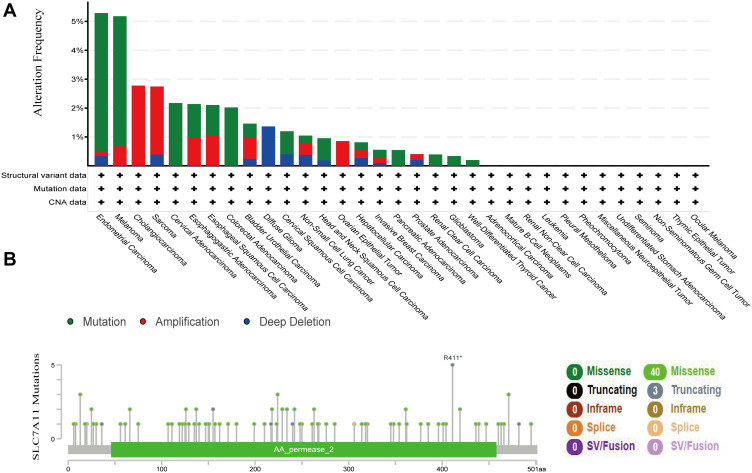
Given that genetic changes play a critical role in regulating cancer oncogenesis and development, genetic alterations in SLC7A11 were further explored based on cBioPortal. A total of 1602 cases retrieved from three tissue studies (10,945 samples from MSKCC, Nat Med 2017; 500 cases from Umich, Nature 2017; 249 cases from Broad/Dana-Farber, Nat Genet 2018) and three cell-line studies (1739 samples from Broad 2019; 1020 samples form Novartis/Broad, Nature 2012; 67 samples NCI, Cancer Res 2012) were analyzed. We found that SLC7A11 mutations were one of the most important single factors for its deregulated expression. The overall mutation rate of SLC7A11 in cancers was relatively low (less than 8%). A total of 43 mutations (40 missense and 3 truncation mutations) were found in SLC7A11. These mutations affect the protein region of amino acids 0 to 501 (Figure 6B). Among the 20 types of cancer, endometrial carcinoma demonstrated the highest rate of SLC7A11 mutation followed by Melanoma (Figure 6A). These results implied that genetic alterations in SLC7A11 played vital roles in tumorigenesis of specific types of cancer, such as endometrial cancer, lymphoma, and colorectal cancer.
Figure 6.
Aberrant expression and mutations of SLC7A11 in the investigated types of cancer. (A) A total of 43 mutation sites were detected, which are located between amino acids 0 to 501 of the SLC7A11 protein. (B) The alteration frequency of SLC7A11 was determined using cBioportal. The alterations included deletions (blue), amplifications (red), and mutations (green); according to the HGVS (Human Genome Variation Society) gene mutation naming rule R411* indicates the arginine at position 411 becomes a termination code (nonsense mutation); an asterisk (*) indicates the termination code.
SLC7A11 Expression was Correlated with TMB and Immune Cell Infiltration in Tumor Microenvironment in Our Pan-Cancer Cohorts
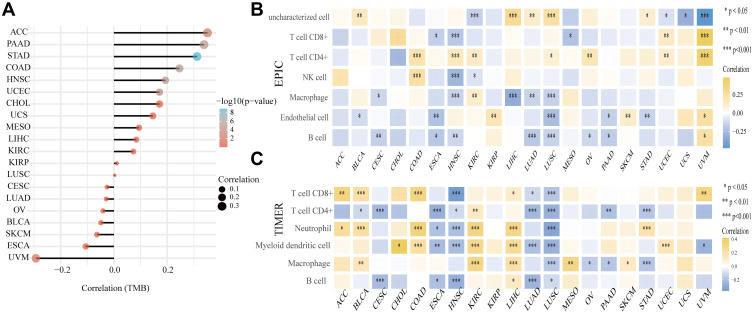
Emerging evidence has highlighted that targeting ferroptosis-associated metabolisms could improve the efficacy of cancer immunotherapy.32 Therefore, we explored the relationship between SLC7A11 expression and immune cell infiltration in tumor microenvironment. According to a previous study, an immunosuppressive tumor microenvironment lacking T lymphocyte infiltration is associated with low tumor mutational burden (TMB).33 Therefore, we explored whether SLC7A11 expression influenced TMB in our pan-cancer cohorts and found that SLC7A11 expression was positively correlated with TMB in most of the studied cancer types, especially in ACC, PAAD, STAD, COAD, and HNSC, but was negatively correlated with the TMB of UVM. Furthermore, by using the EPIC and TIMER algorithms, we found that SLC7A11 expression correlated with the level of immune cell infiltration in tumor microenvironment in different types of cancer, especially in LUSC, LUAD, and LIHC (Figure 7B). Consistent with our findings, immunotherapy was found to exhibit almost identical significant curative effects on hepatocellulr carcinoma (HCC) and LUSC clinically.34–36 These results revealed that SLC7A11 expression was strongly correlated with TMB and immune cell infiltration in tumor microenvironment, suggesting that SLC7A11 may be a predictor of responses of cancers to immunotherapy.
Figure 7.
(A) Correlation coefficients between SLC7A11 expression and TMB in different cancers. The size of the dots in the figure represents the value of the correlation coefficients, and the different colors represent different levels of significance (reflected by P values). The closer the color is to blue, the smaller the P value. The vertical axis represents tumor types, while the horizontal axis represents correlation coefficient between SLC7A11 expression and TMB; a positive relationship is represented by a positive value, and the stronger the correlation, the darker the color (*P < 0.05, **P < 0.01, ***P < 0.001). (B and C) Spearman correlation analysis heat maps showing the correlations between immune scores of microenvironment infiltrating immune cells (CD8+ T cells, CD4+ T cells, NK cells, macrophages, endothelial cells, B cells, and myeloid dendritic cells) and SLC7A11 gene expression in 20 tumor tissues. Two latest algorithms, including TIMER (B) and EPIC (B) were used to evaluate immune scores. The horizontal axis represents different tumor tissues, the vertical axis represents different immune scores, and different colors represent correlation coefficients. A positive value represents a positive correlation and a negative value represents a negative correlation. The stronger the correlation, the darker the color, and the asterisk represents the importance (*p) (*p < 0.05, **p < 0.01, ***p < 0.001). The significance of the two groups of samples passed the Wilcox test.
Drug Responses
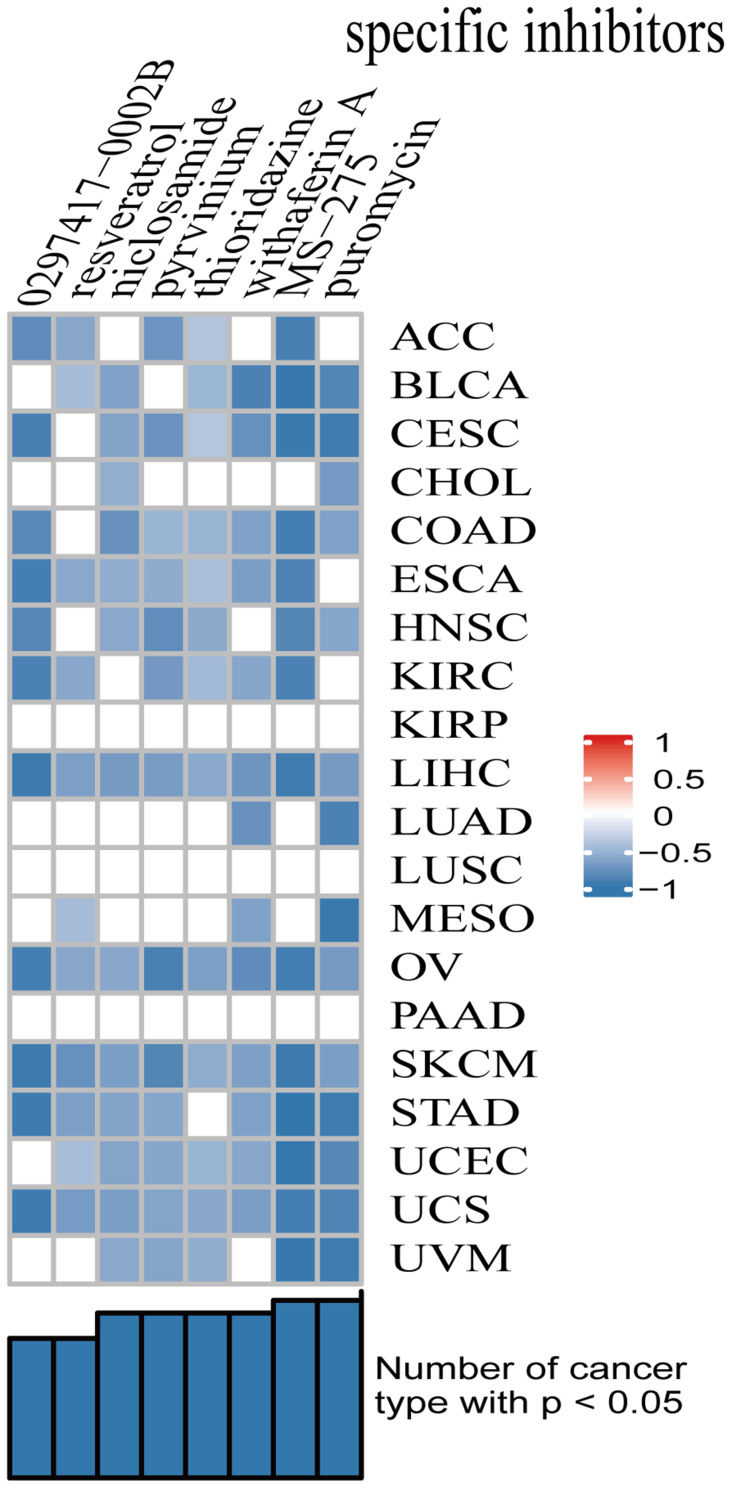
Based on the results of CMap database inquiry, a total of 8 small molecular agents were obtained (Figure 8). The eight small molecules showed negative scores in the analyses, meaning that they have the potential to treat corresponding cancers. Among the small molecules, MS-275 had the highest absolute value of score, meaning that the drug has the potential to treat the 20 types of cancer. These results suggested that MS-275 might be a potential broad-spectrum anti-cancer drug. The therapeutic effects of MS-275 on cancers warrant further investigation.
Figure 8.
Small molecule drugs with significant correlations as revealed by the results of CMap analysis.
Discussion
In this research, we revealed the roles of SLC7A11, a ferroptosis-related gene, as a predictor of survival and a potential regulator of responses to ICIs in 20 types of cancer. Our findings suggested that SLC7A11 expression was up-regulated in a variety of cancers, especially in solid tumors, and that its high expression could be used as a prognostic indicator. We also found that genetic changes and expression level of SLC7A11 had potential effects on prognosis, immune cell infiltration in tumor microenvironment, TMB, and responses to putative small-molecule drugs. To the best of our knowledge, this is the first pan-cancer analysis of SLC7A11 to comprehensively investigate its potential role in underlying tumor prognosis and treatment in cancers.
Ferroptosis is a newly identified form of regulated cell death, which was found to be triggered by the extensive accumulation of iron-dependent reactive oxygen species and lipid peroxides in cancers.7,37,38 In addition, researchers have found that ferroptosis-related genes play increasingly important roles in predicting prognosis and shaping tumor microenvironment in cancers.39–41 Specifically, induction of ferroptosis is a new therapeutic strategy for advanced cancers. SLC7A11 is a critical ferroptosis regulator. Previous studies have identified SLC7A11 as a new UFMylation substrate and revealed that targeting the UFM1/SLC7A11 pathway is a promising cancer treatment strategy.42 Besides, Hagiwara et al found that SLC7A11 was regulated by the PBAF (SWI/SNF) complex, a subunit of which (PBRM1) formed a complex with MUC1-C and NRF2 to increase chromatin accessibility and induce SLC7A11 expression.43 SLC7A11 has also been found to be associated with drug resistance. Lei et al reported that p53 deficiency promoted radiotherapy resistance in cancer cells or tumor tissues at least partly through inhibition of SLC7A11-mediated ferroptosis. Collectively, these data motivated us to focus on SLC7A11-related mechanisms and carry out in-depth explorations of a variety of cancers.
SLC7A11 has been suggested to play an oncogenic role in various tumors. In order to understand the potential influence of SLC7A11 on the prognosis of various types of cancer, we performed pan-cancer analyses of the expression patterns of SLC7A11 across 6313 tumor samples from 20 cancer types deposited by different research groups in TCGA. We observed that SLC7A11 was highly expressed in various tumors, which is consistent with the observations from previous studies.43–46 Notably, by looking at the genetic alterations of SLC7A11, we found that the overall mutation rate of SLC7A11 in cancers was less than 8% and that the main mutation type was missense mutation. We speculate that the mutations in SLC7A11 are responsible for its aberrant expression in cancers. In addition, by exploring the pathways enriched in cancers with high SLC7A11 expression, we found that most of the enriched pathways were metabolic, tumor microenvironment-related and tumorigenesis-related pathways. These findings suggested that aberrant expression of SLC7A11 induced diverse biological effects of tumorigenesis and cancer development. Subsequently, survival analysis revealed that the overexpression of SLC7A11 was associated significantly with reduced OS, indicating that the expression level of SLC7A11 may be a valuable biomarker for the prognosis of cancer patients.
Accumulating evidence suggests that ferroptosis and lipid metabolism play critical roles in modulating tumor immunity.17,47 The mobilization of immune cells to (or within) tumor microenvironment is controlled by various chemokines, cytokines, and pathways, such as ferroptosis.48 To clarify the association between SLC7A11 expression and tumor immunity, we performed Pearson’s correlation analysis and observed a strong positive correlation between SLC7A11 expression and the level of immune cell infiltration in tumor microenvironment in various cancers. Our findings suggest that SLC7A11 expression may be a valuable biomarker for predicting which cancer patients will benefit from immunotherapy. Consistent with our findings, Dingshan Li and Yongsheng Li pointed out that a combination of inducing ferroptosis and blocking tumor immune escape may become the basis for effective antitumor therapies in clinics, and that the expression of SLC7A11 may be a valuable predictive biomarker for the prognosis and responses to immunotherapy in patients with breast carcinoma.47 These findings need to be further confirmed to examine if SLC7A11 could be a potential therapeutic target for cancers.
In addition, we utilized the CMap database to identify several small-molecule drugs with potential therapeutic efficacy against cancers with abnormal SLC7A11 expression. Some of the drugs were shown to have potential anti-cancer effects, such as MS-275/entinostat (an HDAC inhibitor)49 and puromycin,50 which also exhibited inhibitory effects on most types of cancer investigated in our study. Entinostat, combined with exemestane, has shown encouraging efficacy in patients with HR+/HER2-metastatic breast cancer.51 Notably, epigenetic modulators were demonstrated to increase the number of CD8+ effector T cells and improve the efficacy of ICIs. A Phase I clinical study (ETCTN-9844) exhibited that the combination of entinostat, nivolumab and ipilimumab was safe and tolerable with expected rates of immune-related adverse events.52 Thus, these identified small-molecule drugs could be potential therapeutic drugs for cancers with abnormal SLC7A11 expression.
Limitations
Our study has some limitations. First, this is a retrospective study, the data of which come entirely from open databases and the results have not been verified experimentally. Second, the mechanisms underlying SLC7A11ʹs prognostic value in cancers have not been explored. Third, although SLC7A11 expression exhibited correlations with TMB, the level of immune cell infiltration in tumor microenvironment, and drug responses in cancers, the underlying molecular mechanisms must be investigated by further in vivo and in vitro experiments before these results can be generalized to clinical practice.
Conclusions
Through comprehensive pan-cancer analyses using various databases and algorithms, we found that SLC7A11 was highly expressed in various cancers, and this high expression was associated with poor overall survival in most of the cancers investigated. In addition, SLC7A11 expression exhibited significant correlations with TMB, the level of immune cell infiltration in tumor microenvironments, and drug responses in cancers. In summary, these results confirmed the significance of SLC7A11 expression in cancer prognosis and as a potential predictor for the responses to ICIs.
Acknowledgments
This work was supported by Beijing CSCO Clinical Oncology Research Foundation (Y-2019AZMS-0055); the Natural Science Foundation of Liaoning Province (No. 20180550781); the Fundamental Research Funds for the Central Universities (LD202031).
Data Sharing Statement
The datasets analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval
The study protocol was approved by the Ethics Committee of the First affiliated Hospital of Dalian Medical University (YJ-KY-FB-2021-21).
Author Contributions
All authors made a significant contribution to the conception, study design, execution, acquisition of data, analysis and interpretation of data of this work; they also took part in drafting, revising or critically reviewing the manuscript; gave approval of the final version to be published; had agreed on the journal to which the article had been submitted; and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33(17):1974–1982. doi: 10.1200/JCO.2014.59.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378(24):2288–2301. [DOI] [PubMed] [Google Scholar]
- 3.André T, Shiu KK, Kim TW, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura Y, Namikawa K, Yoshino K, et al. Anti-PD1 checkpoint inhibitor therapy in acral melanoma: a multicenter study of 193 Japanese patients. Ann Oncol. 2020;31(9):1198–1206. [DOI] [PubMed] [Google Scholar]
- 5.Wang RF. A special issue on cancer immunotherapy. Cell Res. 2017;27(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Tong Z, Zhang W, et al. FDA-Approved and Emerging Next Generation Predictive Biomarkers for Immune Checkpoint Inhibitors in Cancer Patients. Front Oncol. 2021;7(11):683419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad M, Angeli JP, Vandenabeele P, et al. Regulated necrosis: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2016;15(5):348–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Cheng Y, Mao C, et al. Emerging mechanisms and targeted therapy of ferroptosis in cancer. Mol Ther. 2021;3:S1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Lin D, Yu Q, et al. A promising future of ferroptosis in tumor therapy. Front Cell Dev Biol. 2021;9(9):629150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, Sun S, Johnson T, et al. The glutathione peroxidase Gpx4 prevents lipid peroxidation and ferroptosis to sustain Treg cell activation and suppression of antitumor immunity. Cell Rep. 2021;35(11):109235. [DOI] [PubMed] [Google Scholar]
- 12.Dierge E, Debock E, Guilbaud C, et al. Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell Metab. 2021. [DOI] [PubMed] [Google Scholar]
- 13.Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T, Jiang L, Tavana O, et al. The Deubiquitylase OTUB1 Mediates Ferroptosis via Stabilization of SLC7A11. Cancer Res. 2019;79(8):1913–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato H, Tamba M, Ishii T, et al. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J Biol Chem. 1999;274(17):11455–11458. [DOI] [PubMed] [Google Scholar]
- 16.Koppula P, Zhang Y, Zhuang L, et al. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 2018;38(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Shi J, Liu X, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20(10):1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang X, Green MD, Wang W, et al. Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer Discov. 2019;9(12):1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Green M, Choi JE, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei G, Zhang Y, Koppula P, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30(2):146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye LF, Chaudhary KR, Zandkarimi F, et al. Radiation-Induced Lipid Peroxidation Triggers Ferroptosis and Synergizes with Ferroptosis Inducers. ACS Chem Biol. 2020;15(2):469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173(2):400–416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;2:166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;171(1):243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo JG, Wasser M, Kumar P, et al. Publisher Correction: the Extended Polydimensional Immunome Characterization (EPIC) web-based reference and discovery tool for cytometry data. Nat Biotechnol. 2020;38(6):757. [DOI] [PubMed] [Google Scholar]
- 31.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Green M, Choi JE, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chida K, Kawazoe A, Kawazu M, et al. A Low Tumor Mutational Burden and PTEN Mutations Are Predictors of a Negative Response to PD-1 Blockade in MSI-H/dMMR Gastrointestinal Tumors. Clin Cancer Res. 2021;27(13):3714–3724. [DOI] [PubMed] [Google Scholar]
- 34.Yang JC, Gadgeel SM, Sequist LV, et al. Pembrolizumab in Combination With Erlotinib or Gefitinib as First-Line Therapy for Advanced NSCLC With Sensitizing EGFR Mutation. J Thorac Oncol. 2019;14(3):553–559. [DOI] [PubMed] [Google Scholar]
- 35.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–1833. [DOI] [PubMed] [Google Scholar]
- 36.Kudo M, Matilla A, Santoro A, et al. CheckMate 040 Cohort 5: a phase I/II study of nivolumab in patients with advanced hepatocellular carcinoma and Child-Pugh B cirrhosis. J Hepatol. 2021;1:34. [DOI] [PubMed] [Google Scholar]
- 37.Xu Z, Peng B, Liang Q, et al. Construction of a Ferroptosis-Related Nine-lncRNA Signature for Predicting Prognosis and Immune Response in Hepatocellular Carcinoma. Front Immunol. 2021;17(12):719175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Qiu X, Yan Y, et al. Evaluation of Ferroptosis-related Gene AKR1C1 as a Novel Biomarker Associated with the Immune Microenvironment and Prognosis in Breast Cancer. Int J Gen Med. 2021;28(14):6189–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He F, Chen Z, Deng W, et al. Development and validation of a novel ferroptosis-related gene signature for predicting prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int Immunopharmacol. 2021;12(98):107789. [DOI] [PubMed] [Google Scholar]
- 40.You Y, Fan Q, Huang J, et al. Ferroptosis-Related Gene Signature Promotes Ovarian Cancer by Influencing Immune Infiltration and Invasion. J Oncol. 2021;26(2021):9915312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K, Ping L, Du T, et al. A Ferroptosis-Related lncRNAs Signature Predicts Prognosis and Immune Microenvironment for Breast Cancer. Front Mol Biosci. 2021;7(8):678877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Zhou Y, Xie S, et al. Metformin induces Ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J Exp Clin Cancer Res. 2021;40(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagiwara M, Fushimi A, Yamashita N, et al. MUC1-C activates the PBAF chromatin remodeling complex in integrating redox balance with progression of human prostate cancer stem cells. Oncogene. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bader S, Wilmers J, Pelzer M, et al. Activation of anti-oxidant Keap1/Nrf2 pathway modulates efficacy of dihydroartemisinin-based monotherapy and combinatory therapy with ionizing radiation. Free Radic Biol Med. 2021;20(168):44–54. [DOI] [PubMed] [Google Scholar]
- 45.Hong T, Lei G, Chen X, et al. PARP inhibition promotes ferroptosis via repressing SLC7A11 and synergizes with ferroptosis inducers in BRCA-proficient ovarian cancer. Redox Biol. 2021;42:101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M, Jiang Y, Sun Y. KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma. Biochem Biophys Res Commun. 2021;23(550):77–83. [DOI] [PubMed] [Google Scholar]
- 47.Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther. 2020;5(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elliott MR, Ravichandran KS. The Dynamics of Apoptotic Cell Clearance. Dev Cell. 2016;38(2):147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Z, Zou J, Li S, et al. Epigenetic therapy inhibits metastases by disrupting premetastatic niches. Nature. 2020;579(7798):284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh R, Williams J, Vince R. Puromycin based inhibitors of aminopeptidases for the potential treatment of hematologic malignancies. Eur J Med Chem. 2017;20(139):325–336. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Zhang Q, Li Q, et al. Phase I Study and Pilot Efficacy Analysis of Entinostat, a Novel Histone Deacetylase Inhibitor, in Chinese Postmenopausal Women with Hormone Receptor-Positive Metastatic Breast Cancer. Target Oncol. 2021;16:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roussos Torres ET, Rafie CI, Wang C, et al. Phase 1 Study of Entinostat and Nivolumab with or without Ipilimumab in Advanced Solid Tumors (ETCTN-9844). Clin Cancer Res. 2021;16(clincanres.5017.2021):746. [DOI] [PMC free article] [PubMed] [Google Scholar]