Abstract
This review serves as an introduction to a Special Issue of Comparative Biochemistry and Physiology, focused on using non-human models to study biomedical physiology. The concept of a model differs across disciplines. For example, several models are used primarily to gain an understanding of specific human pathologies and disease states, whereas other models may be focused on gaining insight into developmental or evolutionary mechanisms. It is often the case that animals initially used to gain knowledge of some unique biochemical or physiological process finds foothold in the biomedical community and becomes an established model. The choice of a particular model for biomedical research is an ongoing process and model validation must keep pace with existing and emerging technologies. While the importance of non-mammalian models, such as Caenorhabditis elegans, Drosophila melanogaster, Danio rerio and Xenopus laevis, is well known, we also seek to bring attention to emerging alternative models of both invertebrates and vertebrates, which are less established but of interest to the comparative biochemistry and physiology community.
1. Introduction
The term physiology has roots dating back to the mid-1500s. The original meaning inferred the study of natural science (with physio, meaning “nature”), and at the time, the terms physics and physiology were used interchangeably. By the 1600s, the two terms diverged to become focused on either the physical world (physics) or the biological world (physiology).
Some of the earliest advances in the understanding of animal physiology and anatomy came from Claudius Galenus (Galen) in Rome ca. 120 CE. As a doctor for wounded gladiators, he had the opportunity to examine humans suffering from various degrees of trauma. He was primarily interested in human function but there was a 2-century long prohibition on defiling the human body. As a result, his exploratory work focused on non-humans, first monkeys, then pigs and other mammals. By the time studying humans in detail expanded in the mid-16th century, Galen’s fundamentals of anatomy and function had persisted for 1400 years. In the centuries that followed, physiologists routinely studied both humans and non-human animals to uncover fundamental principle of function, and a great deal of this work used frogs as a convenient, tractable model (see Leonelli and Ankeny, 2013). For example, Luigi Galvani (1737–1798) used frogs to demonstrate the presence of ‘animal electricity’ that was an intrinsic feature of muscle (see Franzini-Armstrong, 2018).
Most of the more familiar physiologists of the late-1800s to mid-1900s have portfolios that include nonhuman animals to study physiological processes. Ivan Pavlov’s work on conditioning in dogs led to principles applied in the treatment of behavioural therapies. He was awarded the 1904 Nobel Prize in Medicine for his work studying the control of digestion in dogs. August Krogh was awarded the 1920 Nobel Prize in Physiology or Medicine for studies on capillaries of frog tongue and skin. Studies of Andrew Huxley and Alan Hodgkin on squid giant axons elucidated how nerve impulses are transmitted (Hodgkin and Huxley, 1952), and garnered the 1963 Nobel Prize. Hugh Huxley’s work on skeletal muscle of rabbits and frogs led to the sliding filament model of skeletal muscle (Huxley, 1953).
A schism between biomedical physiologists and comparative physiologists arose in the mid- to late-1900s and largely coincided with the growth and enrichment of funding opportunities through governmental agencies. There was also a transition toward more targeted research, with the bulk of funds going toward research with direct relevance to improving human health. The research funding models currently used by many countries make a clear demarcation between basic research and that which is health-oriented and translational. However, even under these conditions, many non-traditional models have gained a foothold in the biomedical research community, essentially by achieving critical mass in the decision-making process. As a result, it is common to see medical research funds directed toward researchers studying genetically tractable systems such as the fruit fly (Drosophila melanogaster), the African clawed frog (Xenopus laevis), the nematode Caenorhabditis elegans), and zebrafish (Danio rerio) among others. However, with the exponential growth in the repertoire of tools available to physiologists, reductions in cost, and increases in technical expertise within the community, the barriers between traditional and non-traditional models have greatly diminished. This Special Issue is devoted to exploring the background and merits of what have historically been comparative physiology models, and which are now in use to study biomedical questions.
2. The nature of models
The term model is frequently used to describe an experimental system, but the term means different things to different people. In a general, literal sense, a model is something that resembles something else, often created or used to make projections or inferences that can be applied more generally. That sort of definition would apply equally well to a model train or a mathematical model.
In physiology, a model is something studied in an experimental setting that can apply more broadly in other organisms, identifying commonalities that are highly conserved across taxa. This sort of definition would exclude, for example, studies focused on a particular species of fish because of a special interest in that animal, perhaps because of its economic, cultural, or management value. Such studies might have broader ramifications, but the goal of the work is truly to better understand that specific organism.
Although most would agree on the significance of broader relevance for a model, there are also nuances in the discipline about exactly what else is denoted by the term. Though many more specific terms are used to describe models, the terminology is not used with any degree of precision, and often means quite different things.
2.1. Model organisms
The term model organism is used by some in the literal sense: an organism used as a model. For many, the term “model organism” has a rather restricted interpretation that is understood within select fields to refer to specific organisms chosen decades ago as a focus of an international effort to promote that experimental system, such as Xenopus, Drosophila, and C. elegans. While these are certainly powerful experimental systems, there is nothing about the term “model” that merits such a restrictive definition.
The original model organisms (or systems) were promoted as a means of focusing international, collaborative efforts to advance the field. People shared information and tools as a means of overcoming barriers. For example, sequences of genes of yeast, C. elegans, and Drosophila were circulated within their respective research communities long before ubiquitous ‘omics platforms were developed. This enabled researchers to access genetic information, genetic lines, and molecular tools (e.g., antibodies, nucleic acid probes) to facilitate their own studies. Many of the logistical barriers that existed into the late 20th century are almost unimaginable now. It has been argued that with an exponential increase in genomic information, many more organisms can become an “ideal model organism”, especially now that tools for targeted mutagenesis are commonplace and accessible.
For many years, as editors and reviewers we would oversee arguments with authors who promoted their species of interest as the proverbial “ideal model” for some research problem. The impression seemed to be that it was essential that the experimental system be tagged as both “ideal” and “model” for it to gain broader relevance. In comparative physiology, there is a distinction between models chosen for their unique properties versus those selected because their properties are presumed universal. Many such discussions revolve around the concept of an August Krogh model. Although most comparative physiologists can faithfully provide some rendition of the Krogh Principle, the implications for adopting such models are actually more complicated than is apparent.
2.2. Conservation and divergence in Krogh models
The Krogh principle is an approach widely adopted among physiologists (see Lindstedt and Nishikawa, 2015). First articulated in 1929 by August Krogh and later formally named by Hans Krebs (Krebs, 1975), the principle was stated as: For a large number of problems there will be some animal of choice or a few such animals on which it can be most conveniently studied (Krogh, 1929). Krogh’s experience at that time was based on work with whole animals – Banting and Best’s work with beagles to study diabetes, Christian Bohr’s work on respiration in tortoises – highlighting convenience in a model. But Krogh was best known for his work on the structure and function of capillaries, using frog muscle as an experimental model (see Kissane et al., 2021; Grassi et al., 2021; Poole et al., 2021). Krogh was adept at identifying specific problems (e.g., O2 diffusion vs O2 secretion) and finding models to best study the problem (Larsen et al., 2021). Jorgensen (2001) suggested that Krogh was influenced by his contemporary, Claude Bernard, who took advantage of an atypical arrangement of cervical sympathetic nerves in frogs, which enabled him to interrupt vascular nerves specifically, and which led to the discovery of vasomotor control (Bernard, 1927). In his reflections on Krogh, Sir Hans Krebs summarized those systems that epitomized Krogh models in animal physiology, and most fall in the category of reductionist models (Treberg et al., 2020). Krebs’ own work on muscle metabolism benefited from the unexpected mitochondrial integrity of pigeon muscle. Remarkable insights into fundamental properties of neurons were made possible by the study of electric organs (Nachmansohn, 1959) and the giant axons of squid (Young, 1938), followed by Hodgkin and Huxley’s elucidation of the nature of the action potential (Hodgkin and Huxley, 1952). Amphibian epithelium (toad bladder, frog skin) was also important in exploring the basic properties of transport epithelia (Krogh, 1938, Ussing, 1952: Larsen et al., 2021). Each of these Krogh models takes advantage of a peculiar biological property of an animal feature to study a fundamental property, and most also use the model in a reductionist approach, scaling observations to infer properties of the more complex system.
It should be noted that a model is a system chosen because it appears similar to other systems of interest, but the inherent assumption in a Krogh model is that it differs from the system of interest, albeit in useful ways (see Green et al., 2018). Biomedical researchers choose models based on the assumption that their general properties are conserved and conclusions broadly applicable to human biology. For instance, biomedical research performed on rodents depends on that system faithfully reflecting the homologous system in humans. This creates something of a paradox among comparative and evolutionary physiologists, who may study a species with the expectation that it will differ in important ways from even closely related species. These physiologists search for evolutionary novelty. The concept of a Krogh model has also been applied to study the exceptions- organisms with unique solutions to specific physiological challenges, such as ‘extreme’ environments that often require unique physiological or morphological adaptations. Lauder et al. (1995) argued that these models are useful specifically because they are different, offering an alternative perspective on otherwise conserved relationships between structure, function, and environment. It should also be noted that within humans, there are unique ‘experiments of nature’ – rare groups of humans including patients with unusual conditions – that can be conveniently studied and elucidate key questions in physiological research (Joyner et al., 2021). This includes many examples of populations that show genetic differences that predispose them to atypical responses to stressors (e.g., high altitude hypoxia) and diseases (e.g., obesity).
2.3. Evolutionary models
Comparative physiology models have clear potential to expand our understanding of pathophysiology for two main reasons. First, there is a growing appreciation for the role of evolution in determining human traits in the discipline of evolutionary medicine. Rühli et al. (2016) explore the many pathological conditions for which there is an evolutionary explanation, arguing that the modern condition is best understood through an evolutionary-based approach. Looking at modern humans with knowledge of their evolutionary history better explains the origins, and potentially treatment, of pathological conditions (Stearns and Koella, 2007).
Second, the proliferation of information and analytical tools has made it much easier for physiologists to bring phylogenetic analyses into their research. The speed of data acquisition allows for study of multiple species in parallel that once would have taken an entire career to complete. More species are also accessible to researchers due to increased access to regions of the globe that have been historically difficult to reach. Many studies focus on a collection of related organisms and employ phylogenies to infer evolutionary trajectories in genes, and thereby processes. Many biomedical models are animals that were studied at first because of their unique biology rather than their potential to gain insight into biomedical phenomena. There are many examples of successful cross-over, where research intended for an evolutionary audience gains attention from the biomedical community. An evolution-oriented comparative perspective can enrich the biomedical community through exposure to the range of possible solutions for physiological problems.
3. Comparative models in biomedical research
In this Special Issue, one goal is to focus attention on and promote opportunities to use comparative models to study questions with some direct relevance to biomedical research. In most cases (though see above), this is work done explicitly to better understand human physiology and pathophysiology. Naturally, the species that are most closely related to humans are the most likely models to use to explore human diseases. In some cases, these models are chosen based on practical parameters such as housing and handling. However, there is a growing awareness of the importance of phylogenetic relatedness and its implications, though the specific relationships may not be well known. We have also included simplified phylogenetic trees to show the relationships between the major groups of models. In comparing models, the branch lengths reflect time since common ancestors, and the longer the branch length, the greater the opportunities for evolutionary divergence between the experimental animal and the species it is intended to model. Fig. 1 provides a recent rendering of the phylogenetic relationships between many of the common models used for medical-oriented research. Chimpanzees and humans last shared a common ancestor about 16 mya. The distance between rats and mice is about 3× the distance between chimpanzees and humans; primates and rodents last shared a common ancestor about 80 mya. The distances and times are more striking when considering other mammals, other vertebrates, and invertebrates. The vertebrate lineage diverged echinoderms almost 700 mya, and despite this distance, sea urchins were an early model to study embryonic development. In subsequent sections, we selectively survey the history of the many models used to study biomedical physiology, and implicit in the discussion is the influence of evolutionary-relatedness and the significance of the tremendous periods of time for differences to accrue.
Fig. 1.
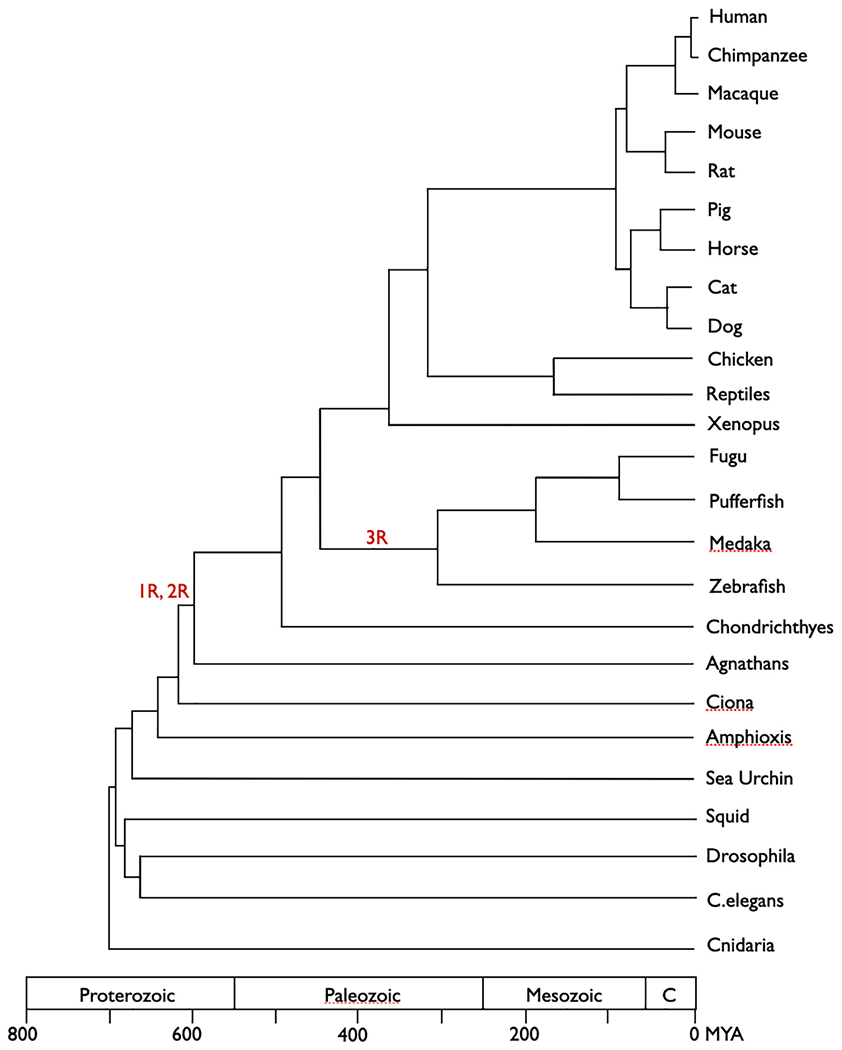
Phylogenetic distances between animal models used in biomedical research. Whole-genome duplication events are represented by 1R, 2R, and 3R, respectively. Source: adapted from Miralles et al. (2019).
3.1. Primates
Non-human primates remain the gold standard for biomedical testing but the ethical considerations are complex (Phillips et al., 2014). As a result, research in non-human primates must meet a very high bar for experimental necessity and is usually late-stage and translational, occurring only after significant validation and testing in lower vertebrates. Nonetheless, non-human primates and humans share many important physiological similarities and cognitive capabilities that simply cannot be matched in other study species, and which are critical to the development of safe and novel treatments for a wide variety of diseases and disorders. For example, non-human primates are critical as late-stage test subjects in evaluating the safety of new vaccines to a developing fetus; tests which clearly could not be performed in human volunteers.
Perhaps the most common use of non-human primates is in the study of neurological function and dysfunction. The human brain is highly complex and specialized, with extensive gyrification of the cerebral cortex to support a larger cortical surface area and greater cognitive power within a limited cranial space (Gautam et al., 2015). Gyrification primarily occurs in primates, ungulates, cephalopods, and large mammals, but is largely absent in rodents (Zilles et al., 2013). Research into neuroprotective mechanisms against ischemic stroke in rodent brain are therefore fundamentally limited by the relatively simple brain system under study and are not sufficient to support advancing of testing of novel therapeutics directly into human patients. Non-human primates play a key role here, bridging the gap between rodent models and human trials. For example, although the efficacy of post-synaptic density 95 (PSD95) inhibitors as a novel treatment for ischemic stroke had been conclusively demonstrated in mice and other lower models, it was not until these results were successfully validated in non-human primates (Cook et al., 2012), that a novel treatment, which offers the first major effective new intervention for stroke since thrombolytics, could be successfully trialed on human patients (Hill et al., 2020). Similarities between non-human primate and human physiology are also critical to the study of neurodegenerative disorders for which animal models are incomplete, such as Alzheimer’s and Parkinson’s.
Non-human primates are the most robust study subjects for diseases that are difficult to assess in lower models, such as depression. They also provide near-human subjects for researching novel treatments for diseases that affect development or can be transmitted to the fetus (e.g., Zika and HIV, among many other examples) (Friedman et al., 2017). Interestingly, non-human primates are probably the only “comparative model” that is studied not because of a novel attribute or unique ability within their environment, but because they are most similar to humans; they are used primarily because research with these animals is less morally problematic than research with human subjects.
3.2. Rats and mice
Mice (usually Mus musculus), and to a lesser extent rats (usually Rattus norvegicus domestica), have been essential in biomedical research for a number of very good reasons. The ease of breeding and holding permits the rearing of large numbers of animals for research. There is also a plethora of immortalized cell culture models available from countless rodent tissues in support of cell biology research. It has been common to interchange rats and mice, trading off the advantages of mice with the size of rats (Ehler et al., 2013). For example, a single preparation of neonatal cardiomyocytes requires hearts from about 10 rat pups per culture (Salameh and Dhein, 1990), whereas 100 s may be needed to get the same yield in mice. Mice were one of the first vertebrates to be used for transgenic studies following the development of transgenic animals through embryonic stem cell injections. Sir Martin Evans was the first to isolate and culture embryonic stem cells from mice (Evans and Kaufman, 1981), leading to the 2007 Nobel Prize in Physiology or Medicine. The use of embryonic cells as recipients of transgenes enabled the development of genetically modified mouse models. The commercial availability of hundreds of strains of transgenic mice enables researchers to use these models to study virtually any biomedical problem.
However, there is an abundance of literature identifying ways in which inherent differences complicate translation of rodent data to humans. Crusio et al. (2009) laments the high number of transgenic mouse studies that demonstrate fundamental flaws associated with how the animals are bred and the appropriate choice of control strains. Bourdi et al. (2011) surveyed a series of contradictory studies related to liver damage and traced the variation in results to the substrains of the C57BL/6 mouse that are typically used as controls. It is somewhat ironic to see significantly flawed studies arising because of comparisons of deeply inbred mouse strains that may differ subtly in genetics yet exhibit pronounced phenotypic differences. Indeed, key phenotypic and behavioural differences between inbred laboratory mice and wild mice exist. For example, domestication impacts female mice to a greater extent than it does males and erases typical sexually dimorphic social behaviors in lab-raised strains (Chalfin et al., 2014). At the molecular level, there are notable differences between inbred and wild-caught mice. For example, wild-caught mice have significantly more active immune function (Abolins et al., 2017), which may confound their use as models for immunity or inflammation, or diseases impacting these pathways. Thus, studies in lab mice may not even be readily extrapolated to wild mice, let alone relevant to humans. These findings also raise questions about the far-reaching impacts of subtle variation in genetic backgrounds.
In addition to erroneous results from lab rodents due to breeding anomalies, there are also examples of sound conclusions demonstrated in rodents that are simply not applicable to humans. Researchers commonly use genetically obese rats and mice that lack either the leptin receptor or the capacity to produce leptin. These genetic models enable the exploration of leptin signaling and metabolic homeostasis and were originally thought to provide ground-breaking insights into obesity in humans. However, hyperphagia due to deficient leptin signaling is the main driver of the obese state in these models. As reviewed by Wang et al. (2014), these rodent models may have limited utility as models for human metabolic diseases associated with obesity, particularly type 2 diabetes, because the etiologies of obesity, metabolic syndrome, and type 2 diabetes rarely involve leptin or leptin receptor deficiencies.
3.3. Rodents: in search of a better mouse
Beyond mice and rats, there are a number of lesser-studied rodents that have proven to be intriguing models. Cohen (2020) summarizes the use of alternative models in the study of SARS-CoV-2. When SARS-CoV-2/COVID-19 appeared in late 2019, there was a rush to identify rodents that were suitable models for studying the disease. However, as has been known for many years, highlighted by SARS, coronavirus did not affect mice in ways analogous to the human immune response. When SARS appeared, one viral target was found to be ACE2, with the viral spike protein binding to the pocket that normally binds angiotensin II. Since mice differ from humans in their sensitivity to coronaviruses, it was noteworthy that ACE2 in mice and humans differ in 11 of the 29 residues that comprise the interaction domain. When McCray Jr. et al. (2006) created transgenic mice that possessed the human version of ACE2, the mice became susceptible to the virus, which manifested symptoms more closely aligned with the human disease. Their model has been resurrected with the appearance of SARS-CoV-2, but others have promoted alternative rodent models where evolutionary differences with humans are minimal. For instance, Syrian hamsters were found to be highly susceptible to SARS-CoV-2. This has been attributed in large part to the similarities in ACE2 sequence, which differs from humans in only 4 residues (Chan et al., 2020). In this particular case, the utility of the Syrian hamster was discovered through serendipity, but now that more genomic information is available, it is feasible to use comparative analysis to identify differences that might impart specific properties based upon unexpected patterns.
While these models are mainly touted as superior versions of a lab mouse, many rodents in nature display traits that make them conducive to studying biomedical questions (Buffenstein, 2005; Smith et al., 2015). For example, in recent years, naked mole-rats (Heterocephalus glaber) have captured the attention of researchers and the general public because of their unusual physiology and appearance. Naked mole-rats are a rare (among mammals) eusocial and subterranean species who, presumably as a result of frequent exposure to harsh environmental challenges throughout their evolutionary history, have proven to be remarkably tolerant to a wide range of clinically relevant stressors. Some examples include tolerance to levels of hypoxia and hypercapnia that would be sufficient to induce loss of consciousness or death in most adult mammals (Chung et al., 2016; Park et al., 2017). Hypoxia is a key component of many clinically relevant pathologies, including stroke, heart attack, anemia, and chronic pulmonary disorders, among others. Similarly, environmental hypercapnia has putatively driven astounding pain-tolerance and insensitivity to acid-based pain sensation at the cutaneous or respiratory surfaces in this species, and also dampened physiological responses to hypercarbia (Clayson et al., 2020; Smith et al., 2020), which can be an important clinical challenge.
Beyond adaptations that seem obviously related to evolution in an “extreme” environment, naked mole-rats are also the longest-lived rodent (up to >30 years). They are a well-established mammalian model of not just longevity (Ruby et al., 2018), but also prolonging healthspan, as they retain reproductive fecundity and general good health into the 95th percentile of their lives. Furthermore, naked mole-rats seemingly have a very low incidence of major aging-related diseases and degeneration, including a remarkably low incidence of cancer of all types (Seluanov et al., 2009; Seluanov et al., 2018). These and other studies describe a range of novel adaptations with important biomedical and translational implications and have driven a surge of interest in this organism: the annual rate of publication of studies investigating naked mole-rats have increased 20-fold over the past 2 decades.
While naked mole-rats have received a disproportionate share of public attention, there are numerous species that live similar social and subterranean lifestyles and that may have the same or other novel adaptations of biomedical interest. Indeed, emerging studies suggest that some of the remarkable adaptations ascribed to naked mole-rats may be broadly conserved, or even surpassed, in related African mole-rat species, and in particular in social species of this lineage (Ivy et al., 2020; Logan et al., 2020; Smith et al., 2020). Similarly, many other rodents living in unique and challenging environments have proven to be valuable study systems. For example, many subterranean, high-altitude dwelling, and/or hibernating rodents are studied for their remarkable hypoxia-tolerance, longevity, metabolic plasticity, and other attributes (Carey et al., 2003; Ballinger and Andrews, 2018; Bhowmick and Drew, 2019; Larson et al., 2014; Storz et al., 2019; Storz and Scott, 2021), which may be informative to not just clinical medicine related to aging and disease, but also to emerging biomedical fields such as space medicine (Feng et al., 2019).
3.4. Non-rodent mammals
Beyond rodents, there are a number of common models to study questions related to human medical concerns. Gurda et al. (2017) summarize the canid (dogs) and felid (cats) genetic models that are available to study genetic diseases found in humans. Typically, mutations are identified as disorders in the pet population, with veterinarians bringing the animals into clinical research. While such diseases are relatively rare in natural populations of mammals, the breeding practices themselves may contribute to spontaneous mutations, which fortuitously align with those seen in humans. Inherited feline diseases have remarkable similarity to analogous genetic diseases in humans (Griffin and Baker, 2002). The same influence of domestication is seen in pigs, another large animal model for human disease. Many studies on pigs focus on renal and digestive physiology because of the similarities with human systems (Gutierrez et al., 2015).
Although cats, dogs, and pigs are used in research, it is the peculiarities of their domestication that have played a major role in their utility as models. Of particular interest in this Special Issue are animals that have evolved in the natural world in ways that offer a different type of mammalian model. In many cases, it is the differences from humans that make them an intriguing model. Of course, the further apart the evolutionary branches between humans and the respective model, the greater care must be used in extrapolating between systems. For example, the enzyme cytochrome oxidase possesses a subunit that exists in vertebrates as two paralogs: COX4-1 and COX4-2. In studies on humans and rodents, COX4-2 is known to be responsive to hypoxia and the enzyme with COX4-2 demonstrates a different relationship with allosteric regulators that is thought to provide protection in hypoxia (Kadenbach and Hüttemann, 2015). However, the regulatory sequence of the gene that confers oxygen sensitivity and the amino acid sequence that imparts the unique allosteric regulation are both traits found in the lineage of mammals the encompasses humans, rodents and lagomorphs. While the paralog exists in other mammals, it appears to have differences in both the gene and protein (Kocha et al., 2015; Porplycia et al., 2017) that may influence responses to oxygen levels in other mammals.
Among the most specious of mammalian orders, bats have long been recognized for their potential as biomedical models for aging (Trapido, 1946) and hypoxia-tolerance (Hiestand et al., 1950). Since these early studies, bats have received considerable attention in the field of aging and have emerged as important models in virus transmission and tolerance (Gorbunova et al., 2020), but their hypoxia-tolerance has been largely underappreciated until very recently. Many species of bats live in small and poorly ventilated spaces, often in large groups and in warm environments. These environmental conditions may present a severe intermittent hypoxic challenge to bats and thus potential molecular and physiological adaptations to hypoxia in this species warrant attention. Marsupials are another order of mammals that have demonstrated significant benefit as a comparative model. Marsupials are born in an immature state and undergo most of their development within their mothers’ pouch (Lillegraven, 1975). This unique arrangement readily enables experimental manipulation of the developing animal without impacting the mother. This is a useful model in the study of mammalian development in general and of developmental disorders in particular (Fukami et al., 2013; Mark and Marotte, 1992; Sharp et al., 2017; Wilson et al., 2003).
3.5. Birds
Non-mammalian tetrapods have been important models for fundamental and biomedical research. In more recent years, nutritional physiology research has used birds both to explore the evolution of metabolic physiology but also as models for metabolic diseases. Many researchers have touted bird models to study aging (Holmes and Ottinger, 2003), taking advantage of practical aspects, such as external fertilization and housing, as well as evolutionary diversity in life span (Austad, 1997; Ottinger et al., 2003). Birds typically have blood glucose levels that are approximately double that of mammals of similar size and are generally insensitive to the regulation of blood glucose by insulin (Braun and Sweazea, 2008). Sustained, elevated hyperglycemia in mammals has well-known deleterious effects on tissues, including cardiovascular disease, neuropathy, nephropathy and retinopathy, owing to the production of reactive oxygen species (ROS) caused by hyperglycemia. In mammals, chronic hyperglycemia and ROS contributes to vascular damage and hypertension (Taniyama and Griendling, 2003). Birds, however, appear to mitigate the negative effects of oxidative stress, including glycation of proteins (Beuchat and Chong, 1998), and have greater longevity compared with mammals of similar size (Holmes and Ottinger, 2003). Thus, birds may be good models for understanding and treating diabetes-related pathologies in humans as well as aging (Austad, 1997). Likewise, birds respond to hyperphagy by stimulating liver hypertrophy and hyperplasia, and greatly increasing lipid content. Though these changes are also seen in humans with fatty liver disease, birds are able to reverse the changes without incurring necrosis or cirrhosis (Wei et al., 2021). Recent work in birds illustrates that neither a refined carbohydrate diet nor a high fat diet has significant effects on plasma glucose, metabolic or vascular physiology (Basile et al., 2020, 2021).
Cardiac disease and cardiac arrhythmias are a leading cause of mortality in developing countries, and the majority of research in this area relies heavily on rodent models. Repolarization of the heart depends largely of a variety of K+ ionic currents, and defects in K+ channels is associated with a variety of cardiac arrhythmias. Identifying animal models with that mimic the currents found in human heart myocardium has proven difficult. Development and testing of anti-arrhythmic drugs for human cardiac disease depends on finding animal models that express cardiac currents that are similar to human myocardial currents. Many of the standard rodent models do not possess the human electrophysiological phenotype and therefore are not ideal models for development of anti-arrhythmic drugs (Janse et al., 1998). Birds have generally not been considered attractive biomedical models for studying cardiac electrophysiology despite being used extensively in developmental cardiovascular physiology owing to the ease of accessibility to an independently developing embryo (Brotto and Creazzo, 1996; Creazzo et al., 2004). However, few studies have examined the electrophysiology of adult bird hearts, despite the similarity with mammals in having a four-chambered heart, high heart rates and endothermy. In this light, a recent study has shown that the Japanese quail (Coturnix japonica) has repolarizing potassium currents that are very similar to the human heart and may prove to be a viable translational model for studying cardiac electrophysiology (Filatova et al., 2021).
3.6. Reptiles
Reptiles were the first terrestrial amniote tetrapods, and the ancestors of birds and mammals. They represent a diverse group that includes the testudines (turtles and tortoises), squamates (lizards and snakes), crocodilians (alligators and crocodiles) and the tuatara (Sphenodon). Reptiles have not been widely used as models for biomedical research, but many investigators have pointed out some unique features that make reptiles attractive to solve serious biomedical problems. For example, some species of turtles of the genera Chrysemys and Trachemys are among the few vertebrates that are capable of withstanding total oxygen deprivation (anoxia). Mammalian neurons and myocardial cells are among the most oxygen-sensitive tissues in the body, and most adult mammals, including humans, cannot survive extended periods of ischemia and hypoxia which occurs during stroke and/or myocardial infarction. The ability of some turtle species to survive anoxia depends on a number of factors including conserving liver glycogen stores through anaerobic metabolic depression; increased ability to buffer lactic acid generated by anaerobic metabolism through elevated carbonate in bone (Jackson, 2004) and preventing tissue oxidative by ROS upon reoxygenation in heart and brain tissue (Pamenter et al., 2007; Bickler and Buck, 2007; Reiterer and Milton, 2020; Bundgaard et al., 2019a; Bundgaard et al., 2019b; Bundgaard et al., 2021). Thus, mechanisms that mitigate the effects of oxygen deprivation on these oxygen-sensitive tissues is of great interest to clinicians and the biomedical community. Additional protective mechanisms include a reduction in channel conductance (‘channel arrest’) (Hochachka, 1986) reflected in reduced glutamatergic excitatory currents, but also increased GABAergic receptor Cl-currents which do not fit the definition of channel arrest, thus the concept of ‘synaptic arrest’ is a better description of the mechanisms that protect the turtle brain from anoxia (Buck and Pamenter, 2018).
Exposure to hypoxia during development in humans is typically maladaptive and results in deleterious long-term consequences for the cardiovascular system (Barker, 2000). However, hypoxic and/or hypercarbic exposure during development is routine for many reptile embryos, thus adaptive responses to developmental exposure to hypoxia and/or hypercarbia may provide insights into the long-term cardiovascular consequences of developmental hypoxia in humans. Similar to birds, reptiles are excellent models for examining the cardiovascular development from embryo to adult owing to the ease of accessing the developing embryo. Recent work with the alligator (Alligator mississippienesis) and snapping turtles (Chelydra serptentina) reveals that developmental exposure to hypoxia and hypercarbia results in immediate (i.e. post-hatch) and long-term (>2 year) changes in cardiovascular function (Marks et al., 2013; Tate et al., 2015; Tate et al., 2016; Filogonio and Crossley 2nd., 2019). This developmental plasticity of the cardiovascular system produces beneficial traits that persist in juvenile animals (Wearing et al., 2016).
Similar to amphibians (see below), some reptile tissues are capable of undergoing complete regeneration. Many lizard species can ‘self-amputate’, or autotomize, their tails in response to a predatory threat with subsequent regeneration of the autotomized limb (Tokuyama et al., 2018). The de novo regeneration of tail tissue in the green anole (Anolis carolinensis) involves the entire peripheral neuromuscular system of the tail within a few weeks (Tokuyama et al., 2018). Although the mechanisms of de novo nerve and neuromuscular junction regeneration appear similar to the development of these structures during development, there are differences in neuromuscular junction number and muscle differentiation that suggest some processes may be specific to regeneration in adult tissue. Recent work in the leopard gecko (Eublepharis macularius) indicates that constitutive cardiomyocyte proliferation occurs routinely and some populations of cardiac cells exhibit slow cycling (Jacyniak and Vickaryous, 2018). Tail autotomy normally prioritizes tail regeneration over somatic growth; however, in leopard geckos, tail autotomy had no effect on proliferation of cardiomyocytes indicating that cardiac homeostasis is not compromised by regeneration of tail tissue (Jacyniak and Vickaryous, 2018).
3.7. Amphibians
Amphibians include a collection of extraordinarily diverse tetrapods, and they have been important models in muscle biology, cardiorespiratory physiology, neurophysiology, developmental biology, and ion and water balance (reviewed by Burggren and Warburton, 2007). They represent the oldest living vertebrate group that made the transition from water to land. The diversity in physiology and life history has stimulated studies to explore the evolution of physiological function, with many examples of lineage-specific solutions to environmental challenges. Amphibians tissues, particularly from frogs, were used as Krogh models to study basic physiology, such as vascular function (Krogh, 1938) and epithelial transport (Ussing, 1952; Larsen, 2021). External fertilization made them convenient as a source of zygotes to study embryonic development, organogenesis and morphogenesis, most commonly in Xenopus laevis. For example, Xenopus oocytes have also been used as a neutral background to study transporter function.
However, in addition to the traits that permit study of the evolution of development and basic physiology, amphibians also have novel characteristics that make them intriguing models to study biomedical questions. Many anurans have the capacity to survive extracellular freezing (Storey and Storey, 1984), which has made them an attractive model to study cryopreservation in the context of tissue preservation (Luu and Storey, 2018). Several caudates have a capacity to regenerate tissues following injury (Grigoryan, 2021). Bettencourt-Diaz et al. (2003) reported that newt (Notophthalmus viridescens) adult heart, as well as the leopard gecko mentioned above, possesses populations of cardiomyocytes that retained the ability to proliferate, which offers some promise for exploring mechanisms to reactivate human cardiomyocytes to facilitate repair following cardiac damage. Recent evidence suggests that the loss of cardiac tissue regenerative capacity in mammals may be linked to increased thyroid hormones associated with the evolution of endothermy (Hirose et al., 2019). Amphibians are also very sensitive to environmental quality and declines in amphibian populations in impacted areas is a harbinger of more subtle effects on humans sharing the same biosphere.
Limb and organ regeneration among salamanders has attracted much attention in the biomedical research community. The neotenic axolotl (Ambystoma) is capable of limb and spinal cord regeneration, which has made it an attractive model for understanding the underlying molecular mechanisms for tissue regeneration in vertebrates, and also how mammals and other vertebrates have lost the ability to regenerate tissues during the course of evolution. The ability to regenerate limb and spinal cord tissue has obvious implications for improvement of function in patients suffering from spinal cord injury (Katoh et al., 2019).
Compared with mammals, and other vertebrates, anuran amphibians (frogs and toads) are extraordinarily tolerant of dehydration and thus are good comparative models for understanding differential dehydration tolerance in vertebrates (Hillman, 2018). The unique ability of anurans to tolerate extreme levels of dehydration that would be lethal for other vertebrates, including humans, lies in their ability to maintain adequate blood volume despite the effects of hypovolemia and hyperviscosity caused by dehydration (Hillman, 2018; Hillman et al., 2021). Mechanisms for maintaining blood volume have typically invoked hydrostatic and oncotic transcapillary forces in mammals (Starling, 1896), but this does not explain how anurans are capable of maintaining blood volume (Hillman et al., 2021) owing to their high rates of transcapillary flux and inability to return plasma from the interstitial to the vascular space via oncotic forces (Hillman, 2018). Anurans have a unique lymphatic system, including lymphatic hearts under feedback regulation from arterial barorecptors, that is capable of returning plasma to the vascular space, thus maintaining blood volume during dehydrational and hemorrhagic stresses that would be lethal to humans (Hillman, 2018). Interestingly, many birds and reptiles have greater dehydration tolerance than mammals, and have similarities with the amphibian lymphatic system (e.g., lymph hearts), yet very little is known about how these groups maintain plasma volume homeostasis (Hedrick et al., 2013). Lymphatic regulation in humans is an important biomedical problem since abnormalities or damage to lymphatic vessels causes lymphedema; lymphatic drainage is often a serious post-surgical complication with few therapeutic solutions; and blocked lymphatic vessels by a parasitic filarial nematode causes ‘elephantiasis’ that affects millions of people worldwide. Thus, greater understanding of the comparative physiology of the lymphatic system of vertebrates may provide unique solutions to a variety of biomedical problems.
3.8. Fishes
Fishes have long held promise as models for biomedical research. Muscle physiologists have benefitted from the anatomical separation of muscle fibre-types (Hidaka and Toida, 1969) and cardiovascular/respiratory physiologists from accessible gills as a surface of gas-exchange (Johansen, 1971). The growth of fish models in recent decades can be attributed to a combination of ethical, logistic, experimental, and technological considerations. Nearing the end of the 20th century, there was increasing ethical pressure to shift to non-primate and nonmammalian biomedical models (Morrison, 2002; Goodman and Check, 2002). In addition to ethical considerations, small fishes are less costly and require much smaller housing footprints (per individual) than their mammalian counterparts. Generally, small fishes also possess a number of traits desirable for experimentation, including year-round breeding windows, rapid generations times (~3 months for medaka and zebrafish), large clutch sizes, transparent embryos and larvae, and relatively easy uptake of many pharmacological agents. Since the late 1990s, the growing genomic databases for Fugu rubripes (see Christoffels et al., 2004 for review), medaka (see Kasahara et al., 2007 for review), and zebrafish (see Varshney et al., 2015 for review) made PCR-based genotyping and gene expression analyses more broadly accessible in various fishes. Recently, the emergence of novel gene-knockdown and gene-editing technologies has catapulted zebrafish to the forefront of biomedical research (Hwang et al., 2013; Varshney and Burgess, 2014). Morpholinos and CRISPR/Cas9, in particular, have helped transform zebrafish into better models of human disease, including cancers, cardiovascular and metabolic diseases, and neurological disorders (Rubbini et al., 2020).
Generally, it was the unexpected fidelity between the fish and human genomes (~ 70% shared genes) that made zebrafish, medaka, and fugu the holy trinity of biomedical fish models. On the other hand, however, several species have emerged as important biomedical models owing to their stark physiological differences. The Mexican tetra (Astyanax mexicanus), for instance, consists of an eyed surface population and more than 30 eyeless cave-dwelling populations (see Rohner, 2018; Krishnan and Rohner, 2019; McGaugh et al., 2020 for review). The unique life-history adaptations of these cave-dwelling populations have made them important vertebrate benchmarks for biomedical research, including insulin resistance (Riddle et al., 2018), social behavior (Kowalko et al., 2013), and circadian dysregulation (Mack et al., 2020). The annual killifish (Austrofundulus limnaeus) has also become an important vertebrate model for a unique life-history trait — its capacity for embryonic diapause (Podrabsky and Hand, 1999). In the past two decades, this system has advanced our understanding of developmental plasticity and bet-hedging (Furness et al., 2015), extreme anoxia tolerance (Podrabsky et al., 2007), and maternal influences (Podrabsky et al., 2010), among others.
3.9. Chordates and echinoderms
Sea squirts of the subphylum Tunicata (tunicates) may bear little resemblance to humans (or animals in general for that matter) but have represented an important biomedical model for embryonic development since the mid 1800’s (see Christiaen et al., 2009 for review). Their close relatives the lancelets (Order: Amphioxiformes) are the only other major group of invertebrate chordates (subphylum cephalochordata). Owing to their unique evolutionary positions (i.e., at the base of the chordate tree), tunicates and amphioxus represent important evolutionary benchmarks for studies in vertebrate evolution. Comparative genomic analyses between the three chordate subphyla (i.e., amphioxus, tunicates, and vertebrates), for instance, revealed that whole-genome duplication events in early vertebrate ancestors likely provided the genomic complexity necessary to derive the distinguishing synapomorphies of the vertebrate subphylum (e.g., homeobox genes and vertebrae) (see Schubert et al., 2006). In addition to this foundational role in genomics (Satoh et al., 2003), tunicates and amphioxus have also become important models for cardiac development (Davidson, 2007), regeneration (Dahlberg et al., 2009), and microbiome research (Corey et al., 2019).
Echinoderms are close relatives of the chordates and represent one of the first comparative models for immunity in the late 1800’s (see Gross et al., 1999 for review). Today, the biomedical community are still leveraging echinoderm models to study immune response. The free-swimming and -feeding purple sea urchin (Strongylocentrotus purpuratus) larva, for instance, has become an important model to understand gut-associated immunity, owing to the easy morphological delineation of the circuitry that regulates this response (Buckley and Rast, 2017). In particular, the cytokine interleukin 17 (IL-17) appears to represent one of the most conserved immune responses between the gut epithelia of echinoderms and mammals (Buckley and Rast, 2019). Echinoderm systems are also increasingly used as biomedical models for their regenerative capacities (Thorndyke et al., 2001). While muscle (e.g., García-Arrarás and Dolmatov, 2010) and neural tissues (e.g., Mashanov et al., 2013) represent the primary tissues of interest in this field, regenerative properties of connective tissues (e.g., collagen networks) also hold great promise for biomaterial development (Ovaska et al., 2017).
3.10. Flies and worms
Among the invertebrates, the most commonly used models are the fruit fly D. melanogaster and the free-living nematode C. elegans. These organisms offer numerous advantages for performing large-scale experiments in the lab, including their small body sizes and large brood sizes, rapid life cycles, and inexpensive maintenance. In addition, the efforts that have gone into delineating aspects of their biology provide useful jumping-off points for further research. For instance, the entire C. elegans cell lineage (Sulston and Horvitz, 1977) and the connectivity of every neuron in C. elegans (White et al., 1986; Jarrell et al., 2012) has been mapped, and the Drosophila central brain (hemibrain) represents the largest reconstructed synaptic-level connectome to date (Scheffer et al., 2020). C. elegans was the first multicellular organism to have a fully sequenced genome (C. elegans Sequencing Consortium, 1998), with the D. melanogaster genome sequenced shortly afterward (Adams et al., 2000), and these models are amenable to genetic manipulation and such approaches as high-throughput genetic screens.
C. elegans and D. melanogaster models enable the study of biochemical and physiological changes across the lifetime of an intact organism with single cell- and tissue-resolution, which facilitates explorations into fundamental biological processes that are relevant to human health. For instance, the human oxygen sensor (prolyl hydroxylases) that modifies HIF was first discovered in C. elegans by Peter Ratcliffe’s group at Oxford (Hon et al., 2002). Ratcliffe shared the 2019 Nobel Prize in Physiology or Medicine for this discovery. Sydney Brenner, Robert Horvitz, and John Sulston were awarded the 2002 Nobel Prize in Physiology or Medicine for using C. elegans to uncover key insights into the genetic regulation of tissue and organ development and apoptosis (e.g., Ellis and Horvitz, 1986). MicroRNAs (miRNAs), a class of non-coding RNA molecules involved in post-transcriptional gene regulation, were first discovered in C. elegans (Lee et al., 1993; Wightman et al., 1993) and are now known to have significant biomedical relevance. Ground-breaking C. elegans experiments also led to the discovery that mutating single genes could extend lifespan (Friedman and Johnson, 1988; Kenyon et al., 1993), which opened up an avenue of research into signaling pathways and mechanisms that have evolutionarily conserved effects on aging.
Along similar lines, the 2017 Nobel Prize in Physiology or Medicine was awarded to Michael Rosbash, Jeff Hall and Michael Young for their D. melanogaster work in identifying and characterizing the role of key genes (period, timeless and doubletime) in transcription and translation-based feedback loops that control the 24-h biological clock. We now know that all multicellular organisms utilize a similar mechanism to control circadian rhythms.
In nature, C. elegans feed on bacteria in soil environments that are rich in organic matter, and are frequently exposed to low oxygen levels, extreme oscillations in nutrient levels, and pathogenic bacteria (Frézal and Félix, 2015). Thus, beyond their utility in the fields of genetics, neurobiology, and developmental and aging biology, their natural history renders them well suited for studies in hypoxia-tolerance, nutrient-sensing, and host-pathogen interactions, D. melanogaster have an increased complexity of organ systems and behaviors, coupled with 60% homology at the level of genes (Wangler et al., 2015) and a comprehensive toolkit to manipulate genes, cells and systems. This makes fruit flies an important system to study the underpinnings of genetics and inheritance, sensory processing, embryonic development, learning, behavior (e.g., feeding, sleep, courtship, grooming etc.) and aging. These models will continue to reveal aspects of physiology and behavior at multiple levels of organization in a relatively short time frame, which has and will continue to aid in understanding human health and disease.
3.11. Other invertebrates
Flies and worms represent the best-established invertebrate models. However, many other species have been used to explore questions of direct relevance to human health. While the studies of Andrew Huxley and Alan Hodgkins on squid giant axons helped elucidate how nerve impulses are transmitted (Hodgkin and Huxley, 1952), cephalopods have more recently been leveraged as a model for learning and memory (Young, 1991; Mather, 1995). Cephalopods represent an especially interesting model because they have evolved high capacity for synaptic plasticity convergent with insects and vertebrates but underscored by drastically different proximate mechanisms (Brown and Piscopo, 2013). Another mollusc, Aplysia, has also become a popular and important model for learning and memory (Roberts and Glanzman, 2003), often in the context of aging (Bailey et al., 1983). Recent work using RNA-seq in Aplysia, for instance, suggests that broad declines in energy metabolism underlie age-associated impairment in memory and learning (Kron et al., 2020). Eric Kandel shared the Nobel Prize (2000) for studies using Aplysia to understand memory.
Invertebrates have also become important models to understand stress response (see Ketchesin et al., 2017). Perhaps none, however, have garnered this role more emblematically than the tardigrades (see Goldstein and Blaxter, 2002). Tardigrades are master tolerators of environmental stress (Møbjerg et al., 2011; Møbjerg and Neves, 2021) and can therefore thrive in every corner of the biosphere, from deep sea tardigrades in submarine caves (Villora-Moreno, 1996) to glacier-dwelling tardigrades in the Himalayas (Dastych, 2019), and terrestrial and freshwater tardigrades in the arctic circle (Pugh and McInnes, 1998). While tardigrades have served as important biomedical models for radiation tolerance (Schill et al., 2009), freeze tolerance, and supercooling (Hengherr et al., 2009) to name a few, their current role in astrobiology perhaps represents the most compelling and imaginative stress model to date (Weronika and Łukasz, 2017).
The lack of a centralized nervous system distinguishes cnidarians from most other animal clades. This unique position (i.e., as an outgroup to the bilateria) makes them an important comparative model to understand the evolution and development of centralized nervous systems (Kelava et al., 2015). The starlet anemone (Nematostella vectensis), in particular, has emerged as an important biomedical model for neural development. Despite more than 500 million years since they shared a common ancestor, there are many striking similarities between neural networks in the starlet anemone and the more centralized neural systems of bilaterians (Kelava et al., 2015). Owing to their relatively easy husbandry, sequenced genome, and arguably the most impressive feats of regeneration in the animal kingdom, the starlet anemone is quickly becoming an important benchmark model in biomedical research (Layden et al., 2016).
4. Final thoughts
The goal of this overview is to frame the Special Issue in a way that promotes the use of alternative models for studying questions related to biomedical physiology. Many comparative researchers study animals for their own traits but recognize that they may also represent opportunities to study questions of direct relevance to human health. While this review is far from comprehensive, we hope that it lays the foundation for more specific papers focused on either experimental models or specific biomedical questions.
References
- Abolins S, King EC, Lazarou L, Weldon L, Hughes L, Drescher P, Raynes JG, Hafalla JCR, Viney ME, Riley EM, 2017. May 3. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat. Commun 8, 14811. 10.1038/ncomms14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Austad SN, 1997. Birds as models of aging in biomedical research. ILAR J. 28, 137–140. [DOI] [PubMed] [Google Scholar]
- Bailey CH, Castellucci VF, Koester J, Chen M, 1983. May. Behavioral changes in aging Aplysia: a model system for studying the cellular basis of age-impaired learning, memory, and arousal. Behav. Neural Biol 38 (1), 70–81. 10.1016/s0163-1047(83)90399-0. [DOI] [PubMed] [Google Scholar]
- Ballinger MA, Andrews MT, 2018. Nature’s fat burning machine: brown adipose tissue in a hibernating mammal. J. Exp. Biol 221 jeb162586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, 2000. In utero programming of cardiovascular disease. Theriogenology 53, 555–574. [DOI] [PubMed] [Google Scholar]
- Basile AJ, Jasbi P, Clark W, Shi X, Gu H, Deviche P, Sweazea KL, 2020. A four-week white bread diet does not alter plasma glucose concentrations, metabolic or vascular physiology in mourning doves, Zenaida macuroura. Comp. Biochem. Physiol. A 247, 110718. [DOI] [PubMed] [Google Scholar]
- Basile AJ, Mohr AE, Jasbi P, Gu H, Deviche P, Sweazea KL, 2021. A four-week high fat diet does not alter glucose or metabolic physiology in wild-caught mourning doves (Zenaida macroura). Comp. Biochem. Physiol. A 251, 110820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, 1927. An Introduction to the Study of Experimental Medicine. Reprint, 1957. Dover Publications, New York. [Google Scholar]
- Bettencourt-Diaz M, Mittnacht S, Brockes JP, 2003. Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. J. Cell Sci 116, 4001–4009. [DOI] [PubMed] [Google Scholar]
- Beuchat CA, Chong CR, 1998. Jul. Hyperglycemia in hummingbirds and its consequences for hemoglobin glycation. Comp. Biochem. Physiol. A Mol. Integr. Physiol 120 (3), 409–416. 10.1016/s1095-6433(98)10039-9. [DOI] [PubMed] [Google Scholar]
- Bhowmick S, Drew KL, 2019. Jun. Mechanisms of innate preconditioning towards ischemia/anoxia tolerance: lessons from mammalian hibernators. Cond. Med 2 (3), 134–141. [PMC free article] [PubMed] [Google Scholar]
- Bickler PE, Buck LT, 2007. Hypoxia tolerance in reptiles, amphibians and fishes: life with variable oxygen availability. Annu. Rev. Physiol 69, 145–170. [DOI] [PubMed] [Google Scholar]
- Bourdi M, Davies JS, Pohl LR, 2011. Jun 20. Mispairing C57BL/6 substrains of genetically engineered mice and wild-type controls can lead to confounding results as it did in studies of JNK2 in acetaminophen and concanavalin A liver injury. Chem. Res. Toxicol 24 (6), 794–796. 10.1021/tx200143x. Epub 2011 May 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EJ, Sweazea KL, 2008. Sep. Glucose regulation in birds. Comp. Biochem. Physiol. B Biochem. Mol. Biol 151 (1), 1–9. 10.1016/j.cbpb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Brotto MA, Creazzo TL, 1996. Feb. Ca2+ transients in embryonic chick heart: contributions from Ca2+ channels and the sarcoplasmic reticulum. Am. J. Phys 270 (2 Pt 2), H518–H525. 10.1152/ajpheart.1996.270.2.H518. [DOI] [PubMed] [Google Scholar]
- Brown ER, Piscopo S, 2013. Jun. Synaptic plasticity in cephalopods; more than just learning and memory? Invertebr. Neurosci 13 (1), 35–44. 10.1007/S10158-013-0150-4. Epub 2013 Apr 3. [DOI] [PubMed] [Google Scholar]
- Buck LT, Pamenter ME, 2018. The hypoxia-tolerant vertebrate brain: arresting synaptic activity. Comp. Biochem. Physiol. B 224, 61–70. [DOI] [PubMed] [Google Scholar]
- Buckley KM, Rast JP, 2017. Oct 23. An organismal model for gene regulatory networks in the gut-associated immune response. Front. Immunol 8, 1297. 10.3389/fimmu.2017.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley KM, Rast JP, 2019. Sep. Immune activity at the gut epithelium in the larval sea urchin. Cell Tissue Res. 377 (3), 469–474. 10.1007/s00441-019-03095-7. Epub 2019 Aug 28. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, 2005. The naked mole-rat: a new long-living model for human aging research. J. Gerontol 60 (11), 1369–1377. [DOI] [PubMed] [Google Scholar]
- Bundgaard A, Qvortrup K, Rasmussen LJ, Fago A, 2019a. Turtles maintain mitochondrial integrity but reduce mitochondrial respiratory capacity in the heart after cold acclimation and anoxia. J. Exp. Biol 222, 1–9. [DOI] [PubMed] [Google Scholar]
- Bundgaard A, James AM, Gruszczyk AV, Martin J, Murphy MP, Fago A, 2019b. Metabolic adaptations during extreme anoxia in the turtle heart and their implications for ischemia-reperfusion injury. Sci. Rep 9, 2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundgaard A, Jensen BS, Jensen FB, Fago A, 2021. Exploring pathways of NO and H2S signaling in metabolic depression: the case of anoxic turtles. Comp. Biochem. Physiol 253, 110857. [DOI] [PubMed] [Google Scholar]
- Burggren WW, Warburton S, 2007. Amphibians as animal models for laboratory research in physiology. ILAR J. 48, 260–269. 10.1093/ilar.48.3.260. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Dec 11. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282 (5396), 2012–2018. 10.1126/science.282.5396.2012. Erratum in: Science 1999 Jan 1; 283(5398):35. Erratum in: Science 1999 Mar 26;283(5410):2103. Erratum in: Science 1999 Sep 3;285(5433):1493. [DOI] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL, 2003. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev 83, 1153–1181. [DOI] [PubMed] [Google Scholar]
- Chalfin L, Dayan M, Levy DR, Austad SN, Miller RA, Iraqi FA, Dulac C, Kimchi T, 2014. Aug 5. Mapping ecologically relevant social behaviours by gene knockout in wild mice. Nat. Commun 5, 4569. 10.1038/ncomms5569. [DOI] [PubMed] [Google Scholar]
- Chan JF-W, Zhang J, Yuan S, Poon VK-M, Chan CC-S, Lee AC-Y, Chan W-M, Fan Z, Tsoi H-W, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai J-P, Kok K-H, Chu H, Chan K-H, Sridhar S, Chen Z, Chen H, To KK-W, Yuen K-Y, 2020. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis ciaa325. 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L, Wagner E, Shi W, Levine M, 2009. Dec. The sea squirt Ciona intestinalis. Cold Spring Harb Protoc 2009 (12). 10.1101/pdb.emo138pdb.emo138. [DOI] [PubMed] [Google Scholar]
- Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B, 2004. Jun. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol 21 (6), 1146–1151. 10.1093/molbev/msh114. Epub 2004 Mar 10. [DOI] [PubMed] [Google Scholar]
- Chung D, Dzal YA, Seow A, Milsom WK, Pamenter ME, 2016. Mar 30. Naked mole rats exhibit metabolic but not ventilatory plasticity following chronic sustained hypoxia. Proc. Biol. Sci 283 (1827), 20160216. 10.1098/rspb.2016.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayson MS, Devereaux MEM, Pamenter ME, 2020. Apr 1. Neurokinin-1 receptor activation is sufficient to restore the hypercapnic ventilatory response in the Substance P-deficient naked mole-rat. Am. J. Phys. Regul. Integr. Comp. Phys 318 (4), R712–R721. 10.1152/ajpregu.00251.2019. Epub 2020 Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J, 2020. From mice to monkeys, animals studied for coronavirus answers. Science 368 (6488), 221–222. [DOI] [PubMed] [Google Scholar]
- Cook DJ, Teves L, Tymianski M, 2012. Feb 29. Treatment of stroke with a PSD-95 inhibitor in the gyrencephalic primate brain. Nature. 483 (7388), 213–217. 10.1038/nature10841. [DOI] [PubMed] [Google Scholar]
- Corey S, Kvederis L, Kingsbury C, Bonsack B, Sanberg PR, Castelli V, Lee JY, Borlongan CV, 2019. Gut microbiome: lactation, childbirth, lung dysbiosis, animal modeling, stem cell treatment, and CNS disorders. CNS Neurol. Disord. Drug Targets 18 (9), 687–694. 10.2174/1871527318666191021145252. [DOI] [PubMed] [Google Scholar]
- Creazzo TL, Burch J, Godt RE, 2004. Calcium buffering and excitation-contraction coupling in developing avian myocardium. Biophys. J. 86, 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusio WE, Goldowitz D, Holmes A, Wolfer D, 2009. Feb. Standards for the publication of mouse mutant studies. Genes Brain Behav. 8 (1), 1–4. 10.1111/j.1601-183X.2008.00438.X. Epub 2008 Sep 6. [DOI] [PubMed] [Google Scholar]
- Dahlberg C, Auger H, Dupont S, Sasakura Y, Thorndyke M, Joly JS, 2009. Refining the Ciona intestinalis model of central nervous system regeneration. PLoS One 4 (2), e4458. 10.1371/journal.pone.0004458. Epub 2009 Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastych H, 2019. Cryobiotus roswithae gen. n., sp. n., a new genus and species of glacier-dwelling tardigrades from Northern Norway (Tardigrada, Panarthropoda). Entomologie heute 31, 95–111. [Google Scholar]
- Davidson B, 2007. Feb. Ciona intestinalis as a model for cardiac development. Semin. Cell Dev. Biol 18 (1), 16–26. 10.1016/j.semcdb.2006.12.007. Epub 2006 Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, Moore-Morris T, Lange S, 2013. Sep 6. Isolation and culture of neonatal mouse cardiomyocytes. J. Vis. Exp 79, 50154. 10.3791/50154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HM, Horvitz HR, 1986. Mar 28. Genetic control of programmed cell death in the nematode C. elegans. Cell 44 (6), 817–829. 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH, 1981. Jul 9. Establishment in culture of pluripotential cells from mouse embryos. Nature 292 (5819), 154–156. 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Feng NY, Junkins MS, Merriman DK, Bagriantsev SN, Gracheva EO, 2019. Sep 23. Osmolyte depletion and thirst suppression allow hibernators to survive for months without water. Curr. Biol 29 (18) 10.1016/j.cub.2019.07.038.3053-3058.e3. Epub 2019 Sep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatova TS, Abramochkin DV, Pavlova NS, Pustovit KB, Konovalova OP, Kuzmin VS, Dobrzynski H, 2021. Repolarizing potassium currents in working myocardium of Japanese quail: a novel translational model for cardiac electrophysiology. Comp. Biochem. Physiol 255, 110919. [DOI] [PubMed] [Google Scholar]
- Filogonio R, Crossley DA 2nd., 2019. Aug. Long term effects of chronic prenatal exposure to hypercarbia on organ growth and cardiovascular responses to adrenaline and hypoxia in common snapping turtles. Comp. Biochem. Physiol. A Mol. Integr. Physiol 234, 10–17. 10.1016/j.cbpa.2019.04.009. Epub 2019 Apr 18. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong C, 2018. Feb 5. The relationship between form and function throughout the history of excitation-contraction coupling. J. Gen. Physiol 150 (2), 189–210. 10.1085/jgp.201711889. Epub 2018 Jan 9. Erratum in: J Gen Physiol. 2018 Jan 22; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frézal L, Félix MA, 2015. Mar 30. C. elegans outside the Petri dish. Elife 4, e05849. 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE, 1988. Jan. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118 (1), 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman H, Ator N, Haigwood N, Newsome W, Allan JS, Golos TG, Kordower JH, Shade RE, Goldberg ME, Bailey MR, Bianchi P, 2017. The critical role of nonhuman primates in medical research. Pathog. Immun 2 (3), 352–365. 10.20411/pai.v2i3.186. Epub 2017 Aug 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami M, Homma K, Hasegawa T, Ogata T, 2013. Apr. Backdoor pathway for dihydrotestosterone biosynthesis: implications for normal and abnormal human sex development. Dev. Dyn 242 (4), 320–329. 10.1002/dvdy.23892. Epub 2012 Nov 19. [DOI] [PubMed] [Google Scholar]
- Furness AI, Lee K, Reznick DN, 2015. Jun. Adaptation in a variable environment: phenotypic plasticity and bet-hedging during egg diapause and hatching in an annual killifish. Evolution 69 (6), 1461–1475. 10.1111/evo.12669. Epub 2015 May 12. [DOI] [PubMed] [Google Scholar]
- García-Arrarás JE, Dolmatov IY, 2010. Echinoderms: potential model systems for studies on muscle regeneration. Curr. Pharm. Des 16 (8), 942–955. 10.2174/138161210790883426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam P, Anstey KJ, Wen W, Sachdev PS, Cherbuin N, 2015. Cortical gyrification and its relationships with cortical volume, cortical thickness, and cognitive performance in healthy mid-life adults. Behav. Brain Res 287, 331–339. 10.1016/j.bbr.2015.03.018. Epub 2015 Mar 21. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Blaxter M, 2002. Jul 23. Tardigrades. Curr. Biol 12 (14), R475. 10.1016/s0960-9822(02)00959-4. [DOI] [PubMed] [Google Scholar]
- Goodman S, Check E, 2002. Jun 13. The great primate debate. Nature 417 (6890), 684–687. 10.1038/417684a. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Kennedy BK, 2020. Jul 7. The world goes bats: living longer and tolerating viruses. Cell Metab. 32 (1), 31–43. 10.1016/j.cmet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi B, Hogan MC, Gladden LB, 2021. Microvascular O2 delivery and O2 utilization during metabolic transitions in skeletal muscle. One-hundred years after the pioneering work by august Krogh. Comp. Biochem. Physiol. Part A 252, 110842. [DOI] [PubMed] [Google Scholar]
- Green S, Dietrich MR, Leonelli S, Ankeny RA, 2018. ‘Extreme’ organisms and the problem of generalization: interpreting the Krogh principle. Hist Philos Life Sci 40, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B, Baker HJ, 2002. Domestic cats as laboratory animals. Lab. Anim. Med 459–482. 10.1016/B978-012263951-7/50015-6. [DOI] [Google Scholar]
- Grigoryan EN, 2021. Jan 18. Study of natural longlife juvenility and tissue regeneration in caudate amphibians and potential application of resulting data in biomedicine. J. Dev. Biol 9 (1), E2 10.3390/jdb9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross PS, Al-Sharif WZ, Clow LA, Smith LC, 1999. Jun–Jul. Echinoderm immunity and the evolution of the complement system. Dev. Comp. Immunol 23 (4–5), 429–442. 10.1016/s0145-305x(99)00022-1. Erratum in: Dev Comp Immunol 1999 Sep;23(6):533. [DOI] [PubMed] [Google Scholar]
- Gurda BL, Bradbury AM, Vite CH, 2017. Canine and feline models of human genetic diseases and their contributions to advancing clinical therapies. Yale J. Biol. Med 90 (3), 417–431. Published 2017 Sep 25. [PMC free article] [PubMed] [Google Scholar]
- Gutierrez K, Dicks N, Glanzner WG, Agellon LB, Bordignon V, 2015. Efficacy of the porcine species in biomedical research. Front. Genet 6, 293. Published 2015 Sep 16. 10.3389/fgene.2015.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick MS, Hillman SS, Drewes RC, Withers PC, 2013. Lymphatic regulation in nonmammalian vertebrates. J. Appl. Physiol 115, 297–308. [DOI] [PubMed] [Google Scholar]
- Hengherr S, Worland MR, Reuner A, Brümmer F, Schill RO, 2009. Mar. Freeze tolerance, supercooling points and ice formation: comparative studies on the subzero temperature survival of limno-terrestrial tardigrades. J. Exp. Biol 212 (Pt 6), 802–807. 10.1242/jeb.025973. [DOI] [PubMed] [Google Scholar]
- Hidaka T, Toida N, 1969. Mar. Biophysical and mechanical properties of red and white muscle fibres in fish. J. Physiol 201 (1), 49–59. 10.1113/jphysiol.1969.sp008741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiestand WA, Rockhold WT, Stemler FW, Stullken DE, Wiebers JE, 1950. Jul. The comparative hypoxic resistance of hibernators and nonhibernators. Physiol. Zool 23 (3), 264–268. 10.1086/physzool.23.3.30152084. [DOI] [PubMed] [Google Scholar]
- Hill MD, Goyal M, Menon BK, Nogueira RG, McTaggart RA, Demchuk AM, Poppe AY, Buck BH, Field TS, Dowlatshahi D, van Adel BA, Swartz RH, Shah RA, Sauvageau E, Zerna C, Ospel JM, Joshi M, Almekhlafi MA, Ryckborst KJ, Lowerison MW, Heard K, Garman D, Haussen D, Cutting SM, Coutts SB, Roy D, Rempel JL, Rohr AC, Iancu D, Sahlas DJ, AYX, Yu, Devlin TG, Hanel RA, Puetz V, Silver FL, BCV, Campbell, Chapot R, Teitelbaum J, Mandzia JL, Kleinig TJ, Turkel-Parrella D, Heck D, Kelly ME, Bharatha A, Bang OY, Jadhav A, Gupta R, Frei DF, Tarpley JW, McDougall CG, Holmin S, Rha JH, Puri AS, Camden MC, Thomalla G, Choe H, Phillips SJ, Schindler JL, Thornton J, Nagel S, Heo JH, Sohn SI, Psychogios MN, Budzik RF, Starkman S, Martin CO, Burns PA, Murphy S, Lopez GA, English J, Tymianski M, ESCAPE-NA1 Investigators, 2020. Mar 14. Efficacy and safety of nerinetide for the treatment of acute ischaemic stroke (ESCAPE-NA1): a multicentre, double-blind, randomised controlled trial. Lancet 395 (10227), 878–887. 10.1016/S0140-6736(20)30258-0. Epub 2020 Feb 20. [DOI] [PubMed] [Google Scholar]
- Hillman SS, 2018. Anuran amphibians as comparative models for understanding extreme dehydration tolerance: a unique negative feedback lymphatic mechanism for blood volume regulation. Am. J. Phys 315, R790–R798. [DOI] [PubMed] [Google Scholar]
- Hillman SS, Drewes RC, Hedrick MS, 2021. Control of blood volume following hypovolemic challenge in vertebrates: transcapillary versus lymphatic mechanisms. Comp. Biochem. Physiol. A 254, 110878. [DOI] [PubMed] [Google Scholar]
- Hirose K, Payumo AY, Cutie S, Hoang A, Zhang H, Guyot R, Lunn D, Bigley RB, Yu H, Wang J, Smith M, Gillett E, Muroy SE, Schmid T, Wilson E, Field KA, Reeder DM, Maden M, Yartsev MM, Wolfgang MJ, Grutzner F, Scanlan TS, Szweda LI, Buffenstein R, Hu G, Flamant F, Olgin JE, Huang GN, 2019. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364, 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, 1986. Defense strategies against hypoxia and hypothermia. Science 231, 234–241. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF, 1952. Aug. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol 117 (4), 500–544. 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DJ, Ottinger MA, 2003. Birds as long-lived animal models for the study of aging. Exp. Gerontol 38, 1365–1375. 10.1016/j.exger.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY, 2002. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417 (6892), 975–978. [DOI] [PubMed] [Google Scholar]
- Huxley HE, 1953. Nov. Electron microscope studies of the organisation of the filaments in striated muscle. Biochim. Biophys. Acta 12 (3), 387–394. 10.1016/0006-3002(53)90156-5. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK, 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol 31 (3), 227–229. 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy CM, Sprenger RJ, Bennett NC, van Jaarsveld B, Hart DW, Kirby AM, Yaghoubi D, Storey KB, Milsom WK, Pamenter ME, 2020. The hypoxia tolerance of eight related African mole-rat species rivals that of naked mole-rats, despite divergent ventilatory and metabolic strategies in severe hypoxia. Acta Physiol. 228 (4), e13436 10.1111/apha.13436. [DOI] [PubMed] [Google Scholar]
- Jackson DC, 2004. Surviving extreme lactic acidosis: the role of calcium lactate formation in the anoxic turtle. Respir. Physiol. Neurobiol 144, 173–178. [DOI] [PubMed] [Google Scholar]
- Jacyniak K, Vickaryous MK, 2018. Constitutive cardiomyocyte proliferation in the leopard gecko (Eublepharis macularius). J. Morphol 2018, 1355–1367. [DOI] [PubMed] [Google Scholar]
- Janse MJ, Opthof T, Kléber AG, 1998. Animal models of cardiac arrhythmias. Cardiovasc. Res 39 (1), 165–177. [PubMed] [Google Scholar]
- Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW, 2012. Jul 27. The connectome of a decision-making neural network. Science 337 (6093), 437–444. 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- Johansen K, 1971. Comparative physiology: gas exchange and circulation in fishes. Annu. Rev. Physiol 33, 569–612. 10.1146/annurev.ph.33.030171.003033. [DOI] [PubMed] [Google Scholar]
- Jorgensen CB, 2001. August Krogh and Claude Bernard on basic principles in experimental physiology. BioScience 51 (1), 59–61. [Google Scholar]
- Joyner MJ, Baker SE, Senefeld JW, Klassen SA, Wiggins CC, 2021. Experiments of nature and within species comparative physiology. Comp. Biochem. Physiol. Part A 253, 110864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B, Hüttemann M, 2015. Sep. The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion. 24, 64–76. 10.1016/j.mito.2015.07.002. Epub 2015 Jul 17. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y, 2007. Jun 7. The medaka draft genome and insights into vertebrate genome evolution. Nature 447 (7145), 714–719. 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Katoh H, Yokota K, Fehlings MG, 2019. Jun 6. Regeneration of spinal cord connectivity through stem cell transplantation and biomaterial scaffolds. Front. Cell. Neurosci 13, 248. 10.3389/fncel.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I, Rentzsch F, Technau U, 2015. Dec 19. Evolution of eumetazoan nervous systems: insights from cnidarians. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 370 (1684), 20150065. 10.1098/rstb.2015.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R, 1993. Dec 2. A C. elegans mutant that lives twice as long as wild type. Nature. 366 (6454), 461–464. 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Ketchesin KD, Stinnett GS, Seasholtz AF, 2017. Sep. Corticotropin-releasing hormone-binding protein and stress: from invertebrates to humans. Stress 20 (5), 449–464. 10.1080/10253890.2017.1322575. Epub 2017 May 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissane RWP, Al-Shammari AA, Egginton S, 2021. Apr. The importance of capillary distribution in supporting muscle function, building on Krogh’s seminal ideas. Comp. Biochem. Physiol. A Mol. Integr. Physiol 254, 110889. 10.1016/j.cbpa.2020.110889. Epub 2021 Jan 11. [DOI] [PubMed] [Google Scholar]
- Kocha KM, Reilly K, Porplycia DS, McDonald J, Snider T, Moyes CD, 2015. Feb 15. Evolution of the oxygen sensitivity of cytochrome c oxidase subunit 4. Am. J. Phys. Regul. Integr. Comp. Phys 308 (4), R305–R320. 10.1152/ajpregu.00281.2014. Epub 2014 Dec 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko JE, Rohner N, Rompani SB, Peterson BK, Linden TA, Yoshizawa M, Kay EH, Weber J, Hoekstra HE, Jeffery WR, Borowsky R, Tabin CJ, 2013. Oct 7. Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr. Biol 23 (19), 1874–1883. 10.1016/j.cub.2013.07.056. Epub 2013 Sep 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA, 1975. Oct. The august Krogh principle: “for many problems there is an animal on which it can be most conveniently studied”. J. Exp. Zool 194 (1), 221–226. 10.1002/jez.1401940115. [DOI] [PubMed] [Google Scholar]
- Krishnan J, Rohner N, 2019. Sweet fish: fish models for the study of hyperglycemia and diabetes. J. Diabetes 11 (3), 193–203. [DOI] [PubMed] [Google Scholar]
- Krogh A, 1929. The progress of physiology. Science 70, 200–204. [DOI] [PubMed] [Google Scholar]
- Krogh A, 1938. The active absorption of ions in some freshwater animals. Zeitschr. vergl. Physiol. 25, 335–350. [Google Scholar]
- Kron NS, Schmale MC, Fieber LA, 2020. Sep 15. Changes in metabolism and proteostasis drive aging phenotype in Aplysia californica sensory neurons. Front. Aging Neurosci 12, 573764. 10.3389/fnagi.2020.573764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen EH, 2021. Dual skin functions in amphibian osmoregulation. Comp. Biochem. Physiol. Part A 253, 110869. [DOI] [PubMed] [Google Scholar]
- Larsen EH, Hoffmann E, Hedrick MS, Wang T, 2021. August Krogh’s contribution to the rise of physiology during the first half the 20th century. Comp. Biochem. Physiol. A 256, 110931. [DOI] [PubMed] [Google Scholar]
- Larson J, Drew KL, Folkow LP, Milton SL, Park TJ, 2014. Apr 1. No oxygen? No problem! Intrinsic brain tolerance to hypoxia in vertebrates. J. Exp. Biol 217 (Pt 7), 1024–1039. 10.1242/jeb.085381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder GV, Huey RB, Monson RK, Jensen RJ, November 1995. Systematics and the study of organismal form and function: advances in systematics are defining new directions for functional morphology and comparative physiology. BioScience 45 (10), 696–704. 10.2307/1312675. [DOI] [Google Scholar]
- Layden MJ, Rentzsch F, Röttinger E, 2016. Jul. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip. Rev. Dev. Biol 5 (4), 408–428. 10.1002/wdev.222. Epub 2016 Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V, 1993. Dec 3. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (5), 843–854. 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leonelli S, Ankeny RA, 2013. What makes a model organism? Endeavor 37 (4), 209–212. [DOI] [PubMed] [Google Scholar]
- Lillegraven JA, 1975. Dec. 1975. Biological considerations of the marsupial-placental dichotomy. Evolution 29 (4), 707–722. 10.1111/j.1558-5646.1975.tb00865.x. [DOI] [PubMed] [Google Scholar]
- Lindstedt SL, Nishikawa KC, 2015. Jun 15. From Tusko to Titin: the role for comparative physiology in an era of molecular discovery. Am. J. Phys. Regul. Integr. Comp. Phys 308 (12), R983–R989. 10.1152/ajpregu.00405.2014. Epub 2015 Apr 8. [DOI] [PubMed] [Google Scholar]
- Logan SM, Szereszewski KE, Bennett NC, Hart DW, van Jaarsveld B, Pamenter ME, Storey KB, 2020. May 11. The brains of six African mole-rat species show divergent responses to hypoxia. J. Exp. Biol 223 (Pt 9), jeb215905. 10.1242/jeb.215905. [DOI] [PubMed] [Google Scholar]
- Luu BE, Storey KB, 2018. Oct. Solving donor organ shortage with insights from freeze tolerance in nature: activating endogenous antioxidant systems with non-coding RNA to precondition donor organs. Bioessays 40 (10), e1800092. 10.1002/bies.201800092. Epub 2018 Aug 27. [DOI] [PubMed] [Google Scholar]
- Mack KL, Jaggard JB, Persons JL, Passow CN, Stanhope BA, Ferrufino E, Tsuchiya D, Smith SE, Slaughter BD, Kowalko J, Rohner N, 2020. Repeated evolution of circadian clock dysregulation in cavefish populations. bioRxiv. 10.1101/2020.01.14.906628.2020.01.14.906628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark RF, Marotte LR, 1992. Feb. Australian marsupials as models for the developing mammalian visual system. Trends Neurosci. 15 (2), 51–57. 10.1016/0166-2236(92)90026-5. [DOI] [PubMed] [Google Scholar]
- Marks C, Eme J, Elsey RM, Crossley DA 2nd., 2013. Oct. Chronic hypoxic incubation blunts thermally dependent cholinergic tone on the cardiovascular system in embryonic American alligator (Alligator mississippiensis). J. Comp. Physiol. B 183 (7), 947–957. 10.1007/s00360-013-0755-2. Epub 2013 Apr 30. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Zueva OR, García-Arrarás JE, 2013. Apr 18. Radial glial cells play a key role in echinoderm neural regeneration. BMC Biol. 11, 49. 10.1186/1741-7007-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather JA, 1995. Cognition in cephalopods. Adv. Study Behav 24, 317. [Google Scholar]
- McCray PB Jr., Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Peng Jia H, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S, 2006. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol 81 (2), 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, Kowalko JE, Duboué E, Lewis P, Franz-Odendaal TA, Rohner N, Gross JB, Keene AC, 2020. Nov. Dark world rises: the emergence of cavefish as a model for the study of evolution, development, behavior, and disease. J. Exp. Zool. B Mol. Dev. Evol 334 (7–8), 397–404. 10.1002/jez.b.22978. Epub 2020 Jul 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles A, Raymond M, Lecointre G, 2019. Empathy and compassion toward other species decrease with evolutionary divergence time. Sci. Rep 9 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møbjerg N, Neves RC, 2021. New insights into survival strategies of tardigrades. Comp. Biochem. Physiol. A 254, 110890. [DOI] [PubMed] [Google Scholar]
- Møbjerg N, Halberg KA, Jørgensen A, Persson D, Bjørn M, Ramløv H, Kristensen RM, 2011. Jul. Survival in extreme environments - on the current knowledge of adaptations in tardigrades. Acta Physiol (Oxford) 202 (3), 409–420. 10.1111/j.1748-1716.2011.02252.x. Epub 2011 Mar 22. [DOI] [PubMed] [Google Scholar]
- Morrison AR, 2002. Making choices in the laboratory. Society 39 (6), 16–23. [Google Scholar]
- Nachmansohn D, 1959. Chemical and Molecular Basis of Nerve Activity. Academic Press, Inc., New York. [Google Scholar]
- Ottinger MA, Reed E, Wu J, Thompson N, French JB Jr., 2003. Jul. Establishing appropriate measures for monitoring aging in birds: comparing short and long lived species. Exp. Gerontol 38 (7), 747–750. 10.1016/s0531-5565(03)00102-5. [DOI] [PubMed] [Google Scholar]
- Ovaska M, Bertalan Z, Miksic A, Sugni M, Di Benedetto C, Ferrario C, Leggio L, Guidetti L, Alava MJ, La Porta CAM, Zapperi S, 2017. Jan. Deformation and fracture of echinoderm collagen networks. J. Mech. Behav. Biomed. Mater 65, 42–52. 10.1016/j.jmbbm.2016.07.035. Epub 2016 Aug 5. [DOI] [PubMed] [Google Scholar]
- Pamenter ME, Richards MD, Buck LT, 2007. Anoxia-induced changes in reactive oxygen species and cyclic nucleotides in the painted turtle. J. Comp. Physiol. B 177, 473–481. [DOI] [PubMed] [Google Scholar]
- Park TJ, Reznick J, Peterson BL, Blass G, Omerbašić D, Bennett NC, Kuich PHJL, Zasada C, Browe BM, Hamann W, Applegate DT, Radke MH, Kosten T, Lutermann H, Gavaghan V, Eigenbrod O, Bégay V, Amoroso VG, Govind V, Minshall RD, Smith ESJ, Larson J, Gotthardt M, Kempa S, Lewin GR, 2017. Apr 21. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 356 (6335), 307–311. 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, Hart BA, Hopkins WD, Hu S-L, Miller LA, Nader MA, Nathanielsz PW, Rogers J, Shively CA, Voytko ML, 2014. Why primate models matter. Am. J. Primatol 76 (9), 801–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podrabsky JE, Hand SC, 1999. Oct. The bioenergetics of embryonic diapause in an annual killifish, Austrofundulus limnaeus . J. Exp. Biol 202 (Pt 19), 2567–2580. [DOI] [PubMed] [Google Scholar]
- Podrabsky JE, Lopez JP, Fan TW, Higashi R, Somero GN, 2007. Jul. Extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus: insights from a metabolomics analysis. J. Exp. Biol 210 (Pt 13), 2253–2266. 10.1242/jeb.005116. [DOI] [PubMed] [Google Scholar]
- Podrabsky JE, Garrett ID, Kohl ZF, 2010. Oct 1. Alternative developmental pathways associated with diapause regulated by temperature and maternal influences in embryos of the annual killifish Austrofundulus limnaeus. J. Exp. Biol 213 (Pt 19), 3280–3288. 10.1242/jeb.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole DC, Kano Y, Koga S, Musch TI, 2021. August Krogh: muscle capillary function and oxygen delivery. Comp. Biochem. Physiol. Part A 253, 110852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porplycia D, Lau GY, McDonald J, Chen Z, Richards JG, Moyes CD, 2017. May 1. Subfunctionalization of COX4 paralogs in fish. Am. J. Phys. Regul. Integr. Comp. Phys 312 (5), R671–R680. 10.1152/ajpregu.00479.2016. Epub 2017 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh PJ, McInnes SJ, 1998. The origin of Arctic terrestrial and freshwater tardigrades. Polar Biol. 19 (3), 177–182. [Google Scholar]
- Reiterer M, Milton SL, 2020. Induction of foxo3a protects turtle neurons against oxidative stress. Comp. Biochem. Physiol. A 243, 110671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle MR, Aspiras AC, Gaudenz K, Peuβ R, Sung JY, Martineau B, Peavey M, Box AC, Tabin JA, McGaugh S, Borowsky R, Tabin CJ, Rohner N, 2018. Mar 29. Insulin resistance in cavefish as an adaptation to a nutrient-limited environment. Nature 555 (7698), 647–651. 10.1038/nature26136. Epub 2018 Mar 21. Erratum in: Nature. 2020 Dec;588(7836):E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AC, Glanzman DL, 2003. Dec. Learning in Aplysia: looking at synaptic plasticity from both sides. Trends Neurosci. 26 (12), 662–670. 10.1016/j.tins.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Rohner N, 2018. “Out of the dark” cavefish are entering biomedical research. In: Zebrafish, Medaka, and Other Small Fishes. Springer, Singapore, pp. 253–268. [Google Scholar]
- Rubbini D, Cornet C, Terriente J, Di Donato V, 2020. Jul. CRISPR meets Zebrafish: accelerating the discovery of new therapeutic targets. SLAS Discov. 25 (6), 552–567. 10.1177/2472555220926920. Epub 2020 May 28. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Smith M, Buffenstein R, 2018. Jan 24. Naked Mole-Rat mortality rates defy gompertzian laws by not increasing with age. Elife 7, e31157. 10.7554/eLife.31157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühli F, van Schaik K, Henneberg M, 2016. Evolutionary medicine: The ongoing evolution of human physiology and metabolism. Physiology 31, 392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salameh A, Dhein S, 1990. Culture of neonatal cardiomyoeytes. In: Dhein S, Mohr FW, Delmar M (Eds.), Practical Methods in Cardiovascular Research. Springer, New York, pp. 568–576. [Google Scholar]
- Satoh N, Satou Y, Davidson B, Levine M, 2003. Jul. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 19 (7), 376–381. 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, Clements J, Hubbard PM, Katz WT, Umayam L, Zhao T, Ackerman D, Blakely T, Bogovic J, Dolafi T, Kainmueller D, Kawase T, Khairy KA, Leavitt L, Li PH, Lindsey L, Neubarth N, Olbris DJ, Otsuna H, Trautman ET, Ito M, Bates AS, Goldammer J, Wolff T, Svirskas R, Schlegel P, Neace E, Knecht CJ, Alvarado CX, Bailey DA, Ballinger S, Borycz JA, Canino BS, Cheatham N, Cook M, Dreher M, Duclos O, Eubanks B, Fairbanks K, Finley S, Forknall N, Francis A, Hopkins GP, Joyce EM, Kim S, Kirk NA, Kovalyak J, Lauchie SA, Lohff A, Maldonado C, Manley EA, McLin S, Mooney C, Ndama M, Ogundeyi O, Okeoma N, Ordish C, Padilla N, Patrick CM, Paterson T, Phillips EE, Phillips EM, Rampally N, Ribeiro C, Robertson MK, Rymer JT, Ryan SM, Sammons M, Scott AK, Scott AL, Shinomiya A, Smith C, Smith K, Smith NL, Sobeski MA, Suleiman A, Swift J, Takemura S, Talebi I, Tarnogorska D, Tenshaw E, Tokhi T, Walsh JJ, Yang T, Horne JA, Li F, Parekh R, Rivlin PK, Jayaraman V, Costa M, Jefferis GS, Ito K, Saalfeld S, George R, Meinertzhagen IA, Rubin GM, Hess HF, Jain V, Plaza SM, 2020. Sep 7. A connectome and analysis of the adult Drosophila central brain. Elife 9, e57443. 10.7554/eLife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill RO, Mali B, Dandekar T, Schnölzer M, Reuter D, Frohme M, 2009. Jul-Aug. Molecular mechanisms of tolerance in tardigrades: new perspectives for preservation and stabilization of biological material. Biotechnol. Adv 27 (4), 348–352. 10.1016/j.biotechadv.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Schubert M, Escriva H, Xavier-Neto J, Laudet V, 2006. May. Amphioxus and tunicates as evolutionary model systems. Trends Ecol. Evol 21 (5), 269–277. 10.1016/j.tree.2006.01.009. Epub 2006 Feb 17. [DOI] [PubMed] [Google Scholar]
- Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, Catania KC, Gorbunova V, 2009. Hypersensitivty to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci 106 (46), 19352–19357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Gladyshev VN, Vijg J, Gorbunova V, 2018. Jul. Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer 18 (7), 433–441. 10.1038/s41568-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JA, Wanyonyi S, Modepalli V, Watt A, Kuruppath S, Hinds LA, Kumar A, Abud HE, Lefevre C, Nicholas KR, 2017. Apr 1. The tammar wallaby: a marsupial model to examine the timed delivery and role of bioactives in milk. Gen. Comp. Endocrinol 244, 164–177. 10.1016/j.ygcen.2016.08.007. Epub 2016 Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ESJ, Schuhmacher L, Husson Z, 2015. The naked mole-rat as an animal model in biomedical research: current perspectives. Open Access Anim. Physiol 7, 137–148. [Google Scholar]
- Smith ESJ, Park TJ, Lewin GR, 2020. May. Independent evolution of pain insensitivity in African mole-rats: origins and mechanisms. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol 206 (3), 313–325. 10.1007/s00359-020-01414-w. Epub 2020 Mar 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starling EH, 1896. On the absorption of fluid from the connective tissue spaces. J. Physiol. Lond 19, 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC, Koella JC, 2007. Evolution in Health and Disease, Second ed. Oxford University Press. 374 pp. [Google Scholar]
- Storey KB, Storey JM, 1984. Biochemical adaptation for freeze tolerance in the wood frog, Rana sylvatica. J. Comp. Physiol. 155, 29–36. [Google Scholar]
- Storz JF, Scott GR, 2021. Phenotypic plasticity, genetic assimilation, and genetic compensation in hypoxia adaptation of high-altitude vertebrates. Comp. Biochem. Physiol. A 253, 110865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Cheviron ZA, McClelland GB, Scott GR, 2019. May 23. Evolution of physiological performance capacities and environmental adaptation: insights from high-elevation deer mice (Peromyscus maniculatus). J. Mammal 100 (3), 910–922. 10.1093/jmammal/gyy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR, 1977. Mar. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol 56 (1), 110–156. 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Taniyama Y, Griendling KK, 2003. Reactive oxygen species in the vasculature: molecular and cellular mechanisms. Hypertension 42, 1075–1081. [DOI] [PubMed] [Google Scholar]
- Tate KB, Kohl ZF, Eme J, Rhen T, Crossley DA 2nd., 2015. Mar-Apr. Critical windows of cardiovascular susceptibility to developmental hypoxia in common snapping turtle (Chelydra serpentina) embryos. Physiol. Biochem. Zool 88 (2), 103–115. 10.1086/677683. Epub 2014 Sep 8. [DOI] [PubMed] [Google Scholar]
- Tate KB, Rhen T, Eme J, Kohl ZF, Crossley J, Elsey RM, Crossley II DA, 2016. Jun 1. Periods of cardiovascular susceptibility to hypoxia in embryonic american alligators (Alligator mississippiensis). Am. J. Phys. Regul. Integr. Comp. Phys 310 (11), R1267–R1278. 10.1152/ajpregu.00320.2015. Epub 2016 Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorndyke MC, Chen WC, Beesley PW, Patruno M, 2001. Dec 15. Molecular approach to echinoderm regeneration. Microsc. Res. Tech 55 (6), 474–485. 10.1002/jemt.1192. [DOI] [PubMed] [Google Scholar]
- Tokuyama MA, Xu C, Fisher RE, Wilson-Rawls J, Kusumi K, Newbern JM, 2018. Developmental and adult-specific processes contribute to de novo neuromuscular regeneration in the lizard tail. Dev. Biol 433, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapido H, 1946. Aug. Observations on the vampire bat with special reference to longevity in captivity. J. Mammal 27, 217–219. [PubMed] [Google Scholar]
- Treberg JR, Martyniuk CJ, Moyes CD, 2020. Dec. Getting the most out of reductionist approaches in comparative biochemistry and physiology. Comp. Biochem. Physiol. B Biochem. Mol. Biol 250, 110483. 10.1016/j.cbpb.2020.110483. Epub 2020 Jul 24. [DOI] [PubMed] [Google Scholar]
- Ussing HH, 1952. Some aspects of the application of tracers in permeability studies. Adv. Enzymol. Relat. Subj. Biochem 13, 21–65. [DOI] [PubMed] [Google Scholar]
- Varshney GK, Burgess SM, 2014. Mar. Mutagenesis and phenotyping resources in zebrafish for studying development and human disease. Brief Funct. Genom 13 (2), 82–94. 10.1093/bfgp/elt042. Epub 2013 Oct 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Sood R, Burgess SM, 2015. Understanding and editing the Zebrafish genome. Adv. Genet 92, 1–52. 10.1016/bs.adgen.2015.09.002. Epub 2015 Nov 12. [DOI] [PubMed] [Google Scholar]
- Villora-Moreno S, 1996. A new genus and species of the deep-sea family Coronarctidae (Tardigrada) from a submarine cave with a deep-sea like condition. Sarsia 81 (4), 277–283. [Google Scholar]
- Wang B, Chandrasekera PC, Pippin JJ, 2014. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr. Diabetes Rev 10 (2), 131–145. 10.2174/1573399810666140508121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler MF, Yamamoto S, Bellen HJ, 2015. Mar. Fruit flies in biomedical research. Genetics 199 (3), 639–653. 10.1534/genetics.114.171785. Epub 2015 Jan 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearing O, Eme J, Rhen T, Crossley II DA, 2016. Phenotypic plasticity in the common snapping turtle (Chelydra serpentina): long-term physiological effects of chronic hypoxia during embryonic development. Am. J. Phys. Regul. Integr. Comp. Phys 310, R176–R184. [DOI] [PubMed] [Google Scholar]
- Wei R, Han C, Deng D, Ye F, Gan X, Liu H, Li L, Xu H, Wei S, 2021. Feb. Research progress into the physiological changes in metabolic pathways in waterfowl with hepatic steatosis. Br. Poult. Sci 62 (1), 118–124. 10.1080/00071668.2020.1812527. Epub 2020 Oct 2. [DOI] [PubMed] [Google Scholar]
- Weronika E, Łukasz K, 2017. Dec. Tardigrades in space research - past and future. Orig. Life Evol. Biosph 47 (4), 545–553. 10.1007/s11084-016-9522-1. Epub 2016 Oct 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S, 1986. Nov 12. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 314 (1165), 1–340. 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G, 1993. Dec 3. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75 (5), 855–862. 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilson JD, Leihy MW, Shaw G, Renfree MB, 2003. Dec 15. Unsolved problems in male physiology: studies in a marsupial. Mol. Cell. Endocrinol 211 (1–2), 33–36. 10.1016/j.mce.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Young JZ, 1938. The functioning of the giant nerve fibres of the squid. J. Exp. Biol 15, 170–185. [DOI] [PubMed] [Google Scholar]
- Young JZ, 1991. Apr. Computation in the learning system of cephalopods. Biol. Bull 180 (2), 200–208. 10.2307/1542389. [DOI] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Amunts K, 2013. May. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 36 (5), 275–284. 10.1016/j.tins.2013.01.006. Epub 2013 Feb 14. [DOI] [PubMed] [Google Scholar]


