Abstract
The effectiveness of SARS-CoV-2 vaccines and therapeutic antibodies have been limited by the continuous emergence of viral variants and by the restricted diffusion of antibodies from circulation into the sites of respiratory virus infection. Here, we report the identification of two highly conserved regions on the Omicron variant receptor-binding domain recognized by broadly neutralizing antibodies. Furthermore, we generated a bispecific single-domain antibody that was able to simultaneously and synergistically bind these two regions on a single Omicron variant receptor-binding domain as revealed by cryo-EM structures. We demonstrated that this bispecific antibody can be effectively delivered to lung via inhalation administration and exhibits exquisite neutralization breadth and therapeutic efficacy in mouse models of SARS-CoV-2 infections. Importantly, this study also deciphered an uncommon and highly conserved cryptic epitope within the spike trimeric interface that may have implications for the design of broadly protective SARS-CoV-2 vaccines and therapeutics.
Keywords: SARS-CoV-2, single-domain antibody, bispecific antibody, broad neutralization, inhalation
Graphical abstract
Therapeutic effects against SARS-CoV-2 infection are seen in mice after inhalation of an engineered, bispecific, single-domain antibody that simultaneously targets two points on the spike protein receptor-binding domain.
Introduction
Since the emergence of the original strain in late 2019, five SARS-CoV-2 variants, namely Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and the newly identified Omicron (B.1.1.529) have been defined as variants of concern (VOCs) by the World Health Organization, inducing waves of prevalence. The Omicron variant, bearing over 30 mutations in the viral spike protein and 15 mutations in the receptor-binding domain (RBD), which is the major target for neutralizing antibodies, is distinct from other VOCs that only possess no more than three RBD mutations. Remarkable resistance of the Omicron variant against neutralization by antibodies and serum has been reported, challenging the protective efficacy of vaccines and therapeutic antibodies (Cameroni et al., 2022; Cao et al., 2022; Cele et al., 2022; Dejnirattisai et al., 2022; Liu et al., 2021).
It is notable that the majority of RBD mutations (9 out of 15) found in Omicron were located in the angiotensin converting enzyme 2 (ACE2) receptor-binding motif (RBM) and the other 6 mutations resided in the ridge side of the RBD core, leaving a part of lateral surface, as well as the cryptic site hidden inside the spike protein trimer, essentially unchanged (Figure 1 A). Previous studies have also revealed the presence of conserved epitopes located on SARS-CoV-2 RBDs (Li et al., 2021; Tian et al., 2020; Yuan et al., 2020). Thus, it is practical to identify the highly conserved epitopes across SARS-CoV-2 variants, which could be important for the design of broadly neutralizing antibodies and universal vaccines.
Figure 1.
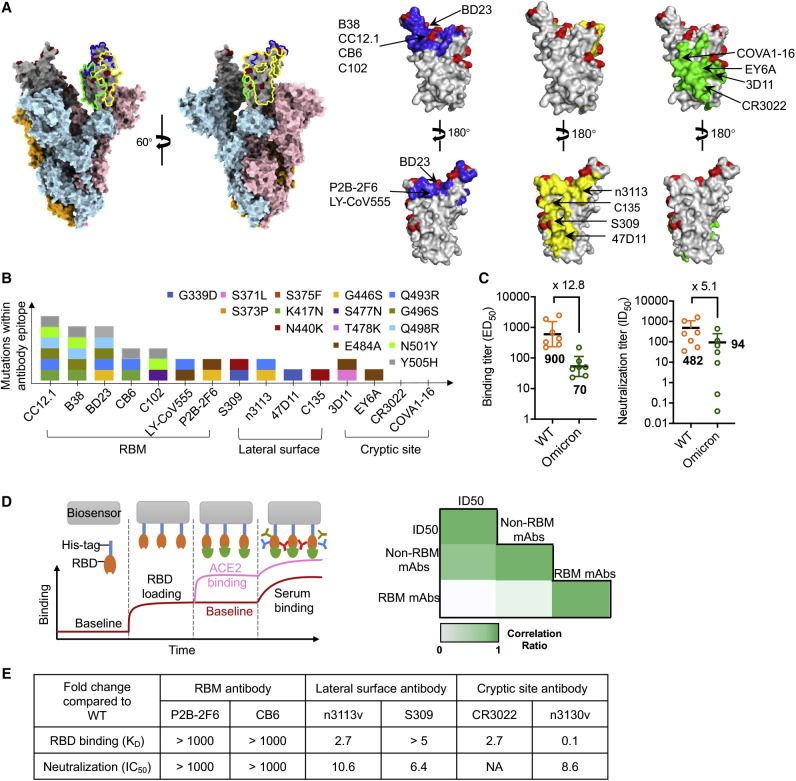
Non-RBM antibodies confer resistance against the SARS-CoV-2 Omicron variant
(A) Epitope clustering of RBD-targeting antibodies on the spike protein and RBD. The Omicron S was shown as surface, with RBD colored in gray; mutations are highlighted in red. The three classes of epitope are circled in S and highlighted in RBD as blue, green, and yellow, respectively.
(B) The amino acid mutations in the RBD region of the Omicron variant are shown as colored boxes. The key mutations found in binding sites of three classes of antibody, such as RBM-binding site, lateral surface, and cryptic site, are displayed.
(C) Binding and neutralization of vaccinee plasma against SARS-CoV-2 WT and Omicron variant. Values denote the geometric mean titer ± SD. Three independent experiments were performed in triplicate.
(D) Illustration of RBD mAbs, RBM mAbs, and non-RBM mAbs in plasma, as measured by BLI (left). Correlation coefficient between plasma neutralization ID50 against the Omicron variant with RBM mAbs or non-RBM mAbs (right), analyzed by Spearman’s rank correlation test.
(E) The binding affinity and neutralization of six antibodies. Values indicate the fold change relative to WT.
See also Figures S1, S2, and S3.
Here, we report the design of a bispecific antibody composed of two different human single-domain antibodies, each directed against a distinct highly conserved region on the Omicron variant RBD. It can be efficiently delivered to lung via inhalation and exhibited superior efficacy in mouse models of SARS-CoV-2 infection. The two arms of this bispecific antibody were found to simultaneously and synergistically target two epitopes of a single RBD, providing unique binding properties and broad neutralizing potency. These results also highlight the importance of cryptic epitopes outside the RBM in neutralization of SARS-CoV-2 Omicron and other variants.
Results
Broad neutralization of Omicron variant by non-RBM antibodies
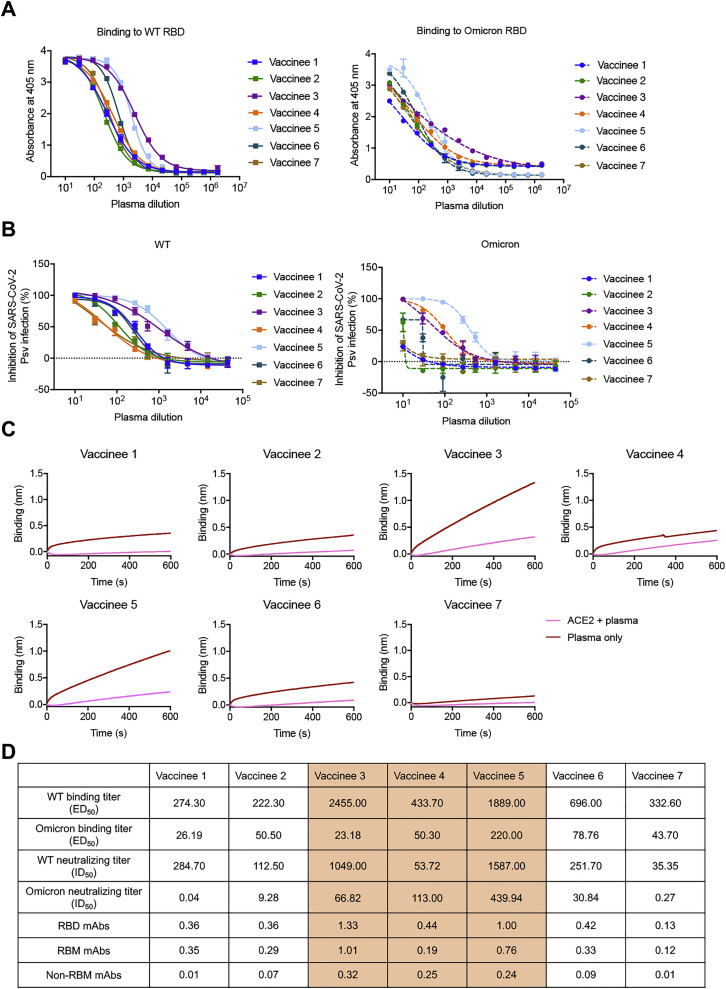
To assess the extent of immune escape of the SARS-CoV-2 Omicron variant, we first mapped its mutations on the RBD and compared these regions with the binding epitopes of some previously reported antibodies (Lu et al., 2021; Yu et al., 2020; Figure S1 ). According to the spatial overlapping of antibody epitopes and viral mutations, the RBD-targeting antibodies can be grouped into three types: those that bind to the RBM, those that bind the cryptic epitopes hidden or partially hidden inside the trimeric interface, and antibodies that bind to lateral surface epitopes outside the trimeric interface (Figure 1A). This classification was comparable to a study of a large panel of SARS-CoV-2 antibodies, which classified epitopes into RBM, inner face, and outer face (Hastie et al., 2021), corresponding to our groups of RBM, cryptic epitopes, and lateral surface epitopes. Evidently, more mutations were involved in the epitopes of apex RBM-binding antibodies than the other two clusters (Figure 1B). To confirm this finding, we measured the binding and neutralization titer of plasma collected from 7 vaccine recipients who received three doses of inactivated SARS-CoV-2 vaccine. The average median binding titer (ED50) of serum declined by 12.8-fold for Omicron RBD, while the neutralizing potency (average median neutralizing titer, ID50) decreased by 5.1-fold against Omicron pseudovirus (Figures 1C, S2 A, and S2B). Interestingly, we found that some vaccinees had high binding and neutralizing antibody titers against the wild-type (WT) SARS-CoV-2 but relatively low neutralization titers against the Omicron variant. To understand the underlying mechanism, we performed epitope binning by saturating the RBM of the SARS-CoV-2 RBD with ACE2, and then loaded plasma to allow the binding of non-RBM antibodies (Figures 1D, S2C, and S2D). The Spearman’s rank correlation test was applied to measure the potential relationship between ID50 with RBM- or non-RBM-binding antibodies. This analysis revealed a high correlation between the binding of non-RBM antibodies in plasma and the Omicron neutralizing potency, implying the functional importance of non-RBM antibodies.
Figure S1.
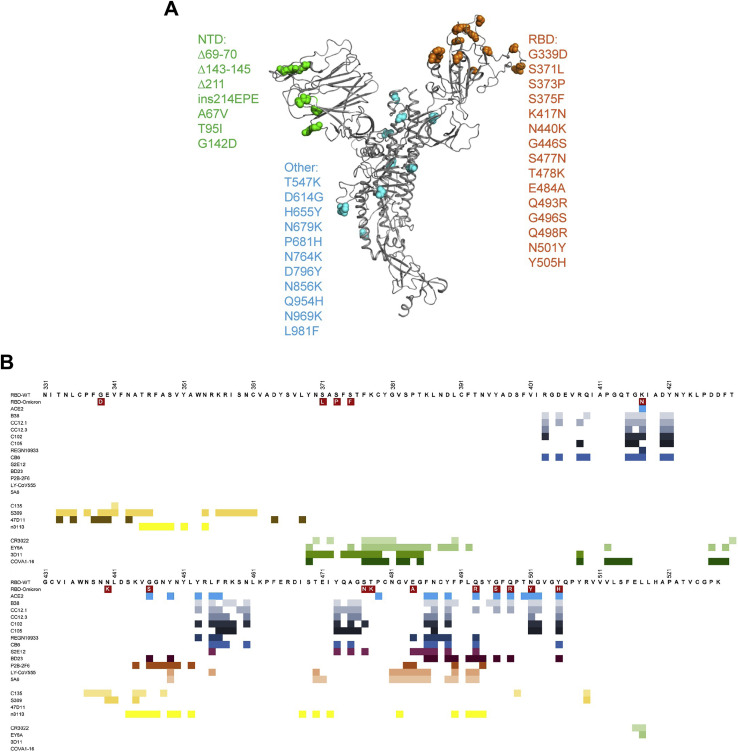
Illustration of mutations on spike of Omicron variant and epitopes for RBD-targeting antibodies, related to Figure 1
(A) The mutations in Omicron variant are shown on spike. The monomeric spike is shown as gray cartoon. The mutations are depicted as spheres in indicated colors. (B) Epitopes for ACE2 and three classes of RBD-targeting antibodies. The residues involved in interaction with antibodies were calculated by ePISA (Proteins, Interfaces, Structures, and Assemblies) and COOT (Crystallographic Object-Oriented Toolkit) and are depicted in colored squares.
Figure S2.
The Omicron variant reduces the binding activity and neutralizing potency of boosted vaccinee plasma, related to Figure 1
(A) Binding curves for WT or Omicron RBD by individual plasma, as determined by ELISA.
(B) Neutralization for WT or Omicron pseudoviruses by individual plasma.
(C) The RBD mAbs, RBM mAbs, and non-RBM mAbs in boosted vaccinee plasma were measured by competition assay using BLI.
(D) Summary of the ED50, ID50 of boosted vaccinee plasma against WT or Omicron variant. The RBD mAbs, RBM mAbs, and non-RBM mAbs in plasma denoted by binding signal.
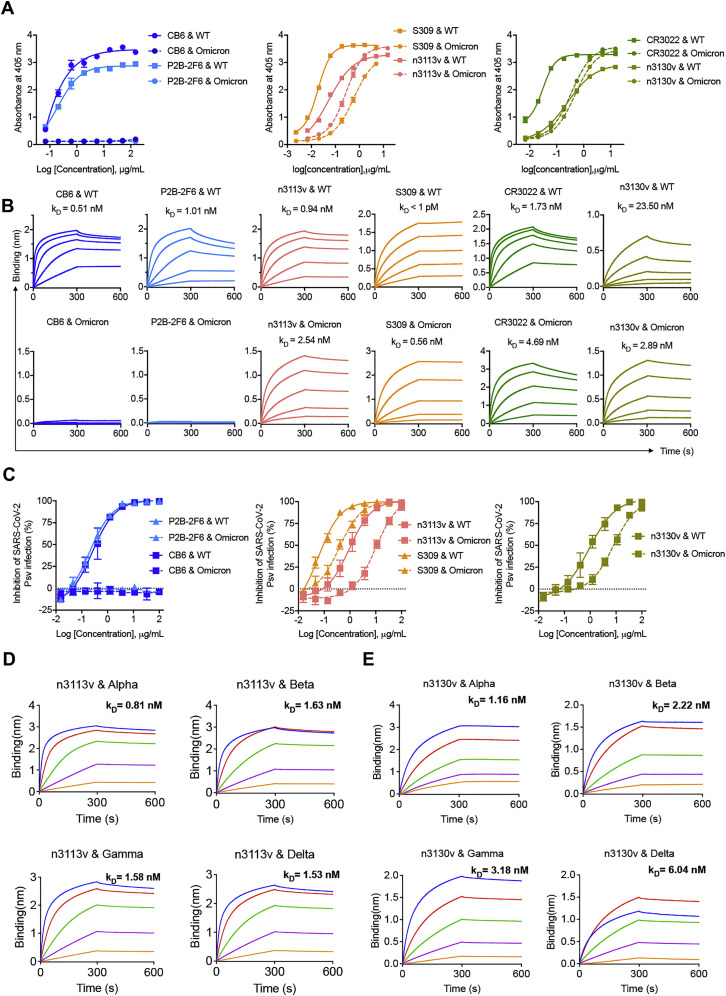
Next, we evaluated the binding ability and neutralizing potency of representative antibodies that occupy three different types of binding sites. Six representative antibodies were cloned, expressed, and purified. We found that CB6 (Shi et al., 2020) and P2B-2F6 (Ju et al., 2020), monoclonal antibodies that bind to RBM and potently block ACE2 from binding to the SARS-CoV-2 RBD, both completely lost Omicron RBD-binding ability and neutralizing potency against Omicron pseudovirus (Figures 1E and S3). In contrast, the potency of some antibodies recognizing the lateral surface epitopes, exemplified by S309 (Pinto et al., 2020) and n3113v, a mutant of previously reported human single-domain antibody n3113 (Wu et al., 2020; Yang et al., 2021), was less affected by Omicron mutations (Figures 1E and S3). Similarly, the antibody CR3022 targets highly conserved cryptic epitopes inside the trimeric interface (Tian et al., 2020; Yuan et al., 2020) and none of the mutations found in Omicron were involved in CR3022 interaction (Figure 1B); therefore, binding of CR3022 to Omicron RBD was only partially diminished (Figures 1E and S3). Such a negligible influence by Omicron on antibody potency was also found in n3130v, a variant of human single-domain antibody n3130 (Wu et al., 2020), which may have epitopes partially overlapping with CR3022, as revealed by its competition with CR3022 for the binding to the SARS-CoV-2 RBD.
Figure S3.
Binding affinity and neutralization of distinct antibody clusters to WT and Omicron variant, related to Figure 1
(A) Binding capacity of antibodies to WT and Omicron RBD, as measured by ELISA.
(B) Binding kinetics of antibodies to WT and Omicron RBD, as measured by BLI.
(C) Neutralization for WT or Omicron pseudoviruses by antibodies.
(D and E) Single-domain antibody n3113v (D) and n3130v (E) exhibited broadly binding capacities against VOCs with high affinity, as measured by BLI.
Taken together, our results suggest that the ACE2-competing antibodies may play a pivotal role in the immune escape of the SARS-CoV-2 Omicron variant. Importantly, two conserved RBD regions targeted by broadly neutralizing antibodies were identified, including a part of the lateral surface (exemplified by n3113v epitope) and the cryptic site inside the trimeric interface (exemplified by n3130v epitope).
Design of bispecific single-domain antibody bn03
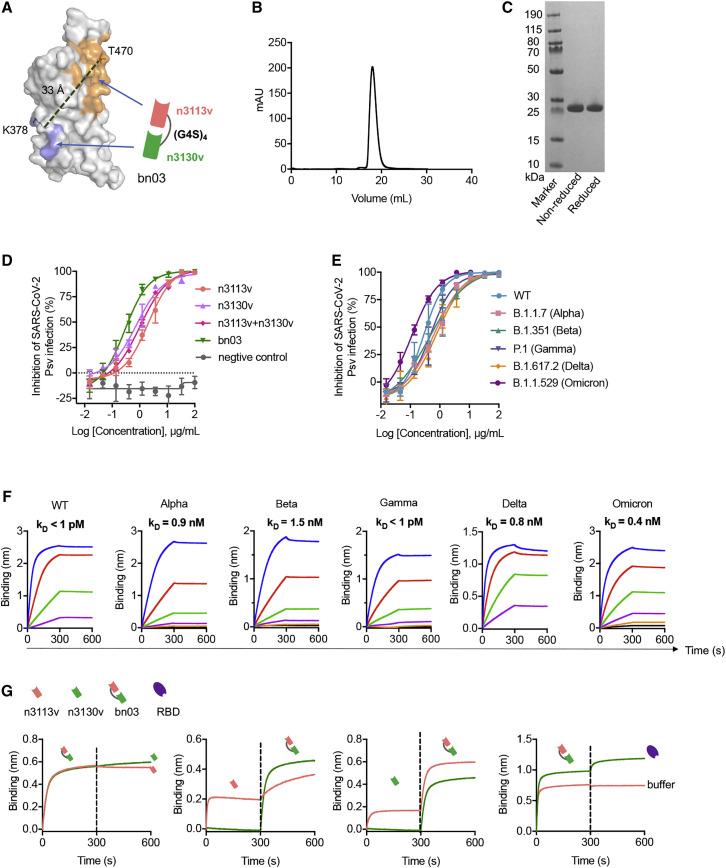
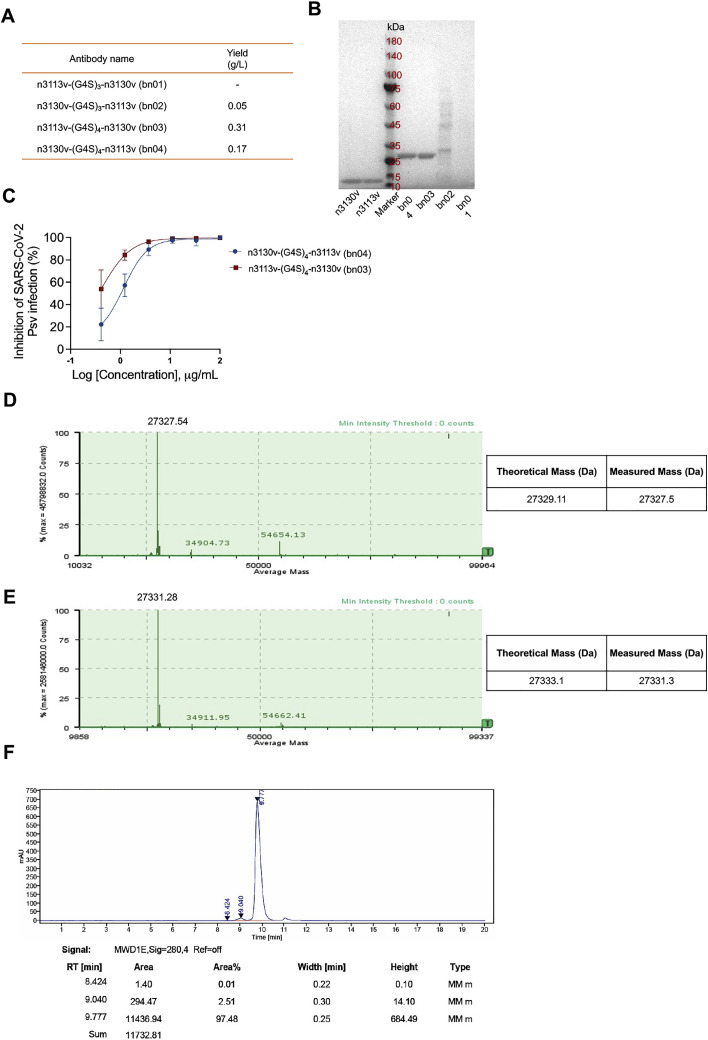
To increase neutralizing breadth for resistance against viral evasion, one strategy is the use of two or more antibodies in combination targeting different epitopes (Baum et al., 2020). An alternative approach is to generate bispecific antibodies, which may have more advantages than the cocktail strategy, such as lower cost and higher efficacy (De Gasparo et al., 2021). We found that both the single-domain antibodies n3113v and n3130v displayed exceptional binding breadth and potency (n3113v, 0.81–2.54 nM; n3130v, 1.16–6.04 nM) to the RBDs of all the five VOCs (Figures S3 B and S3D). To further ensure neutralizing breadth, we connected two single-domain antibodies (n3113v and n3130v) using flexible polypeptide linkers composed of glycine and serine that are widely used in the construction of therapeutic fusion proteins. A panel of bispecific antibodies was generated by combing the structure-based rational design and optimization of linker length, different variants of each arm, and the order of n3113v and n3130v. We hypothesized that n3113v, identified from a phage-displayed library of n3113 mutants, might possess similar binding epitopes to that of n3113. The antibody n3130v was predicted to have partially overlapping epitopes with the antibody CR3022. Therefore, three or four repeats of the GGGGS motif were selected as the linker between n3113v and n3130v in order to fit the measured distance (roughly 30–40 Å) between epitopes of n3113 and CR3022 (Figure 2 A). The bispecific antibodies with (G4S)4 linkers were highly expressed in soluble form in E. coli, while that with (G4S)3 linkers were poorly expressed, possibly due to the misfolding of the antibodies (Figure S4 A). The antibody n3113v-(G4S)4-n3130v, designated as bn03, was chosen for further studies due to its superior biophysical properties and neutralization potency (Figures S4A and S4B). The bispecific antibody bn03 exists in pure monomeric form with a molecular mass of 27 kDa, as demonstrated by size exclusion chromatography (Figure 2B), as well as sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 2C) and mass spectrometry analysis (Figures S4D and S4E) performed under reducing or non-reducing conditions. Furthermore, high-level (grams/liter) production of bn03 was obtained in Chinese hamster ovary (CHO-K1) cells, and a purity of more than 97% was achieved by a single-step downstream process using protein-A chromatography (Figure S4F).
Figure 2.
The design of bispecific single-domain antibody bn03
(A) The bispecific single-domain antibody bn03 contains n3130v and n3113v linked with a linker (GGGGS)4. n3113v and n3130v are colored in pink and green, respectively. The RBD is depicted as gray surface with the epitopes of n3113v and CR3022 (targeting the same epitopes with n3130v) highlighted in orange and blue, respectively. K378 and T470 that are involved in the recognition of CR3022 and n3113, respectively, are shown as sticks. The distance between Cα of K378 and T470 was measured and is labeled in dashed green lines.
(B) Size exclusion chromatography profile of the bispecific antibody bn03.
(C) Reducing and non-reducing SDS-PAGE analysis of bn03.
(D) Neutralization of SARS-CoV-2 pseudoviruses by a panel of single-domain antibodies, including n3113v, n3130v, cocktail of n3113v and n3130v, and bn03. Three independent experiments were performed in triplicate.
(E) Neutralizing potency of bn03 against pseudoviruses of WT and five VOCs. Three independent experiments were performed in triplicate.
(F) Binding affinity of bn03 to RBDs of WT and five VOCs. The KD values are shown.
(G) The bispecific single-domain antibody bn03 simultaneously binds two distinct epitopes on the RBD. The immobilized RBD was incubated with bn03, n3113v (orange), or n3130v (green) until saturation and then incubated with second antibody or the RBD. The binding curves were monitored.
See also Figure S4.
Figure S4.
The properties and neutralization of bispecific single-domain antibodies containing n3113v and n3130v connected with different linkers, related to Figure 2
(A) Four types of bispecific single-domain antibodies (bn01-04) were expressed by E. coli and purified by Ni-NTA resin. The yield was shown. “-” represents no detectable.
(B) SDS-PAGE analysis of n3113v, n3130v and bispecific single-domain antibodies (bn01-04).
(C) Neutralizing activity of bn03 and bn04 against SARS-CoV-2 WT pseudovirus.
(D) Mass spectrometry analysis of intact bn03 using ESI.
(E) Mass spectrometry analysis of reduced bn03 using ESI.
(F) HPLC analysis of bn03.
Binding and neutralization properties of bn03
We found that bn03 exhibited more potent neutralizing efficacy against SARS-CoV-2 than the cocktail of n3113v and n3130v (Figure 2D), denoted by a lower IC50 value of 0.28 μg/mL, which is 4-fold lower than that of the cocktail. Next, we carried out pseudovirus neutralization experiments to characterize the neutralizing breadth of bn03 against WT SARS-CoV-2and all five VOCs. As shown in Figure 2E, bn03 could broadly neutralize all the six viruses, with the IC50 ranging from 0.11–0.76 μg/mL. In alignment with this, we found that bn03 could bind strongly to the RBDs of WT and VOCs with an affinity around 1 nM or lower (Figure 2F). Furthermore, bn03 was still able to bind the RBD that had already been saturated with either n3113v or n3130v (Figure 2G), suggesting that the two arms of bn03 are functional and simultaneously bind to the RBD with no steric hindrance.
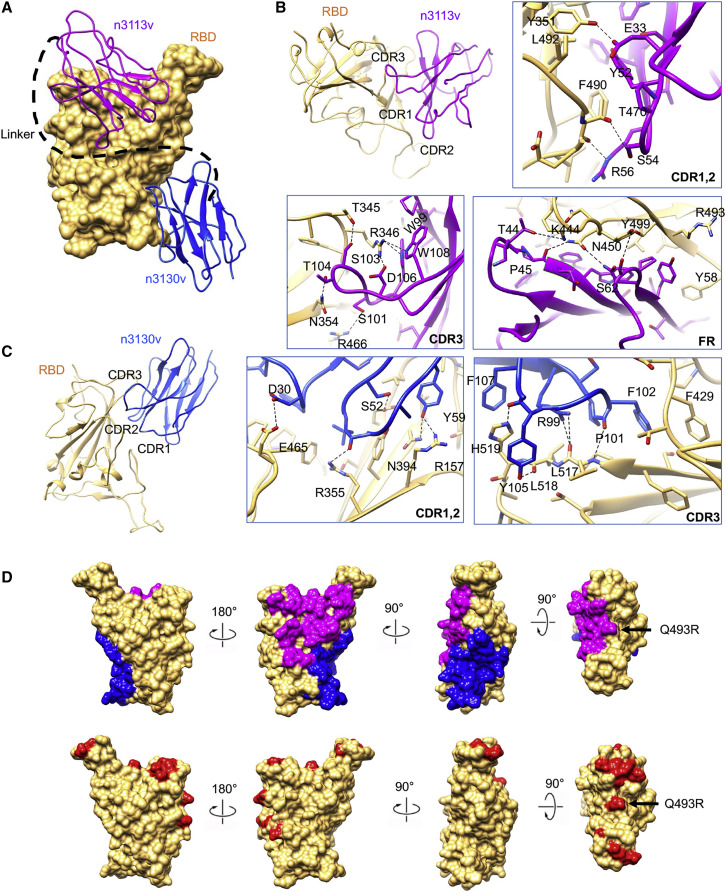
Two arms of bn03 bind simultaneously and synergistically to RBD revealed by cryo-EM structure of Omicron S-bn03
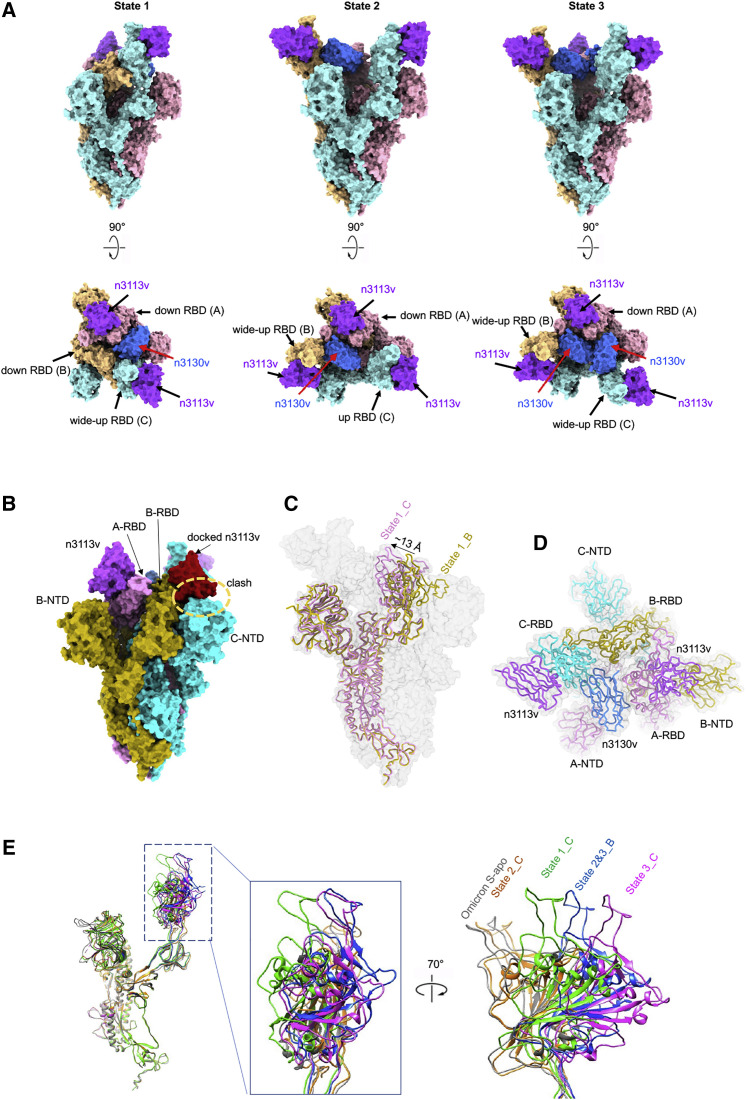
To further investigate the neutralization mechanism of bn03, we determined the cryoelectron microscopy (cryo-EM) structure of the prefusion-stabilized SARS-CoV-2 Omicron S ectodomain trimer complexed with bn03 (Omicron S-bn03). Cryo-EM characterization revealed three conformational states of the complex. The three structures were determined at a resolution of 3.3 (state 1), 2.9 (state 2), and 3.0 Å (state 3), respectively (Figures 3A and S5). In state 1, two arms (n3113v and n3130v) of bn03 simultaneously bind to a single RBD, while one of the other two RBDs is bound by n3113v. The n3113v arm of bn03 occupies a side surface epitope on the RBD, while the n3130v arm recognizes a cryptic epitope located inside the spike protein trimer interface. In state 2, one RBD is bound by the two arms of bn03 and the other two RBDs are bound by two n3113v. In state 3, the spike protein is able to accommodate two bn03 by two RBDs, and the remaining RBD is bound by n3113v (Figure 3 A).
Figure 3.
Cryo-EM structures of Omicron S trimer in complex with bispecific single-domain antibody bn03
(A) Bispecific single-domain antibody bn03 binds to Omicron S trimers in 3 states. Two perpendicular views of Omicron S-bn03 are depicted as surface, with n3113v in magenta, n3130v in blue, and the trimeric spike in cyan, pink, and yellow.
(B) Superposition of RBD-bound n3113v on B-RBD of state 1 structure indicates a clash. A-RBD-bound n3113v is shown as magenta surface. The docked n3113v is shown in red surface.
(C) Alignment of down-state spike from molecule C (pink) and molecule B (yellow) of state 1 structure. The down-state spike is shown as cartoon. Protomer A and C in Omicron S-bn03 structure are depicted as gray surface.
(D) Quaternary binding of n3130 and RBDs. Top view of the state 1 structure shown in cartoon representation.
(E) Comparison of the up-RBDs of bn03-bound Omicron S complexes with the up-RBD of Omicron S-apo (gray). The n3113v-bound state-2 C-RBD is colored orange. The n3113v-n3130v-bound state-1-C, state 2&3-B, and state 3-C are colored green, blue, and magenta, respectively.
See also Figure S5.
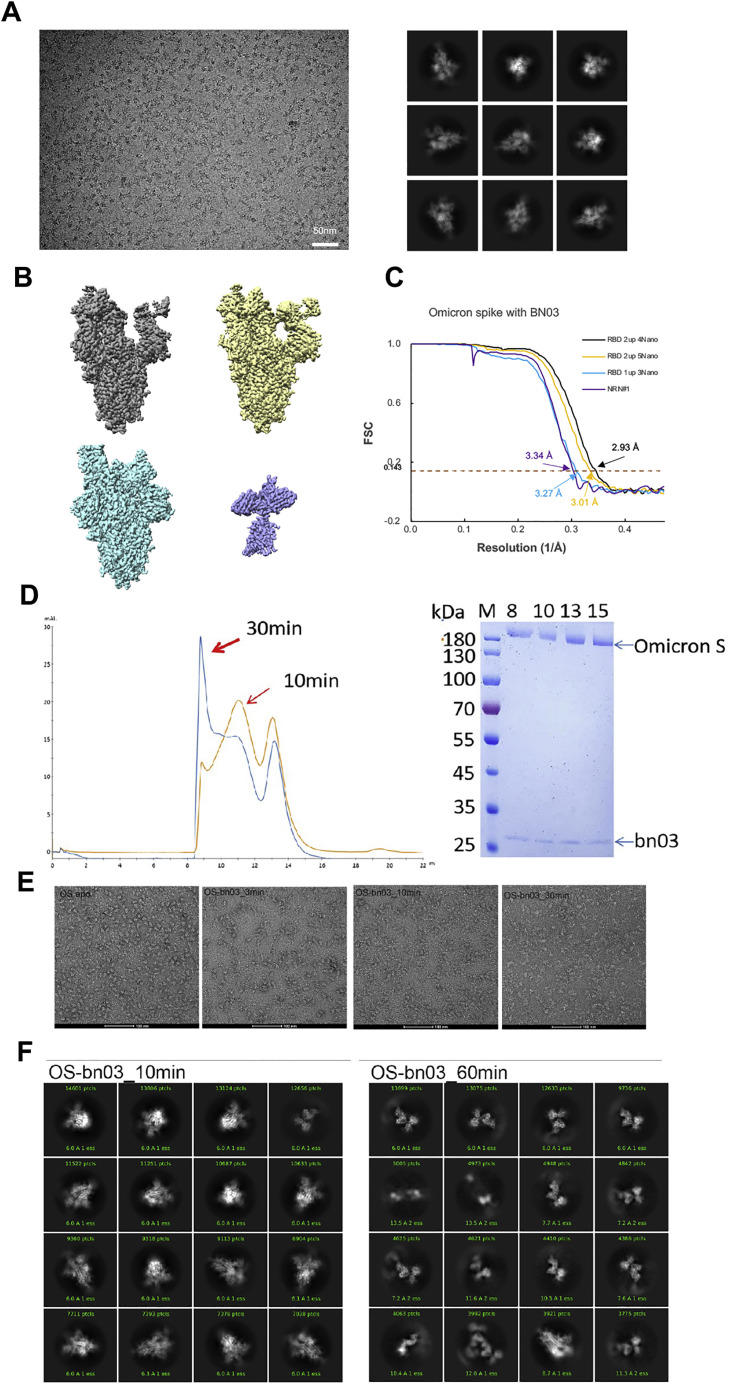
Figure S5.
Cryo-EM data collection and processing of bn03 bound SARS-CoV-2 Omicron S, related to Figure 3
(A) Representative electron micrograph and 2D classification results of XG014 bound SARS-CoV-2 S.
(B) The reconstruction map of the complex structures at three states and one local refinement map. (C) Gold-standard Fourier shell correlation curves for each structure. The 0.143 cut-off is indicated by a horizontal dashed line.
(D) Purification of bn03-Oimcron S complex. The gel-filtration curved showed that incubation of bn03 with Omicron-S-induced aggregation of S protein.
(E) Negative stain images of bn03-Omicron S complex, showing that incubating with bn03 for 30 min leads to Omicron S trimer disassembly. The disassembled monomers tend to aggregate.
(F) The 2D classification result of cryo-EM data collected using 10-min incubation complex and 60-min incubation complex.
In the state 1 complex structure (∼15% of the particles), the bispecific antibody bn03 (n3113v-n3130v) binds on the “up” state RBD and one molecule of n3113v binds on one “down” state RBD (Figure 3A). To simplify the presentation, three protomers of the spike protein were defined counterclockwise as A, B, and C. Interestingly, among the two “down” state RBDs (A-RBD and B-RBD), only A-RBD binds n3113v. Superimposition of the A-RBD-n3113v to the B-RBD demonstrated that n3113v would clash with the neighborhood C-N-terminal domain (NTD) (Figure 3B). Moreover, structural alignments of A and B revealed a ∼13 Å inward shift of A-RBD toward the trimeric axis (Figure 3C). The inward movement of RBD expanded the space between RBD and the neighboring NTD, allowing the accommodation of n3113v. While the complementarity-determining regions (CDRs) of n3130v interact with the C-RBD, the frame regions of n3130v interact with the adjacent A-RBD (Figure 3D). It is reasonable to speculate that the domain shift of A-RBD was induced by n3130v through its quaternary interaction with the A- and C-RBD, indicating that the binding of n3130v synergistically promoted the interaction between n3113v and “down” state RBD.
The bispecific antibody induces Omicron S trimer to unstable wide-up states
In the state 1 structure, binding of n3130v to the RBD induced an obvious movement of the RBD, including a clockwise rotation of 26° and an outward shift of ∼30 Å of the C-RBD when compared with the “up” state RBD of Omicron S-apo structure (Mannar et al., 2021), so-called “wide-up” state RBD (Figure 3A).
The majority of S-bn03 particles are in the state-2 conformation (∼55% particles). In this conformation, only A-RBD is in the down position, which binds an n3113v. Both promoter B and C have their RBD open but at different angles, resulting from the different binding modes of single-domain antibodies. C-RBD only binds n3113v, while B-RBD binds bispecific n3113v-n3130v (Figure 3A). The n3113v-bound up-C-RBD stays at a similar conformation as the up-RBD in Omicron S-apo structure. The bispecific n3113v-n3130v-bound B-RBD opens up more widely than the wide-up C-RBD in state 1, by rotating ∼10° away from the axis of the trimer (Figure 3E).
State 3 (∼30% particles) retains almost identical protomer A and B structures. The major difference between state 2 and state 3 is located at the protomer C-RBD region. Other than the B-RBD, C-RBD also binds with n3113v and n3130v simultaneously, resulting in a dramatic conformational change of the RBD. When compared with the up-RBD of Omicron S-apo structure, the RBD rotates ∼40° and shifts ∼45 Å at the tip. The binding of the second n3130v to the RBD requires more space and pushes the RBD open very widely.
In summary, the binding of n3130v induces RBDs to the wide-up state and introduces instability in RBDs. This is consistent with our observation that incubation of bn03 and Omicron S trimers would result in the gradual disassembly of the trimers into monomers (Figure S5 ). Intriguingly, fusion of the human IgG1 Fc to the single-domain antibodies improved the neutralization activity of n3113v but not the n3130v (Figure S6 A). These findings underscore the advantages of the small-sized n3130v (13 kDa) and bn03 (27 kDa) to penetrate deeply into the cryptic spike protein interface and effectively neutralize SARS-CoV-2.
Figure S6.
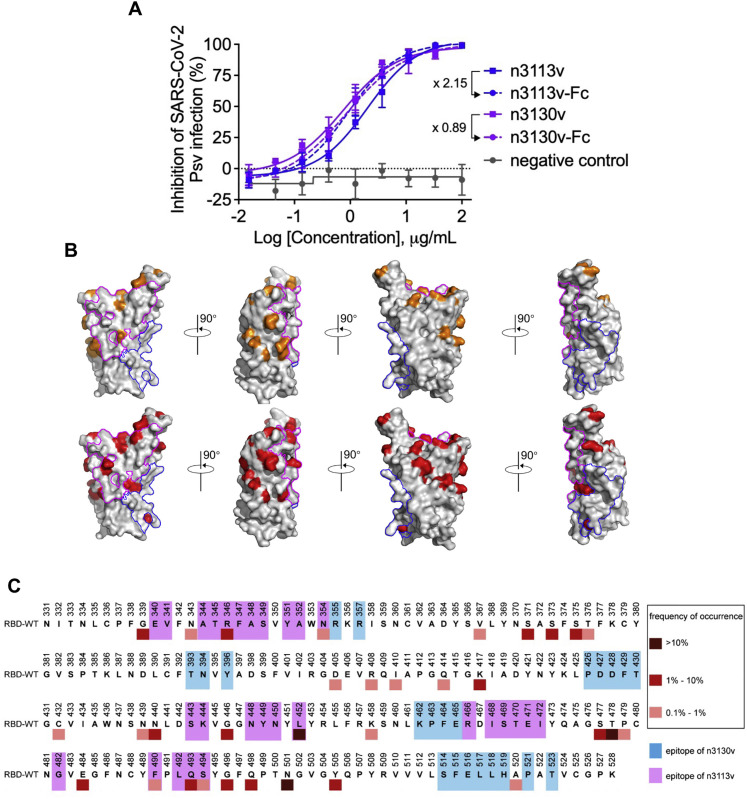
Neutralizing activity of n3113v, n3113v-Fc, n3130v, and n3130v-Fc against SARS-CoV-2 WT pseudovirus, and illustration of epitopes of single-domain antibodies on RBD, related to Figure 4
(A) Fold change in IC50 values reflects the increase or decrease of neutralizing activity post fusing to Fc region.
(B) The RBD was represented as gray surface. Mutations found in VOCs (Alpha, Beta, Gamma, Delta, and Omicron) were highlighted in orange in the upper row. Mutations in RBD with high frequency (occurred in over 0.1% of the total reported sequences) from public database (https://ngdc.cncb.ac.cn/ncov/variation/spike) were indicated in red in the lower row. The epitope of n3130v and n3113v was circled in blue and magenta lines, respectively.
(C) Reported mutations in RBD from public database (https://ngdc.cncb.ac.cn/ncov/variation/spike) were marked by squares in different color. Residues involved in the binding to n3130v and n3113v were highlighted in blue magenta, respectively.
Highly conserved epitopes on Omicron spike protein revealed by bn03
Both n3113v and n3130v target epitopes outside the RBM region (Figure 4 A). The binding of n3113v with S-RBD buries a 973 Å2 surface area. A total of 30 residues from RBD are involved in the interaction (Figure 4B). The interactions of n3113v are mainly mediated by hydrogen bonds between the CDRs and RBDs. Residues R346, T345, N354, Y351, and T470 from the RBD participate in the interaction with the CDRs, forming eight pairs of hydrogen bonds and one pair of salt bridges. The R346 is engaged in two hydrogen bonds (with W99 and W108) and a salt bridge (with D106). Other than the CDR, the framework region 44–48 also forms tight interactions with the RBD by forming hydrogen bonds and through hydrophobic interactions.
Figure 4.
Two conserved epitopes recognized by bn03
(A) Close-up view of the interactions between bn03 and Omicron RBD. The Omicron RBD is displayed in yellow surface. n3113v and n3130v are shown as cartoon colored in magenta and blue, respectively.
(B and C) The interaction of n3113v (B) and n3130v (C) with Omicron RBD. The residues involved in interactions are represented as sticks. Polar interactions are indicated as dotted lines.
(D) The epitopes of n3113v and n3130v on Omicron RBD. Omicron RBD is shown as yellow surface. Four degrees of surface representation of Omicron RBD with epitopes of n3113v and n3130v highlighted in magenta and blue, respectively, are shown. The mutations in Omicron are colored in red.
See also Figure S6.
The binding epitope of n3130v is uncommonly inside the trimeric interface of the spike protein, with the only similar binding mode reported for a broadly neutralizing antibody S2H97 (Starr et al., 2021; Figure 4C). In total, 20 residues from the RBD are involved in the interactions, burying an 842 Å2 surface area. All three CDRs are involved in the binding by forming hydrogen bonds and through hydrophobic interactions. CDR3 forms 5 hydrogen bonds with the loop region of the RBD (515–519). H519 is clapped by F107 and Y105 by π-π stacking. F102 also forms π-π stacking with F429. Besides, The hydrogen bonds formed by residue D30 on CDR1 with E465, as well as by Y59 on CDR2 with R157 and N394, further strengthen the interaction.
In total, 49 out of 50 residues involved in n3113v and n3130v interactions remain unchanged between the WT RBD and Omicron RBD (Figure 4D). R493 is the only residue that was mutated from glutamine. R493 is involved in a π-cation interaction with Y58 from n3113v. The mutation from glutamine and arginine did not affect the interaction, thus not affecting the binding affinity of n3113v to the RBD. The well-conserved epitopes explain the broad neutralization activity of bn03.
Furthermore, we found that the epitopes of n3130v, buried deep inside the trimeric interface of the spike protein, were conserved not only among all the VOCs (Figure S6B) but also the naturally occurring SARS-CoV-2 variants (Figure S6C). We retrieved 3,868,697 full-length SARS-CoV-2 nucleotide sequences uploaded in the public database (https://ngdc.cncb.ac.cn/ncov/variation). These sequences account for 29,624 mutational events in the spike protein and 1,034 in RBDs. Among the mutations in RBDs, those with an occurrence frequency >0.1% were defined as “high frequency”. Notably, the epitope of n3130v was found to be highly conserved among all the variants of high frequencies that have emerged since the outbreak of SARS-CoV-2 (Figures S6B and S6C).
Inhalation as an effective route for delivery of bn03 to lung
It has been reported that camelid-derived nanobodies can be delivered via inhalation for the treatment of pulmonary disease due to their favorable properties of small size, high stability, and solubility (Cunningham et al., 2021; Muyldermans, 2013). Because of the similarities in structure and biophysical properties between nanobodies and human single-domain antibodies, here we explored the potential of delivering human single-domain antibodies via inhalation.
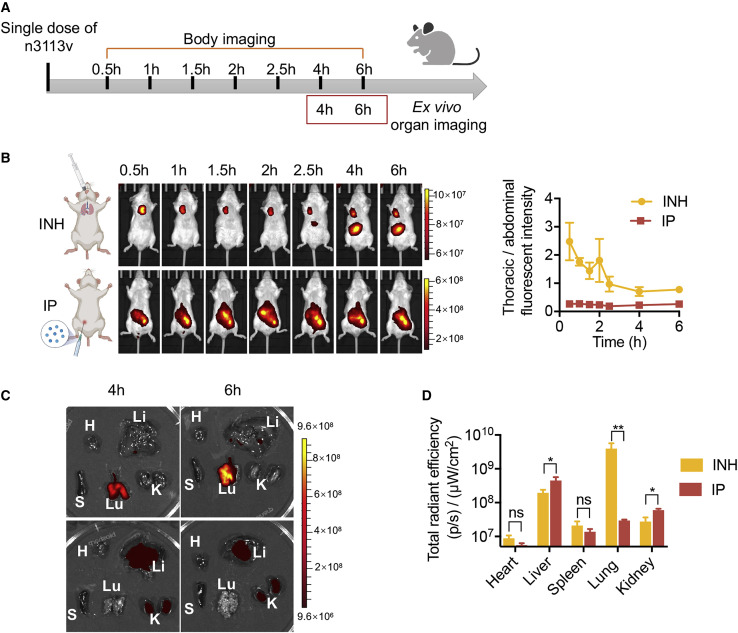
To compare the performance of inhalation administration (INH) with the conventional intraperitoneal (IP) delivery of antibodies in mice, we first conjugated the single-domain antibody n3113v with DyLight 800 to enable in vivo imaging. We employed a previously described high-pressure microsprayer for the quantitative and uniform intratracheal dosing of therapeutics to the lung of mice as a fine aerosolized spray (Kormann et al., 2011; Sakagami, 2020). To assess tissue distribution, BALB/c mice were treated with n3113v through INH or IP delivery (Figure 5 A). Bio-imaging of the mice at different time points indicated that INH effectively delivered the single-domain antibodies to the chest cavity, while in the IP delivery group, the antibodies were largely retained within the abdomen (Figure 5B). We also dissected the mice and measured the fluorescence in different organs at 4 and 6 h post antibody administration (Figure 5C). Most of the antibodies were detected in lung in the INH group. In contrast, the antibodies concentrated mainly in liver and kidney but very few in lung in the IP delivery group (Figure 5D). Therefore, we further quantified the antibody concentrations in lung and plasma and confirmed that a substantially larger number of antibodies could be delivered to lung via INH than IP administration, resulting in significantly higher therapeutic efficacy in animals (Figure 6 ).
Figure 5.
Effective delivery of single-domain antibody to lung via inhalation
(A) Schematic diagram of n3113v biodistribution in mice by inhalation and intraperitoneal injection.
(B) Bio-imaging of mice body at different time points (left) and fluorescence intensity ratio of thoracic cavity against abdomen (right, n = 3).
(C) Mice were sacrificed at 4 and 6 h post antibody administration and dissected organs were imaged (n = 3). Lu, lung; K, kidney; H, heart; S, spleen; Li, liver.
(D) Quantification of organ tissue fluorescence signal.
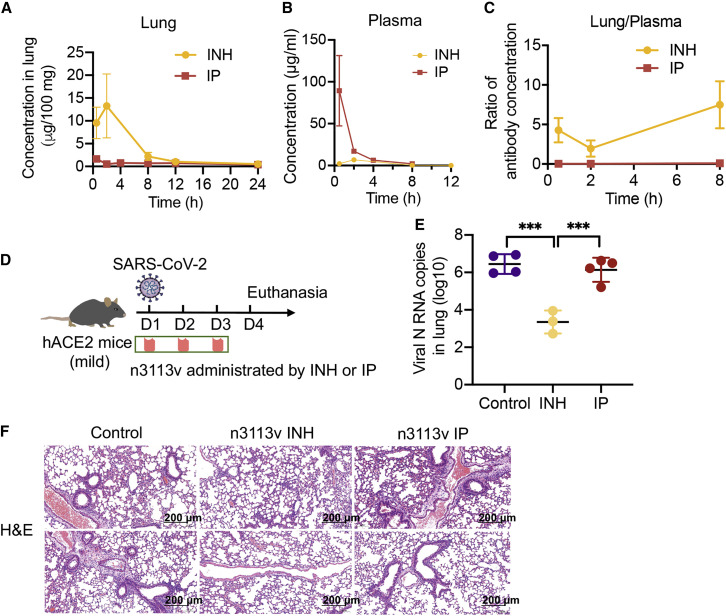
Figure 6.
Inhalation of single-domain antibody exhibits effective therapeutic effects
(A–C) Concentration of n3113v in lung (A), plasma (B), and the ratio of n3113v in lung relative to plasma (C), n = 2.
(D) Experimental design of therapeutic evaluation of n3113v by inhalation and intraperitoneal injection in authentic SARS-CoV-2-infected hACE2 transgenic mice.
(E and F) Lung viral load (E) and lung histology (F) in infected mice with indicated treatments.
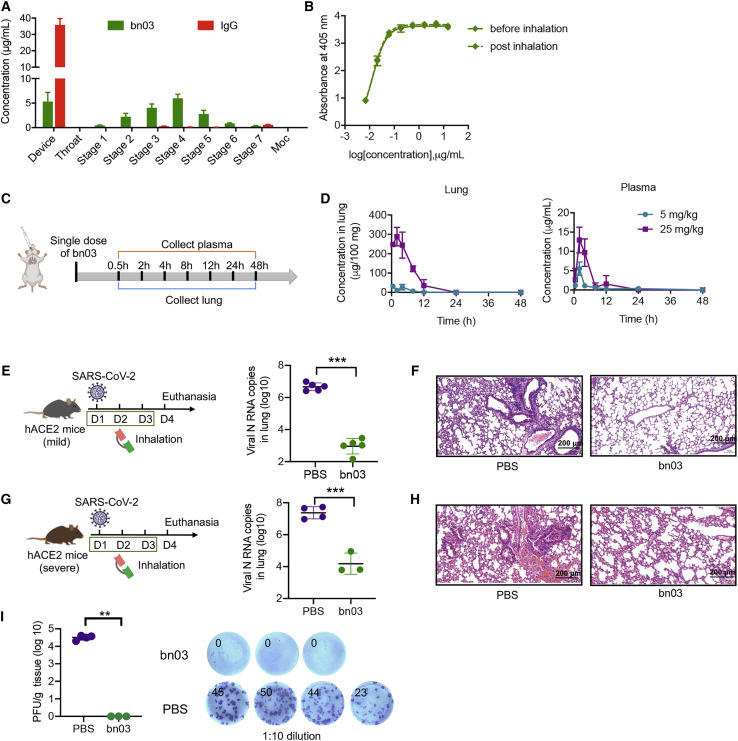
Next, we compared the inhalation potential of the bispecific antibody bn03 with the conventional IgG antibody. For this purpose, we used the next generation impactor (NGI), a high-performance cascade impactor with seven stages, to determine the concentration of bn03 on each cup and to investigate the aerosol performance of bn03 by using a vibrating mesh nebulizer (Figure 7 A). The bn03 displayed good drug deposition on stage 2 through stage 6, indicating that the cut-off diameter of bispecific single-domain antibody post inhalation was within a range of 3–5 μm. The deposition of inhaled particles in the human lung mainly depends on particle size, which should typically be <5 μm (Patton and Byron, 2007). It is widely accepted that aerosol particles 1–5 μm in size are deposited in the small airways and alveoli, larger particles (>5 μm) in the upper respiratory tract, and the smaller particles (<∼0.5 μm) generally fail to deposit and are expelled during exhalation (Bianco et al., 2021). Therefore, bn03 exhibited the potential for aerosol deposition within the respiratory tract. In contrast, most of the IgG antibodies remained in the device and barely distributed to the impactor stages under the same conditions. These results indicate that the bispecific single-domain antibody is more inhalable than IgG.
Figure 7.
Inhalation of bn03 effectively treats SARS-CoV-2 infection in mice
(A) The aerosol performances of bn03 and IgG (S309) by NGI. The concentration of aerosolized particles in different stages is quantified. Three independent experiments were performed in triplicate.
(B) Comparison of the binding ability of bn03 to RBD before or after inhalation using the high-pressure microsprayer. Three independent experiments were performed in triplicate.
(C) Experimental design for evaluation of bn03 in mice by inhalation. BALB/c mice received bn03 via inhalation at a dose of 5 or 25 mg/kg, and blood and lung samples were collected at indicated time points.
(D) Antibody concentration of bn03 in lung (left, n = 2) and plasma (right, n = 2).
(E–H) Therapeutic efficacy of bn03 via inhalation in a hACE2 transgenic mouse model with mild (E) or severe symptoms (G) after authentic SARS-CoV-2 infection. Lung viral load (E and G) and lung histology (F and H) in mice with indicated treatments.
(I) Determination of focus-forming units (FFUs) with lung homogenate in 1:10 dilutions. Lung homogenate of each mouse was serially diluted 10-fold and incubated with Vero E6 cells. The FFUs of each well were counted. The total FFUs of lung tissue were determined and represented (n = 3–4). The right panel shows representative FFUs. Ordinary one-way ANOVA was used in the statistical analysis. Statistical significance is represented as ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; ns, no significance.
See also Figure S7.
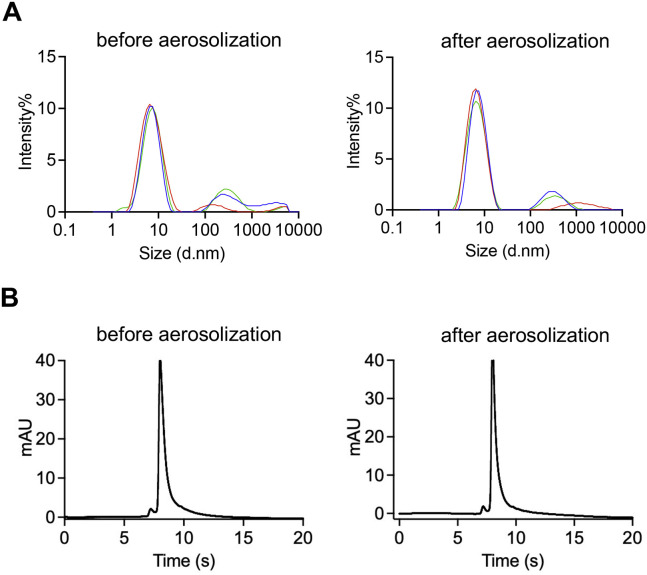
To test whether INH affects the performance of bn03, we collected bn03 before or after aerosolization by the high-pressure microsprayer. Neither aggregation nor degradation of bn03 after aerosolization was observed by dynamic light scattering and high-performance liquid chromatography (HPLC) (Figure S7 ). Furthermore, bn03 bound strongly to the RBD before and after inhalation and displayed unchanged binding efficacy (Figure 7B).
Figure S7.
The properties of bn03 were determined by DLS and HPLC before and after aerosolization by microsprayer aerosolizer, related to Figure 7
(A) Properties of bn03 determined by DLS and (B) properties of bn03 determined by HPLC.
Finally, a single-dose in vivo pharmacokinetics study by inhalation route was carried out in mice using the high-pressure microsprayer (Figure 7C). After inhalation, two mice per group were euthanized at indicated time points for collection of blood and lung. The antibody concentration in plasma and lung was quantified. The results showed that bn03 concentrations in lung were much higher than in circulating blood following inhalation (Figure 7D). The peak concentration of bn03 in lung was 22-fold higher than that of plasma. These results suggest that human single-domain antibodies can be directly delivered to lung by inhalation.
Inhalation of bn03 effectively treats SARS-CoV-2 infection in hACE2 mice
To evaluate the therapeutic effect of inhaled bn03 in vivo, two hACE2 transgenic mouse models of mild and severe symptoms were used. The mouse model of mild COVID-19 was generated using chimeric transgenic mice of hACE2 and mACE2 in which the extracellular region of mACE2 was replaced with hACE2 via CRISPR-Cas9 (Ma et al., 2020). No visible clinical signs were seen in mice after SARS-CoV-2 infection but viral RNA was detected in lung (Figure 7E). INH of bn03 resulted in a significant 3.6-log reduction in viral RNA copies compared to the PBS control group (Figure 7E) and alleviated lung injury (Figure 7F). Another mouse model of severe symptoms was developed by infecting transgenic mice that are highly susceptible to SARS-CoV-2 infection due to the high-level expression of hACE2 in many organs. Indeed, the lungs of these mice had a higher viral titer (Figure 7G) and experienced evident infection-induced pathology, including inflammatory cell infiltration and bronchiolar epithelium disruption (Figure 7H). We found a 2.9-log reduction in lung viral titer in mice treated with bn03 compared to PBS control mice (Figure 7G). H&E staining of lung tissue further revealed that lung injury had been ameliorated through bn03 inhalation therapy (Figure 7H). To evaluate whether the virus in lung tissue could still possess infectious capacity, the lung homogenates of mice from the severe symptom group were incubated with Vero E6 cells. We found that inhalation of bn03 almost entirely eliminated the live virus in the lungs (Figure 7I), reflecting the efficiency of the bispecific antibody.
Discussion
The continuous emergence of new SARS-CoV-2 variants highlights the urgent need to develop broad-spectrum vaccines and antibodies. We showed that the mutations of the Omicron variant were able to diminish the titer of vaccinated sera and abolish antibody binding, especially when their recognizing epitopes overlap with the ACE2-binding sites. We also evaluated the binding ability and neutralizing potency of representative antibodies that occupy different types of binding sites, and identified two highly conserved regions on the RBD that can be recognized by broadly neutralizing antibodies. It was previously found that the ACE2-competing antibodies constitute the majority of RBD-specific antibodies elicited by natural infection or vaccination (Piccoli et al., 2020). It is possible that such antibodies have driven the immune escape of SARS-CoV-2, resulting in the vast number of mutations in the Omicron variant. In this context, the cryptic epitope revealed by n3130v may have advantages in evading viral escape, as very few antibodies were reported to recognize such sterically hindered epitopes.
Intriguingly, novel epitopes hidden in the trimeric interface of influenza A virus hemagglutinin have also been discovered recently (Yu et al., 2017) and a number of antibodies targeting these epitopes are reportedly capable of neutralizing a broad range of influenza isolates (Bangaru et al., 2019; Turner et al., 2019; Watanabe et al., 2019; Wu and Gao, 2019). These findings support the importance of such cryptic epitopes, which could facilitate the development of universal vaccines and antiviral drugs. Indeed, the epitopes of n3130v were found to be highly conserved not only among all the VOCs but also among all the high-frequency (>0.1%) variants that have emerged since the outbreak of SARS-CoV-2 (Figure S6). Notably, we previously reported that a neutralizing monoclonal antibody against SARS-CoV, CR3022, showed cross-reactive binding to the spike protein of SARS-CoV-2 (Tian et al., 2020). The antibody CR3022, as the first reported monoclonal antibody that was able to bind SARS-CoV-2, was also found to bind the cryptic epitope within the spike protein trimeric interface (Yuan et al., 2020). However, CR3022 could only bind to the three-up “open” state of the SARS-CoV-2 spike protein, and thus exhibited strong binding but negligible neutralization. In contrast, n3130v could bind to all the states of SARS-CoV-2 spike protein, penetrate deeply inside the “sterically hidden” trimeric interface, and neutralize the virus by strongly inducing the Omicron S trimer to unstable wide-up states. These findings could provide important insights into the effects of antiviral antibodies targeting these cryptic and highly conserved epitopes.
Recently, several studies revealed that the Fc domain could augment the protection efficacy of neutralizing antibodies in SARS-CoV-2 infected animals by recruiting immune cells such as monocytes, neutrophils, and natural killer cells through engagement of FcγR (Ullah et al., 2021; Yamin et al., 2021). We employed the lung delivery route to improve the in vivo viral neutralizing potency of the single-domain antibody bn03, but the advantage of additional non-neutralizing effector functions like antibody-dependent cell-mediated cytotoxicity (ADCC) may still be missed, when considering the small size without Fc for better aerosol delivery. Therefore, the bn03-Fc fusion protein administrated via the intravenous route may be another promising strategy for enhancing the virus clearance activity of this single-domain antibody.
Our study demonstrated that human single-domain antibodies can be efficiently delivered to lung and can effectively treat pulmonary disease via INH. These antibodies are derived from fully human sequences and thus may have a better safety profile and clinic efficacy compared to camelid-derived nanobodies. Importantly, the bispecific single-domain antibody, bn03, was able to bind two distinct epitopes simultaneously and synergistically on a single RBD of the SARS-CoV-2 Omicron variant. Its small molecular size (27 kDa) not only confers inhalable properties but also enables the deep penetration of bn03 into the trimeric interface of the spike protein to target the highly conserved cryptic epitopes. Because of its unique epitope profile, we propose that such an antibody could possess exceptional neutralization breadth against current and future SARS-CoV-2 variants.
Limitations of the study
This study has some limitations. Due to resource limitations, plasma samples collected from only 7 vaccine recipients were used in this study. Although a high correlation was observed between the percentage of non-RBM antibodies in plasma of the study participants and the neutralizing potency against the Omicron variant, it is still necessary to test samples from more vaccine recipients to confirm such correlation. Furthermore, we have not tested the dose-dependent therapeutic efficacy of bn03 in hACE-2 transgenic mice. The dose we used in this study was based on previous research, which may provide little guidance for selecting a suitable dosage range in clinical trials.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody | Sigma-Aldrich | Cat# A8592-1MG; RRID:AB_439702 |
| Anti-Human IgG (Fab specific)-Peroxidase antibody produced in goat | Sigma-Aldrich | Cat# A0293-1ML; RRID:AB_257875 |
| Polyclonal rabbit anti-SARS Nucleocapsid protein antibody | Rockland | Cat# 200-402-A50; RRID:AB_828403 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP | ThermoFisher | Cat# 31460; RRID:AB_228341 |
| CB6 IgG | This paper | N/A |
| P2B-2F6 IgG | This paper | N/A |
| S309 IgG | This paper | N/A |
| CR3022 IgG | This paper | N/A |
| Bacterial and virus strains | ||
| DH5α Competent Cells | CWBIO | Cat# CW0808 |
| HB2151 Escherichia coli Strains | This paper | N/A |
| SARS-CoV-2 strain nCoV-SH01 | Fudan University | N/A |
| Biological samples | ||
| Plasma, donor Vac01 | This paper | N/A |
| Plasma, donor Vac02 | This paper | N/A |
| Plasma, donor Vac03 | This paper | N/A |
| Plasma, donor Vac04 | This paper | N/A |
| Plasma, donor Vac05 | This paper | N/A |
| Plasma, donor Vac06 | This paper | N/A |
| Plasma, donor Vac07 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate Buffered Saline solution | HyClone | Cat# SH30256.01B |
| Bovine Serum Albumin (BSA) | Yeasen | Cat# 36101ES25 |
| EZ-Trans | Life-iLab | Cat# AC04L098 |
| Dulbecco's Modified Eagle Medium, high glucose | Meilunbio | Cat# PWL035-1 |
| BirA500 Kit | Avidity | N/A |
| Protein Standard Mix 15 -600 kDa | Sigma-Aldrich | Cat# 69385 |
| HisSep Ni-NTA Agarose Resin 6FF | Yeasen | Cat# 20503ES10 |
| High Fidelity PCR Master | Roche | Cat# 12140314001 |
| IPTG | Yeasen | Cat# 10902ES60 |
| SfiI | New England Biolabs | Cat# R0123L |
| Protein A Resin | GenScript | Cat# L00210 |
| Protein G Resin | GenScript | Cat# L00209 |
| Oxoid™ Tryptone | ThermoFisher | Cat# LP0042T |
| EZ-Link™ Sulfo-NHS-LC-LC-Biotin | ThermoFisher | Cat# A35358 |
| Oxoid™ Yeast Extract | ThermoFisher | Cat# LP0021 |
| Tween 20 | Sigma-Aldrich | Cat# 9005-64-5 |
| TRIzol® Reagent | ThermoFisher | Cat# 335912 |
| 2-Propanol | Sigma-Aldrich | Cat# 67-63-0 |
| Ethyl Alcohol | Sigma-Aldrich | Cat# 64-17-5 |
| ABTS | ThermoFisher | Cat# 002024 |
| TrueBlue™ Peroxidase Substrate | Seracare | Cat# 5510-0050 |
| 0.25% Trypsin EDTA phenol red | Meilunbio | Cat# MA0233 |
| SARS-CoV-2 Spike RBD-His (Alpha) | Sino Biological Inc. | Cat# 40592-V08H18 |
| SARS-CoV-2 Spike RBD-His (Beta) | Sino Biological Inc. | Cat# 40592-V08H85 |
| SARS-CoV-2 Spike RBD-His (Gamma) | Sino Biological Inc. | Cat# 40592-V08H86 |
| SARS-CoV-2 Spike RBD-His (Delta) | Sino Biological Inc. | Cat# 40592-V08H90 |
| SARS-CoV-2 Spike RBD-His (Omicron) | ACROBiosystems | Cat# SPD-C522e |
| SARS-CoV-2 Spike RBD-His | Sino Biological Inc. | Cat# 40592-V08B |
| ACE2-His Protein | Novoprotein Scientific Inc. | Cat# C419 |
| Critical commercial assays | ||
| Luciferase Assay System | Promega | Cat# E1501 |
| Dylight 800 Antibody Labeling Kit | ThermoFisher | Cat# 53062 |
| FastKing RT Kit (With gDNAase) | TianGen | Cat# KR116-01 |
| SuperReal PreMix Plus (SYB Green) | TianGen | Cat# FP205 |
| VigoFect | Vigorous Biotechnology | Cat# T001 |
| MultiS one step cloning kit | Vazyme | Cat# C113-01 |
| Experimental models: Cell lines | ||
| Expi293 Expression System | ThermoFisher | Cat# A14635 |
| Vero E6 cells | ATCC | Cat# CRL-1586 |
| Huh-7 cells | Cell Bank of the Chinese Academy of Sciences | N/A |
| 293T cells | ATCC | Cat# CRL-3216 |
| Experimental models: Organisms/strains | ||
| hACE2 mice (B6/JGpt-Ace2em1Cin(hACE2-stop)/Gpt) | GemPharmatech | Strain NO.T037630 |
| hACE2 mice C57BL/6J-Tgtn (CAG-human ACE2-IRES-Luciferase-WPRE-polyA) Smoc | Shanghai Model Organisms Center | Cat# NM-TG-200002 |
| Deposited data | ||
| Cryo-EM map and the coordinated of SARS-CoV-2 Omicron S complexed with bn03(two down and 1 up RBDs) | This paper | EMD-32501 & PDB 7WHJ |
| Cryo-EM map and the coordinated of SARS-CoV-2 Omicron S complexed with bn03(one down and 1 half-up RBDs) | This paper | EMD-32500 & PDB 7WHI |
| Cryo-EM map and the coordinated of SARS-CoV-2 Omicron S complexed with bn03(two up and one down RBDs) | This paper | EMD-32503 & PDB 7WHK |
| Recombinant DNA | ||
| S ectodomain gene: SARS-CoV-2 (strain B.1.1.529) | Genscript | Cat# MC_0101274 |
| RBD gene: SARS-CoV-2 (strain WIV04, GISAID accession no. EPI_ISL_402124) | Genscript | N/A |
| VL and VH gene of S309, CR3022, CB6, P2B-2F6 | Genscript | N/A |
| pComb3x vector | Addgene | Cat# 63891 |
| pSecTag2B expression vector | ThermoFisher | Cat# V90020 |
| Software and algorithms | ||
| UCSF Chimera | UCSF Software | N/A |
| DeepEMhancer | python package | N/A |
| RELION v3.0 | https://www3.mrc-lmb.cam.ac.uk/relion//index.php/Download_&_install | N/A |
| COOT | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot | N/A |
| cryoSPARC | Structura Biotechnology Inc. | N/A |
| MotionCor2 | UCSF Software | https://docs.google.com/forms/d/e/1FAIpQLSfAQm5MA81qTx90W9JL6ClzSrM77tytsvyyHh1ZZWrFByhmfQ/viewform |
| PHENIX | https://phenix-online.org/ | N/A |
| Living Image® Software | PerkinElmer | N/A |
| ForteBio Data Analysis software | Pall ForteBio LLC | N/A |
| Prism 8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| PyMol | PyMol | N/A |
| PDBePISA | Europea Bioinformatics Institute | https://www.ebi.ac.uk/pdbe/prot_int/pistart.html |
Resource availability
Lead contact
Further information and requests for resources and reagents may be directed to and will be fulfilled by the lead contact, Tianlei Ying (tlying@fudan.edu.cn).
Materials availability
All requests for resources and reagents should be directed to the lead contact author. This includes mice and viruses. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Human Specimen
This study included seven volunteers from Fudan University. The participants aged from 24 to 37 consist of 6 females and one male. All participants have received three doses of SARS-CoV-2 inactivated vaccine. Blood samples were collected three weeks after the third dose. All volunteers signed informed consent forms and protocols were approved by the Ethics Committee of the School of Basic Medical Sciences at Fudan University.
Animals
Pathogen-free, male hACE2 mice were purchased from GemPharmatech (B6/JGpt-Ace2em1Cin(hACE2-stop)/Gpt) to mimic the mild infection model. Pathogen-free, male CAG-hACE2-IRES-Luc-Tg transgenic mice, expressing high level of hACE2 and more sensitive to SARS-CoV-2 infection were purchased from Shanghai Model Organisms Center to mimic sever infection model. The mice used in this study were 8-10 weeks old and well fed in BSL-3 labs for several days to adapt to the environment before performing experiments. Mice were randomly allocated to each group and all animal studies were performed in BSL-3 lab of Fudan University.
Cells
Vero E6 cells and 293 T cells were obtained from the American Type Culture Collection (ATCC), Huh-7 cells were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in 37°C, 5% CO2 atmosphere.
Authentic SARS-CoV-2 virus
The authentic SARS-CoV-2 virus hCoV-SH01 used in the focus-forming assay was obtained from Shanghai Medical College and stored at -80°C in a BSL-3 laboratory (Fudan University).
Ethics statements
All the procedures related to animal handling, care, and the treatment were performed and approved by the Ethics Committee of the School of Basic Medical Sciences at Fudan University in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of Fudan University. The authentic SARS-CoV-2 infection and sample collection were performed in BSL-3 lab of Fudan University.
Method details
Antibody footprint analysis
The SARS-CoV-2 spike structure from PDB (PDB entry 7VND) was used for displaying mutations within Omicron and epitope footprints of antibodies. The structures of RBD in complex with CB6 (PDB entry 7C01), B38 (PDB entry 7BZ5), CC12.1 (PDB entry 6XC2), CC12.3 (PDB entry 6XC4), C102 (PDB entry 7K8M), REGN10933 (PDB entry 6XDG), P2B-2F6 (PDB entry 7BWJ), LY-CoV555 (PDB entry 7KMG), S2E12 (PDB entry 7K45), n3113 (PDB entry 7VNB), COVA1-16 (PDB entry 7JMW), CR3022 (PDB entry 6W41) and spike in complex with C105 (PDB entry 6XCM), BD23 (PDB entry 7BYR), 5A6 (PDB entry 7KQB), S309 (PDB entry 6WS6), C135 (PDB entry 7K8Z), 47D11 (PDB entry 7KAJ), EY6A (PDB entry 6ZDH), 3D11 (PDB entry 7KQE) were used to analyze the antibody epitopes. Interface residues were identified by ePISA (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) using default parameters and checked in COOT. The graphs were drawn using PyMoL or ChimeraX.
Protein expression and purification
The sequences of single-domain antibody n3130v, n3113v and bn03 were cloned into pComb3x vector with N-terminal OmpA signal peptide (MKKTAIAIAVALAGFATVAQA) and C-terminal hexahistidine and Flag tag and was expressed in E.coli HB2151. Bacteria transformed with expression plasmid was amplified to express antibody at 30°C for 12 h under the induction of IPTG. The bacteria were pelleted, resuspended in phosphate buffered saline (PBS) buffer and disrupted by ultrasonication, followed by centrifugation at 17,000 rpm for 30 minutes. The supernatant of n3113v was purified by Ni-NTA (Yeasen) and n3130v, and bispecific single-domain antibodies (bn01-04) were purified by Protein A resin (GenScript) following the manufacture’s instruction. The light chain and heavy chain of S309, CB6, P2B-2F6, CR3022 were subcloned into pTT expression vector digested by SfiI in IgG1 format. These IgG antibodies were expressed by Expi293 cells and purified by Protein G (GenScript). Protein integrity was analyzed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Wildtype RBD (WT) was produced and stored in our laboratory. His-tagged Omicron RBD was purchased from AcroBiosystems and other RBD variants (include Alpha, Beta, Gamma, Delta) were purchased from Sino Biological Inc.
Enzyme-linked immunosorbent assay (ELISA)
His-tagged WT and Omicron RBD at 100 ng per well was coated in 96 well half-area microplate (Corning #3690) over night at 4 °C. The antigen coated plate was blocked with PBS containing 5% bovine serum albumin (BSA) for 1 h at 37 °C and washed by three times of PBST (PBS with 0.05% Tween 20). 50 μL of three-fold serially diluted antibody in PBS or plasma from vaccinees at a dilution of 1:10 was added and incubated at 37 °C for 1.5 h. The plate was washed with PBST for three times. The anti-Flag-HRP (Sigma-Aldrich) was used for detection of n3130v and n3113v and anti-Fab-HRP (Sigma-Aldrich) for IgG antibody (S309, CB6, P2B-2F6, CR3022) and plasma. The plate was washed with PBST for five times and the absorbance at 405 nm was measured after incubation with ABTS substrate (Invitrogen) for 10 min. The data was plotted using Graphpad Prism and the antibody concentration or plasma dilution was transformed into log[concentration]/[dilution] for four parameters nonlinear regression fitting. The EC50 (concentration for 50% of maximal effect) and ED50 (median effective dose) were calculated.
Bio-layer interferometry (BLI) binding assay
The binding kinetics of antibodies to RBD were measured by BLI on an Octet-RED96 (ForteBio). The RBD with an AviTag on C terminal was biotinylated by BirA biotin-protein ligase following the manufacturer’s protocol (Avidity). The his-tagged RBD and biotinylated RBD at 10 μg/mL was loaded onto Ni-NTA and streptavidin-coated (SA) biosensors, respectively. Then the antigen immobilized sensors were incubated with three-fold serially diluted antibodies starting at 333 nM in PBST for 300 s for association, and then immersed into PBST for another 300 s at 30 °C. All the curves were fitted by 1:1 binding model using the Data Analysis software 10.0. KD values were determined with R2 values of greater than 95% confidence level.
For the binding-competition assay, the RBD immobilized sensors were incubated with the first antibody until saturation, and then the biosensors were immersed into the second antibody for the same time.
Measurement of RBD binding antibodies, RBM binding antibodies, and non-RBM binding antibodies in vaccinee plasma by BLI
The SA biosensors immobilized with biotin-labeled RBD were incubated with plasma and the signal represented RBD binding antibodies. To further determine the RBM binding antibodies and non-RBM binding antibodies, the competition assay was performed. The SA biosensors loaded with RBD were immersed into 200 nM ACE2 (Novoprotein) until the binding curve reaching saturation, confirming that the RBM of RBD was occupied by ACE2. After that, the complex was incubated 1:50 diluted plasma mixed with 200 nM ACE2. The binding signal indicated that antibodies bind to other regions on RBD except RBM region. The RBM antibodies were determined by RBD antibodies minus non-RBM antibodies. Spearman’s rank correlation test was applied to measure the correlation between plasma neutralization ID50 against Omicron with the RBM antibodies or non-RBM antibodies.
Construction of plasmids encoding SARS-CoV-2 spike protein of various variants for pseudovirus neutralization assay
The gene encoding Omicron S protein was purchased from Genscript and then subcloned into pcDNA3.1 vector. To generate the plasmid encoding SARS-CoV-2 spike protein of various variants (Alpha, Beta, Gamma, and Delta), the site-directed mutagenesis was performed by a QuickMutation™ Site-Directed Mutagenesis Kit (Beyotime Biotechnology). The vector pcDNA3.1 encoding S protein of SARS-CoV-2 from Wuhan-Hu-1 strain (GenBank: MN_908947) was generated and served as template. Firstly, two complementary primers containing the desired mutation were designed. Following site-directed mutagenesis PCR, mutated plasmid was amplified with staggered nicks and DpnI restriction endonuclease (NEB) was added to digest the parental vector for about 3 hours at 37°C. Afterwards, the PCR products including nicked vector with the desired mutations were directly transformed into DH5α competent cells. The mutation was verified by DNA sequencing.
Establishment and validation of a panel of SARS-CoV-2 pseudovirus
Plasmids encoding spike protein of WT and various variants in pcDNA3.1 vector and luciferase reporter-expressing HIV-1 backbone in pNL4-3.luc.RE were 1:1 co-transfected into 293 T cells. After 8 hours, the culture medium was replaced with fresh DMEM medium and the cells were cultured for additional 48 hours. The supernatant containing pseudoviruses were harvested by centrifugation at 3500 rpm/min for 5 minutes and filtered by 0.45 μm pore size filter before stored at −80°C in aliquots for use. Ten-fold serially diluted pseudoviruses were used to infect Huh-7 cells for 12 hours. The supernatant was refreshed 12 hours post-infection and cells were cultured by an additional 48 hours. The luciferase activity was recorded to determine the infectivity of pseudovirus.
Pseudovirus neutralization assay
Huh-7 cells at density of 10,000 per well were coated in 96-well cell culture plate overnight to form monolayer adhered cells. According to the pseudo-viral infectivity determine assay mentioned above, viral dilution of relative light unit (RLU) at around 30,000 was used. Three-fold serially diluted antibody or vaccinated plasma was mixed with pseudovirus at ratio of 1:1 for 1 h at 37 °C. The mixture was added and incubated with Huh-7 cells for 12 hours, followed by refresh of the culture medium with fresh DMEM supplied with 10% FBS. The cells were incubated for another 48 hours, washed with PBS for two times, and subsequently lysed with 50 μL of lysis reagent (Promega). 30 μL of cell lysates were added into the substrate of Firefly Luciferase Assay Kit (Promega) and the RLU readout was detected. Percent of inhibition was calculated as relative reduction of RLU compared with the control well (add cells and pseudovirus, without antibody). Data were non-linear fitted and ID50 was calculated by the equation of four parameters regression using GraphPad Prism.
Determination of antibody homogeneity by analytical size exclusion chromatography (SEC)
100 μg of antibodies in 500 μL of injection volume was chromatographed onto the Superdex™ 200 10/300 GL on AKTA purifier (GE Healthcare). The flow rate was 0.5 mL/min and the system alarm pressure was 3 MPa. Absorbance at 280 nm was monitored. Molecular mass determination was calculated by reference to a protein standard mix (Sigma-Aldrich).
Reduced and non-reduced PAGE
For denaturing SDS-PAGE, 10 μL of protein sample (25 μg protein) were mixed with 2.5 μL of 5× sample loading buffer (Yeasen) and heated at 100 °C for 10 min. For non-reduced PAGE, 25 μg of protein samples were mixed with 2.5 μL of 5× no denaturing protein loading buffer (Yeasen). These samples were then loaded into precast 4%-12% gradient gels (Yeasen) with 8 μL of pre-stained SDS-PAGE Standards. Electrophoresis was performed at room temperature for approximately 60 min using a constant voltage (150 V) in running buffer until the dye front reached the end of the gel. The gels were stained using Coomassie Brilliant Blue (Yeasen) and imaged.
Dynamic light scattering (DLS)
For comparison of aggregation tendency of bn 03 before and after aerosolization by microsprayer aerosolizer (YUYANBIO), the bn03 was filtered through a 0.22-μm filter (Millipore) and adjusted to the concentration (1 mg/mL). After aerosolization, the aerosol was collected and measured using a Zetasizer Nano ZSZEN 3600 (Malvern Instruments Limited) to determine the size of protein particles. The samples (0.2 mL) were analyzed in polystyrene cuvettes at 25°C. Each sample was recorded three times with seven sub runs of 10 s.
Ultra-high-performance liquid chromatography electrospray ionization quadrupole time of flight mass spectrometry (HPLC-ESI-Q-TOF MS) analysis
The molecular size of bn03 was analyzed using an ultra-high performance liquid chromatography (UHPLC) system coupled with a quadrupole time-of-flight mass spectrometry (QTOF-MS) Xevo G2-XS QTOF from Waters equipped with an electrospray ionization (ESI) source operating in positive mode. Briefly, deglycosylated bn03 at a volume of 5 μL was injected by an auto sampler on a ACQUITY UPLC Protein BEH SEC column (200Å, 1.7 μm, 4.6 × 300 mm) for separation. Solvent A and B were water and acetonitrile with 0.1% formic acid, respectively. The gradient expressed as the solvent A ratio was as follows: 0 min, 95% A; 2 min, 95% A; 5 min, 5% A;9 min, 5% A; and 9.1 min, 95% A. The flow rate was 0.4 mL/min. Thereafter, bn03 was identified by ESI-MS under the following conditions: capillary voltage, 3 KV; nebulizer pressure, 40 psi; drying gas flow rate, 10 L/min; gas temperature, 450 °C; fragmentor voltage, 200 V.
Size-exclusion-high-performance liquid chromatography (SEC-HPLC)
Fifty μg of bn03 were applied to a TSK-Gel Super SW3000 (TSK-GEL) using Waters AQUITY UPLC H-class system. The mobile phase was PBS buffer (pH 7.4) run at a flow rate of 0.4 mL/min. Absorbance was monitored at 280 nm. The UV trace was analyzed and integrated by area under the curve to determine percent aggregation, monomer and degradants.
Expression and purification of SARS-CoV-2 Omicron Spike
The human-optimized codon gene encoding SARS-CoV-2 Omicron S ectodomain was purchased from GenScript. HexaPro mutations, “GSAS” substitution at furin cleavage site (residues 682-285) and a C-terminal T4 fibritin trimerization motif were introduced into the gene by MultiS one step cloning kit (Vazyme). The gene was then inserted into the mammalian expression vector pcDNA3.1 with a TwinStrepTag, and an 8×HisTag at C-terminal. The expression plasmid was transiently transfected into suspension HEK293F by polyethylenimine. After 72 hours, the supernatants were harvested and filtered for affinity purification by Histrap HP (GE Healthcare). The protein was then further purified by gel filtration using Superose 6 increase 10/300 column (GE Healthcare) in 20 mM Tris pH 8.0, 200 mM NaCl.
Cryo-EM sample preparation
Purified SARS-CoV-2 Omicron S at 0.5 mg/mL was mixed with bn03 antibody by a molar ratio of 1:2 and incubated for 10 min on ice. A 3 μL aliquot of the sample was loaded onto a freshly glow-discharged holey amorphous nickel-titanium alloy film supported by 400 mesh gold grids, with microarray pattern similar like the commercial Quantifoil 1.2/1.3 grid. The sample was vitrified in liquid ethane using Vitrobot IV (FEI/ThermoFisher), with 2 s blot time and -3 blot force and 10 s wait time.
Cryo-EM data collection and image processing
Cryo-EM data were collected on a Titan Krios microscope (ThermoFisher) operated at 300 kV, equipped with K3 summit direct detector (Gatan) and GIF energy filter (Gatan BioQuantum 967) setting to a slit width of 20 eV. Automated data acquisition was carried out with SerialEM software through beam-image shift method.
Movies were taken in the super-resolution mode at a nominal magnification 81,000×, corresponding to a physical pixel size of 1.064 Å, and a defocus range from -1.2 μm to -2.5 μm. Each movie stack was dose-fractionated to 40 frames with a total exposure dose of about 58 e−/Å2 and exposure time of 3 s.
All the data processing was carried out using either modules on, or through, RELION v3.0 and cryoSPARC. A total of 4,336 movie stacks was binned 2 × 2, dose weighted, and motion corrected using MotionCor2 within RELION. Parameters of contrast transfer function (CTF) were estimated by using Gctf. All micrographs then were manually selected for further particle picking upon ice condition, defocus range and estimated resolution.
Remaining 3,529 good images were imported into cryoSPARC for further patched CTF-estimating, blob-picking and 2D classification. Several good 2D classes were used as templates for template-picking. After 2D classification of particles from template-picking was finished, all good particles from blob-picking and template-picking were merged and deduplicated, subsequently being exported back to RELION through pyem package and re-extracted with binning by 2 (2.128 Å/pixel). After getting an initial-model from cryoSPARC as reference, all 682,853 particles were carried on 1 round of 3D classification in RELION. At this step, two kinds of conformational change could be observed. Then different classes were selected separately by conformational change and last 4 iterations of particles were selected, merged and deduplicated. Another round of 3D classification was carried out to get more certain particles of their own conformation. At last, 534,947 particles of spike protein with two RBD domains up (2-up) were re-extracted unbinned (1.064 Å/pixel) and auto-refined, then CTF-refined and polished, yielding a map at 2.81 Å. Meanwhile, 281,917 particles of spike protein with only one RBD domains up (state1) yielded a map at 2.99 Å through the same procedure. In the 2-up structure, one of the up RBDs was well-resolved, the other up-RBD is more flexible and less well-resolved relative to the rest of the spike protein. Thus, we carried out no-alignment 3D classification with the mask of this up-RBD and its neighboring NTD and nanobody (NTD_RBD_Nanobody, shortly for NRN). We got one class that contains two single-domain antibodies on this up-RBD and another two good classes contains one single-domain antibody on the RBD. These three classes of two conformations were refined with the mask of entire density. Finally, 294,267 particles yielded a 2-up map at 2.93 Å with 5 single-domain antibodies on it, and 170,469 particles yielded a 2-up map at 3.01 Å with 4 single-domain antibodies on it. Furthermore, to get clearer insight of two single-domain antibodies combining on the well-resolved up RBD, we used local-refine strategy to further improve the density of the two single-domain antibodies. That is, the certain particle signal of NRN from this up RBD was subtracted from these 534,947 refined particles. Then local 3D-classification was executed and one good class containing 150,802 particles was local-refined, yielding a 3.34 Å map, which includes two rather improved single-domain antibodies.
In the 1-up structure, the density of bn03 was missed too much. Thus, we carried out no-alignment 3D classification with the mask of entire density to further classify the particles. We got one good class that contains relatively complete single-domain antibodies. This class containing 78,485 particles went on auto-refinement and yielded a 1-up map at 3.27 Å with 3 bn03 on it.
The reported resolutions above are all based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion. All the visualization and evaluation of 3D density maps were performed with UCSF Chimera. These sharpened maps were generated by DeepEMhancer and then “vop zflip” to get the correct handedness in UCSF Chimera for subsequent model building and analysis.
Model building and refinement
For model building of SARS-CoV-2 Omicron S trimer-bn03 complex, the SARS-CoV-2 D614G S trimer model and the nanobody model generated by swiss-model were fitted into the map using UCSF Chimera and then manually adjusted with COOT. COOT was used to introduce the mutations and adding glycans at N-linked glycosylation sites. Several iterative rounds of real-space refinement were further carried out in PHENIX. The RBD domain bounded with two single-domain antibodies was refined against the local refinement map and then docked back into the into global refinement trimer maps. Model validation was performed using MolProbity. Figures were prepared using UCSF Chimera and UCSF ChimeraX.
Aerodynamic particle size measured by Next Generation Impactor
To determine the size distribution of the aerosol antibodies, we used the NGI (Copley Scientific) to analyze the aerodynamic parameters of antibodies according the USP monograph. There are seven-stage droplets collectors representing different cutoff diameters of collected particles in the NGI located in its bottom frame. 2 mg/mL bispecific single-domain antibody bn03 and IgG were aerosolized and deposited on different collection cups at ambient room conditions within 5 s. Specifically, after the assembly was set up and airtight checked followed by vacuum pump running at the constant flow rates of 15 L/min, the antibody solution was added and fired into the cascade impactor immediately. Droplets of each collection plate was washed and collected by PBS, and then the components were dried up before next experiment.
The concentration of IgG in collected solution was quantified by ELISA according to the corresponding standard curve fitted by four parameter nonlinear regression. For the bispecific antibody bn03, the concentration was determined by GatorPlus (Gator Bio). In brief, Ni-NTA biosensors immobilized with his-tagged RBD was immersed into serially diluted purified bn03 to obtain a dose-dependent binding curves and the standard curve is conducted by binding signal versus antibody concentration utilizing linear fitting. Then, the antibody concentrations were calculated from the standard curve.
Bio-distribution of single-domain antibody by inhalation and intraperitoneal injection
Single-domain antibody n3113v labeled by DyLight 800 antibody Labeling Kit (ThermoFisher) at 12 mg/kg was administrated through inhalation or intraperitoneal injection. The single-domain antibody was administrated through intratracheal route using microsprayer aerosolizer (YUYANBIO). Briefly, before performing trachea cannula, mice mouth was opened up by the laryngoscope to visualize the porch of the trachea. The tip syringe of high-pressure microsprayer was gently fed into the main trachea, and the aerosols were delivered quickly. Mice were imaged at different time-points (0.5 h, 1 h, 1.5 h, 2 h, 2.5 h, 4 h, 6 h) and the fluorescence radiance was captured by IVIS Lumina K Series III (PerkinElmer) at Ex/Em 780 nm/845 nm. At 4 hours and 6 hours post antibody administration, mice were sacrificed and organs were collected for fluorescence imaging.
The pharmacokinetic of single-domain antibody in mice by inhalation and intraperitoneal injection
For bn03, 5 mg/kg and 25 mg/kg bn03 were inhaled and administrated into mice using microsprayer aerosolizer. The single-domain antibody n3113v were administrated by inhalation and intraperitoneal injection at a dose of 12 mg/kg. After antibody administration, two mice were sacrificed to collect lung and plasma at indicated time-points (Figure 6C). The lung was weighed, homogenized and then centrifuged at 12,000 rpm. The supernatant was harvested and stored at -80 °C for further quantified.
Antibody concentration in plasma and lung was determined by ELISA. In brief, SARS-CoV-2 RBD at 100 ng per well was coated in 96 well half-area microplate (Corning #3690) over night at 4 °C. The antigen coated plate was blocked with PBS containing 5% BSA for 1 h at 37 °C and washed by three times of PBST (PBS with 0.05% Tween 20). 50 μL of mice plasma or lung homogenate in PBS at a dilution of 1:100 was added for binding at 37 °C for 1.5 h. The plate was washed with PBST for three times and incubated with anti-Flag-HRP (Sigma-Aldrich) for 45 min at 37 °C. The plate was washed with PBST for five times and the enzyme activity was measured by recording the absorbance at 405 nm after incubation with ABTS substrate (Invitrogen) for 10 min. Gradient serially diluted purified antibodies were used to generate quantitative standard curve and fitted by a four-parameter logistic model. The antibody concentration in lung and plasma was calculated from the standard curve.
Therapeutic efficacy of antibodies in the mouse model of SARS-CoV-2 infection
Twelve hACE2 transgenic mice (B6/JGpt-Ace2em1Cin(hACE2-stop)/Gpt) were purchased from GemPharmatech and randomly divided into three groups, inhalation group (INH), intraperitoneal group (IP) and negative control group (Control), respectively. All mice were inoculated intranasally with 1.16 × 105 plaque-forming unit (PFU) SARS-CoV-2 viruses under anesthesia to minimize animal suffering. Two hours post infection, mice were intraperitoneally treated with 12.5 mg/kg n3113v, or inhaled with 12.5 mg/kg n3113v once a day for three days. Animals were sacrificed at 4 dpi (days post infection) and lung tissues were harvested for viral load and histology analysis.
To evaluated the efficacy of bispecific single-domain antibody bn03 in the mouse model of SARS-CoV-2 infection, two types of hACE2-transgenic mice with different hACE2 expressing level were used. The hACE2 mice purchased from GemPharmatech (B6/JGpt-Ace2em1Cin(hACE2-stop)/Gpt) were inoculated intranasally with 1.16 × 105 PFU SARS-CoV-2 viruses to mimic the mild infection model. In contrast, CAG-hACE2-IRES-Luc-Tg transgenic mice (Shanghai Model Organisms Center), expressing high level of hACE2 and more sensitive to SARS-CoV-2 infection, were used as the severe infection model and inoculated intranasally with 1.16 × 104 PFU SARS-CoV-2 viruses two hours before treatment. Viral challenge was operated under anesthesia to minimize animal suffering. Mice were randomly divided into two groups, of which one group received intranasal inhalation of bn03 at 12.5 mg/kg (bn03 group) and the other one received PBS as control (PBS group) for three days. Animals were sacrificed at 4 dpi and lung tissues were harvested for viral load and histology analysis.
Determination of viral titer by qRT-PCR
Briefly, the lung tissue was collected and homogenized by electric homogenizer in Trizol. After centrifugation, the total RNA was extracted from the supernatant by chloroform and isopropanol and reverse-transcribed into cDNA. RNAs were quantitated by qRT-PCR kit (TianGen) in triplicates utilizing SARS-CoV-2 specific primers that target a conserved region in necleocapsid (N) gene of SARS-CoV-2. The SARS-CoV-2 N gene was cloned into a pcDNA3.1 expression plasmid and in vitro transcribed to obtain RNAs for standards. Indicated copies of N standards were 10-fold serially diluted and proceeded to qRT-PCR to obtain standard curves.
Focus-forming assay
10-fold serially diluted lung homogenate was inoculated with monolayer Vero E6 cells in 96-well plate for 1.5 hours followed by the overlay of methylcellulose for 48 h at 37 °C. Cells were fixed with 4% paraformaldehyde for 30 min at 4 °C. The supernatant was removed and the plate was immersed into perm wash buffer with saponin. The cells were permeabilized by 0.2% triton for 15 min, followed by adding of 1:2000 diluted rabbit anti-SARS-CoV nucleocapsid antibody (Rockland) for 3 h at room temperature. The plate was washed by PBS for 2 times and the secondary goat anti-rabbit-HRP antibody was added and incubated for 3 h at room temperature. After washed with PBS for 2 times, TrueBlue substrate was added and the foci was visualized presented as the blue spot.
Quantification and statistical analysis
The results on binding ability of antibody to RBD and antibody concentration are presented as the mean values from two independent experiments. Neutralization titer is presented as the geometric mean of the IC50 values calculated using four-parameter logistic regression from three independent experiments. The viral load is presented as the mean values from three duplicates on a single experiment. Error bars are defined as standard deviation (SD). All data were conducted using Prism software (version 8, GraphPad Software). Statistical significance was analyzed by Student’s t-test, one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test and spearman’s rank correlation test using Prism software (version 8, GraphPad Software).
Acknowledgments
We are grateful to Qi Shao and Zhixin Jing from Sinepharm for the support in NGI experiments and Qian Wang, Yiwei Zhong, and other staff from Core Facility of Microbiology and Parasitology, Shanghai Medical College, for help with experiments. This work was supported by grants from the National Key R&D Program of China (2019YFA0904400), National Natural Science Foundation of China (81902108, 82041003, 81822027, and 32070938), Chinese Academy of Medical Sciences (2019PT350002), Shanghai Municipal Education Commission "Chenguang program” (C620297), Shanghai Municipal Health Commission (GWV-10.2-XD01 and GWV-10.2-YQ06), and Science and Technology Commission of Shanghai Municipality (20411950402, 20XD1401200, 18DZ2210200, 20DZ2254600, and 20DZ2261200).
Author contributions
T.Y., Y.W., L.S., and C.T. initiated, planned, and supervised the project. C.L. performed most of the experiments with assistance from K.H., Y.K., Y.Z., Y.X., Q.D., Y.S., and L.L. S.J., W.Z., X.Z., M.Z., Q.M., Z.C., and L.S. collected cryo-EM data and solved the structures. L.S. and Z.Y. analyzed the structures. G.H., S.D., Y.Z., and X.G. performed animal studies in BSL-3 lab. W.S. performed pseudovirus neutralization assay. The manuscript was written by T.Y., Y.W., L.S., C.L, and Z.Y. and was reviewed, commented on, and approved by all the authors.
Declaration of interests
C.L., Y.W., and T.Y. are listed as inventors on two patent applications related to this work.
Published: March 10, 2022
Data and code availability
The cryo-EM map and the coordinates of SARS-CoV-2 Omicron S complexed with bn03 have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under the accession codes under accession code PDB: 7WHJ, 7WHI, and 7WHK and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bangaru S., Lang S., Schotsaert M., Vanderven H.A., Zhu X., Kose N., Bombardi R., Finn J.A., Kent S.J., Gilchuk P., et al. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell. 2019;177:1136–1152.e18. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco F., Salomone F., Milesi I., Murgia X., Bonelli S., Pasini E., Dellacà R., Ventura M.L., Pillow J. Aerosol drug delivery to spontaneously-breathing preterm neonates: lessons learned. Respir. Res. 2021;22:71. doi: 10.1186/s12931-020-01585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham S., Piedra P.A., Martinon-Torres F., Szymanski H., Brackeva B., Dombrecht E., Detalle L., Fleurinck C., RESPIRE Study Group Nebulised ALX-0171 for respiratory syncytial virus lower respiratory tract infection in hospitalised children: a double-blind, randomised, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2021;9:21–32. doi: 10.1016/S2213-2600(20)30320-9. [DOI] [PubMed] [Google Scholar]
- De Gasparo R., Pedotti M., Simonelli L., Nickl P., Muecksch F., Cassaniti I., Percivalle E., Lorenzi J.C.C., Mazzola F., Magrì D., et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature. 2021;593:424–428. doi: 10.1038/s41586-021-03461-y. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., Liu X., Lambe T., Crook D., Stuart D.I., et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2022;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- Li C., Tian X., Jia X., Wan J., Lu L., Jiang S., Lan F., Lu Y., Wu Y., Ying T. The impact of receptor-binding domain natural mutations on antibody recognition of SARS-CoV-2. Signal Transduct. Target. Ther. 2021;6:132. doi: 10.1038/s41392-021-00536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- Lu L., Su S., Yang H., Jiang S. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184:1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.e9. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez C., Sekirov I., Subramaniam S. SARS-CoV-2 omicron variant: ACE2 binding, cryo-EM structure of spike ProteinACE2 complex and antibody evasion. Preprint at bioRxiv. 2021 doi: 10.1101/2021.12.19.473380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Patton J.S., Byron P.R. Inhaling medicines: delivering drugs to the body through the lungs. Nat. Rev. Drug Discov. 2007;6:67–74. doi: 10.1038/nrd2153. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Sakagami M. In vitro, ex vivo and in vivo methods of lung absorption for inhaled drugs. Adv. Drug Deliv. Rev. 2020;161–162:63–74. doi: 10.1016/j.addr.2020.07.025. [DOI] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner H.L., Pallesen J., Lang S., Bangaru S., Urata S., Li S., Cottrell C.A., Bowman C.A., Crowe J.E., Jr., Wilson I.A., Ward A.B. Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PLoS Biol. 2019;17:e3000139. doi: 10.1371/journal.pbio.3000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158.e15. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., McCarthy K.R., Kuraoka M., Schmidt A.G., Adachi Y., Onodera T., Tonouchi K., Caradonna T.M., Bajic G., Song S., et al. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell. 2019;177:1124–1135.e16. doi: 10.1016/j.cell.2019.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Gao G.F. Breathing" hemagglutinin reveals cryptic epitopes for universal influenza vaccine design. Cell. 2019;177:1086–1088. doi: 10.1016/j.cell.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y., et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898.e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamin R., Jones A.T., Hoffmann H.H., Schäfer A., Kao K.S., Francis R.L., Sheahan T.P., Baric R.S., Rice C.M., Ravetch J.V., Bournazos S. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021;599:465–470. doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wang Y., Jin Y., Zhu Y., Wu Y., Li C., Kong Y., Song W., Tian X., Zhan W., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct. Target. Ther. 2021;6:378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Song H., Wu Y., Chang S.Y., Wang L., Li W., Hong B., Xia S., Wang C., Khurana S., et al. A potent germline-like human monoclonal antibody targets a pH-sensitive epitope on H7N9 influenza hemagglutinin. Cell Host Microbe. 2017;22:471–483.e5. doi: 10.1016/j.chom.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Xiang R., Deng X., Wang L., Yu Z., Tian S., Liang R., Li Y., Ying T., Jiang S. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:212. doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The cryo-EM map and the coordinates of SARS-CoV-2 Omicron S complexed with bn03 have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) under the accession codes under accession code PDB: 7WHJ, 7WHI, and 7WHK and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.