Abstract
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in millions of deaths and seriously threatened public health and safety. Despite COVID-19 vaccines being readily popularized worldwide, targeted therapeutic agents for the treatment of this disease remain very limited. Here, we studied the inhibitory activity of the scutellarein and its methylated derivatives against SARS-CoV-2 main protease (Mpro) by the fluorescence resonance energy transfer (FRET) assay. Among all the methylated derivatives we studied, 4′-O-methylscutellarein exhibited the most promising enzyme inhibitory activity in vitro, with the half-maximal inhibitory concentration value (IC50) of 0.40 ± 0.03 μM. Additionally, the mechanism of action of the hits was further characterized through enzyme kinetic studies and molecular docking. Overall, our results implied that 4′-O-methylscutellarein could be a primary lead compound with clinical potential for the development of inhibitors against the SARS-CoV-2 Mpro.
Keywords: Scutellarein, COVID-19, SARS-CoV-2, Mpro inhibitor
1. Introduction
Coronavirus disease 2019 (COVID-19) pandemic is caused by the emergence coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is still spreading at an alarming speed globally after two years of breakout [1,2]. According to the World Health Organization (WHO) dashboard report, till 1st March 2022, over 400 million positive cases have been reported from over 200 countries and territories. Meanwhile, nearly six million death cases have been confirmed [3]. Although there are multiple effective COVID-19 vaccines available, reinfections and breakthrough infections are frequently reported, meaning that the current vaccines fail to provide long-term steady protection. In addition, the virus continues to evolve and a variety of mutations in the SARS-CoV-2 genome have been found [4]. A new variant named Omicron (PANGO lineage B.1.1.529) enables the virus to evade the immune protective barrier owing to a large number of mutation sites in the receptor binding domain [5,6]. In summary, human beings are likely to coexist with this virus, therefore the development of more effective drugs and specific therapeutics are urgently needed.
As the seventh member of the coronavirus families reported to infect humans [1], SARS-CoV-2 is a highly infectious, enveloped positive-sense single stranded RNA virus with approximately 79.5% genomic identical with SARS-CoV [7,8]. In the SARS-CoV-2 genome, two overlapping polyproteins (pp1a and pp1ab) and four structural proteins are translated upon entry into host cells [9,10]. The polyproteins (PPs) are cleaved by the critical SARS-CoV-2 main protease (Mpro, also known as 3-chymotrypsin-like cysteine protease or 3CLpro) at a minimum of eleven different sites to yield essential viral non-structural proteins (NSPs) required for virus replication and invasiveness [[11], [12], [13]]. Notably, the substrate specificity of the Mpro is highly conserved among different Coronavirus (CoVs) and no Mpro homolog was found in humans, hence reducing the risk of off-target side-effects by Mpro inhibitors. In summary, Mpro is an ideal target for the development of anti-COVID-19 inhibitors [14,15].
Throughout the ages, plants are the richest host for novel drug discovery and development [16]. Many drugs developed from natural lead compounds have indications ranging from coughs to parasitic infection and inflammation [17,18]. Traditional Chinese medicines herbs and formulae have been used in treating viral diseases for a long time. Some of them have been clinically tested to treat COVID-19 [19,20]. Meanwhile, several phytochemicals have been investigated for their therapeutic potentials against SARS-CoV-2. Scutellarin (4′,5,6- trihydroxyflavone-7-O-glucuronide), the main bioactive component extracted from Erigeron breviscapus, has a role as an antineoplastic agent and a proteasome inhibitor [21]. Moreover, Liu et al. demonstrated scutellarein and its flavonoid analogs such as baicalin have the potential as broad-spectrum anti-coronavirus drugs [22]. The binding mode of baicalein with SARS-CoV-2 Mpro determined by X-ray diffraction has been reported [23]. Notably, in the pharmacokinetic studies, scutellarin was hydrolyzed to scutellarein (4′,5,6,7-Tetrahydroxyflavone or 6-Hydroxyapigenin), then absorbed and converted into methylated, sulfated or glucuronidated forms. Among them, 6-O-methylscutellarein is one of the major metabolites in vivo, suggesting the therapeutic effect of scutellarin may be related to methylation [24,25].
In this study, we synthesized scutellarein and its methylated derivatives. Subsequently, we used the fluorescence resonance energy transfer (FRET) to detect the inhibition rates of different compounds against recombinant SARS-CoV-2 Mpro and compared their IC50 values according to the experiment. Finally, 4′-O-methylscutellarein (4), which has an IC50 value of 0.40 ± 0.03 μM, was selected as an excellent lead for further optimization and development of non-covalent, non-peptidomimetic anti-coronavirus drugs [26].
2. Materials and methods
2.1. Chemistry and reagents
Scutellarin and baicalein were purchased from Dingxintong Pharmaceutical Company (Wuhan, China). Chemical syntheses of scutellarein and its methylated derivatives were performed based on our previous studies [29]. Syntheses and characterizations of compounds are in the Supporting Information.
2.2. Protein expression and purification
The cDNA of SARS-CoV-2 Main protease (Wuhan-Hu-1 isolate; accession number: MN908947) was cloned into the pET-28a (+) vector. The SARS-CoV-2 Mpro construct contains both N and C-terminal His tag. The N-terminal Hexa-histidine tag followed with Mpro protease site (SAVLQ↓SGFRK, arrow indicates the cleavage site), was auto-cleaved by SARS-CoV-2 Mpro protease during expression to generate the authentic N-terminus. At the C-terminal, four amino acids (GPLE) were inserted between the Hexa-histidine tag and the full-length SARS-CoV-2 Mpro. The pET-28a (+) plasmid with SARS-CoV-2 Mpro was then transformed into competent E. coli BL21 (DE3) cells and a single colony was picked and inoculated in 5 ml LB medium supplemented with 50 g/ml kanamycin and grown to an optical density at 600 nm of 0.8, and then induced using 0.5 mM IPTG. Induced cultures were incubated at 25 °C for an additional 16 h and then harvested, resuspended, and lysed in buffer A (20 mM HEPES, pH 7.5, 500 mM NaCl, 20 mM Imidazole). The cell debris was removed by centrifugation at 15000 rpm for 90 min at 4 °C. Ni-resin was loaded on a gravity column and after 15 column volumes wash in Buffer A, the protein was eluted in 20 mM HEPES, pH 7.5, 500 mM NaCl, 300 mM Imidazole. As shown in Supplementary Fig. 1, the resulting protein sample was further passed through a size-exclusion chromatography (Superdex™ 200 Increase 10/300 GL, GE Healthcare). The eluted protein samples were stored in a solution (20 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM EDTA) for the enzymatic inhibition assay, protein crystallization, etc.
2.3. The enzymatic inhibition assay
The enzyme activity assays have previously been described [27]. In brief, A fluorescence resonance energy transfer (FRET) protease assay was applied to measure the inhibitory activity of compounds against the SARS-CoV-2 Mpro. We designed a fluorophore-labeled short peptide substrate (Dabcyl-KLSAVLQ↓SGFRKME-Edans) using the sequence between viral polypeptide NSP4-NSP5 junction from SARS-CoV-2. The peptide substrate was synthesized by GL Biochem Ltd (Shanghai, China). The recombinant SARS-Cov-2 Mpro (500 nM at a final concentration) was mixed with 3-fold serial diluted compounds in 50 μL assay buffer (20 mM Tris-HCl, pH 7.5, 10 mM EDTA, 150 mM NaCl) and incubated at 25 °C for 15 min. The reaction was initiated by adding 50 μL fluorogenic substrate with a final concentration of 20 μM. After that, the fluorescence signal at 340 nM (excitation)/490 nM (emission) was immediately measured every 30s for 10 min with a Tecan's Spark multimode reader. The V max of reaction added with compounds at various concentrations compared to the reaction added with assay buffer were calculated and used to generate IC50 curves. For each compound, IC50 values against SARS-CoV-2 Mpro were measured at 10 different concentrations and three independent experiments were performed. All the experimental data were analyzed using GraphPad Prism 8.4.3 software.
2.4. Molecular docking
In order to gain an insight into the binding interaction of investigated 4′-O-methylscutellarein (4) with SARS-CoV-2 Mpro enzyme, we performed molecular docking studies based on the crystal structures of Mpro in complex with the peptide-like inhibitor N3 (PDB ID: 6LU7). The structure of 4′-O-methylscutellarein (4) used for docking was drawn by ChemDraw 19.0 software. The docking of 4′-O-methylscutellarein (4) onto SARS-CoV-2 Mpro was performed with the genetic algorithm approach by Autodock 4.2. Computational molecular docking results were analyzed by PyMOL 2.3 and Chimera 1.15.
3. Results
3.1. Chemistry
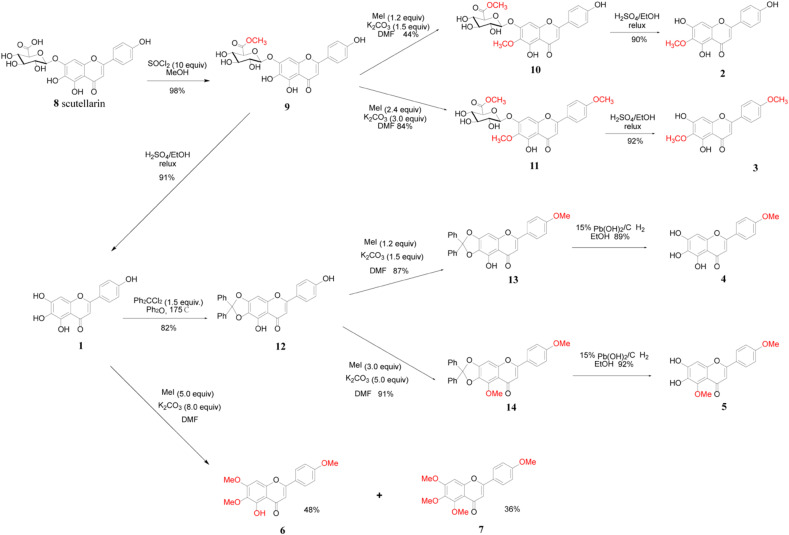
The synthetic route for scutellarein and its methylated derivatives was outlined in Scheme 1 . The scutellarin methyl ester 9 was obtained in 98% yield by treating scutellarin with 10 equiv. amount of SOCl2 in the solution of methanol according to our previously reported method [29]. Scutellarein 1 was obtained in 91% yield by hydrolysis of the sugar moiety of compound 9 in a mixed solvent consisting of H2SO4/EtOH/H2O at reflux. Also, with compound 9 in hands, scutellarein methylated derivatives 2 and 3 could be obtained easily according to our previously reported method [29].
Scheme 1.
Synthesis of scutellarein and its methylated derivatives.
Treatment of scutellarein 1 with 1.5 equiv. of dichlorodiphenylmethane in diphenyl ether at 175 °C led to the desired intermediate 12 in 82% yield after purification by column chromatography. Then the reaction of intermediate 12 with 1.2 equiv. of iodomethane in the presence of K2CO3 afforded compound 13 in 87% yield with a methyl group at the C-4′ phenol hydroxyl position. Under the hydrogenolysis conditions, the diphenylmethylene ketal could be cleaved with 15% hydroxide palladium on carbon as a catalyst in EtOH. Thus, 4′-O-methylscutellarein 4 was obtained in the yield of 89% and no side product was detected by TLC analysis. Similarly, using intermediate 12 as starting material, scutellarein methylated derivative 5 could be obtained in an overall yield of 83.7% for 2 steps. Furthermore, the methylation of scutellarein 1 with iodomethane (5.0 equiv.) afforded the scutellarein methylated derivatives 6 and 7 in 48% and 36% respectively.
3.2. 4′-O-methylscutellarein strongly inhibits SARS-CoV-2 Mpro
We tested the inhibitory activity of the prepared scutellarein and its methylated derivatives against SARS-CoV-2 Mpro and evaluated according to the reported procedures [26]. Firstly, we tested the enzymatic inhibition rate of scutellarein, the 6 methylated derivatives, and compound 9 (PF-07321332) at the concentration of 50 μM. The compound 9 (PF-07321332), identified as one of the strongest Mpro inhibitors authorized for emergency use for the treatment of high-risk COVID-19 patients [30], was employed as the positives control. It demonstrated potent inhibitory effects towards the target enzyme at the concentration of 50 μM. The results from primary screening indicated the inhibition rate of 4 compounds less than 50% at 50 μM (Supplementary Table S1). Then, the remaining 4 compounds with considerable inhibitory effect at 50 μM entered the IC50 value evaluation.
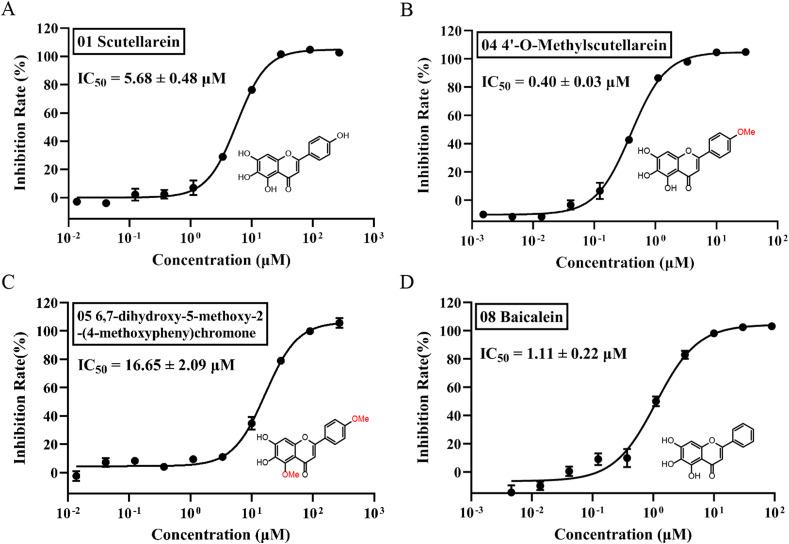
As shown in Fig. 1 , 4′-O-methylscutellarein (4) was characterized as the most potent inhibitor against the target enzyme with IC50 value of 0.40 ± 0.03 μM, which has significant improvement compared with scutellarein (1, IC50 = 5.68 ± 0.48 μM) before structural modification. Moreover, there was no remarkable difference in the inhibitory effect between compound 4 and 8 (IC50 = 0.16 ± 0.02 μM)(Supplementary Fig. 2). Baicalein (8) and 6,7-dihydroxy-5-methoxy-2-(4-methoxyphenyl) chromone (5) have also been identified as inhibitors with IC50 values of 1.11 ± 0.22 μM and 16.65 ± 2.09 μM respectively.
Fig. 1.
Peptide cleavage FRET assay reveals the compounds inhibit the activity of SARS-CoV-2 Mpro. A-D, the hydrolytic activity of SRAS-CoV-2 Mpro was measured in the presence of increasing concentrations of the drug candidates. A. Scutellarein (1). B, 4′-o-Methylscutellarein (4). C, 6,7-dihydroxy-5-methoxy-2-(4-methoxypheny) chromone (5). D, Baicalein. Dose-response curves for IC50 values were determined by nonlinear regression. All date is shown as mean ± SD, n = 3 biological replicates.
3.3. Structure-activity relationship studies
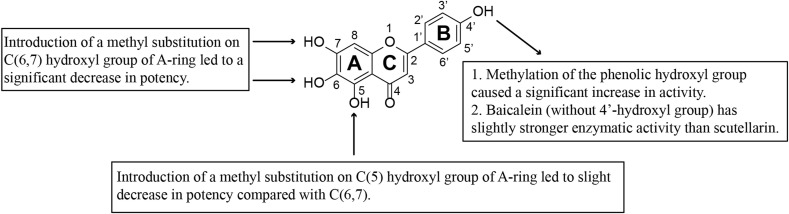
As shown in Table S1 and Fig. 2 , the natural compound scutellarein (1) and Baicalein (10) exhibited potent inhibitory effects against the target SARS-CoV-2 Mpro with inhibition rates of 101.91% and 103.01% respectively at the concentration of 50 μM. By contrast, compound with 7-methoxyl group and/or 6- methoxyl group exhibit reduced rates of inhibition against Mpro under the same concentration,such as compound 2, 3, 6 and 7. Meanwhile, the substitution of 5-hydroxyl group on the A-ring with methoxy also resulted in a decrease in inhibitory activity, which is reflected by the IC50 value of compound 5 (IC50 = 16.65 ± 2.09 μM). The sharp decrease in the activity should be ascribed to the substitution of the hydroxyl group on the A-ring with methoxy. These results are consistent with previous studies conducted by Su et al., the phenolic hydroxyl groups on baicalein made multiple hydrogen bonds with the residues in the binding pocket [23]. In summary, results above demonstrate that any substitution of hydroxyl groups on the A-ring with methoxy has detrimental effects on the enzyme inhibitory potency.
Fig. 2.
SAR analysis of Scutellarein and its methylated derivatives as SARS-CoV-2 Mpro inhibitors.
4′-O-methylscutellarein (4) is a methylated product of 4′hydroxyl group of scutellarein (1). Compound 4 has a dramatically increased inhibitory effect than scutellarein (Fig. 1), as reflected by the change of IC50 value (from 5.68 ± 0.48 μM to 0.40 ± 0.03 μM). Meanwhile, baicalein (8) without B-ring substituents has a higher inhibitory activity (IC50 = 1.11 ± 0.22 μM) than scutellarein (1), which have monohydroxy substitution. It is speculated that the hydrophobicity of B-ring played a vital role on the enzyme inhibitory potency.
3.4. Molecular docking
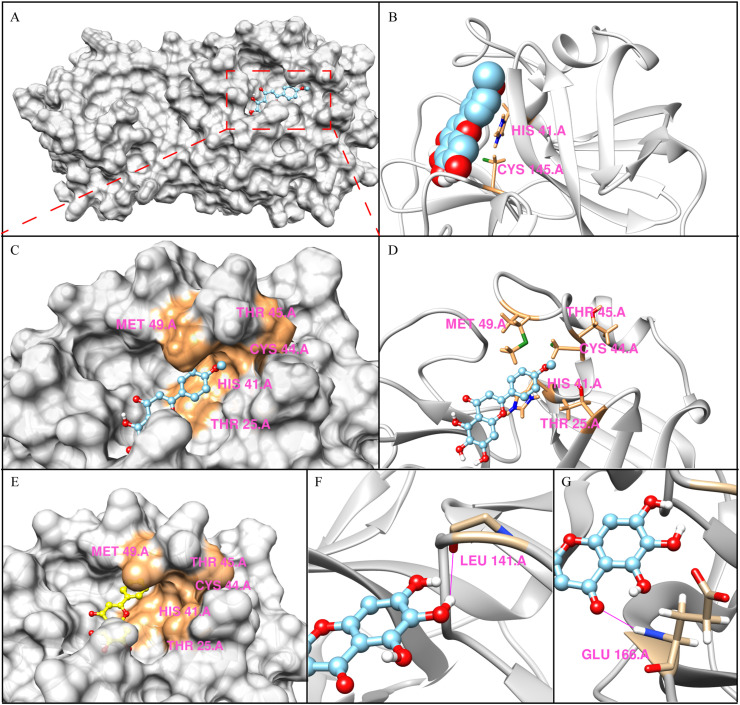
The results of the 20 runs showed that the binding pockets of 4′-O-methylscutellarein (4) were roughly constant with slight changes in the conformation of SARS-CoV-2 Mpro, and they all exhibited low binding free energy, which means the docking results stay rational [28]. As shown in Fig. 3 , 4′-O-methylscutellarein (4) fitted well into the substrate binding site of Mpro enzyme with the calculated binding energy of −17.1544 kJ/mol. Previous studies indicated that SARS-CoV-2 Mpro catalyzes the cleavage of the protein substrate using the catalytic dyad His41 and Cys145 [31]. The docking result indicated that the 4′-O-methylscutellarein (4) is perfectly fitted into the core of the substrate-binding pocket and the catalytic dyad is sequestered from the solvent exposed area to prevent the peptide substrate approaching the active site (Fig. 3B), which is similar to the crystal structure of Mpro in complex with baicalein (PDB: 6M2N) [23]. The 6-phenolic hydroxyl groups of A-ring made a hydrogen bond with the carbonyl group of Leu141 with a distance of 2.5 Å. (Fig. 3F). The only carbonyl group on the A-ring formed a hydrogen bond with Glu166 backbone amide with a distance of 2.3 Å (Fig. 3G). Apart from the hydrophobic interactions, the catalytic Cys145 and His41 also formed S-π and π-π interactions with the aromatic ring of 4′-O-methylscutellarein (4) (Fig. 3B). Notably, compared with the structure of baicalein (8) complexed with SARS-CoV-2 Mpro (PDB: 6M2N), 4′-O-methylscutellarein (4) was well embedded into the active pocket and effectively enhanced the enzymatic inhibitory effect by hydrophobic interactions between the p-methoxyphenyl of 4′-O-methylscutellarein (4) and the pocket formed by hydrophobic side chain of residues Thr15/His41/Cys44/Thr45/Met49 (Fig. 3C, D, 3E).
Fig. 3.
The predicted binding modes of 4′-O-Methylscutellarein (4) in the active site cavity of Mpro. A, Solvent-accessible surface representations of the predicted binding modes of 4′-O-Methylscutellarein (4) with Mpro. B, Ribbon diagram of the predicted binding modes of 4′-O-Methylscutellarein (4) with Mpro. Compound is shown as spheres with carbons in sky blue, oxygen in red, hydrogen in white and sulphur in green. C, Close-up view of the active pocket. D, Stereo view of the acceptor binding pocket in the modeled Mpro structure. E, The structure of SARS-CoV-2 Mpro in complex with baicalein (PDB code 6M2N). F and G, Close-up view of hydrogen bonds between 4′-O-Methylscutellarein (4) and Leu141 and Glu166 in the Mpro binding pocket. H-bond is shown in deep pink. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
The two polyproteins translated from the SARS-CoV-2 genome are cleaved at three sites by papain-like protease (PLpro) and 11 sites by Mpro to produced NSPs of the virus [32]. Since Mpro plays a crucial role at the beginning of viral replication, it has undoubtedly become an ideal drug target [33]. According to current studies, most of the rapidly developed COVID-19 therapeutic drugs from the evaluated compounds target this protein. A peptide-based, covalent oral Mpro inhibitor (PF-07321332) developed by Pfizer was approved by FDA in December 2021, which is dosed with ritonavir as a pharmacokinetic (PK) booster [30]. Meanwhile, there are currently SARS-CoV-2 Mpro inhibitors including FB2001, SIM0417, vv116 and RAY003 in China, which are in the development stage. However, most of the SARS-CoV-2 Mpro inhibitors under study are peptide-based, covalent inhibitors, leading to great challenges with their target selectivity and PK profiles [34,35]. It is necessary to develop safer non-covalent, non-peptidomimetic anti-coronavirus drugs. Shionogi recently published the 2/3 phase clinical trial results (phase 2a) of its oral drug S-217622, which is the first non-peptidic, non-covalent SARS-CoV-2 Mpro inhibitor for treating COVID-19 clinically. Compare with PF-07321332, S-217622 displayed favorable drug metabolism and pharmacokinetic (DMPK) profiles [36].
Scutellarein has been shown to provide a variety of specific bioactivities and health benefits [37,38]. Previous investigations demonstrated that the Scutellarein and baicalein might serve as SARS-CoV and SARS-CoV-2 Mpro [22,39]. In our studies, Scutellarein was characterized as a potent Mpro inhibitor, its methylated derivatives substituted at different positions showed significant changes on the inhibitory effect. Our study provides an ideal lead compound (4′-O-methylscutellarein) for further development of potent non-covalent, non-peptidomimetic inhibitors. However, the IC50 value of enzyme inhibitory activity in vitro may not reflect the results in vivo. The antiviral effect of 4′-O-methylscutellarein against SARS-CoV-2 in vivo should be further evaluated [40,41].
Acknowledgements
This project was funded by National Key Research and Development Program of China [2018YFA0902000]; National Science Foundation of China [31970547]; Natural Science Foundation of Jiangsu Province [BK20190552]; Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University [SKLNMZZ202014]. Research and application of New Coronavirus rapid detection and virus blocking therapy [No. 2020SK3035] supported by Department of Science and Technology of Hunan Province. Q. Wu, S. Yan conceived the research. Y. Li and Y. Xiao designed the project. S. Yan and Y. Wang conducted chemical experiments. Q. Wu and M. Li conducted the experiments on SARS-CoV-2 Mpro, Q. Wu and S. Yan wrote the manuscripts. All authors read and approved the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2022.03.052.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X., Yu Y., Xu J., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study, the Lancet. Respir. Med. 2020;8(5):475–481. doi: 10.1016/s2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO, Coronavirus disease (COVID-19) February 2022. Weekly Epidemiological Update and Weekly Operational Update.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [Google Scholar]
- 4.Gupta D., Sharma P., Singh M., et al. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cell. Mol. Life Sci. 2021;78(24):7967–7989. doi: 10.1007/s00018-021-04008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Q. Si, C. Mengnan, S. Siqi, et al., Genome characterization and potential risk assessment of the novel SARS-CoV-2 variant Omicron (B.1.1.529), Zoonoses 1 (1), 10.15212/ZOONOSES-2021-0024. [DOI]
- 6.Dai L., Gao G.F. Viral targets for vaccines against COVID-19, Nature reviews. Immunology. 2021;21(2):73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt N., Lareau C.A., Keshishian H., et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nature microbiology. 2021;6(3):339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh A.K., Brindisi M., Shahabi D., et al. Drug development and medicinal chemistry efforts toward SARS-coronavirus and covid-19 therapeutics. ChemMedChem. 2020;15(11):907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee R., Perera L., Tillekeratne L.M.V. Potential SARS-CoV-2 main protease inhibitors. Drug Discov. Today. 2021;26(3):804–816. doi: 10.1016/j.drudis.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rut W., Groborz K., Zhang L., et al. SARS-CoV-2 M(pro) inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 12.Anand K., Ziebuhr J., Wadhwani P., et al. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science (New York, N.Y.) 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Lin D., Kusov Y., et al. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63(9):4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., Lin D., Sun X., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.) 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steuten K., Kim H., Widen J.C., et al. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect. Dis. 2021;7(6):1457–1468. doi: 10.1021/acsinfecdis.0c00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J., Jia J. Natural COX-2 inhibitors as promising anti-inflammatory agents: an update. Curr. Med. Chem. 2021;28(18):3622–3646. doi: 10.2174/0929867327999200917150939. [DOI] [PubMed] [Google Scholar]
- 17.Bernardini S., Tiezzi A., Laghezza Masci V., et al. Natural products for human health: an historical overview of the drug discovery approaches. Nat. Prod. Res. 2018;32(16):1926–1950. doi: 10.1080/14786419.2017.1356838. [DOI] [PubMed] [Google Scholar]
- 18.Dias D.A., Urban S., Roessner U. A historical overview of natural products in drug discovery. Metabolites. 2012;2(2):303–336. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D.Y.W., Li Q.Y., Liu J., et al. Traditional Chinese herbal medicine at the forefront battle against COVID-19: clinical experience and scientific basis. Phytomedicine : Int. J.Phytother.Phytopharm. 2021;80:153337. doi: 10.1016/j.phymed.2020.153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du A., Zheng R., Disoma C., et al. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int. J. Biol. Macromol. 2021;176:1–12. doi: 10.1016/j.ijbiomac.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y., Zha H., Rangarajan P., et al. Anti-inflammatory effects of Edaravone and Scutellarin in activated microglia in experimentally induced ischemia injury in rats and in BV-2 microglia. BMC Neurosci. 2014;15:125. doi: 10.1186/s12868-014-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H., Ye F., Sun Q., et al. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzym. Inhib. Med. Chem. 2021;36(1):497–503. doi: 10.1080/14756366.2021.1873977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su H., Yao S., Zhao W., et al. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro, bioRxiv. 2020:2020. doi: 10.1101/2020.04.13.038687. 04.13.038687. [DOI] [Google Scholar]
- 24.Wang L., Ma Q. Clinical benefits and pharmacology of scutellarin: a comprehensive review. Pharmacol. Therapeut. 2018;190:105–127. doi: 10.1016/j.pharmthera.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Shi X., Hu Y., Jiang Y., et al. Scutellarein protects against cardiac hypertrophy via suppressing TRAF2/NF-κB signaling pathway. Mol. Biol. Rep. 2022 doi: 10.1007/s11033-021-07026-0. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman R.L., Kania R.S., Brothers M.A., et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63(21):12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai W., Zhang B., Jiang X.M., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (New York, N.Y.) 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A., Lee K., Kim D. Using reverse docking for target identification and its applications for drug discovery. Expet Opin. Drug Discov. 2016;11(7):707–715. doi: 10.1080/17460441.2016.1190706. [DOI] [PubMed] [Google Scholar]
- 29.Yan S., Xie M., Wang Y., et al. Semi-synthesis of a series natural flavonoids and flavonoid glycosides from scutellarin. Tetrahedron. 2020;76(8):130950. doi: 10.1016/j.tet.2020.130950. [DOI] [Google Scholar]
- 30.Owen D.R., Allerton C.M.N., Anderson A.S., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science (New York, N.Y.) 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira J.C., Fadl S., Villanueva A.J., et al. Catalytic dyad residues His41 and Cys145 impact the catalytic activity and overall conformational fold of the main SARS-CoV-2 protease 3-chymotrypsin-like protease. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.692168. 692168-692168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., He S., Deng W., et al. Comprehensive insights into the catalytic mechanism of Middle East respiratory syndrome 3C-like protease and severe acute respiratory syndrome 3C-like protease. ACS Catal. 2020;10(10):5871–5890. doi: 10.1021/acscatal.0c00110. [DOI] [PubMed] [Google Scholar]
- 33.Jin Z., Du X., Xu Y., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 34.Mengist H.M., Mekonnen D., Mohammed A., et al. Potency, safety, and pharmacokinetic profiles of potential inhibitors targeting SARS-CoV-2 main protease. Front. Pharmacol. 2020;11:630500. doi: 10.3389/fphar.2020.630500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelly S., Liotta D. Potent SARS-CoV-2 direct-acting antivirals provide an important complement to COVID-19 vaccines. ACS Cent. Sci. 2021;7(3):396–399. doi: 10.1021/acscentsci.1c00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unoh Y., Uehara S., Nakahara K., et al. Discovery of S-217622, a non-covalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. bioRxiv. 2022:2022. doi: 10.1101/2022.01.26.477782. 01.26.477782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao K., Pan T., Mou Y., et al. Scutellarein inhibits BLM-mediated pulmonary fibrosis by affecting fibroblast differentiation, proliferation, and apoptosis. Ther. Adv.Chronic Dis. 2020;11 doi: 10.1177/2040622320940185. 2040622320940185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao R., Zhu B.H., Tang S.B., et al. Scutellarein inhibits hypoxia- and moderately-high glucose-induced proliferation and VEGF expression in human retinal endothelial cells. Acta Pharmacol. Sin. 2008;29(6):707–712. doi: 10.1111/j.1745-7254.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu M.S., Lee J., Lee J.M., et al. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22(12):4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayama K. Vitro and animal models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020;41(8):513–517. doi: 10.1016/j.tips.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41(6):363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.