Abstract
The TAR DNA Binding Protein (TARDBP) gene has become relevant after the discovery of its several pathogenic mutations. The lack of evolutionary history is in contrast to the amount of studies found in the literature. This study investigated the evolutionary dynamics associated with the retrotransposition of the TARDBP gene in primates. We identified novel retropseudogenes that likely originated in the ancestors of anthropoids, catarrhines, and lemuriformes, i.e. the strepsirrhine clade that inhabit Madagascar. We also found species-specific retropseudogenes in the Philippine tarsier, Bolivian squirrel monkey, capuchin monkey and vervet. The identification of a retropseudocopy of the TARDBP gene overlapping a lncRNA that is potentially expressed opens a new avenue to investigate TARDBP gene regulation, especially in the context of TARDBP associated pathologies.
Subject terms: Evolutionary genetics, Molecular evolution
Introduction
The availability of whole-genome sequences has accelerated research on the evolution of different genetic elements. Together with genomic DNA-based gene duplication, an important source of evolutionary innovation are the events of RNA retrotranscription and its insertion into the genome1,2. In mammals, retrotranscription depends on the long interspersed nuclear element 1 (L1 or LINE1) enzymatic machinery encoded by retrotransposable elements, which generate an intronless gene duplicate that could produce a protein similar to the parental counterpart3. However, most retrotranscribed sequences are inserted at a random position in the genome, lacking all necessary transcription elements and becoming a pseudogene, a phenomenon called “dead on arrival”3. However, because a significant number of retrocopies are located in introns of other genes, they have potential to regulate their host genes functioning as antisense transcripts4–6. On the other hand, in the human genome, a number of retrocopies overlap with long noncoding RNAs (lncRNAs)7, which are regulatory noncoding RNAs of > 200 nucleotides8. lncRNAs can establish specific interactions with nucleic acids and proteins, acting in diverse fashions as critical regulators of gene expression in several biological processes, including pathological conditions such as cancer and neurodegenerative disorders9.
Retrocopies are abundant in placental mammals, especially in primates10. Extensive evidence indicates that their presence is related to several types of diseases, including neurodegenerative disorders11. Because retrocopies have potential to produce harmful effects on genomes and transcriptomes, silencing mechanisms seem to have evolved to restrict retrotransposition. Intriguingly, during the life of healthy humans the brain is the only known somatic tissue where retrotransposition is de-repressed12. Therefore, identifying the presence of retrocopies/retropseudogenes is not only an important piece of information to have a complete picture of the evolution of any particular gene, but also is necessary to fully understand human health.
The TAR DNA Binding Protein (TARDBP) gene, which encodes the Transactive response DNA-binding protein 43 kDa (TDP-43), has gained considerable attention after the initial discovery that its mutations can cause familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), two major forms of neurodegenerative disorders13,14, with ALS being the most frequent motor neuron disorder in adults15. Up to date, more than 50 pathogenic missense mutations have been characterized13,14. TDP-43 is an RNA-binding protein with a variety of RNA metabolism functions, including transcription, mRNA transport and stabilization, miRNA biogenesis, lncRNA processing, and translation16. More recent findings indicate that TDP-43 participates in the pathogenesis of other neurodegenerative disorders of several other proteinopathies, such as Parkinson’s disease and Alzheimer’s disease, which are conditions characterized by toxic protein aggregation17. In human cells, under physiological conditions, TDP-43 mainly localizes in the nucleus, but in neurons and glial cells of ALS and FTD patients it shuttles and accumulates in the cytoplasm where eventually aggregates and contribute to the onset and progression of these diseases18–22.
The TARDBP gene is conserved in species that share a common ancestor deep in time23, suggesting that this gene carries out essential functions. This gene underwent an event of positive selection in the ancestor of mammals24, suggesting functional adaptations for the group. More recently in evolutionary time, it has been shown that during the evolution of humans, genes related to diseases like Alzheimer's also underwent positive selection25. Although events of positive selection are seen as conferring selective advantage, as a by-product, they can also have adverse effects26. In this regard, it is proposed that human susceptibility to neurodegenerative disorders could be a consequence of improving our cognitive function27,28. Besides these studies, not much is known regarding the evolution of TARDBP in primates. Understanding the evolutionary history of genes represents a critical piece of information, among other things, to perform meaningful comparisons and to understand the variation in function in different species. However, evolutionary studies have been primarily directed to the functional copy, while much less is known of other phenomena that could potentially impact the functions associated with the gene. In this regard, the evolution of primates is characterized by a peak of retrotransposition activity in the anthropoid ancestor29,30, which left a signature of intronless copies, functional or not, of a number of genes in the genome.
The aim of this study is to investigate the retrotransposition dynamics associated with the TARDBP gene in primates. According to our phylogenetic and synteny analyses, we identified retropseudogenes that originated at different times during the evolution of primates. TARDBP retropseudogenes originated in the anthropoid ancestor, between 67 and 43.2 million years ago, in the ancestor of catarrhines, between 43.2 and 29.4 million years ago, and in the ancestor of lemuriformes, i.e. the strepsirrhine clade that inhabit Madagascar, between 59.3 and 55 million years ago. We also found species-specific retropseudogenes in the Philippine tarsier (Carlito syrichta), Bolivian squirrel monkey (Saimiri boliviensis), capuchin monkey (Cebus capucinus imitator) and vervet (Chlorocebus sabaeus). Although annotated sequences are not putatively functional, the identification of a retropseudocopy overlapping a lncRNA opens a new avenue to investigate TARDBP gene regulation.
Results
Multiple retropseudogenes lineages characterize the evolution of the TARDBP gene in primates
According to our phylogenetic and synteny analyses, we identified retropseudogenes of the TARDBP gene that originated at different times during the evolution of primates. We identified retropseudogenes originated in the ancestor of anthropoids, between 67 and 43.2 million years ago, in the ancestor of catarrhines, between 43.2 and 29.4 million years ago, and in the ancestor of lemuriformes, i.e. the strepsirrhine clade that inhabit Madagascar, between 59.3 and 55 million years ago (Fig. 1). More recently in evolutionary time, we found species-specific retropseudogenes in the Philippine tarsier (Carlito syrichta), Bolivian squirrel monkey (Saimiri boliviensis), capuchin monkey (Cebus capucinus imitator) and vervet (Chlorocebus sabaeus). All of them did not have intron sequences and were identified on a different autosome in comparison to the chromosomal location of the functional copy (Fig. 1). Our gene tree did not significantly deviate from the most updated phylogenetic hypotheses for the main group of primates31–33, suggesting that the functional copy of the TARDBP gene was present as a simple copy gene in the ancestor of the group (Fig. 1).
Figure 1.
Maximum likelihood tree showing sister group relationships between the functional copy of TARDBP and primate retropseudogenes. Numbers on the nodes correspond to support values, i.e. the confidence of each node. TARDBP sequences from the African elephant (Loxodonta africana), blue whale (Balaenoptera musculus) and red fox (Vulpes vulpes) were used as outgroups (not shown). The scale denotes substitutions per site and colors represent lineages. The pink lineage represents the TARDBP functional copy. Synteny information is provided for each lineage at the right side of the figure.
We recovered three highly supported monophyletic groups containing representative species of all major groups of anthropoids i.e. apes, Old World monkeys and New World monkeys (light blue, brown and green lineages, Fig. 1), indicating that these retropseudogenes originated in the ancestor of the group, between 67 and 43.2 millions of years ago, and were maintained in representative species of all descendant primate groups. The retropseudogene lineage depicted with the purple shading (Fig. 1), although it was not recovered monophyletic, our synteny analyses suggest that it indeed belongs to a single lineage (Fig. 1). Representative species of the three purple clades possess the same flanking genes, DIAPH3 at the 5´ side and TDRD3 at the 3´ side of the retropseudogene, strongly suggesting that the lack of monophyly could be attributed to a phylogenetic artifact (Fig. 1). The small number of changes, as illustrated by the short branches that define the sister group relationships of the main clades, could be the main cause (Fig. 1). We also found a retropseudogene lineage that according to our phylogenetic tree originated in the ancestor of catarrhine primates, the group that includes apes and Old World monkeys (blue lineage, Fig. 1), between 43.2 and 29.4 million years ago. In this case, we recovered a clade containing the functional copy of the TARDBP gene in catarrhines (upper pink lineage, Fig. 1), sister to a group containing a retropseudogene in the same primate group (blue lineage, Fig. 1). The clade containing TARDBP functional sequences from New World monkeys was recovered sister to the above mentioned clade (Fig. 1). In this clade in addition to the functional TARDBP copy, we found New World monkey specific retropseudogenes for which the evolutionary history is difficult to resolve given the shortness of the branches (Fig. 1). We identified three retropseudogenes, two in the capuchin monkey (Cebus capucinus imitator) and one in the Bolivian squirrel monkey (Saimiri boliviensis). Finally, we recovered a sequence from the Angola colobus (Colobus angolensis)(yellow branch, Fig. 1), which was recovered sister to a clade containing the TARDBP functional copy (pink lineage, Fig. 1) and three retropseudogenes lineages (blue, purple and green clades, Fig. 1). The phylogenetic position of this branch in our gene tree suggests that it represents a retropseudogene originated in the anthropoid ancestor, but only conserved in this species. In support of this claim, the single flanking gene (PRMT2) found in the genomic piece containing the TARDBP retropseudogene in the Angola colobus is not shared with any other gene lineage described in this study (Fig. 1). In all cases, the identified retropseudogenes during the evolutionary history of anthropoid primates have premature stop codons, insertions and/or deletions (supplementary Figs. 1–5).
We also identified retropseudogenes in tarsiers and strepsirrhines (Fig. 1). We found a single retropseudogene in the Philippine tarsier (Carlito syrichta), which shows the hallmark of a sequence free from selective constraints, i.e., a long branch as a signal of an accelerated rate of evolution in comparison to the functional copy (Fig. 1). In the strepsirrhine clade we identified a highly supported lineage containing the TARDBP functional copy in three species, greater bamboo lemur (Prolemur simus), coquerel's sifaka (Propithecus coquereli) and the mouse lemur (Microcebus murinus), which in turn was recovered sister to an also highly supported clade containing retropseudogenes in the greater bamboo lemur (Prolemur simus) and coquerel's sifaka (Propithecus coquereli) (Fig. 1). This tree topology suggests that this retrocopy originated in the ancestor of lemuriformes, i.e. the strepsirrhine clade that inhabits Madagascar, between 59.3 and 55 million years ago, and it has been maintained in the genome of descendant species. Finally, the functional copy of the bushbaby (Otolemur garnettii) was recovered sister to the lemuriformes clade. Similar to the case of anthropoids, all retropseudogenes identified in tarsiers and strepsirrhines have premature stop codons, insertions and/or deletions (supplementary Figs. 6 and 7).
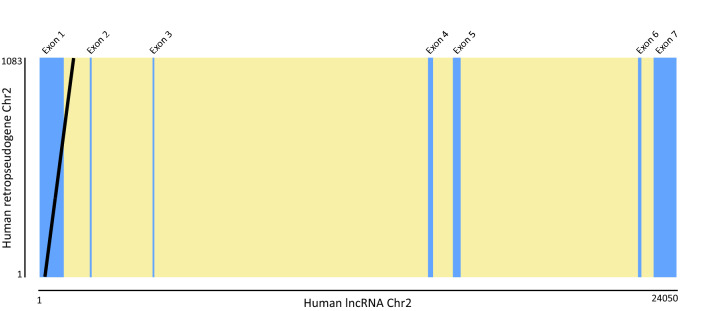
Regarding the location of the retropseudocopies, most of them are within intergenic regions (Table 1). The exception is the one located on chromosome 2, which overlaps with a lncRNA gene (Fig. 2). Specifically, the retropseudocopy starts on position 198 of the first lncRNA exon and finishes on position 375 of the first lncRNA intron.
Table 1 .
Information regarding the genomic location of the TARDBP functional copy and retropseudocopies in humans.
| Chromosome | Type | Genomic coordinates | Region type | Flanking genes | Orientation |
|---|---|---|---|---|---|
| 1 | Functional copy | 11012344–11030528 | Intergenic | C1orf127-TARDBP-MASP2 | Forward |
| 2 | Retropseudocopy | 238231881–238232964 | lncRNA* | ILKAP-RPC-HES6 | Forward |
| 6 | Retropseudocopy | 102453,143–102454069 | Intergenic | GRIK2-RPC-HACE1 | Reverse |
| 8 | Retropseudocopy | 63136407–63138084 | Intergenic | TTPA-RPC-THDF3 | Reverse |
| 13 | Retropseudocopy | 6274779–60276011 | Intergenic | DIAPH3-RPC-TDRD3 | Forward |
| 20 | Retropseudocopy | 6200989–6202191 | Intergenic | FERMT1-RPC-BMP2 | Reverse |
*The ID of the lncRNA is ENSG00000225057; RPC, retropseudocopy.
Figure 2.
Graphical representation of the alignment between the lncRNA (ENSG00000225057)(x-axis) and the retropseudogene (y-axis) sequences of humans located on chromosome 2. Light blue and light yellow vertical rectangles denote exons and introns, respectively. The black diagonal line indicates a locally alignable region of high sequence identity.
Discussion
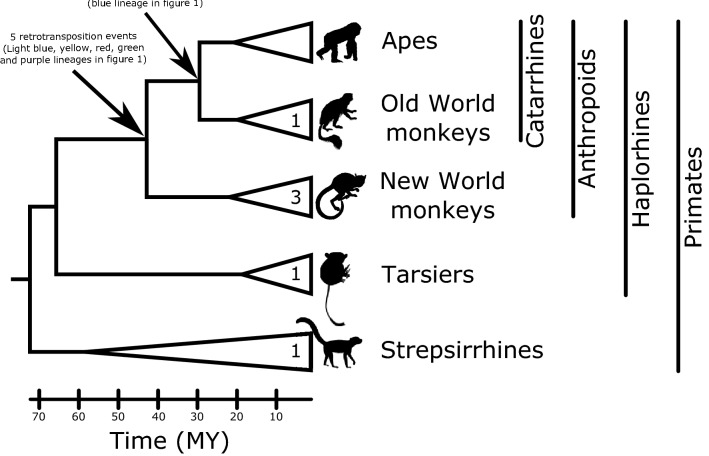
In this study we revealed that the evolutionary history of TARDBP, a gene that in humans encodes TDP-43, an RNA-binding protein involved in several neurodegenerative disorders13,14, is characterized by the presence of retropseudogenes that originated at different ages during the evolutionary history of primates. An important fraction of the retropseudogenes originated in the anthropoid ancestor, between 67 and 43.2 million years ago, and has remained in the genome of the species (Fig. 3). This phenomenon fits the expectation of a peak of retrocopy formation around 40 million years ago, which coincides with an increased activity of L1 retroelements that produced an increment in SINE/Alu retrocopy repeat amplification29,30. Interestingly, this period of time represents a key moment during the evolutionary history of primates, the radiation of the anthropoid lineage, where significant morphological and physiological traits arose34. Thus, this period of Vesuvian mode of evolution could be seen as a source of evolutionary novelty that fueled the origin of the phenotypes that define the anthropoid lineage2,35,36. Other retropseudogenes originated in the catarrhine ancestor, between 43.2 and 29.4 million years ago, and in other primate groups (Fig. 3).
Figure 3.
Time calibrated primate phylogeny showing the origin of the different retropseudogene lineages depicted in Fig. 1. Triangles represent the diversification of each primate group, where the left side angle defines the ancestor of each group. Numbers on the triangles correspond to retropseudogenes originated within a particular group of primates. Silhouette images were obtained from PhyloPic (http://phylopic.org/). Divergence times were obtained from timetree (http://www.timetree.org/).37.
In agreement with the literature, and given the nature of the process originating retrocopies, all of them seem to be non-functional as canonical TARDBP3, which can be verified by the presence of insertions, deletions and/or premature stop codons (supplementary Figs. 1 and 7). The identification of several retropseudogenes for the TARDBP gene in primates appears to be not a surprise as this gene complies with all the requisites to be a gene with multiple retropseudogenes38–40, i.e., short transcripts (coding for 61 to 414 amino acids)41, widely and highly expressed42, low GC-content (47%, average among 23 primate species) and highly conserved (3.4%, maximal divergence among primates). Furthermore, in agreement with the slow rate of pseudogene length shortening over time, the identified retropseudogenes possess a length (mean 1128 bp, median 1193 bp) similar to the functional TARDBP gene (1245 bp).
Among apes, the number of TARDBP retrotransposition events appear to be higher in comparison to the average number of retrocopies per parental gene in their genomes43. On average, ape genomes possess 2.9 retrocopies per parental gene43, however in our study we identified five TARDBP retropseudogenes in each examined ape species. Coincident with previous evidence, we also found a higher number of retropseudogenes in New World monkeys43. Although it is not clear why this group of primates has more retrocopies compared to catarrhines, it is suggested that a specific lineage expansion of L1PA1 and L1P3 subelements could be related to the observed pattern2,43.
TDP-43 binds to long clusters of GU-rich RNA sequences, which in humans are found in one-third of transcribed genes44. This allows TDP-43 to regulate the processing of thousands of transcripts, including that of its own transcript45. In fact, TDP-43 establishes a tightly regulated feedback loop46. It has been demonstrated that a twofold increase or decrease in TDP-43 levels is sufficient to promote neurodegeneration45. Thus, TARDBP retropseudogenes could represent an additional layer of regulation of TDP-43 levels and activity. In this regard, it seems interesting that one of the retropseudocopies is located in a lncRNA (Fig. 2), a pattern that appears not unusual in the human genome47. This fact opens the possibility that this retropseudocopy regulates the expression of the functional copy of TARDBP48–50. In fact, blast searches against the expressed sequence tags (est) database, which represent a snapshot of genes expressed in a given tissue and/or at a specific developmental stage, show at least one record (BI825397), which possesses an identity value of 90.5% with the retropseudocopy located on chromosome 2 and is expressed in medulla in an adult male. In contrast, with the TARDBP functional copy, the identity value is 70.6%. It will be important to determine whether in humans the levels of TDP-43 are affected by the levels of this lncRNA, in particular in the brain of patients suffering the aforementioned neurodegenerative disorders.
In conclusion, in this work, we demonstrate that the TARDBP gene in primates has an evolutionary history characterized by the presence of multiple retropseudogene lineages. In the ancestor of anthropoids occurred a significant increment of retrotransposition activity, which led to intronless sequences that cannot give rise to functional proteins. However, the fact that one of the retropseudocopies is present in a lncRNA and is transcribed opens the opportunity to investigate further its role in regulating the expression of the functional TARDBP gene copy, and its influence in the outcome or fate of the associated neurodegenerative disorders.
Methods
DNA sequences
DNA sequences and phylogenetic analyses
We performed searches for TAR DNA Binding Protein (TARDBP) genes in primate genomes in Ensembl v.10241. We retrieved primate orthologs, using the human (Homo sapiens) entry, based on the ortholog prediction function of Ensembl v.10241. We identified TARDBP retropseudogenes in primate species by performing BLASTN searches51, against the whole genome sequence in Ensembl v.10241 using default settings. In each case the query sequence (TARDBP) was from the same species of the genome in which retropseudogenes were looking for. In our searches, a retropseudogene was recognized as a sequence containing all exons together and found in a different chromosome in comparison to the functional copy. Genomic fragments containing retropseudogenes were extracted and manually annotated by comparing the coding sequence of the same species using the program Blast2seq v2.552 with default parameters. Accession numbers and details about the taxonomic sampling are available in Supplementary Table S1.
Nucleotide sequences were aligned using MAFFT v.753, allowing the program to choose the alignment strategy (FFT-NS-i). We used the proposed model tool of IQ-Tree v.1.6.1254 to select the best-fitting model of nucleotide substitution, which selected GTR + F + R3. We used the maximum likelihood method to obtain the best tree using the program IQ-Tree v1.6.1255. We assessed support for the nodes using three strategies: a Bayesian-like transformation of aLRT (aBayes test)56, SH-like approximate likelihood ratio test (SH-aLRT)57 and the ultrafast bootstrap approximation58. We carried out 20 independent runs to explore the tree space, and the tree with the highest likelihood score was chosen. TARDBP sequences from the African elephant (Loxodonta africana), blue whale (Balaenoptera musculus) and red fox (Vulpes vulpes) were used as outgroups.
Assessment of conserved synteny
We examined genes found upstream and downstream of functional copies and retropseudogenes. We used the estimates of orthology and paralogy derived from the Ensembl Compara database59; these estimates are obtained from a pipeline that considers both synteny and phylogeny to generate orthology mappings. These predictions were visualized using the program Genomicus v100.0160. Our assessments were performed in representative species for each lineage.
Supplementary Information
Acknowledgements
This work was supported by Fondo Nacional de Desarrollo Científico y Tecnológico from Chile (FONDECYT 1210471) and Millennium Nucleus of Ion Channel Associated Diseases (MiNICAD), Iniciativa Científica Milenio, Ministry of Economy, Development and Tourism from Chile to JCO, Fondo Nacional de Desarrollo Científico y Tecnológico from Chile (FONDECYT 1180957) to FJM and LVC and Fondo Nacional de Desarrollo Científico y Tecnológico from Chile (FONDECYT 1211481) to GAM.
Author contributions
J.C.O. and G.A.M. designed the study. K.Z., J.C.O. collected and analyzed data. J.C.O. and G.A.M. wrote the manuscript. L.V.C., F.J.M. reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Juan C. Opazo, Email: jopazo@gmail.com
Gonzalo A. Mardones, Email: gonzalo.mardones@uach.cl
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07908-8.
References
- 1.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: Mechanistic and evolutionary insights. Nat. Rev. Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casola C, Betrán E. The genomic impact of gene retrocopies: What have we learned from comparative genomics, population genomics, and transcriptomic analyses? Genome Biol. Evol. 2017;9:1351–1373. doi: 10.1093/gbe/evx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- 4.Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genom. Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 7.Kubiak MR, Szcześniak MW, Makałowska I. Complex analysis of retroposed genes’ contribution to human genome. Proteome Trans. Genes. 2020;11:542. doi: 10.3390/genes11050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nie L, et al. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 9.Aliperti V, Skonieczna J, Cerase A. Long non-coding RNA (lncRNA) roles in cell biology, neurodevelopment and neurological disorders. Noncoding RNA. 2021;7:36. doi: 10.3390/ncrna7020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- 11.Ciomborowska-Basheer J, Staszak K, Kubiak MR, Makałowska I. Not So dead genes-retrocopies as regulators of their disease-related progenitors and hosts. Cells. 2021;10:912. doi: 10.3390/cells10040912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terry DM, Devine SE. Aberrantly high levels of somatic LINE-1 expression and retrotransposition in human neurological disorders. Front. Genet. 2019;10:1244. doi: 10.3389/fgene.2019.01244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 15.Chiò A, et al. Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology. 2013;41:118–130. doi: 10.1159/000351153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson KA, Kim SH, Tibbetts RS. RNA-binding proteins in neurodegenerative disease: TDP-43 and beyond. Wiley Interdiscip. Rev. RNA. 2012;3:265–285. doi: 10.1002/wrna.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klim JR, Pintacuda G, Nash LA, Juan IGS, . & Eggan, K. K. Connecting TDP-43 Pathology with Neuropathy. Trends Neurosci. 2021 doi: 10.1016/j.tins.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 19.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 20.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 21.Heyburn L, Moussa CE-H. TDP-43 in the spectrum of MND-FTLD pathologies. Mol. Cell. Neurosci. 2017;83:46–54. doi: 10.1016/j.mcn.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinarbasi ES, et al. Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci. Rep. 2018;8:7083. doi: 10.1038/s41598-018-25008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H-Y, Wang I-F, Bose J, Shen C-KJ. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, et al. TDP-43 facilitates milk lipid secretion by post-transcriptional regulation of Btn1a1 and Xdh. Nat. Commun. 2020;11:341. doi: 10.1038/s41467-019-14183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vamathevan JJ, et al. The role of positive selection in determining the molecular cause of species differences in disease. BMC Evol. Biol. 2008;8:273. doi: 10.1186/1471-2148-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt RD, Nesse RM, Williams GC. Why we get sick: The new science of Darwinian medicine. Ecology. 1996;77:983. [Google Scholar]
- 27.Gearing M, Rebeck GW, Hyman BT, Tigges J, Mirra SS. Neuropathology and apolipoprotein E profile of aged chimpanzees: Implications for Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1994;91:9382–9386. doi: 10.1073/pnas.91.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: Which evolutionary genetic models work best? Behav. Brain Sci. 2006;29:385–404. doi: 10.1017/S0140525X06009095. [DOI] [PubMed] [Google Scholar]
- 29.Ohshima K, et al. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pozzi L, et al. Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 2014;75:165–183. doi: 10.1016/j.ympev.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finstermeier K, et al. A mitogenomic phylogeny of living primates. PLoS One. 2013;8:e69504. doi: 10.1371/journal.pone.0069504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kay RF, Ross C, Williams BA. Anthropoid origins. Science. 1997;275:797–804. doi: 10.1126/science.275.5301.797. [DOI] [PubMed] [Google Scholar]
- 35.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 36.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Stecher G, Suleski M, Hedges SB. Timetree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr. Opin. Genet. Dev. 2004;14:328–335. doi: 10.1016/j.gde.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 39.McDonell L, Drouin G. The abundance of processed pseudogenes derived from glycolytic genes is correlated with their expression level. Genome. 2012;55:147–151. doi: 10.1139/g2012-002. [DOI] [PubMed] [Google Scholar]
- 40.Gonçalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yates AD, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlén M, et al. Proteomics: Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 43.Navarro FCP, Galante PAF. A genome-wide landscape of retrocopies in primate genomes. Genome Biol. Evol. 2015;7:2265–2275. doi: 10.1093/gbe/evv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rengifo-Gonzalez JC, El Hage K, Clément M-J, Steiner E, Joshi V, Craveur P, Durand D, Pastré D, Bouhss A. The cooperative binding of TDP-43 to GU-rich RNA repeats antagonizes TDP-43 aggregation. eLife. 2021 doi: 10.7554/eLife.67605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weskamp K, Barmada SJ. TDP43 and RNA instability in amyotrophic lateral sclerosis. Brain Res. 2018;1693:67–74. doi: 10.1016/j.brainres.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayala YM, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milligan MJ, et al. Global intersection of long non-coding RNAs with processed and unprocessed pseudogenes in the human genome. Front. Genet. 2016;7:26. doi: 10.3389/fgene.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: Emerging regulators of gene expression. Front. Genet. 2014;5:476. doi: 10.3389/fgene.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nam J-W, Choi S-W, You B-H. Incredible RNA: Dual Functions of Coding and Noncoding. Mol. Cells. 2016;39:367–374. doi: 10.14348/molcells.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Y, et al. Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat. Commun. 2018;9:903. doi: 10.1038/s41467-018-03311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 53.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anisimova M, Gil M, Dufayard J-F, Dessimoz C, Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SMJ, Amode R, Brent S, Spooner W, Kulesha E, Yates A, Flicek P. Ensembl comparative genomics resources. Database. 2016;2016:bav096. doi: 10.1093/database/bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen NTT, Vincens P, Roest Crollius H, Louis A. Genomicus 2018: Karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 2018;46:D816–D822. doi: 10.1093/nar/gkx1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.