Abstract
This study aimed to investigate the prevalence of foodborne pathogenic bacteria in bovine milk, their antibiogram phenotype, and the carriage of antibiotic resistance genes. Raw bovine milk samples (n = 100) were randomly collected from different suppliers in the northwest of Iran. Antibiotic-resistant patterns and the presence of antibiotic resistance genes were evaluated in the isolates. Escherichia coli, Listeria monocytogenes, Staphylococcus aureus, and Salmonella spp. were isolated from 78%, 47%, 25%, and 21% of samples, respectively. All isolates showed high rates of resistance to amoxicillin, penicillin, and cefalexin. The blaTEM and blaSHV genes were detected in 50.0% and 6.4% of E. coli isolates, respectively. Also, 28.5% and 19.0% of Salmonella isolates were positive for blaTEM and blaSHV. The frequency of mecA and blaZ in S. aureus isolates was 20.0% and 12.0%, respectively. The high prevalence of bovine milk contamination with antimicrobial-resistant species in this study necessitates precise control on antibiotic prescription in veterinary medicine.
Subject terms: Antimicrobials, Bacteria
Introduction
The burden of foodborne diseases in humans remains largely unknown1. During the past decade, the incidence of foodborne microbial diseases has considerably increased in most countries2.
Milk and dairy products, as common food products in many countries, provide favorable environments for the growth of many microorganisms because of their nutrient composition3. Many studies have been performed to improve raw milk quality to reduce the risk of microbial contamination and to increase the chemical nutritional quality of dairy products3,4. In recent years, the consumption of raw milk has been increasingly welcomed due to its potential benefits such as having high nutritional content and beneficial bacteria as well as the prevention of lactose intolerance. However, due to the potential presence of pathogens and their toxins, the consumption of raw milk can pose a serious risk of foodborne disease to public health5–8. Staphylococcus aureus, Salmonella spp., Listeria monocytogenes, and Escherichia coli are the most common pathogens that can be found in raw milk and dairy products made from raw milk4,7. Also, S. aureus, L. monocytogenes, and Salmonella spp. can contribute to bovine mastitis and be excreted directly into the milk8–10.
Inappropriate use of antibiotics is a common problem in medical and veterinary medicine, which may result in the development of multidrug-resistant microorganisms11. The antibiotic resistance in pathogenic bacteria is known as a big challenge for public health worldwide12–14. One of the most important enzymes involved in antibiotic resistance of bacteria is beta-lactamase, especially extended-spectrum beta-lactamase (ESBL), which deactivate the beta-lactam antibiotics through hydrolysis of beta-lactam ring15. The most common ESBL-producing genes are SHV (blaSHV), TEM (blaTEM), and CTX-M genes (blaCTX-M)16.
Over the last decades, mecA has been detected in S. aureusisolates17. The mecA gene is responsible for resistance to methicillin and other β-lactam antibiotics. This gene encodes a penicillin-binding protein (PBP2A) with a low affinity for β-lactam antibiotics18,19. Also, blaZ has been reported as the main gene in S. aureus responsible for resistance against several antibiotics20. TEM and SHV-type β-lactamases are reported as the main causes of resistance in E. colistrains21. Also, numerous beta-lactamases such as TEM, SHV, PER, OXA, and CTX enzymes have been identified in different Salmonella species22. Therefore it seems important to investigate the antibiotic resistance patterns of pathogenic bacteria and the presence of associated encoding genes as the key elements of antibiotics resistance.
This study aimed to evaluate the prevalence of pathogenic foodborne bacteria in raw bovine milk through culture-based techniques, their antibiogram phenotype, and the presence of antibiotic resistance genes among the isolates using multiplex-PCR.
Material and methods
Sampling
Raw bovine milk samples (n = 100) were collected aseptically from different retail sellers in the northwest of Iran. At the seller level, all milk samples were stored in the refrigerator (≤ 4 °C). Samples were transported to the laboratory in an icebox at a temperature less than 4 °C. They were kept in a refrigerator at 4 ± 1 °C before analysis. The microbiological experiments were performed immediately. All microbiological culture mediums were provided by Merck Company (Darmstadt, Germany).
Total bacterial count
Serial tenfold dilutions of raw milk samples were prepared using the tubes containing 9 ml of sterile % 0.1 peptone water (up to 1:10,000 dilutions)23. Then, 0.1 mL of each sample dilutions was cultured on Nutrient agar. The total mesophilic bacterial count was calculated after the plates were incubated aerobically at 37 °C for 48 h24.
Isolation and detection of pathogenic bacteria
Eecherichia coli was isolated from samples according to the method of Feng et al.25and Ombarak et al.26 Three to five presumptive colonies (dark centered and flat colonies with metallic green sheen) from Levine’s Eosin Methylene Blue (L-EMB) agar plates were selected, transferred on tryptic soy agar (TSA), and incubated at 37 °C for 24 h. Biochemical confirmatory tests were performed according to the method of Feng et al.25 and Quinn et al.27.
Staphylococcus aureus was detected in the samples using Baird-parker agar. After incubation of plates at 37 °C for 48 h, typical black colonies with a clear zone were considered as presumptive S. aureus. The isolates were confirmed by biochemical tests such as coagulase, catalase, DNase, lecithinase, oxidase, Lysostaphin sensitivity, VP, urease, glucose, and mannitol fermentation28.
For isolation and detection of L. monocytogenes, samples were enriched in Buffered Listeria enrichment broth (BLEB) at 30 °C for 48 h. The bacterial suspension was streaked onto PALCAM agar and incubated at 35 °C for 48 h. The isolates were confirmed by motility test, gram staining, and biochemical tests such as catalase, oxidase, hemolysis, nitrate reduction, carbohydrate fermentation, Christie-Atkins-Munch-Peterson test (CAMP), methyl red, and Voges-Proskauer (MR/VP)29.
For isolation and detection of Salmonella spp., the raw milk samples were cultured on Bismuth Sulphite agar (BSA), Brilliant Green, and Phenol-Red agar (BGA) for 24 h (BGA)/48 h (BSA) at 37 °C. The suspected colonies were transferred to Samonella-Shigella agar plates and incubated at 37 °C for another 24 h. The presumptive colonies on the plates were subjected to biochemical tests using Lysine Iron agar, Triple Sugar Iron Agar, Sulfide-Indole-Motility medium, and Christensen’s Urea agar30.
Antimicrobial susceptibility test
Antibiotic susceptibility tests of isolates were performed by the Kirby-Bauer disk diffusion method according to the guidelines of clinical laboratory standards31. Isolates were included in the study based on isolation rank (time criterion). Based on this criterion, the first isolate of a particular species isolated from a single sample was included in the analysis32. Briefly, bacterial suspensions were prepared in tubes containing 0.9% (w/v) phosphate-buffered saline with turbidity adjusted to 0.5 McFarland standard. Using a sterile cotton swab, bacterial suspension was streaked uniformly on the surface of Muller-Hinton agar. Antibiotic disks (Padtan Teb, Iran) including amoxicillin (25 μg/disk), azithromycin (15 μg), penicillin (10 IU), cephalexin (30 μg), ceftriaxone (30 μg), gentamicin(10 μg), chloramphenicol (30 μg), and tetracycline (30 μg) were placed on the surface of cultures. The selected antimicrobials were representative of the major classes of antibiotics commonly used in veterinary and human medicine in Iran. Finally, the diameter of the inhibition zone around the disks was measured after incubation of plates at 37 °C for 24 h.
Detection of blaTEM, blaSHV, mecA, and blaZ genes using multiplex-PCR
The genomic DNA was extracted by boiling method33. The primers used for the detection of target genes are listed in Table 1. The reaction contents for each 25 μL PCR consisted of 5.5 μL of deionized water, 12.5 μL RED-Extract-N-Amp master mix 2 × (containing buffer, salts, dNTPs, Taq polymerase, REDTaq dye, and JumpStart Taq antibody) (Sigma-Aldrich, USA), 1 μL of each primer and 3 μL of template DNA. The PCR program for blaTEM and blaSHV genes included initial denaturation for 5 min at 94 °C followed by 32 cycles of denaturation at 94 °C for 30 s, annealing step at 54 °C for 30 s, extension step at 72 °C for 60 s, and a final extension step at 72 °C for 10 min. The PCR condition for mecA and blaZ were as follows: initial denaturation at 95 °C for 4 min, 30 cycles of denaturation at 95 °C for 60 s, annealing step at 58 °C for 60 s, extension step at 72 °C for 60 sand final extension step at 72 °C for 4 min. PCR products were subjected to electrophoresis using 1.5% (w/v) agarose gel. The gel was stained with ethidium bromide. Ultraviolet transillumination (Biorad, USA) was applied for the visualization of DNA.
Table 1.
PCR sets used for detection of target antibiotic-resistance genes in the selected foodborne bacterial isolated from bovine milk.
| Genes | Primer sequence (5' → 3') | Accession number (GenBank) | Annealing (°C) | Amplicon size (bp) | References |
|---|---|---|---|---|---|
| blaTEM | F:ATC AGC AAT AAA CCA GC | NG_068216.1 | 54 | 516 | Eid and Samir34 |
| R: CCC CGA AGA ACG TTT TC | |||||
| blaSHV | F: AGG ATT GAC TGC CTT TTTG | NG_068212.1 | 54 | 392 | Yukawa et al.35 |
| R: ATT TGC TGA TTT CGCTCG | |||||
| mecA | F: AAA ATC GAT GGT AAA GGT TGG C | MK659556.1 | 58 | 532 | Kim et al.36 |
| R: AGT TCT GCA GTA CCG GAT TTG C | |||||
| blaZ | F: TGA CCA CTT TTA TCA GCA ACC | MN689952.1 | 58 | 700 | Meroni et al.37 |
| R: GCC ATT TCA ACA CCT TCT TTC |
Results and discussion
Several studies have revealed that food products such as raw milk and dairy products made from raw milk may be the main sources for the outbreak of antibiotic-resistance pathogens which are known as a challenge for the safety of food products38. This problem is common in developing countries such as Iran, because of the poor food handling practices, inadequate food safety regulations, weak hygienic practices, insufficient financial resources to invest in food safety, weak regulatory systems, and inadequate education for food handlers. In the countries with outbreaks of foodborne diseases, the importance of pathogens like S. aureus, E. coli, L. monocytogenes, and Salmonella spp. has been reported as major causes39.
Numerous researchers previously reported the antimicrobial resistance of E. coli and Salmonella isolates from raw milk to the most common antibiotics in their studies39–42. Also, methicillin-resistant S. aureus as an emerging pathogen has become an important challenge for public health that has been isolated from raw milk11,43. The multidrug-resistant of L. monocytogenes isolates from raw milk to some commonly used antibiotics is reported in various countries such as Ethiopia44, Turkey45, Egypt46, and Pakistan47. So, the present study was designed to study the occurrence of the most common antibiotic-resistant foodborne pathogens from raw milk in Iran.
Totalmesophilic bacterial count, isolation, and identification of bacterial species
The mean total mesophilic bacterial count of the examined raw milk samples in this study was 5.75 ± 0.85 log10 cfu mL−1 which was exceeded the permitted maximum value of raw milk contamination (5 log10 cfu mL−1)48. Our findings of the high rate of contamination in raw milk are in agreement with that of the previous study conducted in Tabriz, indicating the poor microbial quality of raw milk delivered to pasteurized milk plants4. In another study which was conducted in Allahabad city (India), the total bacterial count of examined milk samples was reported between 4.79 log10 cfu mL−1 by Yadav et al.48. Even, a higher level of contamination of about 6.32 ± 0.03 log10 cfu mL−1 was found for the raw milk samples from the collection centers of Guwahati city in India49. In general, the total bacterial count of more than 6 log10 cfu mL−1 reported by many countries is not desirable for raw milk supplies and is not usable for human consumption50.
The increased total bacterial count can be caused by the use of unsanitary equipment for milking, contamination of cow’s udders, inadequate cooling of milk, and occasionally by the milking of cows with mastitis51.
In the present study, 78% of samples were contaminated with E. coli with a mean count of 3.41 ± 0.41 log10 cfu mL−1. High rates of raw milk contamination with E. coli have been reported in many developing and developed countries. It has been reported that 90.67% of raw milk samples in Arusha, Tanzania were contaminated with E. coli52 as well as 76.4% of samples in Egypt26.
In our study, 25% of the raw milk samples were contaminated with S. aureus at an average level of 2.91 ± 0.80 log10 cfu mL−1. In agreement with our study, a study in California showed that 25.3% of the raw milk samples were contaminated with S. aureus5. In another study in Mansoura City, Egypt, the mean S. aureus counts were found to be 3.49 log10 cfu g−1 in raw milk samples43, and 70.4% of raw milk samples in Brazil were contaminated with S. aureus53. These results indicate the different quality of milk samples in different regions of the world.
According to the results of the present study, L. monocytogenes was isolated from 47% of the raw milk samples. Over 70% of positive samples contained L. monocytogenes at a level of less than 10 cfu ml−1. The mean count of this bacterium was detected at 0.60 ± 0.51 log10 cfu mL−1. Many studies in different countries reported the occurrence of L. monocytogenes by various rates of contamination in their raw milk supplies and related products. The occurrence of L. monocytogenes in raw milk has been reported in Kars city (Turkey)45. However, in research in Antakya, Turkey, L. monocytogenes was not detected in any of the raw milk samples54.
In the present study, Salmonella spp. was detected in 21% of the raw milk samples. After enrichment of samples followed by plating, the mean count of Salmonella spp. in the positive samples was detected at 0.26 ± 0.27 log10 cfu mL−1. Similar results have also been reported in different countries. The prevalence of Salmonella spp. in raw milk has also been reported in Arusha, Tanzania (37.33%)52, Egypt(44.44%)55, and Dhaka Metropolis, Bangladesh (25.71%)56.
Antimicrobial susceptibility of isolates to the used antibiotics and detection of blaTEM,blaSHV, mecA, and blaZ genes in the isolates
In this study, it was shown that all strains of E. coli were highly resistant to penicillin (88.46%), cefalexin (82.05%), and amoxicillin (70.51%) (Table 2). Fifty percent (50%) of E. coli isolates had blaTEM and 6.41%of them were positive for blaSHV (Table 3). Consistent with our study, another study reported that 83.1% of isolates of highly antibiotic-resistant E. coli strains, with 100% resistance to acetyl spiramycin, 100% to penicillin, 98.8% to lincomycin, 98.8% to oxacillin, 32.5% to cephalosporin, and 30.1% to ampicillin. The blaTEM was the most frequently detected resistance gene (83.1%)42.
Table 2.
Antibiotic resistance profile of Escherichia coli isolates (n = 78) from bovine milk samples.
| Antimicrobial agent | Disk content | Interpretive categories and zone diameter breakpoints (nearest whole mm)* | No. of isolates (%) | ||||
|---|---|---|---|---|---|---|---|
| R** | I | S | R | I | S | ||
| Azithromycin | 15 μg | ≤ 12 | – | ≥ 13 | 53 (67.94) | – | 25 (32.05) |
| Chloramphenicol | 30 μg | ≤ 12 | 13–17 | ≥ 18 | 15 (19.23) | 2 (2.56) | 61 (78.20) |
| Ceftriaxone | 30 μg | ≤ 19 | 20–22 | ≥ 23 | 17 (21.79) | 14 (17.94) | 47 (60.25) |
| Penicillin | 10 IU | ≤ 14 | – | ≥ 15 | 69 (88.46) | – | 9 (11.53) |
| Gentamicin | 10 μg | ≤ 12 | 13–14 | ≥ 15 | 6 (7.69) | 3 (3.84) | 69 (88.46) |
| Amoxicillin | 25 μg | ≤ 13 | 14–16 | ≥ 17 | 55 (70.51) | 3 (3.84) | 20 (25.64) |
| Tetracycline | 30 μg | ≤ 11 | 12–14 | ≥ 15 | 20 (25.64) | 9 (11.53) | 49 (62.82) |
| Cephalexin | 30 μg | ≤ 14 | – | ≥ 15 | 64 (82.05) | – | 14 (17.94) |
*From CLSI31.
**S susceptible, I intermediate, R resistant.
Table 3.
Distribution of resistance genes in the selected foodborne bacterial isolated from bovine milk.
| Target genes | No of isolates (%) | |||
|---|---|---|---|---|
| E. coli | Salmonella spp. | L. monocytogenes | S. aureus | |
| blaSHV | 5 (6.41%) | 4 (19.04) | – | – |
| blaZ | – | – | – | 3 (12.00) |
| blaTEM | 39 (50%) | 6 (28.57) | – | – |
| mecA | – | – | – | 5 (20.00) |
In the present study, blaTEM was the most common resistance gene in E. coli isolates. However, only 50% of the resistant isolates to both penicillin and amoxicillin harbored this gene. Also, blaSHV was present in five isolates of E. coli. All isolates containing this gene showed resistance to cephalexin, penicillin, and amoxicillin in phenotypic experiments.
The isolated strains of L. monocytogenes in our study were highly resistant to penicillin, cefalexin, and amoxicillin (97.87%) (Table 4). Since ampicillin is an important first-choice antibiotic for the treatment of listeriosis57, the isolates of L. monocytogenes were evaluated for the presence of known genes responsible for resistance to beta-lactam antibiotics (blaTEM, blaSHV, mecA, blaZ) using the specific primers. However, none of the resistance genes were detected in L. monocytogenes (Table 3). Similar results were found by Marian et al.58 that showed 100% of L. monocytogenes strains in their study were resistant to ampicillin and penicillin, with no involvement of blaZ and mecA genes in their resistance. Also, Bertsch et al.57 examined the antimicrobial susceptibility and antibiotic resistance genes in foodborne, clinical, and environmental isolates of L. monocytogenes that were negative for the presence of blaZ and mecA genes.
Table 4.
Antibiotic resistance profile of Listeria monocytogenes isolates (n = 47) from bovine milk samples.
| Antimicrobial agent | Disk content | Interpretive categories and zone diameter breakpoints (nearest whole mm)* | No. of isolates (%) | ||||
|---|---|---|---|---|---|---|---|
| R** | I | S | R | I | S | ||
| Azithromycin | 15 μg | ˂ 17 | 17–21 | ≥ 22 | 12 (25.53) | 14 (29.78) | 21 (44.68) |
| Chloramphenicol | 30 μg | ˂ 18 | 18–20 | ≥ 21 | 22 (46.80) | 13 (27.65) | 12 (25.53) |
| Ceftriaxone | 30 μg | ˂ 15 | 15–20 | ≥ 21 | 17 (36.17) | 18 (38.29) | 12 (25.53) |
| Penicillin | 10 IU | < 8 | 8–28 | ≥ 29 | 46 (97.87) | 0 (0) | 1 (2.12) |
| Gentamicin | 10 μg | < 18 | 18–20 | ≥ 21 | 24 (51.06) | 21 (44.68) | 2 (4.25) |
| Amoxicillin | 25 μg | ˂ 14 | 14–24 | ≥ 25 | 46 (97.87) | 1 (2.12) | 0 (0) |
| Tetracycline | 30 μg | ˂ 22 | 22–24 | ≥ 25 | 23 (48.93) | 2 (4.25) | 22 (46.80) |
| Cephalexin | 30 μg | ˂ 12 | 12–17 | ≥ 18 | 46 (97.87) | 1 (2.12) | 0 (0) |
The results of antimicrobial resistance tests showed that the isolated strains of Salmonella were highly resistant to penicillin (100%), cefalexin (100%), and amoxicillin (71.42%) (Table 5). High rates of antibiotic resistance for Salmonella spp. have been reported by many studies. In a study by Obaidat and Stringer (2019), more than 50% of S. enterica isolates in raw milk were resistant to kanamycin, streptomycin, amoxicillin, and tetracycline. In another study, the highest rate of antibiotic resistance for Salmonella was detected to ampicillin, chloramphenicol, streptomycin, sulfonamide, tetracycline, amoxicillin, ceftiofur, and ceftriaxone41. These results were consistent with the results obtained from the present study.
Table 5.
Antibiotic resistance profile of Salmonella spp. isolates (n = 21) from bovine milk samples.
| Antimicrobial agent | Disk content | Interpretive categories and zone diameter breakpoints (nearest whole mm)* | No. of isolates (%) | ||||
|---|---|---|---|---|---|---|---|
| R** | I | S | R | I | S | ||
| Azithromycin | 15 μg | ≤ 12 | – | ≥ 13 | 8 (38.09) | – | 13 (61.90) |
| Chloramphenicol | 30 μg | ≤ 12 | 13–17 | ≥ 18 | 6 (28.57) | 1 (4.76) | 14 (66.66) |
| Ceftriaxone | 30 μg | ≤ 19 | 20–22 | ≥ 23 | 5 (23.80) | 3 (14.28) | 13 (61.90) |
| Penicillin | 10 IU | ≤ 14 | – | ≥ 15 | 21 (100) | – | 0 (0) |
| Gentamicin | 10 μg | ≤ 12 | 13–14 | ≥ 15 | 0 (0) | 0 (0) | 21 (100) |
| Amoxicillin | 25 μg | ≤ 13 | 14–16 | ≥ 17 | 15 (71.42) | 3 (14.28) | 3 (14.28) |
| Tetracycline | 30 μg | ≤ 11 | 12–14 | ≥ 15 | 5 (23.80) | 7 (33.33) | 9 (42.85) |
| Cephalexin | 30 μg | ≤ 14 | – | ≥ 15 | 21 (100) | – | 0 (0) |
*From CLSI31.
**S susceptible, I intermediate, R resistant.
In this study, six (28.57%) and 4 (19.04%) isolates of Salmonella spp. were positive for blaTEM and blaSHV, respectively (Table 3). Four isolates with multidrug resistance to penicillin, ceftriaxone, amoxicillin, and cephalexin, carried both blaTEM and blaSHV genes. In a study by Ranjbar et al.62 the frequency of Salmonella spp. with blaTEM and blaSHV genes was 29.9% and 2.89%, while the prevalence of these two genes in Salmonella in another study was reported 15.38% and 12.82%, respectively63. The results of these studies were in agreement with the present study.
Staphylococcus aureus isolates were highly resistant to amoxicillin (100%), cephalexin (100%), and penicillin (84.00%), respectively (Table 6). Antimicrobial resistance in S. aureus species is very common in raw milk samples, as reported by many researchers. Li et al.64 indicated that 80.5% of S. aureus isolates were resistant to penicillin and ampicillin. The resistance of S. aureus isolates to penicillin G (87.9%), cloxacillin (75.9%), and amoxicillin (55.6%) was also reported by Al-Ashmawyet al.43 in Mansoura City, Egypt.
Table 6.
Antibiotic resistance profile of Staphylococcus aureus isolates (n = 25) from bovine milk samples.
| Antimicrobial agent | Disk content | Interpretive categories and zone diameter breakpoints (nearest whole mm)* | No. of isolates (%) | ||||
|---|---|---|---|---|---|---|---|
| R** | I | S | R | I | S | ||
| Azithromycin | 15 μg | ≤ 13 | 14–17 | ≥ 18 | 8 (32.00) | 8 (32.00) | 9 (36.00) |
| Chloramphenicol | 30 μg | ≤ 12 | 13–17 | ≥ 18 | 6 (24.00) | 13 (52.00) | 6 (24.00) |
| Ceftriaxone | 30 μg | ≤ 13 | 14–20 | ≥ 21 | 6 (24.00) | 12 (48.00) | 7 (28.00) |
| Penicillin | 10 IU | ≤ 28 | – | ≥ 29 | 21 (84.00) | – | 4 (16.00) |
| Gentamicin | 10 μg | ≤ 12 | 13–14 | ≥ 15 | 3 (12.00) | 21 (84.00) | 1 (4.00) |
| Amoxicillin | 25 μg | ≤ 28 | – | ≥ 29 | 25 (100) | – | 0 (0) |
| Tetracycline | 30 μg | ≤ 14 | 15–18 | ≥ 19 | 7 (28.00) | 11 (44.00) | 7 (28.00) |
| Cephalexin | 30 μg | ≤ 21 | – | ≥ 22 | 25 (100) | – | 0 (0) |
The presence of the mecA gene was found in five (20%) isolates of S. aureus and the blaZ gene was positive in three (12.00%) isolates of S. aureus (Table 3). Notably, S. aureus isolates with phenotypic resistances to penicillin, amoxicillin, ceftriaxone, and cephalexin always harbored mecA and blaZ either individually or concurrently. These two genes are common genes involved in the antibiotic resistance of S. aureus strains. The electrophoresis pattern of the PCR products of the resistance genes in the bacteria under this study is shown in Figs. 1, 2 and 3.
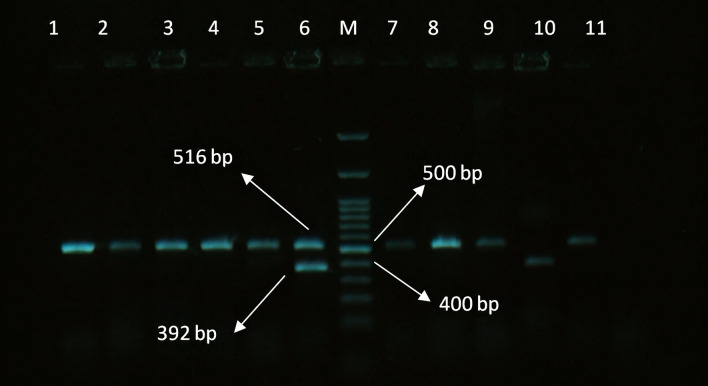
Figure 1.
Electrophoresis pattern obtained by multiplex PCR for detection of blaTEM and blaSHV genes in E. coli isolates. M: marker 100 bp; lane 1, 2, 3, 4, 5, 6, 7, 8, 9, 11: amplification of blaTEM gene at 516 bp; lane 6, 10: amplification of blaSHV gene at 392 bp.
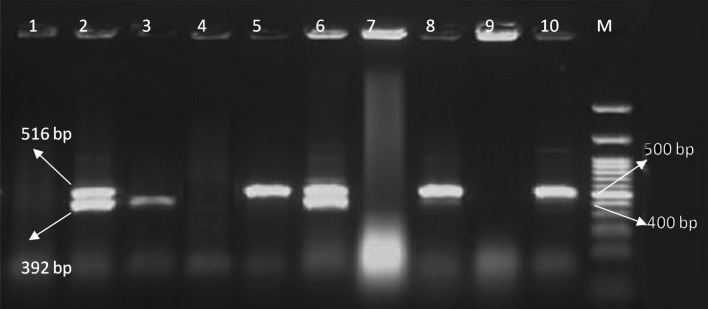
Figure 2.
Electrophoresis pattern obtained by multiplex PCR for detection of blaTEM and blaSHV genes in Salmonella isolates. M: marker 100 bp; Lanes 2, 5, 6, 8, and 10: amplification of blaTEM gene at 516 bp; lane 2, 3, 6: amplification of blaSHV gene at 392 bp.
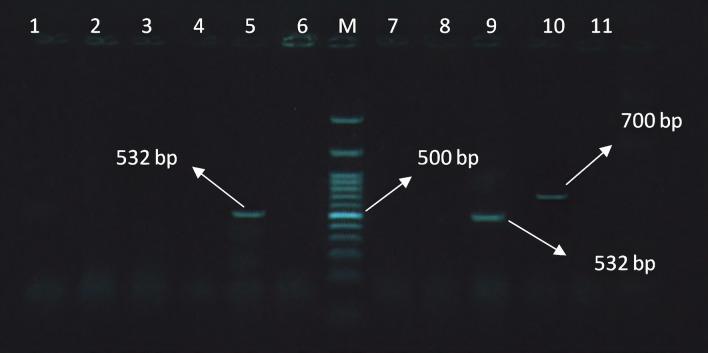
Figure 3.
Electrophoresis pattern obtained by multiplex PCR for detection of mecA and blaZ genes in S. aureus isolates. M: marker 100 bp; Lanes 5 and 9: amplification of mecA gene at 532 bp; lane 10: amplification of blaZgene at 700 bp.
The blaZ and mecA were identified as resistance genes in S. aureus isolated from subclinical mastitis in Egypt65. In another study that investigated the genomic profile of S. aureus isolates from bulk tank milk and dairy cows with clinical mastitis, the prevalence of blaZ gene was detected in 17.2% of isolates66.
Resistance to penicillin, amoxicillin, cephalexin, and ceftriaxone was more prevalent than the associated antibiotic resistance genes between isolates. The discrepancies between the phenotypic resistances and associated resistance genes in this study may be due to the fact that the entire suite of resistant genes, which could result in phenotypic resistance, was not evaluated in this study. Also, it is possible that the antibiotic-resistant genes detected may be mutated and/or non-functional, inducible or not expressed. Other mechanisms of resistance such as multidrug efflux pumps, mutations in outer membrane porins, or other unknown resistance genes may be involved in the phenotypic resistance67,68.
In the present study, high resistance levels and multidrug resistances against up to 7 antibiotics were detected between the evaluated isolates, with a high proportion for beta-lactams. Since beta-lactams are the most commonly used antibiotics in veterinary medicine, the emergence of beta-lactam-resistant pathogenic bacteria can be a serious threat to the wide use of these drugs69.
The occurrence of antibiotic-resistance pathogens in raw milk can be directly affected by farm management and practices. Regular cleaning of the farm can decrease the prevalence of antibiotic resistance pathogens70. The types of animal breeding (intensive, semi-intensive, or free-ranging) can influence the occurrence of antibiotic resistance pathogens due to the inappropriate administration of antibiotics. Excessive use of antibiotics in therapeutic and sub-therapeutic levels in dairy cattle farms can result in the presence of antibiotic-resistant pathogens in raw milk. So, if raw milk is not heat-treated, the presence of antibiotic-resistant foodborne pathogens in raw milk may pose food safety hazards to humans70,71.
Conclusion
Our results show that raw milk has a great potential for transmission of antibiotic-resistant pathogens such as E. coli, S. aureus, L. monocytogenes, and Salmonella spp. In the present study, high levels of resistance were observed among the screened isolates to the most common beta-lactams such as amoxicillin, penicillin, and cefalexin. Also, the prevalence of beta-lactamase genes in E. coli, S. aureus, and Salmonella spp. provided evidence on the high risk of resistant food-borne pathogens to humans through raw milk.
Since antibiotics have extensive applications in dairy cattle farms in developing countries such as Iran; the microbiota of raw milk may contain relatively high levels of antibiotic-resistance bacteria. Therefore, enhancing the safety of milk and implementing good manufacturing practices are extremely important for the health of consumers. Pasteurization of raw milk, prevention of cross-contamination, storage of raw milk in cold temperature, appropriate authority supervision, and regulatory monitoring on the use of antibiotics in dairy cattle farms are necessary to ensure the safety of milk and dairy products.
The main route for the contamination of raw milk with resistant bacteria can be the subject of future studies to determine whether these bacteria get into the milk via cow’s udder or mixed into the milk during or after milking. Since phylogenetic assays can be used to ensure the genetic variations of resistant bacteria; it is recommended that these assays be performed on foodborne pathogenic isolates in future studies.
Acknowledgements
This article is provided from the thesis of Mrs. Sima Hassani entitled “Microbiological quality, antimicrobial resistance and genomic investigation of pathogenic bacteria isolated from raw milk in Iran” in the master’s degree in the University of Tabriz, Iran. The authors are grateful to Vice-Chancellery for Research and Technology, University of Tabriz, for their financial support in performing this research [grant number 43.8641].
Author contributions
S.H.: Data curation, funding, investigation, project administration, data analysis; M.-H.M.: Conceptualization, design of methodology, supervision for the research activities, verification of data; writing of the original draft, writing and editing of the manuscript; S.N.G.: Data curation, Design of methodology, supervision for the research activities, investigation, project administration, analysis and verification of data; S.A.K.: investigation, editing of the manuscript; A.H.: editing of the manuscript; Z.B.: editing of the manuscript. All authors have reviewed and approved the final manuscript.
Funding
This work was supported by the vice-chancellery for research and technology, University of Tabriz.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H, Opsteegh M, Langelaar M, Threfall J, Scheutz F, Van der Giessen J. Food-borne diseases: The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010;139:S3–15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. General Information Related to Microbiological Risks in Food. http://www.who.int/foodsafety/micro/general/en/index.html (2012)
- 3.Moosavy MH, Kordasht HK, Khatibi SA, Sohrabi H. Assessment of the chemical adulteration and hygienic quality of raw cow milk in the northwest of Iran. Qual. Assur. Saf. Crops Foods. 2019;11:491–498. [Google Scholar]
- 4.Moosavy M, Mahmoudi R, Ghorbanpour E, Khatibi SA. Evaluation of microbial and physicochemical characteristics of raw cow milk delivered to pasteurized milk plants in Tabriz city, Iran. J. Food Res. 2018;28:183–196. [Google Scholar]
- 5.Heidinger JC, Winter CK, Cullor JS. Quantitative microbial risk assessment for Staphylococcus aureus and Staphylococcus enterotoxin A in raw milk. J. Food Prot. 2009;72:1641–1653. doi: 10.4315/0362-028x-72.8.1641. [DOI] [PubMed] [Google Scholar]
- 6.Moosavy MH, Hallaj Salahipor M, Mostafavi E, Khatibi SA. Risk factors for human brucellosis in Mianeh, Iran. J. Zoonotic Dis. 2018;3:10–21. [Google Scholar]
- 7.Sugrue I, Tobin C, Ross RP, Stanton C, Hill C. In: Foodborne Pathogens and Zoonotic Diseases. Nero LA, De Carvalho AF, editors. Academic Press; 2019. pp. 259–272. [Google Scholar]
- 8.Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implication. Foodborne Pathogen Disease. 2005;2:115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- 9.Ding T, Suo Y, Zhang Z, Liu D, Ye X, Chen S, Zhao YA. Multiplex RT-PCR assay for S. aureus, L. monocytogenes, and Salmonella spp. detection in raw milk with pre-enrichment. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobirka M, Tancin V, Slama P. Epidemiology and classification of mastitis. Animals. 2020;10:2212. doi: 10.3390/ani10122212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asiimwe BB, Baldan R, Trovato A, Cirillo DM. Prevalence and molecular characteristics of Staphylococcus aureus, including methicillin-resistant strains, isolated from bulk can milk and raw milk products in pastoral communities of South-West Uganda. BMC Infect. Dis. 2017;17:422. doi: 10.1186/s12879-017-2524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frieri M, Kumar K, Boutin A. Antibiotic resistance. J. Infect. Public Health. 2017;10:369–378. doi: 10.1016/j.jiph.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MKF, Baloch Z. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018;2:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abadi ATB, Rizvanov AA, Haertlé T, Blatt NL. World Health Organization report: Current crisis of antibiotic resistance. BioNano Sci. 2019;9:778–788. [Google Scholar]
- 15.Thenmozhi S, Moorthy K, Sureshkumar B, Suresh M. Antibiotic resistance mechanism of ESBL producing Enterobacteriaceae in clinical field: A review. Int. J. Pure Appl. Biosci. 2014;2:207–226. [Google Scholar]
- 16.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 17.Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao X, Cullen PJ, Liu D, Muhammad AI, Chen S, Ye X, Wang J, Ding T. Combating Staphylococcus aureus and its methicillin resistance gene (mecA) with cold plasma. Sci. Total Environ. 2018;645:1287–1295. doi: 10.1016/j.scitotenv.2018.07.190. [DOI] [PubMed] [Google Scholar]
- 19.Elhassan MM, Ozbak HA, Hemeg HA, Elmekki MA, Ahmed LM. Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. BioMed Res. Int. 2015;2015:1–5. doi: 10.1155/2015/895860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen JE, Christensen H, Aarestrup FM. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 2006;57:450–460. doi: 10.1093/jac/dki492. [DOI] [PubMed] [Google Scholar]
- 21.Elumalai S, Muthu G, Selvam REM, Ramesh S. Detection of TEM, SHV and CTX-M-type β-lactamase production among clinical isolates of Salmonella species. J. Med. Microbiol. 2014;63:962–967. doi: 10.1099/jmm.0.068486-0. [DOI] [PubMed] [Google Scholar]
- 22.Na SH, Moon DC, Kang HY, Song HJ, Kim SJ, Choi JH, Yoon JW, Yoon SS, Lim SK. Molecular characteristics of extended-spectrum β-lactamase/AmpC-producing Salmonella enterica serovar Virchow isolated from food-producing animals during 2010–2017 in South Korea. Int. J. Food Microbiol. 2020;322:1–9. doi: 10.1016/j.ijfoodmicro.2020.108572. [DOI] [PubMed] [Google Scholar]
- 23.Weldaragay H, Yilma Z, Tekle-Giorgis Y. Hygienic practices and microbiological quality of raw milk produced under different farm size in Hawassa, southern Ethiopia. Wudpecker J. Agric. Res. Rev. 2012;4:132–142. [Google Scholar]
- 24.Lianou DT, Michael CK, Vasileiou NG, Petinaki E, Cripps PJ, Tsilipounidaki K, Katsafadou AI, Politis AP, Kordalis NG, Ioannidi KS, Gougoulis DA. Extensive countrywide field investigation of somatic cell counts and total bacterial counts in bulk-tank raw milk in sheep flocks in Greece. Foods. 2021;10:1–18. doi: 10.3390/foods10020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng, P., Weagant, SD., Grant, M. A., Burkhardt, W., Shellfish, M. & Water, B. BAM: Enumeration of Escherichia coli and the Coliform Bacteria. https://www.fda.gov/food/laboratory-methods-food/bam-chapter-4-enumeration-Escherichia-coli-and-coliform-bacteria (2002).
- 26.Ombarak RA, Hinenoya A, Awasthi SP, Iguchi A, Shima A, Elbagory ARM, Yamasaki S. Prevalence and pathogenic potential of Escherichia coli isolates from raw milk and raw milk cheese in Egypt. Int. J. Food Microbiol. 2016;221:69–76. doi: 10.1016/j.ijfoodmicro.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Quinn PJ, Markey BK, Leonard FC, Fitzpatrick ES, Fanning S, Hartigan PJ. In: Veterinary Microbiology and Microbial Disease. Sayers M, editor. Blackwell Science; 2011. pp. 143–148. [Google Scholar]
- 28.Tallent, S., Hait, J., Bennett, R. W. & Lancette, G. A. Bam Chapter12: Staphylococcus aureus. https://www.fda.gov/food/laboratory-methods-food/bam-chapter-12-Staphylococcus-aureus (2019).
- 29.Hitchins, A. D., Jinneman, K. & Chen, Y. BAM Chapter 10: Detection of Listeria Monocytogenes in Foods and Environmental Samples, and Enumeration of Listeria Monocytogenes in Foods. https://www.fda.gov/food/laboratory-methods-food/bam-chapter-10-detection-Listeria-monocytogenes-foods-and-environmental-samples-and-enumeration (2017).
- 30.Andrews WH, Flowers RS, Siliker J, Bailey JS, Labbe RG. In: Compendium of Methods for the Microbiological Examination of Foods. Downes FP, Ito K, editors. American Public Health Association; 2001. pp. 357–376. [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100 (31th ed.). https://clsi.org/standards/products/microbiology/documents/m100 (2021).
- 32.Cornaglia G, Hryniewicz W, Jarlier V, Kahlmeter G, Mittermayer H, Stratchounski L, Baquero F. European recommendations for antimicrobial resistance surveillance. Clin. Microbiol. Infect. 2004;10:349–383. doi: 10.1111/j.1198-743X.2004.00887.x. [DOI] [PubMed] [Google Scholar]
- 33.Nayak R, Stewart TM, Nawaz MS. PCR identification of Campylobacter coli and Campylobacter jejuni by partial sequencing of virulence genes. Mol. Cell. Probes. 2005;19:187–193. doi: 10.1016/j.mcp.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Eid S, Samir AH. Extended-spectrum beta-lactamase and class 1 integrons in multidrug-resistant Escherichia coli isolated from turkeys. Vet. World. 2019;12:1167–1174. doi: 10.14202/vetworld.2019.1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yukawa S, Uchida I, Tamura Y, Ohshima S, Hasegawa T. Characterisation of antibiotic resistance of Salmonella isolated from dog treats in Japan. Epidemiol. Infect. 2019;147:1–6. doi: 10.1017/S0950268819000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YH, Kim HS, Kim S, Kim M, Kwak HS. Prevalence and characteristics of antimicrobial-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus aureus from retail meat in Korea. Food Sci. Anim. Resour. 2020;40:758–771. doi: 10.5851/kosfa.2020.e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meroni G, Soares Filipe JF, Drago L, Martino PA. Investigation on antibiotic-resistance, biofilm formation and virulence factors in multi drug resistant and non multi drug resistant Staphylococcus pseudintermedius. Microorganisms. 2019;7:1–11. doi: 10.3390/microorganisms7120702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulusoy BH, Chirkena K. Two perspectives of Listeria monocytogenes hazards in dairy products: The prevalence and the antibiotic resistance. Food Qual. Saf. 2019;3:233–241. [Google Scholar]
- 39.Tadesse HA, Gidey NB, Workelule K, Hailu H, Gidey S, Bsrat A, Taddele H. Antimicrobial resistance profile of E. coli isolated from raw cow milk and fresh fruit juice in Mekelle. Tigray. Ethiopia. Vet. Med. Int. 2018;2018:1–7. doi: 10.1155/2018/8903142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obaidat MM, Stringer AP. Prevalence, molecular characterization, and antimicrobial resistance profiles of Listeria monocytogenes, Salmonella enterica, and Escherichia coli O157:H7 on dairy cattle farms in Jordan. J. Dairy Sci. 2019;102:8710–8720. doi: 10.3168/jds.2019-16461. [DOI] [PubMed] [Google Scholar]
- 41.Van Kessel JS, Sonnier J, Zhao S, Karns JS. Antimicrobial resistance of Salmonella enterica isolates from bulk tank milk and milk filters in the United States†. J. Food Prot. 2013;76:18–25. doi: 10.4315/0362-028X.JFP-12-263. [DOI] [PubMed] [Google Scholar]
- 42.Yu ZN, Wang J, Ho H, Wang YT, Huang SN, Han RW. Prevalence, antimicrobial-resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J. Glob. Antimicrob. Resist. 2020;22:94–101. doi: 10.1016/j.jgar.2019.12.016. [DOI] [PubMed] [Google Scholar]
- 43.Al-Ashmawy MA, Sallam KI, Abd-Elghany SM, Elhadidy M, Tamura T. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathog. Dis. 2016;13:156–162. doi: 10.1089/fpd.2015.2038. [DOI] [PubMed] [Google Scholar]
- 44.Girma Y, Abebe B. Isolation, identification and antimicrobial susceptibility of Listeria species from raw bovine milk in Debre-Birhan Town, Ethiopia. J. Zoonotic Dis. Public Health. 2018;2:4. [Google Scholar]
- 45.Aksoy A, Sezer Ç, Vatansever L, Gülbaz G. Presence and antibiotic resistance of Listeria monocytogenes in raw milk and dairy products. Kafkas Univ. Vet. Fak. Dergis. 2018;24:415–421. [Google Scholar]
- 46.Tahoun AB, Abou Elez RM, Abdelfatah EN, Elsohaby I, El-Gedawy AA, Elmoslemany AM. Listeria monocytogenes in raw milk, milking equipment and dairy workers: Molecular characterization and antimicrobial resistance patterns. J. Glob. Antimicrob. Resist. 2017;10:264–270. doi: 10.1016/j.jgar.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Gohar S, Abbas G, Sarfraz M, Ali S, Ashraf M, Aslam R, Yaseen K. Prevalence and antimicrobial resistance of Listeria monocytogenes isolated from raw milk and dairy products. Matrix Sci. Med. 2017;1:10–14. [Google Scholar]
- 48.Yadav J, Paul S, Peter JK, Kumar Y, Singh AK, Masih F, Masih H. Comparative evaluation of pathogenic bacterial incidence in raw and pasteurized milk. Int. J. Eng. Sci. 2014;3:11–20. [Google Scholar]
- 49.Dinki N, Balcha E. Detection of antibiotic residues and determination of microbial quality of raw milk from milk collection centres. Adv. Anim. Vet. Sci. 2013;1:80–83. [Google Scholar]
- 50.Kivaria F, Noordhuizen J, Kapaga A. Evaluation of the hygienic quality and associated public health hazards of raw milk marketed by smallholder dairy producers in the Dar es Salaam region, Tanzania. Trop. Anim. Health Prod. 2006;38:185–194. doi: 10.1007/s11250-006-4339-y. [DOI] [PubMed] [Google Scholar]
- 51.Vietoris V, Zajac P, Zubrická S, Čapla J, Čurlej J. Comparison of total bacterial count (TBC) in bulk tank raw cow's milk and vending machine milk. Carpathian J. Food Sci. Technol. 2016;8:184–191. [Google Scholar]
- 52.Lubote R, Shahada F, Matemu A. Prevalence of Salmonella spp. and Escherichia coli in raw milk value chain in Arusha, Tanzania. Am. J. Res. Commun. 2014;2:1–13. [Google Scholar]
- 53.Rall VLM, Vieira FP, Rall R, Vieitis RL, Fernandes A, Candeias JMG, Cardoso KFG, Araújo JP. PCR detection of staphylococcal enterotoxin genes in Staphylococcus aureus strains isolated from raw and pasteurized milk. Vet. Microbiol. 2008;132:408–413. doi: 10.1016/j.vetmic.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 54.Aygun O, Pehlivanlar S. Listeria spp. in the raw milk and dairy products in Antakya, Turkey. Food Control. 2006;17:676–679. [Google Scholar]
- 55.Elafify M, Darwish WS, Al-Ashmawy M, Elsherbini M, Koseki S, Kawamura S, Abdelkhalek A. Prevalence of Salmonella spp. in Egyptian dairy products: Molecular, antimicrobial profiles and a reduction trial using d-tryptophan. J. Consum. Prot. Food Saf. 2019;14:399–407. [Google Scholar]
- 56.Yasmin S, Parveen S, Munna MS, Noor R. Detection of Salmonella spp. and microbiological analysis of milk and milk based products available within Dhaka Metropolis, Bangladesh. Microbiol. Res. J. Int. 2015;5:474–480. [Google Scholar]
- 57.Bertsch D, Muelli M, Weller M, Uruty A, Lacroix C, Meile L. Antimicrobial susceptibility and antibiotic resistance gene transfer analysis of foodborne, clinical, and environmental Listeria spp isolates including Listeria monocytogenes. Microbiol. Open. 2014;3:118–127. doi: 10.1002/mbo3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marian M, Aminah SS, Zuraini M, Son R, Maimunah M, Lee H, Wong W, Elexson N. MPN-PCR detection and antimicrobial resistance of Listeria monocytogenes isolated from raw and ready-to-eat foods in Malaysia. Food Control. 2012;28:309–314. [Google Scholar]
- 59.CA-SFM Comitéde l’Antibiogramme de la Socie´te´ Franc¸aise de Microbiologie Report 2003. Int. J. Antimicrob. Agents. 2003;21:364–391. doi: 10.1016/s0924-8579(03)00021-9. [DOI] [PubMed] [Google Scholar]
- 60.Hansen JM, Gerner-Smidt P, Bruun B. Antibiotic susceptibility of Listeria monocytogenes in Denmark 1958–2001. APMIS. 2005;113:31–36. doi: 10.1111/j.1600-0463.2005.apm1130105.x. [DOI] [PubMed] [Google Scholar]
- 61.Soussy CJ, Cluzel R, Courvalin P. Definition and determination of in vitro antibiotic susceptibility breakpoints for bacteria in France. Eur. J. Clin. Microbiol. Infect. Dis. 1994;13:238–246. doi: 10.1007/BF01974543. [DOI] [PubMed] [Google Scholar]
- 62.Ranjbar R, Ardashiri M, Samadi S, Afshar D. Distribution of extended-spectrum β-lactamases (ESBLs) among Salmonella serogroups isolated from pediatric patients. Iran. J. Microbiol. 2018;10:294–299. [PMC free article] [PubMed] [Google Scholar]
- 63.Aljanaby AAJ, Medhat AR. Research article prevalence of some antimicrobials resistance associated-genes in Salmonella typhi isolated from patients infected with typhoid fever. J. Biol. Sci. 2017;17:171–184. [Google Scholar]
- 64.Li L, Zhou L, Wang L, Xue H, Zhao X. Characterization of methicillin-resistant and-susceptible staphylococcal isolates from bovine milk in northwestern China. PLoS ONE. 2015;10:e0116699. doi: 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdeen EE, Walid M, Hussien H, Roshdy S. PCR for detection of virulence and antibiotic resistance genes of coagulase-positive Staphylococcus aureus from clinical mastitis in Egypt. Int. J. Basic Appl. Sci. 2015;4:315–319. [Google Scholar]
- 66.Ronco T, Klaas IC, Stegger M, Svennesen L, Astrup LB, Farre M, Pedersen K. Genomic investigation of Staphylococcus aureus isolates from bulk tank milk and dairy cows with clinical mastitis. Vet. Microbiol. 2018;215:35–42. doi: 10.1016/j.vetmic.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Davis MA, Besser TE, Orfe LH, Baker KN, Lanier AS, Broschat SL, New D, Call DR. Genotypic-phenotypic discrepancies between antibiotic resistance characteristics of Escherichia coli isolates from calves in management settings with high and low antibiotic use. Appl. Environ. Microbiol. 2011;77:3293–3299. doi: 10.1128/AEM.02588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith M, Do TN, Gibson JS, Jordan D, Cobbold RN, Trott DJ. Comparison of antimicrobial resistance phenotypes and genotypes in enterotoxigenic Escherichia coli isolated from Australian and Vietnamese pigs. J. Glob. Antimicrob. Resist. 2014;2:162–167. doi: 10.1016/j.jgar.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Ghazaei C. Phenotypic and molecular detection of Beta-Lactamase enzyme produced by Bacillus cereus isolated from pasteurized and raw milk. J. Med. Bacteriol. 2019;8:1–7. [Google Scholar]
- 70.Kamaruzzaman EA, Abdul Aziz S, Bitrus AA, Zakaria Z, Hassan L. Occurrence and characteristics of extended-spectrum β-Lactamase-producing Escherichia coli from dairy cattle, milk, and farm environments in peninsular Malaysia. Pathogens. 2020;9:1–10. doi: 10.3390/pathogens9121007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wallensten A, Hernandez J, Ardiles K, Gonzalez-Acuna D, Drobni M, Olsen B. Extended spectrum beta-lactamases detected in Escherichia coli from gulls in Stockholm, Sweden. Infect. Ecol. Epidemiol. 2011;1:1–4. doi: 10.3402/iee.v1i0.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.