Abstract
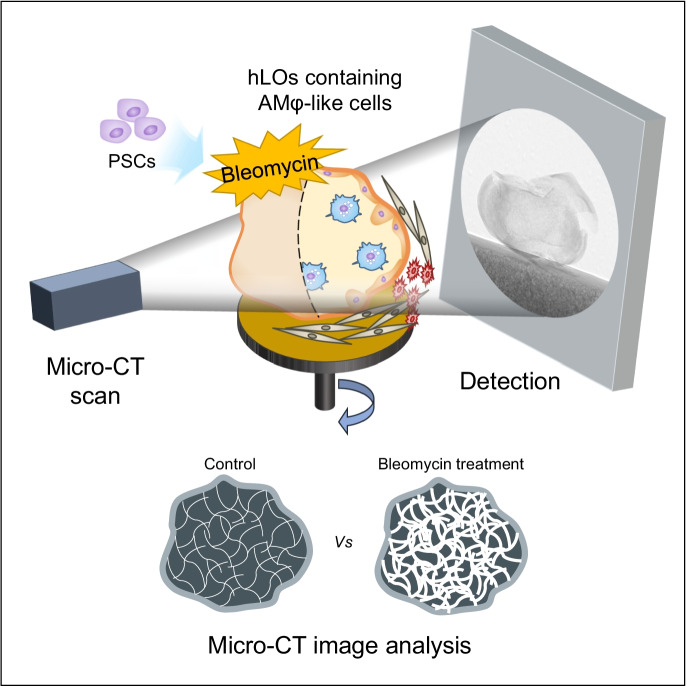
Human lung organoids (hLOs) are useful for disease modelling and drug screening. However, a lack of immune cells in hLOs limits the recapitulation of in vivo cellular physiology. Here, we generated hLOs containing alveolar macrophage (AMφ)–like cells derived from pluripotent stem cells (PSC). To bridge hLOs with advanced human lung high-resolution X-ray computed tomography (CT), we acquired quantitative micro-CT images. Three hLO types were observed during differentiation. Among them, alveolar hLOs highly expressed not only lung epithelial cell markers but also AMφ-specific markers. Furthermore, CD68+ AMφ-like cells were spatially organized on the luminal epithelial surface of alveolar hLOs. Bleomycin-treated alveolar hLOs showed upregulated expression of fibrosis-related markers and extracellular matrix deposits in the alveolar sacs. Alveolar hLOs also showed structural alterations such as excessive tissue fraction under bleomycin treatment. Therefore, we suggest that micro-CT analyzable PSC-derived alveolar hLOs are a promising in vitro model to predict lung toxicity manifestations, including fibrosis.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s10565-022-09698-1.
Keywords: Human alveolar organoid, Fibrosis model, Micro-CT, Alveolar macrophage
Introduction
Patients with respiratory diseases, such as chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis (IPF), have been increasing. These diseases are caused by environmental pollution, chemical exposure, smoking, and unknown factors (Daba et al. 2004; Thannickal et al. 2004; Zhang et al. 2020). Pulmonary fibrosis is a chronic respiratory disease characterized by the distortion of structure and loss of function of the lungs, due to the proliferation of fibroblasts and myofibroblasts and excessive deposition of disassembled collagen and extracellular matrix (ECM) after progressive damage to the lung tissue (Thannickal et al. 2004). Pulmonary fibrosis is a potential adverse effect of exposure to drugs such as penicillamine, amiodarone, and procainamide or environmental factors such as gases, asbestos, silica, and pesticides (Daba et al. 2004; Kim et al. 2018). Bleomycin is used to treat several malignancies; however, it is known to cause IPF (Tashiro et al. 2008). Additionally, polyhexamethylene guanidine phosphate, a humidifier disinfectant, accounts for 80.3% of cases of humidifier disinfectant-associated lung injuries (Paek et al. 2015; Song et al. 2014).
In vivo animal models have been used in cellular and molecular epidemiological studies, pathogenesis assessment, and drug development for IPF; specifically, bleomycin has been widely used to establish lung fibrosis models (Carrington et al. 2018; Della Latta et al. 2015). The characteristics of pulmonary fibrosis have been confirmed by a comparative analysis of accumulation of ECM, expression of inflammatory cytokines, structural changes in the lung tissue, and expression of fibrosis-related markers (Spagnolo et al. 2021; Thannickal et al. 2004). With recent advances in medical imaging and image processing technologies, the use of high-resolution X-ray computed tomography (CT) for the diagnosis of chronic lung diseases has rapidly increased (Hartley et al. 1994; Ohkubo et al. 2018). CT is a non-destructive technique to visualize the internal features of objects and is an important tool for disease diagnosis (Colombi et al. 2015; Ohkubo et al. 2018). For example, in routine clinical settings, IPF can be diagnosed by CT imaging without performing invasive lung biopsy. In animal models, micro-CT imaging has been used. Recently, Laura et al. developed a new micro-CT-guided densitometric approach for in vivo lung fibrosis staging in a bleomycin mouse model (Mecozzi et al. 2020).
Although animal models have been widely used for research purposes, they have an inherent limitation of the physiological differences between humans and animals. Three-dimensional (3D) organoids, composed of organ-specific cells and endowed with in vitro self-organization capability, have emerged in the field of stem cell research. Organoids are considered potential alternatives to animal models for disease modelling, regenerative medicine, physiological and pathological studies, and drug screening (Chen et al. 2018; Neal et al. 2018). Several protocols have been reported to establish human lung organoids (hLOs) derived from lung tissues or pluripotent stem cells (PSCs) (Chen et al. 2017; Miller et al. 2019; Neal et al. 2018). Patient tissue–derived hLOs have been used to model experimental infections, study pathogenesis, and screen candidate coronavirus disease (COVID-19) therapeutics (Han et al. 2020a, b; Youk et al. 2020). However, organoids lacking blood vessels and immune cells present in human organs are limited in their ability to mimic responses in vivo. To our knowledge, there have been no reports on brain, intestinal, cardiac, kidney, or lung organoids containing immune cells, except when they are co-cultured. However, infiltrating immune cells (i.e., B cells, T cells, natural killer cells, and macrophages) have been identified in adenocarcinoma patient–derived organoids (Neal et al. 2018).
Among different types of immune cells in the lungs, tissue-resident alveolar macrophages (AMφs) play a crucial role as the first line of defense against pathogens via phagocytosis, cytokine secretion, and antigen presentation to helper T cells (Heo et al. 2018; Kotton and Morrisey 2014). Moreover, AMφs are crucial regulators of fibrosis and secrete numerous pro-fibrotic soluble mediators, chemokines, and matrix metalloproteases (Zhang et al. 2018). Furthermore, in a normal injury repair response, macrophages readily acquire a phenotype that promotes fibroproliferation (Byrne et al. 2016). Mouse AMφs are identified by surface markers CD68+Siglec F+CD11bloCD64+ and CD206+, and human AMφs express CD68+CD11c+CD206+ surface markers (Byrne et al. 2016; Misharin et al. 2013). Recent studies have demonstrated that AMφs derived from embryonic precursors seed the lungs before birth, self-maintain locally throughout adult life, and self-renew in most adult tissues in a steady state (Roszer 2018; Yao et al. 2020). Interestingly, McGrath et al. reported that hematopoietic progenitors emerge before hematopoietic stem cells and generate functional blood cells in the mammalian embryo (McGrath et al. 2015). They also showed that embryonic stem cell–derived hematopoiesis recapitulates early yolk sac hematopoiesis and gives rise to primitive definitive erythro-myeloid progenitors (McGrath et al. 2015; Perdiguero et al. 2015). These progenitors first emerge in the yolk sac of human embryos, and then, they are found in the fetal liver (Frame et al. 2013). The origin of AMφs is controversial (Joshi et al. 2018), and it has long been thought that circulating monocytes differentiate into all tissue-resident macrophages including AMφs. Recently, it has been reported that macrophages originating from the fetal liver develop tissue-specific macrophages, such as those of the lungs (AMφs), brain (glial cell), and skin (Langerhans cell), on exposure to niche signals (Ginhoux and Guilliams 2016; Mowat et al. 2017; T'Jonck et al. 2018). Several researchers have attempted to generate tissue-specific macrophage-derived PSCs in vitro, but without a tissue-specific niche in vivo; it has remained a challenge (Nadkarni et al. 2016; Takata et al. 2017). Therefore, by incorporating an in vivo lung niche, Ginhoux et al. generated mouse iPSC–derived primitive macrophages that were intranasally transferred to the lungs of Csf2ra−/− mice, wherein they differentiated into functional AMφ-like cells, demonstrating that a lung niche is essential for the development of AMφs (Takata et al. 2017).
Here, we demonstrate PSC-derived alveolar hLOs containing AMφ-like cells that can be structurally and quantitatively analyzed by micro-CT. This model, which closely recapitulates the human lung, can be widely used as an in vitro model to predict lung toxicity manifestations such as fibrosis, assess infectious diseases, and drug screening.
Methods
Cell culture
Human PSCs, ES cell line (WA09), was obtained from WiCell Research Institute. The Korea institutional review board approved all experiments involving the use of PSCs (#P01-202,005–41-001). The PSCs were cultured in mTeSR™-1 medium (Cat# 85,850; STEMCELL Technologies, CA, USA) on Matrigel® hESC-qualified matrix, LDEV-free (Cat# 354,277; Corning, NY, USA) coated plates. The cells were cultured in an incubator at 37 °C with humidified air containing under 5% CO2.
Differentiation of hLOs
Before the initiation of hLO differentiation, human PSCs were harvested with 0.5 mM EDTA, seeded on Matrigel® hESC-qualified matrix, LDEV-free (Cat# 354277; Corning), and cultured using mTeSR™-1 medium (Cat# 85850; STEMCELL Technologies) for 3 days. On differentiation day 0, the medium of PSCs was removed, and then STEMdiff™ Endoderm basal medium supplemented with 1 × MR and 1 × CJ (Cat#05,110; STEMCELL Technologies) was added. After 24 h, the basal medium was replaced with fresh DE medium every day. On day 4, the culture medium was aspirated and washed with 1 × DPBS. The differentiating cells were incubated with TrypLE™ Express Enzyme (1 ×), without phenol red (Cat#12604013; Gibco, MA, USA) for 3 min at 37 °C in a 5% CO2 incubator. After harvesting the cells with DPBS, the differentiated cells were centrifuged at 1000 rpm for 3 min. The harvested multiple cell types were plated in Matrigel-coated plate containing DMEM/F12 (Cat#11320033; Gibco) medium supplemented with 1 × N2 (Cat#17502048; Gibco), 1 × B27 (Cat#17504044; Gibco), 1 × penicillin–streptomycin (P/S) (Cat#15140122; Gibco), 10 μM SB431542 (Cat#S1067; Selleckchem, TX, USA), and 100 ng/mL Noggin (Cat#120-10C; Peprotech, CT, USA), instead of CXCR4+ cell type only. After 24 h, the medium was replaced with DMEM/F12 containing 1 × N2, 1 × B27, 1 × P/S, 1 μM IWP4 (Cat#04–0036; Stemgent, MA, USA), and 10 μM SB431542. On day 7, anterior foregut endoderm medium (DMEM/F12 containing 1 × N2, 1 × B27, 1 × P/S, 3 μM CHIR99021 (Cat#S2924; Selleckchem), 10 ng/mL BMP4 (Cat#120–05; Peprotech), 10 ng/mL FGF7 (Cat#100–19; Peprotech), 10 ng/mL FGF10 (Cat#100–26; Peprotech), 50 nM all-trans retinoic acid (Cat#0695; R&D System, MN, USA), and Matrigel were used to replace the medium every 2 days until day 17. On days 15–17, the cells were harvested by pipetting and then suspension cultured on ultra-low attachment plates (Cat#3471, Corning). That medium was changed half and half every other day until the termination of culture. The hLOs were observed daily using a Leica DMi8 Microscope, and images were captured and the size of the hLOs was calculated using the LAS X software (Leica, Wetzlar, DE). The volume of hLOs was calculated under the assumption that the hLO was perfectly spherical.
Flow cytometry
The differentiating cells and hLOs on days 4, 10, 20, 30, and 60 were dissociated into single cells with 0.25% trypsin/EDTA (Cat#25200,114; Gibco) and 10 mg/mL Collagenase type II (Cat#17101015; Gibco). After passing the suspension through a 40-μm cell strainer (BD, NJ, USA), the dissociated cells were stained directly with phycoerythrin (PE), allophycocyanin (APC), Alexa Fluor-647, and fluorescein (FITC)-conjugated specific antibodies for 15 min at 4 °C. The list of antibodies is presented in Table S1. FITC-conjugated antibodies used were as follows: EpCAM (Cat#324204; Biolegend, CA, USA), CD68 (Cat#333806; Biolegend), and MUC1 (Cat#ab187833; Abcam, Cambridge, UK). PE-conjugated antibodies used were as follows: CXCR4 (Cat#341604; Biolegend) and PDGFRα (Cat#sc-338; Santa Cruz, TX, USA). APC-conjugated antibodies used were c-KIT (Cat#313206; Biolegend), SOX17 (Cat#IC1924A; R&D systems), and SOX9 (Cat#565493; BD Pharmingen, NJ, USA). The following PerCP-Cy5.5-conjugated antibody was used: CD11b (Cat#301418; Biolegend). BV421-conjugated antibody used was CD11c (Cat#301628; Biolegend). The cells were stained with TTF-1 (Cat#ab76013; Abcam) and PDPN (Cat#sc-59347; Santa Cruz) primary antibodies, and then with Alexa Fluor 488-, 594-, and 647-conjugated secondary antibodies. All stained cells were analyzed on CytoFLEX (Beckman Coulter, CA, USA), and the results were analyzed using NovoExpress software (ACEA Biosciences, CA, USA) and FlowJo software (BD). For t-SNE analysis, live cells were concatenated by downsampling each individual sample as the same proportion for subsequent high-dimensional data analysis to normalize between samples under different differentiation days. Samples were analyzed in parallel by t-SNE and overlaid onto the t-SNE map using FlowJo 10.6.2.
Immunofluorescence staining
For immunofluorescence, the hLOs were fixed with 4% paraformaldehyde. After washing two times with PBS, the fixed hLOs were treated with permeabilization solution. After 5 min, the hLOs were washed with washing buffer (PBS containing 0.1% Triton X-100) three times, followed by a blocking step with 2% BSA in washing buffer for 30 min. Subsequently, the hLOs were incubated with primary antibodies at 4 °C for 16–24 h. The primary antibodies used are listed in Table S2. After washing three times with washing buffer, the hLOs were incubated with appropriate Alexa Fluor 488- or 594- or 647-conjugated anti-rabbit IgG or anti-mouse IgG secondary antibodies (1:1000, Alexa 488-conjugated anti-mouse, Cat#A-21202, Invitrogen, MA, USA; Alexa 594-conjugated anti-mouse, Cat#A-11032, Invitrogen; Alexa 488-conjugated anti-rabbit, Cat#A-11034, Invitrogen; Alexa 594-conjugated anti-rabbit, Cat#A-21207, Invitrogen) for 1 h. The nuclei are stained with NucBlue™ Fixed Cell Stain ReadyProbes™ (Cat# R37606; Invitrogen). Bronchial (B)-type hLOs in the Matrigel block were prepared for cryosection as detailed below. B-type hLOs were fixed with 4% PFA and then embedded in optimal cutting temperature solution before being flash frozen. Sections of thickness 8 μm were prepared and air-dried for 2 h. The frozen sections were washed in PBS, blocked with 2% BSA and 0.1% Triton X-100 containing PBS for 1 h, and then covered with VECTASHIELD® (Cat# H-1200; Vector Laboratories, Inc., CA, USA). Both stained hLOs and cryosections of hLOs were visualized under a confocal microscope (LSM 800, Carl Zeiss, Berlin, DE), and images were analyzed using ZEN software (Carl Zeiss).
Scanning electron microscopy
The samples were fixed in 2.5% paraformaldehyde-glutaraldehyde mixture buffered with 0.1 M phosphate (pH 7.2) for 2 h, post-fixed in 1% osmium tetroxide in the same buffer for 1 h, dehydrated in graded series of ethanol, and substituted with isoamyl acetate. Thereafter, the samples were dried at the critical point under CO2. Finally, the samples were sputtered with gold in a sputter coater (Polaron SC502; Fison Instruments, Glasgow, UK) and observed using a scanning electron microscope (FEI Quanta 250 FEG, FEI Company, OR, USA).
Transmission electron microscopy
The hLOs (on day 85) were fixed with 2.5% glutaraldehyde for 3 h at room temperature. The samples were further processed, sectioned, and stained by Korea Basic Science Institute, Electron Microscope Facility (Cheongju, KR). Briefly, the sample was dehydrated with EtOH (50% to 100%) and then incubated in propylene oxide/Epon812 medium (EMS) (3:1 ratio) for 5 h. The medium was replaced with propylene oxide/Epon812 medium (EMS) (1:1 ratio) and incubated overnight. Finally, the samples were transferred into propylene oxide/Epon812 medium (EMS) (1:3 ratio) and allowed to stand for 5 h at room temperature; subsequently, the samples were transferred into new Embed 812 and incubated overnight at room temperature, after which the samples were embedded in fresh Embed 812 and polymerized overnight at 70 °C. Plastic-embedded samples were sectioned at 70 nm, and grids were stained in 4% aqueous uranyl acetate for 10 min at 70 °C followed by lead citrate for 5 min at room temperature. The sections were examined under Bio-TEM at 120 kV (Tecnai G2 Spirit, FEI Company). Images were acquired using US4000 software (Gatan, CA, USA).
Morphogenesis of hLOs
The upper part of the upside-down 24-well trans-well inserts were coated with growth factor reduced Matrigel (Cat#354230; Corning). The inverted upper portion of the 6.5-mm diameter insert (Cat#3415, Corning) was coated with 50 µL of growth factor reduced Matrigel. After air-drying, the hLOs (on days 13–15) were embedded in growth factor reduced Matrigel in 24 trans-well inserts and incubated at 37 °C in CO2 incubators until the Matrigel solidified. The hLO medium was added into the wells, and the trans-well inserts were placed as well. The medium was changed every other day. The morphology of hLO-derived branching tree-like clusters in the Matrigel was observed daily using a Leica DMi8 Microscope and images were captured using LAS X software.
Hematoxylin and eosin staining and immunohistochemistry
The hLO samples were fixed with 10% buffered formaldehyde overnight at room temperature with rocking, embedded in paraffin, and sectioned at 5-µm thickness. After de-paraffinization and epitope retrieval, the sections were stained with H&E to visualize histologic characteristics of the hLOs. These stained sections were observed and photographed under a microscope (Carl Zeiss). For immunohistochemistry, after the blocking step, the epitope-retrieved sections were stained with anti-α-SMA antibody (cat# ab150301; Abcam, 1: 100) or anti-VIMENTIN antibody (cat# ab20346; Abcam, 1: 100) overnight at 4 °C. After the washing step, the sections were treated with anti-rabbit-IgG-HRP conjugated secondary antibody, and DAB was used for HRP substrate.
Quantitative real-time PCR
The total RNA was isolated from cells (A549, lung cancer cell line; ATCC, BEAS-2B, human immortalized bronchial epithelial cell line; ATCC, HPAEpic, human primary alveolar epithelial cells; ScienCell Research Laboratories, Inc.) and hLOs on days 0, 4, 10, 20, 30, and 40 of differentiation. The cDNA was generated by reverse transcription of 1 μg of RNA with random hexamers and PrimeScript™ First Strand cDNA Synthesis kit (Cat#6110A; Takara, Kyoto, JP) following the manufacturer’s instructions. Real-time qPCR was performed on a SimpliAmp Thermal cycler (Applied Biosystems, MA, USA) using ABI Power SYBR Green PCR Master Mix (Applied Biosystems). The real-time qPCR conditions were as follows: 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min, with dissociation/melt curves obtained for each of the genes. Absolute quantification of each gene was normalized to the housekeeping gene GAPDH. The qPCR for each sample was performed in triplicates. The qPCR primer sequences are listed in Table S3.
Total RNA sequencing and bioinformatic analysis
The total RNA from three types of hLOs was purified using TRIzol (Cat#15596018, Invitrogen). RNA concentration and RNA integrity number (RIN) were determined using an Agilent 2100 BioAnalyzer (Agilent Technologies, CA, USA), and samples with RIN > 7 were used for RNA sequencing. mRNA was isolated using magnetic beads–based poly (A) selection, and the first and second strand cDNA fragments were synthesized using random hexamers. Subsequently, the cDNA fragments were purified and PCR-amplified to create the final cDNA libraries using the Illumina TruSeq RNA prep kit (Illumina, CA, USA). The libraries were then sequenced using Illumina HiSeq2500 (Illumina) to generate more than 30 million paired-end 101-bp reads at TheragenEtex BiO Institute. All raw sequence data have been deposited in the NCBI database with accession number PRJNA596053.
All raw sequence reads by RNA-Seq were pre-processed using Trimmomatic (v0.36) to trim the adaptor sequences and remove low-quality sequences (Bolger et al. 2014). The remaining clean reads were mapped to the human genome (hg38) using Hisat2 with default parameters. The alignment results were then uploaded to Cufflinks (v2.2.1) and the relative expression levels in the three types of hLOs were calculated as fragment per kilobase of exon per million of mapped fragments (FPKM) using CuffDiff of Cufflinks. We defined the differentially expressed genes (DEG) based on the false discovery rate (FDR) < 0.01 and fold change > 2 of the mean expression values. Subsequently differences in the enrichment of gene ontology (GO) categories and KEGG pathways for the DEGs were analyzed using Gorilla (http://cbl-gorilla.cs.technion.ac.il/) and DAVID (https://david.ncifcrf.gov) tools. To perform a principal component analysis (PCA) of the transcriptome profiles in the three hLO types, the “prcomp” function in “stats” package in R (version 3.4.3) was used. For comparative transcriptome analysis, lung RNA-seq data in the early developmental stage were obtained from LungMAP database (https://lungmap.net) including sequencing data from tissues of infants and children who died shortly after birth due to brain injury. After excluding patient samples with known lung disease, a total of 18 lung transcriptome data were used and we selected for further analysis genes whose expression level of at least 5 FPKM.
Phagocytosis assay
Differentiated alveolar macrophage dissociated from the hLOs with 1 mL of 0.25% trypsin EDTA were plated on Matrigel-coated μ-Slide 8-well chambers (Cat#80826; ibidi, Martinsried, DE) and then incubated in a 5% CO2 incubator. Attached AMφs were activated with PMA (10 ng/mL, Cat#P8139; Sigma-Aldrich) for 18 h, and then Fluoresbrite Polychromatic Red 1.0 μm Latex beads (Cat#18660, Polyscience Inc, NY, USA) were added to the activated or non-activated AMφs. After 4 h, the cells were analyzed by laser confocal microscopy (Carl Zeiss).
Bleomycin-induced fibrosis modeling using alveolar hLO
Suspension-cultured alveolar hLOs on day 85 were treated with 10 µg/mL bleomycin for 48 h. As a control, PBS-treated alveolar hLOs were used. Each sample was harvested and analyzed for the expression of fibrosis-related genes and proteins.
Micro-CT imaging and quantitative image analysis
The organoid samples were fixed in 2.5% paraformaldehyde-glutaraldehyde mixture buffered with 0.1 M phosphate (pH 7.2) for 2 h, postfixed in 1% osmium tetroxide in the same buffer for 1 h, dehydrated in a graded series of ethanol, and substituted with isoamyl acetate. The samples were then dried to the critical point under CO2. Micro-CT scans of the hLO samples were acquired using a SkyScan 1272 high-resolution micro-CT scanner (Bruker microCT, Kontich, BEL). The scanning parameters were as follows: 35 kV source voltage, 190 A source current, 0.100 exposure rotation step, and 1.300004 μm image pixel size. Images were reconstructed using scanner software (NRecon 1.7.0.4; Bruker) into 16-bit TIFF format. A water phantom (a PCR tube containing water) with the same parameters were scanned and used to convert the TIFF files to the DICOM format, based on the air and water intensities as Hounsfield units (HUs) in the DICOM format; air 1000 HU and water 0 HU. Regions of hLOs were then semi-automatically segmented from the DICOM images using ITK-SNAP 3.6.0 software (Moorefield et al. 2018) and 3D geometry of hLOs were obtained.
Statistical analysis
Evaluation of experimental data for significant differences was performed using the Student’s t-test, which was conducted using the Prism software package (GraphPad Software). Results with *p < 0.05, **p < 0.01, and ***p < 0.001 were considered significant for these studies.
Results
Differentiation of PSCs into hLOs
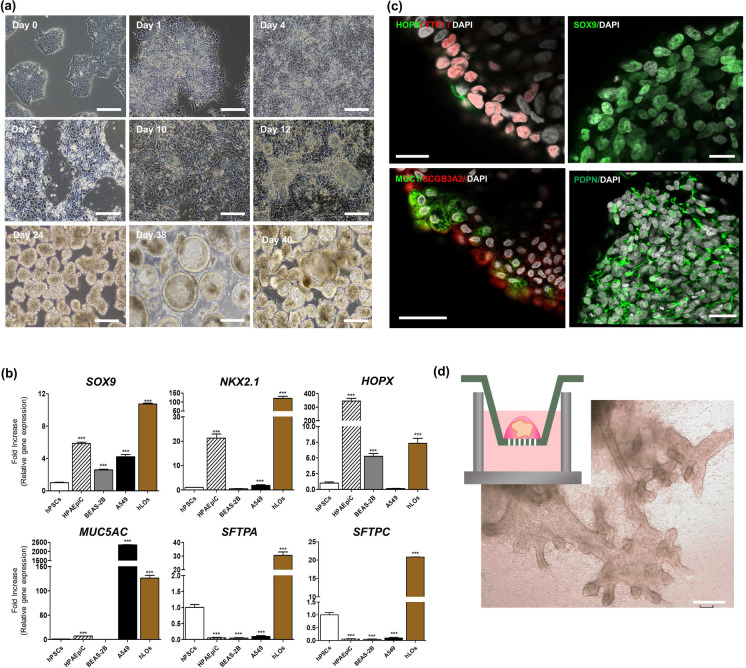
We modified existing protocols (Chen et al. 2017; Jacob et al. 2017; Strikoudis et al. 2019) to generate hLOs containing AMφ-like cells derived from differentiated PSCs (Supplementary Fig. 1a and Fig. 1a). Briefly, undifferentiated CD326+ PSCs differentiated into definitive endoderm lineage on a Matrigel-coated plate. After 4 days, these differentiated cells comprised diverse cell populations such as CXCR4+ EpCAM+ cells (93.49%), CXCR4− EpCAM+ cells (5.34%), SOX17+ PDGFR-α+ cells (4.9%), and CXCR4+c-KIT+ cells (85.74%), as confirmed by flow cytometry (Supplementary Fig. 1). Unlike the existing protocols, for further differentiation of these cells into hLOs containing AMφ-like cells, all the differentiated cells in this stage were differentiated into the anterior foregut endoderm on the Matrigel-coated plates in a medium containing NOGGIN, SB431542, and IWP4 without performing an artificial CXCR4+-sorting step. After 2 days, the medium was changed to a medium containing BMP4, FGF10, FGF7, ATRA, and CHIR99021 for differentiating PSCs into hLOs. Consequently, budding clumps started to form on days 10–12. On days 13–15, the cells were harvested by mechanical pipetting; subsequently, the harvested cell clumps were cultured in a suspension. Under this condition, the 3D clumps of heterogeneous sizes and morphologies were differentiated into hLOs (Fig. 1a). During the differentiation of hLOs, we performed quantitative PCR (qPCR) and flow cytometry to characterize these hLOs in a time-dependent manner. As shown in Supplementary Fig. 2a, NKX2-1 (lung epithelial primordial progenitor marker), SOX9 (epithelial progenitor marker/bud tip progenitor marker), HOPX (epithelial type I and II cell marker), and SFTPA expression was increased in hLOs on day 30. We also compared the expression levels of lung-specific genes (SOX9, NKX2-1, HOPX, TP63, MUC5AC, and SFTPC) between the hLOs and positive controls, which were HPAEpiC cells (primary alveolar epithelial type I cell line), BEAS-2B cells (human bronchial epithelial cell line), and A549 cells (human adenocarcinoma lung cell line). Interestingly, HPAEpiC cells expressed SOX9, NKX2.1, and HOPX but not MUC5AC, SFTPA, and SFTPC. A549 cells expressed high levels of MUC5AC, whereas BEAS-2B cells expressed SOX9 and HOPX. Overall, the expression of most lung-specific genes was higher in hLOs than in the positive controls (Fig. 1b), indicating that hLOs comprised diverse lung cell types. The immunofluorescence analysis also showed that hLOs expressed the alveolar epithelial cell markers HOPX, TTF1, and PDPN; the secretory cell marker SCGB3A2; the distal bud tip progenitor marker SOX9; and the alveolar epithelial type II cell marker MUC1, on day 68 (Fig. 1c). The tSNE dot plots represent the significant changes in the expression of the definitive endoderm markers (EpCAM and CXCR4) and lung-specific markers (MUC1 and NKX2-1) over time (Supplementary Fig. 2b, c). Both EpCAM+/CXCR4+ and EpCAM+/c-KIT+ cell populations were dominant on day 4, but were not detected on day 30 (Supplementary Fig. 2b). Conversely, MUC1+ or TTF1+ cell populations were detected on day 40 in hLOs but not on day 4 (Supplementary Fig. 2c), showing that PSC-derived definitive endoderm stage cells successfully differentiated into hLOs expressing lung-specific markers in a time-dependent manner. Moreover, we investigated the potential of branching morphogenesis of hLOs and observed that budding tips began to appear during days 27–30 (Supplementary Fig. 3) and branching airway tree-like structures around day 50 (Fig. 1d). These results collectively demonstrate that hLOs have lung-specific characteristics.
Fig. 1.
Generation of PSC-derived hLOs. a Bright-field images show the morphology of budding and differentiating 3D hLOs derived from PSCs in a time-dependent manner. Scale bar = 250 μm.) Lung-specific gene expression of the PSC-derived hLOs. The expression of SOX9, NKX2-1, HOPX, MUC5AC, SFTPA, and SFTPC in hLOs (brown bar). As controls, HPAEpiC cells (human primary alveolar epithelial cell line, hatched bar), BEAS-2B cells (human immortalized bronchial epithelial cell line, grey bar), and A549 cells (human lung cancer cell line, black bar) were used. The overall expression is relative to that in undifferentiated hPSCs on day 0. *p < 0.05, **p < 0.01, ***p < 0.001. All error bars represent SD. c Immunofluorescence data show that the hLOs express MUC1+ (green)/SCGB3A2+ (red): HOPX+ (green)/ NKX2-1+ (red): SOX9+ (green): PDPN+ (green) cells in the hLOs. The nucleus was visualized by DAPI staining (white). Scale bars = 20 μm. d Several new branches of hLOs in the Matrigel-coated Trans-well expanding over time were observed. Images of the hLOs on day 65 were captured by bright field microscopy. Scale bar = 200 μm
Alveolar hLOs express high levels of immune-associated genes
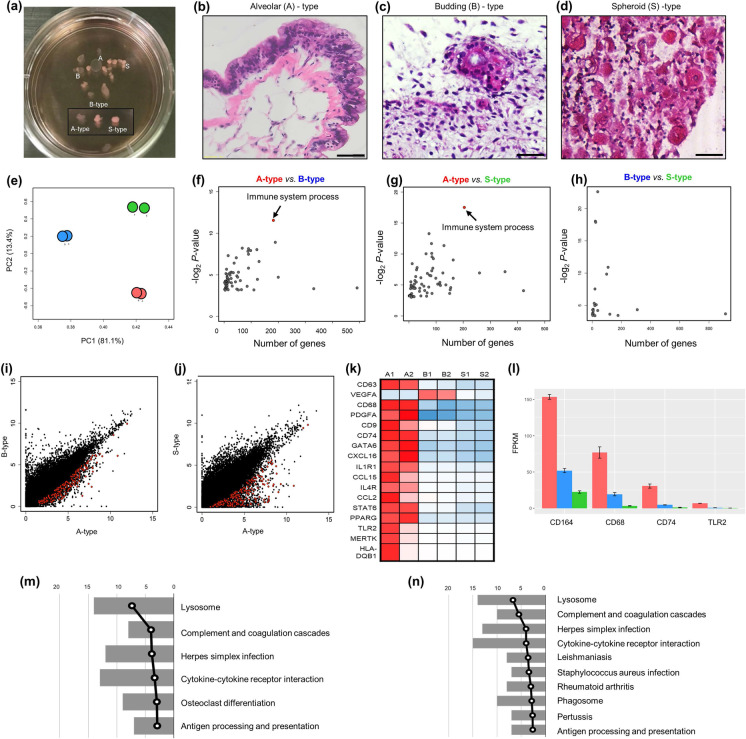
Interestingly, during long-term suspension culture, hLOs with a heterogeneous morphology, including transparent cyst-like, budding, and solid structures, were observed from day 30 (Fig. 2a). The cyst-like hLOs had a transparent air-sac structure, similar to the alveolus; the budding-type hLOs had branching tips; and the solid-type hLOs were spheroid. To evaluate the histological differences among these hLOs, each type of hLO was stained with H&E on day 85. The analysis of alveolar hLOs with transparent cyst-like morphological characteristics showed that they had an outer structure surrounded by smooth muscle cells and bronchial epithelial cells as well as a structure comprising thin alveolar epithelial cells that resembled an alveolar sac (Fig. 2b). Conversely, the irregular budding-type hLOs consisted of bronchial epithelial cells (Fig. 2c), whereas the spheroid-like cell clusters (spheroid-type) did not have distinctive defining characteristics (Fig. 2d). To further analyze the characteristics of the three hLO types, we performed genome-wide expression analysis by RNA-Seq on day 85 and obtained highly reproducible results (Spearman’s correlation coefficient, mean p = 0.9716 ± 0.0003). The subsequent principal component analysis of the RNA-Seq results categorized the transcriptome profiles of the hLOs into three distinct phases (Fig. 2e). Notably, certain genes were expressed in specific hLO types (Supplementary Fig. 4). In alveolar hLOs, “immune system processes” was the most significant GO term in the biological processes category (Fig. 2f–h). These immune cell–related genes were more commonly upregulated in alveolar hLOs than in bronchoalveolar or spheroid hLOs (Fig. 2i, j). Specifically, in our transcriptome profiles, alveolar hLOs exhibited upregulated expression of macrophage-specific genes (e.g., CD68, TLR2, CD74, and CD164; Fig. 2k, l) compared with other type of hLOs. Additionally, the KEGG pathway analysis of the immune system process-specific genes of alveolar hLOs revealed altered immune pathways, indicating a high expression of genes involved in lysosomal activity, complement and coagulation cascades, cytokine-cytokine receptor interactions, and antigen processing and presentation (Fig. 2m, n). These findings suggest that alveolar hLOs comprise immune cells. Furthermore, to analyze which developmental stage of human lung tissues represents our hLOs, we collected 18 lung transcriptomes from LungMAP database, which includes molecular maps of the neonatal, pediatric, and early adult human lung and compared those to our six hLOs’ transcriptome data using UPGMA dendrogram analysis. Two major transitions in expression patterns were observed as expected between the human ES cell–derived hLOs and the human lung samples, but intriguingly the hLOs transcriptomes are relatively closer to those of neonatal lung (D005 and DP02 samples corresponding to day 1) than the others including pediatric and early adult lung. A similar pattern was so observed when immune cell–related genes were assessed. These results indirectly indicate that our hLOs are functionally similar to the early stage of lung tissue (Supplementary Fig. 5).
Fig. 2.
Large-scale transcriptome analyses of alveolar hLOs. a A photograph of floating 3D hLOs showing three different types of 3D hLOs. b–d Hematoxylin and eosin staining displaying the hLO structure. b Alveolar (A) hLOs possessing sac-like structures with thin layer alveolar epithelial type I cells resembling the alveolus inside and smooth muscle (pink) and epithelial cells on the outside. Scale bar = 50 μm. c Budding-type hLOs showing bronchial epithelial cells and bronchiole morphology. d Specific structure was not detected in spheroid (S)-type hLOs. Scale bar = 250 μm. e PCA analysis of three types of hLOs; red, blue, and green colors indicate A-, B-, and S-type hLOs, respectively. f–h Biological process gene ontology analysis of differentially expressed genes identified between (f) A- and B-types, g A- and S-types, and h B- and S-types. i–j Scatter plots of gene expression levels in (I) A- and B-types and (J) A- and S-types; 182 and 204 immune-related genes from (I) and (J) indicated with red dots. k Heatmap of macrophage-related genes. Rows represent individual genes and columns represent individual samples. The high (red), average (white), and low (blue) expression levels are depicted with a color scale. All gene expression levels were normalized by FPKM. (l) The average expression levels of macrophage-specific genes; error bars represent standard errors (n = 2). m–n KEGG pathway analysis of immune-related genes in I (m) and J (n); the bars represent the number of genes and broken lines represent the transformed P values
Alveolar hLOs contain CD68+ cells
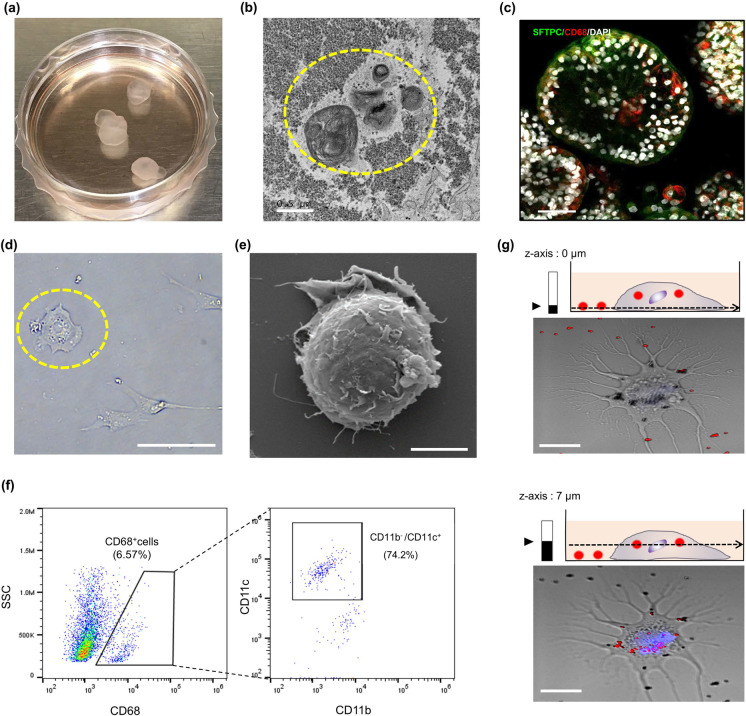
Around differentiation day 60, alveolar hLOs resembling a cystic air sac were observed; their average area and volume were 0.25 mm2 and 96 mm3, respectively (Supplementary Fig. 6). Our observation of the immune cell–specific gene expression in alveolar hLOs prompted us to examine whether these transparent cyst-like alveolar hLOs also contained AMφ-like cells using immunofluorescence analysis (Fig. 3a). Firstly, we identified alveolar hLOs through transmission electron microscopy (TEM) by verifying whether they comprised epithelial type II cells with laminar bodies, which are secretory organelles, containing surfactants. As shown in Fig. 3b, the TEM analysis verified the presence of laminar bodies in the alveolar hLOs. Furthermore, the immunofluorescence analysis showed that SFTPC+ (green color, Fig. 3c) epithelial type II cells were dense outside the alveolar sacs, whereas cells expressing the AMφ-specific marker CD68 were located inside the hollow sacs (red color, Fig. 3c, Supplementary Fig. 7). This demonstrated that the spatial organization of CD68+ AMφ-like cells within the alveolar lumen was similar to that within the human alveolar lumen. Upon reviewing single cells dissociated from alveolar hLOs by bright field and scanning electron microscopy, AMφ-specific morphology, round shape, and pseudopods (Fig. 3d, e) were observed, similar to AMφs in the human lung tissue. These data indicate that a population of CD68+ cells is present in the transparent alveolar hLOs. Next, to further validate the presence of CD68+ cells inside alveolar hLOs, we performed flow cytometry. As shown in Fig. 3f, 6.57% of the total cell population of alveolar hLOs were CD68+ cells, which were negative for CD11b. Moreover, 74.2% of the CD68+ cell population expressed CD11c+, indicating that CD68+CD11c+CD11b−AMφ-like cells are present inside alveolar hLOs (Fig. 3f). However, these CD68+ cell population were not detected until day 40 of differentiation, and these were detected after day 55 of differentiation (Supplementary Fig. 8). Furthermore, we performed a phagocytosis assay of AMφ-like cells dissociated from alveolar hLOs and activated by phorbol 12-myristate 13-acetate using Fluoresbrite Polychromatic Red 1.0 μm Latex beads. These latex beads in the cytosol of activated AMφ-like cell were observed in a 7-μm z-position (Fig. 3g), demonstrating that AMφ-like cells in alveolar hLOs exhibit the AMφ-specific function of phagocytosis. This finding, combined with the results of the marker expression analysis, suggests that PSC-derived alveolar hLOs endogenously contain functional AMφ-like cells with phagocytic activity. Furthermore, to validate the expression of CD68 and AMφ-like cell differentiation–related genes in hLOs, we performed the qPCR analysis in a time-dependent manner. As shown in Supplementary Fig. 9, the expression of GM-CSF and PPAR-γ, which are known as inducers for AMφ differentiation, began to increase during days 10–20 and continued to increase significantly throughout hLO differentiation up to day 60. CD68 expression was also significantly increased on day 60. Furthermore, CD68, GM-CSF, and PPAR-γ were highly expressed in the alveolar-type LOs compared with those in the others (Supplementary Fig. 10).
Fig. 3.
AMφ-like cells exist inside alveolar hLOs. a Visualization of the floating alveolar hLOs on day 120. b Representative image of transmission electron microscopy revealing that PSC-derived hLOs possess tightly packed, concentric membrane sheets (lamellae) forming lamellar bodies (yellow circle). Scale bar = 0.5 μm. c Whole 3D alveolar hLOs stained with CD68 (red), SFTPC (green), and DAPI (white), and imaged by confocal microscopy on day 85. Scale bar = 100 μm. d The morphology of dissociated cells from the hLOs on day 90. The yellow circle shows a round AMφ-like cell (bright field image). Scale bar = 100 μm. e Scanning electron microscopy images of round AMφ-like cells. Scale bar = 5 μm. f Dot plots represent the CD68+ cell population of dissociated alveolar hLOs expressing CD11c, but not CD11b, in hLOs on day 60. g Confocal microscopy images focused at the bottom (z = 0 μm, lower) and center (z = 7 μm, upper) of AMφ-like cells. Blue color indicates nucleus stained with Hoechst 33,342. Data are representative of independent experiments replicated four times. Scale bar = 25 μm
Fibrotic characteristics of bleomycin-treated alveolar hLOs
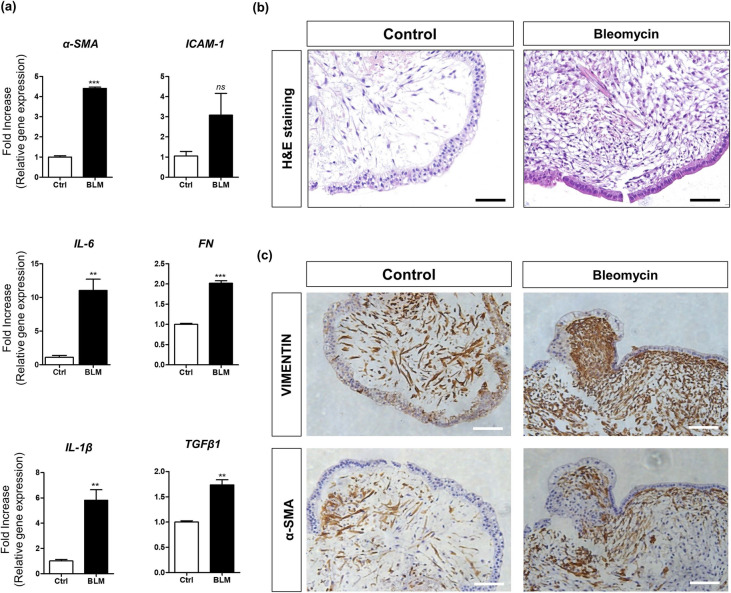
To confirm whether alveolar hLOs containing AMφ-like cells can be used as a predictive toxicity assessment model for pulmonary fibrosis, isolated alveolar hLOs were treated with bleomycin, a pulmonary fibrosis inducer. After 48 h, we harvested these hLOs and analyzed fibrotic gene expression using qPCR. Interestingly, bleomycin-treated alveolar hLOs showed a significantly upregulated expression of the following lung fibrosis-related genes: α-SMA, FN, IL-1β, IL-6, and TGF-β1 (Fig. 4a). Furthermore, as shown in Supplementary Fig. 11, the expression levels of α-SMA (1.95 ± 0.008-fold) and TGF-β1 (1.21 ± 0.023-fold) were significantly increased in the BLM-treated bronchoalveolar (B)-type hLOs compared with those in the PBS-treated control. The expression of α-SMA (1.24 ± 0.023-fold) and TGF-β1 (1.33 ± 0.040-fold) in the spheroid-type hLOs treated with BLM was slightly increased compared with that in the control. In contrast, the expression of α-SMA (2.8 ± 0.004-fold) and TGF-β1 (6.82 ± 0.792-fold) in alveolar-type hLOs treated with BLM was considerably increased (Supplementary Fig. 11). Here, we suggest that the high expression of fibrosis genes by bleomycin treatment in this alveolar hLOs might attributed to various reasons including the presence of AMφ-like cells and growth factors induced by various cells constituting alveolar hLO other than the presence of AMφ hLOs.
Fig. 4.
Bleomycin induces an increase in fibrotic marker expression in alveolar hLOs. a Quantitative PCR analysis of fibrosis-related marker expression in alveolar hLOs treated with bleomycin. Bar graphs show relative gene expression (fold increase) of α-SMA, ICAM-1, IL-1β, IL-6, FN, and TGF-β1 in alveolar hLOs after bleomycin treatment (10 µg/ml) for 48 h. Black bar indicates alveolar hLOs treated with bleomycin, and white bar indicates alveolar hLOs treated with PBS (control). Data are presented as mean ± SD. (*p ≤ 0.05). b Histology of bleomycin-treated alveolar hLOs and control. H&E staining data show that fibroblast-like cells increased in bleomycin-treated alveolar hLOs. Scale bar = 100 μm. c IHC analysis showed the expression of vimentin and α-SMA in bleomycin-treated alveolar hLOs (left) and control (right). Scale bar = 100 μm
Additionally, H&E staining images showed higher fibroblast proliferation and larger fibrotic area in the bleomycin-treated alveolar hLOs than in the PBS-treated controls (Fig. 4b). The immunohistochemistry analysis showed higher α-SMA and Vimentin expression in the bleomycin-treated alveolar hLOs than in the PBS-treated controls (Fig. 4c).
Micro-CT analysis of bleomycin-treated alveolar hLOs
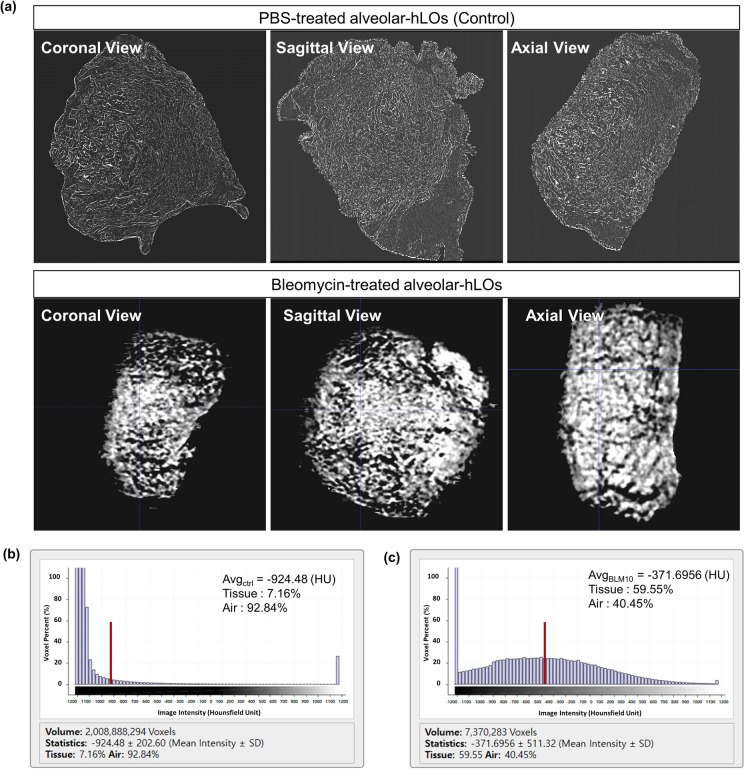
The overall scheme of quantitative micro-CT imaging is described in Fig. 5. CT density distribution per voxel (volume elements, 1 µm3) of an alveolar hLO sample can be shown as a histogram. As shown in Fig. 6a, the control hLO sample (upper) depicted air spaces (darker regions) in the alveoli. However, in the bleomycin-treated hLO sample (lower), a substantial portion of the sample was covered by a high attenuation area, reflecting that tissues refer to non-air components. The histogram in Fig. 6b shows marked changes in the average CT density (from − 905 ± 207 HU to − 336 ± 493 HU) and tissue fraction (from 9 to 62.9%) between the control and bleomycin-treated samples. The bleomycin-induced tissue fraction and standard deviation increased to 63% and 493 HU, respectively, indicating the characteristics of lung fibrosis. These observations are consistent with in vivo CT values for the human lungs that ranged between − 950 HU and − 701 HU for normal regions and between − 700 HU and 0 HU for fibrotic regions (Moorefield et al. 2018). We also found that the alveolus size in alveolar hLOs was consistent between the H&E staining and micro-CT analysis (Supplementary Fig. 12).
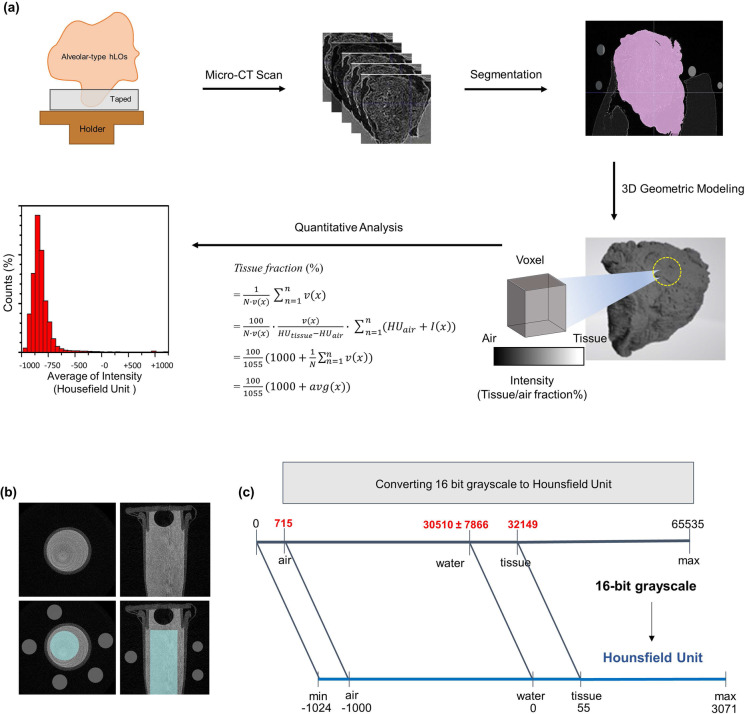
Fig. 5.
Schematic workflow of quantitative micro-CT imaging analysis of alveolar hLOs. a Workflow of micro-CT imaging of alveolar hLOs. The workflow shows alveolar hLO sampling, micro-CT scanning, segmentation, 3D geometric modelling, and quantitative analysis in sequence. The results are presented as the distribution of voxel's HU values. b Axial (left) and coronal (right) view micro-CT images of the water phantom (water in a PCR tube, gray regions) in the upper panels were used to compute mean intensities of water (cyan region inside the tube in the lower panels) and air (small grey circles in the lower panels) for unit conversion from the TIFF to DICOM format. c Chart for converting 16-bit grayscale TIFF to DICOM in HU. The HU values of blank sample (air), water, and tissue are − 1000, 0, and 55, respectively
Fig. 6.
Micro-CT image analysis shows an increase in tissue fraction in the alveolar hLOs treated with bleomycin. a 3D views of reconstructed micro-CT images of normal control (upper) and bleomycin-treated (lower) alveolar hLOs. Images were visualized with a central level of − 500 HU and a window of 1500 HU. b CT density (intensity in HU) distribution of the PBS control-treated alveolar hLO micro-CT image. c CT density (intensity in HU) distribution of the bleomycin (10 µg/ml)-treated alveolar hLO micro-CT image. The red bars represent the average HU values for each model
Discussion
We established alveolar hLOs containing AMφ-like cells derived from human PSCs using a modified protocol and long-term suspension culture method. RNA-Seq data and qPCR analysis revealed that macrophage-related genes are highly expressed in alveolar hLOs compared with those in other organoids; the TEM images showed the morphology of AMφ-like cells. We confirmed by immunofluorescence analysis that AMφ-like cells expressing CD68, an AMφ marker, were located within these alveolar-organoids and had phagocytotic activity. We also successfully acquired and quantitatively analyzed micro-CT images of human alveolar hLOs, which, to the best of our knowledge, has not been achieved before. Through this, we confirmed that fibrosis substantially increased the tissue fraction of alveolar hLOs, as observed in human lung CTs. This was consistent with the histological changes and expression patterns of the fibrosis-related markers in the fibrosis-induced alveolar-organoids, suggesting that quantitative micro-CT imaging analysis of the alveolar hLO model is a promising tool for the precise assessment of lung fibrosis.
Several groups have succeeded in generating hLOs (Chen et al. 2017; Strikoudis et al. 2019; Yamamoto et al. 2017); however, there have been no reports of hLOs containing endogenously derived AMφ-like cells from PSCs. Enrichment of hLOs with AMφs has been achieved by co-culturing organoids with iPS-derived primitive macrophages (iPCs) after differentiation, but AMφs are difficult to obtain in the absence of a live human lung niche (Happle et al. 2017; Takata et al. 2017). The currently established protocols for hLO differentiation only use definitive endodermal CXCR4+ cells to generate hLOs (Chen et al. 2017; Strikoudis et al. 2019). In contrast to previously reported methods, we differentiated all cells derived from PSCs in the definitive endoderm stage (EpCAM+/− cells, CXCR4−PDGFRα− cells, CXCR4−PDGFRα+ cells, and SOX17+/− cells) into hLOs without any sorting step, and obtained three types of hLOs. Additionally, RNA-Seq of these hLO types provided an important clue, that is, immune cell–related genes are more highly expressed in alveolar hLOs than in others (Fig. 2).
CXCR4+PDGFRα+ mesendodermal cells can differentiate into mesodermal or endodermal cells via epigenetic regulation (Chu et al. 2016; Wang and Chen 2016). Regarding the stimuli that trigger the differentiation of mesendodermal cells into AMφs, several studies have reported that granulocyte–macrophage colony-stimulating factor (GM-CSF) produced by type II pneumocytes is necessary for the development of AMφs from precursor cells in the fetal liver or yolk sac (Cohen et al. 2018; Mass et al. 2016; T'Jonck et al. 2018). Moreover, the TGF-β1 and GM-CSF pathways are known to upregulate PPAR-γ expression, stimulating AMφ development (Yu et al. 2017). As shown in Supplementary Fig. 9a, GM-CSF and PPAR-γ expression was upregulated during alveolar hLO differentiation. Therefore, we suggest that AMφ-like cells originate from these PSC-derived EpCAM+CXCR4+SOX17−/+c-KIT+/PDGRFα+ mesendodermal cells and that GM-CSF and PPAR-γ secreted by epithelial cells during hLOs differentiation induced their differentiation into AMφ-like cells, although further studies are needed to incorporate cell lineage tracking.
Alveolar hLOs containing AMφs generated through long-term suspension culture were shaped like a balloon; their structure was similar to that of the alveolar sac of the lung tissues. The lung consists of air space and tissue components that can be decomposed solely by a single local intensity value in each voxel. This anatomical nature of the lungs enables lung structural and functional analyses by conventional and micro-CT imaging. We successfully conducted quantitative micro-CT imaging of alveolar hLOs and analyzed the structure of the hLO models showing reasonable CT density distribution (Fig. 6). To the best of our knowledge, this is the first quantitative micro-CT imaging study of alveolar hLOs containing AMφ-like cells. However, the preparation and fixation of alveolar hLO samples for micro-CT imaging are highly challenging for successful imaging, owing to the small size of alveolar hLOs compared with that of the lung tissue and the thin sac structure of 3D organoids consisting only cells without scaffolds. Nevertheless, we revealed that the tissue fraction analyzed by micro-CT in the bleomycin-treated alveolar hLOs was higher than that in the control. The micro-CT finding of increased tissue fraction in the bleomycin-treated alveolar hLOs correlated with the findings of H&E staining, fibrotic gene expression analysis, and immunohistochemistry analysis (Supplementary Fig. 12). Differences between the fibrotic alveolar hLOs and control models in the CT density map (Fig. 6) and in the air and tissue fractions were associated with the changes observed in histological imaging, providing improved pathophysiological insights. Therefore, our study presents a highly innovative convergence of research technologies, integrating organoid, medical imaging, and biomedical engineering methods.
Conclusions
Overall, this study is the first to successfully establish alveolar hLOs containing AMφ-like cells derived from human PSCs. Additionally, it is the first to conduct micro-CT imaging of alveolar hLOs and analyze their structure. The bleomycin-treated alveolar hLOs showed upregulated expression of fibrosis-associated markers and increased tissue fraction following quantitative analysis of micro-CT images, suggesting that alveolar hLOs can be used as useful preclinical fibrosis quantitative evaluation models. Furthermore, these alveolar hLOs, which can be analyzed by micro-CT imaging, have the potential to be widely used for various research purposes, such as infectious disease modelling, inflammatory response studies, pathogenesis assessment, drug screening, and drug toxicity testing. Therefore, our 3D alveolar hLO model has potential to bridge the gap between structural and functional relationships among 2D cell models, human quantitative CT imaging studies, and preclinical animal studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
Eun-Mi Kim, Seri Choi, Jihong Song, and Gohar Manzar designed the experiments and Eun-Mi Kim and Seri Choi performed most of the experiments. Seri Choi, Seung-Yeon Kim, and Eun-Mi Kim performed generation of hLOs and Flow cytometry analysis. Seongmin Cheon, Chungoo Park, and Eun-Kee Park performed bioinformatics analysis. Dae-Hwan Nam, Su-Man Kim, Hyung-Sik Kang, Sang-Hun Jeong, and Ju-Han Lee performed H&E staining. Ji-eun Kim and Ki-Suk Kim performed the long-term culture. Hyang-Ae Lee performed real-time qPCR. Seri Choi, Seung-Yeon Kim, Han-Jin Park, Won Keun Oh, and Kee K.Kim generated fibrosis model and performed Immunofluorescence. Chang Hyun Lee, Jiwoong Choi, and Minseob Lee analyzed micro-CT imaging. Jihong Song, Gohar Manzar, and Kee K.Kim provided comments. Eun-Mi Kim, Jihong Song, Gohar Manzar, Chungoo Park, and Jiwoong Choi wrote the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant, which is funded by the Korea government (MSIT) (No. 2018R1D1A1B07045689), the Korea Institute of Toxicology (1711133839 and 1711133843), and the Korea Environment Industry & Technology Institute (KEITI) grants (2018001360003 and 2018001360001) through the Environment Health Action Program, funded by the Korea Ministry of Environment (MOE).
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Seri Choi and Jiwoong Choi contributed equally to this work.
Contributor Information
Chungoo Park, Email: chungoo@jnu.ac.kr.
Chang Hyun Lee, Email: changhyun.lee@snu.ac.kr.
Eun-Mi Kim, Email: eunmi.kim@kitox.re.kr.
References
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (oxford, England) 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AJ, Maher TM, Lloyd CM. Pulmonary macrophages: a new therapeutic pathway in fibrosing lung disease? Trends Mol Med. 2016;22:303–316. doi: 10.1016/j.molmed.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Carrington R, Jordan S, Pitchford SC, Page CP. Use of animal models in IPF research. Pulm Pharmacol Ther. 2018;51:73–78. doi: 10.1016/j.pupt.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Chen Y, Feng J, Zhao S, Han L, Yang H, Lin Y, Rong Z. Long-term engraftment promotes differentiation of alveolar epithelial cells from human embryonic stem cell derived lung organoids. Stem Cells and Development. 2018;27:1339–1349. doi: 10.1089/scd.2018.0042. [DOI] [PubMed] [Google Scholar]
- Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LF, Leng N, Zhang J, Hou Z, Mamott D, Vereide DT, Choi J, Kendziorski C, Stewart R, Thomson JA. Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol. 2016;17:173. doi: 10.1186/s13059-016-1033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Giladi A, Gorki A-D, Solodkin DG, Zada M, Hladik A, Miklosi A, Salame T-M, Halpern KB, David E, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175:1031–1044.e1018. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- Colombi D, Dinkel J, Weinheimer O, Obermayer B, Buzan T, Nabers D, Bauer C, Oltmanns U, Palmowski K, Herth F, et al. Visual vs fully automatic histogram-based assessment of idiopathic pulmonary fibrosis (IPF) progression using sequential multidetector computed tomography (MDCT) PLoS ONE. 2015;10:e0130653. doi: 10.1371/journal.pone.0130653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daba MH, El-Tahir KE, Al-Arifi MN, Gubara OA. Drug-induced pulmonary fibrosis. Saudi Med J. 2004;25:700–706. [PubMed] [Google Scholar]
- Della LV, Cecchettini A, Del Ry S, Morales MA. Bleomycin in the setting of lung fibrosis induction: From biological mechanisms to counteractions. Pharmacol Res. 2015;97:122–130. doi: 10.1016/j.phrs.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Frame JM, McGrath KE, Palis J. Erythro-myeloid progenitors: "definitive" hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol Dis. 2013;51:220–225. doi: 10.1016/j.bcmd.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- Han Y., Yang L., Duan X., Duan F., Nilsson-Payant B.E., Yaron T.M., Wang P., Tang X., Zhang T., Zhao Z. et al., Identification of candidate COVID-19 therapeutics using hPSC-derived Lung Organoids. In: bioRxiv. 2020a.
- Han Y., Yang L., Duan X., Duan F., Nilsson-Payant B.E., Yaron T.M., Wang P., Tang X., Zhang T., Zhao Z. et al. 2020b. Identification of candidate COVID-19 therapeutics using hPSC-derived lung organoids. 2020b.2005.2005.079095. 10.1101/2020b.05.05.079095%JbioRxiv.
- Happle C., Lachmann N., Ackermann M., Wetzke M., Mirenska A., Mucci A., Glomb T., Oliver D.-B., Moritz T., Hansen G., Induced pluripotent stem cell derived macrophages differentiate into AM like cells in the lungs of humanized PAP mice. In: Paediatric Bronchology. 2017.
- Hartley P.G., Galvin J.R., Hunninghake G.W., Merchant J.A., Yagla S.J., Speakman S.B., Schwartz D.A. 1994. High-resolution CT-derived measures of lung density are valid indexes of interstitial lung disease. 76. 271-277. 10.1152/jappl.1994.76.1.271. [DOI] [PubMed]
- Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, Boonekamp KE, Bowden G, Hendrickx APA, Willems RJL, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol. 2018;3:814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A, Morley M, Hawkins F, McCauley KB, Jean JC, Heins H, Na C-L, Weaver TE, Vedaie M, Hurley K, et al. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell. 2017;21:472–488.e410. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Walter JM, Misharin AV. Alveolar macrophages. Cell Immunol. 2018;330:86–90. doi: 10.1016/j.cellimm.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Kim HR, Shin DY, Chung KH. A review of current studies on cellular and molecular mechanisms underlying pulmonary fibrosis induced by chemicals. Environ Anal Health Toxicol. 2018;33:e2018014–e2018010. doi: 10.5620/eht.e2018014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotton DN, Morrisey EE. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E., Ballesteros I., Farlik M., Halbritter F., Günther P., Crozet L., Jacome-Galarza C.E., Händler K., Klughammer J., Kobayashi Y. et al. 2016. Specification of tissue-resident macrophages during organogenesis. Science (New York, NY) 353. 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed]
- McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, Palis J. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 2015;11:1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecozzi L, Mambrini M, Ruscitti F, Ferrini E, Ciccimarra R, Ravanetti F, Sverzellati N, Silva M, Ruffini L, Belenkov S, et al. In-vivo lung fibrosis staging in a bleomycin-mouse model: a new micro-CT guided densitometric approach. Sci Rep. 2020;10:18735. doi: 10.1038/s41598-020-71293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Dye BR, Ferrer-Torres D, Hill DR, Overeem AW, Shea LD, Spence JR. Generation of lung organoids from human pluripotent stem cells in vitro. Nat Protoc. 2019;14:518–540. doi: 10.1038/s41596-018-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GRS, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield EC, Blue RE, Quinney NL, Gentzsch M, Ding S. Generation of renewable mouse intestinal epithelial cell monolayers and organoids for functional analyses. BMC Cell Biol. 2018;19:15. doi: 10.1186/s12860-018-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat AM, Scott CL, Bain CC. Barrier-tissue macrophages: functional adaptation to environmental challenges. Nat Med. 2017;23:1258–1270. doi: 10.1038/nm.4430. [DOI] [PubMed] [Google Scholar]
- Nadkarni RR, Abed S, Draper JS. Organoids as a model system for studying human lung development and disease. Biochem Biophys Res Commun. 2016;473:675–682. doi: 10.1016/j.bbrc.2015.12.091. [DOI] [PubMed] [Google Scholar]
- Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972-1988.e1916. 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed]
- Ohkubo H, Nakagawa H, Niimi A. Computer-based quantitative computed tomography image analysis in idiopathic pulmonary fibrosis: A mini review. Respir Investig. 2018;56:5–13. doi: 10.1016/j.resinv.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Paek D, Koh Y, Park DU, Cheong HK, Do KH, Lim CM, Hong SJ, Kim YH, Leem JH, Chung KH, et al. Nationwide study of humidifier disinfectant lung injury in South Korea, 1994–2011. Incidence and Dose-Response Relationships. Ann Am Thorac Soc. 2015;12:1813–1821. doi: 10.1513/AnnalsATS.201504-221OC. [DOI] [PubMed] [Google Scholar]
- Perdiguero EG, Klapproth K, Schulz C, Busch K, de Bruijn M, Rodewald HR, Geissmann F. The origin of tissue-resident macrophages: when an Erythro-myeloid progenitor is an erythro-myeloid progenitor. Immunity. 2015;43:1023–1024. doi: 10.1016/j.immuni.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Roszer T. 2018. Understanding the biology of self-renewing macrophages. Cells 7. 10.3390/cells7080103. [DOI] [PMC free article] [PubMed]
- Song JA, Park HJ, Yang MJ, Jung KJ, Yang HS, Song CW, Lee K. Polyhexamethyleneguanidine phosphate induces severe lung inflammation, fibrosis, and thymic atrophy. Food Chem Toxicol. 2014;69:267–275. doi: 10.1016/j.fct.2014.04.027. [DOI] [PubMed] [Google Scholar]
- Spagnolo P, Kropski JA, Jones MG, Lee JS, Rossi G, Karampitsakos T, Maher TM, Tzouvelekis A, Ryerson CJ. Idiopathic pulmonary fibrosis: disease mechanisms and drug development. Pharmacol Ther. 2021;222:107798. doi: 10.1016/j.pharmthera.2020.107798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikoudis A, Cieślak A, Loffredo L, Chen Y-W, Patel N, Saqi A, Lederer DJ, Snoeck H-W. Modeling of fibrotic lung disease using 3D organoids derived from human pluripotent stem cells. Cell Rep. 2019;27:3709–3723.e3705. doi: 10.1016/j.celrep.2019.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- T'Jonck W, Guilliams M, Bonnardel J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell Immunol. 2018;330:43–53. doi: 10.1016/j.cellimm.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, et al. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity. 2017;47(183–198):e186. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Tashiro M, Izumikawa K, Yoshioka D, Nakamura S, Kurihara S, Sakamoto N, Seki M, Kakeya H, Yamamoto Y, Yanagihara K, et al. Lung fibrosis 10 years after cessation of bleomycin therapy. Tohoku J Exp Med. 2008;216:77–80. doi: 10.1620/tjem.216.77. [DOI] [PubMed] [Google Scholar]
- Thannickal VJ, Toews GB, White ES, Lynch JP, 3rd, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med. 2004;55:395–417. doi: 10.1146/annurev.med.55.091902.103810. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen YG. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J Mol Biol. 2016;428:1409–1422. doi: 10.1016/j.jmb.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, Sone N, Nagasaki T, Matsumoto H, Muro S, et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods. 2017;14:1097–1106. doi: 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- Yao Y, Liu Q, Adrianto I, Wu X, Glassbrook J, Khalasawi N, Yin C, Yi Q, Dong Z, Geissmann F, et al. Histone deacetylase 3 controls lung alveolar macrophage development and homeostasis. Nat Commun. 2020;11:3822. doi: 10.1038/s41467-020-17630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J, Kim T, Evans KV, Jeong YI, Hur Y, Hong SP, Kim JH, Yi K, Kim SY, Na KJ, et al. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–919.e910. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Buttgereit A, Lelios I, Utz SG, Cansever D, Becher B, Greter M. The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity. 2017;47(903–912):e904. doi: 10.1016/j.immuni.2017.10.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang Y, Wu G, Xiong W, Gu W, Wang CY. Macrophages: friend or foe in idiopathic pulmonary fibrosis? Respir Res. 2018;19:170. doi: 10.1186/s12931-018-0864-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. 2020. Identifying airborne transmission as the dominant route for the spread of COVID-19. 117. 14857–14863. 10.1073/pnas.2009637117 %J Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Not applicable.