Abstract
The durable responses and favorable long-term outcomes are limited to a proportion of advanced melanoma patients treated with immune checkpoint inhibitors (ICI). Considering the critical role of antitumor immunity status in the regulation of ICI therapy responsiveness, we focused on the immune-related gene profiles and aimed to develop an individualized immune signature for predicting the benefit of ICI therapy. During the discovery phase, we integrated three published datasets of metastatic melanoma treated with anti-PD-1 (n = 120) and established an immune-related gene pair index (IRGPI) for patient classification. The IRGPI was constructed based on 31 immune-related gene pairs (IRGPs) consisting of 51 immune-related genes (IRGs). The ROC curve analysis was performed to evaluate the predictive accuracy of IRGPI with AUC = 0.854. Then, we retrospectively collected one anti-PD-1 therapy dataset of metastatic melanoma (n = 55) from Peking University Cancer Hospital (PUCH) and performed the whole-transcriptome RNA sequencing. Combined with another published dataset of metastatic melanoma received anti-CTLA-4 (VanAllen15; n = 42), we further validated the prediction accuracy of IRGPI for ICI therapy in two datasets (PUCH and VanAllen15) with AUCs of 0.737 and 0.767, respectively. Notably, the survival analyses revealed that higher IRGPI conferred poor survival outcomes in both the discovery and validation datasets. Moreover, correlation analyses of IRGPI with the immune cell infiltration and biological functions indicated that IRGPI may be an indicator of the immune status of the tumor microenvironment (TME). These findings demonstrated that IRGPI might serve as a novel marker for treating of melanoma with ICI, which needs to be validated in prospective clinical trials.
Keywords: immune-related gene pair index (IRGPI), immune checkpoint inhibitor (ICI), melanoma, prediction, immune infiltration
Introduction
Malignant melanoma is an aggressive malignant tumor with a poor clinical prognosis, the incidence of which is increasing globally (1–3). With the development of immunotherapy, immune checkpoint inhibitors (ICI) therapy has been approved as the standard treatment for melanoma (4–8). According to the reports from multiple clinical trials (9–12), the overall response rate (ORR) of PD-1 blockade with nivolumab or pembrolizumab ranged from 26% to 44%, thus indicating almost 50% of patients with severely progressed melanoma do not obtain complete or partial response, with roughly 24% reach a stable disease only. Notably, as the main subtypes of Asian patients with melanoma, only 10~20% of acral and mucosal cases can benefit from ICI therapy (13–15). Therefore, development of novel biomarkers in the hope of better prediction of the response to ICI therapy are urgently required.
Several biomarkers have been developed for predicting the benefit of ICI therapy for melanoma patients, including PD-L1 expression (16), tumor mutation burden (TMB) (17), interferon-γ signal (18, 19), and tumor infiltrating lymphocytes (TILs) (20). However, the immunohistochemistry analysis of PD-L1 varies significantly among different antibodies (21), thereby making it difficult to define the positive threshold of PD-L1 expression. In addition, the whole-genome sequences from 183 melanoma samples revealed that the burden of mutations is more frequent in cutaneous compared with acral and mucosal melanoma (22). Thus, the widespread detection value of TMB in acral and mucosal subtypes are limited.
The past decade has witnessed rapid progress in tumor genomics. Some studies utilized RNA sequencing data to establish immune-related gene signatures for the evaluation of immune response and prognosis in melanoma (23, 24). Unfortunately, none has been confirmed to be translated into clinical application owing to the small size of discovery data and lack of sufficient validation (25). Nowadays, a series of immunotherapy data regarding PD-1/PD-L1 or CTLA-4 blockade in melanoma patients have been reported all over the world. Integrated analyses may provide a complete picture of ICI therapy in different populations and summarize more superior predictive biomarkers. However, the information of gene expression profiling (GEP) was measured using different sequence platforms, which is not applied to normalizing gene expression levels through traditional approaches (26). Furthermore, the potential biological heterogeneity across datasets was also a challenge. Recently one method based on the construction of immune-related gene pairs (IRGPs) from GEP can be an excellent choice, which calculates the relative ranking of gene expression levels without the requirement for data preprocessing and has been demonstrated to establish robust models for the application of cancer classification (27–29). Hence, it is imperative to identify novel biomarkers based on IRGPs for guiding ICI therapy.
In this study, we integrated three published datasets of metastatic melanoma treated with anti-PD-1 (n = 120) and constructed an immune-related gene pair index (IRGPI). The IRGPI was constructed based on 31 immune-related gene pairs (IRGPs) consisting of 51 immune-related genes (IRGs), which may be a promising biomarker for predicting the response of ICI therapy and survival outcomes in melanoma patients. The predictive performance of IRGPI was also validated in Peking University Cancer Hospital (PUCH, n = 55) and VanAllen15 (n = 42) datasets treated with PD-1 or CTLA-4 blockade. Furthermore, the analyses of the TME, the immune cell infiltration, and biological functions of different IRGPI groups were also performed, which demonstrated that IRGPI may be an indicator of the immune status of the TME.
Materials and Methods
Patients and GEP
From March 2016 to March 2019, 55 melanoma patients treated with anti-PD-1 therapy were recruited for this study from PUCH. Formalin-fixed, paraffin-embedded pretreatment tumor samples were obtained from all patients. We separated all the clinical and pathological data by medical record review, including sex, age, primary site, metastasis status, and clinical efficacy. Tumor responses were evaluated using the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, including complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). CR and PR were regarded as responders, while PD and SD were regarded as non-responders. In this study, overall survival (OS) and progression-free survival (PFS) were used as the primary and secondary survival endpoints, respectively. Gene expression data for the PUCH cohort was based on the Illumina NovaSeq 6000 platform. The details of processing the GEP of the PUCH cohort have been described in our previous study (30). This study was conducted according to the Declaration of Helsinki Principles and approved by the Medical Ethics Committee of PUCH. Informed consent for the use of material in medical research was obtained from all participants.
External Data Acquisition
We obtained RNA-seq and clinical data from four publicly available cohorts of melanoma patients treated with anti-PD-1 or anti-CTLA-4 therapy, including Gide19 (n = 41) (31), Hugo16 (n = 28) (32), Riaz17 (n = 51) (33), and VanAllen15 (n = 42) (34). Data of Gide19 cohort (PRJEB23709) were downloaded from the European Nucleotide Archive database (ENA; https://www.ebi.ac.uk/ena). Data of Hugo16 cohort (GSE91061) and Riaz17 cohort (GSE78220) were downloaded from the Gene Expression Omnibus database (GEO; https://www.ncbi.nlm.nih.gov/geo/). Data of VanAllen15 cohort (phs000452.v2.p1) were downloaded from the database of Genotypes and Phenotypes (dbGap; http://www.ncbi.nlm.nih.gov/gap). The treatment response to immunotherapy consisted of CR/PR/SD/PD according to RECIST 1.1 guidelines, which were used in our analysis.
Moreover, we downloaded the RNA-sequencing data of all available cutaneous melanoma samples from The Cancer Genome Atlas (TCGA) database through the GDC tool (http://portal.gdc.cancer.gov/). The survival data of these patients were extracted from cbioPortal (http://www.cbioportal.org). Patients with OS less than one month were excluded from our analysis. In addition, we separated the TMB data of melanoma patients in the TCGA-SKCM cohort from The Cancer Immunome Atlas (http://tcia.at/home) (35).
Construction of the IRGPI
We constructed a predictive signature based on IRGs gathered from the ImmPort Web portal (https://www.immport.org/home) (36). Two IRGs constituted one IRGP and formed as “IRG-A|IRG-B”. The score of IRGPI was generated through pairwise comparison of gene expression levels in specific samples. When the expression level of IRG-A was higher than IRG-B, the IRGP was assigned a score of 1; otherwise, the IRGP score was 0. IRGPs with score of 0 or 1 in over 80% of the specimens were regarded as IRGPs with constant values, which does not contribute to the difference of patient survival (37). Therefore, we excluded these IRGPs with constant values from our analysis.
The Gide19, Hugo16, and Riaz17 cohorts merged into the meta cohort, which was used for the construction of IRGPI. Firstly, we used the log-rank test to investigate the correlation of each IRGP to patients’ OS in the meta cohort. According to the analysis results, IRGPs with a false discovery rate (FDR) < 0.001 were candidates to build the IRGPI. Then, the multivariate Cox regression analyses were performed to obtain the hub IRGPs and the respective coefficients. Finally, the IRGPI formula was defined as follows:
Validation of the IRGPI
The predictive and prognostic values of IRGPI for immunotherapy were validated in PUCH and VanAllen15 cohorts. Based on the treatment response to immunotherapy, we conducted the receiver operating characteristic (ROC) curve analysis to estimate the prediction accuracy of IRGPI. Using the cut-off value that generated the maximum Youden index (38), the patients were divided into IRGPI-high and IRGPI-low groups. Then, the log-rank tests were conducted for comparison of the survival outcomes between two IRGPI groups.
Tumor Immune Microenvironment Analysis
The transcriptomic data of TCGA-SKCM (skin cutaneous melanoma) cohort were used for analyzing the association of IRGPI with immune-related features. Using the IRGPI formula, we calculated the IRGPI score of each patient in the TCGA-SKCM cohort. The cut-off value for the IRGPI was determined on the basis of the association with patients’ OS by using X-tile software (version 3.6.1) (39). Based on two bioinformatic analyses of GEP data in the TCGA-SKCM cohort, we calculated the enrichment of immune cells between two IRGPI groups. Briefly, we used Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) method to calculate the immune score and ESTIMATE score of patients (40). CIBERSORT was further used to distinguish 22 immune cells, such as T cell types, B cell types, NK cells, and myeloid cell types (41). In the signaling analysis, we conducted gene set enrichment analysis (GSEA) to distinguish which immune-related pathways were markedly different between IRGPI-high and IRGPI-low groups.
To further characterize the tumor immune microenvironment between two IRGPI groups, we performed single simple GSEA on some previously published immune-related signatures (19, 42–45) and compared the score between IRGPI-high and IRGPI-low groups.
Statistical Analysis
All statistical analyses were performed using the R software (version 3.6.3) and Prism 8. Survival analyses were performed using the R packages “survival” and “survminer”. The signature of IRGPs was obtained using the R package “glmnet”. Univariate and multivariate Cox regression analyses were conducted using the R package “survival”. ROC curve analyses were performed using the R package “survivalROC”. ESTIMATE analysis was conducted using the R package “estimate”. CIBERSORT analysis was processed using the R packages “e1701”, “preprocessCore”, and “limma”. All statistical analyses were two-sided, and P < 0.05 was considered as statistically significant.
Results
Patients Characteristics
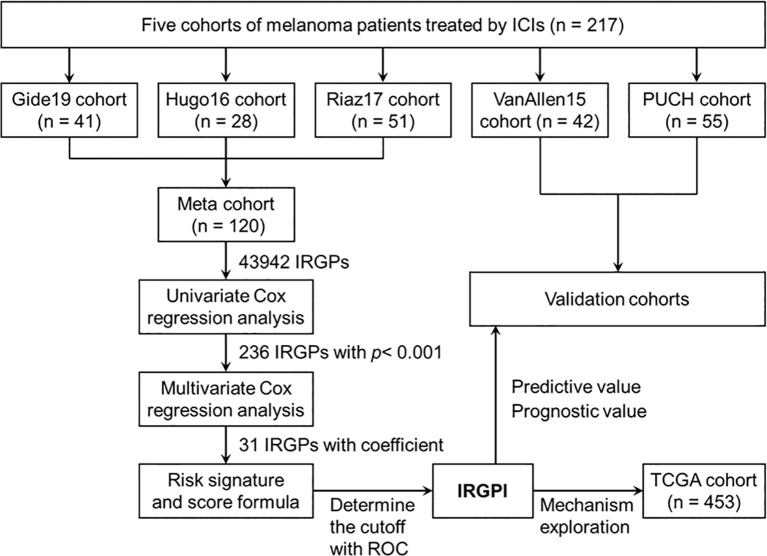
The flowchart of this study design is presented in Figure 1 . A total of 217 patients treated with ICI from five cohorts were included in this study. We constructed the IRGPI based on the meta cohort (n = 120), which consisted Gide19 (n = 41), Hugo16 (n = 28), and Riaz17 (n = 51) cohorts. The PUCH (n = 55) and VanAllen15 (n = 42) cohorts were used for validation the prediction model. The clinicopathological characteristics are summarized in Table 1 . The median follow-up is 13.1~32.7 months in five cohorts. Notably, 43.6% of patients in PUCH cohort were acral melanomas, which have been reported to be the main subtype of melanoma in Asians. However, the vast majority of patients in other cohorts were cutaneous melanoma.
Figure 1.
The flowchart showing the scheme of this study.
Table 1.
Clinicopathological characteristics of five immunotherapy cohorts included in this study.
| Patient characteristics | Training cohorts | Validation cohorts | |||
|---|---|---|---|---|---|
| Gide19 | Hugo16 | Riaz17 | VanAllen15 | PUCH | |
| No. of patients | 41 | 28 | 51 | 42 | 55 |
| Median age in yrs (range) | 66 (37-90) | 61 (19-84) | – | 61 (22-83) | 51 (27-72) |
| Sex, n (%) | |||||
| Male | 26 (63.4) | 20 (71.4) | – | 28 (66.7) | 17 (30.9) |
| Female | 15 (36.6) | 8 (28.6) | – | 14 (33.3) | 38 (69.1) |
| Primary site, n (%) | |||||
| Acral | – | – | 1 (2.0) | – | 24 (43.6) |
| Mucosal | – | 3 (10.7) | 7 (13.7) | 2 (4.8) | 8 (14.5) |
| Cutaneous | – | 21 (75.0) | 32 (62.7) | 37 (88.1) | 18 (32.7) |
| Ocular | – | 4 (7.9) | 3 (7.1) | – | |
| Unknown | – | 4 (14.3) | 7 (13.7) | – | 5 (9.1) |
| Metastasis status, n (%) | |||||
| M0 | – | 1 (3.6) | 1 (1.9) | 1 (2.4) | 10 (18.2) |
| M1a | – | 2 (7.1) | 11 (21.6) | 3 (7.1) | 16 (29.1) |
| M1b | – | 3 (10.7) | 8 (15.7) | 7 (16.7) | 18 (32.7) |
| M1c | – | 22 (78.6) | 23 (45.1) | 31 (73.8) | 11 (20.0) |
| Unknown | – | – | 8 (15.7) | – | – |
| BRAF V600, n (%) | – | 12 (42.9) | 14 (27.4) | – | – |
| Prior MAPKi, n (%) | – | 12 (42.9) | – | 4 (9.5) | – |
| Treatment, n (%) | |||||
| Anti-PD-1 | 41 (100) | 28 (100) | 51 (100) | – | 55 (100) |
| Anti-CTLA-4 | – | – | – | 42 (100) | |
| Best overall response, n (%) | |||||
| CR | 4 (9.8) | 5 (17.9) | 3 (5.9) | – | 1 (1.8) |
| PR | 15 (36.6) | 10 (35.7) | 7 (13.7) | – | 13 (23.6) |
| CR/PR | – | – | 19 (45.2) | ||
| SD | 6 (14.6) | – | 16 (31.4) | – | 6 (10.9) |
| PD | 16 (39.0) | 13 (46.4) | 25 (49.0) | 23 (54.8) | 35 (63.6) |
| Median PFS (months) | 9.0 | – | – | 2.8 | 3.9 |
| Median OS (months) | 29.3 | 32.7 | 21.1 | 13.1 | 28.1 |
MAPKi, MAPK pathway inhibitors; Anti-PD-1, anti-programmed death-1; Anti-CTLA-4, anti-cytotoxic T lymphocyte antigen-4; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; PFS, progression-free survival; OS, overall survival.
Construction and Definition of the IRGPI
Among the 2487 IRGs from the ImmPort database, 1138 IRGs commonly occurred in the GEP of five cohorts and 1295044 IRGPs were calculated. We excluded 1251102 IRGPs (96.6%) with constant values in any cohort and 43942 IRGPs remained for subsequent construction of the IRGPI ( Table S1 ). According to the univariate Cox regression analyses of the correlation between each IRGP and patients’ OS in the meta cohort, 236 IRGPs with adjusted P < 0.001 were selected as prognostic IRGPs ( Table S2 ). Then, we performed the multivariate Cox regression analyses to filtrate IRGPs to construct the IRGPI. Finally, 31 IRGPs were filtrated to define the IRGPI ( Table 2 and Figure S1A ). The IRGPI consisted of 51 unique IRGs, most of which encoded molecules involved in antimicrobials, cytokines, and cytokine receptors.
Table 2.
Model information of IRGPI.
| IRG-A | Full name | Immune pathway | IRG-B | Full name | Immune pathway | Coefficient |
|---|---|---|---|---|---|---|
| CD1B | CD1b molecule | Antigen Processing and Presentation | AMHR2 | anti-Mullerian hormone receptor type 2 | Cytokine Receptors, TGFb Family Member Receptor | -0.133719837 |
| CD1C | CD1c molecule | Antigen Processing and Presentation | GDNF | glial cell derived neurotrophic factor | Cytokines, TGFb Family Member | -0.006407801 |
| CD1E | CD1e molecule | Antigen Processing and Presentation | NGF | nerve growth factor | Cytokines | -0.118650926 |
| HLA-C | major histocompatibility complex, class I, C | Antigen Processing and Presentation, NaturalKiller Cell Cytotoxicity | SPP1 | secreted phosphoprotein 1 | Cytokines | -0.000146032 |
| HSPA6 | heat shock protein family A (Hsp70) member 6 | Antigen Processing and Presentation | PI15 | peptidase inhibitor 15 | Antimicrobials | -0.006568346 |
| IFNG | interferon gamma | Antigen Processing and Presentation, Antimicrobials, Cytokines, Interferons, NaturalKiller Cell Cytotoxicity, TCR Signaling Pathway | NTS | neurotensin | Cytokines | -0.208747404 |
| RELB | RELB proto-oncogene, NF-kB subunit | Antigen Processing and Presentation | NFATC4 | nuclear factor of activated T cells 4 | BCR Signaling Pathway, NaturalKiller Cell Cytotoxicity, TCR Signaling Pathway | -0.001379582 |
| CXCL13 | C-X-C motif chemokine ligand 13 | Antimicrobials, Chemokines, Cytokines | PLAU | plasminogen activator, urokinase | Antimicrobials, Chemokines, Cytokines | -0.042607265 |
| XCL1 | X-C motif chemokine ligand 1 | Antimicrobials, Chemokines, Cytokines | FABP6 | fatty acid binding protein 6 | Antimicrobials | -0.179318494 |
| SFTPD | surfactant protein D | Antimicrobials | CR2 | complement C3d receptor 2 | BCR Signaling Pathway | 0.248588976 |
| MMP9 | matrix metallopeptidase 9 | Antimicrobials | NOX4 | NADPH oxidase 4 | Antimicrobials | -0.126050935 |
| RBP7 | retinol binding protein 7 | Antimicrobials | PRF1 | perforin 1 | NaturalKiller Cell Cytotoxicity | 0.040708869 |
| IFIH1 | interferon induced with helicase C domain 1 | Antimicrobials | VAV3 | vav guanine nucleotide exchange factor 3 | BCR Signaling Pathway, NaturalKiller Cell Cytotoxicity, TCR Signaling Pathway | -0.601441422 |
| IDO1 | indoleamine 2,3-dioxygenase 1 | Antimicrobials | CD72 | CD72 molecule | BCR Signaling Pathway | -0.238320598 |
| IDO1 | indoleamine 2,3-dioxygenase 1 | Antimicrobials | SECTM1 | secreted and transmembrane 1 | Cytokines | -0.16744787 |
| IRF1 | interferon regulatory factor 1 | Antimicrobials | HMOX1 | heme oxygenase 1 | Antimicrobials | -0.00086748 |
| IRF1 | interferon regulatory factor 1 | Antimicrobials | IL1R1 | interleukin 1 receptor type 1 | Cytokine Receptors, Interleukins Receptor | -0.115025852 |
| ZYX | zyxin | Antimicrobials | IRF9 | interferon regulatory factor 9 | Antimicrobials | 0.117305566 |
| TNFAIP3 | TNF alpha induced protein 3 | Antimicrobials | IL1R1 | interleukin 1 receptor type 1 | Cytokine Receptors, Interleukins Receptor | -0.209151382 |
| HMOX1 | heme oxygenase 1 | Antimicrobials | IL32 | interleukin 32 | Cytokines | 0.199502086 |
| CCR7 | C-C motif chemokine receptor 7 | Antimicrobials, Chemokine Receptors, Cytokine Receptors | IL11 | interleukin 11 | Cytokines, Interleukins | -0.036970847 |
| PTGDR | prostaglandin D2 receptor | Antimicrobials, Cytokine Receptors | EGF | epidermal growth factor | Cytokines | -0.088054831 |
| RAC3 | Rac family small GTPase 3 | BCR Signaling Pathway, NaturalKiller Cell Cytotoxicity | NR1D1 | nuclear receptor subfamily 1 group D member 1 | Cytokine Receptors | 0.174898131 |
| CD19 | CD19 molecule | BCR Signaling Pathway | EGF | epidermal growth factor | Cytokines | -0.012058023 |
| INPP5D | inositol polyphosphate-5-phosphatase D | BCR Signaling Pathway | IL1R1 | interleukin 1 receptor type 1 | Cytokine Receptors, Interleukins Receptor | -0.014592558 |
| CXCR3 | C-X-C motif chemokine receptor 3 | Chemokine Receptors, Cytokine Receptors | IL11 | interleukin 11 | Cytokines/Interleukins | -0.12999067 |
| EGF | epidermal growth factor | Cytokines | TNFRSF11A | TNF receptor superfamily member 11a | Cytokine Receptors, TNF Family Members Receptors | 0.254354052 |
| IL33 | interleukin 33 | Cytokines, Interleukins | RARG | retinoic acid receptor gamma | Cytokine Receptors | -0.27380975 |
| IL7 | interleukin 7 | Cytokines, Interleukins | PRF1 | perforin 1 | NaturalKiller Cell Cytotoxicity | 0.062199575 |
| IL20RB | interleukin 20 receptor subunit beta | Cytokine Receptors, Interleukins Receptor | TNFRSF10C | TNF receptor superfamily member 10c | Cytokine Receptors, NaturalKiller Cell Cytotoxicity, TNF Family Members Receptors | 0.049115859 |
| TEK | TEK receptor tyrosine kinase | Cytokine Receptors | CD28 | CD28 molecule | TCR Signaling Pathway | 0.046180545 |
Evaluation of the Prediction Accuracy of IRGPI for the Efficacy of ICI Therapy
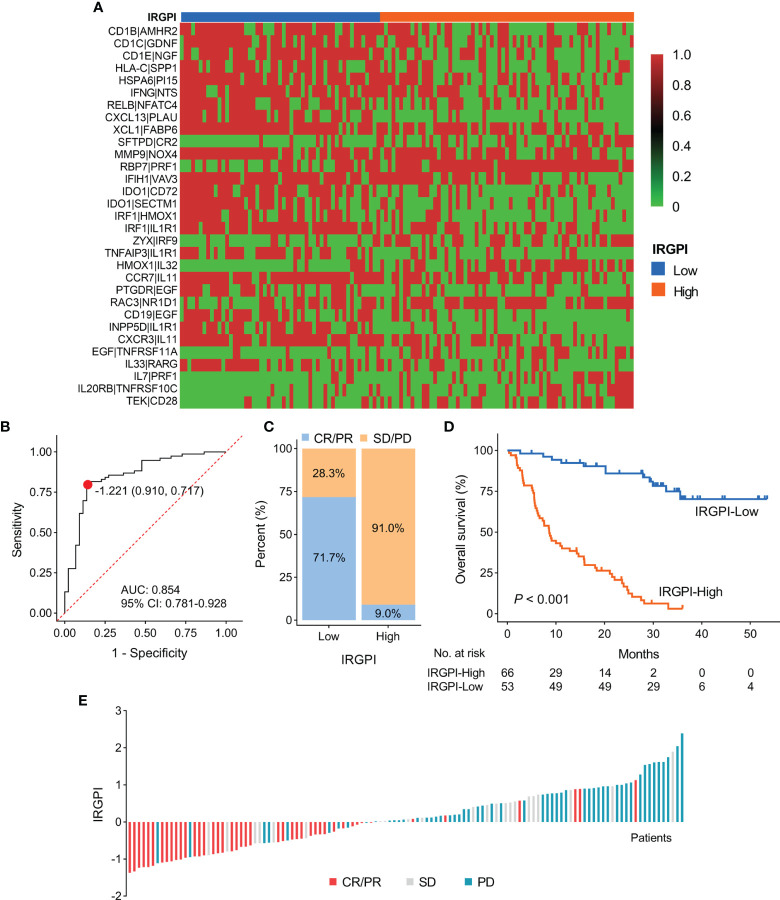
Based on the IRGPI formula, we calculated each patient’s IRGPI score in the meta cohort and exhibited the result in a heatmap ( Figure 2A ). We conducted ROC curve analysis to evaluate the prediction accuracy of IRGPI for the efficacy of ICI therapy in the meta cohort. With a cut-off value based on the Youden index of -1.221, we found that IRGPI-low group patients were correctly classified as CR/PR with a sensitivity of 71.7% (38/53). Further, IRGPI-high group patients were successfully classified as SD/PD with a specificity of 91.0% (61/67). The overall accuracy of IRGPI was 82.5% (99/120) with AUC of 0.854 ( Figure 2B ). IRGPI-low group showed a higher ORR than IRGPI-high group (71.7% vs. 9.0%; Figures 2C, E ). Moreover, we evaluated the relationship between the IRGPI score and OS/PFS in the meta cohort. Kaplan-Meier survival analysis revealed that IRGPI-low group patients had significantly longer OS (P < 0.001; Figure 2D ). The median PFS for IRGPI-high group patients was markedly shorter than IRGPI-low group patients in the Gide19 cohort (P < 0.001; Figure S2A ). The pooled hazard ratio (HR) along with 95% confidence interval (CI) for the association between high IRGPI score and OS in 119 cases of patients was 8.02 (3.91~15.19), and no significant heterogeneity among the three datasets was observed (I2 = 0%, P = 0.98, Figure S1B ). Overall, the IRGPI showed a superior prediction for the benefit of ICI therapy in the meta cohort.
Figure 2.
Construction and evaluation of IRGPI in the discovery cohort. (A) A heatmap of the identified 31 IRGPs with corresponding IRGPI groups. (B) ROC curve for the predictive performance of IRGPI. (C) The rate of durable clinical response for patients with high and low IRGPI scores. (D) Kaplan-Meier plots of overall survival segregated by IRGPI score with cut-off points selected according to the Youden index. (E) Waterfall plot of IRGPI for distinct clinical response groups. IRGPI, immune-related gene pair index; IRGPs, immune-related gene pairs; ROC, receiver operating characteristic; AUC, area under curve; CI, confidence interval.
Validation of the Robustness of IRGPI in Predicting the Efficacy of ICI Therapy
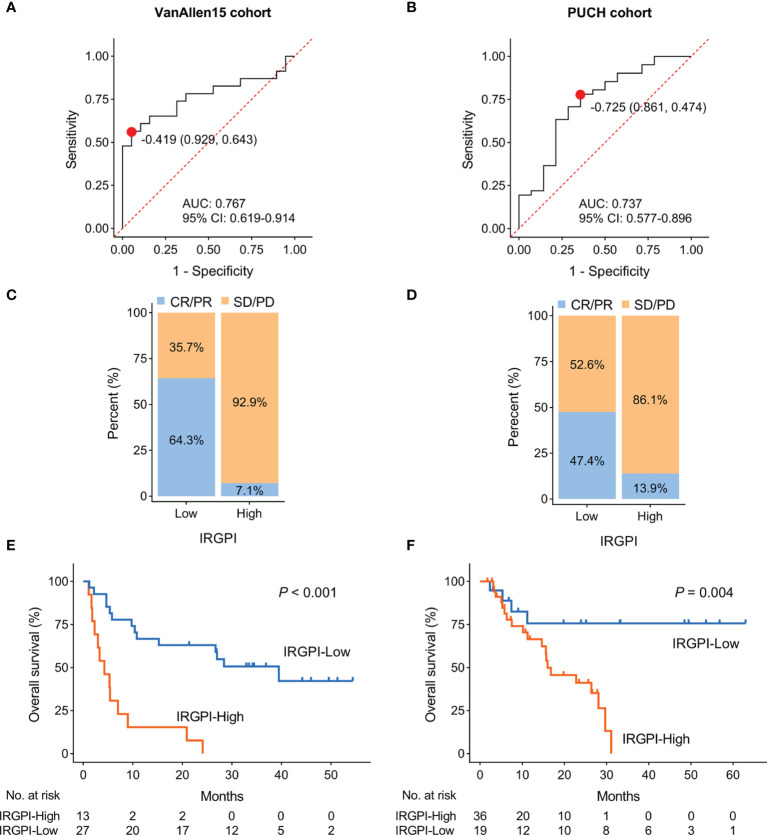
To verify the robustness of IRGPI in predicting the efficacy of ICI therapy, we assessed the correlation of IRGPI score with overall response rate and survival outcomes in VanAllen15 and PUCH cohorts. In the VanAllen15 cohort, the IRGPI successfully identified 31 of 42 patients with an overall accuracy of 73.8% and an AUC of 0.767 ( Figure 3A ). Similarly, in the PUCH cohort, the IRGPI demonstrated an overall accuracy of 72.7% (40/55) and AUC of 0.737 ( Figure 3B ). The ORR of IRGPI-high group was lower than IRGPI-low group in VanAllen15 cohort (64.3% vs. 7.1%; Figures 3C and S3A ) and PUCH cohort (47.4% vs. 13.9%; Figures 3D and S3B ), respectively. As expected, higher IRGPI conferred poor survival outcomes in VanAllen15 cohort (OS: P < 0.001, PFS: P < 0.001; Figures 3E and S2B ) and PUCH cohort (OS: P = 0.004, PFS: P = 0.015; Figures 3F and S2C ), respectively. These results confirmed that the IRGPI is reliable for the prediction of ICI therapy responsiveness in VanAllen15 and PUCH cohorts.
Figure 3.
Validation the performance of IRGPI in two cohorts. (A, B) ROC curves for the predictive performance of IRGPI in VanAllen15 and PUCH cohorts, respectively. (C, D) The rate of durable clinical response for patients with high and low IRGPI scores in VanAllen15 and PUCH cohorts, respectively. (E, F) Kaplan-Meier plots of overall survival segregated by IRGPI score with cut-off points selected according to the Youden index in VanAllen15 and PUCH cohorts, respectively. IRGPI, immune-related gene pair index; ROC, receiver operating characteristic; AUC, area under curve; CI, confidence interval.
Association of IRGPI With Tumor Immune Microenvironment in Melanoma
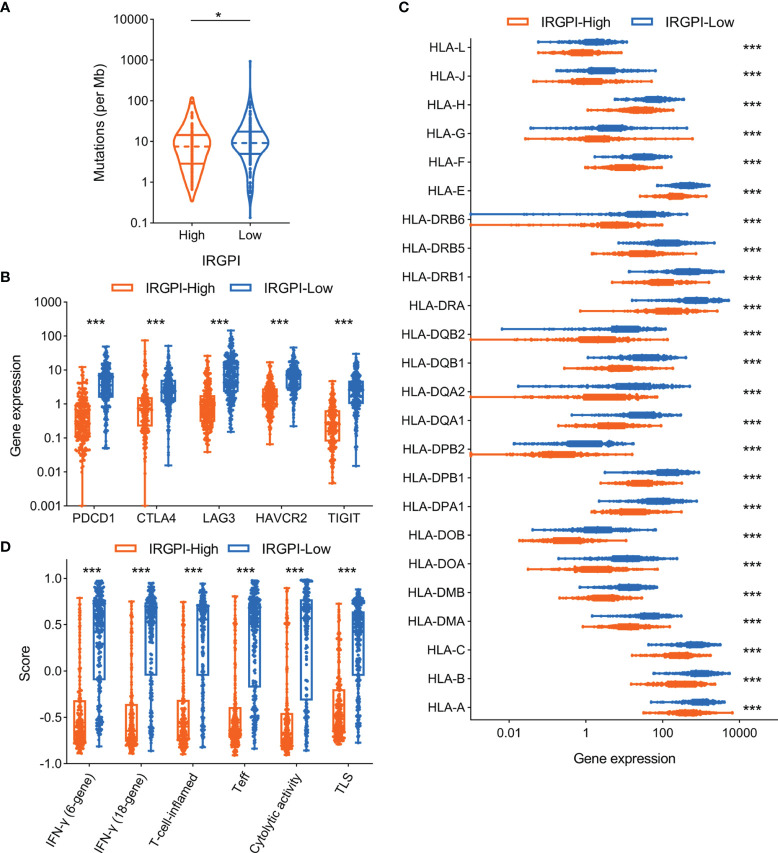
Reportedly, the infiltration of immune cells, especially CD8+ T cells, is associated with immunotherapy response in many types of cancer (46). Based on the above results, we further investigated the relationship between IRGPI and tumor immune microenvironment features in melanoma patients. The TCGA-SKCM cohort was stratified into IRGPI-high and IRGPI-low groups using X-tile software ( Table S3 and Figure S4 ). Firstly, we calculated the immune score and ESTIMATE score of patients in TCGA-SKCM cohort by ESTIMATE algorithm. The data showed that both the immune score and ESTIMATE score were considerably increased in IRGPI-low group compared with IRGPI-high group ( Figures 4A, B ). Secondly, the CIBERSORT analytical tool was used to estimate the proportions of 22 types of immune cells in each SKCM sample. The results revealed that the infiltration levels of CD8+ T cells, activated memory CD4+ T cells, naive B cells, and NK cells in IRGPI-high group were lower than that in IRGPI-low group, while resting memory CD4+ T cells, M0 and M2 macrophages showed the opposite trend ( Figure 4C ). Finally, we performed GSEA to identify which pathways were enriched at specific IRGPI levels. As shown in Figure 4D , the pathways of inflammatory response, interferon response, antigen processing and presentation, and T cell receptor signaling were markedly upregulated in IRGPI-low group.
Figure 4.
Comparison of immune microenvironment characteristics according to IRGPI status. (A, B) ESTIMATE algorithm revealed the ImmuneScore and ESTIMATEScore between IRGPI-high and IRGPI-low groups. (C) Evaluation of 22 immune cell infiltrating using the CIBERSORT method. (D) GSEA plots of immune-related pathways in comparison between IRGPI-high and IRGPI-low groups. IRGPI, immune-related gene pair index; GSEA, gene set enrichment analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
Correlation of IRGPI to Other Potential Immunotherapy Biomarkers in Melanoma
A series of potential biomarkers have been developed to predict the response of ICI therapy in malignant tumors, such as TMB, immune inhibitory receptor expression levels. We analyzed the relationship between IRGPI and TMB in TCGA-SKCM cohort and the results showed that higher IRGPI conferred lower TMB ( Figure 5A ). As expected, the expression levels of immune inhibitory receptors (including PD-1, CTLA-4, LAG3, TIM-3, and TIGIT) showed the same trend as TMB between IRGPI-high and IRGPI-low groups ( Figure 5B ). Moreover, the deficiency of human leukocyte antigen (HLA) could impair antigen presentation and initiate antitumor immunity, which consequently resulting in primary resistance to immunotherapy (47). We then investigated the correlation of IRGPI to the expression levels of HLA members and the data indicated most HLA members were substantially upregulated in IRGPI-low group compared with IRGPI-high group ( Figure 5C ).
Figure 5.
Association of IRGPI to other potential biomarkers in melanoma. (A) Comparison of tumor mutation burden level according to IRGPI status. (B) Correlation of IRGPI to immune inhibitory receptors, including PDCD1, CTLA4, LAG3, HAVCR2, TIGIT. (C) The profile of HLA member expression levels between IRGPI-high and IRGPI-low groups. (D) Box plot of the immune-related signatures in comparison of the IRGPI-high and IRGPI-low groups. IRGPI, immune-related gene pair index; HLA, human leukocyte antigen; IFN, interferon; Teff, effective T cells; TLS, tertiary lymphoid structure. *P < 0.05, ***P < 0.001.
Some immune-related GEP signatures have been described to predict the benefit of ICI therapy in melanoma ( Table S4 ). We therefore compared these GEP signature scores between IRGPI-high and IRGPI-low groups. Consistent with other biomarkers, these GEP signature scores were significantly downregulated in IRGPI-high group compared with IRGPI-low group ( Figure 5D ).
Discussion
Over the past decades, the incidence of malignant melanoma has continued to increase, but the mortality has decreased, largely due to the rapid development of ICI and targeted therapies (48). Compared with the excellent clinical efficacy of ICI therapy in melanoma, the investigations of its biomarkers are relatively insufficient. The data from the real world revealed that durable responses and favorable long-term outcomes are limited a proportion of melanoma (12). Thus, more attention should be paid to the discovery and establishment of novel biomarkers for selecting patients who may benefit from ICI therapy.
In this study, we integrated the data of ICI therapy of melanoma patients from our center and other four Western cohorts (31–34), and constructed an individualized immune predictive signature (IRGPI). The rate of durable clinical response for IRGPI-low patients in the discovery cohort, VanAllen15 and PUCH cohorts were 71.7%, 64.3% and 47.4%, respectively. Further, the percentage of non-responder in IRGPI-high group in discovery cohort, VanAllen15 and PUCH cohorts were 91%, 92.9% and 86.1%, respectively. The AUCs of ROC curve were all more than 0.7 in the discovery and validation cohorts. This reflected the good prediction accuracy and sensitivity of IRGPI for ICI therapy. The patient classification based on IRGPI also showed significantly different survival outcomes. Meanwhile, it is noted that the patients from our center are mainly acral and mucosal subtypes, while the patients from four Western cohorts are mainly cutaneous melanomas, which is consistent with the previous studies (49, 50). Considering the differential subtypes of melanoma and robust prediction accuracy across five cohorts, we reasonably assume that IRGPI is a reliable biomarker for guiding ICI therapy in both Western and Eastern patients with melanoma.
The IRGPI consisted of 51 unique IRGs, of which 36 encoded molecules involved in antimicrobials, cytokines, and cytokine receptors, which play vital roles in the regulation of the response to tumor immune microenvironment. Meanwhile, many of these IRGs have been demonstrated to be correlated with PD-L1 signaling and anti-PD-1 therapy, such as MMP9 (matrix metallopeptidase 9) and EGF (epidermal growth factor). Zhao et al. found that TGFβ pathway inhibition promoted the proliferation expansion of stromal fibroblasts, thereby facilitating MMP9-dependent cleavage of PD-L1 surface expression, leading to PD-1 blockade resistance in melanoma models (51). Furthermore, inhibition of MMP9 promoted the therapeutic efficacy of PD-1 blockade, with a marked reduction of tumor burden and extension of survival time (52). Li et al. discovered that the immunosuppressive activity of PD-L1 was tightly regulated by ubiquitination and N-glycosylation, in which glycogen synthase kinase 3β (GSK3β) could induce phosphorylation-dependent proteasome degradation of PD-L1 (53). In addition, EGF could stabilize PD-L1 via GSK3β inactivation in basal-like breast cancer (53). Therefore, blocking of EGF signaling using gefitinib resulted in the destabilization of PD-L1, enhancing antitumor T cell immunity and the treatment response of PD-1 blockade in syngeneic mouse models. What’s more, some IRGs (including IFNG, PRF1, IDO1, CXCL13) were included in an IFN-γ-related T cell-inflamed GEP, which have been developed into a clinical-grade assay for evaluating the treatment efficacy of pembrolizumab in pan-tumors (19). As expected, the GSEA results of our study showed that lower IRGPI conferred upregulated interferon response and inflammatory signaling. The above data and analyses indicated that IRGPI could predict T cell inflammation in melanoma and explain the relationship between IRGPI and ICI therapy responsiveness to some extent.
TILs, especially CD8+ T cells, can be used for predicting ICI therapy responsiveness and survival outcomes (46). In our study, we calculated the relative proportion of 22 types of immune cells based on the CIBERSORT algorithm and the results revealed that melanoma samples with high IRGPI harbored more infiltration of CD8+ T cells, activated memory CD4+ T cells, naive B cells, and NK cells, which further elucidated the reason why IRGPI-high patients with melanoma can benefit from ICI therapy. However, the infiltration levels of regulatory T cells (Tregs) in IRGPI-high patients were also significantly higher compared with that in IRGPI-low patients, which was contradictory with the previous report of Tregs with an immunosuppressive role in TME (54). Further investigations are required to evaluate the infiltration levels of Tregs using immunohistochemistry or flow cytometry.
There are some limitations, unresolved concerns, and potential perspectives in our study. First, the current study combined the data from different datasets, which can sometimes present a selection bias, due to various therapy settings, different pre-existing mutations, and baseline patient characteristics. Although this gene-pair based approach we used in this study does not require normalization of GEP, this bias across cohorts is inevitable. Second, for the IRGPI, there are still some genes whose function are not fully elucidated. Further studies, such as knockdown or overexpression of IRGs in melanoma cell lines, are required to verify the role of these genes. Moreover, the basic experiments were also lacking to examine the immune cell infiltrating and PD-L1 expression of patients treated with ICI therapy. Finally, the patients in PUCH cohort were treated with different anti-PD-1 antibodies from various pharmaceutical companies, which may lead to drug bias. Compared with two previous studies (NCT02821000 and NCT02836795) of PD-1 blockade for treating melanoma patients, the data showed the ORR of two types of anti-PD-1 antibodies in mucosal subtype were 13.3% and 0, respectively (14, 55). Thus, further studies, preferably in a prospective setting, are required to stringently evaluate the correlation of IRGPI to the immunotherapy response and survival outcomes.
Conclusions
In summary, we constructed an individualized immune predictive signature (IRGPI), which could robustly predict the ICI therapy responsiveness and long-term survival outcomes. In addition, IRGPI may be an indicator of the immune characteristics of the TME in melanoma patients. These findings indicated that IRGPI might serve as a novel marker for treating of melanoma with ICI, which needs to be validated in prospective clinical trials.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/search/?dbId=&q=HRA000524.
Ethics Statement
The studies involving human participants were reviewed and approved by Peking University Cancer Hospital & Institute. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SC and YK were involved in conception and design of the study. JuY performed and evaluated the experiment. XW and JiY helped to analyze the results. YK and XW provided materials or patients. JuY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Natural Science Foundation of China (82002906, 81902789, 82002897) and CSCO-Roche Cancer Research Fund 2019 (Y-Roche2019/2-0028).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.839901/full#supplementary-material
Forest plot of different IRGPI groups. (A) Multivariate Cox analysis of 31 hub immune-related gene pairs. (B) Forest plot of high IRGPI score with poor OS in patients from three datasets.
The performance of the IRGPI in predicting progression-free survival in three cohorts.
Waterfall plot of IRGPI for distinct clinical response groups in VanAllen15 and PUCH cohorts.
X-tile plots of the IRGPI scores in TCGA-SKCM cohort.
Abbreviations
AUC, Area Under Curve; CR, complete response; ESTIMATE, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data; FDR, false discovery rate; GEP, gene expression profiling; CI, confidence interval; GSEA, gene set enrichment analysis; HLA, human leukocyte antigen; HR, hazard ratio; ICI, immune checkpoint inhibitors; IRGs, immune-related genes; IRGPs, immune-related gene pairs; IRGPI, immune-related gene pair index; ORR, overall response rate; OS, overall survival; PD, progressive disease; PFS, progression-free survival; PUCH, Peking University Cancer Hospital; PR, partial response; RECIST, Response Evaluation Criteria in Solid Tumors; ROC, receiver operating characteristic SD, stable disease; TCGA, The Cancer Genome Atlas; TILs, tumor infiltrating lymphocytes; TME, tumor microenvironment; TMB, tumor mutation burden; Tregs, regulatory T cells.
References
- 1. Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a Molecular Classification of Melanoma. J Clin Oncol (2007) 25(12):1606–20. doi: 10.1200/JCO.2006.06.0442 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer Statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4. Robert C, Ribas A, Schachter J, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab Versus Ipilimumab in Advanced Melanoma (KEYNOTE-006): Post-Hoc 5-Year Results From an Open-Label, Multicentre, Randomised, Controlled, Phase 3 Study. Lancet Oncol (2019) 20(9):1239–51. doi: 10.1016/S1470-2045(19)30388-2 [DOI] [PubMed] [Google Scholar]
- 5. Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, et al. Nivolumab and Ipilimumab Versus Ipilimumab in Untreated Melanoma. N Engl J Med (2015) 372(21):2006–17. doi: 10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 Pathway Blockade for Cancer Therapy: Mechanisms, Response Biomarkers, and Combinations. Sci Transl Med (2016) 8(328):328rv4. doi: 10.1126/scitranslmed.aad7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eggermont A, Blank CU, Mandala M, Long GV, Atkinson V, Dalle S, et al. Adjuvant Pembrolizumab Versus Placebo in Resected Stage III Melanoma. N Engl J Med (2018) 378(19):1789–801. doi: 10.1056/NEJMoa1802357 [DOI] [PubMed] [Google Scholar]
- 8. Rozeman EA, Hoefsmit EP, Reijers I, Saw R, Versluis JM, Krijgsman O, et al. Survival and Biomarker Analyses From the OpACIN-Neo and OpACIN Neoadjuvant Immunotherapy Trials in Stage III Melanoma. Nat Med (2021) 27(2):256–63. doi: 10.1038/s41591-020-01211-7 [DOI] [PubMed] [Google Scholar]
- 9. Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C, et al. Pembrolizumab Versus Investigator-Choice Chemotherapy for Ipilimumab-Refractory Melanoma (KEYNOTE-002): A Randomised, Controlled, Phase 2 Trial. Lancet Oncol (2015) 16(8):908–18. doi: 10.1016/S1470-2045(15)00083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and Tumor Responses With Lambrolizumab (Anti-PD-1) in Melanoma. N Engl J Med (2013) 369(2):134–44. doi: 10.1056/NEJMoa1305133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, et al. Survival, Durable Tumor Remission, and Long-Term Safety in Patients With Advanced Melanoma Receiving Nivolumab. J Clin Oncol (2014) 32(10):1020–30. doi: 10.1200/JCO.2013.53.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mao L, Qi Z, Zhang L, Guo J, Si L. Immunotherapy in Acral and Mucosal Melanoma: Current Status and Future Directions. Front Immunol (2021) 12:680407. doi: 10.3389/fimmu.2021.680407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura Y, Namikawa K, Yoshino K, Yoshikawa S, Uchi H, Goto K, et al. Anti-PD1 Checkpoint Inhibitor Therapy in Acral Melanoma: A Multicenter Study of 193 Japanese Patients. Ann Oncol (2020) 31(9):1198–206. doi: 10.1016/j.annonc.2020.05.031 [DOI] [PubMed] [Google Scholar]
- 14. Si L, Zhang X, Shu Y, Pan H, Wu D, Liu J, et al. A Phase Ib Study of Pembrolizumab as Second-Line Therapy for Chinese Patients With Advanced or Metastatic Melanoma (KEYNOTE-151). Transl Oncol (2019) 12(6):828–35. doi: 10.1016/j.tranon.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang B, Chi Z, Chen Y, Liu X, Wu D, Chen J, et al. Safety, Efficacy, and Biomarker Analysis of Toripalimab in Previously Treated Advanced Melanoma: Results of the POLARIS-01 Multicenter Phase II Trial. Clin Cancer Res (2020) 26(16):4250–9. doi: 10.1158/1078-0432.CCR-19-3922 [DOI] [PubMed] [Google Scholar]
- 16. Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, et al. Host Expression of PD-L1 Determines Efficacy of PD-L1 Pathway Blockade-Mediated Tumor Regression. J Clin Invest (2018) 128(2):805–15. doi: 10.1172/JCI96113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational Landscape and Significance Across 12 Major Cancer Types. Nature (2013) 502(7471):333–9. doi: 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M, et al. Conserved Interferon-γ Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell (2021) 39(1):122. doi: 10.1016/j.ccell.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-Related mRNA Profile Predicts Clinical Response to PD-1 Blockade. J Clin Invest (2017) 127(8):2930–40. doi: 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zito Marino F, Ascierto PA, Rossi G, Staibano S, Montella M, Russo D, et al. Are Tumor-Infiltrating Lymphocytes Protagonists or Background Actors in Patient Selection for Cancer Immunotherapy. Expert Opin Biol Ther (2017) 17(6):735–46. doi: 10.1080/14712598.2017.1309387 [DOI] [PubMed] [Google Scholar]
- 21. Sunshine JC, Nguyen PL, Kaunitz GJ, Cottrell TR, Berry S, Esandrio J, et al. PD-L1 Expression in Melanoma: A Quantitative Immunohistochemical Antibody Comparison. Clin Cancer Res (2017) 23(16):4938–44. doi: 10.1158/1078-0432.CCR-16-1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-Genome Landscapes of Major Melanoma Subtypes. Nature (2017) 545(7653):175–80. doi: 10.1038/nature22071 [DOI] [PubMed] [Google Scholar]
- 23. Zhang JA, Zhou XY, Huang D, Luan C, Gu H, Ju M, et al. Development of an Immune-Related Gene Signature for Prognosis in Melanoma. Front Oncol (2020) 10:602555. doi: 10.3389/fonc.2020.602555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan J, Wu X, Yu J, Zhu Y, Cang S. Prognostic Role of Tumor Mutation Burden Combined With Immune Infiltrates in Skin Cutaneous Melanoma Based on Multi-Omics Analysis. Front Oncol (2020) 10:570654. doi: 10.3389/fonc.2020.570654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian J, Simon R. Gene Expression-Based Prognostic Signatures in Lung Cancer: Ready for Clinical Use. J Natl Cancer Inst (2010) 102(7):464–74. doi: 10.1093/jnci/djq025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the Widespread and Critical Impact of Batch Effects in High-Throughput Data. Nat Rev Genet (2010) 11(10):733–9. doi: 10.1038/nrg2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heinäniemi M, Nykter M, Kramer R, Wienecke-Baldacchino A, Sinkkonen L, Zhou JX, et al. Gene-Pair Expression Signatures Reveal Lineage Control. Nat Methods (2013) 10(6):577–83. doi: 10.1038/nmeth.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li B, Cui Y, Diehn M, Li R. Development and Validation of an Individualized Immune Prognostic Signature in Early-Stage Nonsquamous Non-Small Cell Lung Cancer. JAMA Oncol (2017) 3(11):1529–37. doi: 10.1001/jamaoncol.2017.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xue YN, Xue YN, Wang ZC, Mo YZ, Wang PY, Tan WQ. A Novel Signature of 23 Immunity-Related Gene Pairs Is Prognostic of Cutaneous Melanoma. Front Immunol (2020) 11:576914. doi: 10.3389/fimmu.2020.576914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cui C, Xu C, Yang W, Chi Z, Sheng X, Si L, et al. Ratio of the Interferon-γ Signature to the Immunosuppression Signature Predicts Anti-PD-1 Therapy Response in Melanoma. NPJ Genom Med (2021) 6(1):7. doi: 10.1038/s41525-021-00169-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell (2019) 35(2):238–55.e6. doi: 10.1016/j.ccell.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 32. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell (2016) 165(1):35–44. doi: 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution During Immunotherapy With Nivolumab. Cell (2017) 171(4):934–949.e16. doi: 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science (2015) 350(6257):207–11. doi: 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-Cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep (2017) 18(1):248–62. doi: 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 36. Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: Disseminating Data to the Public for the Future of Immunology. Immunol Res (2014) 58(2-3):234–9. doi: 10.1007/s12026-014-8516-1 [DOI] [PubMed] [Google Scholar]
- 37. Kim S, Lin CW, Tseng GC. MetaKTSP: A Meta-Analytic Top Scoring Pair Method for Robust Cross-Study Validation of Omics Prediction Analysis. Bioinformatics (2016) 32(13):1966–73. doi: 10.1093/bioinformatics/btw115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Youden WJ. Index for Rating Diagnostic Tests. Cancer (1950) 3(1):32–5. doi: [DOI] [PubMed] [Google Scholar]
- 39. Camp RL, Dolled-Filhart M, Rimm DL. X-Tile: A New Bio-Informatics Tool for Biomarker Assessment and Outcome-Based Cut-Point Optimization. Clin Cancer Res (2004) 10(21):7252–9. doi: 10.1158/1078-0432.CCR-04-0713 [DOI] [PubMed] [Google Scholar]
- 40. Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the Population Abundance of Tissue-Infiltrating Immune and Stromal Cell Populations Using Gene Expression. Genome Biol (2016) 17(1):218. doi: 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells With CIBERSORT. Methods Mol Biol (2018) 1711:243–59. doi: 10.1007/978-1-4939-7493-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spranger S, Bao R, Gajewski TF. Melanoma-Intrinsic β-Catenin Signalling Prevents Anti-Tumour Immunity. Nature (2015) 523(7559):231–5. doi: 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 43. McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, et al. Clinical Activity and Molecular Correlates of Response to Atezolizumab Alone or in Combination With Bevacizumab Versus Sunitinib in Renal Cell Carcinoma. Nat Med (2018) 24(6):749–57. doi: 10.1038/s41591-018-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanchez A, Furberg H, Kuo F, Vuong L, Ged Y, Patil S, et al. Transcriptomic Signatures Related to the Obesity Paradox in Patients With Clear Cell Renal Cell Carcinoma: A Cohort Study. Lancet Oncol (2020) 21(2):283–93. doi: 10.1016/S1470-2045(19)30797-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Messina JL, Fenstermacher DA, Eschrich S, Qu X, Berglund AE, Lloyd MC, et al. 12-Chemokine Gene Signature Identifies Lymph Node-Like Structures in Melanoma: Potential for Patient Selection for Immunotherapy. Sci Rep (2012) 2:765. doi: 10.1038/srep00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu C, Jiang A. Dendritic Cells and CD8 T Cell Immunity in Tumor Microenvironment. Front Immunol (2018) 9:3059. doi: 10.3389/fimmu.2018.03059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins T, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell (2017) 171(6):1259–71.e11. doi: 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Curti BD, Faries MB. Recent Advances in the Treatment of Melanoma. N Engl J Med (2021) 384(23):2229–40. doi: 10.1056/NEJMra2034861 [DOI] [PubMed] [Google Scholar]
- 49. Chi Z, Li S, Sheng X, Si L, Cui C, Han M, et al. Clinical Presentation, Histology, and Prognoses of Malignant Melanoma in Ethnic Chinese: A Study of 522 Consecutive Cases. BMC Cancer (2011) 11:85. doi: 10.1186/1471-2407-11-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McLaughlin CC, Wu XC, Jemal A, Martin HJ, Roche LM, Chen VW. Incidence of Noncutaneous Melanomas in the U. S Cancer (2005) 103(5):1000–7. doi: 10.1002/cncr.20866 [DOI] [PubMed] [Google Scholar]
- 51. Zhao F, Evans K, Xiao C, DeVito N, Theivanthiran B, Holtzhausen A, et al. Stromal Fibroblasts Mediate Anti-PD-1 Resistance via MMP-9 and Dictate Tgfβ Inhibitor Sequencing in Melanoma. Cancer Immunol Res (2018) 6(12):1459–71. doi: 10.1158/2326-6066.CIR-18-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye Y, Kuang X, Xie Z, Liang L, Zhang Z, Zhang Y, et al. Small-Molecule MMP2/MMP9 Inhibitor SB-3CT Modulates Tumor Immune Surveillance by Regulating PD-L1. Genome Med (2020) 12(1):83. doi: 10.1186/s13073-020-00780-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and Stabilization of Programmed Death Ligand-1 Suppresses T-Cell Activity. Nat Commun (2016) 7:12632. doi: 10.1038/ncomms12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jacobs JF, Nierkens S, Figdor CG, de Vries IJ, Adema GJ. Regulatory T Cells in Melanoma: The Final Hurdle Towards Effective Immunotherapy. Lancet Oncol (2012) 13(1):e32–42. doi: 10.1016/S1470-2045(11)70155-3 [DOI] [PubMed] [Google Scholar]
- 55. Tang B, Yan X, Sheng X, Si L, Cui C, Kong Y, et al. Safety and Clinical Activity With an Anti-PD-1 Antibody JS001 in Advanced Melanoma or Urologic Cancer Patients. J Hematol Oncol (2019) 12(1):7. doi: 10.1186/s13045-018-0693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of different IRGPI groups. (A) Multivariate Cox analysis of 31 hub immune-related gene pairs. (B) Forest plot of high IRGPI score with poor OS in patients from three datasets.
The performance of the IRGPI in predicting progression-free survival in three cohorts.
Waterfall plot of IRGPI for distinct clinical response groups in VanAllen15 and PUCH cohorts.
X-tile plots of the IRGPI scores in TCGA-SKCM cohort.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/search/?dbId=&q=HRA000524.