Abstract
Fluoroquinolone efflux was studied in 47 Staphylococcus aureus clinical strains with MICs of ciprofloxacin (CFX) of ≤2 μg/ml. Forty-three strains were wild type for gyrA, gyrB, and grlA quinolone resistance-determining regions and for norA and its promoter region. Forty of these strains (MICs of CFX, 0.1 to 0.2 μg/ml) did not show efflux of fluoroquinolones. Three strains (MICs of CFX, 1 to 2 μg/ml) showed efflux. These results suggest that efflux can appear in S. aureus clinical strains in the absence of mutations in norA and its promoter.
Fluorinated quinolones (FQ) appeared in the 1980s as synthetic broad-spectrum antibiotics that were very active mainly against gram-negative bacteria. Mutations in grlA, the gene encoding topoisomerase IV subunit A, seem to be the main factor in Staphylococcus aureus resistance to older FQ, such as ciprofloxacin and ofloxacin (1, 2, 10). Mutations in DNA gyrase (gyrA mainly) usually appear in the presence of previous mutations in grlA, leading to additional increases in MICs (10). Some newer FQ have a different behavior. The main target for some FQ, in gram-positive bacteria such as Streptococcus pneumoniae is DNA gyrase, instead of topoisomerase IV (11). Nevertheless, this characteristic has not been shown for other gram-positive bacteria.
Another mechanism involved in quinolone resistance in S. aureus is overexpression of norA. This gene encodes a multidrug efflux protein (NorA) capable of transporting FQ outside the bacteria (4–6, 16, 17). Overexpression of norA has been related to mutations 89 bp upstream from the putative ATG start codon (4, 9). NorA-mediated resistance has been described in the apparent absence of mutations in topoisomerase genes (6). Moreover, NorA-mediated resistance can appear both in the presence (4, 9) and in the absence of promoter mutations (5). S. aureus strains derived from a single strain (SA-1199) that can overexpress norA in a constitutive or inducible manner have been reported. Inducible strains lack promoter mutations (6).
NorA has been shown to play a role even in quinolone-susceptible strains, since norA disruption leads to MICs eightfold lower than for the parent strain (16). We have studied the presence of mutations in the genes encoding DNA gyrase and topoisomerase IV and in the norA gene and its promoter in FQ-sensitive and borderline strains. When strains with the same genotype had different FQ susceptibilities, efflux activity was studied.
Bacterial strains.
Forty-seven S. aureus clinical strains with ciprofloxacin MICs of ≤2 μg/ml were studied.
Antimicrobial susceptibility.
MICs of ofloxacin, ciprofloxacin, sparfloxacin, and trovafloxacin were determined by the agar dilution method, according to National Committee for Clinical Laboratory Standards guidelines (8). Drugs were obtained from their respective manufacturers as standard powders. MIC determinations were also performed in the presence of 20 μg of reserpine per ml.
PCR procedures.
gyrA (13), grlA (1), and gyrB (3) quinolone resistance-determining regions (QRDRs) and norA (4) and its promoter region were amplified and sequenced (12) according to previously described methods.
Uptake of quinolones.
Uptake and accumulation of fluoroquinolones was determined by the method described by Takenouchi et al. Fluorescence was used as a means of quantifying FQ concentrations (15).
The results of sequencing of the four genes appear in Table 1. Among the 47 strains with ciprofloxacin MICs of ≤2 μg/ml, four strains showed a mutation in grlA leading to a Ser80-to-Ile substitution (group B), and 43 strains were wild type for gyrA, gyrB, grlA, and norA. The four strains with mutations in grlA showed MICs of ciprofloxacin of 1 to 2 μg/ml. Among the 43 wild-type strains, we found two groups of strains. Forty strains showed ciprofloxacin MICs of around 0.1 to 0.2 μg/ml (group A1). Three strains showed MICs similar to grlA mutant strains (1 to 2 μg/ml) (group A2).
TABLE 1.
In vitro activities of four quinolones, alone and combined with reserpine, against the three groups of S. aureus strains studied
| Group | No. of strains | MIC range (μg/ml) of quinolone alone/quinolone plus reserpine (20 μg/ml)
|
|||
|---|---|---|---|---|---|
| Ciprofloxacin | Ofloxacin | Sparfloxacin | Trovafloxacin | ||
| A1 | 40 | 0.1–0.2/0.1–0.2 | 0.1–0.2/0.1–0.2 | 0.01–0.03/0.01–0.03 | ≤0.008/≤0.008 |
| A2 | 3 | 1–2/0.1–0.2 | 1–2/0.1–0.2 | 0.06/0.01–0.03 | 0.03/≤0.008 |
| B | 4 | 1–2/1–2 | 1–2/1–2 | 0.1/0.1 | 0.03/0.03 |
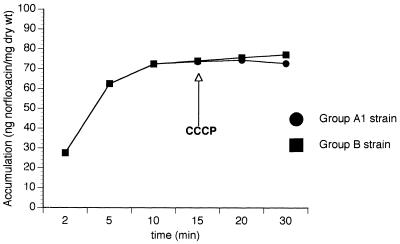
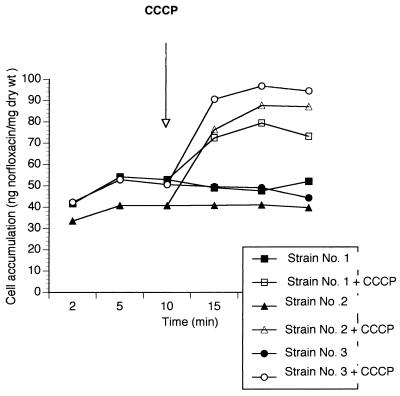
The study of FQ uptake by the wild-type strains in group A1 and the three grlA mutant strains (group B) led to results similar to the uptake curve appearing in Fig. 1 (which corresponds to one of the strains). Carbonyl cyanide m-chlorophenylhydrazone (CCCP) did not modify their behavior. When FQ uptake was studied in the three wild-type strains with higher ciprofloxacin MICs (group A2), we observed the behavior shown in Fig. 2. FQ uptake increased in the presence of CCCP.
FIG. 1.
Example of norfloxacin accumulation in strains of groups A1 and B.
FIG. 2.
Norfloxacin accumulation in strains of group A2.
NorA-mediated quinolone efflux has been extensively studied in S. aureus. Previous studies have shown that norA overexpression can lead to FQ resistance, both in the presence and in the absence of topoisomerase alterations (4, 5). Quinolone resistance due to norA overexpression was first related to mutations in the norA promoter region (4, 9). Previous studies suggested that the thymine-to-adenine mutation in the norA promoter region might be responsible for increased norA transcription (4, 9), but norA overexpression happens independently of this mutation (5), suggesting that norA regulation can be located elsewhere in the chromosome (6). Recent studies suggest that this mutation might be necessary for constitutively increased norA expression, but it is not necessary when overproduction is inducible (6). Efflux-pump mediated quinolone-resistance has not been described in gyrA or grlA wild-type clinical strains. Our results show that efflux-mediated resistance might be not so infrequent in this kind of strain. The three S. aureus strains in group A2 were wild type for norA and its promoter region. Results obtained in this study suggest that resistance in these strains is directly related to efflux. We did not find any mutation in gyrA, gyrB, and grlA QRDRs and norA and its promoter region. grlB mutations have been described in S. aureus only following gyrA and grlA mutations (14). Moreover, MICs of ciprofloxacin decreased by eightfold in the three strains when ciprofloxacin was combined with reserpine (Table 1). One possible explanation is that mutations in areas other than the norA promoter region might be involved in constitutive norA overexpression. Nevertheless, since norA expression has not been determined, we cannot know certainly the origin of the efflux system, and the possibility of efflux pumps other than NorA cannot be disregarded. In other species, such as Pseudomonas aeruginosa, the presence of several efflux systems able to efflux quinolones with different levels of efficacy has been described (7).
In the three strains in group A2, efflux was associated with MICs similar to those caused by grlA mutations in the strains in group B. Changes were not the same for all of the quinolones tested (Table 1). Ciprofloxacin and ofloxacin are hydrophilic quinolones, and MICs of these compounds ranged from 0.1 to 0.2 μg/ml in wild-type strains without efflux activity. MICs of these FQ against strains with efflux activity ranged between 1 and 2 μg/ml and reverted to 0.1 to 0.2 μg/ml when tested in presence of reserpine. MICs of sparfloxacin, a more hydrophobic compound, were less affected. MICs ranged between 0.01 and 0.03 μg/ml for strains without efflux activity, and MICs for the three strains with efflux activity were 0.06 μg/ml. MICs for trovafloxacin remained very low for all strains. MICs for strains with efflux activity were 0.03 μg/ml, while MICs for strains without efflux activity were ≤0.008 μg/ml (four- to eightfold lower).
These results show that efflux can appear in clinical strains, even in the absence of mutations in the genes usually involved in quinolone resistance, particularly in strains with to MICs at or above the breakpoint. The mechanisms controlling this efflux, and the possibility that these strains might even increase their efflux activity in an inducible way, are now being studied.
Acknowledgments
This study was supported in part by a grant from the FIS (Ministry of Health, Spain) (96/18).
REFERENCES
- 1.Ferrero L, Cameron B, Manse B, Lagneaux D, Creuzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 2.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2016–2023. doi: 10.1128/aac.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaatz G W, Seo S M, Ruble C R. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaatz G W, Seo S M. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:2650–2655. doi: 10.1128/aac.39.12.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaatz G W, Seo S M. Mechanisms of fluoroquinolone resistance in genetically related strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2733–2737. doi: 10.1128/aac.41.12.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köhler T, Michea-Hamzepour M, Plesiat P, Kahr A L, Pechere J C. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. Approved standard M7-A3. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 9.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan X S, Fisher L H. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sreedharan S, Oram M, Jensen B, Peterson L R, Fisher L M. DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J Bacteriol. 1990;172:7260–7262. doi: 10.1128/jb.172.12.7260-7262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi H, Kikuchi T, Shoji S, Fujimura S, Lutfor A B, Tokue Y, Nukiwa T, Watanabe A. Characterization of gyrA, gyrB, grlA and grlB mutations in fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:49–58. doi: 10.1093/jac/41.1.49. [DOI] [PubMed] [Google Scholar]
- 15.Takenouchi T, Tabata F, Iwata Y, Hanzawa H, Sugawara M, Ohya S. Hydrophilicity of quinolones is not an exclusive factor for decreased activity in efflux-mediated resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1835–1842. doi: 10.1128/aac.40.8.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada H, Kurose-Hamada S, Fukuda Y, Mitsuyama J, Takahata M, Minami S, Watanabe Y, Narita H. Quinolone susceptibility of norA-disrupted Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2308–2309. doi: 10.1128/aac.41.10.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Komo N. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]