Abstract
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype. TNBC progression is sustained by recruitment of a strong tumor microenvironment (TME) mainly composed of cancer-associated fibroblasts (CAFs) able to endorse tumor hallmarks. Increasing evidences demonstrate that exosomes mediate the crosstalk between cancer cells and the TME. We examined TNBC-derived exosomes and their microRNA (miRNA) cargo in activation of normal fibroblasts (NFs) toward CAFs. We demonstrated that TNBC cell-derived exosomes increased NF collagen contraction and migration alongside CAF molecular markers. Exosome-activated fibroblasts promoted the invasion potential of normal breast epithelial cells, as assessed by an organotypic co-culture assay that resembled the in vivo context. We also investigated TNBC cell-derived exosome cargo in activating NFs to CAFs by performing small RNA sequencing. We found that the synergistic action of miR-185-5p, miR-652-5p, and miR-1246 boosted fibroblast migration and contraction, promoting specific CAF subspecialization toward a pro-migratory functional state. These data highlight the role of breast cancer cells in re-education of the TME and their contribution to tumor evolution.
Keywords: CAF, exosomes, miRNA, TME, TNBC, cancer, breast cancer, BC, fibroblast, cross-talk
Graphical Abstract

Scognamiglio et al. study the mechanisms involved in crosstalk between breast cancer cells and stromal fibroblasts in the tumor microenvironment and identify microRNAs from breast cancer cell-derived exosomes able to synergistically activate stromal fibroblasts toward a cancer-associated, fibroblasts-related, pro-migratory phenotype.
Introduction
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype, and there is no molecular targeted therapy available.1 TNBC is characterized by high intra-tumoral heterogeneity and plasticity, which dictate multiple malignant signs, including treatment resistance and metastasis. Rapid spread of metastases remains the main obstacle for a positive clinical outcome.2 This aggressive spread is encouraged by the tumor microenvironment (TME), which actively participates in all stages of tumor progression and is now recognized as a cancer hallmark.3,4 In breast cancer, the TME is composed mainly of cancer-associated fibroblasts (CAFs), which are generally derived from the surrounding stroma and can be recruited by cancer cells to endorse different tumor functions, in particular invasion and metastasis.5, 6, 7, 8
Research on CAFs has grown in recent years and has been focused in particular on the distinct subspecializations CAFs acquire during cancer.9 CAFs are the most effective cells in the TME in extracellular matrix (ECM) remodeling because of their capacity for enzyme-mediated ECM degradation and force-dependent ECM reshuffling.10, 11, 12, 13 In this scenario, extracellular vesicles, such as exosomes, have been found to be important mediators, particularly in the crosstalk between cancer and the TME through transfer of biological molecules, such as proteins, lipids, mRNAs, and microRNAs (miRNAs or miRs), that sustain cancer development and progression.14, 15, 16, 17, 18
Dysregulation of miRNAs in cancer has been shown extensively to influence cell proliferation, metastasis, angiogenesis, stem phenotype, and resistance to therapies. Therefore, miRNA profiling has been employed for cancer diagnosis, prognosis, and drug response prediction in affected individuals.19, 20, 21
For example, the tumor suppressor miR-34 family is recognized as one of the most significant in cancer and is investigated in numerous pre-clinical and clinical studies.22, 23, 24, 25, 26, 27, 28, 29 A miR-34a mimic-based drug was employed in the MRX34 clinical trial (NCT01829971), which was the first-in-human phase 1 study of miRNA-based cancer therapy.30 Even though the clinical trial terminated prematurely, it was the first concrete application of miRNAs in the clinic and a solid proof of concept for miRNA-based cancer therapy.
MiRNAs can be loaded selectively into the exosomes released by cancer cells and directed to their extracellular target compartments, contributing to regulation of different tumor processes, including recruitment of cellular components from the TME.31,32
Here we investigated whether TNBC-derived exosomes and their miRNA cargo may affect conversion of fibroblasts into a CAF-like phenotype in the TME. Our results show that exosomal miR-185-5p, miR-652-5p, and miR-1246 synergistically activate stromal fibroblasts by promoting a specific pro-migratory functional state that could serve for TNBC cell migration and invasion.
Results
Breast cancer-derived exosomes are transferred to normal fibroblasts
To study whether the crosstalk between breast cancer cells and surrounding fibroblasts is also mediated by exosomes, we isolated exosomes from MDA-MB-231 cells through a polymeric precipitation method (ExoQuickTC, System Biosciences [SBI]). The tumoral exosomes were then characterized by Nanoparticle Tracking Analysis (NTA), transmission electron microscopy (TEM), and western blot analysis.
NTA, performed with the Nanosight NS300 system equipped with a blue laser (405 nm), revealed that there was a major particle size peak at approximately 105 nm, corresponding to the mean diameter of exosomes (Figure 1A). Western blotting revealed expression of the typical exosomal protein markers CD63, CD81, CD9, and Tsg101 and absence of the endoplasmic reticular protein Calnexin, indicating no cellular contamination (Figure 1B). Then the morphology of exosomes was analyzed by TEM (Figure 1C). These data confirmed the exosomal origin of isolated vesicles.
Figure 1.
. MDA-MB-231 cell-derived exosomes: characterization and fibroblast uptake
(A) Results from the NTA performed on MDA-MB-231 cell-derived exosomes. The peak (at 105 nm) indicates the mean ratio of vesicle size and concentration derived from three measurements. (B) Western blot showing expression of exosome-specific markers (CD63, CD81, CD9, and Tsg101) in MDA-MB-231 cells and exosomes and the absence of the endoplasmic reticulum (ER) protein Calnexin. (C) Representative TEM images of exosomes from MDA-MB-231 cells (scale bar, 500 nm). A yellow square indicates a larger magnification (scale bar, 100 nm). (D) Immunofluorescence assay performed on NFs (pt. #1) incubated with MDA-MB-231 cell-derived exosomes labeled with PKH26 dye. The images from confocal microscopy show co-localization of the red signal derived from PKH26-labeled exosomes and the green signal from FITC-conjugated anti-β-actin (merged), indicative of exosome uptake by NFs. Magnification, 63×.
To determine whether there was a transfer of breast cancer-derived exosomes to normal fibroblasts, we took advantage of primary stromal fibroblasts derived from surgical resection of normal breast tissues and/or fibroadenomas. Because of the benign origin of fibroadenoma, we chose to define fibroblasts isolated from this tumor by convention as normal fibroblasts (NFs). Primary fibroblasts were derived from three female individuals (pt. #1, #2, and #3) between 21 and 44 years of age and presenting with various clinicopathological features, including two fibroadenomas (pt. #1 and #2) and one mammary reduction (pt. #3) without any pathological signs. PKH26-labeled MDA-MB-231 cell-derived exosomes were then incubated for 12 h with NFs (pt. #1) and subjected to immunofluorescence analysis. We observed that PKH26-labeled exosomes were distinctly taken up by NFs. Confocal microscopy indicated co-localization of PKH26 derived from exosomes and β-actin from NFs (Figure 1D), confirming that NFs actively took up breast cancer-derived exosomes.
MDA-MB-231 cell-derived exosomes promote a CAF-like phenotype in NFs
Cell contractility and motility are regarded as major hallmarks of activated CAFs in the TME to foster cancer cell invasion.33, 34, 35 Therefore, we assessed the effect of breast cancer-derived exosomes on fibroblast-mediated collagen contraction. NFs (pt. #1, #2, and #3) were plated in a type 1 collagen matrix and incubated with MDA-MB-231 cell-derived exosomes or PBS (control) to examine their contraction ability. After 24 h, we observed that NFs cultured in the presence of exosomes reduced the collagen plug area compared with control NFs, indicating increased cell contractility mediated by exosomes (Figures 2A and 2B).
Figure 2.
. MDA-MB-231 cell-derived exosomes promote a CAF-like phenotype in NFs
(A) Collagen contraction assay. Shown are representative pictures of collagen plugs containing NFs cultured with MDA-MB-231 cell-derived exosomes (+ exosomes) or PBS (no treatment [NT]) after 24 h. (B) Histogram of mean collagen plug areas for NFs + exosomes over NT, measured with ImageJ. Standard deviations were calculated on replicates from two independent experiments performed with three different NF cells (pt. #1, #2, and #3). The p value was calculated using two-tailed unpaired t test; ∗∗p = 0.0028. (C) Migration assay performed with NFs (pt. #1, #2, and #3) cultured in the presence of MDA-MB-231 cell-derived exosomes for 24h (+exosomes) or PBS (NT). Shown are representative bright-field images of NFs migrated through the Transwell chamber and colored with crystal violet. Bars indicate size expressed in micrometers (pt. #2 and #3) or pixels (pt. #1). Magnification, 5×. (D) Histogram of mean absorbance of crystal violet eluted from NFs + exosomes over NT. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cells (pt. #1, #2, and #3). The p value was calculated using two-tailed unpaired t test; ∗p = 0.039. (E) Histogram of quantitative real-time PCR showing FAP, Caveolin-1, SLC16A3, and SLC2A1 mRNA relative expression in NFs + exosomes over NT. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cells (pt. #1, #2, and #3). The p values were calculated using multiple t test with false discovery rate (FDR) adjustment (FAP and SLC2A1, ∗∗p = 0.0014; SLC16A3, ∗∗∗p = 0.00064; Caveolin-1, ∗∗∗p = 0.00014).
To verify whether breast cancer-derived exosomes affected the migration potential of fibroblasts, we pre-treated NFs (pt. #1, #2, and #3) with MDA-MB-231 cell-derived exosomes or PBS for 48 h and then performed a Transwell migration assay. We found improved migration when NFs were cultured in the presence of exosomes (Figures 2C and 2D). Then, to assess the progression of NFs toward a CAF-like phenotype, the expression of molecular markers usually associated with the CAF phenotype was examined. We performed quantitative real-time PCR to measure the levels of FAP, Caveolin-1, SLC16A3, and SLC2A1 mRNAs in NFs cultured with MDA-MB-231 cell-derived exosomes for 48h. The presence of exosomes was associated with upregulation of these genes, supporting the notion of fibroblasts being converted to a CAF-like phenotype (Figure 2E).
Exosome-activated fibroblasts induce invasion in normal breast epithelial cells
To investigate the effects of fibroblast activation in the TME, we carried out three-dimensional cell-cell interaction modeling by setting up an in vitro invasion organotypic assay, as reported previously.36 NFs (pt. #3) were seeded on a type 1 collagen matrix and then cultured in the presence of MDA-MB-231 cell-derived exosomes or PBS for 48 h to ensure fibroblast activation. Subsequently, normal breast epithelial cells (cell line MCF10A) were seeded on top of the fibroblast-containing collagen matrix for 14 days, which was transferred to an invasion grid and finally paraffin embedded for analysis. Activation of NFs induced by the presence of exosomes increased the number of MCF10A cells invading the matrix, as demonstrated by the positive pan-cytokeratin signal from cells stained in the collagen section (Figures 3A and 3B). These data suggest that MDA-MB-231-derived exosomes have a pro-tumor stromal function triggering NFs to a CAF-like phenotype. Our organotypic assay outlined how the effect of cancer-derived exosomes on fibroblasts contributes to malignant transformation promoting the invasion capacity of non-tumorigenic breast epithelial cells.
Figure 3.
3D Organotypic invasion assay of normal breast epithelial cells in a matrix containing exosome-activated fibroblasts
(A) Left panel: representative phase-contrast images of a collagen matrix-embedded section (10-μm slides) showing MCF10A cell invasion through the matrix when three-dimensionally co-cultured with NFs (pt. #3) incubated with MDA-MB-231-derived exosomes (+ exosomes) or PBS (NT). Right panel: confocal microscopy images of collagen matrix sections showing MCF10A cells stained with DAPI for nuclei (blue signal) and Alexa 594-conjugated anti-pan-cytokeratin antibody (red signal) as an epithelial marker. (B) Histogram of the number of invading MCF10A cells (NFs + exosomes and NFs_NT), calculated for different fields of confocal microscopy images. Standard deviations were calculated on technical replicates. The p value was calculated using two-tailed unpaired t test; ∗∗∗∗p < 0.0001.
miRNA expression profile of breast cancer exosomes
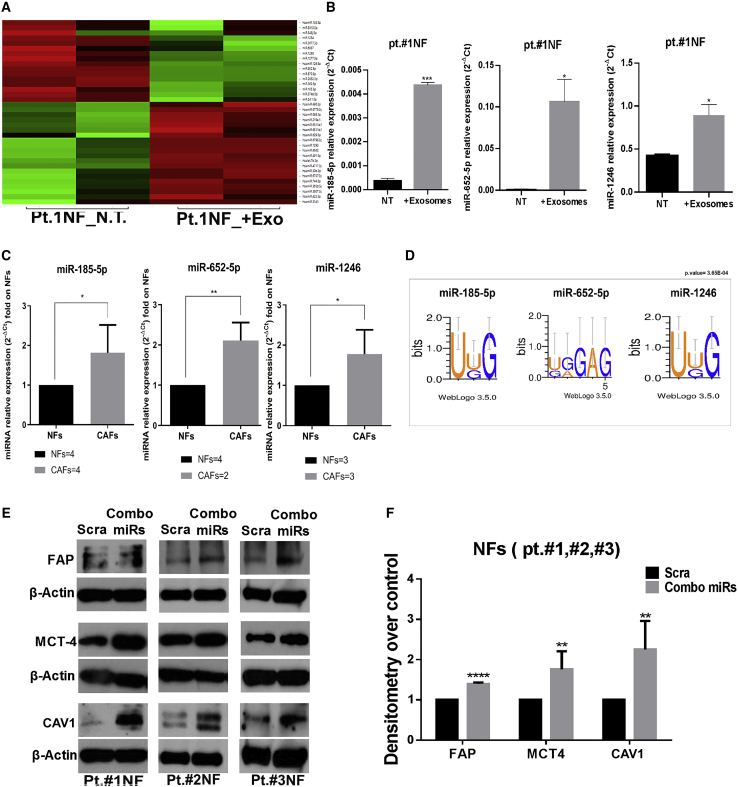
To investigate whether the breast cancer exosomal cargo was involved in fibroblast activation in the TME, we performed small RNA sequencing to identify miRNAs differentially expressed in NFs cultured with MDA-MB-231 cell-derived exosomes or PBS for 24 h. We found that 14 miRNAs were upregulated significantly in NFs (pt. #1) when exposed to exosomes (Figure 4A). We then focused our attention on miR-185-5p, miR-652-5p, and miR-1246 because they have been demonstrated to be upregulated in NFs upon MDA-MB-231-derived exosome incubation (Figure 4B). The basal expression levels of these three miRNAs were higher in CAFs compared with NFs (Figure 4C), indicating that they are involved in fibroblast activation. We found that miR-185-5p, miR-652-5p, and miR-1246 were predicted to be actively sorted into exosomes by the Motif Discovery on Short Nucleotide Sequences (MDS2) bioinformatic tool (http://cse-jcui-08.unl.edu:7000/input), as described previously.37 We found short-sequence motifs (EXOmotifs) in the three miRNAs that predicted their loading into exosomes (Figure 4D). Based on the prediction results, we investigated whether the three miRNAs were loaded into exosomes by adopting an overexpression model. We transfected miR-185-5p, miR-652-5p, miR-1246, and a scramble sequence (as a control) in MDA-MB-231 cells for 48 h. Then we isolated exosomes and performed qRT-PCR to check the expression levels of the three miRNAs in cells and in the relative exosomes. We found that consistent portions of miR-185-5p, miR-652-5p, and miR-1246 were overexpressed in MDA-MB-231-derived exosomes (Figure S1), indicating that they were loaded into exosomes. These findings directed us to study these three miRNAs among the other upregulated miRNAs identified by small RNA sequencing. We then investigated whether the miRNAs shuttled via breast cancer-derived exosomes trigger NF conversion to CAFs. We found that transfection of the individual miRNAs had no significant biological effects on fibroblast activation, as shown by western blotting for FAP, MCT4, and Caveolin-1, known markers associated with the CAF phenotype (Figures S2A and S2B). Therefore, we hypothesized that miR-185-5p, miR-652-5p, and miR-1246 might work synergistically, as already reported for miR-185-5p.38 We found that combined transfection of the three miRNAs (combo miRs) was associated with upregulated CAF markers (FAP, MCT4, and Caveolin-1) in NFs derived from all three individuals (Figures 4E and 4F).
Figure 4.
Differentially expressed miRNAs in fibroblasts upon exposure to MDA-MB-231 cell-derived exosomes
(A) Heatmap of data from small RNA sequencing analysis, showing miRNAs differentially expressed in NFs (pt. #1) incubated with MDA-MB-231 cell-derived exosomes (+exosomes) and PBS (NT). Upregulated miRNAs are represented in green and downregulated miRNAs in red. The results are based on technical duplicates (p < 0.05). (B) Histograms of qRT-PCR results showing the expression levels of miR-185-5p, miR-652-5p, and miR-1246 in NFs (pt. #1) incubated with exosomes compared with NT. Data are shown as relative expression of the miRNAs. Standard deviations were calculated on technical duplicates. The p values were calculated using unpaired t test (∗p = 0.0317; ∗∗∗p = 0.0008). (C) Histograms of qRT-PCR results showing the basal expression levels of miR-185-5p, miR-652-5p, and miR-1246 in NFs compared with CAFs. Data are shown as relative expression of miRNAs folded on NFs. Standard deviations were calculated with replicates from two independent experiments performed on four different primary NF cells (pt. #1, #2, #3, and #4) and four different primary CAF cells (pt. #81, #82, #87, and #89). The p values were calculated using unpaired t test (∗p = 0.0210; ∗∗p = 0.0027). (D) Representative images of the EXOmotif analysis with MDS2 software showing a short motif (3–5 nt) in miR-185-5p, miR-652-5p, and miR-1246 sequences, predictive of their active loading into exosomes. (E) Western blot showing overexpression of FAP, Caveolin-1, and MCT4 proteins in NFs (pt. #1, #2, and #3) transfected with combo miRs compared with control (scrambled [Scra]) after 72 h. (F) Histogram of densitometric measurement of bands, obtained with ImageJ. Quantification of protein expression is represented as the mean of folded densitometry from NFs transfected with combo miRs over Scra. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cells (pt. #1, #2, and #3). The p values were calculated using multiple t test with FDR adjustment (FAP, ∗∗∗∗p < 0.0001; MCT4, ∗∗p = 0.0019; Caveolin-1, ∗∗p = 0.0016).
Exosomal miR-185-5p, miR-652-5p, and miR-1246 synergistically activate fibroblasts toward a pro-migratory functional phenotype
The main ability acquired by CAFs in the TME is ECM remodeling, a process promoting permissive tracks for cancer cell invasion.34 Our results so far demonstrated that the combo miRs mediated activation of NFs by primarily boosting their motility in the ECM. To explore this, we set up a collagen contraction assay, plating NFs pre-transfected with combo miRs or a scrambled control onto type 1 collagen matrices. We found that the synergistic action of the three miRNAs promoted fibroblast collagen contraction, as shown by reduction of the collagen plug area in combo miR-transfected NFs (Figures 5A and 5B).
Figure 5.
Combo miRs promote fibroblast-mediated ECM remodeling
(A) Representative pictures of collagen plugs containing NFs transfected with combo miRs or Scra taken after 24 h. (B) Histogram of mean collagen plug area for NFs transfected with combo miRs, fold over Scra at 24 h (ImageJ). Standard deviations were calculated on replicates from two independent experiments performed with three different NF cells (pt. #1, #2, and #3). The p value was calculated using two-tailed unpaired t test; ∗∗p = 0.0011. (C) Transwell invasion assay with NFs (pt. #2 and #3) transfected with combo miRs or Scra (control). Shown are representative bright-field images of NFs that invaded the matrix and migrated through the Transwell chamber, fixed and colored with crystal violet. Bars indicate size, expressed in micrometers or pixels. Magnification, 5×. (D) Histogram of mean absorbance values for crystal violet eluted from NFs transfected with combo miRs folded on Scra. Standard deviations were calculated on replicates from two independent experiments performed on two different NF cells (pt. #2 and #3). The p value was calculated using two-tailed unpaired t test; ∗∗p = 0.0018.
(E) Western blot showing overexpression of MMP1, MMP2, and MMP3 proteins together with ITGB1 and ITGA5 in NFs (pt. #1, #2, and #3) transfected with combo miRs and Scra after 48 and 72 h. (F) Histograms of mean densitometric measurement of bands (ImageJ) for NFs transfected with combo miRs over Scra. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cells (pt. #1, #2, and #3). The p values were calculated using multiple t test with FDR adjustment (MMP1, ∗p = 0.023; MMP2, ∗∗∗p = 0.00087; MMP3, ∗∗p = 0.0035; ITGB1, ∗∗∗p = 0.00014; ITGA5, ∗∗p = 0.0054).
We then investigated the invasion potential of fibroblasts, specifically their ability to move within the ECM as a result of its degradation. We transfected NFs (pt. #1, #2, and #3) with combo miRs and performed a Transwell invasion assay, seeding the cells in a Matrigel solution plated on top of the migration chamber, simulating the three-dimensional ECM. We observed increased invasion by NFs transfected with combo miRs, as demonstrated by the higher absorbance values of crystal violet eluted from migrated cells (Figures 5C and 5D).
Considering that ECM remodeling depends on force-mediated and protease-dependent mechanisms,34 we investigated integrin (ITG) and matrix metalloprotease (MMP) expression in NFs after combo miR overexpression. We conducted western blot analysis for expression of ITGB1 and ITGA5 proteins, major players in mechanical force-mediated cell invasion and ECM modeling, especially during CAF transformation.39, 40, 41, 42 We found that ITGB1 and ITGA5 were increased in NFs transfected with combo miRs after 72 h (Figures 5E and 5F), suggesting that there was a higher driving force exerted by fibroblasts to contract the collagen matrix. However, the protease activity involved in matrix degradation is arbitrated by different members of the MMP protein family. In particular, MMP1, MMP2, MMP3, and MMP9 are mainly upregulated in breast cancer stroma under the influence of cancer cells.43, 44, 45 We found that, after 48 h of transfection, combo miRs led to increased levels of MMP1, MMP2, and MMP3, justifying the boost in fibroblast invasion potential (Figures 5E and 5F).
We found that combo miRs promoted a pro-migratory phenotype in NFs, as demonstrated by Transwell migration and scratch assays (Figures 6A–6D). To corroborate these results, we investigated expression of FAK, the main regulator of focal adhesion turnover responsible for cell movement.46 We found upregulation of the activated form of FAK protein (phosphorylated at Y576/577) using western blot analysis (Figures 6E and 6F) in NFs (pt. #1, #2, and #3) transfected with combo miRs, indicating activation of the molecular pathway involved in cell migration. These data demonstrated that miR-185-5p, miR-652-5p, and miR-1246 synergistically foster a pro-migratory functional state in NFs in the TME.
Figure 6.
Combo miRs promote migration of NFs
(A) Wound healing assay performed with NFs transfected with combo miRs or Scra. Shown are representative images of NFs in bright fields 0 and 24 h from scratch. Bars indicate size, expressed in pixels. (B) Histogram of mean scratch area at 24 h, normalized on 0 h, of NFs transfected with combo miRs over Scra. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cells (pt. #1, #2, and #3). The p value was calculated using two-tailed unpaired t test; ∗∗p = 0.0061. (C) Transwell migration assay performed with NFs (pt. #1, #2, and #3) transfected with combo miRs and Scra (control). Shown are representative images of NFs in bright fields migrating through the Transwell chamber and colored with crystal violet. Bars on the images indicate size, expressed in micrometers (pt. #1 and #2) or pixels (pt. #3). (D) Histogram of mean absorbance values of crystal violet eluted from migrated cells. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cells (pt. #1, #2, and #3). The p value was calculated using two-tailed unpaired t test; ∗∗p = 0.0049. (E) Western blot showing overexpression of phosphorylated FAK (Y576/577) protein in NFs transfected with combo miRs compared with Scra after 72 h. (F) Histogram of mean densitometric measurement of bands (ImageJ) for NFs transfected with combo miRs over Scra. Standard deviations were calculated on replicates from two independent experiments performed on three different NF cell lines (pt. #1, #2, and #3). The p value was calculated using two-tailed unpaired t test; ∗p = 0.029.
Discussion
The data presented here shed light on the role of exosomes in the crosstalk between the TME and breast cancer cells. Our results strengthen knowledge of the mechanisms adopted by breast cancer cells to potentiate the oncogenic phenotype of neighboring cells in the TME. We demonstrate that TNBC-derived exosomes and the miRNA cargo they hold activate stromal fibroblasts to obtain a specific pro-migratory functional phenotype, potentially enabling tumor invasion and metastasis.
It is known that fibroblasts are recruited by cancer cells in the TME to support different tumor traits.7,47 However, the major obstacle when studying CAFs is high functional heterogeneity and lack of specific molecular markers defining their status, even considering occasional antitumorigenic roles.9 However, some researchers have tried to associate precise gene signatures with CAF subtypes.48 In our case, fibroblast activation mediated by cancer cell exosomal miRNAs was associated with upregulation of MCT4, FAP, and Caveolin-1, among other canonical CAF markers. The role of Caveolin-1 in CAF activation is controversial. In fact, some studies have demonstrated that loss of Caveolin-1 expression is a trait associated with CAF transformation.49, 50, 51 In contrast, our data are in line with those of Goetz et al.13 showing that the presence of Caveolin-1-enriched CAFs correlates with tumor invasion and metastasis by promoting biomechanical remodeling of tumoral stroma.
In the context of the breast cancer TME, intracellular and extracellular miRNAs have been reported widely to mediate the crosstalk between CAFs and cancer cells.52, 53, 54, 55, 56, 57, 58 In this study, we provide evidence supporting the notion that breast cancer cell exosomal miRNAs induce a CAF-related pro-migratory phenotype rather than a proliferative pro-survival one. No significant effect on activation of proliferative and survival pathways mediated by combo miRs was observed (Figures S3A and S3B), as shown by cell viability assay and western blotting for phosphorylated AKT serine/threonine kinase (S473) and phosphorylated β-catenin (S33/37/T41), known markers of cell proliferation, self-renewal, and survival in cancer.59, 60, 61, 62 In contrast, delivery to fibroblasts of the breast-cancer-cell exosomal combo miRs induced the former to increase migration, contraction, and invasion, characteristics acquired by specific CAF subtypes. Otherwise, incubation of NFs with MCF10A-derived exosomes, which presented lower levels of two of the three miRNAs, showed no effect on fibroblast migration, indicating the presence of a specific mechanism related to the combined exosomal miRNA function (Figures S4A and S4B). Overexpression in fibroblasts of combo miRs induced expression of MMPs (types 1, 2, and 3) and ITGs (α5 and β1 subunits) and increased FAK phosphorylation (Y576/577), components of major pathways involved in cancer cell invasion and motility, because they operate in protease-dependent ECM remodeling and cellular movement.40,54,63
Although breast cancer prognosis has improved with development of molecular targeted therapies, treatment of TNBC is still a challenge because of its highly invasive nature and relatively low response rate. These adverse clinicopathological aspects are often caused by CAFs populating the activated tumoral stroma and the exosomal cargo shed by cancer cells into the surrounding TME.64, 65, 66 Thus, discovering new molecular targets for TNBC prognosis and drug response prediction has become fundamental. Regarding this point, our study shows that three exosomal miRNAs (miR-185-5p, miR-652-5p, and miR-1246) act synergistically to promote fibroblast transformation in the context of TNBC.
In individuals with severe active alopecia, miR-185-5p has already been reported to work in synergy with other miRNAs rather than alone,38 a finding coherent with our initial hypothesis. Upregulation of this miRNA has been observed specifically in lymph nodes with metastases from breast cancer; similarly, it has emerged as a prognostic factor of radiation-related toxicity in the serum of individuals with oropharyngeal cancer67 and as a predictive biomarker of chemotherapy response and metastasis formation in colorectal and gastric cancer.68,69
miR-652-5p has been reported to be upregulated in breast malignancies. However, decreased expression of this miRNA is correlated with esophageal carcinoma progression and recurrence.70,71 This difference could be explained by the fact that miRNAs can operate in different ways depending on the biological system in which they are acting.
miR-1246 is a well-known master regulator in cancer. In particular, its upregulation has been associated with tumor growth, metastasis, and drug resistance in different types of cancer.72, 73, 74 In breast cancer, exosomal miR-1246 has been used as diagnostic biomarker because of its high expression specificity.
Given the considerable role of these miRNAs in cancer, the novelty of our study lies in the combined effect exerted by these exosomal miRNAs on fibroblast activation in the TME. This could reflect the existence of a specific miRNA profile in the tumor cells’ exosomal cargos, with a well-defined scope of action.75
Our results demonstrate that exosomes encourage breast cancer development by delivering specific miRNAs that stimulate formation of a singular and aggressive TME. These findings may aid development of novel, alternative strategies for TNBC theragnostics. Better comprehension of the mechanisms underlying the behavior of CAFs in the context of a tumor may help to adapt them for specific clinical benefits.
Materials and methods
Primary and continuous cells cultures
Primary cultures of fibroblasts (NFs) were obtained from individuals undergoing breast reduction surgery at Mediterranea Cardiocentro (Naples, Italy). Informed consent was obtained before sample collection. This study was conducted according to the criteria set by the declaration of Helsinki and approved by the Research Ethics Committees of the University of Naples Federico II (119/15ES1) and A.S.L. Napoli 1 (247/C.E.-20/2021Oss). Briefly, human breast specimens were cut by mechanical fragmentation with sterile blades and tongs. The ECM was digested with collagenase (Sigma-Aldrich, Code(Cod.) C5138) for 2 h under continuous agitation (200 rpm) at 37°C. Then the cellular suspension was centrifuged to separate epithelial cells from the fibroblast population (200 rpm for 2 min to obtain a pellet of epithelial cells and 1,300 rpm for 5 min to obtain the fibroblast population). Fibroblasts were grown in Dulbecco’s modified Eagle’s medium/nutrient F12-Ham (DMEM-F12; Sigma-Aldrich, Cod. D8437, lot RNBG9065) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, Cod. F7524, lot BCBW0228), 1% penicillin/streptomycin (A/A; Gibco, Cod. 15240-062, lot 2321085), and 1% amphotericin B (Gibco, Cod. 15290-026, lot 2244434) at 37°C with 5% CO2. The breast cancer continuous cell line MDA-MB-231 (ATCC) was grown in RPMI 1640 medium (Sigma-Aldrich, Cod. R8758, lot RNBF0094) supplemented with 10% heat-inactivated FBS, 1% A/A. The normal breast epithelial cell line MCF-10A (ATCC) was cultured in DMEM-F12 supplemented with 5% heat-inactivated fetal horse serum, 1% A/A, 1% amphotericin B, and all hormones and factors needed for their growth: epidermal growth factor (EGF; 1 μg/μL), hydrocortisone (1 μg/μL), cholera toxin (100 μg/μL), and insulin (20 μg/μL). All media and supplements were from Sigma-Aldrich (Milan, Italy).
Exosome isolation
Exosomes were isolated from cell culture media of MDA-MB-231 cells. 4 × 106 cells were plated in 150 × 25 mm cell culture dishes (Corning, 430599) with their usual growth medium (described in the previous section) to allow plate attachment. After 24 h, cells were washed twice with PBS (Dulbecco’s PBS, Sigma-Aldrich, Cod. D8537, lot RNBH3372), and 12 mL of RPMI medium (Sigma-Aldrich) supplemented with 10% FBS depleted of exosomes (Exo-FBS, SBI, Cod. EXO-FBS-250A-1, lot 161004-002), 1% A/A, and 1% amphotericin B was added. After 48 h, culture media were collected and centrifuged at 3,000 × g for 15 min at room temperature (RT) to remove cellular debris. The supernatants were transferred into new sterile tubes, and the appropriate volume of the ExoQuick-TC exosome isolation reagent (SBI, Cod. EXOTC50-A) was added according to the manufacturer’s instructions. Then the tubes were gently mixed until the separation between the two phases was no longer visible. The tubes were kept standing at 4°C overnight (O/N). The following day, the tubes were centrifuged first at 1,500 × g for 30 min and then at 1,500 × g for 5 min to ensure complete removal of the ExoQuick-TC solution. Last, exosome pellets were resuspended in 300 μL of PBS solution.
NTA
Exosome size and particle number were analyzed using the NS300 nanoparticle characterization system (NanoSight; Alfatest, Rome, Italy) equipped with a blue laser (405 nm). In brief, 40 μL of exosome isolation (see “Exosome Isolation”) was diluted with PBS to a final volume of 400 μL and loaded into the instrument. For the measurement, the instrument’s software (NTA 3.1 Build 3.1.54) was used. The capture settings were as follows: camera type, scientific CMOS camera (sCMOS); camera level, 15; slider shutter, 1,206; slider gain, 366; Frame Per Second (FPS), 25.0; number of frames, 1,498; temperature, 24.6°C–24.7°C; viscosity (water), 0.895–0.897 centipoises (cP); dilution factor, 2 × 10 × 102; syringe pump speed, 20. The analysis settings were as follows: detection threshold, 5; blur size, auto; max jump distance, auto (12.3–12.9 pixels). Sample measurement was performed in triplicate.
TEM
TEM imaging was carried out at the Department of Radiology of Leiden University Medical Center (Leiden, the Netherlands). Carbon-coated grids (formvar/carbon on 200-mesh copper; AGS162; Van Loenen Instruments, Zaandam, the Netherlands) were glow discharged for 1 min at 2 × 10−1 mbar and 20 mA using the Emitech K950X Turbo Evaporator (Quorum Technologies, Ashford, UK). Then 3 μL of sample solution was transferred to the glow-discharged grid and left for 1 min to adhere. Excess liquid was blotted onto filter paper, and 3 μL of 2% uranyl acetate in distilled water was applied to the grid for negative staining. Excess uranyl acetate was removed by blotting after 1 min, and the sample was air dried for 10 min. Grids were placed on an RT holder and observed at a voltage of 120 kV with a Tecnai 12 Twin (FEI; OR, USA) fitted with a OneView camera, model 1095 (Gatan; Pleasanton, CA, USA). DigitalMicrograph 3.4 was used to capture and save digital images (Gatan).
Cell transfection and exosome treatment
NFs (2.5 × 105) were seeded in 60 × 15 mm cell culture dishes (Corning, 353004), and a combination of miRNA precursor (pre-miR)-185-5p, pre-miR-652-5p, and pre-miR-1246 (combo miRs) as well as pre-miRNA-scramble (Ambion, Life Technologies, Milan, Italy) was transiently transfected at a final concentration of 150 μM for each transfection point using Oligofectamine reagent (Invitrogen, Thermo Fisher Scientific, Milan, Italy; Cod. 12252-011, lot 2030861) under reduced serum conditions (Opti-MEM, Gibco, Cod. 31985-047, lot 2091581). After 4 h, the cell medium was supplemented with a final concentration of 10% FBS to restore optimal cell growth conditions. Cells were collected at 24, 48, and/or 72 h of transfection for downstream analysis or applications. Exosomes isolated from MDA-MB-231 cells were quantized using Bradford reagent (a protein assay dye, Bio- Rad, Cod. 5000006, lot 64254929), and a total amount of 40 μg was used for NF treatments. Briefly, NFs (2.5 × 105) were seeded in 60-mm dishes in 10% FBS-DMEM-F12 as long as they get attached, then washed twice with PBS solution and kept in DMEM-F12 medium supplemented with 10% Exo-FBS for the exosome treatment. Last, NFs were collected after 24 and/or 48 h for downstream analysis.
Exosome labeling and immunofluorescence assay
Exosomes isolated from MDA-MB-231cells (see “Exosome isolation and characterization”) were labeled with the red fluorescent cell membrane linker PKH26 (Sigma-Aldrich, SLBT6344). Briefly, exosomes (40 μg) were stained with PKH26 (0.33 μL) for 5 min in the dark at RT in a final reaction volume of 2 mL. Then the same amount of 1% BSA (2 mL) was added to stop the labeling reaction. Finally, samples were ultracentrifuged (Beckman Coulter, Optima MAX) twice at 100,000 × g for 70 min at 4°C, and pellets were resuspended in 500 μL of PBS. For the immunofluorescence assay, 5 × 105 NFs were plated on glass coverslips in a 24-well plate. The following day, NFs were treated with PKH26-labelled exosomes for 12 h, washed three times with PBS, and finally fixed with MetOH/acetone 1:1 for 10 min at −20°. After 3 washes in PBS, cells were blocked in 1% PBS-BSA with 0.3% Triton X-100 (Sigma-Aldrich, catalog number 9002-93-1) solution at RT for 15 min. Subsequently, cells were stained with anti- β-actin primary antibody (1:1,000) diluted in blocking solution for 1 h at RT for cytoskeleton detection. After 3 washes in PBS, the secondary antibody, goat anti-mouse immunoglobulin G (IgG)-fluorescein isothiocyanate (FITC) (Santa Cruz Biotechnology, F0211, 1:300 in PBS), was added for 30 min at RT. Last, cells were incubated with DAPI (BD Pharmingen, catalog number 564907, 1:1,000 in PBS) for 10 min at RT in the dark for nucleus visualization. Coverslips were washed and mounted with 2μL of 1:1 glycerol (Sigma-Aldrich, 114K0183V) in PBS on a microscope slide, and images taken from confocal microscopy (Leica LSM700) were analyzed to check exosome uptake.
Protein isolation and western blotting
Cells were washed twice in ice-cold PBS and exosomes isolated previously were lysed in JS buffer (50 mM HEPES [pH 7.5] containing 150 mM NaCl, 1% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 5 mM EGTA, 1 mM Na3VO4, and 1× protease inhibitor cocktail). Protein concentration was determined by Bradford reagent (BioRad, Cod. 5000006, lot 64254929) using BSA as the standard, and equal amounts of proteins were analyzed by SDS-PAGE (12.5% acrylamide, Bio-Rad, Cod. 1610158, lot 64269544). First, gels were electroblotted into nitrocellulose membranes (GE Healthcare Life Science, catalog 10600002). Then membranes were blocked for 1 h with 5% blotting-grade blocker (Bio-Rad, 1706404) in Tris-buffered saline (TBS; Bio-Rad, 1706435) containing 0.1% Tween 20 (Sigma-Aldrich, P1379-1L) and finally incubated at 4°C O/N with the primary antibodies. Signal detection was performed by peroxidase-conjugated secondary antibodies using the enhanced chemiluminescence system (Thermo Fisher Scientific, Milan Italy). Primary antibodies used were as follows: anti-FAP (Abcam, ab53066, 1:1,000 in 5% TBS-milk), anti-Caveolin-1 (Santa Cruz Biotechnology, MA, USA; sc-53564, 1:500 in 5% TBS-BSA), anti-MCT4 (Santa Cruz Biotechnology, sc-376140, 1:500 in 5% TBS-BSA), anti-β-actin (Sigma, A5441, 1:15,000 in 5% TBS-milk), anti-phospho-FAK(Y576/577) (Cell Signaling Technology, 3281, 1:500 in 5% TBS-BSA), anti-FAK (Cell Signaling Technology, 71433, 1:1,000 in 5% TBS-milk), anti-CD63 (Santa Cruz Biotechnology, sc-15363, 1:500 in TBS-milk), anti-TAPA1 (Abcam, ab35026, 1:1,000 in 5% TBS-milk), anti-Tsg101 (Abcam, ab83, 1:1,000 in 5% TBS-milk), anti-Hsp70 (Santa Cruz Biotechnology, sc-32239, 1:500 in 5% TBS-milk), anti-Calnexin (Abcam, ab10286, 1:1,000 in 5% TBS-milk), anti MMP1 (Santa Cruz Biotechnology, sc-21731, 1:500 in 5% TBS-milk), MMP2 (Santa Cruz Biotechnology, sc-21731, 1:250 in 5% TBS-milk), MMP3 (Santa Cruz Biotechnology, sc-21731, 1:500 in 5% TBS-milk), anti-ITG α5 (Santa Cruz Biotechnology, sc-13547, 1:500 in 5% TBS-milk), anti-ITG β1 (Santa Cruz Biotechnology, sc-13547, 1:1,000 in 5% TBS-milk), anti-AKT (Cell Signaling Technology, 9272, 1:1,000 in 5% TBS-milk), anti-phospho-AKT (Ser473) (Cell Signaling Technology, 9271, 1:1,000 in 5% TBS-milk); anti-β-Catenin (Cell Signaling Technology, Inc, #9582 1:1000 in 5% TBS-BSA), and anti-phospho-β-Catenin (Ser33/37/Thr41) (Cell Signaling Technology, 9561, 1:1,000 in 5% TBS-BSA).
RNA extraction and real-time PCR
Total RNA (miRNAs and mRNA) was extracted using TRIzol reagent (Life Technologies, 15596018). Reverse transcription was performed starting from an equal volume of total RNA/sample (150–300 ng) using the miScript Reverse Transcription Kit (QIAGEN, catalog number 218161) for total miRNAs and SuperScript III First-Strand (Invitrogen, catalog number 18080051) for mRNAs. Quantitative analysis of miR-185-5p, miR-652-5p, miR-1246, and RNU6A (as an internal reference) was performed by real-time PCR using the miScript SYBR Green PCR Kit (QIAGEN, catalog number 218075) and miScript Primer Assays (QIAGEN, catalog number 3406126). The reaction for detection of miRNAs was performed as follows: 95°C for 15 min, 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. For mRNA amplification of FAP, Caveolin-1, SLC16A3, SLC2A1, and β-actin as the internal normalizer gene, we performed real-time PCR with iTaq Universal SYBR Green Supermix (Bio-Rad, catalog number 1725124) and custom-made primers for mRNAs (Integrated DNA Technologies, Milan, Italy). The reaction for detection of mRNAs was performed as follows: 95°C for 15 min, 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. All reactions were run in triplicate.
Collagen contraction assay
The collagen contraction assay was performed with NFs in 35 × 10 mm cell culture dishes (Corning, 430165) for exosome treatment and in 12-well plates (Corning, 3513) for combo miRNA transfection. Type 1 collagen (Corning, 354236, 33 ng/μL) was resuspended in an acidic environment composed of acetic acid (5 mM) and minimum essential Eagle’s medium (Sigma, MO275, 10×). Then NaOH (1 M) was added drop by drop to restore the neutral pH for cell resuspension. For the contraction assay performed with combo miRNAs, NFs were first transfected with combo miRNAs (scrambled for control) for 48 h and then used for the contraction assay; for exosomes, NFs were first plated in the collagen plug and then treated with exosomes. In both cases, 1.5 × 105 NFs resuspended in 250 μL of FBS were added to type 1 the collagen mix prepared before. All steps during collagen handling must be performed on ice to avoid early collagen solidification. Then plates containing collagen plugs and cells were taken at 37°C with 5%CO2 for 3 h to allow collagen solidification. Subsequently, collagen plugs were detached from plate walls to allow cell contraction, and DMEM-F12 FBS-free medium (with or without exosomes, depending on the experiment) was added. Images of the entire collagen plugs were taken after 24 h with the camera tool of a mobile phone held in a fixed position. Then plug areas were calculated with ImageJ software and analyzed for NF contraction ability after exosome treatment or combo miR transfection.
3D organotypic co-culture assay
NFs were starved in DMEM-F12 medium without FBS for 24 h and then seeded in 35 × 10 mm cell culture dishes (Corning, 430165) in a neutralized matrix made of type 1 collagen treated with MDA-MB-231-derived exosomes (or PBS as a control) to ensure their activation (the same procedure as described under “Contraction assay”). 1 × 105 MCF10A cells were seeded on top of the collagen plug for 48 h. Then the plugs were transferred to an invasion grid (screens for CD-1 size 40 mesh, Sigma-Aldrich) in a 60-mm plate, and complete growth medium was added underneath to create an air/liquid interface to trigger epithelial cell invasion. After 14 days, matrices were fixed, paraffin embedded, and cut into 10-μm sections. Organotypic matrices were stained with anti-pan-cytokeratin (Santa Cruz Biotechnology, sc-8018, 1:400 in blocking solution) O/N at 4°C and then with the secondary antibody, Alexa 594-conjugated goat anti-mouse (Abcam, ab150116, 1:400 in blocking solution) for 1 h at RT in the dark. Images were taken with an inverted microscope and with a fluorescent one. Pan-cytokeratin-positive cells were counted using ImageJ software in different fields of the images to quantify the number of invading cells.
Transwell migration assay
The migration assay was carried out with 8.0 μm polycarbonate-membrane-permeable, 6.5-mm Transwell inserts (Corning, Cod. 3422, lot 11619021). NFs pretreated with exosomes (40 μg) or transfected with miRNAs (as described under “Cell transfection and exosome treatment”) were harvested with a trypsin-EDTA solution (Sigma-Aldrich, Cod. T4D49, lot SLCH3365) and counted with a Neubauer chamber. Then 1 × 105 cells were washed with PBS to remove any FBS residue, resuspended in DMEM-F12 FBS-free medium, and seeded in the upper part of the Transwell chamber. The lower part of the chamber was filled with 600 μL of DMEM-F12 medium supplemented with 10% FBS, 1% A/A, and 1% amphotericin B to create the chemical gradient needed for cell migration. Cells were incubated at 37°C with 5% CO2 for 24 h. Then the Transwell chambers were stained and fixed with 0.1% crystal violet in 25% methanol for 20 min at RT in the dark. The reaction was stopped with water, and non-migrated cells were scraped off the top of the chamber with a cotton swab. Representative images were taken with the phase-contrast microscope (Leica DMI3000 B). The percentage of migrated cells was calculated by eluting crystal violet with 600 μL of 1% SDS for each well and measuring the respective absorbance at 490 nm with a Multiskan FC plate reader (Thermo Scientific).
Invasion assay
The in vitro invasion assay was performed in Corning 24-well plates with 8.0 μm polycarbonate-membrane-permeable 6.5 mm Transwell inserts (Corning, Cod. 3422, lot 11619021). NFs (1.2 × 105). NFs transfected previously with combo miRs for 24 h were resuspended in a mix containing Matrigel Matrix Basement Membrane (Corning, Cod. 354230, lot 6207017) diluted 1:5 in DMEM-F12, FBS free. The lower part of the chamber was filled with 600 μL of DMEM-F12 medium supplemented with 10% FBS, % antibiotic-antimycotics, and 1% amphotericin B to create the chemical gradient for cell movement. Cells were incubated at 37°C with 5% CO2 for 72 h. The Transwell supports were stained and fixed with 0.1% crystal violet in 25% methanol for 20 min at RT in the dark. The reaction was stopped with water, and non-migrated cells together with residual Matrigel solution were scraped off the top of the Transwell with a cotton swab. Representative images were taken with a phase-contrast microscope (Leica DMI3000 B). The percentage of migrated cells was evaluated by eluting crystal violet with 600 μL of 1% SDS for each well and measuring the respective absorbance at 490 nm with a Multiskan FC plate reader (Thermo Scientific).
Scratch assay
NFs (5 × 104) were seeded in a 12-well plate (Corning, 3513) and, on the following day, transfected with miR-185-5p, miR-652-5p, miR-1246, or a scrambled sequence as a control. After 48 h, cells were starved for 3 h in DMEM-F12 FBS-free culture medium. Next, a scratched wound was made with a 200-μL tip in each well, and then cells were grown continuously in DMEM-F12 culture medium complemented with 10% FBS and 1% A/A for 24 h. Microscopy images were taken in different fields of the wound at the scratch moment (t0) and after 24 h (t24) using a 5× objective of an inverted microscope (DMI3000 B, Leica, Milan, Italy). Scratch area was calculated with ImageJ software and analyzed to measure the wound-healing ability of NFs after combo miR transfection.
Small RNA sequencing
NFs (5 × 105) were plated in 100-mm dishes with DMEM-F12 culture medium supplemented with 10% Exo-FBS (SBI) and 1% A/A and treated with 120 μg of MDA-MB-231-derived exosomes and the same volume of PBS solution as a control. After 24 h, cells were collected, and RNA was extracted using TRIzol reagent (Life Technologies, 15596018). Samples were shipped to Genomix4Life (Baronissi, Salerno, Italy), who performed small RNA sequencing using Illumina HiSeq2500 (SmallRNA 1 × 20M Cod. G4L1630 – iMir, Cod. G4L15055) and bioinformatics analysis (PCA component and differential expression analysis). Two biological replicates for each experimental point were analyzed. For the statistical analysis, p < 0.05 alone was considered for experimental significance; no p value adjustment was performed because of the small sample size (NFs from pt. #1).
In vitro cell viability assay
Cell viability was evaluated with the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Milan, Italy) according to the manufacturer’s protocol. After 30 min of incubation, the plates were analyzed on a multilabel counter (Bio-Rad) to measure the absorbance values used for the analysis.
Data availability
The RNA sequencing data discussed in this publication have been deposited in database: NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE185654 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185654).
Acknowledgments
This work was supported by grants from Associazione Italiana Ricerca sul Cancro AIRC (IG 2016 N. 18473), POR Campania FESR 2014-2020 “SATIN”, and the Earlier Foundation (to G.C.). This work has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie projects cONCReTE (872391), PRISAR2 (872860), CANCER (777682), PAVE (861190), and CAST (857894). G.R. is the recipient of a European GRANT BARRICADE (IF_MSCA n: 895151). A.A. is the recipient of European GRANT GlEXO (IF MSCA n:891551).
Author contributions
Conceptualization, I.S., L.C., I.P., and C.Q.; data curation, I.S. and L.C.; formal analysis, I.S., L.C., and C.Q.; funding acquisition, G.C.; investigation, I.S. and L.C.; methodology, I.S., L.C., I.P., F.P., F.I., M.P.S., S.B., A.A., G.R., S.N., R.V.C., and T.S.; project administration, G.C. and I.S.; resources, G.T.; software, I.S., L.C., C.Q., and A.A.; supervision, G.C.; validation, I.S., L.C., and I.P.; visualization, I.S. and L.C.; writing – original draft, I.S. and L.C.; writing – review & editing, G.C., F.I., F.P., G.R., M.P.S., and A.A. All authors have read and approved the manuscript and its content.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.02.013.
Supplemental information
References
- 1.Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 2.Karaayvaz M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., Specht M.C., Bernstein B.E., Michor F., Ellisen L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018;9:3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Balkwill F.R., Capasso M., Hagemann T. The tumor microenvironment at a glance. J. Cell Sci. 2012;125:5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- 5.Orimo A., Gupta P.B., Sgroi D.C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V.J., Richardson A.L., Weinberg R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 6.Toullec A., Gerald D., Despouy G., Bourachot B., Cardon M., Lefort S., Richardson M., Rigaill G., Parrini M.C., Lucchesi C., et al. Oxidative stress promotes myofibroblast differentiation and tumour spreading. EMBO Mol. Med. 2010;2:211–230. doi: 10.1002/emmm.201000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdogan B., Webb D.J. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem. Soc. Trans. 2017;45:229–236. doi: 10.1042/BST20160387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiro N., Gonzalez L.O., Fraile M., Cid S., Schneider J., Vizoso F.J. Breast cancer tumor stroma: cellular components, phenotypic heterogeneity, intercellular communication, prognostic implications and therapeutic opportunities. Cancers. 2019;11:664. doi: 10.3390/cancers11050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahai E., Astsaturov I., Cukierman E., DeNardo D.G., Egeblad M., Evans R.M., Fearon D., Greten F.R., Hingorani S.R., Hunter T., et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kechagia J.Z., Ivaska J., Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 2019;20:457–473. doi: 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- 11.Hooper S., Gaggioli C., Sahai E. A chemical biology screen reveals a role for Rab21-mediated control of actomyosin contractility in fibroblast-driven cancer invasion. Br. J. Cancer. 2010;102:392–402. doi: 10.1038/sj.bjc.6605469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeltz C., Primac I., Erusappan P., Alam J., Noel A., Gullberg D. Cancer-associated fibroblasts in desmoplastic tumors: emerging role of integrins. Semin. Cancer Biol. 2020;62:166–181. doi: 10.1016/j.semcancer.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Goetz J.G., Minguet S., Navarro-Lerida I., Lazcano J.J., Samaniego R., Calvo E., Tello M., Osteso-Ibanez T., Pellinen T., Echarri A., et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y., Luo X., Lv W., Hu W., Zhao C., Xiong M., Yi Y., Wang D., Wang Y., Wang H., et al. Tumor-derived exosomal components: the multifaceted roles and mechanisms in breast cancer metastasis. Cell Death Dis. 2021;12:547. doi: 10.1038/s41419-021-03825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 16.Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu D.D., Wu Y., Shen H.Y., Lv M.M., Chen W.X., Zhang X.H., Zhong S.L., Tang J.H., Zhao J.H. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015;106:959–964. doi: 10.1111/cas.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giordano C., La Camera G., Gelsomino L., Barone I., Bonofiglio D., Ando S., Catalano S. The biology of exosomes in breast cancer progression: dissemination, immune Evasion and metastatic colonization. Cancers. 2020;12:2179. doi: 10.3390/cancers12082179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y.S., Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio M.V., Croce C.M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 22.Palma F., Affinito A., Nuzzo S., Roscigno G., Scognamiglio I., Ingenito F., Martinez L., Franzese M., Zanfardino M., Soricelli A., et al. miR-34c-3p targets CDK1 a synthetic lethality partner of KRAS in non-small cell lung cancer. Cancer Gene Ther. 2021;28:413–426. doi: 10.1038/s41417-020-00224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silen W., Machen T.E., Forte J.G. Acid-base balance in amphibian gastric mucosa. Am. J. Physiol. 1975;229:721–730. doi: 10.1152/ajplegacy.1975.229.3.721. [DOI] [PubMed] [Google Scholar]
- 24.Siemens H., Jackstadt R., Hunten S., Kaller M., Menssen A., Gotz U., Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 25.Roy S., Levi E., Majumdar A.P., Sarkar F.H. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J. Hematol. Oncol. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L., Hermeking H. miR-34a and miR-34b/c Suppress Intestinal Tumorigenesis. Cancer Res. 2017;77:2746–2758. doi: 10.1158/0008-5472.CAN-16-2183. [DOI] [PubMed] [Google Scholar]
- 27.Liang J., Li Y., Daniels G., Sfanos K., De Marzo A., Wei J., Li X., Chen W., Wang J., Zhong X., et al. LEF1 targeting EMT in prostate cancer invasion is regulated by miR-34a. Mol. Cancer Res. 2015;13:681–688. doi: 10.1158/1541-7786.MCR-14-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Wang L., Dong D., Wang Z., Ji W., Yu M., Zhang F., Niu R., Zhou Y. MiR-34b/c-5p and the neurokinin-1 receptor regulate breast cancer cell proliferation and apoptosis. Cell Prolif. 2019;52:e12527. doi: 10.1111/cpr.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahlhut C., Slack F.J. Combinatorial action of MicroRNAs let-7 and miR-34 effectively synergizes with Erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. 2015;14:2171–2180. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong D.S., Kang Y.K., Borad M., Sachdev J., Ejadi S., Lim H.Y., Brenner A.J., Park K., Lee J.L., Kim T.Y., et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingenito F., Roscigno G., Affinito A., Nuzzo S., Scognamiglio I., Quintavalle C., Condorelli G. The role of Exo-miRNAs in cancer: a focus on therapeutic and diagnostic applications. Int. J. Mol. Sci. 2019;20:4687. doi: 10.3390/ijms20194687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotani A., Ito M., Kudo K. Non-coding RNAs and lipids mediate the function of extracellular vesicles in cancer cross-talk. Semin. Cancer Biol. 2021;74:121–133. doi: 10.1016/j.semcancer.2021.04.017. [DOI] [PubMed] [Google Scholar]
- 33.Sanz-Moreno V., Gaggioli C., Yeo M., Albrengues J., Wallberg F., Viros A., Hooper S., Mitter R., Feral C.C., Cook M., et al. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 2011;20:229–245. doi: 10.1016/j.ccr.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 34.Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi H., Sakai R. Direct interaction between carcinoma cells and cancer associated fibroblasts for the regulation of cancer invasion. Cancers. 2015;7:2054–2062. doi: 10.3390/cancers7040876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timpson P., McGhee E.J., Erami Z., Nobis M., Quinn J.A., Edward M., Anderson K.I. Organotypic collagen I assay: a malleable platform to assess cell behaviour in a 3-dimensional context. JoVE. 2011;13:e3089. doi: 10.3791/3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sanchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng Y., Qi S., Hu R., Zhao J., Rui W., Miao Y., Ma J., Yang Q. Identification of blood microRNA alterations in patients with severe active alopecia areata. J. Cell. Biochem. 2019;120:14421–14430. doi: 10.1002/jcb.28700. [DOI] [PubMed] [Google Scholar]
- 39.Maschler S., Wirl G., Spring H., Bredow D.V., Sordat I., Beug H., Reichmann E. Tumor cell invasiveness correlates with changes in integrin expression and localization. Oncogene. 2005;24:2032–2041. doi: 10.1038/sj.onc.1208423. [DOI] [PubMed] [Google Scholar]
- 40.Jang I., Beningo K.A. Integrins, CAFs and mechanical forces in the progression of cancer. Cancers. 2019;11 doi: 10.3390/cancers11050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheppard D. Epithelial integrins. BioEssays : News Rev. Mol. Cell. Dev. Biol. 1996;18:655–660. doi: 10.1002/bies.950180809. [DOI] [PubMed] [Google Scholar]
- 42.Kumaravel S., Abbey C.A., Bayless K.J., Chakraborty S. The beta1-integrin plays a key role in LEC invasion in an optimized 3-D collagen matrix model. Am. J. Physiol. Cell Physiol. 2020;319:C1045–C1058. doi: 10.1152/ajpcell.00299.2020. [DOI] [PubMed] [Google Scholar]
- 43.Eiro N., Fernandez-Garcia B., Vazquez J., Del Casar J.M., Gonzalez L.O., Vizoso F.J. A phenotype from tumor stroma based on the expression of metalloproteases and their inhibitors, associated with prognosis in breast cancer. Oncoimmunology. 2015;4 doi: 10.4161/2162402X.2014.992222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eck S.M., Cote A.L., Winkelman W.D., Brinckerhoff C.E. CXCR4 and matrix metalloproteinase-1 are elevated in breast carcinoma-associated fibroblasts and in normal mammary fibroblasts exposed to factors secreted by breast cancer cells. Mol. Cancer Res. 2009;7:1033–1044. doi: 10.1158/1541-7786.MCR-09-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito A., Nakajima S., Sasaguri Y., Nagase H., Mori Y. Co-culture of human breast adenocarcinoma MCF-7 cells and human dermal fibroblasts enhances the production of matrix metalloproteinases 1, 2 and 3 in fibroblasts. Br. J. Cancer. 1995;71:1039–1045. doi: 10.1038/bjc.1995.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K. FAK-dependent cell motility and cell Elongation. Cells. 2020;9:192. doi: 10.3390/cells9010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi R.S., Kanugula S.S., Sudhir S., Pereira M.P., Jain S., Aghi M.K. The role of cancer-associated fibroblasts in tumor progression. Cancers. 2021;13:1399. doi: 10.3390/cancers13061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Busch S., Andersson D., Bom E., Walsh C., Stahlberg A., Landberg G. Cellular organization and molecular differentiation model of breast cancer-associated fibroblasts. Mol. Cancer. 2017;16:73. doi: 10.1186/s12943-017-0642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Outschoorn U.E., Lisanti M.P., Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin. Cancer Biol. 2014;25:47–60. doi: 10.1016/j.semcancer.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Sotgia F., Martinez-Outschoorn U.E., Pavlides S., Howell A., Pestell R.G., Lisanti M.P. Understanding the Warburg effect and the prognostic value of stromal caveolin-1 as a marker of a lethal tumor microenvironment. Breast Cancer Res. 2011;13:213. doi: 10.1186/bcr2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercier I., Casimiro M.C., Wang C., Rosenberg A.L., Quong J., Minkeu A., Allen K.G., Danilo C., Sotgia F., Bonuccelli G., et al. Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 downregulation and RB tumor suppressor functional inactivation: implications for the response to hormonal therapy. Cancer Biol. Ther. 2008;7:1212–1225. doi: 10.4161/cbt.7.8.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roscigno G., Cirella A., Affinito A., Quintavalle C., Scognamiglio I., Palma F., Ingenito F., Nuzzo S., De Micco F., Cuccuru A., et al. miR-216a acts as a negative regulator of breast cancer by modulating stemness properties and tumor microenvironment. Int. J. Mol. Sci. 2020;21:2313. doi: 10.3390/ijms21072313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J.E., Kim B.G., Jang Y., Kang S., Lee J.H., Cho N.H. The stromal loss of miR-4516 promotes the FOSL1-dependent proliferation and malignancy of triple negative breast cancer. Cancer Lett. 2020;469:256–265. doi: 10.1016/j.canlet.2019.10.039. [DOI] [PubMed] [Google Scholar]
- 54.Wu H.J., Hao M., Yeo S.K., Guan J.L. FAK signaling in cancer-associated fibroblasts promotes breast cancer cell migration and metastasis by exosomal miRNAs-mediated intercellular communication. Oncogene. 2020;39:2539–2549. doi: 10.1038/s41388-020-1162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baroni S., Romero-Cordoba S., Plantamura I., Dugo M., D'Ippolito E., Cataldo A., Cosentino G., Angeloni V., Rossini A., Daidone M.G., Iorio M.V. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7:e2312. doi: 10.1038/cddis.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen B., Sang Y., Song X., Zhang D., Wang L., Zhao W., Liang Y., Zhang N., Yang Q. Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics. 2021;11:3932–3947. doi: 10.7150/thno.53412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dou D., Ren X., Han M., Xu X., Ge X., Gu Y., Wang X. Cancer-associated fibroblasts-derived exosomes suppress immune cell function in breast cancer via the miR-92/PD-L1 pathway. Front. Immunol. 2020;11:2026. doi: 10.3389/fimmu.2020.02026. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Donnarumma E., Fiore D., Nappa M., Roscigno G., Adamo A., Iaboni M., Russo V., Affinito A., Puoti I., Quintavalle C., et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget. 2017;8:19592–19608. doi: 10.18632/oncotarget.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duchartre Y., Kim Y.M., Kahn M. The Wnt signaling pathway in cancer. Crit. Rev. 2016;99:141–149. doi: 10.1016/j.critrevonc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida T., Sopko N.A., Kates M., Liu X., Joice G., McConkey D.J., Bivalacqua T.J. Three-dimensional organoid culture reveals involvement of Wnt/beta-catenin pathway in proliferation of bladder cancer cells. Oncotarget. 2018;9:11060–11070. doi: 10.18632/oncotarget.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ediriweera M.K., Tennekoon K.H., Samarakoon S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin. Cancer Biol. 2019;59:147–160. doi: 10.1016/j.semcancer.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y., Wang T., Du J., Li Y., Wang X., Zhou Y., Yu X., Fan W., Zhu Q., Tong X., Wang Y. The critical role of PTEN/PI3K/AKT signaling pathway in shikonin-induced apoptosis and proliferation inhibition of chronic myeloid leukemia. Cell Physiol. Biochem. 2018;47:981–993. doi: 10.1159/000490142. [DOI] [PubMed] [Google Scholar]
- 63.Nagase H., Visse R., Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu T., Di G. Role of tumor microenvironment in triple-negative breast cancer and its prognostic significance. Chin. J. Cancer Res. 2017;29:237–252. doi: 10.21147/j.issn.1000-9604.2017.03.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dong X., Bai X., Ni J., Zhang H., Duan W., Graham P., Li Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020;11:987. doi: 10.1038/s41419-020-03189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomasik B., Papis-Ubych A., Stawiski K., Fijuth J., Kedzierawski P., Sadowski J., Stando R., Bibik R., Graczyk L., Latusek T., et al. Serum MicroRNAs as xerostomia biomarkers in patients with oropharyngeal cancer undergoing radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2021;111:1237–1249. doi: 10.1016/j.ijrobp.2021.07.008. [DOI] [PubMed] [Google Scholar]
- 68.Sur D., Balacescu L., Cainap S.S., Visan S., Pop L., Burz C., Havasi A., Buiga R., Cainap C., Irimie A., Balacescu O. Predictive Efficacy of MiR-125b-5p, MiR-17-5p, and MiR-185-5p in liver metastasis and chemotherapy response among advanced stage colorectal cancer patients. Front. Oncol. 2021;11:651380. doi: 10.3389/fonc.2021.651380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun J., Zhao J., Yang Z., Zhou Z., Lu P. Identification of gene signatures and potential therapeutic targets for acquired chemotherapy resistance in gastric cancer patients. J. Gastrointest. Oncol. 2021;12:407–422. doi: 10.21037/jgo-21-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsui D., Zaidi A.H., Martin S.A., Omstead A.N., Kosovec J.E., Huleihel L., Saldin L.T., DiCarlo C., Silverman J.F., Hoppo T., et al. Primary tumor microRNA signature predicts recurrence and survival in patients with locally advanced esophageal adenocarcinoma. Oncotarget. 2016;7:81281–81291. doi: 10.18632/oncotarget.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao P., Wang D., Liu M., Chen S., Yang Z., Zhang J., Wang H., Niu Y., Wang W., Yang J., Sun G. DNA methylation-mediated repression of exosomal miR-652-5p expression promotes oesophageal squamous cell carcinoma aggressiveness by targeting PARG and VEGF pathways. Plos Genet. 2020;16:e1008592. doi: 10.1371/journal.pgen.1008592. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Torii C., Maishi N., Kawamoto T., Morimoto M., Akiyama K., Yoshioka Y., Minami T., Tsumita T., Alam M.T., Ochiya T., et al. miRNA-1246 in extracellular vesicles secreted from metastatic tumor induces drug resistance in tumor endothelial cells. Sci. Rep. 2021;11:13502. doi: 10.1038/s41598-021-92879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ueta E., Tsutsumi K., Kato H., Matsushita H., Shiraha H., Fujii M., Matsumoto K., Horiguchi S., Okada H. Extracellular vesicle-shuttled miRNAs as a diagnostic and prognostic biomarker and their potential roles in gallbladder cancer patients. Sci. Rep. 2021;11:12298. doi: 10.1038/s41598-021-91804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen S., Fu Z., Wen S., Yang X., Yu C., Zhou W., Lin Y., Lv Y. Expression and diagnostic value of miR-497 and miR-1246 in hepatocellular carcinoma. Front. Genet. 2021;12:666306. doi: 10.3389/fgene.2021.666306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kruger S., Abd Elmageed Z.Y., Hawke D.H., Worner P.M., Jansen D.A., Abdel-Mageed A.B., Alt E.U., Izadpanah R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer. 2014;14:44. doi: 10.1186/1471-2407-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data discussed in this publication have been deposited in database: NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE185654 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185654).